Abstract
Xenarthra (armadillos, sloths, and anteaters) constitutes one of the four major clades of placental mammals. Despite their phylogenetic distinctiveness in mammals, a reference phylogeny is still lacking for the 31 described species. Here we used Illumina shotgun sequencing to assemble 33 new complete mitochondrial genomes, establishing Xenarthra as the first major placental clade to be fully sequenced at the species level for mitogenomes. The resulting data set allowed the reconstruction of a robust phylogenetic framework and timescale that are consistent with previous studies conducted at the genus level using nuclear genes. Incorporating the full species diversity of extant xenarthrans points to a number of inconsistencies in xenarthran systematics and species definition. We propose to split armadillos into two distinct families Dasypodidae (dasypodines) and Chlamyphoridae (euphractines, chlamyphorines, and tolypeutines) to better reflect their ancient divergence, estimated around 42 Ma. Species delimitation within long-nosed armadillos (genus Dasypus) appeared more complex than anticipated, with the discovery of a divergent lineage in French Guiana. Diversification analyses showed Xenarthra to be an ancient clade with a constant diversification rate through time with a species turnover driven by high but constant extinction. We also detected a significant negative correlation between speciation rate and past temperature fluctuations with an increase in speciation rate corresponding to the general cooling observed during the last 15 My. Biogeographic reconstructions identified the tropical rainforest biome of Amazonia and the Guiana Shield as the cradle of xenarthran evolutionary history with subsequent dispersions into more open and dry habitats.
Keywords: mammals, Xenarthra, shotgun Illumina sequencing, molecular phylogenetics, mitochondrial genomes, molecular dating
Introduction
Xenarthra (armadillos, anteaters, and sloths), with only 31 living species currently recognized (Wetzel 1985; Gardner 2008), is the least diversified of the 4 major groups of placental mammals (Meredith et al. 2011). As old, South American endemics, their evolutionary history has been shaped by their geographical isolation for the greater part of the Cenozoic until the Great American Biotic Interchange (GABI) triggered by the formation of the Isthmus of Panama (Marshall et al. 1982; Bacon et al. 2015). Xenarthrans have been major components during this interchange, with many taxa successfully dispersing into Central and North America (Patterson and Pascual 1968; McDonald 2005). The reduced number of extant species is likely the result of extinctions, especially during the terminal Pleistocene extinctions that occurred around 11,000 years ago. This major extinction event seems to have preferentially affected the largest terrestrial forms, such as giant ground sloths and glyptodonts (Lyons et al. 2004). Surviving xenarthran species can be regarded as unique witnesses of the oldest South American endemic radiation of placental mammals (Delsuc et al. 2004). They thus represent an ideal model for understanding the biogeographical patterns and diversification processes at work during South America’s “splendid isolation” (Simpson 1980; Moraes-Barros and Arteaga 2015).
The last decade has seen much progress in elucidating xenarthran phylogeny, thanks to new molecular data. Most of these studies have focused on their position within placental mammals because morphological studies placed them as a sister group of other placentals, referred to as the Epitheria hypothesis (Novacek 1992). The seminal molecular phylogenetic studies of placentals have shown convincingly that armadillos, anteaters, and sloths (Xenarthra) constitute one of the four major placental clades, establishing them as an essential component of the early placental radiation alongside Afrotheria, Laurasiatheria, and Euarchontoglires (Madsen et al. 2001; Murphy, Eizirik, Johnson, et al. 2001; Murphy, Eizirik, O'Brien, et al. 2001). Yet, despite studies using multigene data (Delsuc et al. 2002; Amrine-Madsen et al. 2003; Meredith et al. 2011), retroposon insertions (Churakov et al. 2009; Nishihara et al. 2009) and genome-wide data (McCormack et al. 2012; Song et al. 2012; Romiguier et al. 2013), their exact position within placentals remains contentious.
Within Xenarthra, molecular studies have converged upon a robust phylogeny of the 14 recognized genera (Delsuc et al. 2002, 2003, 2012; Möller-Krull et al. 2007). This phylogenetic framework has served for specifying the timing of their diversification in South America during the Cenozoic (Delsuc et al. 2004). These studies have also helped refine xenarthran taxonomy with, for instance, the recognition of two distinct families within anteaters (Delsuc et al. 2001; Barros et al. 2008) to reflect the deep divergence (about 40 Ma) estimated between the pygmy anteater (Cyclopes didactylus; Cyclopedidae) and the giant anteater and tamanduas (Myrmecophagidae). Molecular data have also recently revealed an ancient divergence of fairy armadillos, supporting the classification of the two living species in two distinct genera (Chlamyphorus and Calyptophractus) and grouped into the distinct subfamily Chlamyphorinae (Delsuc et al. 2012).
Despite these significant advances, a fully resolved species-level phylogeny is still lacking for Xenarthra. The reason for this includes the rarity of many of its constitutive species and a dated taxonomy with persistent uncertainty on species delimitations and distributions due to a lack of basic field data (Superina et al. 2014). Establishing a good reference framework would be critical for the conservation of this peculiar placental group, which includes a number of endangered species. According to the last IUCN Red list Assessment (IUCN Red List of Threatened Species 2015), 6 of the 31 living species (19%) were considered threatened, while the conservation status of 5 species could not be assessed due to lack of data. One particularly striking example is the iconic Brazilian three-banded armadillo for which no molecular data currently exist and for which only scarce biological information can be found in the literature (Superina et al. 2014; Feijó et al. 2015). More generally, 9 species among the 31 currently recognized xenarthrans still have not been investigated via molecular means are thus not represented in GenBank.
The scarcity of molecular data for this group is perhaps best reflected by the fact that only four xenarthran complete mitochondrial genomes are available, only two of which have been published (Arnason et al. 1997, 2002). With current advances in DNA sequencing, mitogenomes have become a standard for estimating well-sampled species-level phylogenies in numerous mammalian groups (Hassanin et al. 2012; Finstermeier et al. 2013; Guschanski et al. 2013; Mitchell et al. 2014). In facilitating access to museum specimens and recently extinct species, next-generation sequencing techniques promise the development of museomics (Mason et al. 2011; Rowe et al. 2011; Fabre et al. 2014) and a rebirth of ancient DNA studies based on complete mitogenomes (Enk et al. 2011; Paijmans et al. 2013). Here we have used an Illumina shotgun sequencing approach to obtain and assemble 33 mitogenomes representing all living xenarthran species using both modern tissues and historic museum samples, including a type specimen. This allowed us to establish a robust phylogenetic framework and timescale that we have used as a reference to evaluate the systematics and species delineation within xenarthrans, and to study their diversification history and biogeography with respect to the paleoenvironmental changes that took place in South America throughout the Cenozoic.
Results and Discussion
Shotgun Mitogenomics and Phylogenetics
Using shotgun Illumina sequencing of genomic DNA and a combination of de novo assembly and mapping methods, we were able to successfully assemble 33 new complete mitochondrial genomes from individuals representing all 31 living xenarthran species (table 1). Using ancient DNA laboratory conditions to avoid potential contaminations, and cross-contamination, we extracted DNA from a variety of modern (ear biopsies, internal organs, and blood) and historical (dried skins and bones) tissue samples, whose collection dates ranged from 1896 to 2011 and have been stored in different collections since that time (table 1). Not surprisingly, the quality and quantity of DNA varied dramatically among samples. Nevertheless, shotgun Illumina sequencing from each total genomic DNA extract allowed the successful assembly of the mitogenome, with mean depth of coverage varying from 2,319X for the pygmy sloth (Bradypus pygmaeus) to 7X for the six-banded armadillo (Euphractus sexcinctus). The total number of reads required to obtain reasonable coverage was highly variable, as was also the proportion of mitochondrial reads recovered from each sample. At the two extremes of the range stand the pink fairy armadillo (Chlamyphorus truncatus) with 11.76% of mitochondrial reads from which we obtained a 244X coverage depth with only 570,194 total reads, and one of the two hairy long-nosed armadillo (Dasypus pilosus) samples that contained only 0.02% of mitochondrial reads and required the use of 44,847,476 total reads to reach 23X depth of coverage (table 1). These results confirmed that there is no a priori predictor of the final mitogenome coverage that may be obtained for a given sample, because it appears to be mainly dependent on the mitochondrial/nuclear cell ratio in the sampled tissue (Tilak et al. 2015). Also, predicting the amount of final mitogenome coverage from shotgun data is further complicated for museum specimens by the fraction of endogenous versus exogenous DNA in the sample. Our results nevertheless illustrate the utility of a shotgun approach for mammalian mitogenomics (Enk et al. 2011; Botero-Castro et al. 2013) enabling the efficient use of museum specimens for full mitogenome assembly (Rowe et al. 2011).
Table 1.
Sample Details and Assembly Statistics of the 33 New Xenarthran Mitochondrial Genomes.
| Species | Common Name | Collection Number | Origin | Collection Date | Sample Type | Total Reads | Mito Reads | MtDNA (%) | Mean Coverage Depth | Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|
| Bradypus pygmaeus | Pygmy sloth | USNM 579179 | Panama | 1991 | Internal organ | 31,262,238 | 383,944 | 1.23 | 2,319X | KT818523 |
| Bradypus torquatus | Maned sloth | BA449/11 | Brazil | 2011 | Skin biopsy | 39,749,058 | 61,728 | 0.16 | 375X | KT818524 |
| Bradypus tridactylus | Pale-throated three-toed sloth | ISEM T-5013 | French Guiana | 2006 | Ear biopsy | 2,016,278 | 11,916 | 0.59 | 42X | KT818525 |
| Bradypus variegatus | Brown-throated three-toed sloth | MVZ 155186 | Peru | 1978 | Internal organ | 726,173 | 30,043 | 4.14 | 121X | KT818526 |
| B. variegatus | Brown-throated three-toed sloth | NC_006923 | ||||||||
| Choloepus didactylus | Southern two-toed sloth | MNHN 1998-1819 | French Guiana | 1997 | Internal organ | 670,499 | 7,289 | 1.09 | 26X | KT818537 |
| Cho. didactylus | Southern two-toed sloth | NC_006924 | ||||||||
| Choloepus hoffmanni | Hoffmann's two-toed sloth | ISEM T-6052 | Panama | 2001 | Kidney | 1,258,442 | 3,174 | 0.25 | 11X | KT818538 |
| Cyclopes didactylus | Pygmy anteater | MNHN 1998-234 | French Guiana | 1995 | Internal organ | 1,832,592 | 5,630 | 0.31 | 21X | KT818539 |
| Myrmecophaga tridactyla | Giant anteater | ISEM T-2862 | French Guiana | 2001 | Internal organ | 1,102,261 | 7,215 | 0.65 | 26X | KT818549 |
| Tamandua mexicana | Northern tamandua | MVZ 192699 | Mexico | 1977 | Internal organ | 1,552,407 | 4,167 | 0.27 | 16X | KT818551 |
| Tamandua tetradactyla | Southern tamandua | ISEM T-6054 | French Guiana | 2001 | Ear biopsy | 14,370,168 | 6,373 | 0.04 | 30X | KT818552 |
| Ta. tetradactyla | Southern tamandua | NC_004032 | ||||||||
| Dasypus kappleri | Greater long-nosed armadillo | ISEM T-3365 | French Guiana | 2001 | Internal organ | 1,059,009 | 46,167 | 4.36 | 166X | KT818541 |
| Dasypus hybridus | Southern long-nosed armadillo | ZVC M2010 | Uruguay | 1976 | Tail bone | 30,663,498 | 38,967 | 0.13 | 123X | KT818540 |
| Dasypus septemcinctus | Seven-banded armadillo | ISEM T-3002 | Argentina | 2000 | Ear biopsy | 1,878,745 | 2,174 | 0.12 | 8X | KT818546 |
| Dasypus novemcinctus | Nine-banded armadillo | ISEM T-1863 | French Guiana | 1995 | Ear biopsy | 4,525,766 | 2,104 | 0.05 | 8X | KT818542 |
| Da. novemcinctus | Nine-banded armadillo | USA | NC_001821 | |||||||
| Dasypus pilosus | Hairy long-nosed armadillo | MSB 49990 | Peru | 1980 | Dried skin | 21,384,760 | 11,901 | 0.06 | 54X | KT818544 |
| Da. pilosus | Hairy long-nosed armadillo | LSUMZ 21888 | Peru | 1978 | Dried skin | 44,847,476 | 8,525 | 0.02 | 23X | KT818543 |
| Dasypus sabanicola | Northern long-nosed armadillo | USNM 372834 | Venezuela | 1966 | Dried skin | 32,354,212 | 10,323 | 0.03 | 46X | KT818545 |
| Dasypus yepesi | Yunga's long-nosed armadillo | MLP 30.III.90.2 | Argentina | 1988 | Rib bone | 1,518,470 | 4,203 | 0.28 | 13X | KT818547 |
| Chaetophractus nationi | Andean hairy armadillo | ISEM T-LP1 | Bolivia | 2008 | Blood | 790,237 | 2,453 | 0.31 | 9X | KT818534 |
| Chaetophractus vellerosus | Screaming hairy armadillo | ISEM T-CV1 | Argentina | 2005 | Internal organ | 1,212,063 | 9,552 | 0.79 | 34X | KT818533 |
| Chaetophractus villosus | Large hairy armadillo | ISEM T-NP390 | Argentina | 2001 | Ear biopsy | 978,540 | 4,889 | 0.50 | 18X | KT818535 |
| Euphractus sexcinctus | Six-banded armadillo | ISEM T-1246 | NA | NA | Dried skin | 2,885,506 | 2,029 | 0.07 | 7X | KT818548 |
| Zaedyus pichiy | Pichi | ISEM T-6060 | Argentina | 2005 | Internal organ | 1,939,442 | 3,202 | 0.17 | 12X | KT818555 |
| Calyptophractus retusus | Greater fairy armadillo | ZSM T-Bret | Bolivia | 1974 | Internal organ | 1,766,903 | 3,063 | 0.17 | 10X | KT818532 |
| Chlamyphorus truncatus | Pink fairy armadillo | ISEM T-CT1 | Argentina | 2005 | Internal organ | 570,194 | 67,049 | 11.76 | 244X | KT818536 |
| Priodontes maximus | Giant armadillo | ISEM T-2353 | Argentina | 2000 | Skin biopsy | 13,662,361 | 5,123 | 0.04 | 19X | KT818550 |
| Tolypeutes matacus | Southern three-banded armadillo | ISEM T-2348 | Argentina | 2000 | Ear biopsy | 1,356,101 | 6,051 | 0.45 | 20X | KT818553 |
| Tolypeutes tricinctus | Brazilian three-banded armadillo | JB21 | Brazil | 2007 | Ear biopsy | 13,307,971 | 20,076 | 0.15 | 97X | KT818554 |
| Cabassous centralis | Northern naked-tailed armadillo | AMNH MO-10752 | Costa Rica | 1896 | Skull bone | 35,461,781 | 14,828 | 0.04 | 51X | KT818527 |
| Cabassous chacoensis | Chacoan naked-tailed armadillo | ISEM T-2350 | Argentina | 2000 | Tail biopsy | 910,714 | 4,848 | 0.53 | 18X | KT818528 |
| Cabassous tatouay | Greater naked-tailed armadillo | ZVC M365 | Uruguay | 1966 | Dried skin | 40,034,772 | 14,614 | 0.04 | 39X | KT818529 |
| Cabassous unicinctus | Southern naked-tailed armadillo | MNHN 1999-1068 | French Guiana | 1995 | Internal organ | 2,099,343 | 19,302 | 0.92 | 66X | KT818531 |
| Cab. unicinctus | Southern naked-tailed armadillo | ISEM T-2291 | French Guiana | 2000 | Internal organ | 1,021,049 | 4,205 | 0.41 | 16X | KT818530 |
| Dugong dugon | Dugong | NC_003314 | ||||||||
| Loxodonta Africana | African elephant | NC_000934 | ||||||||
| Orycteropus afer | Aardvark | NC_002078 |
Note.—NA: not available; USNM: National Museum of Natural History, Washington, USA; ISEM: Institut des Sciences de l'Evolution, Montpellier, France; MVZ: Museum of Vertebrate Zoology, Berkeley, USA; MNHN: Museum National d'Histoire Naturelle, Paris, France; ZVC: Colección de Vertebrados de la Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay; MSB: Museum of Southwestern Biology, Albuquerque, USA; LSUMZ: Louisiana State University Museum of Natural Science, Baton Rouge, USA; MLP: Museo de La Plata, La Plata, Argentina; AMNH: American Museum of Natural History, New York, USA; ZSM: Zoologische Staatssammlung München, Munich, Germany.
