Abstract
Genetically distinct hantaviruses have been identified in five species of fossorial moles (order Eulipotyphla, family Talpidae) from Eurasia and North America. Here, we report the isolation and partial characterization of a highly divergent hantavirus, named Nova virus (NVAV), from lung tissue of a European mole (Talpa europaea), captured in central Poland in August 2013. Typical hantavirus-like particles, measuring 80–120 nm in diameter, were found in NVAV-infected Vero E6 cells by transmission electron microscopy. Whole-genome sequences of the isolate, designated NVAV strain Te34, were identical to that amplified from the original lung tissue, and phylogenetic analysis of the full-length L, M and S segments, using maximum-likelihood and Bayesian methods, showed that NVAV was most closely related to hantaviruses harbored by insectivorous bats, consistent with an ancient evolutionary origin. Infant Swiss Webster mice, inoculated with NVAV by the intraperitoneal route, developed weight loss and hyperactivity, beginning at 16 days, followed by hind-limb paralysis and death. High NVAV RNA copies were detected in lung, liver, kidney, spleen and brain by quantitative real-time RT-PCR. Neuropathological examination showed astrocytic and microglial activation and neuronal loss. The first mole-borne hantavirus isolate will facilitate long-overdue studies on its infectivity and pathogenic potential in humans.
A paradigm-disruptive shift in hantavirus host diversity has emerged from the recent discovery of genetically distinct hantaviruses (family Bunyaviridae, genus Hantavirus) in multiple species of shrews and moles (order Eulipotyphla)1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19 and insectivorous bats (order Chiroptera)20,21,22,23,24 in widely separated geographic regions in Europe, Asia, Africa and North America. Moreover, phylogenetic analyses, based on partial and full-length hantavirus genomes, suggest that ancestral shrews, moles and/or bats probably predated rodents as the reservoir hosts of primordial hantaviruses25,26,27. However, despite being referred to as novel viruses, nearly all of the 36 newfound non-rodent-borne hantaviruses have not been isolated in cell culture or animals and exist only as viral sequences. As such, almost nothing is known about their biology and transmission dynamics, as well as infectivity and pathogenic potential in humans.
Hantaviruses possess a negative-sense, tripartite RNA genome, designated as large (L), medium (M) and small (S) segments, which encode an RNA-dependent RNA polymerase (RdRp), envelope glycoproteins (Gn and Gc) and a nucleocapsid (N) protein, respectively28. Some hantaviruses cause diseases of varying clinical severity, known as hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome (HPS)29. Until recently, rodents (order Rodentia) of the Muridae and Cricetidae families were believed to serve as the exclusive reservoir hosts. The recent flurry of descriptions of newfound non-rodent-borne hantaviruses was presaged by reports, published two to three decades ago which went largely unnoticed, of HFRS antigens in tissues of the Eurasian common shrew (Sorex araneus), Eurasian water shrew (Neomys fodiens) and European mole (Talpa europaea), captured in Russia and the former Yugoslavia30,31,32.
Among the five hantaviruses discovered to date in moles (family Talpidae) from Europe, Asia and North America, Nova virus (NVAV), originally detected in archival liver tissue of a single European mole (Talpa europaea) captured in 1999 in Zala County, Hungary18, and subsequently shown to be widespread in France33 and Poland34, is the most divergent25,26. Here, we report the isolation and partial characterization of NVAV. Since hantaviruses are notoriously difficult to isolate, the long-awaited isolation of NVAV in cell culture is a noteworthy milestone that will accelerate acquisition of new knowledge about its transmission dynamics and facilitate long-overdue studies to ascertain its infectivity and pathogenic potential in humans.
Results
Prevalence of NVAV infection
Of 22 European moles (Fig. 1A), captured in Huta Dłutowska between June 21, 2013 and August 26, 2013, NVAV RNA was detected by RT-PCR in lung tissue from 11 (50%). Based on the intensity of the RT-PCR amplicon on ethidium bromide-stained gels, lung tissue homogenates were prepared from four European moles and used for virus isolation attempts.
Figure 1.
(A) European mole (Talpa europaea) captured in Huta Dłutowska, in central Poland, in August 2013. (B) Thin-section transmission electron micrograph, showing hantavirus particles in Vero E6 cells inoculated with lung homogenate from a NVAV-infected European mole. Bar = 100 nm.
Isolation and propagation of NVAV
Initially, NVAV RNA was detected in Vero E6 cells at 17 days following inoculation with a 1% lung tissue homogenate from one of four European moles. Viral RNA was also detected at day 14 of the first passage, but the virus isolate was lost on subsequent passages. After several other failed attempts, NVAV RNA was detected in cells and culture media at 34 days and on subsequent inoculation of fresh Vero E6 cells with culture media, indicating virus replication. At 120 days, the NVAV infectivity in culture media was 8 × 103 plaque-forming units (pfu)/mL. Like all other rodent- and shrew-borne hantaviruses isolated to date, the NVAV isolate did not produce cytopathic effect (or CPE) in Vero E6 cells. Typical hantavirus-like particles, measuring 80–120 nm in diameter, were found by thin-section transmission electron microscopy (Fig. 1B). No other virus-like particles were observed.
Sequence analysis of NVAV
The full-length S-, M- and L-genomic segments of the cell culture grown NVAV isolate, designated NVAV strain Te34, and the full-length S segment of the NVAV from the original lung tissue, were sequenced. While the overall genomic organization of NVAV was indistinguishable from other hantaviruses, the deduced-amino acid sequence similarities were significantly different from that of rodent-, shrew- and mole-borne hantaviruses, differing by more than 50% for the N protein and Gn/Gc envelope glycoproteins and 40% for the RdRp (Table 1).
Table 1. Nucleotide and amino acid sequence similarity (%) between NVAV strain Te34 and other representative rodent-, shrew-, mole- and bat-borne hantaviruses.
