Abstract
Background. Human rhinoviruses (HRVs) are frequently detected in children with acute respiratory illnesses (ARIs) but also in asymptomatic children. We compared features of ARI with HRV species (A, B, C) and determined genotypes associated with repeated HRV detections within individuals.
Methods. We used clinical data and respiratory samples obtained from children <3 years old during weekly active household-based surveillance. A random subset of samples in which HRV was detected from individuals during both ARI and an asymptomatic period within 120 days of the ARI were genotyped. Features of ARI were compared among HRV species. Concordance of genotype among repeated HRV detections within individuals was assessed.
Results. Among 207 ARI samples sequenced, HRV-A, HRV-B, and HRV-C were detected in 104 (50%), 20 (10%), and 83 (40%), respectively. Presence of fever, decreased appetite, and malaise were significantly higher in children with HRV-B. When codetections with other viruses were excluded (n = 155), these trends persisted, but some did not reach statistical significance. When 58 paired sequential HRV detections during asymptomatic and ARI episodes were sequenced, only 9 (16%) were identical genotypes of HRV.
Conclusions. Clinical features may differ among HRV species. Repeated HRV detections in young children frequently represented acquisition of new HRV strains.
Keywords: children, human rhinovirus, Peru, respiratory illness
Human rhinoviruses (HRVs) are frequently detected during acute respiratory illnesses (ARIs) in children [1–4]. Some HRV infections have been associated with severe lower respiratory tract infection (LRTI), including bronchiolitis and pneumonia [5, 6]. Human rhinoviruses are divided into 3 species, HRV-A, HRV-B, and HRV-C, with over 150 distinct genotypes currently described within those species [7]. The pathogenic potential of HRV may vary by species. In prior studies, HRV-C has been associated with more severe manifestations of wheezing and asthma exacerbations [8–12]. In addition to ARI in which HRV is the sole pathogen detected, HRVs are also frequently codetected with other pathogens and may contribute to disease associated with other viral or bacterial infections [5, 6]. Whether other environmental, geographic, or host factors influence the pathogenicity of HRV is unknown, supporting the need for assessment of these factors in diverse settings.
Detection of HRV in a patient with ARI does not always confirm an etiologic role for the virus. Human rhinoviruses are also frequently detected in asymptomatic children [13–19], complicating the clinical interpretation of HRV detections in children with ARI. Longitudinal assessments and molecular characterization of HRV detections in the same individual during asymptomatic and symptomatic periods are needed to clarify the clinical significance of HRV detection in young children over time. However, very few longitudinal studies have evaluated the molecular epidemiology of HRV detections, and most have been restricted to selected urban, high population-density areas with surveillance performed in healthcare facilities [14, 18, 20–22].
Our prospective household-based cohort study of Respiratory Infections in Andean Peruvian Children (RESPIRA-PERU) [23, 24] is uniquely suited to provide information about the detection of HRV during symptomatic ARI and asymptomatic periods from sequential respiratory samples obtained from the same child during prospective household surveillance in a rural, high-altitude setting. We sought to compare clinical features among ARI with different HRV species and to determine the concordance of genotypes during repeated HRV detections.
METHODS
Study Design
This study uses data from RESPIRA-PERU, a prospective cohort study designed to evaluate the epidemiology, clinical features, and impact of environmental factors on ARIs in young Andean children <3 years of age [6, 24–28]. The median age at enrollment was 4.6 months (interquartile range, 0.5–17.1). Weekly active household-based surveillance for ARI was conducted from May 1, 2009 through September 30, 2011 by trained field workers, and clinical data and respiratory samples were systematically collected from the participants. This study was approved by the Vanderbilt Institutional Review Board and by the Ethics Committee of the Instituto de Investigacion Nutricional. Written informed consent was provided by caregivers of participants before enrollment or initiation of any study procedures.
Study Setting
The RESPIRA-PERU study was conducted in the Province of San Marcos, Department of Cajamarca, located in the northern highlands of Peru. Study households were located between 1500 and 4000 meters above sea level. The population is primarily rural, with low income, low educational level, and limited access to healthcare services, as previously described [24]. To ensure a representative sample of the communities in the study area, broad selection criteria were used for enrollment, consisting of the following: (1) families with children aged <3 years, and (2) intention to remain in the study area for the next year.
Acute Respiratory Illness Definitions
Acute respiratory illness was defined as the presence of either cough or fever [24, 29, 30]. Acute respiratory illness severity was assessed by trained field workers if the child experienced cough or fever on the visit day or the day prior. Children were considered to have a severe ARI if 1 or more of the general WHO danger signs [31] was present, including signs of LRTI such as tachypnea, audible wheezing, intercostal and subcostal retractions, grunting, nasal flaring, stridor, or cyanosis.