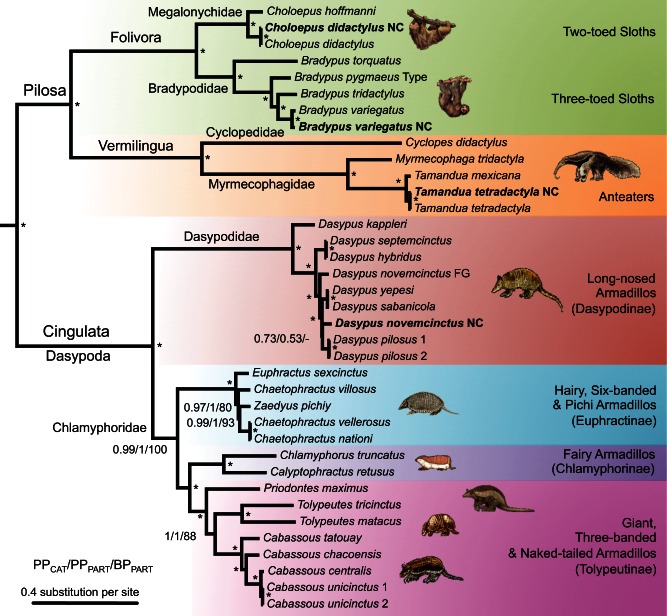
The complete mitogenomes allowed us to construct a highly informative phylogenetic data set containing 15,006 sites for 40 taxa (37 xenarthrans and 3 outgroups). The best way to analyze mitogenomic data continues to be debated (Leavitt et al. 2013; Powell et al. 2013). Despite representing a single linked locus, selection pressures and evolutionary rates are highly heterogeneous across the mitochondrial DNA molecule (Galtier et al. 2006; Nabholz et al. 2012) and particular substitution patterns and base composition biases exist among sites and strands (Reyes et al. 1998). One way to accommodate this rate heterogeneity is to use partitioned models implemented in both maximum likelihood (ML) and Bayesian approaches, which have been shown to improve phylogenetic inference (Chiari et al. 2012; Kainer and Lanfear 2015). However, determining the best partition scheme currently requires the a priori definition of biologically relevant partitions (Lanfear et al. 2012). In our case, the optimal partition scheme selected by PartitionFinder (Lanfear et al. 2012) attributed a GTR+G+I model to four partitions, basically capturing the substitution rate heterogeneity among codon positions of protein-coding genes, RNAs, and ND6 that is the only gene to be encoded on the heavy strand (supplementary table S1, Supplementary Material online). A valuable alternative to partitioned models is provided by the Bayesian site-heterogeneous CAT–GTR (general time reversible) mixture model (Lartillot and Philippe 2004), in which the optimal number of site-specific substitution pattern categories is directly estimated from the data. The application of this model for analyzing mammalian mitogenomic data is only starting, but it has already been rather promising (Hassanin et al. 2012; Botero-Castro et al. 2013; Fabre et al. 2013). In the case of our xenarthran data set, ML and Bayesian implementations of the optimal partitioned model and Bayesian inference under the CAT-GTR mixture model gave exactly the same, fully resolved topology apart from one unsupported node within the genus Dasypus (fig. 1).
Fig. 1.
Phylogenetic relationships of all extant xenarthran species. Bayesian consensus phylogram obtained using PhyloBayes under the CAT-GTR-G mixture model. Values at nodes indicate Bayesian posterior probabilities (PP) obtained under the CAT-GTR-G4 mixture model and maximum-likelihood bootstrap percentages (BP) obtained under the optimal partitioned model, respectively. Asterisks (*) mark branches that received maximal support from both methods. The afrotherian outgroup is not shown (full tree provided as supplementary fig. S1, Supplementary Material online). NC: GenBank reference mitogenomes (in bold); FG: French Guiana. Type: Museum type specimen.
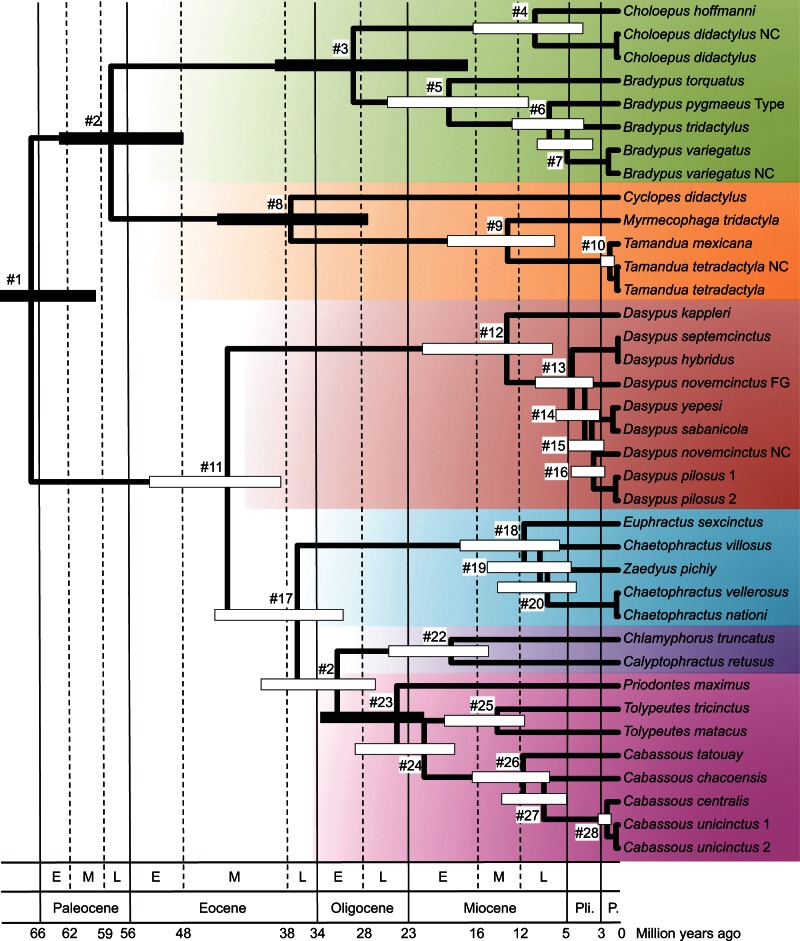
The xenarthran mitogenomic tree shows a fair amount of branch length heterogeneity among groups, with fast evolving clades including anteaters and Dasypodinae, and slow evolving clades such as sloths and hairy armadillos (fig. 1). Lineage-specific variation in evolutionary rates in mammalian mitochondrial genomes has been previously characterized (Gissi et al. 2000). Such variation has been linked to differences in mutation rates that correlate well with longevity in mammals (Nabholz et al. 2008). As a result, the mammalian mitochondrial clock is particularly erratic (Nabholz et al. 2009) and substitution rate variations among lineages should be accounted for in dating analyses by using relaxed clock models (Thorne et al. 1998). The selection of the clock model is, nevertheless, often arbitrary and appears mostly dependent upon the software choice, with an overwhelming majority of studies relying on BEAST (Drummond et al. 2012), thus generally using an uncorrelated gamma (UGAM also known as UCLN) relaxed clock model (Drummond et al. 2006). However, it has been shown that autocorrelated rate models, such as the autocorrelated log-normal model (LN; Thorne et al. 1998), generally offer a better fit, especially with large data sets above the species level (Lepage et al. 2007; Rehm et al. 2011). We thus compared the fit of the UGAM and LN models with a strict molecular clock (CL) model using cross-validation tests. The latter showed that the relaxed clock models both largely outperform the strict clock model (UGAM vs. CL: 14.42 ± 9.12; LN vs. CL: 18.47 ± 4.86), and among relaxed clock models, LN fits our data better than UGAM (LN vs. UGAM: 4.05 ± 7.87). Accordingly, we present and discuss the divergence times obtained with the autocorrelated LN relaxed clock model (fig. 2 and table 2).
Fig. 2.
Molecular timescale for all extant xenarthran species. The Bayesian chronogram was obtained using a rate-autocorrelated LN relaxed molecular clock model using PhyloBayes under the CAT-GTR-G mixture model with a birth–death prior on the diversification process, and six soft calibration constraints. Node bars indicate the uncertainty around mean age estimates based on 95% credibility intervals. Divergence dates less than 0.5 Ma are not represented. Plain black node bars indicate nodes used as a priori calibration constraints. Numbers at nodes refer to table 2. The afrotherian outgroup is not shown (full tree provided as supplementary fig. S2, Supplementary Material online). Vertical lines delimitate the main geological periods of the Cenozoic following the 2012 Geological Time Scale of the Geological Society of America (Gradstein et al. 2012). E = early; M = middle; L = late; Pli. = Pliocene; P. = Pleistocene.
Table 2.
Divergence Time Estimates for All Xenarthran Nodes Inferred Using the Site-Heterogeneous CAT-GTR+G4 Substitution Model and an Autocorrelated LN Relaxed Molecular Clock Model.
| Nodes | This study | Delsuc et al. (2012) | Delsuc et al. (2004) |
|---|---|---|---|
| 1. Xenarthraa | 67.7 ± 3.0 [60.4–71.6] | 67.8 ± 3.4 [61.3–74.7] | 64.7 ± 4.9 [55.3–74.6] |
| 2. Pilosaa (anteaters + sloths) | 58.4 ± 4.1 [48.6–64.7] | 60.1 ± 3.6 [53.1–67.2] | 55.2 ± 4.9 [45.8–65.2] |
| 3. Folivoraa (sloths) | 29.9 ± 6.5 [16.5–39.6] | 28.3 ± 3.4 [22.0–35.2] | 20.8 ± 3.3 [15.0–27.8] |
| 4. Megalonychidae (two-toed sloths) | 9.2 ± 3.5 [3.5–16.7] | N.A. | N.A. |
| 5. Bradypodidae (three-toed sloths) | 19.0 ± 4.7 [9.6–27.0] | N.A. | N.A. |
| 6. Bradypus pygmaeus/others | 7.7 ± 2.4 [3.6–12.6] | N.A. | N.A. |
| 7. Bradypus tridactylus/Bradypus variegatus | 5.7 ± 1.8 [2.6–9.5] | N.A. | N.A. |
| 8. Vermilinguaa (anteaters) | 37.8 ± 4.9 [26.9–46.2] | 45.5 ± 3.7 [38.4–52.8] | 40.0 ± 4.4 [31.8–49.0] |
| 9. Myrmecophaga/Tamandua | 12.7 ± 3.3 [7.0–19.8] | 13.6 ± 2.1 [9.9–18.2] | 10.1 ± 1.8 [6.9–14.1] |
| 10. Tamandua mexicana/Tamandua tetradactyla | 1.0 ± 0.4 [0.4–2.0] | N.A. | N.A. |
| 11. Cingulata (armadillos) | 44.9 ± 3.5 [38.3–52.1] | 42.3 ± 3.8 [35.1–50.0] | 39.7 ± 4.5 [31.3–49.1] |
| 12. Dasypodinae (long-nosed armadillos) | 12.4 ± 3.4 [7.2–20.4] | 11.2 ± 2.0 [7.8–15.6] | 7.3 ± 1.6 [4.6–10.9] |
| 13. Dasypus septemcinctus + Dasypus hybridus/others | 5.1 ± 1.7 [2.7–9.2] | N.A. | N.A. |
| 14. Dasypus novemcinctus FG/others | 3.7 ± 1.2 [2.0–6.8] | N.A. | N.A. |
| 15. Dasypus sabanicola + Dasypus yepesi/others | 2.9 ± 1.0 [1.5–5.4] | N.A. | N.A. |
| 16. Dasypus novemcinctus NC/Dasypus pilosus | 2.8 ± 0.9 [1.5–5.1] | N.A. | N.A. |
| 17. Chlamyphoridae | 37.2 ± 3.4 [31.5–44.7] | 34.5 ± 3.6 [27.8–41.9] | 32.9 ± 4.1 [25.2–41.5] |
| 18. Euphractinae (hairy armadillos) | 11.0 ± 2.8 [6.8–17.8] | 8.3 ± 1.6 [5.5–11.8] | 6.2 ± 1.4 [3.8–9.3] |
| 19. Chaetophractus villosus/others | 9.1 ± 2.4 [5.5–15.1] | 7.1 ± 1.4 [4.7–10.3] | N.A. |
| 20. Zaedyus pichiy/others | 8.2 ± 2.3 [4.9–13.7] | N.A. | N.A. |
| 21. Chlamyphorinae/Tolypeutinae | 32.6 ± 3.1 [27.9–40.0] | 32.9 ± 3.6 [26.3–40.2] | N.A. |
| 22. Chlamyphorinae (fairy armadillos) | 19.4 ± 2.7 [15.2–25.9] | 17.3 ± 2.7 [12.4–23.0] | N.A. |
| 23. Tolypeutinaea | 25.7 ± 2.7 [22.4–32.7] | 26.1 ± 3.2 [20.2–32.9] | 21.8 ± 3.3 [15.8–28.9] |
| 24. Tolypeutes/Cabassous | 22.5 ± 2.6 [19.0–29.0] | 24.2 ± 3.1 [18.5–30.7] | 20.5 ± 3.2 [14.7–27.3] |
| 25. Tolypeutes | 14.1 ± 2.0 [11.0–19.1] | N.A. | N.A. |
| 26. Cabassous | 10.9 ± 1.9 [8.0–15.5] | N.A. | N.A. |
| 27. Cabassous chacoensis/others | 8.6 ± 1.6 [6.0–12.4] | N.A. | N.A. |
| 28. Cabassous centralis/Cabassous unicinctus | 1.3 ± 0.3 [0.8–2.1] | N.A. | N.A. |
Note.—Mean posterior estimates, associated standard errors, and 95% credibility intervals are expressed in Ma (mean date ± SD [95% CredI]).
