Abstract
Increasing evidence has been suggested that microRNA-144 (miR-144) involved in tumor initiation, development and metastasis in various cancers. However, the biological roles and potential mechanisms of miR-144 in laryngeal squamous cell carcinoma (LSCC) remain unclear. In the present study, we discovered that miR-144 expression levels in LSCC tissues were significantly lower than those of adjacent normal tissues. Furthermore, overexpression of miR-144 in LSCC cells inhibited cell proliferation, colony formation, migration, and invasion in vitro. Consistently, stable overexpression of miR-144 suppressed the growth of LSCC cell xenografts in vivo. Bioinformatic algorithms and luciferase reporter assays confirmed that insulin receptor substrate 1 (IRS1) is a direct target of miR-144. Overexpreesion of miR-144 obviously decreased IRS1 expression thereby suppressing phosphatidylinositol 3-kinase (PI3K)/AKT pathway activation. Further functional studies suggested that downregulation of IRS1 had similar effects as that of miR-144 overexpression, and upregulation of IRS1 partially reversed the inhibitory effects of miR-144. These findings suggested that miR-144 functioned as a tumor suppressor in LSCC by targeting IRS1, and that miR-144 might serve as a potential target for LSCC treatment.
Keywords: miR-144, laryngeal squamous cell carcinoma, IRS1, growth, metastasis
Introduction
Laryngeal cancer is the eleventh most common cancer worldwide with high mortality rate [1,2]. Laryngeal squamous cell carcinoma (LSCC), accounting for approximately 85-90% of all malignant tumors of the larynx, is an aggressive malignancy that represents the second most common site of head and neck tumors [1,2]. Despite the improvement of currently available treatment strategies, including surgical resection, radiotherapy, and chemotherapy, the survival rate of patients with LSCC has changed little over the past 10 years [3]. There is very little information about the precise molecular pathways involved in LSCC development and procession. Therefore, there is an urgent need to understand the molecular mechanisms that underlie LSCC development, which may contribute to identification of novel diagnositic marker and novel therapeutic targets.
MicroRNAs (miRNAs, miRs) are endogenous, single-stranded, short (18-24 nucleotides in length), non-coding RNAs that regulate gene expression at the transcriptional or posttranscriptional level by binding to the complementary 3’-untranslated region (3’-UTR) of messenger RNAs (mRNAs) [4,5]. It has been showed that miRNAs involved in many physiological and pathological processes through regulating cellular proliferation, differentiation, metabolism and apoptosis [6]. It is well known that miRNAs play significant roles in various human cancers, functioning as oncogenes by inhibiting tumor suppressor genes or tumor suppressor by downregulating oncogenes [7-9]. Numerous miRNAs had been found to involve in LSCC development and procession [10]. Therefore, miRNAs are presently considered as potential novel targets for LSCC therapy.
Increasing reports were focused on the role of miR-144 carcinogenesis because it is dysregulated and involved in the tumorigenesis of various cancer, such as lung cancer [11], osteosarcoma [12], hepatocellular carcinoma [13], thyroid cancer [14], bladder cancer [15] and colorectal carcinoma [16]. However, the biological functions and underlying molecular mechanism of miR-144 in LSCC are not yet described. Therefore, in the present study, we investigate the biological role and potential mechanism of miR-144 in LSCC by several experiments in vitro and tumor growth of xenograft in vivo.
Materials and method
Tissue sample
Twenty-four pairs of human LSCC and adjacent normal tissues were harvested at Department of Department of Otorhinolaryngology-Head and Neck Surgery, the First Hospital of Jilin University (Changchun, China) from September 2011 to October 2014. The corresponding adjacent noncancerous tissues were obtained from surgically resected tissues that were located over 5 cm away from the tumors. All tissue samples were immediately snap-frozen in liquid nitrogen, and stored in liquid and stored at -80°C until further processing. Informed consent was obtained from all patients. This study was approved by the Ethic Committee of Jilin University (Changchun, China).
Cell culture
The human Hep2 (laryngeal cancer cell line) was obtained from the Cell Biology Institute of Shanghai, Chinese Academy of Science (Shanghai, china) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO, Carlsbad, CA) containing with 10% fetal bovine serum (FBS, Hyclone, Logan, USA), 100 units/ml penicillin or 100 μg/ml streptomycin in a humidified atmosphere at 37°C in 5% CO2.
Cell transfection
miR-144 mimics (miR-144), and corresponding miRNA negative control (miR-NC) were brought from Qiagen (Frederick, MD, USA). siRNA against IRS1 (si-IRS1), siRNA against negative control (si-NC) plasmid were brought from GenePharma (Shanghai, China). Overexpression IRS1 plasmid pCDNA3.1-IRS1) was granted from Doctor Peng Zhang (Changchun, China). These molecular productions were transient transfected into Hep2 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Transfection efficiencies were determined in every experiment at 48 h after transfection.
