Abstract
The blood–brain barrier is composed of cerebrovascular endothelial cells and tight junctions, and maintaining its integrity is crucial for the homeostasis of the neuronal environment. Recently, we discovered that mitochondria play a critical role in maintaining blood–brain barrier integrity. We report for the first time a novel mechanism underlying blood–brain barrier integrity: miR-34a mediated regulation of blood–brain barrier through a mitochondrial mechanism. Bioinformatics analysis suggests miR-34a targets several mitochondria-associated gene candidates. We demonstrated that miR-34a triggers the breakdown of blood–brain barrier in cerebrovascular endothelial cell monolayer in vitro, paralleled by reduction of mitochondrial oxidative phosphorylation and adenosine triphosphate production, and decreased cytochrome c levels.
Keywords: Blood–brain barrier, cerebrovascular endothelial cells, cytochrome c, mir-34a, mitochondria
Introduction
The blood–brain barrier (BBB) is composed of highly specialized cerebrovascular endothelial cells (CECs), separates brain tissue from the circulating blood, and maintains homeostasis of the neuronal environment.1 The CECs are interconnected by tight junctions including cytoplasmic zonula occludens (ZO) proteins, and various transmembrane proteins such as occludin and claudins.2 Disruption of BBB tight junctions has been well documented in cerebrovascular diseases and neurodegenerative disorders and is considered to be a pathological condition of the diseases and plays a key role in disease progression as well.2
A recent study demonstrates that the mitochondrial mechanisms regulate BBB integrity and permeability using oxygen–glucose deprivation and reoxygenation (OGD-R), an in vitro model of ischemic reperfusion injury.3 Our work demonstrates that compromised mitochondria lead to the disruption of tight junctions, opening of the BBB, and exacerbation of stroke outcomes.4 As such, regulation of mitochondrial function may affect BBB openings and could be critical in limiting the pathological progression of cerebrovascular diseases and neurodegenerative disorders.
MicroRNAs (miRNAs) are short non-coding functional RNAs that target certain messenger RNAs (mRNAs) through complementary base-pairing between the miRNAs and its mRNA targets, resulting in the inhibition of mRNA translation or degradation of mRNA.5 It has been documented that miRNAs are involved in mitochondrial structure and function, such as miR-181c which regulates mitochondrial morphology,6 miR-1 which affects mitochondrial mRNA translation,7 and miR-378 which targets mitochondrial enzymes involved in oxidative energy metabolism.8 Additionally, several miRNAs have recently been found to regulate BBB permeability. MiR-155, miR-181c, and miR-29c negatively affect BBB function by targeting tight junction protein genes directly or affecting related signal pathways.9–11 The miR-34 family members were discovered computationally and later verified experimentally as a part of the p53 tumor suppressor network. Recent work demonstrates that miR-34a modulates the expression of synaptic targets and neuronal morphology and function.12 However, little is known regarding the role of miR-34a in mitochondrial function and BBB permeability.
In the present study, we report that the overexpression of miR-34a breaks down the BBB through inhibition of mitochondrial function. Furthermore, cytochrome c (CYC) is experimentally verified as a target of miR-34a in vitro.
Materials and methods
BBB permeability assay in vitro
Cell culture and transfection were detailed in supplementary information (SI). To identify the effect of miR-34a on the BBB, we employed a CEC (bEnd.3 cell line from ATCC) culture model, which is a murine CEC line with established BBB characteristics. We first transfected the CECs with miRNA-Alxea-Fluo®-555 by lipofectomine RNAiMAX reagent and confirmed high transfection efficiency (74.9%) in CECs by flow cytometry (Supplementary Figure 1(a)) and microscopy (Supplementary Figure 1(b)). Then, we constructed a miR-34a expression plasmid and observed an approximate nine-fold increase in miR-34a copies in CECs with the miR-34a plasmid compared to vector control as determined by real-time polymerase chain reaction (PCR) (Supplementary Figure 1(c)). We performed the following experiments under the same transfection condition.