Respiratory Sample Collection
Nasal Swab Collection
During weekly household visits, field workers collected a nasal swab (NS) from any child with ARI following procedures previously reported [30]. In brief, 1 nonflocked polyester-tipped swab was placed into each nostril sequentially and rotated beneath the turbinates to collect epithelial cells and absorb secretions. After each nostril was swabbed, the swab was inserted into a tube with Remel M4RT viral transport medium.
Nasopharyngeal Sample Collection
Nasopharyngeal (NP) swabs were collected monthly from each child under observation, whether or not respiratory symptoms were present, and during complicated ARI, defined by the presence of danger signs established by the World Health Organization (WHO) [32]. Nasopharyngeal samples were processed according to WHO recommendations for identifying pneumococcal colonization [33, 34]. In brief, samples were collected with a nonflocked deep NP Rayon swab and then immediately placed in 1 mL Skim Milk-Tryptone-Glucose-Glycerol transport medium.
Despite slightly different collection methods, prior studies by our group and others have demonstrated a very high agreement (89%–99%) in the detection of respiratory viruses between NS and NP samples collected in this manner [27]. All NP and NS specimens were transported in cold packs to the laboratory within 8 hours of collection and preserved at −70°C before diagnostic testing.
Selection of Samples
All NS samples collected during ARI episodes underwent comprehensive viral testing by real-time reverse transcription-polymerase chain reaction (RT-PCR). A subset of NP samples collected during asymptomatic periods were also randomly selected to undergo comprehensive viral testing by real-time RT-PCR [17]. A subset of HRV-positive ARI samples underwent genetic sequencing for species and genotype identification. In addition, we identified a subset of paired observations in which an asymptomatic NP sample collected within 120 days before or after an HRV ARI (the “index ARI”) tested positive for HRV. We performed HRV genotypic characterization of these within-child paired ARI-asymptomatic samples to determine whether they represented persistent detection of identical strains of HRV or acquisition of new HRV strains.
Laboratory Procedures
RNA was extracted from frozen specimen aliquots using an automated nucleic acid extraction method (MagMax Express 96, Life Technologies/Applied Biosystems), and purified RNA aliquots were stored at −80°C until analysis. Reverse transcription-PCR was performed using primers that amplified a fragment encompassing the VP4/VP2 region. Amplified fragments were sequenced on an ABI 3730 × 1 DNA analyzer. Sequence alignment was performed with published HRV sequences obtained from GenBank using MacVector version 13 (MacVector). Phylogenetic analysis was performed and the final tree generated using the neighbor-joining algorithm in Geneious version 8.1 (Biomatters).
Statistical Analysis
To compare the clinical features of ARI among HRV species, Fisher exact or Kruskal-Wallis tests were used as appropriate for categorical or continuous variables, respectively. Concordance of species detection among ARI-asymptomatic sample pairs was performed using the McNemar's exact test. Human rhinovirus genotypes from ARI and asymptomatic periods within an individual were compared and described.
RESULTS
Clinical Features of Acute Respiratory Illness by Human Rhinovirus Species
Two hundred sixty-three HRV-positive NS samples collected during ARI were randomly selected for genotypic characterization. Of these, 210 (80%) were successfully amplified and sequenced. Three isolates from samples that tested positive for HRV by RT-PCR during ARI were identified as enteroviruses by sequencing. Thus, 207 HRV ARI detections successfully underwent genotypic characterization. In 80 of 207 (39%) episodes of ARI, both cough and fever were present, whereas in 87 of 207 (42%) ARI episodes only cough in the absence of fever was noted, and in 40 of 207 (19%) episodes there was only fever. Of these 207 HRV ARIs, 104 (50.2%) were HRV-A, 20 (9.7%) were HRV-B, and 83 (40.1%) were HRV-C. The median age at time of HRV ARI differed significantly among species, with children with HRV-C ARI significantly older than children with HRV-A or HRV-B ARI. When all HRV ARI were considered, the clinical features of ARI varied significantly between HRV species, with HRV-B more likely to be associated with fever (P = .048), malaise (P = .006), and decreased appetite (P = .009) than HRV-A or HRV-C (Table 1). The prevalence of cough was high and did not differ between HRV species. Acute respiratory illness severity assessments were performed in 123 of 207 (59.4%) at the time of the home visit or the prior day. Only 5 of these 123 (3.4%) HRV-associated ARIs were severe, and they occurred in children 7, 10, 16, 25, and 26 months of age. The prevalence of severe ARI did not differ significantly among HRV species (P = .540). There were no severe ARI associated with HRV-B. Signs of LRTI, including wheezing, were uncommon and did not differ significantly between species. As previously described [6], detections of HRV occurred during each calendar month with no discernible seasonal pattern.