RNA extraction and quantitative reverse-transcription PCR (qRT-PCR)
For detection miR-144 expression, total RNA was extracted from cultured cells and tissue using mirVana miRNA Isolation kit (Ambion) according to the manufacturer’s protocol. The quantitative RT-PCR (qRT-PCR) was performed by the mirVanaTM miRNA detection kit (Ambion, USA) with miR-144 specific primer (Ambion, USA) using ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). U6 RNA was used as internal control.
For detection IRS1 expression, total RNA was extracted from cultured cells and tissue using TRIzol reagent (Life Technologies, Inc, USA) according to manufacturer’s instructions, and was reverse-transcribed into cDNA using M-MLV reverse transcriptase kits (Promega, USA) according to the manufacturer’s instructions. Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa, Dalian, China) under ABI PRISM 7000 Sequence Detection System. The primer of IRS1 and β-actin were used in this study as described previously [17]. β-actin was used as internal control. Relative gene expression levels were calculated using the 2-ΔΔCt method.
Cell proliferation and colony formation assays
For cell proliferation assay, 2 × 103 transfected cells/well were plated into 96-well plates and cultured at the indicated time points (24, 48, 72 and 96 h). Then, 10 μl CCK-8 reagent (DoJinDo, Japan) was added to each well. The OD450 nm value was determined by a microplate reader (Bio-Tek Instruments, Winooski, VT).
For colony formation assay, 1 × 103 transfected cells/well were seeded in 6-well plate and cultured for 2 weeks. Then cells were fixed with 4% paraformaldehyde for 20 min and stained with 1% crystal violet. The total number of colonies was counted under a light microscope (Olympus, Tokyo, Japan).
Wound healing assay
Wound healing assays were performed to assess the motility of indicated cells. In briefly, when transfected cells reached 90-95% confluence, they were scratched with a sterile plastic micropipette tip in the cell monolayer. After wounding, the debris was removed by washing the cells with PBS, and complete DMEM medium with 10% FBS was added, and were cultured for 24 h. Recovery of the wound was observed and captured images by a phase-contrast microscope (Zeiss, Gottingen, Germany). For each image, the area that was uncovered by cells was analyzed by using Image Pro Plus 4.5.1 software.
Transwell assay
The invasive behaviors of indicated cells were analyzed by Transwell chamber (Corning CostarCorp., Cambridge, MA, USA) assay. In briefly, 2 × 105 transfected cells in serum-free medium were added to each upper chamber pre-coated with Matrigel matrix, and 500 μl of DMEM medium containing 10% FBS were added to the lower chamber to serve as chemoattractant. After 24 h incubation, cells on the surface of upper chamber were removed by scraping with a cotton swab, and the invasive cells on the lower membrane surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, photographed, and quantified by counting them in five random fields by a light microscope (Olympus, Tokyo, Japan).
Luciferase assays
The human IRS1 3’UTR oligonucleotides containing the wild-type (Wt) or mutant (Mut) miR-144 binding site of were amplified using PCR and inserted into downstream of the luciferase gene in the pGL3-luciferase reporter plasmid (Ambion, Austin, TX, USA). For luciferase assay, Hep2 cells were added to 24-well plates, and then transfected with 100 ng of luciferase reporter vectors (Wt/Mut IRS1 3’UTR) and 100 nM of miR-144 or miR-NC. Forty-eight hours after transfection, luciferase activities were measured using the dual-luciferase reporter gene assay kit (Promega, Beijing, China), according to the manufacturer’s instructions.
Western blot analysis
The transfected cells and tissues were harvested and lysed with RIPA buffer (Beyotime, Shanghai, China). Total Protein concentrations were measured by the Bradford assay (Bio-Rad, Hercules, CA, USA). Equal amounts of proteins (20 μg each lane) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Richmond, CA, USA). Then the membranes were blocked in 5% skim milk for 1 h at room temperature, followed incubated with specific primary antibodies overnight at 4°C as followed: anti-IRS1, anti-Akt, anti-p-Akt (Ser473) and anti-β-actin (all antibody from Santa Cruz Biotechnology Inc., California, USA; all diluted 1:1000). After extensive washing, they were incubated with secondary horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibodies (Santa Cruz Biotechnology) at room temperature for 2 h. Protein bands were visualized using chemiluminescent detection system (ECL, Thermo Scientific, Rockford, IL, USA).
In vivo tumorigenesis assay
Animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Jilin University (Changchun, China). Twenty male BALB/C nude mice (18-20 g, 6-7 weeks old) were obtained from the Experimental Animal Center of Changchun Institute for Biological Sciences (Changchun, China).
