Abstract
Nanostructured materials exhibit unique magnetic, electrical and catalytic properties. These characteristics are determined by the chemical composition, size and shape of the nanostructured components, which are challenging to modulate on such small size scales and to interface with living cells. To address this problem, we are using a self-assembling filament protein, PrgI, as a scaffold for bottom-up inorganic nanostructure synthesis. PrgI is a small protein (80 amino acids) that oligomerizes to form the type-III secretion system needle of Salmonella enterica. We demonstrate that purified PrgI monomers also spontaneously self-assemble into long filaments and that high-affinity peptide tags specific for attachment to functionalized particles can be integrated into the N-terminal region of PrgI. The resulting filaments selectively bind to gold, whether the filaments are assembled in vitro, sheared from cells or remain attached to live S. enterica cell membranes. Chemical reduction of the gold-modified PrgI variants results in structures that are several micrometres in length and which incorporate a contiguous gold surface. Mutant strains with genomically incorporated metal-binding tags retain the secretion phenotype. We anticipate that self-assembled, cell-tethered protein/metal filamentous structures have applications in sensing and energy transduction in vivo.
Keywords: nanostructures, biomineralization, type-III secretion, self-assembly, microbial electrocatalysis
1. Introduction
Nanomaterials exhibit unique properties that arise from their molecular-level organization, geometry and large surface area to volume ratios. For example, gold (Au) nanoprisms absorb in the near-infrared region and have strong localized surface plasmon resonance due to their sharp corners and edges, while Au nanospheres absorb in the ultraviolet-visible (UV/vis) region [1,2]. Thin Au nanosheets crystallize in hexagonal close-packed structures instead of the face-centred cubic structures that are usually observed in nanoparticles of other geometries [2,3]. As a result of these and other interesting physical properties, metal nanostructures are used in opto-electronics, medicine, energy and catalysis, as well as material reinforcement and water desalination [1]. However, for many applications that require nanoscale control over the system components to confer the unique ensemble properties required for such applications, new methods to enable control over molecular features will be critical to achieve precise organization, assembly timing and targeting of the nanostructured materials. Here, traditional top-down approaches are severely constrained, because they are inherently planar, suffer from limited optical resolution and result in nanostructures that are difficult to manipulate [4].
Biomolecules such as lipids, DNA and proteins are emerging as useful scaffolds for building three-dimensional inorganic materials at sub-millimetre length scales from the bottom up [5–7]. Proteins are especially attractive because they undergo amino acid sequence-specific self-assembly and have sequence-based molecular recognition properties. In addition, complex protein assemblies are often thermodynamically favourable. Finally, the diversity afforded by amino acid biochemistry provides numerous bioorthogonal methods for conjugating proteins to inorganic nanostructures and for enabling protein interactions in specific ways with desired ligands, membranes or surfaces [7]. Thus, engineered self-assembling protein systems provide a route to spontaneous and programmable organization of complex structures with new material properties, at shrinking size scales.
Conjugating inorganic materials to self-assembled peptide backbones is an established strategy for the production of functional materials [8]. For example, filamentous phage coat proteins and clathrin have been used as templates for constructing liquid-crystalline ZnS films and titanium dioxide particles, respectively [9,10]. However, these strategies are limited by the inherent size and shape of the protein structures that are used as templates and lack high-resolution material flexibility. For instance, bacteriophage M13 is 900 nm long, and therefore any M13-based materials are limited to 900 nm in the smallest subunit length dimension [9]. Clathrin, as a vesicle coat protein, is used only with spherical geometries. Other protein scaffolds that have been engineered in a variety of contexts to bind to inorganic materials include tobacco mosaic virus coat proteins [11], apoferritin [12], actin [13], curli [14], microtubule [15] and amyloid and amyloid-like fibres [16,17], each with unique advantages and drawbacks.
The type-III secretion system (T3SS) includes a filamentous structure that has all the characteristics of a promising scaffold for metal templating. The T3SS is a membrane-anchored multi-protein apparatus native to many Gram-negative pathogens, such as the highly genetically tractable Salmonella enterica, and the filament extends 60 nm from the outer membrane into the extracellular space [18]. This filament is a homomultimeric protein assembly comprising approximately 120 copies of the PrgI monomer, which has a molecular weight of less than 10 kDa, affording high resolution over resulting assemblies [19]. The monomers spontaneously assemble into a helical, symmetric filament as they are secreted one by one through the nascent membrane-embedded structure. Recent solid-state nuclear magnetic resonance studies indicate that the structure of PrgI comprises two α helices, with the N-terminal amino acids forming a flexible loop extending away from the polymerized filament [20].