Table 1.
Clinical Features of HRV ARI by Species (HRV-A, HRV-B, HRV-C)
| Characteristics of ARI | All HRV ARI |
HRV ARI Excluding Codetections With Other Viruses |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Any HRV (n = 207) |
HRV-A (n = 104; 50.2%) |
HRV-B (n = 20; 9.7%) |
HRV-C (n = 83; 40.1%) |
P Valuea | Any HRV (n = 155) |
HRV-A (n = 78; 50.3%) |
HRV-B (n = 13; 8.4%) |
HRV-C (n = 64; 41.3%) |
P Valuea | |
| Age (m), median (IQR) | 17 (10–27) | 15.5 (9–25) | 15.5 (9.5–24.5) | 21 (12–29) | .026 | 16 (9–25) | 13 (8–23) | 15 (9–28) | 19.5 (10.5–28.5) | .023 |
| Fever, no. (%) | 120 (58.0) | 53 (51.0) | 16 (80.0) | 51 (61.4) | .040 | 86 (55.5) | 39 (50.0) | 11 (84.6) | 36 (56.2) | .064 |
| Duration fever, median d (IQR) | 2 (1–3) | 2 (1–2) | 2 (1–3) | 2 (1–3) | .464 | 2 (1–2) | 2 (1–2) | 2 (1–3) | 2 (1–3) | .629 |
| Cough, no. (%) | 167 (80.7) | 84 (80.8) | 14 (70.0) | 69 (83.1) | .410 | 124 (80.0) | 61 (78.2) | 8 (61.5) | 55 (85.6) | .124 |
| Duration cough, median d (IQR) | 5 (3–8) | 4 (3–7) | 6.5 (3–8) | 4 (3–8) | .758 | 4 (3–8) | 5 (3–7) | 4.5 (2–7) | 4 (3–8) | .682 |
| Decreased appetite, no. (%) | 41 (19.8) | 24 (23.1) | 8 (40.0) | 9 (10.8) | .007 | 27 (17.4) | 14 (17.9) | 5 (38.4) | 8 (12.5) | .078 |
| Duration decreased appetite, median d (IQR) | 3 (2–6) | 3 (2–6) | 2 (1–3) | 4 (3–10) | .041 | 3 (2–7) | 3.5 (3–6) | 1 (1–2) | 5 (3–10.5) | .025 |
| Malaise, no. (%) | 55 (26.6) | 30 (28.8) | 10 (50.0) | 15 (18.1) | .011 | 40 (25.8) | 20 (25.6) | 7 (53.8) | 13 (20.3) | .055 |
| Duration malaise, median d (IQR) | 2 (1–4) | 2 (1–4) | 2 (1–3) | 2 (1–4) | .750 | 2 (1–3) | 2 (1–3.5) | 2 (1–2) | 2 (1–3) | .571 |
| Rhinorrhea, no. (%) | 101 (48.8) | 50 (48.0) | 7 (35.0) | 44 (53.0) | .344 | 73 (47.0) | 36 (46.1) | 3 (23.0) | 34 (53.1) | .137 |
| Codetection with ≥1 other virus, no. (%) | 52 (25.1) | 26 (25.0) | 7 (35.0) | 19 (22.9) | .533 | NA | NA | NA | NA | NA |
| Taken to a health center | 59 (28.5) | 33 (31.7) | 5 (25.0) | 21 (25.3) | .586 | 40 (25.8) | 25 (32.0) | 2 (15.3) | 13 (20.3) | .210 |
| ARI severity assessed, no. (%) | 123 (59.4) | 58 (55.8) | 10 (50.0) | 55 (66.3) | 88 (56.8) | 40 (51.3) | 4 (30.8) | 44 (68.8) | ||
| ARI severe | 5 (4.1) | 1 (1.7) | 0 (0.0) | 4 (7.3) | .366 | 3 (3.4) | 0 (0.0) | 0 (0.0) | 3 (6.8) | .342 |
| Tachypnea | 1 (0.8) | 0 (0.0) | 0 (0.0) | 1 (1.8) | .533 | 1 (1.1) | 0 (0.0) | 0 (0.0) | 1 (2.3) | 1.000 |
| Stridor | 1 (0.8) | 0 (0.0) | 0 (0.0) | 1 (1.8) | .528 | 1 (1.1) | 0 (0.0) | 0 (0.0) | 1 (2.3) | 1.000 |
| Wheezing | 3 (2.4) | 1 (1.7) | 0 (0.0) | 2 (3.6) | .700 | 1 (1.1) | 0 (0.0) | 0 (0.0) | 1 (2.3) | 1.000 |
Abbreviations: ARI, acute respiratory illness; HRV, human rhinovirus; IQR, interquartile range; NA, not applicable.