SD: standard deviation; 95% CredI: 95% credibility interval; FG: French Guiana; NC: GenBank reference mitogenome (specimen from the USA); N.A.: not applicable
aUsed as a priori calibration constraints.
Phylogenetic Framework and Timescale for Living Xenarthrans
Our analyses provide the first comprehensive phylogeny including all living species of Xenarthra. We obtained a fully resolved tree with high ML bootstrap and Bayesian support values, except for one node within the genus Dasypus (fig. 1). This mitogenomic topology is entirely congruent with previous studies conducted at the genus level using nuclear exons (Delsuc et al. 2002), combinations of mitochondrial and nuclear genes (Delsuc et al. 2001, 2003, 2012), and retroposons and their flanking noncoding sequences (Möller-Krull et al. 2007). Furthermore, the newly estimated timescale (fig. 2 and table 2) is compatible with previous time estimates provided by the analyses of nuclear exons alone (Delsuc et al. 2004) or in combination with mitochondrial genes (Delsuc et al. 2012). The few discrepancies concern nodes for which the species sampling has been substantially increased such as Folivora, Dasypodinae, Euphractinae, and Tolypeutinae (table 2). For these nodes, the newly inferred dates appear older than previous estimates performed at the genus level as expected with a denser species sampling. Such global congruence with previous nuclear-based phylogenetic and dating analyses, allows being confident that ancient introgression and/or hybridization events have not significantly affected the mitogenomic tree of xenarthrans. A number of new surprising and important inferences are to be drawn from our mitogenomic framework with respect to phylogenetic relationships and species delimitation within the different xenarthran groups.
Sloths (Pilosa; Folivora)
The six living species of sloths belong to two genera, with two-toed sloths (genus Choloepus) and three-toed sloths (genus Bradypus) having been placed in two distinct families (Megalonychidae and Bradypodidae, respectively) to reflect their numerous morphological differences and a probably diphyletic origin from two different fossil groups (Webb 1985). Their independent adaptation to the arboreal lifestyle also led to a number of anatomical convergences related to their peculiar suspensory locomotion (Nyakatura 2012). Our results confirm this deep dichotomy with a divergence date between the two genera around 30 Ma (fig. 2 and table 2), which appears more ancient than previously estimated with nuclear data (Delsuc et al. 2004). This difference might stem from our increased taxon sampling, because only a single representative species of each genus was previously considered. Their considerable molecular divergence nevertheless supports the classification of the two modern sloth genera into distinct families.
Within two-toed sloths, the new mitochondrial genome sequence obtained for the Southern two-toed sloth (Choloepus didactylus) appears almost identical to the reference mitogenome (NC_006924) deposited in GenBank (99.8% pairwise identity). As expected, the Hoffmann’s two-toed sloth (Choloepus hoffmanni) is more divergent (pairwise distance of 7.2% with Cho. didactylus). The divergence time between the two toed-sloth species is estimated at about 9 Ma (fig. 2 and table 2). Choloepus hoffmanni presents two disjunct northern and southern populations. A recent study estimated the divergence between northern and southern Cho. hoffmanni mitochondrial lineages at about 7 Ma (Moraes-Barros and Arteaga 2015), but it did not include Cho. didactylus. Obtained from a captive individual most likely coming from Panama, our Cho. hoffmanni mitogenome belongs to the northern population. Therefore, we could not discard the possibility that a southern Cho. hoffmanni sequence will not belong to the Cho. didactylus lineage. The occurrence of hybrids in captivity raises the question of whether hybridization also occurs in natural populations, especially between the southern populations of Cho. hoffmanni and individuals of Cho. didactylus inhabiting north-central Peru and south-western areas of Brazil (Steiner et al. 2010). This evidence coupled with the significant variation in chromosome number observed in South American Choloepus (2n = 53–67; Hayssen 2010) indicates the need for a taxonomic review of both Cho. hoffmanni and Cho. didactylus. This will require the analysis of additional mitochondrial and nuclear data for an extensive sampling, especially along the southern distribution of Cho. hoffmanni, where it is sympatric with Cho. didactylus.
Concerning three-toed sloths, the endangered maned sloth (Bradypus torquatus) is the sister group of the three other described species (fig. 1). This phylogenetic position is in agreement with previous studies based on a few mitochondrial genes (Barros et al. 2003; Moraes-Barros et al. 2011). Our dating estimates confirm the maned sloth as an ancient Atlantic forest endemic, which may have diverged from other sloths as early as 19 Ma (fig. 2 and table 2). Such an old divergence date associated with its distinctive morphological characters would argue for a classification of B. torquatus in its own genus (Scaeopus), as advocated by Barros et al. (2008). As the maned sloth is one of the most threatened xenarthran species (Superina, Plese, et al. 2010), its phylogenetic distinctiveness should be considered in future conservation assessments.
The critically endangered pygmy sloth (B. pygmaeus) is restricted to Isla Escudo de Veraguas, in the islands of Bocas del Toro (Panama). Anderson and Handley (2001) described this insular population as a distinct species on the basis of morphometric analyses showing a reduced body size compared with the mainland and other island populations of the brown-throated three-toed sloth (Bradypus variegatus). Our analyses, based on the sequencing of the type specimen (USNM 579179), show that B. pygmaeus constitutes a distinct lineage within three-toed sloths that is clearly separated from Bradypus tridactylus and B. variegatus (fig. 1) from which it diverged some 8 Ma (fig. 2 and table 2). By sequencing a pale-throated three-toed sloth (B. tridactylus) specimen from French Guiana, where only this species occurs, we were able to confirm that the GenBank reference mitogenome (NC_006923) was originally misidentified as B. tridactylus and in fact belongs to the brown-throated three-toed sloth (B. variegatus), as previously shown by Moraes-Barros et al. (2011). Indeed, our newly sequenced B. variegatus specimen from Peru (MVZ 155186) is 97.3% identical to NC_006923, whereas the pairwise distance with B. tridactylus reaches 9.5%. The divergence date between B. variegatus and B. tridactylus is estimated here around 6 Ma (fig. 2 and table 2). However, similar to the Hoffmann’s two-toed sloth, the brown-throated three-toed sloth includes divergent mitochondrial lineages (Moraes-Barros et al. 2011; Moraes-Barros and Arteaga 2015) and occurs as far north as Honduras. Therefore, we cannot exclude the possibility that individuals from mainland Central America (Panama, Costa Rica, Nicaragua, and Honduras) not included in this study might belong to the distinct pygmy sloth (B. pygmaeus) lineage rather than to B. variegatus.
Anteaters (Pilosa; Vermilingua)
Anteaters are the least diverse xenarthran group with only four described species classified in three genera. The mitogenomic tree (fig. 1) confirms the phylogenetic distinctiveness of the monotypic pygmy anteater (Cyc. didactylus) from the two closely related genera Tamandua and Myrmecophaga. The early divergence of the pygmy anteater is estimated around 38 Ma, whereas Tamandua and Myrmecophaga diverged much more recently around 13 Ma (fig. 2 and table 2). This profound dichotomy appears fully compatible with previous estimates (Delsuc et al. 2004, 2012) and confirms the rationality of dividing anteaters into two distinct families Cyclopedidae (Cyclopes) and Myrmecophagidae (Tamandua and Myrmecophaga), as proposed by Barros et al. (2008). This taxonomic distinction also reflects the numerous morphological differences observed between the two main anteater lineages (Gaudin and Branham 1998). Regarding lesser anteaters, our new sequence of the southern tamandua (Tamandua tetradactyla) from French Guiana confirmed the identification of the GenBank reference mitogenome (NC_004032), the two sequences being 99.4% identical (fig. 1). However, our mitogenomic data revealed a very limited genetic differentiation between northern (Tamandua mexicana) and southern (Ta. tetradactyla) tamanduas, with a pairwise distance of only 2.8% (2.1% on Cytochrome c oxidase subunit 1; COX1). The two tamanduas are considered two distinct species with parapatric distributions separating on each side of the Northern Andes in Venezuela, Colombia, Ecuador, and Peru (Superina, Miranda, et al. 2010). However, the species diagnoses are based on differences in coat coloration, body size, skull characters, and number of caudal vertebrae that can be quite variable within populations (Wetzel 1985). Our whole mitochondrial genome data question the species status of these anteaters and encourage future nuclear studies aiming at delimitating species within the genus Tamandua.
Armadillos (Cingulata; Dasypoda)
Armadillos are the most speciose xenarthran group with 21 extant species and 9 genera. Our phylogenetic results unambiguously support the monophyly of each of the four armadillo subfamilies Dasypodinae, Euphractinae, Chlamyphorinae, and Tolypeutinae (fig. 1), as previously recognized with a concatenation of nuclear and mitochondrial genes (Delsuc et al. 2012). The first dichotomy separates long-nosed armadillos (Dasypodinae) from the other three subfamilies, which form a monophyletic group (PPCAT = 0.99/PPPART = 1.0/BPPART = 100). This deep divergence occurred early in the Cenozoic at an estimated time of about 45 Ma (fig. 2 and table 2). Given this remarkably ancient divergence date relative to other placental families (Meredith et al. 2011), we propose splitting armadillos into two distinct families: Dasypodidae and Chlamyphoridae. The proposed use of Chlamyphoridae is based on the taxonomic rank elevation of the oldest constitutive subfamily that is Chlamyphorinae Bonaparte 1850. Within Chlamyphoridae, the mitogenomic tree (fig. 1) supports the grouping of Chlamyphorinae (fairy armadillos) with Tolypeutinae (giant, three-banded, and naked-tailed armadillos) to the exclusion of Euphractinae (hairy armadillos), in line with previous studies including nuclear noncoding (Möller-Krull et al. 2007) and protein-coding (Delsuc et al. 2012) sequences. The early branching of Euphractinae is estimated around 37 Ma, relatively quickly followed by the separation between Chlamyphorinae and Tolypeutinae, circa 33 Ma (fig. 2 and table 2).
Dasypodinae.
This subfamily includes seven species of long-nosed armadillos belonging to the single genus Dasypus. Species in this genus are characterized by an elongated nose that can be functionally related to the use of the tongue to gather ants, termites, and a diversity of soil invertebrates (Loughry and McDonough 2013). An unusual particularity, thought to be shared by all species belonging to this genus, is their reproduction by obligate polyembryony in which the female systematically gives birth to genetically identical litters (Galbreath 1985). The most common and best studied is the nine-banded armadillo (Dasypus novemcinctus), which has the largest distribution from Argentina to North America as a consequence of its ongoing invasion of the southern United States (Taulman and Robbins 2014; Feng and Papes 2015). The greater long-nosed armadillo (Dasypus kappleri) is the largest of the group, and it has been proposed on morphological grounds to classify this species in its own subgenus Hyperoambon (Wetzel and Mondolfi 1979). The hairy long-nosed armadillo (Da. pilosus), which is an endemic of Peru, is also morphologically distinctive in being the only armadillo possessing a carapace entirely covered with dense fur. This peculiarity has led some authors to propose its taxonomic distinction in the subgenus Cryptophractus (Wetzel and Mondolfi 1979). A recent morphological study, which was mainly based on the analysis of the structure of its osteoderms, even proposed to raise it to the genus level (Castro et al. 2015). The remaining species constitute a complex of morphologically similar taxa with historical taxonomic uncertainty. The southern long-nosed armadillo (Dasypus hybridus) and the seven-banded armadillo (Dasypus septemcinctus) are particularly hard to distinguish, with globally parapatric distributions that might overlap in southern Brazil, northern Argentina, and Paraguay (Abba and Superina 2010). The species status of the northern long-nosed armadillo (Dasypus sabanicola) has also been historically hard to establish (Wetzel 1985), and the Yunga’s lesser long-nosed armadillo (Dasypus yepesi) has only been recently recognized as a distinct species (Vizcaíno 1995).
The phylogenetic tree obtained from the mitogenomes (fig. 1) clearly identifies Da. kappleri as the sister group to all other long-nosed armadillo species. Molecular dating, estimates its early divergence at more than 12 Ma (fig. 2 and table 2). This fairly ancient date, coupled with well-characterized morphological differences such as the presence of unique scutes on the knee, would argue for its placement in the distinct genus Hyperoambon, originally proposed as a subgenus by Wetzel and Mondolfi (1979). Second to diverge within Dasypodinae is a clade composed of Da. hybridus and Da. septemcinctus whose mitogenome sequences appear almost identical (99.3% identity). This observation is consistent with noted morphological similarity and historical synonymy between these two taxa (Abba and Superina 2010). However, given the fact that our Da. septemcinctus species is from northern Argentina and our Da. hybridus is from Uruguay, both in potential areas of sympatry between the two taxa, we cannot exclude the possibility of misidentification, hybridization, and/or introgression being responsible for the observed mitogenomic similarity. Further clarification of the taxonomic status of these two species would require collecting additional mitochondrial and nuclear data for specimens coming from the two extremes of their ranges in central Argentina for Da. hybridus and northern Brazil for Da. septemcinctus.