With this work, we demonstrate that the PrgI filament makes an excellent scaffold for nanostructured materials both in vitro and tethered to the S. enterica cell surface. We first demonstrate that peptide tags can be integrated into the N-terminal region of PrgI without disrupting filament formation. We incorporate the nickel (II) (Ni2+)-binding polyhistidine (6XHis) tag to PrgI in this manner and allow filaments to form either in vitro or in vivo. After incubating with Ni2+-nitrilotriacetic acid-Au (Ni2+-NTA-Au) nanoparticles, we observe Au-bound PrgI filaments and bundles. For the methods involving native PrgI assembly in S. enterica, we use recombination-mediated genetic engineering techniques in order to protect the secretion phenotype of the natural system. Future applications using this bioinorganic system might include the incorporation of peptide tags that selectively bind heavy metals or minerals for cell-powered sensing applications for bioremediation or energy applications. The metal-functionalized secretion system described herein is therefore a first step in a variety of endeavours for interfacing biological metabolism with synthetic extracellular circuits.
2. Material and methods
2.1. Reagents
Reagents were used as received from commercial sources. In all experiments, water was deionized using a MilliQ system (Millipore). Chemicals were obtained from Fisher Chemical except Tris base (VWR). Molecular biology reagents were obtained from New England Biolabs.
2.2. Cloning
Standard restriction enzyme-based molecular cloning techniques were employed for all cloning. Linearized vectors were obtained using polymerase chain reaction (PCR) with Pfu DNA polymerase and S. enterica genomic DNA for the template, followed by DpnI digest of the resulting PCR products. To construct the pTet-6XH-PrgI plasmid, we used the pTet-ColE1-Cam vector backbone and BglII/BamHI as our restriction enzymes for the insertion of 6XH-prgI. To construct the pET28b(+)-6XH-PrgI plasmid, we used the pET28b(+)-ColE1-Kan vector backbone (Novagen) and NdeI/BamHI as our restriction enzymes for the insertion of prgI. The DH export plasmid, used to test secretion in engineered strains, was constructed for a previous study [21]. More details, including a list of plasmids and PCR primers used in this study, are available in the electronic supplementary material, tables SI and SII.
2.3. Strains and growth conditions
All non-T3SS-induced strains were grown in LB-Lennox medium at 225 r.p.m. and 37°C. T3SS-induced strains were grown in T3SS-inducing medium (17 g l−1 NaCl) at 120 r.p.m. and 37°C as described in [21]. Escherichia coli BL21 cultures expressing pET28b(+)-6XH-PrgI were subcultured 1 : 100 from an overnight culture in 500 ml Terrific Broth, in a 1 litre baffled flask. Cultures were grown at 225 r.p.m. and 30°C for 6 h and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) after 2 h. In this study, all engineered PrgI proteins also had the solubility-enhancing mutations V65A and V67A, which do not impact the protein's oligomerization dynamics [22]. All S. enterica strains were ΔflhCD to avoid production of flagella [23].
2.4. Expression and purification of 6XH-PrgI
6XHis-PrgI was overexpressed in E. coli BL21 DE(3) pLysS cells using a pET28b(+) IPTG-inducible vector as described. The cells were centrifuged at 6000g, the cell pellets were resuspended in 30 ml Ni2+ column buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 20 mM imidazole, 500 mM NaCl buffer, pH 7.3), and the resuspended cells were lysed by sonication. The insoluble fraction was removed by centrifugation at 17 000g for 1 h. The supernatant was decanted and passed through a Ni-NTA affinity column (GE Healthcare) at room temperature under native conditions, washed with 60 ml Ni2+ column buffer, then eluted in Ni2+ elution buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 500 mM imidazole, 500 mM NaCl buffer, pH 7.3). The protein solution was buffer-exchanged into 2.5 ml 20 mM HEPES, 50 mM NaCl buffer pH 5.5 using a PD-10 desalting column (GE Healthcare), then concentrated to approximately 1 mg ml−1 using 3 kDa molecular weight cut-off spin concentration columns (Sartorius). Purified 6XH-PrgI monomers were allowed to self-assemble into filamentous structures at 4°C over three weeks.
2.5. Ni2+-NTA-Au conjugation with filaments
Approximately 10 mg of 6XH-PrgI filaments were incubated with an 8 M excess of Ni2+-NTA-Au nanoparticles (Nanoprobes) at 4°C for 16 h in 5 ml 20 mM HEPES, 50 mM NaCl buffer, rocking gently. Excess nanoparticles were removed by size exclusion using a NAP-5 column (GE Healthcare). Labelled filaments were stored in 2.5 ml phosphate-buffered saline.