| Virus strain | S segment |
M segment |
L segment |
|||
|---|---|---|---|---|---|---|
| 1,284 nt | 428 aa | 3,381 nt | 1,127 aa | 6,471 nt | 2,157 aa | |
| NVAV MSB95703 | 86.4 | 96.5 | 82.4 | 97.7 | 85.7 | 96.4 |
| NVAV 1135 | 99.8 | 99.5 | – | – | 99.9 | 99.8 |
| NVAV 2086 | 97.7 | 99.5 | – | – | 96.3 | 100.0 |
| NVAV 2097 | 99.6 | 100.0 | – | – | 95.9 | 99.1 |
| NVAV YA0077 | 85.0 | 97.7 | – | – | 83.5 | 97.0 |
| NVAV YA0087 | 86.1 | 97.4 | – | – | 85.5 | 97.4 |
| NVAV YA0109 | 86.2 | 97.4 | – | – | 84.4 | 97.4 |
| XSV F42682 | 59.3 | 56.4 | 63.2 | 64.3 | 71.0 | 78.8 |
| LBV BT20 | 57.9 | 58.3 | 63.5 | 58.0 | 68.8 | 68.2 |
| LQUV Ra10 | 54.9 | 54.6 | 44.3 | 44.4 | 67.9 | 71.3 |
| HUPV Pa1 | 61.6 | 57.6 | – | – | 71.7 | 79.8 |
| MOYV KB576 | – | – | – | – | 70.4 | 77.8 |
| MGBV 1209 | – | – | – | – | 68.8 | 68.6 |
| MJNV Cl 05-11 | 50.2 | 44.9 | 41.2 | 41.2 | 65.9 | 63.0 |
| TPMV VRC66412 | 49.2 | 47.0 | 42.5 | 42.0 | 66.2 | 62.6 |
| ULUV FMNH158302 | 62.0 | 48.1 | 55.9 | 40.6 | 64.6 | 61.4 |
| KMJV FMNH174124 | 50.8 | 49.1 | 49.7 | 42.1 | 66.7 | 62.7 |
| ASAV N10 | 50.9 | 48.4 | 54.6 | 45.7 | 66.3 | 62.1 |
| OXBV Ng1453 | 51.5 | 50.0 | 54.4 | 45.8 | 63.9 | 61.3 |
| JMSV MSB144475 | 51.4 | 49.5 | 60.0 | 49.8 | 65.1 | 62.8 |
| SWSV mp70 | 51.6 | 50.7 | 54.6 | 60.2 | 62.8 | 58.7 |
| QHSV YN05-284 | 53.0 | 49.3 | 61.2 | 51.1 | 71.2 | 73.8 |
| KKMV MSB148794 | 58.4 | 48.8 | 57.3 | 45.2 | 64.4 | 62.3 |
| CBNV CBN-3 | 57.2 | 50.2 | 46.8 | 44.3 | 66.5 | 61.9 |
| JJUV 10-11 | 50.9 | 50.7 | 52.4 | 45.0 | 64.6 | 60.1 |
| TGNV Tan826 | 64.0 | 47.6 | – | – | 70.3 | 64.2 |
| AZGV KBM15 | 39.4 | 50.0 | 55.6 | 40.6 | 65.6 | 61.4 |
| BOWV VN1512 | 59.1 | 49.1 | 44.9 | 43.5 | 64.8 | 60.6 |
| RKPV MSB57412 | 57.9 | 52.3 | 54.3 | 48.6 | 65.4 | 60.8 |
| HTNV 76-118 | 50.4 | 51.4 | 52.8 | 45.1 | 65.7 | 61.7 |
| SEOV 80-39 | 57.7 | 49.5 | 42.9 | 45.2 | 65.3 | 61.8 |
| SOOV SOO-1 | 56.6 | 50.9 | 50.6 | 44.9 | 65.2 | 61.9 |
| DOBV Greece | 57.5 | 51.9 | 43.4 | 45.3 | 65.0 | 62.5 |
| ANDV Chile9717869 | 53.4 | 51.4 | 42.9 | 46.1 | 64.8 | 60.7 |
| SNV NMH10 | 51.3 | 51.4 | 43.4 | 46.6 | 65.6 | 61.3 |
| PUUV Sotkamo | 58.6 | 53.3 | 45.6 | 46.2 | 65.3 | 61.1 |
| TULV M5302v | 51.6 | 51.6 | 57.2 | 47.5 | 65.4 | 61.4 |
| PHV PH-1 | 50.1 | 50.0 | 59.1 | 47.1 | 64.1 | 60.9 |
Abbreviations: ANDV, Andes virus; ASAV, Asama virus; AZGV, Azagny virus; BOWV, Bowé virus; CBNV, Cao Bang virus; DOBV, Dobrava virus; HTNV, Hantaan virus; HUPV, Huangpi virus; JMSV, Jemez Spring virus; JJUV, Jeju virus; KKMV, Kenkeme virus; KMJV, Kilimanjaro virus; LBV, Laibin virus; LQUV, Longquan virus; MGBV, Magboi virus; MJNV, Imjin virus; MOYV, Mouyassué virus; NVAV, Nova virus; OXBV, Oxbow virus; PHV, Prospect Hill virus; PUUV, Puumala virus; QHSV, Qian Hu Shan virus; RKPV, Rockport virus; SEOV, Seoul virus; SNV, Sin Nombre virus; SOOV, Soochong virus; SWSV, Seewis virus; TGNV, Tanganya virus; TPMV, Thottapalayam virus; TULV, Tula virus; ULUV, Uluguru virus; XSV, Xuan Son virus. nt, nucleotides; aa, amino acids.– indicates that no sequences are available.
The full-length, 1,825-nucleotide S-segment sequence of the NVAV isolate and the NVAV in the original lung tissue were identical, containing a single open reading frame with a predicted N protein of 428 amino acids (nucleotide positions 53 to 1,339), and 52- and 486-nucleotide 5′– and 3′–noncoding regions, respectively. Comparison of the S segment between the NVAV isolate and the prototype NVAV strain MSB95703 from Hungary showed nucleotide and amino acid sequence similarities of 85.6% and 96.5%, respectively. The secondary structure of the predicted N protein of NVAV strain Te34 was virtually identical to that reported earlier for the prototype strain (data not shown).
The entire M-genomic segment of NVAV strain Te34 was 3,590 nucleotides, with a predicted glycoprotein precursor of 1,127 amino acids. The highly conserved WAASA amino acid motif of the M segment was found at amino acid positions 641–645, and the NVAV glycoprotein had six potential N-linked glycosylation sites (five on Gn at amino acid positions 101, 133, 344, 396 and 539, and one on Gc at position 924). The very low degree of amino acid sequence similarity between the GnGc of NVAV and other hantaviruses would predict the absence of antibody cross-neutralization between NVAV and hantaviruses harbored by rodents, shrews, moles and bats (Table 1).
The full-length L-genomic segment of NVAV strain Te34 was 6,563 nucleotides, with a predicted RdRp of 2,157 amino acids, starting at nucleotide position 34 and including 56 nucleotides of the 3′–noncoding region. As with all other hantaviruses, the six conserved motifs (premotif A and motifs A, B, C, D and E) were found. Premotif A had a conserved lysine and two arginine residues. Motifs A, B and D had conserved aspartate, glycine and lysine, respectively. In motif C, there were two conserved aspartic acid residues. The XDD motif, essential for catalytic activity, and motif E, containing the E(F/Y)XS site, were also present. The higher sequence similarity of approximately 60% or more in the L segment probably signified the functional constraints of the RdRp (Table 1).