The next diverging lineage is represented by an individual identified as Da. novemcinctus from French Guiana, which unambiguously represents a distinct branch in the mitogenomic tree (fig. 1). In an early phylogeographic study of nine-banded armadillos based on the mitochondrial D-loop, we observed that individuals from French Guiana were indeed very distant from the ones of the invasive US populations to which the reference mitochondrial genome for Da. novemcinctus belongs (Huchon et al. 1999). The two mitogenomes are indeed fairly divergent with a pairwise distance of 5.6% (5.7% on COX1). The divergence date between the French Guianan lineage and other long-nosed armadillos is estimated around 3.7 Ma (fig. 2 and table 2), which strongly suggests that the French Guianan lineage might represent a previously unrecognized species. This potentially new species is the sister group of a clade regrouping Da. yepesi and Da. sabanicola on one side, and Da. pilosus and Da. novemcinctus on the other side. Only the position of the hairy long-nosed armadillo (Da. pilosus) appears unstable with low statistical support (fig. 1). Although Da. sabanicola and Da. yepesi are restricted to very distinct localities of South America respectively in Venezuela/Colombia and north-eastern Argentina, they possess very similar mitogenomes (98.7% identity). Our results confirm that the taxonomic status of both species is questionable and needs further review (Abba and Superina 2010). Further morphological and molecular species delimitation studies will be needed to fully understand the species boundaries within long-nosed armadillos.
Finally, the hairy long-nosed armadillo (Da. pilosus) appears to constitute a distinct lineage of long-nosed armadillos (fig. 1), but its molecular divergence does not seem to match its morphological distinctiveness. Indeed, its pairwise distance with both Da. novemcinctus and with the Da. sabanicola/Da. yepesi clade is about 5%. The divergence date between Da. pilosus and Da. novemcinctus is estimated around 2.8 Ma (fig. 2 and table 2). Our phylogenetic reconstruction also strongly contradicts the results of Castro et al. (2015) who found Da. pilosus to be the sister group to all other long-nosed armadillos based on cladistic analysis of morphological characters. Based on these results and the peculiar structure of its osteoderms, they proposed resurrecting Cryptophractus as the genus name for the hairy long-nosed armadillo. Our results do not support such a taxonomic reassessment and argue in favor of conserving the hairy long-nosed armadillo in the genus Dasypus. More generally, our mitogenomic topology for long-nosed armadillos reveals many conflicts with the cladistic analysis of Castro et al. (2015). This suggests that osteodermal characters are of limited taxonomic value in being highly homoplastic, and should therefore be used with caution in phylogenetic analyses.
Euphractinae.
Euphractine armadillos constitute an ecologically homogeneous and morphologically similar group with five traditionally recognized species classified in the three genera Chaetophractus, Euphractus, and Zaedyus (Abba and Superina 2010). The genera Euphractus and Zaedyus are monospecific and include the six-banded armadillo (E. sexcinctus) and the pichi (Zaedyus pichiy), respectively. The genus Chaetophractus classically encloses three species, the large hairy armadillo (Chaetophractus villosus), the screaming hairy armadillo (Chaetophractus vellerosus), and the Andean hairy armadillo (Chaetophractus nationi). The interrelationships between the three genera have been difficult to decipher with both morphological (Engelmann 1985; Gaudin and Wible 2006) and molecular data (Delsuc et al. 2002, 2003; Möller-Krull et al. 2007) likely due to their rapid diversification (Delsuc et al. 2004). A recent study investigated the phylogenetic relationships among all five species using an integrative approach based on skull geometric morphometrics and molecular phylogenetics (Abba et al. 2015). It was proposed that Cha. nationi should be considered a synonym of Cha. vellerosus based on shared mitochondrial haplotypes and a close proximity at the nuclear level coupled with a very similar morphology because the two species only differ in size. Moreover, phylogenetic analyses of a combination of six noncoding nuclear markers and two nuclear exons suggested the paraphyly of the genus Chaetophractus, with Cha. vellerosus being more closely related to Z. pichiy than to Cha. villosus. The relative positions of the large hairy armadillo (Cha. villosus) and the six-banded armadillo (E. sexcinctus) nevertheless remained uncertain, as conflicting positions were obtained with the noncoding and protein-coding partitions (Abba et al. 2015).
The complete mitochondrial genome sequences confirm that the threatened and geographically restricted Andean hairy armadillo (Cha. nationi) could not be genetically distinguished from the widespread screaming hairy armadillo (Cha. vellerosus), with 99.8% mitogenomic identity between the two taxa. This result reinforces the proposition of taxonomically synonymizing these two species by retaining only Cha. vellerosus (Abba et al. 2015). Moreover, the mitogenomic tree (fig. 1) offers some additional support for the paraphyly of the genus Chaetophractus caused by the strongly supported sister group relationship of Z. pichiy with Cha. vellerosus/Cha. nationi (PPCAT = 0.99/PPPART = 1/BPPART = 93). As with nuclear data, the early branching of E. sexcinctus is slightly less supported by ML but nevertheless appears quite robust (PPCAT = 0.97/PPPART = 1/BPPART = 80). Molecular dating results (fig. 2 and table 2) also confirm a rapid diversification of Euphractinae, with speciation events occurring within a few million years between 8 and 11 Ma, a period marked by the appearance of more arid areas in the Southern Cone of South America where most of these species are distributed. Definitive resolution of the relationships among hairy armadillos might require large-scale genomic data to account for possible discordances between gene trees and the species tree resulting from incomplete lineage sorting.
Chlamyphorinae.
This recently recognized subfamily (Delsuc et al. 2012) consists of only two species of fairy armadillos or pichiciegos that count among the most elusive mammals due to their nocturnal and subterranean habits. The pink fairy armadillo (Chl. truncatus) is restricted to sandy plains of central Argentina, while the greater fairy armadillo (Calyptophractus retusus) is found in the Gran Chaco of northern Argentina, Paraguay, and eastern Bolivia (Abba et al. 2010). The mitogenomic tree (fig. 1) corroborates earlier results by strongly supporting the monophyly of fairy armadillos (Delsuc et al. 2012) and their sister group relationship with tolypeutines (Möller-Krull et al. 2007; Delsuc et al. 2012). Molecular dating (fig. 2 and table 2) also confirms the considerably old divergence of the two species (ca. 19 Ma) and their ancient split from tolypeutine armadillos (ca. 33 Ma). These results underline again the phylogenetic distinctiveness of the two fairy armadillo species and argue in favor of their classification in two distinct genera within their own subfamily. The phylogenetic uniqueness of fairy armadillos, combined with their scarcity in the wild, make pleads for increased conservation attention of these atypical mammals.
Tolypeutinae.
This subfamily includes seven species classified in three genera. Two tribes are classically recognized (Wetzel 1985; McKenna and Bell 1997): Priodontini grouping the giant armadillo (Priodontes maximus) with naked-tailed armadillos of the genus Cabassous, and Tolypeutini consisting solely of two species of three-banded armadillos (genus Tolypeutes). Giant and naked-tailed armadillos are typically fossorial and are equipped with large anterior claws used for digging. Three-banded armadillos are ground dwelling and morphologically distinctive in being the only armadillos capable of entirely rolling into a ball by locking their carapace as a defensive strategy. In contrast to morphological data that always favored the monophyly of the Priodontes and Cabassous genera on the basis of numerous anatomical similarities (Engelman 1985; Gaudin and Wible 2006) and characteristically spoon-shaped spermatozoa (Cetica et al. 1998), the phylogenetic relationships within the family Tolypeutinae have been notoriously difficult to resolve with molecular data (Delsuc and Douzery 2008). The concatenation of nuclear exons and two mitochondrial genes has basically left the issue unresolved (Delsuc et al. 2002, 2003, 2012), whereas analyses of noncoding retroposon flanking sequences offered some support for a sister group relationship between Cabassous and Tolypeutes (Möller-Krull et al. 2007). The mitogenomic picture (fig. 1) is congruent with noncoding nuclear data in supporting the paraphyly of the tribe Priodontini by grouping Tolypeutes with Cabassous to the exclusion of Priodontes (PPCAT = 1/PPPART = 1/BPPART = 88). This suggests that the morphological characters related to fossoriality used to define this tribe might either have been acquired convergently by giant and naked-tailed armadillos or, more probably, represent symplesiomorphies inherited from a fossorial ancestor.
Concerning Tolypeutini, we collected the first molecular data for the flagship Brazilian three-banded armadillo (Tolypeutes tricinctus). This endangered endemic of the North-Eastern Brazilian Caatinga biome was chosen as a mascot to increase awareness about biodiversity and ecosystem conservation. Our mitochondrial genome data revealed an unexpectedly high sequence divergence with its sister species, the southern three-banded armadillo (Tolypeutes matacus). The pairwise distance between the two mitogenomes of these morphologically and ecologically similar species reaches 12% (11.9% on COX1). Accordingly, molecular dating estimated a deep divergence of circa 14 Ma between the two allopatrically distributed species (fig. 2 and table 2). The considerable phylogenetic distinctiveness revealed for the Brazilian three-banded armadillo reinforces the conservation concerns expressed for a species considered to be one of the most threatened Brazilian mammals (Feijó et al. 2015).
Our mitogenomic study is the first to include all four recognized species of the conspicuous and fossorial naked-tailed armadillos. The greater naked-tailed armadillo (Cabassous tatouay) is the first to diverge, followed by the Chacoan naked-tailed armadillo (Cabassous chacoensis) and the two closely related northern (Cabassous centralis) and southern (Cabassous unicinctus) naked-tailed armadillos (fig. 1). The emergence of Cab. tatouay appears quite ancient (ca. 11 Ma), as is the separation of Cab. chacoensis from Cab. centralis and Cab. unicinctus (ca. 9 Ma), which are estimated to have diverged much more recently, less than 2 Ma (fig. 2 and table 2). The mitogenomes of the northern and southern naked-tailed armadillos appear very similar in sequence (98.0% identity), with a pairwise distance of only 1.9% based on COX1. This situation is reminiscent of the case revealed between the northern and southern tamanduas, with closely related species presenting parapatric distributions in Central and South America only interrupted by the Northern Andes. Additional nuclear studies would be warranted for further defining the taxonomic status of Cab. centralis and Cab. unicinctus that appear only weakly differentiated based on their mitogenomes.
Diversification and Historical Biogeography of Xenarthra
The strongly resolved tree obtained for all living xenarthran species allowed us to derive a reference timescale that can be used to study the patterns and processes underlying their diversification. Seeking the causes of species diversification and extinction by teasing apart the role of abiotic (e.g., physical environmental changes) and/or biotic (e.g., species interactions) factors (Benton 2009), is now made possible by the use of different birth–death models (Morlon 2014). We applied a suite of diversification models to sequentially consider the effects of past environmental changes (Condamine et al. 2013), rate variation through time (Stadler 2011; Rabosky 2014), and diversity-dependent processes (Etienne et al. 2012) on the macroevolutionary history of Xenarthra.
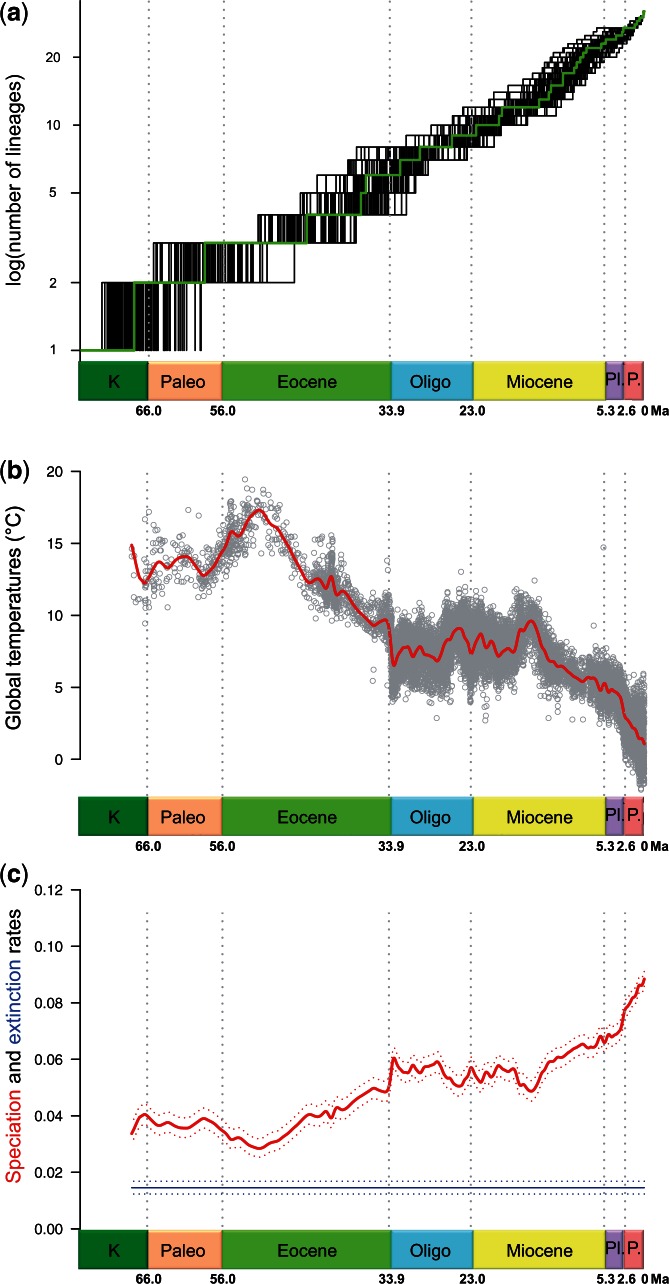
It was previously proposed that xenarthran diversification has been influenced by paleoenvironmental changes triggered by Andean uplift and sea level fluctuations in South America during the Cenozoic (Delsuc et al. 2004). At that time, no models of diversification integrating the effect of environmental variables were available to formally test the synchronicity of some cladogeneses with periods of cooling and Andean uplift. Such explicit models are now available (Condamine et al. 2013), and when applied to our data, the best temperature-dependent model showed that the speciation rate over the entire xenarthran timetree correlates negatively with temperature (fig. 3, table 3, and supplementary table S2, Supplementary Material online). This pattern is the opposite of the one found, for instance, in Cetacea (Condamine et al. 2013), and may be explained by the fact that a number of rapid speciation events in the xenarthran tree and especially within armadillos occurred in the last 10–15 Ma, during a period of intense cooling (Zachos et al. 2001). This continuous drop in temperature since the middle Miocene, followed by the setup of the circum Antarctic current and the last Andean uplift phase (Garzione et al. 2008), caused the aridification of South America and the formation of dry biomes such as Caatinga and Cerrado in the North, and Chaco and pampas in the Southern Cone (Simon et al. 2009; Hoorn et al. 2010).