2.6. Au reduction for growth of contiguous Au structures
6XHis-PrgI filaments were labelled with 17 nM Au nanoparticles as described, and 1 ml of labelled filament solution was incubated with 0.01% HAuCl4, 40 mM NH2OH and 0.05% Triton X-100 (Sigma Aldrich) at room temperature for 5 min in a cuvette. UV/Vis absorbance was measured at 526 nm. Structures were inspected via transmission electron microscopy (TEM).
2.7. Isolation of T3SS needle structures
Cells were grown in T3SS-inducing conditions in 1 litre of T3SS-inducing medium as described and harvested in late log phase, then resuspended in 30 ml TE buffer (10 mM Tris Cl/1 mM EDTA, pH7.5) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Needles were mechanically sheared from cells as described in Kubori et al. [24]. Briefly, the cell suspension was vortexed three times for 1.5 min each and passed six times through a 25-gauge needle. The cells were removed by centrifugation at 9000g at 4°C for 20 min. The supernatant was centrifuged at 100 000g at 4°C for 1 h. The pellet contained the needles, which were resuspended in TE buffer, washed twice by the same method and visualized by TEM.
2.8. SDS-PAGE and western blotting
Protein samples were prepared for sodium dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE) by boiling for 6 min in 4× Laemmli buffer with 8% SDS. The samples were then loaded onto 12.5% polyacrylamide gels and subjected to 130 V for 70 min. For samples analysed by Coomassie staining, the gels were stained according to the standard methods. For samples analysed by western blotting, the samples were then blotted from the gels to polyvinylidene fluoride membranes following standard procedures. Western blots were completed using horseradish peroxidase-conjugated anti-FLAG and anti-histidine antibodies (Sigma) against the incorporated 1× C-terminal FLAG tags and 6× N-terminal His tags on all proteins of interest. Detection was performed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) over a total exposure time of 10 min on a Bio-Rad Chemidoc Imager, and blots were auto-corrected for contrast.
2.9. Creation of ΔflhCD and ΔprgI strains and integration of 6XHis-prgI in the Salmonella enterica genome
The method of Thomason et al. [25] was employed to make a homologous recombination-based flhCD and prgI knockout strain, and then to reintroduce the mutant gene 6XHis-prgI in the prgI genetic locus. The ΔflhCD strain was created first, and subsequent modifications to remove and replace the prgI gene followed. All DNA primers used to make deletions, insertions and confirm correct sequencing are listed in the electronic supplementary material.
2.10. DH secretion assay
Salmonella enterica strains were grown with and without T3SS-inducing conditions for 6–8 h. Cultures were centrifuged and supernatants were collected, treated and probed for the presence of DH by western blotting with an anti-FLAG antibody as described in [21].
2.11. Dynamic light scattering
Dynamic light scattering (DLS) was performed using a Zetasizer Nano (Malvern Instruments). Before analysis samples were filtered through 0.22 µm filtres (Millipore). Measurements were taken in 20 mM HEPES, 50 mM NaCl buffer pH 5.5 at 25°C in backscattering mode with 10 × 10 mm quartz cuvettes.
2.12. Transmission electron microscopy
TEM images were taken using an FEI Technai 12 transmission electron microscope with an accelerating voltage of 120 kV. Samples were desalted using NAP-5 desalting columns (GE Healthcare), concentrated to approximately 50 µM using 100 kDa molecular weight cut-off spin filtres (Millipore), incubated on Formvar-coated copper mesh grids for 5 min and wicked off using filter paper and dried in air briefly. These grids were then quickly washed with water and immediately wicked again. The samples were stained with 1% UO2(OAc)2 for 2 min, and again wicked and dried.