a Fisher exact or Kruskal-Wallis test was used as appropriate for categorical or continuous variables, respectively. P values <.05 were considered statistically significant (indicated in bold).
Viral Codetection During Acute Respiratory Illness
Human rhinovirus was codetected with at least 1 other viral pathogen in 52 of 207 (25.1%) ARI samples. The viruses most frequently codetected with HRV were adenovirus (AdV), parainfluenza virus (PIV), and respiratory syncytial virus (RSV), in 26 of 207 (12.6%), 15 of 207 (7.2%), and 11 of 207 (5.3%) ARI samples, respectively. Of the 5 severe ARI in which HRV was detected, 2 (40%) were associated with codetection of other pathogens (PIV3 and AdV).
When HRV codetections with other viruses were excluded in secondary analyses (n = 155), the trends in differences in the clinical features of ARI among HRV species persisted, but some were no longer statistically significant. The reduction in the sample size when codetections were excluded in this subanalysis limited the power and precision of these assessments.
Asymptomatic Human Rhinovirus Detection
Ninety-nine HRV-positive NP samples collected during asymptomatic periods within 120 days of an HRV ARI were identified for sequencing. Of these, 77 of 99 (78%) were successfully amplified and sequenced. Twenty-nine (38%) of 77 asymptomatic samples sequenced were species A, 22 (29%) were B, and 26 (34%) were C. Other viruses were codetected with HRV in 19 of 99 (17%) of the asymptomatic samples. The majority of these codetections were with AdV (17 of 19; 89%). Detections of other viruses (MPV, PIV, RSV, influenza) were very uncommon during asymptomatic periods [17].
Paired Human Rhinovirus Detections
For this analysis, we focused on paired observations in which HRV was detected in both an index ARI and an asymptomatic sample collected within 120 days either before or after the index ARI. Human rhinovirus genotyping data were available for 58 paired HRV detections, including 5 index ARI samples with 2 NP samples collected during asymptomatic periods. Among the 58 pairs, there were no significant differences in relative viral load concentrations, as approximated by RT-PCR cycle threshold values, between ARI detections (mean, 27.3 seconds; 95% confidence interval [CI], 26.0–28.6) and asymptomatic (mean, 27.6; 95% CI, 26.2–29.0) detections (P = .72).
Of 58 total paired detections, 25 (43%) were concordant by species (eg, the same species was detected during both the ARI and the asymptomatic period) (Table 2). Fifteen of the concordant pairs by species were A-A detections, 5 were B-B detections, and 5 were C-C detections. The probability of detecting the same species during ARI and asymptomatic periods was not statistically different across HRV species (HRV-A, P = .0522; HRV-B, P = .0574; HRV-C, P = .6900). However, when genetic sequences were compared to determine similarity, the majority (49 of 58) of repeated HRV detections within individuals represented acquisition of new strains, rather than persistence of identical genotypes. Only 9 (16%) of the 58 total paired detections represented presence of identical genotypes of HRV during both the ARI and the asymptomatic period. In 8 of these 9 (89%) pairs, concordant genotypes occurred within 30 days of each other, whereas only 15 of 49 (31%) paired detections associated with different genotypes occurred within 30 days.
Table 2.
Patterns of 58 Pairs of HRV Index ARI and Asymptomatic HRV Detections
| Species of HRV in Index ARI and Asymptomatic HRV Detection |
Asymptomatic HRV Detected Before HRV Index ARI |
Asymptomatic HRV Detected After HRV Index ARI |
||||
|---|---|---|---|---|---|---|
| N pairs | Asymptomatic species | Index ARI species | −120 to −31 d | −30 to −8 d | 8 to 30 d | 31 to 120 d |
| 15a | A | A | A,A | (A),A,A | (A),A | (A),A,A,A,A,A,A,A |
| 7 | A | B | B,B | B | B | B,B,B |
| 12 | A | C | C | C,C | C | C,C,C,C,C,C,C,C |
| 1 | B | A | A | |||
| 5a | B | B | (B),(B) | (B),(B),(B) | ||
| 2 | B | C | C | C | ||
| 7 | C | A | A,A,A,A,A | A | A | |
| 4 | C | B | B | B,B,B | ||
| 5a | C | C | C | (C) | C | C,C |
Abbreviations: ARI, acute respiratory illness; HRV, human rhinovirus.