Phylogenetic analysis of NVAV
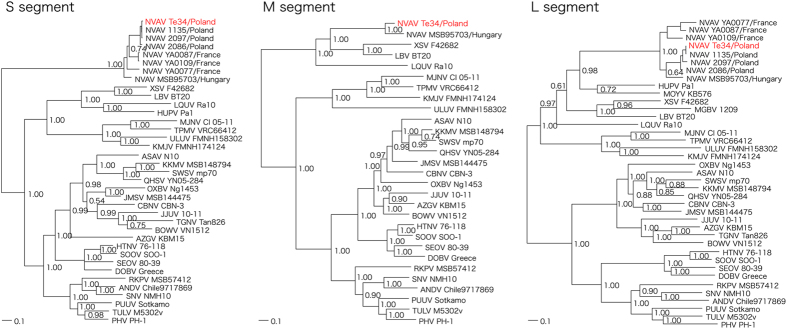
As determined by maximum-likelihood and Bayesian methods, phylogenetic analysis, based on full-length sequences of each genomic segment, demonstrated tree topologies, well supported by bootstrap analysis and posterior node probabilities, showing the NVAV isolate forming a distinctly divergent clade, which comprised other NVAV strains from Hungary, France and Poland (Fig. 2). Similar topologies were found for deduced amino acid sequences of the encoded proteins (data not shown). Collectively, these data strongly supported an ancient hantavirus-host relationship without evidence of host switching.
Figure 2. Phylogenetic trees generated by the Bayesian method, using the GTR+I+Γ model of evolution as estimated from the data, based on the alignment of the coding regions of the full- length S, M and L segments of NVAV strain Te34.
Topologies of the unrooted phylogenetic trees using the maximum-likelihood method were nearly identical. Relationships are shown to the prototype NVAV strain from Hungary (MSB95703, S: FJ539168; M: HQ840957; L: FJ593498) and to representative NVAV strains from Poland (1135, S: JX990924; L: JX990948; 2086, S: KF515970; L: JX990963; and 2097, S: JX990930; L: KF663727) and France (YA0077, S: KF010573; L: KF010538; YA0087, S: KF010571; L: KF010534; YA0109, S: KF010565; L: KF010521). Other hantaviruses harbored by shrews and moles included Thottapalayam virus (TPMV VRC66412), Imjin virus (MJNV Cl 05-11), Uluguru virus (ULUV FMNH158302), Kilimanjaro virus (KMJV FMNH174124), Asama virus (ASAV N10), Oxbow virus (OXBV Ng1453), Rockport virus (RKPV MSB57412), Jemez Springs virus (JMSV MSB144475), Seewis virus (SWSV mp70), Kenkeme virus (KKMV MSB148794), Qian Hu Shan virus (QHSV YN05-284), Cao Bang virus (CBNV CBN-3), Azagny virus (AZGV KBM15), Tanganya virus (TGNV Tan826), Bowé virus (BOWV VN1512), and Jeju virus (JJUV SH42). Bat-borne hantaviruses included Magboi virus (MGBV MGB1209), Mouyassué virus (MOYV KB576), Huangpi virus (HUPV Pa-1), Longquan virus (LQUV Ra-10), Laibin virus (LBV BT20), and Xuan Son virus (XSV F42682). Also shown are representative rodent-borne hantaviruses, including Hantaan virus (HTNV 76-118), Soochong virus (SOOV SOO-1), Dobrava/Belgrade virus (DOB/BGDV Greece), Seoul virus (SEOV 80-39), Tula virus (TULV M5302v), Puumala virus (PUUV Sotkamo), Prospect Hill virus (PHV PH-1), Sin Nombre virus (SNV NMH10) and Andes virus (ANDV Chile9717869). The numbers at each node are posterior node probabilities based on 150,000 trees. The scale bar indicates nucleotide substitutions per site.
Experimental NVAV infection
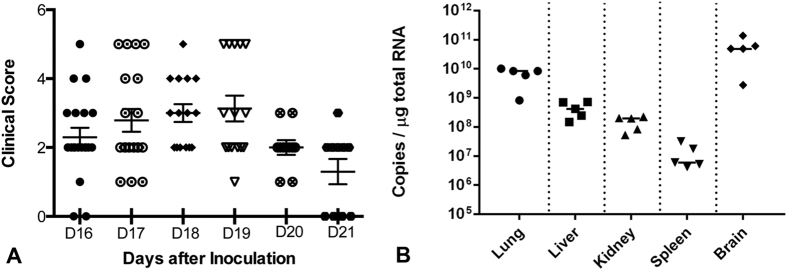
Of 20 infant Swiss Webster mice, inoculated with 200 pfu of NVAV strain Te34 by the intraperitorneal route, all developed weight loss and hyperactivity, beginning at 16 days. In 11 mice, disease progressed to include hind-limb paralysis, during the ensuing four-day period (Fig. 3A). Moribund mice, with clinical scores of 5, were subsequently euthanized, and their sera had antibodies against NVAV, as determined by the indirect immunofluorescent antibody (IFA) test (data not shown). Using conventional RT-PCR and quantitative real-time RT-PCR, NVAV RNA was detected in lung, liver, kidney, spleen and brain from experimentally infected mice. NVAV RNA copies in the brain were 10- to 100-fold higher than in other tissues (Fig. 3B).
Figure 3.
(A) Clinical scores of infant Swiss Webster mice inoculated with NVAV strain Te34 by the intraperitoneal route. Data are combined from two independent experiments, consisting of 10 mice each. Mice were monitored twice daily for disease severity. Clinical scores were designated as: 1, ruffled fur/hunched back; 2, hyperactivity/twitching; 3, paresis/difficulty walking; 4, paralysis; and 5, moribund/euthanized. Scores are shown for each mouse, beginning on day 16 post-inoculation. Error bars represent SEM. (B) Real-time RT-PCR quantification of NVAV RNA in lung, liver, kidney, spleen and brain of Swiss Webster mice inoculated with NVAV strain Te34. NVAV RNA copies per μg of total RNA are shown for moribund mice sacrificed on day 17 post-inoculation. The solid horizontal line for each tissue signifies the median for five moribund mice.
Moreover, brain tissue homogenates from NVAV-infected moribund mice, when inoculated intraperitoneally into new litters of infant mice, produced lethal meningoencephalitis, which was indistinguishable to that observed in infant mice experimentally infected with the NVAV cell-culture isolate. In addition, NVAV sequences amplified from tissues of infected mice were identical to that of the NVAV cell-culture isolate used for experimental infection, as well as to the NVAV originally detected in the European mole lung tissue.