3. Results
3.1. N-terminal modification to PrgI does not disrupt filament assembly in vivo
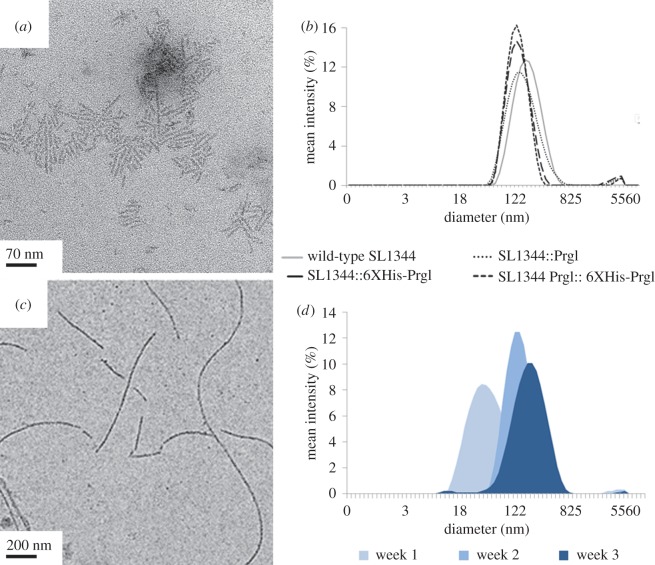
We used the filament-forming PrgI protein of S. enterica as a template for the incorporation of inorganic materials. Incorporation of inorganic materials requires a binding peptide which is not in the wild-type filament. We chose to N-terminally append this peptide because structural studies indicate this end is flexible, minimizing the possibility of steric hindrance (electronic supplementary material, figure S1) [20]. We first set out to ensure the N-terminus of PrgI is indeed amenable to alteration and demonstrate that fusions to the N-terminus would not prevent filament formation. To this end, we created a genetic construct that results in a fusion of the Ni2+-binding polyhistidine tag (HHHHHH) to the flexible N-terminus of PrgI (6X-His PrgI). We knocked out the genomic copy of prgI to make the S. enterica ΔprgI strain and introduced 6XHis-prgI on a plasmid under the control of an anhydrotetracycline (aTc)-inducible promoter (pTet), to ensure that only polyhistidine-modified PrgI would be produced by the ΔprgI stain. After growing under T3SS-inducing conditions and inducing 6X-His PrgI expression with the addition of aTc, we mechanically sheared the needles from cells. We compared the size and morphology of mutant and wild-type needles expressed from the inducible plasmid with wild-type needles expressed from the genome using TEM (figure 1a) and DLS (figure 1b). We confirmed the identity of the protein by anti-polyhistidine western blotting (electronic supplementary material, figure S2). These techniques revealed that episomal expression of wild-type and polyhistidine-tagged PrgI in ΔprgI strains produced needles of the same hydrodynamic radius and geometry as wild-type strains.
Figure 1.
Needle morphology of 6XH-PrgI is similar to wild-type PrgI both in vivo and in vitro. (a) TEM images of 6XHis-PrgI needles mechanically sheared from cells, which are morphologically similar to wild-type needles. (b) DLS traces of T3SS needles mechanically sheared from cells, indicating that the S. enterica wild-type strain with no plasmid, wild-type strains expressing PrgI and 6XHis-PrgI from the pTet promoter, and ΔprgI::pTet-PrgI strains all form needles. (c) TEM images and (d) DLS traces tracking needle growth of recombinant purified 6XHis-PrgI monomers into filaments over three weeks. Image is from the three-week timepoint. Graphs use a log scale on the x-axis. (Online version in colour.)
3.2. Recombinant mutant PrgI monomers form filaments in vitro
The complemented mutant needles were 50–80 nm long, but many applications in energy and sensing require much longer filaments. Therefore, we also constructed an inducible plasmid for the recombinant production of 6XHis-PrgI in E. coli in order to polymerize PrgI-based filaments in vitro. We expressed and purified the protein from cell lysate using nickel chromatography and confirmed its presence by western blotting (electronic supplementary material, figures S2 and S3). Filaments assemble spontaneously in solution and are visible by TEM within 24 h, but acidic buffer conditions slow the assembly and prevent the formation of large aggregates. Filament growth was monitored by DLS over the course of three weeks and the heterogeneous mixture of long needles was observed using TEM (figure 1c,d). The recombinant needles were 1–2 µm in length. Recombinant expression and purification of a non-assembling 6XH-PrgI mutant, 6XH-PrgIQ26A/K50A, which is unable to form the hydrogen bond between the helices of the protein, does not result in visible filaments (electronic supplementary material, figure S4) [26].
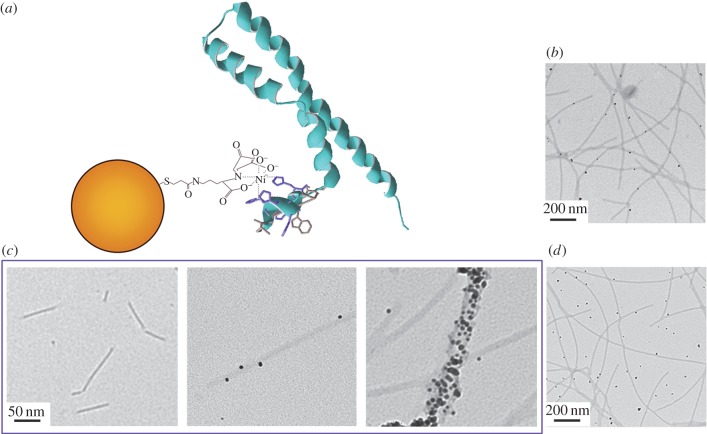
3.3. Building a protein nanowire
The polyhistidine tag is commonly used for affinity chromatography in protein purification, and the 6XHis-PrgI structures were designed to bind to bivalent nickel cations. To test this, we confirmed the conjugation of assembled 6XHis-PrgI filament structures with Ni2+-NTA-functionalized Au nanoparticles (figure 2a). The recombinant needle structures were able to selectively bind these Au particles (figure 2b). Increasing concentrations of Ni2+-NTA-Au nanoparticles resulted in increased binding as observed by TEM, as well as notable aggregation of filaments, which we termed ‘bundling’ (figure 2c). The nanoparticles did not associate with PrgI filaments that did not incorporate a 6XHis-tag (figure 2d).