Concordant HRV species among pairs are indicated by shaded rows.
a Of 58 total paired HRV detections, 9 represented concordant genotypes, indicated by (). Three of the paired A-A species detections represented identical HRV genotypes. Asymptomatic samples were collected 22 days before, 8 days after, and 36 days after the index ARI. All 5 repeated B-B species detections represented identical HRV genotypes between ARI and asymptomatic paired sample (asymptomatic samples collected 15 days before, 14 days before, 8 days after, 21 days after, and 29 days after index ARI). One of the paired C-C detections represented identical HRV genotypes (asymptomatic sample collected 15 days before index ARI).
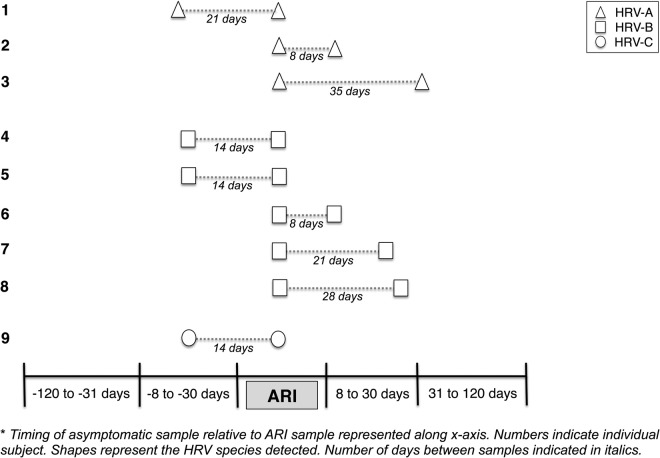
Of 15 paired A-A detections, only 3 pairs were identical genotypes (Figure 1, Table 2). In these 3 pairs, asymptomatic samples were collected 22 days before, 8 days after, and 36 days after the index ARI. All 5 paired B-B detections represented identical genotypes, and each of the asymptomatic detections with identical genotypes to ARI occurred within 30 days before or after the ARI. Only 1 of the 5 C-C paired detections represented identical genotypes, with the asymptomatic detection occurring 15 days before ARI sample collection. In 4 of 9 identical pairs, the asymptomatic HRV detection occurred before ARI sample collection. Human rhinovirus was codetected with another virus in 2 of 4 (1 PIV, 1 AdV) of these pre-ARI asymptomatic detections.
Figure 1.
Repeated detections of identical human rhinovirus (HRV) strains during acute respiratory illness (ARI) paired with asymptomatic period in individual subjects (n = 9 identical pairs).
Human Rhinovirus Diversity
Although several clinical differences were seen among HRV species, there was also substantial genetic diversity among viruses. Phylogenetic analysis was performed with sequences from multiple specimens (133 A, 42 B, 109 C). The phylogenetic tree (Supplementary Figure 1) depicts the wide distribution of HRV genotypes detected during both asymptomatic and ARI periods. In total, more than 100 different putative HRV genotypes were identified, although strict definitions for HRV typing have not been established. There was more genetic diversity among HRV-A and HRV-C than HRV-B. It is interesting to note that a number of clusters of identical or near-identical strains were detected (indicated by arcs in Supplementary Figure 1). These clusters likely represented a single virus spreading through the community during a brief period of a few months, despite the geographically dispersed nature of the rural communities in the study. Moreover, several of these clusters occurred concurrently, indicating that more than 1 virus was simultaneously circulating in the region.
DISCUSSION
We previously reported that HRV was frequently detected from the same children during both ARI (32%) and asymptomatic periods (31%) in the parent RESPIRA-Peru study. Several asymptomatic HRV detections occurred after the HRV ARI, suggesting that detection of HRV in asymptomatic periods may represent the same HRV genotype detected during ARI [17]. The current study extended those early observations using genotypic analysis of HRV detections. We showed circulation of all 3 HRV species, with HRV-B infections associated with more clinical symptoms than HRV-A or HRV-C when all HRV ARI were considered, and that HRV-C ARI occurred in older children relative to ARI with HRV-A or HRV-B. We also demonstrated that the vast majority of repeated detections of HRV-A or HRV-C within individual children were caused by new HRV strains, rather than long-term persistence of a single HRV genotype. In contrast, the same HRV-B genotypes were detected in both HRV-B ARI and asymptomatic samples frequently, suggesting variability in persistence of detections by species.