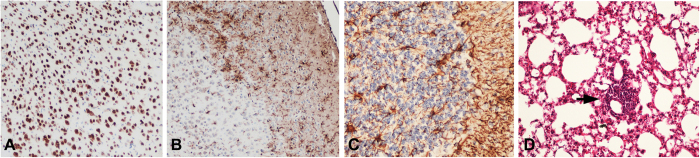
As assessed by light-microscopic examination of hematoxylin and eosin-stained tissues and by immunohistochemical staining, pathological abnormalities in NVAV-infected infant mice were largely confined to the brain. Perivascular and subpial lymphocytic infiltrates were found, particularly in the cerebellum. Minimal focal loss of neurons was evident in the cerebral cortex, using a neuronal nuclei-specific antibody (NeuN) (Fig. 4A). Microglial activation and astrocytosis, as evidenced by staining for a microglia-specific calcium-binding protein (Iba1) and glial fibrillary acidic protein (GFAP), respectively, were also prominent (Fig. 4B,C). In the lung, perivascular lymphocytic infiltrates were conspicuous with mild widening of the alveolar septa and interstitial infiltration, but without evidence of hyaline membranes or intra-alveolar edema (Fig. 4D).
Figure 4. Histopathology in infant mice, 17 days after inoculation by the intraperitoneal route with 200 pfu of NVAV strain Te34.
Immunohistochemical staining, showing (A) neuronal loss and (B) microglial activation in the cerebral cortex and (C) widespread astrogliosis in the cerebellum, using primary antibodies against NeuN, Iba1 and glial fibrillary acidic protein, respectively. (D) Lung tissue, stained with hematoxylin and eosin, showing perivascular lymphocytic infiltration (arrow). Original magnification: (A) and (B), X100; (C), X200; (D), X200.
Materials and Methods
Trapping and specimen processing
Moles were trapped using PVC pipes, equipped with flat aluminum latches at both ends, in Huta Dłutowska (N51°35′49.51, E19°22′46.80), near Łódź in central Poland, during June to August 2013. Live-caught moles were euthanized by cervical dislocation, and stored at 4 °C or −20 °C for several hours or days before harvesting of lung tissues, which were stored at −80 °C until used for RT-PCR and virus isolation.
Ethics statement
Field procedures and protocols, including trapping, euthanasia and tissue processing, were approved by the Łódź Ethical Committee on Animal Testing (14/LB/511/2010 and 29/LB/548/2011) and the Directorate General for Environmental Protection (DOP-OZGiZ.4200/N2732/10/JRO, DOP- OZGiZ.6401.05.25.2011kp.3 and DOP-OZGiZ.6401.05.28.2011kp.1), as well as by the Łódź Regional Directorate for Environmental Protection (WPN-I6631.2010.MS and WPN-L6400.59.2011.MS), in accordance to guidelines of the American Society of Mammalogists35. For experimental NVAV infection in Swiss Webster mice, approval was received from the University of Hawaii Institutional Animal Care and Use Committee (protocol 06-055), which adhered to guidelines and regulations of the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals and the American Veterinary Medical Association. All experiments involving mice were conducted in consultation with veterinary and animal care staff of the University of Hawaii animal biosafety level-3 laboratory.
RNA extraction, cDNA synthesis and RT-PCR amplification
As an initial screening for NVAV infection, total RNA was extracted from 20–50 mg of mole lung tissues, using the PureLink Micro-to-Midi total RNA purification kit (Invitrogen, San Diego, CA), and cDNA was synthesized using the SuperScript III First-Strand Synthesis Systems (Invitrogen), then analyzed for NVAV RNA by RT-PCR, using NVAV-specific oligonucleotide primers33,34.
Virus isolation and propagation
Attempts to isolate hantavirus from lung tissues of moles with genetic evidence of NVAV infection were made in Vero E6 cells (CRL 1586, American Type Culture Collection, Manassas, VA), using previously described methods6. Briefly, subconfluent monolayers of Vero E6 cells, grown in 25-cm2 flasks, were inoculated with 1% (w/v) suspensions of lung tissue homogenates, prepared in Dulbecco’s minimum essential medium (DMEM) containing 30% fetal bovine serum, from four NVAV RNA-positive European moles. Cells were sub-cultured at two- to five-week intervals, at which time aliquots of cells and supernatants were examined for NVAV RNA by RT-PCR. Supernatant from NVAV-infected cultures were then used to inoculate fresh Vero E6 cells to demonstrate virus replication and to determine infectivity titers by plaque assay6.
Thin-section electron microscopy
NVAV-infected Vero E6 cells were pelleted and fixed overnight with 2.5% glutaraldehyde (Ted Pella, Inc., Redding, CA) in 0.1 M sodium cacodylate buffer, pH 7.3, washed twice in 0.1 M cacodylate, followed by post-fixation with 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 hour. Infected cells were then dehydrated in a graded series of ethanol washes (30%, 50%, 70%, 85%, 95%, 100%), substituted with propylene oxide, and embedded in LX112 epoxy resin. Ultrathin sections (60 to 80 nm) were obtained using an RMC Powertome ultramicrotome (Boeckler Instruments, Inc., Tuscon, AZ), double stained with uranyl acetate and lead citrate. Grids were viewed with a Hitachi HT7700 high-contrast/high-resolution transmission electron microscope operating at 100kV (Hitachi, Japan), and images were photographed with an AMT XR41 4 megapixel camera (Advanced Microscopy Techniques, Corp., Woburn, MA).
Whole genome sequencing
Oligonucleotide primers were designed using the MegAlign Clustal W program (DNASTAR Inc., Madison, WI) to obtain the whole genome of NVAV strain Te34 from cDNA prepared from total RNA, extracted from NVAV-infected Vero E6 cells and from lung tissue of the original wild-caught European mole (Table 2). The 5′– and 3′–termini of each segment were amplified using the 3′–Full RACE Core Set (Takara Bio Inc., Otsu, Japan). Nested or hemi-nested PCR was performed in 20 μL reaction mixtures, containing 250 μM dNTP, 2.5 mM MgCl2, 1 U of Takara LA Taq polymerase (Takara) and 0.25 μM of each primer (Table 2). Initial denaturation at 94 °C for 2 min was followed by two cycles each of denaturation at 94 °C for 30 sec, two-degree step-down annealing from 46 °C to 38 °C for 40 sec, and elongation at 72 °C for 1 min, then 30 cycles of denaturation at 94 °C for 30 sec, annealing at 42 °C for 40 sec, and elongation at 72 °C for 1 min, in a GeneAmp PCR 9700 thermal cycler (Perkin-Elmer, Waltham, MA)34. PCR products were separated, using MobiSpin S-400 spin columns (MoBiTec, Goettingen, Germany), and amplicons were sequenced directly using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA)34.