Figure 2.
Protein–ligand interaction of 6XHis-PrgI. (a) Schematic depicting 6XHis-PrgI bound to a Ni2+-NTA-Au nanoparticle of 5 nm diameter. Modified from PrgI structure, PDB code 2LPZ [20]. (b) TEM images showing recombinant 6XHis-PrgI needles bound to 5 nM Ni2+-NTA-Au particles. (c) Binding density is linearly dependent on ligand concentration. In the panels from left to right, the concentration of Ni2+-NTA-Au is 0 nM, 5 nM and 17 nM. (d) Non-histidine-tagged PrgI needles do not associate with Ni2+-NTA-Au nanoparticles. The TEM image was taken before washing the structures, to exhibit the random distribution of Au nanoparticles in the protein solution. (Online version in colour.)
Although the recombinant 6XHis-PrgI structures were able to bind the Ni2+-NTA-Au nanoparticles in a concentration-dependent fashion, the metal coverage was incomplete. We next tested whether the protein–metal conjugates could be reduced to form contiguous metal structures. We reacted HAuCl4 with hydroxylamine, a mild reducing agent, in solution with metal-conjugated filaments. Tween 20, a non-ionic detergent, was added at small concentrations to the reaction to reduce protein aggregation. Soluble Au3+ was deposited and reduced to Au(0) from HAuCl4 to ‘grow’ the Au nanoparticles that were nucleated on the protein filaments [27]. The reaction was monitored by UV/Vis absorption spectroscopy, which revealed the emergence of a surface plasmon resonance at 525 nm, typical of nanoscale Au particles (figure 3a). The reaction resulted in the formation of crude nanowire networks and large Au-containing structures. The morphology and composition of the Au structures was confirmed by energy-dispersive X-ray spectroscopy and TEM (figure 3b; electronic supplementary material, figure S5), and protein filaments which did not include an N-terminal histidine tag did not result in network-like structures (electronic supplementary material, figure S6).
Figure 3.
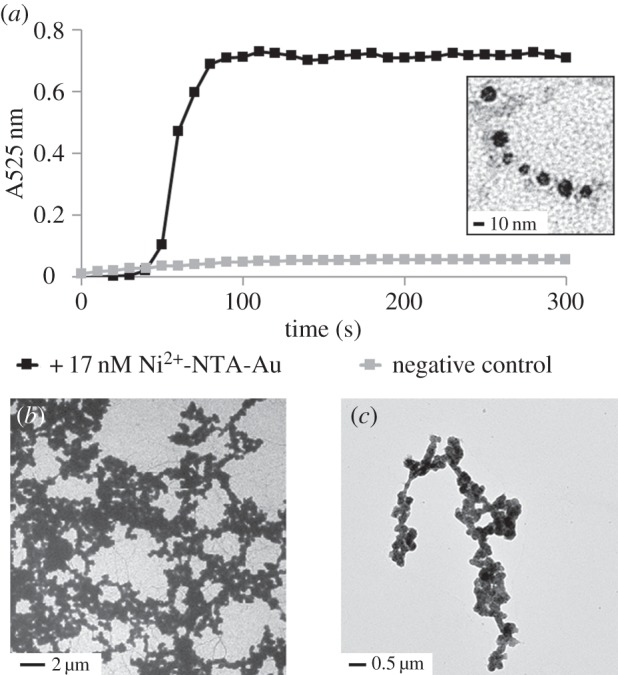
Au reduction to make contiguous nanowires. (a) Au reduction on recombinant 6XHis-PrgI needles nucleated with Au particles can be monitored in solution by UV/Vis spectroscopy at 525 nm. Negative control indicates non-Au-conjugated 6XHis-PrgI in solution with the same reagents. Inset: 6XHis-PrgI filament seeded with Au nanoparticles that have been enlarged due to partial reduction. (b) TEM images of contiguous Au networks and (c) structures.