Fig. 3.
Diversification pattern of Xenarthra. (a) Lineages-through-time plot constructed from 100 Bayesian posterior trees showing a steady accumulation of species through time. (b) Past fluctuations of temperatures over the Cenozoic (data plotted from Zachos et al. 2001, 2008). (c) Speciation (bold curve) and extinction rates through time for xenarthrans obtained from the relationship between diversification and paleotemperatures estimated using the approach of Condamine et al. (2013). The best model indicates a negative correlation between speciation and past temperatures and no dependence on extinction. K = Cretaceous; Paleo. = Paleocene; Oligo. = Oligocene; Pl. = Pliocene; P. = Pleistocene.
Table 3.
Summary of Diversification Analyses Results.
| Type of Birth-Death | Method Used | Reference | Data Used | Settings | Result |
|---|---|---|---|---|---|
| Paleoenvironmental dependence (rates vary “continuously” as a function of time) | RPANDA (fit_env) | Condamine et al. (2013) | 100 posterior chronograms | 7 ML models testing whether rates vary or not (exponential and linear variation) | Speciation is “negatively” linked to past temperatures, and “constant extinction” |
| Among clade and time variation of rates | BAMM | Rabosky (2014) | Bayesian chronogram | Bayesian model testing rate shift(s) among clade and through time (Poison prior = 1.0) | No significant rate shift detected: Speciation increased through time, “constant extinction” |
| Time dependence (rates vary “discretely” as a function of time) | TreePar (bd.shifts.optim) | Stadler (2011) | 100 posterior chronograms | 4 ML models testing from no rate shift to 3 rate shifts | No global rate shift detected, a constant birth-death is supported |
| Diversity dependence (rates vary as a function of the number of species) | DDD (dd_ML) | Etienne et al. (2012) | Bayesian chronogram | 5 ML models testing whether speciation declines with diversity and/or extinction increases with diversity | The clade has reached its carrying capacity, with extinction increasing as diversity increases |
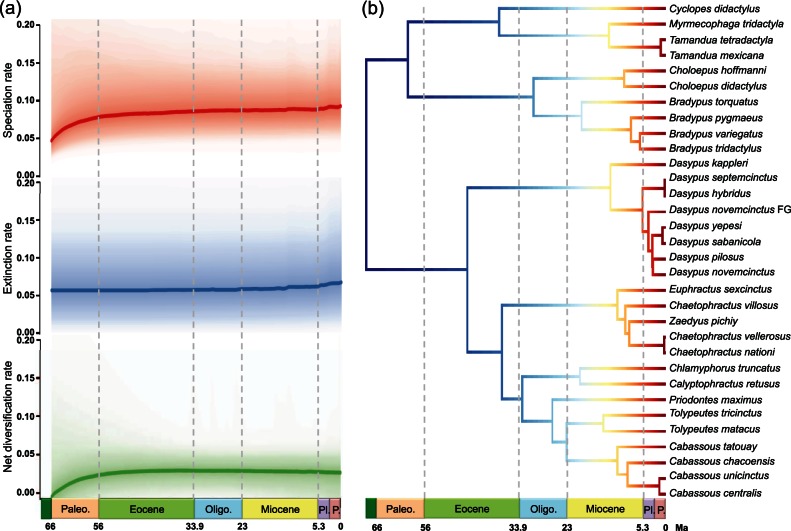
The Bayesian analysis of macroevolutionary mixtures (BAMM, Rabosky 2014) show that a single macroevolutionary rate explains the diversification of the group over time (table 3 and supplementary fig. S3, Supplementary Material online), and that the net diversification rate tends to increase through time driven by a higher speciation rate in the last 15 Ma (fig. 4). This corroborates the results obtained with the paleoenvironmental model. Furthermore, time-dependent diversification analyses (table 3 and supplementary table S2, Supplementary Material online) also portray Xenarthra as an old and species-poor, but nevertheless successful clade with a low diversification rate throughout the Cenozoic characterized by a high species turnover driven by an intermediate, but constant extinction rate. The BAMM and TreePar models are thus congruent on inferring no detectable diversification rate shift and constant extinction through time, but they disagree on the speciation rate inference for which only BAMM estimates an increase over time. This discrepancy in estimates might be explained by the relatively small size of the xenarthran timetree, which includes only 32 tips, but may increase with the addition of extinct xenarthran genomes. The inference of extinction on the xenarthran phylogeny is nevertheless in agreement with the fossil record, which documents a relatively high rate of extinction in this clade (Patterson and Pascual 1968; Simpson 1980). Notably here, extinction is estimated to be high but nevertheless constant, confirming that these models are able to detect extinction signal even in species-poor clades (Morlon et al. 2011; Jansa et al. 2014; Beaulieu and O’Meara 2015).
Fig. 4.
Bayesian analysis of macrevolutionary mixtures in Xenarthra. (a) Bayesian reconstruction of rate variations in speciation, extinction, and net diversification through time. (b) Maximum a posteriori probability shift configuration represented as a phylorate plot showing variations in speciation rates (cool colors = slow, warm = fast) along each branch of the xenarthran phylogeny. Each unique color section of a branch represents the mean of the marginal posterior density of speciation rates on a localized segment of a phylogenetic tree. The rate variation pattern for lineages involves a uniform, although slight, temporal acceleration in speciation rates. “Dasypus novemcinctus FG” denotes the French Guiana lineage. Paleo. = Paleocene; Oligo. = Oligocene; Pl = Pliocene; P = Pleistocene.
Interestingly, xenarthrans do not seem to have been particularly affected by the GABI, with, for instance, giant sloths and glyptodonts successfully colonizing Central and North America. Xenarthra was finally depauperated by the extinction of the largest forms at the terminus of the Pleistocene (Lyons et al. 2004), a very recent event that is hardly detectable with current methods of macroevolutionary analyses. This effect is especially resonant for Pilosa in which the successful northern emigrants such as giant ground sloths were drawn to extinction (McDonald 2005). Ultimately, fossil data would have to be integrated into diversification analyses to adequately model the macroevolutionary history of this species-poor clade (Fritz et al. 2013; Silvestro et al. 2014).
The role of biotic interactions was finally assessed using the diversity-dependent diversification (DDD) model of Etienne et al. (2012). The DDD model depicts Xenarthra as a clade that has reached its carrying capacity, with extinction increasing as diversity increases (table 3 and supplementary table S2, Supplementary Material online). Thus, niche partitioning in the Neotropics may be a dominant factor in shaping the pattern of species richness in Xenarthra. Evidence from current geographical distributions shows that in clades such as Tolypeutinae and Dasypodinae, only little overlap in distribution ranges is observed (Abba and Superina 2010; Superina, Miranda, et al. 2010; Superina, Plese, et al. 2010). In line with these observations, a detailed study has recently identified the tropical rainforest in the Amazon Basin as an area of high ecological diversity for xenarthrans, indicating a high disparity between pairs of coexisting species (Fergnani and Ruggiero 2015).
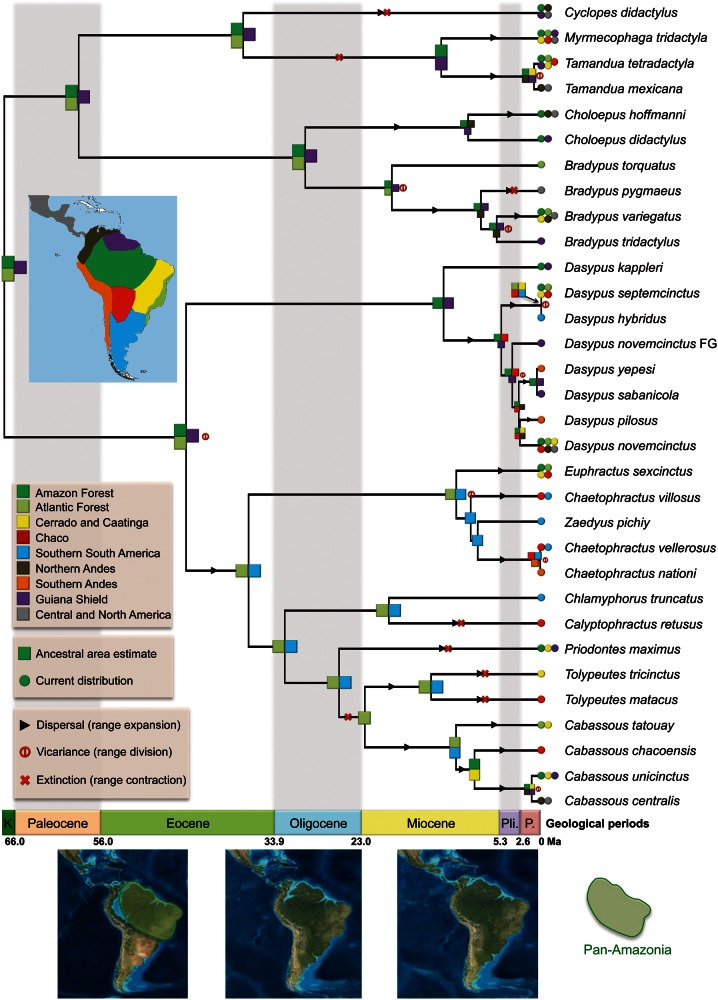
Biogeographic analyses could provide further insight into this question, and help understand the role of niche partitioning in time and the consequences on the resulting current biodiversity. We thus used the Dispersal-Extinction-Cladogenesis (DEC, Ree and Smith 2008) model on the xenarthran timetree to estimate ancestral biogeographic ranges by taking into account the connectivity among areas through time, as well as the dispersal abilities between areas according to the regional biome evolution (fig. 5). The biogeographic analysis identifies Pan-Amazonia (tropical lowland rainforest of Amazonia and Guiana Shield) and Atlantic forest as the cradle of Xenarthra evolution for most of the Paleogene (left map on fig. 5). This fits well with the high phylogenetic and ecological diversities observed for xenarthrans in the Amazonian region (Fergnani and Ruggiero 2015). Nonetheless, the common ancestor of Chlamyphoridae (Euphractinae, Chlamyphorinae, and Tolypeutinae) subsequently dispersed toward the Southern Cone in the late Eocene (central map on fig. 5). We also estimated that different species of armadillos colonized Central America after the closing of the Isthmus of Panama (Pliocene), but not in the middle or late Miocene when land connections were first made (Montes et al. 2015). Interestingly, only 9 of the 31 phylogenetic events are explained by allopatric speciation, 4 of which involved the Andes and 5 other biome divergences, such as tropical forest and savannah. Moreover, most of the allopatric speciation events are recent (last 8 My), indicating an important role of vicariance due to the building of the northern Andes, especially in northern Colombia for species with parapatric distributions in South and Central America, such as Cab. unicinctus/Cab. centralis or Ta. tetradactyla/Ta. mexicana (Moraes-Barros and Arteaga 2015).
Fig. 5.
Historical biogeography of living xenarthrans. The biogeographical range estimation was inferred under the Dispersal-Extinction-Cladogenesis model taking into account the change of connectivity and dispersal ability between areas defined as the main biomes of the American continent. Paleogeographic maps depict the tectonic evolution of South America adapted from Blakey (2008). “Dasypus novemcinctus FG” denotes the French Guiana lineage. K = Cretaceous; Pli. = Pliocene; P. = Pleistocene.
The pattern inferred within Pilosa and Cingulata shows notable differences in the biogeographic processes. Regarding allopatric speciation (vicariance)/range expansion (dispersal)/range contraction (local extinction), we found 3/8/3 events for Pilosa versus 5/17/5 for Cingulata. These results are suggestive of a relatively stable biogeographic history in the clade Pilosa, and a more dynamic history in Cingulata. Indeed anteaters and sloths appeared more stable in time and centered on the Pan-Amazonian region (Amazonia and Guiana Shield) and the Atlantic forest. The formation of the Cerrado and Caatinga isolated Amazonia from the Atlantic Forest about 9 Ma (Simon et al. 2009) and might be associated with the diversification of B. torquatus restricted to the Atlantic forest and its sister group constituted of the other three-toed sloths (B. pygmaeus, B. tridactylus, and B. variegatus) within Pan-Amazonia further expanding into Central America. The final rise of the Northern Andes probably explains the vicariance between the two Tamandua species. These findings corroborate patterns of Xenarthra’s diversification discussed by Moraes-Barros and Arteaga (2015) who proposed a Western South America origin for Bradypus. These authors also pointed to a West to East dispersal through Amazonia for B. variegatus with a later colonization of the Atlantic forest during the Pleistocene.
On the contrary, armadillos display a more dynamic pattern with many inferred events of range expansions within all groups, also compensated by several local extinctions. For instance, we evidenced six independent colonizations of the Chaco region that likely occurred during the middle to late Miocene cooling and the aridification from the southern Amazon region (right map on fig. 5). The Chaco is indeed an area where current taxonomic richness is high in Xenarthra (Fergnani and Ruggiero 2015). The creation of new open habitats (biomes) in response to this general cooling (Simon et al. 2009) probably also promoted the diversification of Chlamyphoridae, especially within Euphractinae, with many species being now restricted to the different biomes: Z. pichiy in the Southern Cone (semiarid steppe grasslands); Chl. truncatus in the Central Desert; Cab. chacoensis, Cal. retusus, and To. matacus in the Chaco; To. tricinctus in the Caatinga; and Cab. tatouay in the Cerrado (Anacleto et al. 2006; Abba et al. 2012). Few species have a large repartition covering different biomes, such as P. maximus, Cab. unicinctus, and E. sexcinctus. The concurrent colonizations of arid areas and positive correlation of xenarthran diversification with cooler temperatures (cooler climate favors drier conditions) indicate that our diversification and biogeographical results are concordant and corroborate the invasion of the Southern cone within the last 10–15 My from a tropical origin. This is consistent with the reported increase in ecological diversity observed for xenarthrans in this region (Fergnani and Ruggiero 2015). Overall, the historical biogeography of Xenarthra is best explained by a progressive biome specialization of species due to the Cenozoic differentiation of biomes toward the present that probably led to more opportunities to disperse and diversify. The diversity-dependence pattern is likely attributed to allopatry within clades and secondary sympatry established between clades.