Our study found a highly heterogeneous and dynamic pattern of HRV acquisition and circulation among young children, with >100 different HRV genotypes identified in our study population. These findings are consistent with the few other longitudinal assessments of genotypic characterization of HRV detections in young children, which have demonstrated that there is high genetic diversity among HRV genotypes identified, and suggested that repeat HRV detections among young children often represent acquisition of new strains [14, 20, 21, 35]. In a recent study from an urban location in the United States, children were monitored from near birth to 12 months of age for serial rhinovirus infections [15]. In the study by Loeffelholz et al [15], persistence of the same genotype occurred in only 8 of 179 (4.5%) ARI, similar to our finding of only 1 of 58 (1.7%) asymptomatic detections in our cohort that persisted more than 30 days after an ARI. Taken together, these findings suggest that persistence of individual HRV genotypes beyond 30 days is infrequent in young children. In addition, the current study expands upon the Loeffelholz et al [15] study by evaluating detections in children up to 3 years of age. There were no differences seen in the features of HRV pathogenicity or patterns of acquisition of new genotypes in children <12 months of age compared with children 12 months or older. Although most of the other studies were mainly conducted in urban, highly populated settings, the similarities in observations with our study, which focused on rural, low-density, and relatively isolated communities [26], are remarkable.
Several studies have reported differences in the clinical features of HRV-C compared with HRV-A ARI, most notably increased wheezing and possibly more LRTI associated with HRV-C [11, 36–38]. Other comparisons of HRV-C and HRV-A have been more variable [39], with increased supplemental oxygen requirement associated with HRV-C in one study [11], but increased proportions of pneumonia or bronchiolitis with HRV-A in another [8]. However, these studies focused primarily on infants hospitalized with ARI. Moreover, comparisons with HRV-B infections are rare because in many previous studies, HRV-B typically comprised only 3%–8.5% of the HRV detections [8, 9, 11, 36]. However, one study reported lower odds of moderate to severe respiratory illness associated with HRV-B compared with HRV-A and HRV-C [40]. In our study, 20 (9.7%) of 207 HRV-associated ARI were HRV-B, enabling comparisons among all 3 species. Although the proportion of severe ARI did not differ significantly between HRV species, the overall prevalence of severe ARI was low in our household-based surveillance study. Fever, decreased appetite, and malaise occurred significantly more frequently with HRV-B ARI than with HRV-A or HRV-C ARI in both the presence and absence of viral codetections. The variability of findings regarding the clinical differences of ARI among HRV species in our study and others suggest that differences in severity or other clinical manifestations may be linked to certain viral genotypes, rather than species, although the substantial genetic HRV diversity in our study precluded further analysis of that point. It is interesting to note that HRV-B represented 29% of HRV detected during asymptomatic periods but only 10% of HRV detected during ARI. It is difficult to compare these observations with previous studies that conducted molecular genotyping of HRV detections, given that the proportions of HRV-B detection during ARI compared with asymptomatic periods were not directly reported [15, 35, 21]. However, this observation warrants further evaluation studies and may suggest different patterns of HRV species detection among symptomatic and asymptomatic periods.
Our findings must be interpreted in light of several limitations. The RESPIRA-PERU study conducted prospective weekly household-based surveillance, and most children were identified early in their disease with mild ARI. Few subjects were captured with severe ARI or LRTI symptoms, or wheezing, precluding comparisons with other studies that have evaluated children presenting to the emergency department or hospitalized with ARI. In addition, this study is restricted to identification of respiratory viruses, with no assessment of the concurrent presence of commensal and pathogenic bacteria in the nasopharynx, which may influence patterns of respiratory viral detection or viral pathogenicity [41–43].
This study also has several important strengths. First, this is one of few intensive prospective evaluations of respiratory illnesses in young children in a household-based surveillance setting, adding information that may more accurately capture the spectrum of HRV disease associated with mild-to-moderate respiratory illness. Second, of few studies that have provided longitudinal assessments of HRV detection in children over time [14–16, 20, 21, 44], ours is one of the only evaluations conducted in a rural, high-altitude setting, and one of few that provided detailed genotypic characterization of HRV detections. In addition, rather than using an external control group for comparison, the study collected prospective data from individual subjects over a period of longitudinal follow-up, allowing within-subject comparisons wherein each subject served as his own control, minimizing the role of intersubject variability.