For in vivo tumorigenesis assay, 2 × 106 Hep2 cells stable expression miR-144 or miR-NC were suspended in 100 μl of serum-free DMEM containing 1% matrigel and then injected subcutaneously in the right posterior flank of BALB/c-nude mice (10 mice in each group), respectively. Ten days later, tumor growth was monitored and recorded by measuring tumor length and width every five day, and tumor volume was calculated according to according to the formula: V = 0.5 × L (length) × W2 (width). After 30 days, mice were sacrificed, and the tumors tissue were dissected and weighted. Part of tumor tissues collected for analysis of the expression of miR-144 and IRS1.
Statistical analysis
All data are expressed as means ± standard deviation (SD) at least from three independent experiments. Statistical analyses were performed using SPSS version 19.0 software (SPSS, Chicago, IL, USA). The differences between groups were analyzed using Student’s t-test or one-way ANOVA. The relationship between IRS1 and miR-144 expressions was tested with two-tailed Pearson’s correlation. Differences were considered significant when the P < 0.05.
Results
miR-144 is down-regulated in human LSCC tissues
The expression status of miR-144 in laryngeal tumor samples and the adjacent normal tissues obtained from the same patients was determined by qRT-PCR. As shown in Figure 1, the expression level of miR-144 was significantly downregulated in tumor samples compared with the corresponding normal tissues (P < 0.01).
Figure 1.
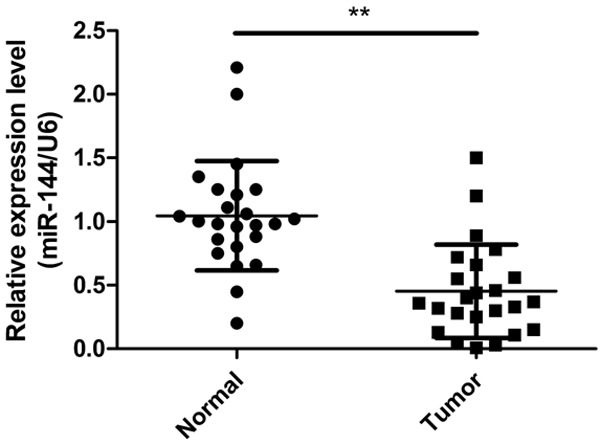
The expression of miR-144 is downregulated in human LSCC tissues. Relative expression levels of miR-144 in 24 LSCC tumor tissues and corresponding normal sample by qRT-PCR. **P < 0.01 versus normal tissues.
miR-144 overexpression inhibits proliferation and colony formation in Hep2 cells
To investigate the role of miR-144 in LSCC tumorigenesis, we analyzed the functional effects of miR-144 upon its ectopic expression by transfection with miR-144 or miR-NC. qRT-PCR assay confirmed that miR-144 expression level significantly increased in Hep2 cells after transfected with miR-144 mimic (Figure 2A). Then we investigated the effects of miR-144 overexpression on cell proliferation and colony formation. Compared to miR-NC group, overexpression miR-144 in Hep2 cells significantly inhibited cell proliferation (Figure 2B) and colony formation (Figure 2C).
Figure 2.
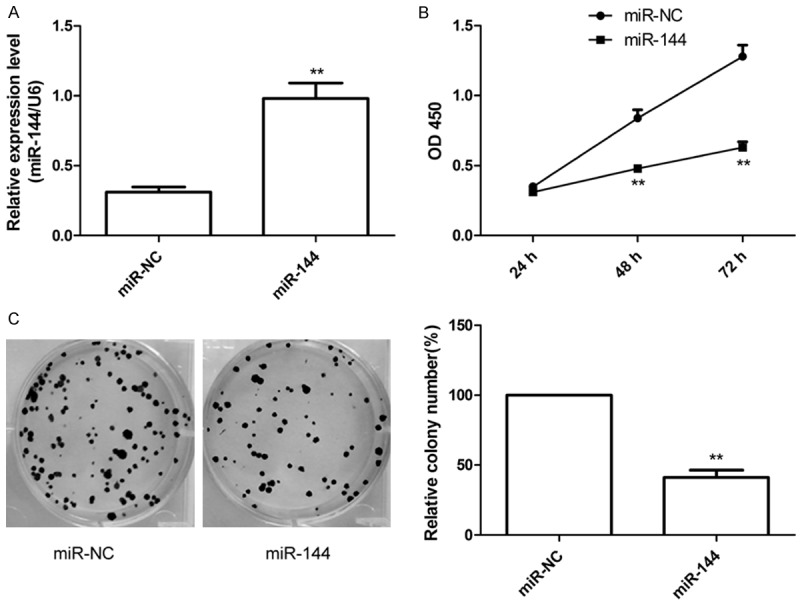
MiR-144 overexpression reduces proliferation and colony formation in Hep2 cells. A. qRT-PCR determined miR-144 expression in Hep2 cells transfected with miR-144 mimic or miR-NC. B. CCK8 assay were performed to determined cell proliferation in Hep2 cells transfected with miR-144 mimic or miR-NC. C. Colony formation was determined in Hep2 cells transfected with miR-144 mimic or miR-NC. *P <0.05, **P < 0.01 versus miR-NC.