Table 2. Oligonucleotide primers for amplification of the full-length S, M and L segments of NVAV strain Te34.
| Primer | Sequence (5′ - 3′) | Segment | Polarity |
|---|---|---|---|
| Han-5′end-EcoRI | CTC GAA TTC TAG TAG TAG AC | S/M/L | + |
| OSM55 | TAG TAG TAG ACT CC | S/M/L | + |
| Shrew-end-EcoRI | CTC GAA TTC TAG TAG T | S/M/L | − |
| Mole-S732R | GRA AKC CDA TVA CTC CCA T | S | − |
| Mole-S712R | CAT HAC AGG ACT WAT CA | S | − |
| Tal-S696F | TGA TNA GYC CTG TNA TGG GAG T | S | + |
| Tal-S710F | ATG GGA GTG ATA GGC TTT CA | S | + |
| HTN-S6 | AGC TCN GGA TCC ATN TCA TC | S | − |
| NVAV-S1143F | ACA TGA GAA GGA CTC AGT C | S | + |
| NVAV-S1169F | ATG CAG ATG GAT CAG CGA | S | + |
| NVAV-3′endR | TAG TAG TAT ACT CCT TGA AAA GC | S | − |
| NVAV-M360R | CAC ATA TGC CAA GTA ATG AAC CGT | M | − |
| NVAV-M330R | AAG TAG TAT CAC CTG CCT GAG TA | M | − |
| NVAV-M1350R | ACA ATG TCT TTA TCA AGC CGT TGA CA | M | − |
| NVAV-M1290R | CTA CCA ATA AGA CAT GTT GGA GA | M | − |
| NVAV-M1440F | ATG TTT CCA TGG ATT GCA CA | M | + |
| NVAV-M1460F | ATG GCT GTA GAA GTT TGT G | M | + |
| HTN-M2400R | ARA TAA ANN CCA CAN GCA GTA CA | M | − |
| HTN-M2355R | CCW GGR CAA TCH VGA GGR TTA CA | M | − |
| NVAV-M2230F | TAG GTC ATT GGA TGG ATG GAG AGC | M | + |
| NVAV-M3200F | GAT GGC TTA AAG GCA AGT GCT CCT C | M | + |
| NVAV-M3260F | CAG ATG CAT TTT CAG ATG GTT CG | M | + |
| NVAV-L2031R | CAC ATA TAT CTC TAT TGC ACT C | L | − |
| Bat-L1929F | ATG AAR NTN TGT GCA YTG TTT GA | L | + |
| Han-L2970R | CCN GGN GAC CAY TTN GTD GCA TC | L | − |
| Tal-L2759F | ATW GAR GAT TAT TAT GAT GC | L | + |
| Tal-L2855F | GAA AGG GCA TTN MGA TGG GCN TCA GG | L | + |
| Han-L2984F | ATG TAT GTN AGT GCW GAT GC | L | + |
| Han-L3588R | GGN ATH GAN ACN GCA CAN CCY TCA AA | L | − |
| Han-L-F1 | ATG TAY GTB AGT GCW GAT GC | L | + |
| Han-L-R1 | AAC CAD TCW GTY CCR TCA TC | L | − |
| Han-L-F2 | TGC WGA TGC HAC NAA RTG GTC | L | + |
| Han-L-R2 | GCR TCR TCW GAR TGR TGD GCA A | L | − |
| Shrew-L3370F | GCH CAY CAY TCW GAT GAT GC | L | + |
| Han-L4330R | TGY TGY TTH GCY TGC AT | L | − |
| Han-L4297R | ADN GGD GAY TGY AWN GTC AT | L | − |
| Han-L5239R | TGN AYR CAR TAW GCA TCA TA | L | − |
| NVAV-L4983F | TGA TGC AGA TGT TAG AGA GTG GTT TC | L | + |
| NVAV-L5080F | GTT GAT CCT GAA ATA CAA TGT GCT GGC | L | + |
Mixed bases: B = G, T, C; D = G, A, T; H = A, T, C; K = G, T; M = A, C; N = A, T, G, C; R = A, G; V = A, G, C; W = A, T; Y = C, T.
Phylogenetic analysis
To assess the evolutionary relationships, phylogenetic analysis was performed on the coding regions of the full-length S-, M- and L-genomic segment sequences of NVAV and other representative rodent-, shrew-, mole- and bat-borne hantaviruses, using maximum likelihood and Bayesian methods, implemented in PAUP* (Phylogenetic Analysis Using Parsimony, 4.0b10)36, RAxML Blackbox webserver37 and MrBayes 3.138, under the best-fit GTR+I+Γ model of evolution selected by hierarchical likelihood-ratio test in MrModeltest v2.339 and jModelTest version 0.140. Two replicate Bayesian Metropolis–Hastings Markov Chain Monte Carlo runs, each comprising six chains of 10 million generations sampled every 100 generations with a burn-in of 25,000 (25%), resulted in 150,000 trees overall. Sequences were aligned using Clustal W41, and each genomic segment was treated separately in phylogenetic analyses. The posterior node probabilities were based on 2 million generations and estimated sample sizes over 100 (implemented in MrBayes).
Experimental NVAV infection
To ascertain if experimental NVAV infection resembled that observed with hantaviruses isolated from murid rodents and crocidurine shrews, two-day old Swiss Webster mice were inoculated with 200 pfu of NVAV strain Te34, by the intraperitoneal route. Mice were monitored for weight loss and neurological signs, and the severity of their clinical condition was scored, using a standardized scale42,43. Terminal sera were tested for antibodies against NVAV by the IFA technique6, using NVAV-infected Vero E6 cells spotted onto 10-well slides (Tekdon, Inc., Myakka City, FL) and Alexa Fluor® 488 conjugated goat anti-mouse IgG (H+L) as the secondary antibody (ThermoFisher, Cat. #A-11001, Waltham, MA). Lung, liver, kidney, spleen and brain tissues were collected from moribund mice for histopathology and immunohistochemistry, as well as for quantitation of NVAV RNA by real-time PCR. In addition, brain tissue homogenates from NVAV-infected moribund mice were inoculated intraperitoneally into new litters of infant mice, and NVAV sequences in tissues were compared to that of the NVAV cell-culture isolate and original lung tissue used in the isolation attempts.