Conclusions
Our exhaustive data set establishes Xenarthra as the first major clade of placental mammals to be completely sequenced at the species level for mitogenomes from both modern and archival tissues. These data provide a reference phylogenetic framework and timescale, setting the scene for studying the diversification and biogeography of xenarthrans. The phylogenetic scaffold defined here will also be particularly useful for phylogenetic analyses based on ancient DNA of the numerous recently extinct taxa contained in this group. Finally, the wealth of molecular data generated here will be important for forthcoming studies on the phylogeography, species delimitation, barcoding, and conservation efforts of this understudied group of placentals.
Materials and Methods
Biological Samples
The complete taxon sampling we used in this study is detailed in table 1. The samples notably include several tissues linked to specimen vouchers deposited in International Natural History Museums including a type specimen (B. pygmaeus; USNM 579179). Most other tissue samples are conserved in the Mammalian Tissue Collection of the Institut des Sciences de l’Evolution de Montpellier (Catzeflis 1991). The maned sloth (B. torquatus) sample was collected under Brazilian permit Ibama/MMA 19267-3/14597869, and the Brazilian three-banded armadillo (To. tricinctus) with permit Ibama/MMA 42354-1.
DNA Extraction, Illumina Library Preparation, and Sequencing
All DNA extractions and library preparations took place in a separate, dedicated clean room (Biobubble) of McMaster Ancient DNA Centre, used strictly for low template samples.
DNA Extractions
For bone samples, up to 50 mg of bone materials were split into small pieces (1–5 mm) that were demineralized with 0.75 ml of 0.5 M EDTA pH 8 at room temperature overnight with agitation, and the supernatants removed following centrifugation. For soft tissue samples, up to 50 mg were cut into small pieces (1–5 mm). The one tissue preserved in Dimethylsulfoxid (DMSO) was washed multiple times with 0.1 × Tris-EDTA buffer (TE) pH 8 and blotted dry to remove this reducing agent before further processing. Pelleted bone and tissue samples were then digested with 0.5 ml of a Tris HCl–based proteinase K solution with 20 mM Tris–Cl pH 8, 0.5% sodium lauryl sarcosine (Fisher Scientific), 5 mM calcium chloride, 1% polyvinylpyrrolidone (Fisher Scientific), 50 mM dithiothreitol, 2.5 mM N-phenacyl thiazolium bromide (Prime Organics), and 250 µg/ml proteinase K. Proteinase digestion was performed at 55 °C for a couple of hours, with agitation. Following centrifugation the digestion supernatants were removed and extracted of organics using phenol:chloroform:isoamyl alcohol (25:24:1), and the resulting postcentrifugation aqueous solution was extracted with chloroform. We then concentrated the final aqueous phase with 30 kDA Amicon Ultra 0.5 ml centrifugal filters (Millipore) at 14,000 × g, with three washes using 0.1 × TE buffer pH 8 plus 0.05% Tween-20 to provide a desalted concentrate of 50 µl.
Genomic DNA Fragmentation
For 26 extracts, DNA fragmentation was done by digestion of 1 µg of DNA extract with NEBNext dsDNA Fragmentase (New England Biolabs). The reactions were purified using the MinElute Polymerase Chain Reaction (PCR) Purification kit (Qiagen) and eluted in 20 µl buffer EB. The remaining seven extracts were sonicated using a Covaris S220 according to manufacturer’s protocol for a median fragment length of 200 bp with a reduced input volume of 50 µl and a maximum of 10 µg of DNA (5 or 10 µl of extract).
Library Preparation and Indexing
We used between 0.1 and 1 µg of sheared DNA in Illumina library preparations as described in Meyer and Kircher (2010) with the modification of a blunt-end repair reaction volume of 40 µl or 70 µl and replacing all SPRI bead clean-ups with MinElute purifications to 20 µl buffer EB. We did not heat deactivate the Bst polymerase following the fill-in step and instead purified the reaction with MinElute to 20 µl buffer EB. A first set of libraries was index amplified using the common P5 and a set of unique P7 indexing primers (Meyer and Kircher 2010) in 50 µl reactions consisting of 1 × PCR buffer II, 2.5 mM MgCl2, 250 µM Deoxynucleotide (dNTP) mix, 200 nM each forward (P5) and reverse (P7) primer, 2.5 U Taq Gold, and 2 µl (100 ng) of template library. Thermal cycling conditions were as follows: initial denaturation at 95 °C for 4 min, 12 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 10 min. Amplifications were performed using a MJ thermocycler (BioRad). A second set of libraries was dual-index-amplified using unique P5 and P7 indexing primers (Kircher et al. 2012) in 50 µl reactions consisting of 1 × AccuPrime Pfx Reaction mix, 0.5 × EvaGreen, 500 nM each forward (P5) and reverse (P7) primer, 1.25 U AccuPrime Pfx DNA Polymerase, and 5 µl (250–2,500 ng) of template library. Thermal cycling conditions were as follows: initial denaturation at 95 °C for 2 min, 13 cycles of 95 °C for 15 s, 60 °C for 30 s, 68 °C for 30 s, and a final extension at 68 °C for 10 min. Amplifications were performed in real time with a CFX96 Real-time PCR platform (BioRad). All indexed libraries were finally purified with MinElute to 50 µl EB.
Illumina Sequencing
The 33 libraries were sequenced on 3 different lanes. The first run including 19 libraries was processed on an Illumina Genome Analyzer IIx using 72 bp single-end reads. The second run of four libraries was also processed on an Illumina Genome Analyzer IIx but using 72 bp paired-end reads. These first two runs were subcontracted to Ambry Genetics (Aliso Viejo, CA, USA). The third run of 10 libraries was run on an Illumina HiSeq 2500 instrument with 100 bp paired-end reads at the Donnelly Sequencing Centre of the University of Toronto (Canada). Initial data processing and base calling, including extraction of cluster intensities, was done using RTA1.8 (SCS version 2.8). Sequence quality filtering script was executed in the Illumina CASAVA software (ver 1.7.0, Illumina, Hayward, CA).
Mitochondrial Genomes Assembly
Raw sequence reads were first trimmed to remove any adapter and index tag sequences using CutAdapt (Martin 2011). De novo assembly of the trimmed reads was performed using the ABySS assembler (Simpson et al. 2009) with default parameters. We used a range of kmers to optimize contig lengths and identical contigs resulting from the use of different kmers were identified and collapsed using CD-HIT (Fu et al. 2012). Mitochondrial contigs were then extracted using BLASTN similarity searches against the closest reference mitogenome. If not already full length, contigs were assembled into complete mitochondrial genomes with Geneious R7 (Kearse et al. 2012). Using Geneious, mitogenomes were checked by mapping the trimmed reads to the mitogenome assemblies with the low sensitivity option, and scanned by eye to confirm appropriate mapping, particularly in regions with repeats. Any gaps in the contig assemblies were progressively filled by extending the contigs using iterations of the mapping procedure on the consensus sequence as implemented in Geneious. Final mitochondrial genomes were annotated by alignment with published xenarthran mitogenomes, and protein-coding regions were checked to confirm no indels or stop codons were present. The 33 new xenarthran mitogenomes have been deposited in GenBank under accession numbers KT818523–KT818555.
Phylogenetic Reconstructions
The 33 new mitochondrial genomes were added to the 4 available xenarthran mitogenomes and to 3 afrotherian species that were used as outgroups: the dugong (Dugong dugon; NC_003314), the African savannah elephant (Loxodonta africana; NC_000934), and the aardvark (Orycteropus afer; NC_002078). Individual mitochondrial genes were aligned with the MAFFT v7.017 plugin (Katoh and Standley 2013) in Geneious, using the amino acid translation for protein-coding genes. Unambiguously aligned sites were then selected by Gblocks (Castresana 2000) with default relaxed settings and codon options for protein-coding genes. The final data set contained 14,917 sites for 40 taxa and is available as supplementary material. PartitionFinder v1.1 (Lanfear et al. 2012) was used to find the optimal partition schemes and models of sequence evolution for RAxML, using the greedy algorithm starting from 42 a priori defined partitions corresponding to the 3 codon positions of each of the 13 protein-coding genes, 12S ribosomal RNA (rRNA), 16S rRNA, and the combined tranfer RNAs (tRNAs). Branch lengths have been unlinked among partitions and the Bayesian information criterion was used for selecting the best-fitting partition scheme.
ML inference was implemented with RAxML v7.8 (Stamatakis et al. 2008) using separate general time-reversible models with gamma distribution for each of the four best-fit partitions selected by PartitionFinder. Statistical reliability of the ML tree was evaluated with nonparametric bootstrapping (100 replications) through the Thorough Bootstrap option of RAxML under the optimal partitioned model to obtained ML bootstrap percentages (BPPART).
Bayesian phylogenetic inference under a mixed model was conducted using the MPI version of MrBayes 3.2.3 (Ronquist et al. 2012) using separate GTR+G8+I models for each of the four selected partitions, as determined by PartitionFinder, with parameters being unlinked across partitions. Two independent runs of four incrementally heated MCMCMC starting from a random tree were performed. MCMCMC was run for 10,000,000 generations, with trees and associated model parameters being sampled every 1,000 generations. The initial 2,500 trees in each run were discarded as burn-in samples after convergence checking. The 50% majority-rule Bayesian consensus tree and the associated posterior probabilities (PPPART) were then computed from the 15,000 combined trees sampled in the 2 independent runs. Bayesian phylogenetic reconstruction under the CAT-GTR-G4 mixture model (Lartillot and Philippe 2004) was conducted using PhyloBayes-MPI 1.5a (Lartillot et al. 2013). Two independent Markov Chain Monte Carlo (MCMC) starting from a random tree was run for 35,000 cycles (2,400,000 tree generations), with trees and associated model parameters being sampled every 10 cycles. The initial 350 trees (10%) sampled in each MCMC run were discarded as burn-in after checking for convergence in both likelihood and model parameters (tracecomp subprogram), and clade posterior probability (bpcomp subprogram). The 50% majority-rule Bayesian consensus tree and the associated posterior probabilities (PPCAT) were then computed from the remaining combined 6,300 (2 × 3,150) trees using bpcomp.
Molecular Datings
Molecular dating analyses were performed under a Bayesian relaxed molecular framework using PhyloBayes 3.3f (Lartillot et al. 2009). In all dating calculations, the tree topology was fixed to the majority-rule consensus tree previously inferred in Bayesian analyses. Dating analyses were conducted using the site-heterogeneous CAT-GTR+G4 mixture model and a relaxed clock model, as recommended by Lartillot et al. (2009) with a birth–death prior on divergence times (Gernhard 2008) combined with soft fossil calibrations (Yang and Rannala 2006). We used the following five, well-justified afrotherian and xenarthran calibration intervals defined by Meredith et al. (2011): 1) Paenungulata (maximum age 71.2 Ma, minimum age 55.6 Ma); 2) Xenarthra (maximum age 71.2 Ma, minimum age 58.5 Ma); 3) Pilosa (maximum age 65.5 Ma, minimum age 31.5 Ma); 4) Folivora (maximum age 40.6 Ma, minimum age 15.97 Ma); and 5) Vermilingua (maximum age 61.1 Ma, minimum age 15.97 Ma). We also added a recently proposed calibration point within armadillos based on the oldest armadillo fossil skull identified as a stem Tolypeutinae and found in the late Oligocene of Deseadean in Bolivia at 26 Ma (Billet et al. 2011). This finding allows us to set the minimum age for Tolypeutinae at a conservative 23.0 Ma, corresponding to the upper boundary of late Oligocene. The maximum age for the origin of Tolypeutinae was set at 37.8 Ma using the lower boundary of the late Eocene, because the oldest fossils of the closest outgroup (Euphractinae) trace back to the Casamayoran, at least 36 Ma (Kay et al. 1999). The prior on the root of the tree (Placentalia) was set at 100 Ma according to Meredith et al. (2011).
Although Lepage et al. (2007) and Rehm et al. (2011) showed that the autocorrelated LN relaxed clock model generally offers the best fit, we compared it with the UGAM model (Drummond et al. 2006) and a strict molecular clock (CL) model. These three clock models were compared against each other using the same prior settings (see above) in a cross-validation procedure as implemented in PhyloBayes. The cross-validation tests were performed by dividing the original alignment in 2 subsets of 13,426 sites (learning set) and 1,491 sites (test set). The overall procedure was repeated over 10 random splits for which a MCMC chain was run on the learning set for a total 1,100 cycles sampling posterior rates and dates every cycle. The first 100 samples of each MCMC were excluded as the burn-in period for calculating the cross-validation scores averaged across the 10 replicates. The final dating calculations were conducted under the best fitting model by running two independent MCMC chains for a total 50,000 cycles, sampling posterior rates and dates every 10 cycles. The first 500 samples (10%) of each MCMC were excluded as the burn-in after checking for convergence in both likelihood and model parameters. Posterior estimates of divergence dates were then computed from the remaining 4,500 samples of each MCMC using the readdiv subprogram. The values reported in table 2 are averages over the 2 independent chains.
Diversification Analyses
We used either the Bayesian chronogram or 100 randomly sampled chronograms obtained from the post burn-in posterior distribution of the PhyloBayes dating analyses to estimate diversification rates with different methods. These trees have been restricted to 32 taxa to better reflect the current xenarthran species diversity by excluding redundant taxa. To visualize the tempo and mode of diversification of the group, we first reconstructed lineages-through-time plots and then used a suite of ML models and a Bayesian model of diversification (Morlon 2014). For each type of ML diversification model, we computed the corrected Akaike information criterion (AICc). We then checked the support for the selected model against all models nested within it using likelihood ratio test (LRT). The scenario supported by LRT with the lowest AICc was considered the best fit. Bayes factors were used to assess model fit in the Bayesian framework.
Paleoenvironment-Dependent Diversification Model
To test the effect that past environmental change might have had on the diversification of Xenarthra, we used a model derived from the one of Morlon et al. (2011) that allows speciation and extinction rates to vary according to an environmental variable, which itself varies through time (Condamine et al. 2013), such as past variations in temperature (Zachos et al. 2001, 2008). Following the approach of Morlon et al. (2011), we designed four models to be tested: 1) BCSTDCST, a time constant birth–death model (null model); 2) BVARDCST, speciation rate is exponentially varying with temperature and extinction rate is constant; 3) BCSTDVAR, speciation rate is constant and extinction rate is exponentially varying with temperature; and 4) BVARDVAR, speciation and extinction rates are both exponentially varying with temperature. We also repeated these three rate-variable models with a linear dependence with temperature (because we have no prior expectation about how speciation or extinction might vary with temperature).