miR-144 overexpression inhibits migration and invasion in Hep2 cells
To study the effect of miR-144 on metastasis ability of LSCC cells, migration and invasion assays were performed in Hep2 cells transfected with miR-144 or miR-NC by wound healing and invasion chamber assay, respectively. It was found that restoration of miR-144 substantially inhibited the migration and invasion capabilities of HepG2 cells (Figure 3A, 3B).
Figure 3.
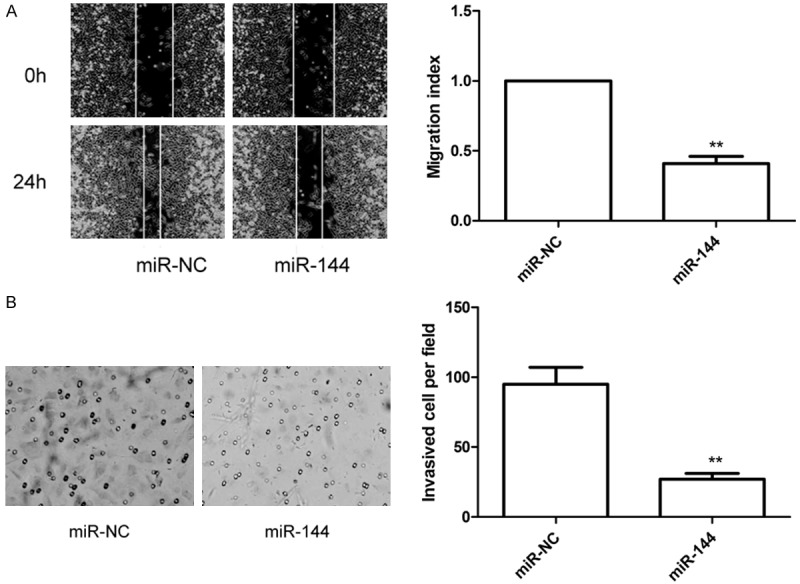
miR-144 overexpression inhibits migration and invasion in Hep2 cells. A. Would healing assay were performed to determined cell migration in Hep2 cells transfected with miR-144 mimic or miR-NC. B. Transwell invasion assay were performed to determined cell invasion in Hep2 cells transfected with miR-144 mimic or miR-NC. *P < 0.05, **P < 0.01 versus miR-NC.
IRS1 was a direct target of miR-144
To explore the target of miR-144 in LSCC, prediction algorithm (Targetscan6.2 and miRanda) was used. IRS1 was predicted to be a potential target of miR-144 (Figure 4A). Luciferase activity assay further revealed that miR-144 overexpression only significantly suppressed the luciferase activity of the Wt- 3’-UTR of IRS1 in Hep2 cells (Figure 4B), while the activity of the Mut -3’ UTR IRS1 site was not change (Figure 4B). Moreover, overexpression of miR-144 remarkably inhibited IRS1 expression on mRNA level (Figure 4C) and protein expression (Figure 4D). We also found that overexpression of miR-144 remarkably inhibited p-AKT expression (Figure 4D), an IRS1 downstream protein, suggesting that overexpression of miR-144 inhibited AKT pathway activation. We also investigated IRS1 mRNA expression in 24 pairs LSCC tissues and corresponding normal tissue by qRT-PCR. It was found that IRS1 expression on mRNA levels were upregulated in LSCC tissues (Figure 4E), and its expression was inversely correlated with miR-144 expression by two-tailed Pearson’s correlation (Figure 4F; r = -0.433, P < 0.05).
Figure 4.