Quantitative Taqman real-time PCR
The primers and probe targeting the NVAV S segment were designed using the Primer Express® software version 3.0 (Applied Biosystems, Foster City, CA). The S-segment forward and reverse primer sequences for real-time PCR were 5′–GCGGTGTTAAGGTCCCAAAA–3′ and 5′–CTTCTGCCTTGATAGCTGATTGAG–3′, respectively, and the probe was 5′–CTTTATGTCTCACTCCCAAC–3′. The probe was labeled with the reporter dye FAM at the 5′–end and quencher dye MGB/non fluorescent at the 3′–end, respectively. Each 20-μL reaction contained 1 μL cDNA, 10 μL 2X TaqMan Gene Expression master mix (Applied Biosystems), 0.5 μL forward and reverse primers (36 μM), 0.5 μL fluorescent probe (10 μM), and 7.5 μL double deionized water. The reaction was performed at 50 °C for 2 min and 90 °C for 10 min, followed by 40 cycles at 95 °C for 15 sec and 60 °C for 1 min, in an ABI 7500 Real-Time PCR system (Applied Biosystems). The standard curve was constructed using RNA from NVAV-infected Vero E6 cells. RNA concentration was measured using a NanoDrop spectrophotometer. After RT-PCR using random primers (10 μM), 10-fold serial dilutions of the cDNA were used in duplicate to generate a standard curve.
Discussion
Prior to this report, only two non-rodent-borne hantaviruses had been isolated. One is the prototype shrew-borne hantavirus, known as Thottapalayam virus (TPMV), isolated initially in infant mice injected with spleen tissue homogenate from an Asian house shrew (Suncus murinus), captured during 1964 in southern India44. The other is Imjin virus (MJNV), isolated from the Ussuri white-toothed shrew (Crocidura lasiura), captured near the demilitarized zone in the Republic of Korea6. This report represents the isolation and partial characterization of the first mole-borne hantavirus, known as NVAV, which is widespread across the geographic distribution of its talpid host, the European mole33,34.
Because most of the newfound hantaviruses detected in shrews, moles and insectivorous bats have relied on archival tissues, poor tissue preservation, suboptimal RNA quality and limited sample quantity have all conspired to thwart intensive efforts at virus isolation and at acquiring full-length genome sequences. In addition, the high sequence diversity of non-rodent-borne hantaviruses has hampered reliable primer design. Thus, only a few of the hantaviruses harbored by shrews, moles and bats have been fully sequenced, preventing definitive phylogenetic analysis.
The availability of NVAV in cell culture has facilitated the acquisition of its whole genome, resulting in robust phylogenetic trees based on all three segments, with each topology demonstrating that NVAV occupies a highly divergent lineage. NVAV was closely related to bat-borne hantaviruses, suggesting a complex evolutionary history, presumably involving host-switching events, as shown previously for Asama virus in the Japanese shrew mole (Urotrichus talpoides)16, Oxbow virus in the American shrew mole (Neurotrichus gibbsii)17 and Rockport virus in the eastern mole (Scalopus aquaticus)19, as well as periodic episodes of host/pathogen co-divergence through deep evolutionary time19,25,26.
The Gn and Gc glycoproteins, encoded by the M segment, are antigenic and serve as determinants of the protective humoral immune response45,46. One of the traditional criteria to assign species status to hantaviruses is the absence of antibody cross neutralization, as determined by plaque-reduction neutralization tests (PRNT)47. Previously, no antibody cross neutralization could be demonstrated between TPMV or MJNV and rodent-borne hantaviruses6,48. In the present report, PRNT were not performed because the less than 50% amino acid sequence similarity between the glycoproteins of NVAV and representative hantaviruses for which isolates are available predicted no antibody cross neutralization.
The N-linked glycosylation sites on the Gn (N134, N235, N347, N399 and N609) and Gc (N928) of prototype Hantaan virus (HTNV) strain 76-118 are relatively well conserved among murid rodent-borne hantaviruses and are critical for the conformation of these proteins for transport, receptor binding and antigenicity49,50,51,52. Overall, the first, third and fourth N-linked glycosylation sites on the Gn are also generally preserved among cricetid rodent-borne hantaviruses, as well as the few shrew- and mole-borne hantaviruses for which full-length M-segment sequences are available6,10,12,16,17,19,52. By contrast, in both NVAV strain Te34, reported here, and NVAV strain BE/Namur/TE/2013/1 from Belgium (GenBank KT004446)53, the glycosylation site of N101 on the Gn is unique among all hantaviruses reported to date, including Laibin virus24, the only bat-borne hantavirus which has a whole M-segment sequence. To what extent this unique N-linked glycosylation site impacts the function of the Gn is unknown.
As mentioned above, the L-protein motifs of NVAV closely resembled that of other rodent-borne hantaviruses54, as well as the six shrew- and one mole-borne hantaviruses for which full-length L segment sequences are available6,7,10,12,14,19. Recently, antibodies directed against the middle portion of the Tula virus L protein were used to study the membrane association of the L protein in infected cells55. A similar approach would allow the detection and monitoring of L protein expression in NVAV-infected cells and tissues. These and other studies are now possible with the isolation of NVAV.
The lack of a small animal model has hampered studies aimed at elucidating the pathogenesis of HFRS. Infant mice, as well as certain strains of adult mice (such as C57BL/6, BALB/c, AKR/J, and SJL/J), develop fatal meningoencephalitis following inoculation with HTNV and other HFRS-causing murid rodent-borne hantaviruses56,57,58,59,60,61,62, as well as TPMV and MJNV from crocidurine shrews63 (Gu et al. unpublished observations). Our data indicate that experimental NVAV infection also resulted in neuroinvasive disease. By contrast, infant mice are not susceptible to infection with cricetid rodent-borne hantaviruses, such as Sin Nombre virus (SNV). The absence of convincing human correlates of acute hantaviral neurological disease and of hantavirus antigen and RNA in brains of HFRS and HPS patients argue against the utility and significance of these infant mouse models. That is, the infectivity of hantaviruses in infant and adult mice does not predict the pathogenic potential of hantaviruses in humans.
On the other hand, Syrian hamsters (Mesocricetus auratus) may serve as a useful experimental host. Because of their susceptibility to experimental hantavirus infection, Syrian hamsters have been used extensively in the testing of candidate vaccines for HFRS64,65. Moreover, a lethal HPS model has been developed in adult Syrian hamsters, which recapitulates the clinical and pathological features of HPS in humans66,67. Recently, we demonstrated that infant and juvenile Syrian hamsters develop a fatal disease, characterized by respiratory distress, weight loss, hind limb paralysis and seizures, following intraperitoneal inoculation with MJNV68. High viral loads were found in tissues, and histopathology included hepatic necrosis, widespread inflammation in lungs without pulmonary edema, and extensive MJNV antigens in microvascular endothelial cells. Adult hamsters, on the other hand, were resistant to MJNV infection. Whether or not Syrian hamsters, which are transiently immunosuppressed with dexamethasone and cyclophosphamide, will be rendered susceptible to MJNV infection, as has been shown for SNV, the HPS-causing prototype hantavirus69, awaits future study.