CONCLUSIONS
Taken together, our findings suggest that the clinical features of ARI associated with HRV vary by HRV species. Repeated detections of HRV in young children frequently represented acquisition of new HRV strains rather than prolonged infection with the same strain.
Supplementary Material
Supplementary material is available online at Open Forum Infectious Diseases (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
We are indebted to the communities of San Marcos, Cajamarca Peru for participation in this study. We also acknowledge the approval and continuous support of the Cajamarca Health Region authorities. We are also indebted to the field workers and field supervisors whose efforts in difficult geographical areas and harsh weather conditions allowed the conduct of this study. The following persons worked on behalf of the study of Respiratory Infections in Andean Peruvian Children: Philip J. Budge and Yuwei Zhu (Vanderbilt University); Jorge E. Vidal and Keith P. Klugman (Emory University); Hector Verastegui and Stella M. Hartinger (Instituto de Investigacion Nutricional).
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was funded by a Vanderbilt University Clinical and Translational Science Award (grant UL1 RR024975) from the National Institutes of Health; an investigator initiated research grant from Pfizer (IIR WS1898786(0887X1-4492); to C. G. G.); the Thrasher Research Fund (grant 02832-9; to C. G. G.); and Early Career Award (to L. M. H.).
Potential conflicts of interest. C. G. G. has served as a consultant to Pfizer in unrelated work. M. R. G. receives grant funding from MedImmune. K. M. E. receives grant funding from Novartis in unrelated work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Collaborators: for the RESPIRA-PERU Group, Marie R. Griffin, John V. Williams, Leigh M. Howard, Kathryn M. Edwards, Philip J. Budge, Yuwei Zhu, Monika Johnson, Carlos G. Grijalva, Jorge E. Vidal, Keith P. Klugman, Hector Verastegui, Stella M. Hartinger, Ana I. Gil, and Claudio F. Lanata
References
- 1.Miller EK, Lu X, Erdman DD et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis 2007; 195:773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller EK, Williams JV, Gebretsadik T et al. Host and viral factors associated with severity of human rhinovirus-associated infant respiratory tract illness. J Allergy Clin Immunol 2011; 127:883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piotrowska Z, Vazquez M, Shapiro ED et al. Rhinoviruses are a major cause of wheezing and hospitalization in children less than 2 years of age. Pediatr Infect Dis J 2009; 28:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieninger E, Fuchs O, Latzin P et al. Rhinovirus infections in infancy and early childhood. Eur Respir J 2013; 41:443–52. [DOI] [PubMed] [Google Scholar]
- 5.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet 2011; 377:1264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budge PJ, Griffin MR, Edwards KM et al. A household-based study of acute viral respiratory illnesses in Andean children. Pediatr Infect Dis J 2014; 33:443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev 2013; 26:135–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwane MK, Prill MM, Lu X et al. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis 2011; 204:1702–10. [DOI] [PubMed] [Google Scholar]
- 9.Linder JE, Kraft DC, Mohamed Y et al. Human rhinovirus C: age, season, and lower respiratory illness over the past 3 decades. J Allergy Clin Immunol 2013; 131:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller EK, Edwards KM, Weinberg GA et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol 2009; 123:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller EK, Khuri-Bulos N, Williams JV et al. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol 2009; 46:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller EK, Mackay IM. From sneeze to wheeze: what we know about rhinovirus Cs. J Clin Virol 2013; 57:291–9. [DOI] [PubMed] [Google Scholar]
- 13.Self WH, Williams DJ, Zhu Y et al. Respiratory viral detection in children and adults: comparing asymptomatic controls and patients with community-acquired pneumonia. J Infect Dis 2016; 213:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Zalm MM, Wilbrink B, van Ewijk BE, Highly frequent infections with human rhinovirus in healthy young children: a longitudinal cohort study. J Clin Virol 2011; 52:317–20. [DOI] [PubMed] [Google Scholar]
- 15.Loeffelholz MJ, Trujillo R, Pyles RB et al. Duration of rhinovirus shedding in the upper respiratory tract in the first year of life. Pediatrics 2014; 134:1144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol 2006; 78:644–50. [DOI] [PubMed] [Google Scholar]
- 17.Howard LM, Johnson M, Williams JV et al. Respiratory viral detections during symptomatic and asymptomatic periods in young Andean children. Pediatr Infect Dis J 2015; 34:1074–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byington CL, Ampofo K, Stockmann C et al. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin Infect Dis 2015; 61:1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monto AS, Malosh RE, Petrie JG et al. Frequency of acute respiratory illnesses and circulation of respiratory viruses in households with children over 3 surveillance seasons. J Infect Dis 2014; 210:1792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Principi N, Zampiero A, Gambino M et al. Prospective evaluation of rhinovirus infection in healthy young children. J Clin Virol 2015; 66:83–9. [DOI] [PubMed] [Google Scholar]