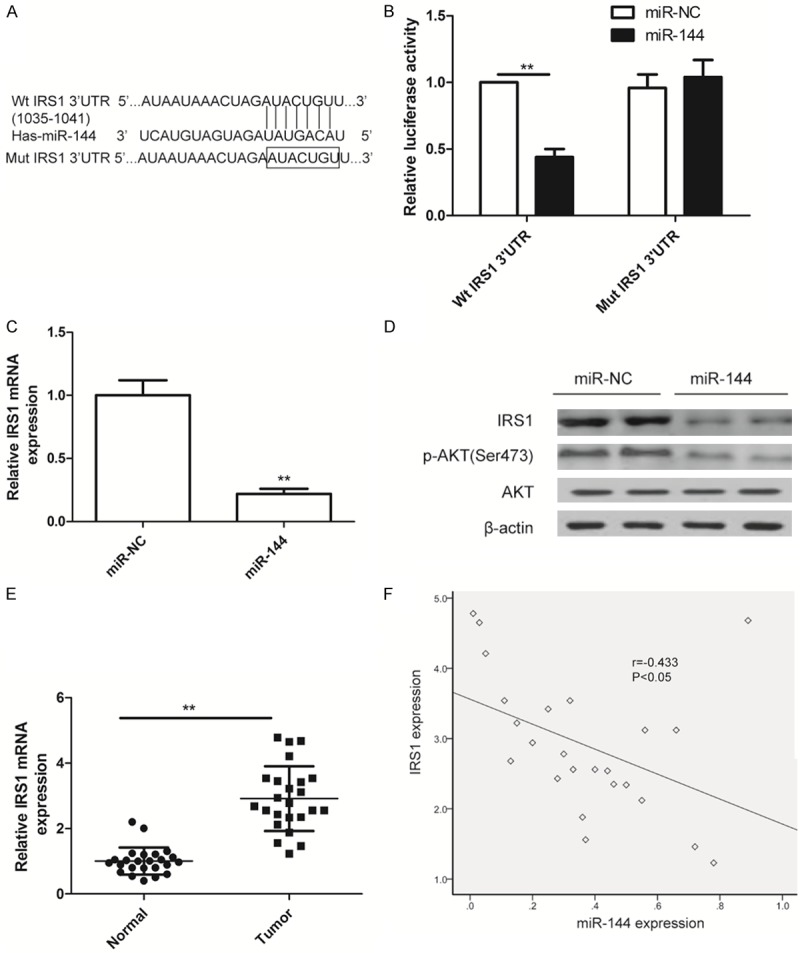
IRS1 is a direct target of miR-144a. A. The predicted binding sites for miR-144 in the 3’UTR of IRS1 and the mutations in the binding sites are shown. B. The luciferase activity of the wild type IRS1 3-UTR (Wt) and mutant IRS1 3’UTR (Mut) co-transfected with miR-144 or miR-NC was determined. C. IRS1 expression on mRNA level in Hep2 cells transfected with miR-144 mimic or miR-NC were determined by qRT-PCR. D. IRS1, AKT and p-AKT protein expression were determined in Hep2 cells transfected with miR-144 mimic or miR-NC by western blot. qRT-PCR and western blot data were normalized to β-actin. *P < 0.05, **P < 0.01 versus miR-NC. E. qRT-PCR determined IRS1 mRNA expression in 24 cases of LSCC tissues and matched normal tissues. The β-actin was used as an internal control. *P < 0.05, **P < 0.01 versus normal tissues. F. The reverse relationship between IRS1 mRNA expression and miR-144 expression was determined by Pearson’s correlation.
Downregulation of IRS1 had similar effects of miR-144 ovexpression in Hep2 cells To investigate the biological functions of IRS1 on LSCC cells, Hep2 cells were transfected with si-IRS1 or the si-NC, and found that knockdown IRS1 by si-IRS1 could obviously decreased IRS1 expression on mRNA level (Figure 5A) and protein level (Figure 5B). We also found that downregulation of IRS1 in Hep2 cells significantly inhibited cell proliferation and colony formation, migration and invasion (Figure 5C-F), suggesting that downregulation of IRS1 mimicked the effect of miR-144 overexpression on LSCC cells.
Figure 5.
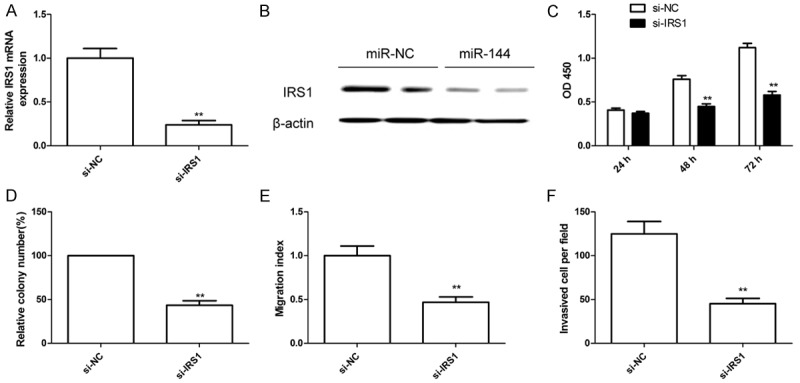
Downregulation of IRS1 had similar effects of miR-144 ovexpression in Hep2 cells. (A and B) IRS1 expression on mRNA level (A) and protein level (B) were detected in Hep2 cells transfected with si-IRS1 or si-NC. β-actin was used as an internal control. (C-F) Cell proliferation (C), colony formation (D), migration (E) and invasion (F) were determined in Hep2 cells transfected with si-IRS1 or si-NC. *P < 0.05, **P < 0.01 versus si-NC group.