3.4. Conjugating Au nanoparticles to a living cell
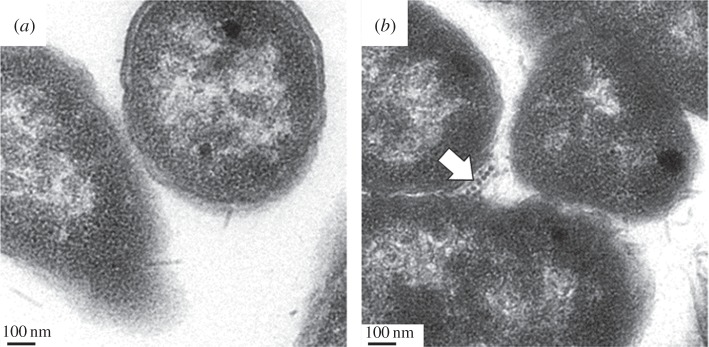
We recognize that, for some applications, it will be useful to interact inorganic materials with protein filaments that are still in contact with the cell. Thus, we next asked whether we could conjugate the Ni2+-NTA-Au to the filaments formed from 6XHis-PrgI while they are still part of the T3SS apparatus embedded in the membranes of living cells. To accomplish this, we grew the S. enterica ΔprgI::pTet-6XHis-prgI strain in T3SS-inducing conditions and added Ni2+-NTA-Au to the cultures. We isolated and washed the cells and then compared them with wild-type cells using TEM. In doing so, we observed several T3SS needles that were labelled with Au nanoparticles (figure 4). We did not observe Au-labelled needles in wild-type strains to which the nanoparticles were added.
Figure 4.
Cell surface-bound Au needles. (a) Wild-type S. enterica cells expressing T3SS. (b) Salmonella enterica pTet:6XHis-prgI cells expressing T3SS conjugated to Au nanoparticles, indicated by arrow.
3.5. Rescuing secretion phenotype for heterologous protein secretion
The native function of the T3SS is protein secretion, and we investigated the ability of the PrgI mutant strains to secrete heterologous proteins. The S. enterica SL1344-derived ΔprgI strain is not secretion-competent because PrgI is necessary for type-III secretion. We tested this using a model protein, the DH domain from human intersectin, fused to the N-terminus of a natively secreted substrate (figure 5) [21]. Complementation of PrgI in this strain does not rescue the secretion phenotype, even though complemented cells do construct T3SS needles that are morphologically indistinguishable from wild-type needles by TEM. Therefore, to restore the secretion phenotype in S. enterica while maintaining its ability to produce metal-binding needles, we used a recombination-mediated genetic engineering strategy to reintroduce the 6XHis-prgI gene back into the ΔprgI strain at the native locus for prgI (electronic supplementary material, figure S7) [25]. This strain, S. enterica 6XHis-prgI, was able to secrete the DH domain at wild-type level titres. It is intriguing that a genomic copy of prgI is required for secretion but a plasmid-complemented copy of the gene is sufficient for needle construction, and understanding of this feature will require further study into the genetic architecture and regulation of the pPrgH operon of Salmonella pathogenicity island-1.
Figure 5.
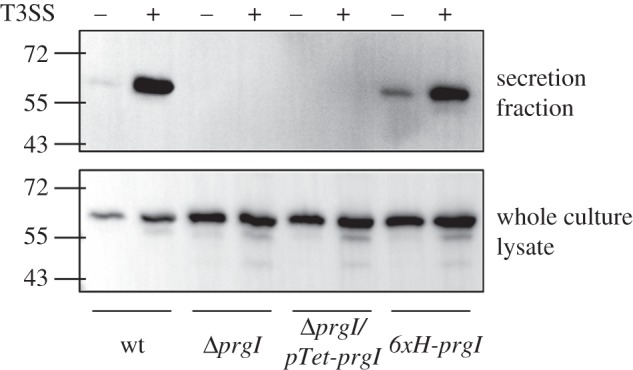
Restoring the secretion phenotype. Western blot comparing DH secretion and expression in S. enterica SL1344 wild-type, ΔprgI, ΔprgI::pTet-6XHis-PrgI and 6XHis-PrgI strains in T3SS-inducing and non-inducing conditions.
4. Discussion and conclusion
The T3SS needle can be used as a flexible template for the production of wire-like nanostructures of variable composition. In this study, we used Au nanoparticles; however, genetic incorporation of other metal- or mineral-binding peptides and subsequent polymerization would allow for templating of materials in a variety of redox states. The high resolution afforded by the small size of the PrgI monomer introduces the possibility of constructing multi-component metallic wires with distinct nanoscale elements, without first anchoring them to a silicon substrate. PrgI monomer size may also support applications in surface plasmon-mediated energy transfer and molecular transport junctions because these systems require precise, high-resolution spacing between device sections. Additionally, we engineered the strain to preserve the native secretion phenotype so that, in the future, it is possible for heterologous electron transfer proteins to be secreted from the system to synergistically complement nanowire-mediated cross-membrane electron transfer in a tuneable fashion.