These models rely on a past environmental variable describing how the environment varied through time. For temperature, we relied on the well-known Cenozoic temperature curves published by Zachos et al. (2001, 2008). We used the R package pspline to reconstruct smooth lines of the paleo-data for the environmental variable. In other words, a smooth line is introduced in the birth–death model to represent the paleoenvironment through time, and at each point in time the model refers to this smooth line to obtain the value of the temperature. Given the dated phylogeny, the model then estimates speciation and extinction rates as a function of this value (Condamine et al. 2013).
Time-Dependent Diversification Models
We assessed whether diversification rates remained constant during the evolutionary history of Xenarthra. We first used BAMM to estimate speciation and extinction rates through time along the xenarthran phylogeny (Rabosky 2014). BAMM allows studying complex evolutionary processes on phylogenetic trees, potentially shaped by a heterogeneous mixture of distinct evolutionary dynamics of speciation and extinction across clades. The method is designed to automatically detect rate shifts and sample distinct evolutionary dynamics that best explain the whole diversification dynamics of the clade. In BAMM, the speciation rate is allowed to vary exponentially through time while extinction is maintained constant (Rabosky, Donnellan, et al. 2014). Subclades in the tree might diversify faster (or slower) than others, and BAMM allows detecting these diversification rate shifts and comparing how many and where these shifts might occur. BAMM is implemented in a C++ command line program and the BAMMtools R package (Rabosky, Grundler, et al. 2014). We set 4 MCMC running for 10 million generations and sampled every 10,000 generations. Other parameters were set to default values except the Poisson process prior that we set to 1.0 following the authors’ recommendation (Rabosky, Grundler, et al. 2014). We performed four independent runs (with a burn-in of 15%) using different seeds, and we used effective sample size to assess the convergence of the runs, considering values above 200 as indicating convergence. The posterior distribution was used to compute the best global rates of diversification through time, to estimate the configuration of the diversification rate shifts by evaluating alternative diversification models as compared by Bayes factors.
The TreePar package (Stadler 2011) was used to assess speciation and extinction rates through time, and to specifically detect potential rapid and global changes in diversification rates that might be due to environmental factors, such as climatic shifts following major geological events. We employed the “bd.shifts.optim” function that allows estimating discrete changes in speciation, extinction rates, and mass extinction events in undersampled phylogenies (Stadler 2011). At each time t, the rates are allowed to change and the species may undergo a shift in diversification. TreePar analyses were run with the following settings: start = 0, end = crown age estimated by dating analyses, grid = 0.1 Myr, and posdiv = FALSE to allow the diversification rate to be negative (i.e., allows for periods of declining diversity).
Diversity-Dependent Diversification Model
We investigated whether lineages diversified rapidly in their early stages and have reached equilibrium (or are being bounded), suggesting that diversity is saturated toward the present, as niches became occupied and diversification rates slowed down. We used the method of Etienne et al. (2012) to explore the effect of diversity on speciation and extinction rates. The function “dd_ML” was used to fit five models: 1) speciation declines linearly with diversity and no extinction (DDL), 2) speciation declines linearly with diversity and extinction (DDL + E), 3) speciation declines exponentially with diversity and extinction (DDX + E), 4) extinction increases linearly with diversity (DD + EL), and 5) extinction increases exponentially with diversity (DD + EX). The initial carrying capacity was set to the current species diversity, and the final carrying capacity was estimated according to the models and parameters.
Biogeographic Reconstructions
The ancestral range estimation was performed using BioGeoBEARS (Matzke 2014). The analyses were carried out using the same time-calibrated phylogeny as used for the diversification analyses. We used the DEC model (Ree and Smith 2008) to conduct local optimizations and estimate the ancestral character state of each node according to the current distribution and biogeographical model the user introduces in the analyses. The root was left unconstrained but optimized by the method. We did not assess whether the inclusion of the founder-event speciation (parameter J) significantly improved the likelihood because DEC-J is appropriate for island-dwelling clades (Matzke 2014).
A geographic model was incorporated to include operational areas that are defined as geographic ranges shared by two or more species and delimited by geological or oceanic features, which may have acted as barriers to dispersal. The distribution of anteaters, armadillos, and sloths ranges from southern North America to southern South America (Abba and Superina 2010; Superina, Plese, et al. 2010; Superina, Miranda, et al. 2010). We further divided these two regions into smaller biogeographic identities to obtain higher resolution in the inference of the ancestral area of origin. Using tectonic reconstructions, notably the evolution of past Amazonian landscapes (e.g., Hoorn et al. 2010), the model comprised nine component areas: 1) Amazonia (Amazon Forest plus Amazon Basin); 2) Mata Atlântica (Atlantic Forest along the Brazilian coast); 3) Cerrado (tropical savannah) and Caatinga (desert and xeric shrublands); 4) Chaco region and Pantanal (semiarid lowland and adjacent wetlands); 5) southern South America (grasslands); 6) Southern and Central Andes (Chile, western Bolivia, and Peru); 7) Northern Andes (Ecuador, Colombia, and western Venezuela); 8); Guiana Shield (eastern Venezuela, Guyana, Suriname, French Guiana), and 9) Central and North America (from Panama to southern USA). The adjacency matrix was designed while taking into account the geological history and the biological plausibility of combined ranges (Hoorn et al. 2010). We discarded ranges larger than six areas in size that were not subsets of observed species ranges (Da. novemcinctus had the largest range with six areas). Distributional data were compiled from monographs and IUCN data (Abba and Superina 2010; Superina, Plese, et al. 2010; Superina, Miranda, et al. 2010).
We did not split our sample into smaller geographic areas for several reasons. The complexity of the geological history of this region makes it difficult to accurately reconstruct past distributions of land and sea at any given point in time. For instance, many uncertainties remain about the appearance and disappearance of the Pebas System, or the exact timing of the rising of the Andes (Hoorn et al. 2010). Moreover, our goal was to investigate the dominant biogeographical processes that shaped the xenarthran distribution pattern. For the latter, we were particularly interested in assessing the relative contributions of vicariance and dispersal.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work largely benefited from the help of the following individuals and institutions who provided tissue samples: François Catzeflis (Institut des Sciences de l’Evolution, Montpellier, France), Jean-François Mauffrey, Philippe Gaucher, Eric Hansen, François Ouhoud-Renoux, Jean-Christophe Vié, Philippe Cerdan, Michel Blanc, and Rodolphe Paowé (French Guiana), Sergio Vizcaíno (Museo de La Plata, La Plata, Argentina), Jorge Omar García and Rodolfo Rearte (Complejo Ecológico Municipal, Presidencia Roque Sáenz Peña, Argentina), Daniel Hernández (Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay), John Trupkiewicz (Philadelphia Zoo, USA), Darrin Lunde (National Museum of Natural History, Washington, USA), Jim Patton and Yuri Leite (Museum of Vertebrate Zoology, Berkeley, USA), Ross MacPhee (American Museum of Natural History, New York, USA), Jonathan Dunnum and Joseph Cook (Museum of Southwestern Biology, Albuquerque, USA), Donna Dittman and Mark Hafner (Louisiana State University Museum of Natural Science, Baton Rouge, USA), Gerhard Haszprunar and Michael Hiermeier (Zoologische Staatssammlung München, Munich, Germany), Géraldine Véron (Museum National d’Histoire Naturelle, Paris, France), Agustín Jiménez-Ruiz, Guido Valverde, and Guillermo Pérez-Jimeno. Our thanks also extend to Trish McLenachan and David Penny who sequenced and made freely available the first sloth mitogenomes, Lionel Hautier and Philippe Gaubert for taxonomical advice, and two anonymous referees for helpful comments. This work was supported by grants from the EU’s Seventh Framework Programme (No 286431) to N.M.-B., the Centre National de la Recherche Scientifique (CNRS), the Scientific Council of Université Montpellier 2 (UM2), and Investissement d’Avenir of the Agence Nationale de la Recherche (CEBA: ANR-10-LABX-25-01) to F.D., and the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chairs program to H.N.P. This is contribution ISEM 2015-227-S of the Institut des Sciences de l’Evolution de Montpellier.
References
- Abba AM, Cassini GH, Valverde G, Tilak MK, Vizcaíno SF, Superina M, Delsuc F. 2015. Systematics of hairy armadillos and the taxonomic status of the Andean hairy armadillo (Chaetophractus nationi). J Mammal. 96:673–689. [Google Scholar]
- Abba AM, Superina M. 2010. The 2009/2010 armadillo Red List assessment. Edentata 11:135–184. [Google Scholar]
- Abba AM, Tognelli MF, Seitz VP, Bender JB, Vizcaíno SF. 2012. Distribution of extant xenarthrans (Mammalia: Xenarthra) in Argentina using species distribution models. Mammalia 76:123–136. [Google Scholar]
- Amrine-Madsen H, Koepfli KP, Wayne RK, Springer MS. 2003. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 28:225–240. [DOI] [PubMed] [Google Scholar]
- Anacleto TCS, Diniz JAF, Vital MVC. 2006. Estimating potential geographic ranges of armadillos (Xenarthra, Dasypodidae) in Brazil under niche-based models. Mammalia 70:202–213. [Google Scholar]
- Anderson RP, Handley CO. 2001. A new species of three-toed sloth (Mammalia: Xenarthra) from Panama, with a review of the genus Bradypus. Proc Biol Soc Wash. 114:1–33. [Google Scholar]
- Arnason U, Adegoke JA, Bodin K, Born EW, Esa YB, Gullberg A, Nilsson M, Short RV, Xu X, Janke A. 2002. Mammalian mitogenomic relationships and the root of the eutherian tree. Proc Natl Acad Sci U S A. 99:8151–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason U, Gullberg A, Janke A. 1997. Phylogenetic analyses of mitochondrial DNA suggest a sister group relationship between Xenarthra (Edentata) and Ferungulates. Mol Biol Evol. 14:762–768. [DOI] [PubMed] [Google Scholar]
- Bacon CD, Silvestro D, Jaramillo C, Smith BT, Chakrabarty P, Antonelli A. 2015. Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc Natl Acad Sci U S A. 112:6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MC, Sampaio I, Schneider H. 2003. Phylogenetic analysis of 16S mitochondrial DNA data in sloths and anteaters. Genet Mol Biol. 26:5–12. [Google Scholar]
- Barros MC, Sampaio I, Schneider H. 2008. Novel 12S mtDNA findings in sloths (Pilosa, Folivora) and anteaters (Pilosa, Vermilingua) suggest a true case of long branch attraction. Genet Mol Biol. 31:793–799. [Google Scholar]
- Beaulieu JM, O'Meara BC. 2015. Extinction can be estimated from moderately sized molecular phylogenies. Evolution 69:1036–1043. [DOI] [PubMed] [Google Scholar]
- Benton MJ. 2009. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323:728–732. [DOI] [PubMed] [Google Scholar]
- Billet G, Hautier L, de Muizon C, Valentin X. 2011. Oldest cingulate skulls provide congruence between morphological and molecular scenarios of armadillo evolution. Proc R Soc Lond B. 278:2791–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakey RC. 2008. Gondwana paleogeography from assembly to breakup—a 500 million year odyssey. In: Fielding CR, Frank TD, Isbell JL, editors. Resolving the late Paleozoic ice age in time and space. Boulder (CO): Geological Society of America Special Paper 441; p. 1–28. [Google Scholar]
- Botero-Castro F, Tilak MK, Justy F, Catzeflis F, Delsuc F, Douzery EJP. 2013. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol Phylogenet Evol. 69:728–739. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Castro MC, Ciancio MR, Pacheco V, Salas-Gismondi RM, Bostelmann JE, Carlini AA. 2015. Reassessment of the hairy long-nosed armadillo “Dasypus” pilosus (Xenarthra, Dasypodidae) and revalidation of the genus Cryptophractus Fitzinger, 1856. Zootaxa 3947:30–48. [DOI] [PubMed] [Google Scholar]
- Catzeflis FM. 1991. Animal tissue collections for molecular genetics and systematics. Trends Ecol Evol. 419:565–571. [DOI] [PubMed] [Google Scholar]
- Cetica PD, Solari AJ, Merani MS, de Rosas JC, Burgos MH. 1998. Evolutionary sperm morphology and morphometry in armadillos. J Submicrosc Cytol Pathol. 30:309–314. [PubMed] [Google Scholar]
- Chiari Y, Cahais V, Galtier N, Delsuc F. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Kriegs JO, Baertsch R, Zemann A, Brosius J, Schmitz J. 2009. Mosaic retroposon insertion patterns in placental mammals. Genome Res. 19:868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine FL, Rolland J, Morlon H. 2013. Macroevolutionary perspectives to environmental change. Ecol Lett. 16:72–75. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Catzeflis FM, Stanhope MJ, Douzery EJP. 2001. The evolution of armadillos, anteaters, and sloths depicted by nuclear and mitochondrial phylogenies: implications for the status of the enigmatic fossil Eurotamandua. Proc R Soc Lond BS. 268:1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Douzery EJP. 2008. Recent advances and future prospects in xenarthran molecular phylogenetics. In: Vizcaĺno SF, Loughry WJ, editors. The Biology of the Xenarthra. Gainesville: University Press of Florida; p. 11–23. [Google Scholar]
- Delsuc F, Scally M, Madsen O, Stanhope MJ, De Jong WW, Catzeflis FM, Springer MS, Douzery EJP. 2002. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol Biol Evol. 19:1656–1671. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Stanhope MJ, Douzery EJP. 2003. Molecular systematics of armadillos (Xenarthra, Dasypodidae): contribution of maximum likelihood and Bayesian analyses of mitochondrial and nuclear genes. Mol Phylogenet Evol. 28:261–275. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Superina M, Tilak MK, Douzery EJP, Hassanin A. 2012. Molecular phylogenetics unveils the ancient evolutionary origins of the enigmatic fairy armadillos. Mol Phylogenet Evol. 62:673–680. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Vizcaíno SF, Douzery EJP. 2004. Influence of Tertiary paleoenvironmental changes on the diversification of South American mammals: a relaxed molecular clock study within xenarthrans. BMC Evol Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann GF. 1985. The phylogeny of the Xenarthra . In: Montgomery GG, editor. The Evolution and Ecology of Armadillos, Sloths and Vermilinguas. Washington and London: Smithsonian Institution Press; p. 51-64. [Google Scholar]