We transfected the CECs in 24-well plates and conducted BBB permeability assays in transwell inserts (Millipore, Darmstadt, Germany) in triplicates. We seeded transfected CECs (1.5 × 105) on inserts for two days. We added 250 µg/ml FITC-labeled dextran FD-4 (4 kDa, Sigma) to the apical side of the filters and then sampled the medium in the basolateral compartment every 15 min for 2 h. We read the mean fluorescent intensity of FD-4 on a plate reader (excitation 490 nm and emission 515 nm) and calculated the apparent permeability coefficient (Papp, cm/s) with an FD-4 standard curve.
Oxygen consumption rate evaluation
We measured oxygen consumption rate (OCR) at 37℃ using an XF96e extracellular analyzer (Seahorse Bioscience, Massachusetts, USA) according to the manufacturer’s instructions. Briefly, we seeded CECs into Seahorse Bioscience XF96e cell culture plates (16,000 cells/well for 48 h culture; 10,000 cells/well for 72 h culture) and cultured overnight. We performed miR-34 plasmid versus vector control transfection with Lipofectamine RNAiMAX reagent (Life Technologies, New York, USA) and cultured CECs for additional 48 h or 72 h. We changed the cell culture medium to un-buffered (pH 7.4) DMEM prior to measurements. We prepared 10 × dilutions of oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), rotenone, and antimycin A (all from Sigma) and implemented the measure protocol. This protocol allowed determination of the basal level of oxygen consumption, the amount of oxygen consumption linked to adenosine triphosphate (ATP) production, maximal respiration capacity, and non-mitochondrial oxygen consumption.
ATP measurement
We cultured cells in 96-well black plates and measured ATP level by ATP bioluminescence assay kit (Promega, Wisconsin, USA) according to the manufacturer’s instruction.
Detect respiratory chain complex I–IV by flow cytometry
We cultured CECs in six-well plates and transfected the cells with miR-34a plasmid or control, then we washed the cells and performed intracellular staining with an intracellular staining set (cat. 72-5775, eBioscience, California, USA). Antibody information is detailed in SI. We acquired data on BD Calibur flow cytometry and analyzed mean fluorescence intensity by Flowjo software.
Statistical analysis
We performed statistical analysis using Prism 5 software (Graphpad software, California, USA). Differences between groups were analyzed by the unpaired Student’s t-test, 1-way analysis of variance (ANOVA), or 2-way ANOVA as indicated in the figure legends.
Results
Overexpression of miR-34a affects BBB permeability and disrupts tight junctions in CECs
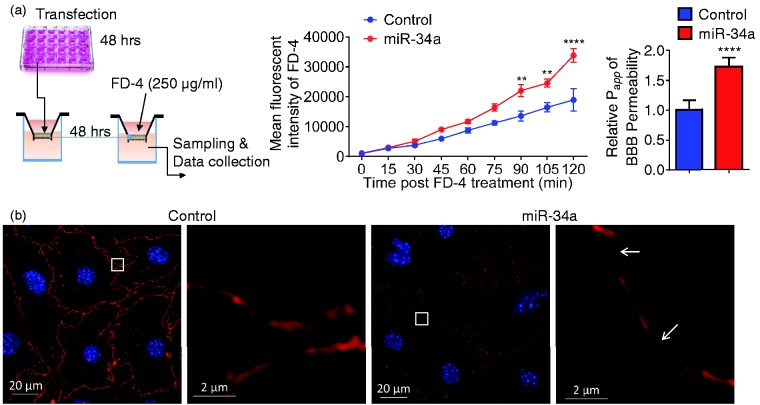
To determine whether miR-34a functionally affected the BBB, we transfected CECs with miR34a plasmid versus vector control in 24-well plates, cultured the cells for 48 h, conducted a BBB permeability assay in a CEC monolayer transwell system in vitro with an additional culture of 48 h, and measured the fluorescent dye FD-4 permeability of each well (Figure 1(a)). As shown in Figure 1(a), FD-4 permeability was significantly increased in wells containing miR-34a overexpression CEC monolayer. Papp, the permeability coefficient, was also significantly higher in CECs overexpressed with miR-34a in comparison to vector controls (Figure 1(a)). Furthermore, immunohis-tochemistry staining of tight junction-related proteins revealed that ZO-1 was continuously distributed in the control, but a discontinuous distribution of ZO-1 was observed in miR-34a overexpressed CEC monolayer (Figure 1(b)). Disruption of tight junctions was not associated with cell viability in CECs transfected with plasmids for 48 h or 96 h (Supplementary Figure 2). Altogether, these data suggest that overexpression of miR-34a increases BBB permeability and compromises BBB tight junctions.