There is tremendous potential to harness metabolic energy from intracellular redox reactions by building a synthetic electronic conduit across membranes. Yet, to date, little work has been done to precisely interface metal-functionalized proteins with cells beyond exploring the natural systems that perform this function, and all biomolecular templates for metal conjugation have been studied in vitro. Natural systems in Shewanella oneidensis MR-1 and Geobacter species construct protein-based, conductive, nanowire-like pili in oxygen-limiting conditions that extend beyond the cell membranes in order to make contact with extracellular electron-accepting solid phases [28]. Yet these systems are poor candidates for engineering well-controlled nanostructure–cell interfaces, because they are necessary for the organisms' central metabolism and are not well characterized or genetically tractable. Furthermore, Shewanella and Geobacter express conductive pili exclusively in strict anaerobic conditions, and neither organism can be easily grown in a fermenter. The T3SS filament of S. enterica offers a tuneable scaffold that retains its interaction with the cellular membrane, thus providing a starting point for mimicking these fascinating natural systems.
Previously, Jensen et al. [29] showed that integrating a protein-based electron transfer chain in a bacterial cell envelope, with the final electron acceptor localized to the outer membrane, caused an eightfold increase in Fe(III) citrate reduction compared with wild-type E. coli. Our efforts to incorporate a metallic T3SS-templated nanowire in the cell membrane present a method for extension of this work; as the rate of reduction does not scale linearly with surface area, linking a membrane-embedded nanowire with a protein-based electron transport chain that is on the same size scale can increase the efficiency of electron transfer out of the cell. Furthermore, metallized protein filaments that are attached to cell surfaces to create nanowire-like electron conduits would allow for a delocalized electron transport system that is more efficient than stepwise electron transport in a protein-based electron transfer chain. In the future, bioenergy applications may benefit from integrating a chain of redox-active proteins with a membrane-embedded nanowire to mediate electron transfer across the membrane.
Supplementary Material
Acknowledgements
The authors thank the Dr Caroline Ajo-Franklin laboratory and Dr Darryl Sasaki for helpful discussions and equipment usage. The authors thank the Tullman-Ercek laboratory for insightful feedback on experimental design and the manuscript.
Authors' contributions
A.A. and D.T.E. designed the project. A.A performed the experiments. A.A. and D.T.E wrote the manuscript and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
The work was funded by the UC Berkeley/Sandia National Laboratories Excellence in Engineering Graduate Fellowship (A.A.).
References
- 1.Fan Z, Huang X, Tan C, Zhang H. 2015. Thin metal nanostructures: synthesis, properties and applications. Chem. Sci. 6, 95–111. ( 10.1039/c4sc02571g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia Y, Xiong Y, Lim B, Skrabalak SE. 2009. Shape-controlled synthesis of metal nanocrystals: simple chemistry meets complex physics? Angew. Chem. Int. Ed. 48, 60–103. ( 10.1002/anie.200802248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, et al. 2011. Synthesis of hexagonal close-packed gold nanostructures. Nat. Commun. 2, 1–6. ( 10.1038/ncomms1291) [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Okazaki S. 2000. Pushing the limits of lithography. Nature 406, 1027–1031. ( 10.1038/35023233) [DOI] [PubMed] [Google Scholar]
- 5.Xu G-K, Li Y, Li B, Feng X-Q, Gao H. 2009. Self-assembled lipid nanostructures encapsulating nanoparticles in aqueous solution. Soft Matter 5, 3977–3983. ( 10.1039/B906918F) [DOI] [Google Scholar]
- 6.Yin P, Choi HM, Calvert CR, Pierce NA. 2008. Programming biomolecular self-assembly pathways. Nature 451, 318–322. ( 10.1038/nature06451) [DOI] [PubMed] [Google Scholar]
- 7.Gradišar H, Jerala R. 2014. Self-assembled bionanostructures: proteins following the lead of DNA nanostructures. J. Nanobiotechnol. 12, 4 ( 10.1186/1477-3155-12-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CL, Rosi NL. 2010. Peptide-based methods for the preparation of nanostructured inorganic materials. Angew. Chem. Int. Ed. 49, 1924–1942. ( 10.1002/anie.200903572) [DOI] [PubMed] [Google Scholar]