- Enk J, Devault A, Debruyne R, King CE, Treangen T, O’Rourke D, Salzberg SL, Fisher D, MacPhee R, Poinar H. 2011. Complete Columbian mammoth mitogenome suggests interbreeding with woolly mammoths. Genome Biol. 12:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne RS, Haegeman B, Stadler T, Aze T, Pearson PN, Purvis A, Phillimore AB. 2012. Diversity-dependence brings molecular phylogenies closer to agreement with the fossil record. Proc R Soc Lond B. 279:1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PH, Jønsson KA, Douzery EJP. 2013. Jumping and gliding rodents: mitogenomic affinities of Pedetidae and Anomaluridae deduced from an RNA-Seq approach. Gene 531:388–397. [DOI] [PubMed] [Google Scholar]
- Fabre PH, Vilstrup JT, Raghavan M, Der Sarkissian C, Willerslev E, Douzery EJP, Orlando L. 2014. Rodents of the Caribbean: origin and diversification of hutias unravelled by next-generation museomics. Biol Lett. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó A, Garbino GST, Campos BATP, Rocha PA, Ferrari SF, Langguth A. 2015. Distribution of Tolypeutes Illiger, 1811 (Xenarthra: Cingulata) with comments on its biogeography and conservation. Zool Sci. 32:77-87. [DOI] [PubMed] [Google Scholar]
- Feng X, Papes M. 2015. Ecological niche modelling confirms potential north-east range expansion of the nine-banded armadillo (Dasypus novemcinctus) in the USA. J Biogeogr. 42:803-807. [Google Scholar]
- Fergnani PN, Ruggiero A. 2015. Ecological diversity in South American mammals: their geographical distribution shows variable associations with phylogenetic diversity and does not follow the latitudinal richness gradient. PLoS One 10:e0128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finstermeier K, Zinner D, Brameier M, Meyer M, Kreuz E, Hofreiter M, Roos C. 2013. A mitogenomic phylogeny of living primates. PLoS One 8:e69504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz SA, Schnitzler J, Eronen JT, Hof C, Böhning-Gaese K, Graham CH. 2013. Diversity in time and space: wanted dead and alive. Trends Ecol Evol. 28:509–516. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbreath GJ. 1985. The evolution of monozygotic polyembryony in Dasypus. In: Montgomery GG, editor. The Evolution and Ecology of Armadillos, Sloths and Vermilinguas. Washington and London: Smithsonian Institution Press; p. 243-245. [Google Scholar]
- Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K. 2006. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 16:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AL. 2008. Magnorder Xenarthra. In: Gardner AL, editor. Mammals of South America: Volume 1 Marsupials, Xenarthrans, Shrews, and Bats. Chicago (IL): University of Chicago Press; p. 127–128. [Google Scholar]
- Garzione CN, Hoke GD, Libarkin JC, Withers S, MacFadden B, Eiler J, Ghosh P, Mulch A. 2008. Rise of the Andes. Science 320:1304–1307. [DOI] [PubMed] [Google Scholar]
- Gaudin TJ, Branham DG. 1998. The phylogeny of the Myrmecophagidae (Mammalia, Xenarthra, Vermilingua) and the relationship of Eurotamandua to the Vermilingua. J Mammal Evol. 5:237-265. [Google Scholar]
- Gaudin TJ, Wible JR. 2006. The phylogeny of living and extinct armadillos (Mammalia, Xenarthra, Cingulata): A craniodental analysis. In: Carrano MT, Gaudin TJ, Blob RW, Wible JR, editors. Amniote Paleobiology. Chicago: The University of Chicago Press; p. 153-198. [Google Scholar]
- Gissi C, Reyes A, Pesole G, Saccone C. 2000. Lineage-specific evolutionary rate in mammalian mtDNA. Mol Biol Evol. 17:1022-1031. [DOI] [PubMed] [Google Scholar]
- Gradstein FM, Ogg JG, Schmitz MD, Ogg GM. 2012. The geologic time scale 2012, volume set. Boston: Elsevier. [Google Scholar]
- Guschanski K, Krause J, Sawyer S, Valente LM, Bailey S, Finstermeier K, Sabin R, Gilissen E, Sonet G, Nagy ZT, et al. 2013. Next-generation museomics disentangles one of the largest primate radiations. Syst Biol. 62:539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT, et al. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 335:32–50. [DOI] [PubMed] [Google Scholar]
- Hayssen V. 2011. Choloepus hoffmanni (Pilosa: Megalonychidae). Mammalian Species 43:37–55. [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez-Meseguer A, Anderson CL, Figueiredo JP, et al. 2010. Amazonia through time: Andean uplift, climate change, landscape evolution and biodiversity. Science 330:927–931. [DOI] [PubMed] [Google Scholar]
- Huchon D, Delsuc F, Catzeflis FM, Douzery EJP. 1999. Armadillos exhibit less genetic polymorphism in North America than in South America: nuclear and mitochondrial data confirm a founder effect in Dasypus novemcinctus (Xenarthra). Mol Ecol. 8:1743–1748. [DOI] [PubMed] [Google Scholar]
- IUCN Red List of Threatened Species. 2015. Version 2014.3. Available from: www.iucnredlist.org. Accessed on 15 April 2015. [Google Scholar]
- Jansa SA, Barker FK, Voss RS. 2014. The early diversification history of didelphid marsupials: a window into South America’s “Splendid Isolation”. Evolution 68:684–695. [DOI] [PubMed] [Google Scholar]
- Kainer D, Lanfear R. 2015. The effects of partitioning on phylogenetic inference. Mol Biol Evol. 2:1611–1627. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RF, Madden RH, Vucetich GM, Carlini AA, Mazzoni MM, Re GH, Heizler M, Sandeman H. 1999. Revised geochronology of the Casamayoran South American land mammal age: climatic and biotic implications. Proc Natl Acad Sci U S A. 96:13235–13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Leavitt JR, Hiatt KD, Whiting MF, Song H. 2013. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: a phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol Phylogenet Evol. 67:494–508. [DOI] [PubMed] [Google Scholar]
- Lepage T, Bryant D, Philippe H, Lartillot N. 2007. A general comparison of relaxed molecular clock models. Mol Biol Evol. 24:2669–2680. [DOI] [PubMed] [Google Scholar]
- Loughry WJ, McDonough CM. 2013. The nine-banded armadillo: a natural history. Norman (OK): University of Oklahoma Press. [Google Scholar]
- Lyons SK, Smith FA, Brown JH. 2004. Of mice, mastodons and men: human-mediated extinctions on four continents. Evol Ecol Res. 6:339–358. [Google Scholar]
- Madsen O, Scally M, Douady CJ, Kao DJ, DeBry RW, Adkins R, Amrine HM, Stanhope MJ, de Jong WW, Springer MS. 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409:610–614. [DOI] [PubMed] [Google Scholar]
- Marshall LG, Webb SD, Sepkoski JJ, Raup DM. 1982. Mammalian evolution and the great American interchange. Science 215:1351–1357. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. [Google Scholar]
- Mason VC, Li G, Helgen KM, Murphy WJ. 2011. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res. 21:1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst Biol. 63:951–970. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Faircloth BC, Crawford NG, Gowaty PA, Brumfield RT, Glenn TC. 2012. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species tree analysis. Genome Res. 22:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HG. 2005. Paleoecology of extinct xenarthrans and the Great American Biotic Interchange. Bull Fla Mus Nat Hist. 45:313–333. [Google Scholar]
- McKenna MC, Bell SK. 1997. Classification of mammals above the species level. New York: Columbia University Press. [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334:521–524. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010: pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pratt RC, Watson LN, Gibb GC, Llamas B, Kasper M, Edson J, Hopwood B, Male D, Armstrong KN, et al. 2014. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol Biol Evol. 31:2322–2330. [DOI] [PubMed] [Google Scholar]
- Möller-Krull M, Delsuc F, Churakov G, Marker C, Superina M, Brosius J, Douzery EJP, Schmitz J. 2007. Retroposed elements and their flanking regions resolve the evolutionary history of xenarthran mammals (armadillos, anteaters, and sloths). Mol Biol Evol. 24:2573–2582. [DOI] [PubMed] [Google Scholar]
- Montes C, Cardona A, Jaramillo C, Pardo A, Silva JC, Valencia V, Ayala VC, Pérez-Angel LC, Rodriguez-Parra LA, Ramirez V, et al. 2015. Middle Miocene closure of the Central American seaway. Science 348:226–229. [DOI] [PubMed] [Google Scholar]
- Moraes-Barros N, Arteaga MC. 2015. Genetic diversity in Xenarthra and its relevance to patterns of Neotropical biodiversity. J Mammal. 96:690–702. [Google Scholar]
- Moraes-Barros N, Silva JA, Morgante JS. 2011. Morphology, molecular phylogeny, and taxonomic inconsistencies in the study of Bradypus sloths (Pilosa: Bradypodidae). J Mammal. 92:86–100. [Google Scholar]
- Morlon H. 2014. Phylogenetic approaches for studying diversification. Ecol Lett. 17:508–525. [DOI] [PubMed] [Google Scholar]
- Morlon H, Parsons TL, Plotkin J. 2011. Reconciling molecular phylogenies with the fossil record. Proc Natl Acad Sci U S A. 108:16327–16332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WJ, Zhang YP, Ryder OA, O'Brien SJ. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409:614–618. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Ellegren H, Wolf JB. 2012. High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol Biol Evol. 30:272–284. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol. 25:120–130. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glémin S, Galtier N. 2009. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol. 9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Maruyama S, Okada N. 2009. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc Natl Acad Sci U S A. 106:5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novacek MJ. 1992. Mammalian phylogeny: shaking the tree. Nature 356:121–125. [DOI] [PubMed] [Google Scholar]
- Nyakatura JA. 2012. The convergent evolution of suspensory posture and locomotion in tree sloths. J Mammal Evol. 19:225–234. [Google Scholar]
- Paijmans JL, Gilbert MTP, Hofreiter M. 2013. Mitogenomic analyses from ancient DNA. Mol Phylogenet Evol. 69:404–416. [DOI] [PubMed] [Google Scholar]
- Patterson B, Pascual R. 1968. The fossil mammal fauna of South America. Q Rev Biol. 43:409–451. [Google Scholar]
- Powell AF, Barker FK, Lanyon SM. 2013. Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: an avian case study. Mol Phylogenet Evol. 66:69–79. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9:e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Donnellan SC, Grundler M, Lovette IJ. 2014. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst Biol. 63:610–627. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, Shi JJ, Brown JW, Huang H, Larson JG. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol. 5:701–707. [Google Scholar]
- Ree RH, Smith SA. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 57:4–14. [DOI] [PubMed] [Google Scholar]
- Rehm P, Borner J, Meusemann K, von Reumont BM, Simon S, Hadrys H, Misof B, Burmester T. 2011. Dating the arthropod tree based on large-scale transcriptome data. Mol Phylogenet Evol. 61:880–887. [DOI] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 15:957–966. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery EJP. 2013. Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol. 30:2134–2144. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe KC, Singhal S, Macmanes MD, Ayroles JF, Morelli TL, Rubidge EM, Bi K, Moritz CC. 2011. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Mol Ecol Res. 11:1082–1092. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Schnitzler J, Liow LH, Antonelli A, Salamin N. 2014. Bayesian estimation of speciation and extinction from incomplete fossil occurrence data. Syst Biol. 63:349–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MF, Grether R, de Queiroz LP, Skema C, Pennington RT, Hughes CE. 2009. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proc Natl Acad Sci U S A. 108:20359–20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. 1980. Splendid isolation: the curious history of mammals in South America. New Haven (CT): Yale University Press. [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Liu L, Edwards SV, Wu S. 2012. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci U S A. 109:14942–14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T. 2011. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci U S A. 108:6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Houck ML, Ryder OA. 2010. Species identification and chromosome variation of captive two-toed sloths. Zoo Biol. 29:1–13. [DOI] [PubMed] [Google Scholar]
- Superina M, Miranda FR, Abba AM. 2010. The 2010 anteater Red List assessment. Edentata 11:96–114. [Google Scholar]
- Superina M, Pagnutti N, Abba AM. 2014. What do we know about armadillos? An analysis of four centuries of knowledge about a group of South American mammals, with emphasis on their conservation. Mammal Rev. 44:69–80. [Google Scholar]
- Superina MT, Plese N, de Moraes-Barros N, Abba AM. 2010. The 2010 sloth red list assessment. Edentata 11:115–134. [Google Scholar]
- Taulman JF, Robbins LW. 2014. Range expansion and distributional limits of the nine-banded armadillo in the United States: an update of Taulman & Robbins (1996). J Biogeogr. 41:1626–1630. [Google Scholar]
- Thorne JL, Kishino H, Painter IS. 1998. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 15:1647–1657. [DOI] [PubMed] [Google Scholar]
- Tilak MK, Justy F, Debiais-Thibaud M, Botero-Castro F, Delsuc F, Douzery EJP. 2015. A cost-effective straightforward protocol for shotgun Illumina libraries designed to assemble complete mitogenomes from non-model species. Conserv Genet Res. 7:37–40. [Google Scholar]
- Vizcaíno SF. 1995. Identificación específica de las “mulitas”, género Dasypus L. (Mammalia, Dasypodidae), del noroeste argentino. Descripción de una nueva especie. Mastozool Neotrop. 2:5–13. [Google Scholar]
- Webb SD. 1985. The interrelationships of tree sloths and ground sloths. In: Montgomery GG, editor. The Evolution and Ecology of Armadillos, Sloths and Vermilinguas. Washington: Smithsonian Institution; p. 105–112. [Google Scholar]
- Wetzel RM. 1985. The identification and distribution of recent Xenarthra (= Edentata). In: Montgomery GG, editor. The evolution and ecology of armadillos, sloths, and vermilinguas. Washington and London: Smithsonian Institution Press; p. 5–21. [Google Scholar]
- Wetzel RM, Mondolfi E. 1979. The subgenera and species of long-nosed armadillos, Genus Dasypus L. In: Eisenberg JF, editor. Vertebrate Ecology in the Northern Neotropics. Washington: The National Zoological Park, Smithsonian Institution; p. 39–63. [Google Scholar]
- Yang Z, Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using fossil calibrations with soft bounds. Mol Biol Evol. 23:212–226. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE. 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451:279–283 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.