Similarly, if NVAV is pathogenic in immunocompetent or immunosuppressed Syrian hamsters, new insights will be gained about hantavirus pathogenesis, and the development of medical countermeasures for hantavirus disease prevention will be accelerated. Alternatively, the development of a chronic NVAV infection model in Syrian hamsters with persistent virus shedding in saliva, urine and/or feces, that presumably resemble natural NVAV infection in European moles, would serve as a convenient model system in which to study NVAV transmission dynamics and genomic reassortment.
Finally, the long-awaited isolation of a highly divergent mole-borne hantavirus will accelerate the acquisition of new knowledge about its infectivity and pathogenic potential in humans. That is, because NVAV infection is highly prevalent in European moles and because European moles often reside near human habitation, clinicians and healthcare personnel should be vigilant for possible NVAV infection in individuals, who develop febrile illnesses or unusual clinical syndromes following known or suspected exposures. The availability of the NVAV isolate will facilitate the development of robust diagnostic tests in search for human infection and disease.
Additional Information
Accession codes: GenBank: NVAV strain Te34 cell-culture isolate: S segment (KR072621); M segment (KR072622); L segment (KR072623); NVAV strain Te34 amplified from lung tissue: S segment (KM403445).
How to cite this article: Gu, S. H. et al. Isolation and partial characterization of a highly divergent lineage of hantavirus from the European mole (Talpa europaea). Sci. Rep. 6, 21119; doi: 10.1038/srep21119 (2016).
Acknowledgments
This research was supported in part by U.S. Public Health Service grant R01AI075057 from the National Institute of Allergy and Infectious Diseases and P20GM103516 and P30GM114737 from the National Institute of General Medical Sciences, National Institutes of Health; N N303 604538 from the State Committee for Scientific Research in Poland; and by the Healthy Ageing Research Centre project (REGPOT-2012-2013-1, 7FP). The expert assistance provided by Tina Carvalho, of the Biological Electron Microscopy Facility, is gratefully acknowledged, as are the services of the Genomics Core Facility, funded partially by the Centers of Biomedical Research Excellence program (P30GM103341). Grant DBI-1040548 from the National Science Foundation provided funding to acquire the Hitachi HT7700 TEM system and RMC Powertome ultramicrotome. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions S.H.G., M.K. and R.Y. conceived and designed the experiments; J.H., J.M. and M.M. conducted the trapping and field collections; S.H.G., M.K., B.S. and P.P.L. performed the experiments; S.H.G., M.K. and R.Y. contributed mouse model expertise; S.H.G., M.K., B.S., P.P.L. and R.Y. analyzed the data and prepared the figures; and all authors prepared and reviewed the manuscript.
References
- Klempa B. et al. Novel hantavirus sequences in shrew, Guinea. Emerg Infect Dis 13, 520–522 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S. et al. Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis 13, 1420–1423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W. et al. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg Infect Dis 13, 1784–1787 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W. et al. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol J 4, 114 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S. et al. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg 78, 348–351 (2008). [PMC free article] [PubMed] [Google Scholar]
- Song J.-W. et al. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura). J Virol 83, 6184–6191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J., Arai S., Hope A. G., Cook J. A. & Yanagihara R. Novel hantavirus in the flat-skulled shrew (Sorex roboratus). Vector Borne Zoonotic Dis 10, 593–597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J. et al. Genetic diversity of Thottapalayam virus, a hantavirus harbored by the Asian house shrew (Suncus murinus) in Nepal. Am J Trop Med Hyg 85, 540–545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J., Kadjo B., Dubey S., Jacquet F. & Yanagihara R. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d’Ivoire. Virol J 8, 373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S. et al. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology 424, 99–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H. et al. Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol J 10, 160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H., Nicolas V., Lalis A., Sathirapongsasuti N. & Yanagihara R. Complete genome sequence analysis and molecular phylogeny of a newfound hantavirus harbored by the Doucet’s musk shrew (Crocidura douceti) in Guinea. Infect Genet Evol 20, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radosa L. et al. Detection of shrew-borne hantavirus in Eurasian pygmy shrew (Sorex minutus) in Central Europe. Infect Genet Evol 19, 403–410 (2013). [DOI] [PubMed] [Google Scholar]
- Kang H. J., Stanley W. T., Esselstyn J. A., Gu S. H. & Yanagihara R. Expanded host diversity and geographic distribution of hantaviruses in sub-Saharan Africa. J Virol 88, 7663–7667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo S. Q. et al. A new hantavirus from the stripe-backed shrew (Sorex cylindricauda) in the People’s Republic of China. Virus Res 184, 82–86 (2014). [DOI] [PubMed] [Google Scholar]
- Arai S. et al. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proc Natl Acad Sci USA 105, 16296–16301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J. et al. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 388, 8–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J. et al. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS One 4, e6149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J., Bennett S. N., Hope A. G., Cook J. A. & Yanagihara R. Shared ancestry between a mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J Virol 85, 7496–7503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumibcay L. et al. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Côte d’Ivoire. Virol J 9, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. et al. Hantavirus in bat, Sierra Leone. Emerg Infect Dis 18, 159–161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S. et al. Novel bat-borne hantavirus, Vietnam. Emerg Infect Dis 19, 1159–1161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.-P. et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores and rodents. PLoS Pathog 9, e1003159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Novel hantavirus identified in black-bearded tomb bats, China. Infect Genet Evol 31, 158–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara R., Gu S. H., Arai S., Kang H. J. & Song J.-W. Hantaviruses: rediscovery and new beginnings. Virus Res 187, 6–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S. N., Gu S. H., Kang H. J., Arai S. & Yanagihara R. Reconstructing the evolutionary origins and phylogeography of hantaviruses. Trends Microbiol 22, 473–482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. & Zhang Y. Z. The evolution and emergence of hantaviruses. Curr Opin Virol 10, 27–33 (2014). [DOI] [PubMed] [Google Scholar]
- Vaheri A. et al. Uncovering the mysteries of hantavirus infections. Nature Rev Microbiol 11, 539–550 (2013). [DOI] [PubMed] [Google Scholar]
- Jonsson C. B., Figueiredo L. T. M. & Vapalahti O. A global perspective on hantavirus ecology, epidemiology and disease. Clin Microbiol Rev 23, 412–441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya I. N. et al. Features of circulation of hemorrhagic fever with renal syndrome (HFRS) virus among small mammals in the European U.S.S.R. Arch Virol 75, 313–316 (1983). [DOI] [PubMed] [Google Scholar]
- Tkachenko E. A. et al. Potential reservoir and vectors of haemorrhagic fever with renal syndrome (HFRS) in the U.S.S.R. Ann Soc Belg Med Trop 63, 267–269 (1983). [PubMed] [Google Scholar]
- Gligic A. et al. Hemorrhagic fever with renal syndrome in Yugoslavia: epidemiologic and epizootiologic features of a nationwide outbreak in 1989. Eur J Epidemiol 8, 816–825 (1992). [DOI] [PubMed] [Google Scholar]
- Gu S. H., Dormion J., Hugot J.-P. & Yanagihara R. High prevalence of Nova hantavirus infection in the European mole (Talpa europaea) in France. Epidemiol Infect 142, 1167–1171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H. et al. Co-circulation of soricid- and talpid-borne hantaviruses in Poland. Infect Genet Evol 28, 296–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Animal Care and Use Committee. Guidelines for the capture, handling and care of mammals as approved by the American Society of Mammalogists. J Mammal 79, 1416–1431 (1998). [Google Scholar]
- Swofford D. L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts (2003).