IRS1 overexpression partially attenuated the tumor suppressive effects of miR-144 in Hep2 cells
To further confirm that the effect of miR-144 on Hep2 cells was dependent on downregulation of IRS1 expression, overexpression plasmid (pcDNA3.1-IRS1) was transfected into the Hep2 cells with high levels of miR-144 or miR-NC to counteract the function of miR-144. qRT-PCR and western blot assay showed that overexpression IRS1 plasmid could increase IRS1 expression on mRNA level (Figure 6A) and protein level (Figure 6B) in Hep2 cells. Then we analyzed the cell proliferation, colony formation, migration and invasion in the above groups, respectively. We found that overexpression of IRS1 in Hep2 cells could partially reverse the suppressive effect of miR-144 on cell proliferation, colony formation, migration and invasion (Figure 6C-F). These data indicated that miR-144 exerts tumor suppressive effects on LSCC cells partially through inhibition of IRS1 expression.
Figure 6.
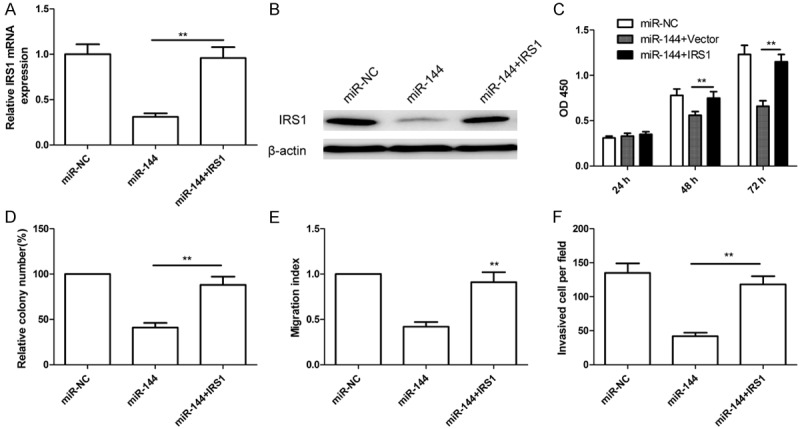
IRS1 overexpression partially attenuated the tumor suppressive effects of miR-144 in Hep2 cells. (A and B) Overexpression IRS1 plasmid (pcDNA3.1-IRS1) were transfected into the Hep2 cells with high levels of miR-144 or miR-NC, IRS1 expression on mRNA level (A) and protein level (B) were determined in above cells. β-actin was used as an internal control. (C-F) Cell proliferation (C), colony formation (D), migration (E) and invasion (F) were determined in above cells. *P < 0.05, **P < 0.01 versus miR-144.
miR-144 suppresses the tumor growth through inhibiting IRS1 in vivo
We also investigated the effects of miR-144 on tumor growth of Hep2 cells through an in vivo tumorigenesis assays. The Hep2 stably expression miR-144 or miR-NC was implanted subcutaneously into nude mice to allow tumor formation, respectively. As shown in Figure 7A-C, the mice inoculated with Hep2 cells with a high level of miR-144 had a significant reduction in the tumor volumes and weight, compared with that inoculated with miR-NC-transfected cells. In addition, the expression level of miR-144 in tumor tissues with Hep2/miR-144 transfection group was higher than that in tumor tissues with Hep2/miR-NC transfection group (Figure 7D), while IRS1 expression was decreased both mRNA level and protein level in Hep2/miR-144 transfection group compared to Hep2/miR-NC transfection group (Figure 7E and 7F). These data suggested that enforced expression of miR-144 can significantly suppressed LSCC cell growth in vivo through inhibiting IRS1 expression.
Figure 7.
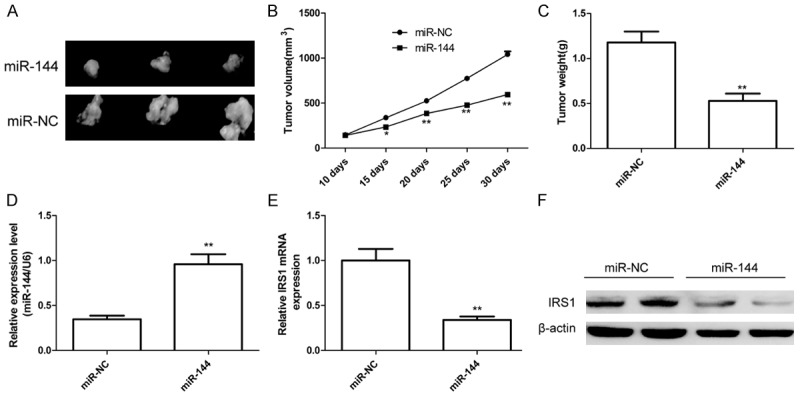
MiR-144 suppresses the tumor growth through inhibiting IRS1 in vivo. (A) Representative photographs of dissected tumors from nude mice. (B) The curve of tumor growth was shown. (C) Tumor weights were measured. (D) qRT-PCR determined miR-144 expression in tumor tissue. (E and F) IRS1 expression on mRNA level (E) and protein level (F) in tumor tissue were determined in tumor tissues. β-actin was used to an internal control. *P < 0.05, **P < 0.01 versus miR-NC.