Figure 1.
Overexpression of miR-34a increases BBB permeability in vitro. (a) A schematic protocol using fluorescein isothiocyanate–dextran-4 (FD-4) to detect BBB permeability in vitro. FD-4 permeability in CECs that overexpressed miR-34a plasmid (0.017 ng) versus control was presented as real-time rate of FD-4 mean fluorescent intensity (2-way ANOVA followed by post hoc Dunnett’s test; n = 3; **, P < 0.01; ****, P < 0.0001). Calculated apparent permeability coefficient Papp (Student’s t-test; ****, P < 0.0001) is expressed as mean ± SD. (b) Confocal fluorescence images of CECs confluent monolayers confirmed microscopically after transfection with miR-34a plasmid versus control. Fluorescent staining: tight junctions ZO-1 (red), cell nuclei (DAPI, blue). Overexpression of miR-34a apparently disrupted tight junctions and resulted in gaps between cells (white arrows). Results are representative of three independent experiments.
MiR-34a affects mitochondrial function by targeting CYC in CECs
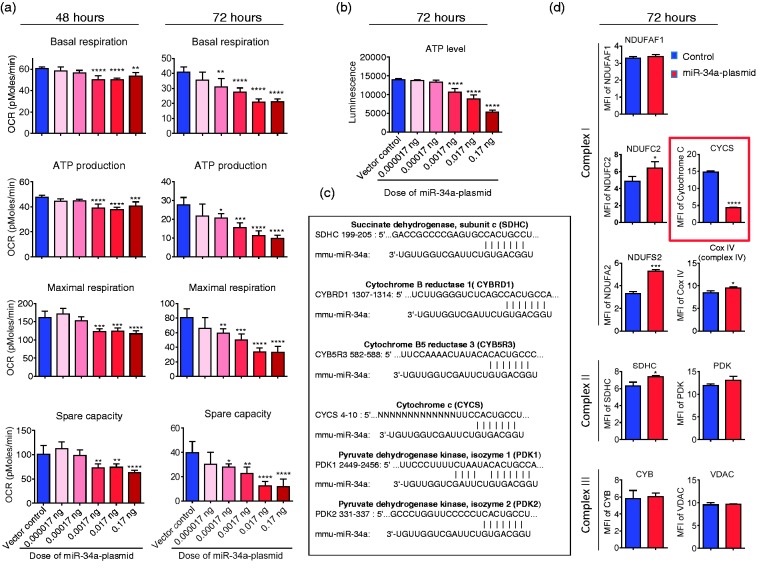
Our recent work demonstrated that mitochondria play a pivotal role in the maintenance of BBB integrity. BBB tight junctions are rapidly disrupted if oxidative phosphorylation is reduced by mitochondrial inhibitors.4 To investigate whether the miR-34a regulates BBB openings via affecting mitochondrial function in CECs, we examined cellular energetic OCRs in CECs transfected with miR-34a plasmid versus vector control. Interestingly, overexpression of miR-34a significantly impaired mitochondrial function in CECs (Figure 2(a) and Supplementary Figure 3). Basal respiration, ATP production, maximal respiration, and spare capacity were all significantly reduced in CECs overexpressing miR-34a for 48 and 72 h (Figure 2(a)). ATP level was also substantially reduced in CECs following overexpression of miR-34a in a dose dependent manner at 72 h (Figure 2(b)).
Figure 2.