- Stamatakis A., Hoover P. & Rougemont J. A rapid bootstrap algorithm for the RAxML Web-Servers. Syst Biol 75, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Posada D. & Crandall K. A. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998). [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanteri M. C. et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest 119, 3266–3277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Roe K., O’Connell M. & Nerurkar V. R. Induction of virus-specific effector immune cell response limits virus replication and severe disease in mice infected with non-lethal West Nile virus Eg101 strain. J Neuroinflammation 12, 178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey D. E., Reuben R., Panicker K. N., Shope R. E. & Myers R. M. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. Indian J Med Res 59, 1758–1760 (1971). [PubMed] [Google Scholar]
- Hooper J. W., Kamrud K. I., Elgh F., Custer D. & Schmaljohn C. S. DNA vaccination with hantavirus M segment elicits neutralizing antibodies and protects against Seoul virus infection. Virology 255, 269–278 (1999). [DOI] [PubMed] [Google Scholar]
- Hooper J. W., Custer D. M., Thompson E. & Schmaljohn C. S. DNA vaccination with the Hantaan virus M gene protects hamsters against three of four HFRS hantaviruses and elicits a high-titer neutralizing antibody response in Rhesus monkeys. J Virol 75, 8469–8477 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A. et al. Bunyaviridae. In: King A. M. Q., Lefkowitz E. J., Adams M. J. & Carstens E. B. (Eds.), Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA, pp. 725–741 (2012). [Google Scholar]
- Chu Y. K., Lee H. W., LeDuc J. W., Schmaljohn C. S. & Dalrymple J. M. Serological relationships among viruses in the Hantavirus genus, family Bunyaviridae. Virology 198, 196–204 (1994). [DOI] [PubMed] [Google Scholar]
- Shi X. & Elliott R. M. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J Virol 78, 5414–5422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepojoki J., Strandin T., Lankinen H. & Vaheri A. Hantavirus structure – molecular interactions behind the scene. J Gen Virol 93, 1631–1644 (2012). [DOI] [PubMed] [Google Scholar]
- Muyangwa M., Martynova E. V., Khaiboullina S. F., Morzunov S. P. & Rizvanov A. A. Hantaviral proteins: structure, functions and role in hantavirus infection. Front Microbiol 6, 1326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P. D., Vincent M. J. & Nichol S. T. Thottapalayam virus is genetically distant to the rodent-borne hantaviruses, consistent with its isolation from the Asian house shrew (Suncus murinus). Virol J 4, 80 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laenen L. et al. Complete genome sequence of Nova virus, a hantavirus circulating in the European mole in Belgium. Genome Announc 3, e00770–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen S. K., Vaheri A. & Plyusnin A. L protein, the RNA-dependent RNA polymerase of hantaviruses. Arch Virol 150, 533–556 (2005). [DOI] [PubMed] [Google Scholar]
- Kukkonen S. K., Vaheri A. & Plyusnin A. Tula hantavirus L protein is a 250 kDa perinuclear membrane-associated protein. J Gen Virol 85, 1181–1189 (2004). [DOI] [PubMed] [Google Scholar]
- Kurata T., Tsai T. F., Bauer S. P. & McCormick J. B. Immunofluorescence studies of disseminated Hantaan virus infection of suckling mice. Infect Immun 41, 391–398 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi T. et al. Experimental infection in newborn mice and rats by hemorrhagic fever with renal syndrome (HFRS) virus. Microbiol Immunol 28, 1345–1353 (1984). [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yanagihara R., Gibbs C. J. Jr. & Gajdusek D. C. Immune spleen cell-mediated protection against fatal Hantaan virus infection in infant mice. J Infect Dis 151, 691–697 (1985). [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yanagihara R., Gibbs C. J. Jr., Amyx H. L. & Gajdusek D. C. Differential susceptibility and resistance of immunocompetent and immunodeficient mice to fatal Hantaan virus infection. Arch Virol 86, 109–120 (1985). [DOI] [PubMed] [Google Scholar]
- Kim G. R. & McKee K. T. Jr. Pathogenesis of Hantaan virus infection in suckling mice: clinical, virologic and serologic observations. Am J Trop Med Hyg 34, 388–395 (1985). [DOI] [PubMed] [Google Scholar]
- Yoo Y. C. et al. Comparison of virulence between Seoul virus strain SR-11 and Hantaan virus strain 76–118 of hantaviruses in newborn mice. Microbiol Immunol 37, 557–562 (1993). [DOI] [PubMed] [Google Scholar]
- Wichmann D. et al. Hantaan virus infection causes an acute neurological disease that is fatal in adult laboratory mice. J Virol 76, 8890–8899 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W., Baek L. J., Schmaljohn C. S. & Yanagihara R. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg Infect Dis 13, 980–985 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C. S., Chu Y.-K., Schmaljohn A. L. & Dalrymple J. M. Antigenic subunits of Hantaan virus expressed by baculovirus and vaccinia virus recombinants. J Virol 64, 3162–3170 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. K., Jennings G. B. & Schmaljohn C. S. A vaccinia virus-vectored Hantaan virus vaccine protects hamsters from challenge with Hantaan and Seoul viruses but not Puumala virus. J Virol 69, 6417–6423 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J. W., Larsen T., Custer D. M. & Schmaljohn C. S. A lethal disease model for hantavirus pulmonary syndrome. Virology 289, 6–14 (2001). [DOI] [PubMed] [Google Scholar]
- Safronetz D., Ebihara H., Feldmann H. & Hooper J. W. The Syrian hamster model of hantavirus pulmonary syndrome. Antiviral Res 95, 282–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H. et al. Lethal disease in infant and juvenile Syrian hamsters experimentally infected with Imjin virus, a newfound crocidurine shrew-borne hantavirus. Infect Genet Evol 36, 231–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato R. L. et al. A lethal disease model for hantavirus pulmonary syndrome in immunosuppressed Syrian hamsters infected with Sin Nombre virus. J Virol 88, 811–819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]