Discussion
Numerous miRNAa have been identified to play significant roles in the pathogenesis of LSCC through regulating tumor cell proliferation, metastasis, invasion and apoptosis [10]. For example, Tian et al. reported that miR-27a acts as an oncogene in laryngeal squamous cell carcinoma through down-regulation of PLK2 [18]. Xu et al. demonstrated that miR-106b could increase the proliferation and invasion of laryngeal carcinoma cells of through targeting RUNX3, suggesting miR-106 acted as oncogene in LSCC [19]. Luo et al. found that miR-375 can act as tumor suppressor, and significantly inhibit LSCC cell proliferation, migration, and invasion and promote apoptosis to suppress tumor via IGF1R-mediated AKT signaling pathway [20]. Li et al. showed that miR-30b significantly promoted p53-mediated tumor cell apoptosis in HEp-2 cells, and increased the anti-tumor and pro-apoptotic effect of Ad-p53 in HEp-2 implanted nude mice [21]. Here, we found that miR-144 expression was downregulated in LSCC tissues, and that miR-144 overexpression inhibited proliferation, colony formation, migration and invasion in vitro, and suppresses tumor growth in vivo by targeting IRS1. These will be helpful for the better understanding of LSCC carcinogenesis and could represent a new therapeutic target for the treatment of LSCC.
Several studies have shown that miR-144 was down-regulated and functioned as a tumor suppressor in lung cancer [11], osteosarcoma [12], hepatocellular carcinoma [13], thyroid cancer [14], bladder cancer [15] and colorectal carcinoma [16]. However, a report demonstrated that miR-144 could promote cell proliferation, migration, and invasion in nasopharyngeal carcinoma (NPC) through targeting phosphatase and tensin homolog (PTEN) expression [22]. These inconsistent results suggested that dysregulation of miR-144 in various cancers may be dependent on the tumor tissue-specific. Here, our result showed that miR-144 expression was downregulated in LSCC tissues, restoration of miR-144 significantly inhibited LSCC growth in vitro and in vivo, suggesting that miR-144 functions as a tumor suppressor in LSCC.
IRS1, a docking protein, can serve as the key molecule in mediating the metabolic and mitogenic effects of insulin in peripheral tissues by transmitting the signals from the insulin receptor to the down-stream enzymes, such as PI3K, Akt, and PDK-1 [23]. It has been reported that IRS1 expression was upregulated in various cancers [24]. Increasing evidence has been showed that IRS1 could act as oncogene, and involve in tumors initiation and progression, tumor angiogenesis and chemosensitivity by regulating downstream pathway, such as PI3K/AKT and MAPK signal pathway [23,25-27]. In addition, it has been showed IRS1 was target gene of several miRNAs, such as miR-126 [28], miR-370 [29], miR-128 [17], miR-23a [30] and miR-145 [31]. In this study, we confirmed IRS1 was a direct target of miR-144 by luciferase reporter assay. Restoration of miR-144 expression inhibited IRS1 expression on mRNA level and protein level in Hep2 cells, as well as decreased downstream protein expression. Our results also showed that IRS1 expression was upregulated in LSCC tissues and its expression inversely correlated with miR-144 expression. Of note, we found that downregulation of IRS1 had similar the effects of miR-144 overexpression, and upregulation of IRS1 partially reversed the inhibitory effects of miR-144. These results might suggest that miR-144 exerted tumor suppressor in laryngeal squamous cell carcinoma through repression of IRS1 expression.