Overexpression of mir-34a reduces mitochondrial function and decreases CYC level in cerebrovascular endothelial cells. (a) Basal respiration, ATP production, maximal respiration, and spare capacity were calculated from the bioenergetics functional assay at post-transfection 48 and 72 h (raw data in Supplementary Figure 3). Data are expressed as mean ± SD (n = 5). 1-way ANOVA followed by post hoc Tukey’s test. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (b) ATP level was measured at 72 h post-transfection. Data are expressed as mean ± SD (n = 5). 1-way ANOVA followed by post hoc Tukey’s test. (****, P < 0.0001). (c) Bioinfomatic analysis of miR-34a-targeting candidates related to mitochondria. (d) Flow cytometry analysis of mitochondrial specific proteins for complex I proteins (NDUFAF1, NDUFC2 and NDUFS2), complex II protein (SDHC), complex III protein (CYB), complex IV protein (CYC oxidase, Cox IV), cytochrome c (CYCS), pyruvate dehydrogenase kinase (PDK), and voltage-dependent anion channel protein (VDAC) at 72 h post-transfection. CYC level was significantly lower in the cells that were transfected with the miR-34a plasmid. Data are presented as mean ± SD (n = 3) and analyzed by Student’s t-test, *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Results are representative of three independent experiments.
To further determine miR-34a targets and uncover the mechanism that is used to affect mitochondria, we performed a bioinformatics analysis of the miR-34a database (miRbase and TargetScan). MiR-34a potentially targets several mitochondria-associated gene candidates including succinate dehydrogenase subunit c (SDHC), cytochrome B reductase 1 (CYBRD1), cytochrome B5 reductase 3 (CYBRD5), cytochrome c (CYCS), pyruvate dehydrogenase kinase isozyme 1 and 2 (PDK1 and PDK2) (Figure 2(c). However, CECs transfected with the miR-34a plasmid had robustly decreased CYCS levels measured by flow cytometry, suggesting that CYCS is one of the miR-34a targets among the potential candidates (Figure 2(d)). Moreover, overexpression of miR-34a slightly increased potential target SDHC but did not change the protein level of CYB and PKD (Figure 2(d)). Off-target genes, NDUFAF1, and VDAC showed no significant change in protein level, but NDUFC2, NDUFS2, and Cox IV were all increased in parallel with overexpression of miR-34a (Figure 2(d)). Taken together, these results experimentally verified CYCS as a miR-34a target, which is associated with the reduction of mitochondrial oxidative phosphorylation in CECs.
Discussion
In the present study, we demonstrated that the overexpression of miR-34a results in an increased BBB permeability and the disruption of tight junctions ZO-1 in CECs. Consistently, overexpression of miR-34a impaired mitochondrial oxidative phosphorylation and reduced ATP production in CECs. Bioinformatics analysis revealed series of potential miR-34a-targeting candidates related to mitochondrial function. We elucidated that CYCS is a miR-34a target, and the overexpression of miR-34a inhibited the CYCS expression and increased with the expression of other mitochondria-associated genes.
The overexpression of miR-34a disrupted tight junction protein ZO-1 (Figure 1). However, bioinformatics analysis indicated that miR-34a did not target the ZO-1 gene or other tight junction related genes, which suggests that the increased BBB permeability is not directly caused by the targeting of tight junction protein genes. The compromised mitochondrial function by overexpression of miR-34a may influence cellular metabolism in a way that is critical to maintain BBB tight junctions. Among several potential mitochondria-associated gene targets (Figure 2(c)), miR-34a initiated the reduction of CYCS level. Interestingly, potential target SDHC and other off-target gene proteins (NDUFC2, NDUFS2, and Cox IV) were concurrently upregulated (Figure 2(d)), which might be due to the compensation for the reduced target gene protein CYCS, or the disturbance of the coordinated gene translation in mitochondria. We therefore concluded that CYCS is a miR-34a target and is responsible for the miR-34a-induced reduction of mitochondrial oxidative phosphorylation.
Protein kinase C (PKC) signaling has also been shown to affect BBB or other endothelial barriers in vitro and in vivo. A recent study reported that miR-34a regulated blood–tumor barrier by targeting PKCɛ using glioma endothelial cells.13 In this study, we did not assess the PKC pathways that could contain additional targets of miR-34a. However, our data do support that miR-34a affects BBB via a mitochondrial mechanism, which is novel and may lead a new direction for designing BBB-related therapeutics.