- 9.Lee SW, Mao C, Flynn CE, Belcher AM. 2002. Ordering of quantum dots using genetically engineered viruses. Science 296, 892–895. ( 10.1126/science.1068054) [DOI] [PubMed] [Google Scholar]
- 10.Schoen AP, Schoen DT, Huggins KN, Arunagirinathan MA, Heilshorn SC. 2011. Template engineering through epitope recognition: a modular, biomimetic strategy for inorganic nanomaterial synthesis. J. Am. Chem. Soc. 133, 18 202–18 207. ( 10.1021/ja204732n) [DOI] [PubMed] [Google Scholar]
- 11.Shenton W, Douglas T, Young M, Stubbs G, Mann S. 1999. Inorganic–organic nanotube composites from template mineralization of tobacco mosaic virus. Adv. Mater. 11, 253–256. () [DOI] [Google Scholar]
- 12.Iwahori K, Yoshizawa K, Muraoka M, Yamashita I. 2005. Fabrication of ZnSe nanoparticles in the apoferritin cavity by designing a slow chemical reaction system. Inorg. Chem. 44, 6393–6400. ( 10.1021/ic0502426) [DOI] [PubMed] [Google Scholar]
- 13.Henry E, Dif A, Schmutz M, Legoff L, Amblard F, Marchi-Artzner V, Artzner F. 2011. Crystallization of fluorescent quantum dots within a three-dimensional bio-organic template of actin filaments and lipid membranes. Nano. Lett. 11, 5443–5448. ( 10.1021/nl203216q) [DOI] [PubMed] [Google Scholar]
- 14.Chen AY, Deng Z, Billings AN, Seker UO, Lu MY, Citorik RJ, Zakeri B, Lu TK. 2014. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523. ( 10.1038/nmat3912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boal AK, Headley TJ, Tissot RG, Bunker BC. 2003. Microtubule-templated biomimetic mineralization of lepidocrocite. Adv. Funct. Mater. 14, 19–24. ( 10.1002/adfm.200304435) [DOI] [Google Scholar]
- 16.Scheibel T, Parthasarathy R, Sawicki G, Lin XM, Jaeger H, Lindquist SL. 2003. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl Acad. Sci. USA 100, 4527–4532. ( 10.1073/pnas.0431081100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acar H, Garifullin R, Guler MO. 2011. Self-assembled template-directed synthesis of one-dimensional silica and titania nanostructures. Langmuir 27, 1079–1084. ( 10.1021/la104518g) [DOI] [PubMed] [Google Scholar]
- 18.Burkinshaw BJ, Strynadka NC. 2014. Assembly and structure of the T3SS. Biochim. Biophys. Acta 1843, 1649–1663. ( 10.1016/j.bbamcr.2014.01.035) [DOI] [PubMed] [Google Scholar]
- 19.Rathinavelan T, Lara-Tejero M, Lefebre M, Chatterjee S, McShan AC, Guo DC, Tang C, Galan JE, De Guzman RN. 2014. NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J. Mol. Biol. 426, 2958–2969. ( 10.1016/j.jmb.2014.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loquet A, et al. 2012. Atomic model of the type III secretion system needle. Nature 486, 276–279. ( 10.1038/nature11079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azam A, Li C, Metcalf K, Tullman-Ercek D. 2015. Type III secretion as a generalizable strategy for the production of full-length biopolymer-forming proteins. Biotech. Bioeng. 9999, 1–8. ( 10.1002/bit.25656) [DOI] [PubMed] [Google Scholar]
- 22.Poyraz O, et al. 2010. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 17, 788–792. ( 10.1038/nsmb.1822) [DOI] [PubMed] [Google Scholar]
- 23.Ikebe T, Iyoda S, Kutsukake K. 1999. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 74, 179–183. ( 10.1266/ggs.74.179) [DOI] [PubMed] [Google Scholar]
- 24.Kubori T, Sukhan A, Aizawa SI, Galán JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl Acad. Sci. USA 97, 10 225–10 230. ( 10.1073/pnas.170128997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 78, 116.1–1.16.24. ( 10.1002/0471142727.mb0116s78) [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Ouellette AN, Egan CW, Rathinavelan T, Im W, De Guzman RN. 2007. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371, 1304–1314. ( 10.1016/j.jmb.2007.06.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown KR, Natan MJ. 1998. Hydroxylamine seeding of colloidal Au nanoparticles in solution and on surfaces. Langmuir 14, 726–728. ( 10.1021/la970982u) [DOI] [Google Scholar]
- 28.Shi L, Squier TC, Zachara JM, Fredrickson JK. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20. ( 10.1111/j.1365-2958.2007.05783.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen HM, Albers AE, Malley KR, Londer YY, Cohen BE, Helms BA, Weigele P, Groves JT, Ajo-Franklin CM. 2010. Engineering of a synthetic electron conduit in living cells. Proc. Natl Acad. Sci. USA 107, 19 213–19 218. ( 10.1073/pnas.1009645107) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.