In conclusion, the present study first showed that miR-144 was significantly decreased in LSCC tissues, and that restoration miR-144 expression inhibits LSCC cell proliferation, colony formation, migration and invasion, as well as suppresses tumor growth in vivo at least partially through inhibiting IRS1 expression. These results suggested that miR-144 functioned as a tumor suppressor in LSCC by targeting IRS1, and that miR-144 might serve as a potential target for laryngeal squamous cell carcinoma treatment.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph E, Dyckhoff G, Becher H, Dietz A, Ramroth H. Effects of tumour stage, comorbidity and therapy on survival of laryngeal cancer patients: a systematic review and a metaanalysis. Eur Arch Otorhinolaryngol. 2011;268:165–179. doi: 10.1007/s00405-010-1395-8. [DOI] [PubMed] [Google Scholar]
- 3.Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;41:673–695. v. doi: 10.1016/j.otc.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNAMediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 6.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–44. [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Li Z. The role of microRNAs expression in laryngeal cancer. Oncotarget. 2015;6:23297–23305. doi: 10.18632/oncotarget.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Li P, Li J, Wang Y, Du Y, Chen X, Zang W, Wang H, Chu H, Zhao G, Zhang G. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem. 2015;35:997–1007. doi: 10.1159/000369755. [DOI] [PubMed] [Google Scholar]
- 12.Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu J, Wang K, Liu D, Zhang X, Yin W. The downregulation of miR-144 is associated with the growth and invasion of osteosarcoma cells through the regulation of TAGLN expression. Int J Mol Med. 2014;34:1565–1572. doi: 10.3892/ijmm.2014.1963. [DOI] [PubMed] [Google Scholar]
- 13.Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumour Biol. 2014;35:10759–10764. doi: 10.1007/s13277-014-2017-7. [DOI] [PubMed] [Google Scholar]
- 14.Guan H, Liang W, Xie Z, Li H, Liu J, Liu L, Xiu L, Li Y. Down-regulation of miR-144 promotes thyroid cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine. 2015;48:566–574. doi: 10.1007/s12020-014-0326-7. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 16.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, Wakabayashi G, Mori M, Mimori K. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Shi B, Huang K, Fan G. MicroRNA-128 suppresses cell growth and metastasis in colorectal carcinoma by targeting IRS1. Oncol Rep. 2015;34:2797–2805. doi: 10.3892/or.2015.4251. [DOI] [PubMed] [Google Scholar]
- 18.Tian Y, Fu S, Qiu GB, Xu ZM, Liu N, Zhang XW, Chen S, Wang Y, Sun KL, Fu WN. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2 in laryngeal carcinoma. BMC Cancer. 2014;14:678. doi: 10.1186/1471-2407-14-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen S, Wang B. MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett. 2013;587:3166–3174. doi: 10.1016/j.febslet.2013.05.069. [DOI] [PubMed] [Google Scholar]
- 20.Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS, Yang W, Fu QL, Lei W. miR-375 suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. Biomed Res Int. 2014;2014:374598. doi: 10.1155/2014/374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Wang B. Overexpression of microRNA-30b improves adenovirus-mediated p53 cancer gene therapy for laryngeal carcinoma. Int J Mol Sci. 2014;15:19729–19740. doi: 10.3390/ijms151119729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL, Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, Fu L. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–463. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- 23.Saltiel AR, Pessin JE. Insulin signaling pathways in time and space. Trends Cell Biol. 2002;12:65–71. doi: 10.1016/s0962-8924(01)02207-3. [DOI] [PubMed] [Google Scholar]
- 24.Reiss K, Del Valle L, Lassak A, Trojanek J. Nuclear IRS-1 and cancer. J Cell Physiol. 2012;227:2992–3000. doi: 10.1002/jcp.24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergmann U, Funatomi H, Kornmann M, Beger HG, Korc M. Increased expression of insulin receptor substrate-1 in human pancreatic cancer. Biochem Biophys Res Commun. 1996;220:886–890. doi: 10.1006/bbrc.1996.0500. [DOI] [PubMed] [Google Scholar]
- 26.Dearth RK, Cui X, Kim HJ, Kuiatse I, Lawrence NA, Zhang X, Divisova J, Britton OL, Mohsin S, Allred DC, Hadsell DL, Lee AV. Mammary tumorigenesis and metastasis caused by overexpression of insulin receptor substrate 1 (IRS1) or IRS-2. Mol Cell Biol. 2006;26:9302–9314. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baserga R. The contradictions of the insulinlike growth factor 1 receptor. Oncogene. 2000;19:5574–5581. doi: 10.1038/sj.onc.1203854. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 29.Chang KW, Chu TH, Gong NR, Chiang WF, Yang CC, Liu CJ, Wu CH, Lin SC. miR-370 modulates insulin receptor substrate-1 expression and inhibits the tumor phenotypes of oral carcinoma. Oral Dis. 2013;19:611–619. doi: 10.1111/odi.12046. [DOI] [PubMed] [Google Scholar]
- 30.Cao M, Li Y, Lu H, Meng Q, Wang L, Cai L, Dong X. MiR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J Cancer Res Clin Oncol. 2014;140:1661–1670. doi: 10.1007/s00432-014-1725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z, Wu J, Zhou Y. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446:1255–1260. doi: 10.1016/j.bbrc.2014.03.107. [DOI] [PubMed] [Google Scholar]