We have noted several limitations in our study. First, we did not examine the effects of knockdown or knockout miR-34a on BBB function, which might fully establish the role of miR-34a in the BBB and mitochondria. Second, this work was conducted in cell culture models, which adequately address the mechanism of effect that miR-34a exerts on the BBB and mitochondria but do not provide evidence of its involvement in cerebrovascular or neurodegenerative conditions. Further studies in relevant experimental models are warranted.
Mitochondria play a pivotal role in cellular bioenergetics and cell survival, participating in a variety of cellular processes, including the generation of ATP, and the regulation of apoptotic signaling and other signaling pathways.14 MiR-34a targets and represses multiple genes involved in cell proliferation, apoptosis, cell cycle, migration, etc.,15 but it is not known if these effects are modulated by the observed mitochondrial effects as well. The present study provides the first description of miR-34a affecting mitochondrial activity, which could lead to a revision of current miR-34a targets and may lead to discovery of new mechanisms. The elucidation of the miR-34a’s role in mitochondrial oxidative phosphorylation and the BBB integrity offers a novel therapeutic strategy for targeting miR-34a to treat cerebrovascular and neurodegenerative diseases such as stroke and Alzheimer’s disease. These neuropathological diseases are known to involve a host of conditions that lead to mitochondrial impairment and BBB disruption. Finally, transient opening of the BBB could prove to be useful for CNS drug delivery.
Supplementary Material
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH (P20 GM109098, P01 AG027956 and U54 GM104942). Imaging experiments were performed at the West Virginia University (WVU) Microscope Imaging Facility supported by the Mary Babb Randolph Cancer Center (MBRCC) and NIH (P20 RR016440, P30 RR032138/GM103488 and P20 RR016477). Flow cytometry was performed at the WVU Flow Cytometry Core Facility supported by MBRCC and NIH (P30 RR032138/GM103488 and GM103434).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
MB, SNS, HH, DQ, and XR acquired data. JWS and XR designed research and wrote the manuscript.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data.
References
- 1.Mehta DC, Short JL, Hilmer SN, et al. Drug access to the central nervous system in Alzheimer's disease: preclinical and clinical insights. Pharm Res 2015; 32: 819–839. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg GA. Neurological diseases in relation to the blood-brain barrier. J Cereb Blood Flow Metab 2012; 32: 1139–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alluri H, Stagg HW, Wilson RL, et al. Reactive oxygen species-caspase-3 relationship in mediating blood-brain barrier endothelial cell hyperpermeability following oxygen-glucose deprivation and reoxygenation. Microcirculation 2014; 21: 187–195. [DOI] [PubMed] [Google Scholar]
- 4.Doll DN, Hu H, Sun J, et al. Mitochondrial crisis in cerebrovascular endothelial cells opens the blood-brain barrier. Stroke 2015; 46: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Li J, Chi H, et al. MicroRNA-181c targets Bcl-2 and regulates mitochondrial morphology in myocardial cells. J Cell Mol Med 2015; 19: 2084–2097. [DOI] [PMC free article] [PubMed]
- 7.Zhang X, Zuo X, Yang B, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014; 158: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrer M, Liu N, Grueter CE, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA 2012; 109: 15330–15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Ramirez MA, Wu D, Pryce G, et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 2014; 28: 2551–2565. [DOI] [PubMed] [Google Scholar]
- 10.Tominaga N, Kosaka N, Ono M, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun 2015; 6: 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalani A, Kamat PK, Familtseva A, et al. Role of microRNA29b in blood-brain barrier dysfunction during hyperhomocysteinemia: an epigenetic mechanism. J Cerebr Blood Flow Metab 2014; 34: 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agostini M, Tucci P, Steinert JR, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA 2011; 108: 21099–21104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Wang P, Ma J, et al. MiR-34a regulates blood-tumor barrier function by targeting protein kinase Cepsilon. Mol Biol Cell 2015; 26: 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol 2011; 11: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget 2014; 5: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.