- 21.Muller L, Mack I, Tapparel C et al. Human rhinovirus types and association with respiratory symptoms during the first year of life. Pediatr Infect Dis J 2015; 34:907–9. [DOI] [PubMed] [Google Scholar]
- 22.Storch GA. Editorial commentary: plethora of respiratory viruses and respiratory virus data. Clin Infect Dis 2015; 61:1225–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huicho L, Trelles M, Gonzales F. National and sub-national under-five mortality profiles in Peru: a basis for informed policy decisions. BMC Public Health 2006; 6:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grijalva CG, Griffin MR, Edwards KM et al. Cohort profile: the study of respiratory pathogens in Andean children. Int J Epidemiol 2014; 43:1021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budge PJ, Griffin MR, Edwards KM et al. Impact of home environment interventions on the risk of influenza-associated ARI in Andean children: observations from a prospective household-based cohort study. PLoS One 2014; 9:e91247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu A, Budge PJ, Williams J et al. Incidence and risk factors for respiratory syncytial virus and human metapneumovirus infections among children in the remote highlands of Peru. PLoS One 2015; 10:e0130233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grijalva CG, Griffin MR, Edwards KM et al. Concordance between RT-PCR-based detection of respiratory viruses from nasal swabs collected for viral testing and nasopharyngeal swabs collected for bacterial testing. J Clin Virol 2014; 60:309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grijalva CG, Griffin MR, Edwards KM et al. The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin Infect Dis 2014; 58:1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanata CF, Rudan I, Boschi-Pinto C et al. Methodological and quality issues in epidemiological studies of acute lower respiratory infections in children in developing countries. Int J Epidemiol 2004; 33:1362–72. [DOI] [PubMed] [Google Scholar]
- 30.Poehling KA, Edwards KM, Weinberg GA et al. The underrecognized burden of influenza in young children. N Engl J Med 2006; 355:31–40. [DOI] [PubMed] [Google Scholar]
- 31.Gove S. Integrated management of childhood illness by outpatient health workers: technical basis and overview. The WHO Working Group on Guidelines for Integrated Management of the Sick Child. Bull World Health Organ 1997; 75:7–24. [PMC free article] [PubMed] [Google Scholar]
- 32.Integrated management of childhood illness: conclusions. WHO Division of Child Health and Development. Bull World Health Organ 1997; 75(suppl 1):119–28. [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien KL, Nohynek H; World Health Organization Pneumococcal Vaccine Trials Carriage Working G. Report from a WHO working group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J 2003; 22:133–40. [DOI] [PubMed] [Google Scholar]
- 34.Satzke C, Turner P, Virolainen-Julkunen A et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013; 32:165–79. [DOI] [PubMed] [Google Scholar]
- 35.Jartti T, Lee WM, Pappas T et al. Serial viral infections in infants with recurrent respiratory illnesses. Eur Respir J 2008; 32:314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Q, Zheng S, Zhou L et al. Impact of human rhinovirus types and viral load on the severity of illness in hospitalized children with lower respiratory tract infections. Pediatr Infect Dis J 2015; 34:1187–92. [DOI] [PubMed] [Google Scholar]
- 37.Cox DW, Bizzintino J, Ferrari G et al. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med 2013; 188:1358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierangeli A, Ciccozzi M, Chiavelli S et al. Molecular epidemiology and genetic diversity of human rhinovirus affecting hospitalized children in Rome. Med Microbiol Immunol 2013; 202:303–11. [DOI] [PubMed] [Google Scholar]
- 39.Lau SK, Yip CC, Lin AW et al. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis 2009; 200:1096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee WM, Lemanske RF Jr, Evans MD et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 2012; 186:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen EK, Koeppel AF, Hendley JO et al. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome 2014; 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babiuk LA, Lawman MJ, Ohmann HB. Viral-bacterial synergistic interaction in respiratory disease. Adv Virus Res 1988; 35:219–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosch AA, Biesbroek G, Trzcinski K et al. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 2013; 9:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Annamalay AA, Khoo SK, Jacoby P et al. Prevalence of and risk factors for human rhinovirus infection in healthy aboriginal and non-aboriginal Western Australian children. Pediatr Infect Dis J 2012; 31:673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.