Abstract
Essential trace elements possess vital functions at molecular, cellular, and physiological levels in health and disease, and they are tightly regulated in the human body. In order to assess variability and potential adaptive evolution of trace element homeostasis, we quantified 18 trace elements in 150 liver samples, together with the expression levels of 90 genes and abundances of 40 proteins involved in their homeostasis. Additionally, we genotyped 169 single nucleotide polymorphism (SNPs) in the same sample set. We detected significant associations for 8 protein quantitative trait loci (pQTL), 10 expression quantitative trait loci (eQTLs), and 15 micronutrient quantitative trait loci (nutriQTL). Six of these exceeded the false discovery rate cutoff and were related to essential trace elements: 1) one pQTL for GPX2 (rs10133290); 2) two previously described eQTLs for HFE (rs12346) and SELO (rs4838862) expression; and 3) three nutriQTLs: The pathogenic C282Y mutation at HFE affecting iron (rs1800562), and two SNPs within several clustered metallothionein genes determining selenium concentration (rs1811322 and rs904773). Within the complete set of significant QTLs (which involved 30 SNPs and 20 gene regions), we identified 12 SNPs with extreme patterns of population differentiation (FST values in the top 5% percentile in at least one HapMap population pair) and significant evidence for selective sweeps involving QTLs at GPX1, SELENBP1, GPX3, SLC30A9, and SLC39A8. Overall, this detailed study of various molecular phenotypes illustrates the role of regulatory variants in explaining differences in trace element homeostasis among populations and in the human adaptive response to environmental pressures related to micronutrients.
Keywords: quantitative trait loci, trace elements, positive selection, proteomics, micronutrient homeostasis
Introduction
Humans depend on at least nine essential trace elements (bromine, cobalt, copper, iodine, iron, manganese, molybdenum, selenium, and zinc) and possibly more trace elements (boron and chromium). In spite of dietary intake fluctuations, the physiological and cellular concentrations of essential trace elements (or inorganic micronutrients) are tightly kept in homeostasis by a number of membrane transport proteins and metal-binding proteins. Such regulation is fundamental to ensure the correct molecular and cellular functions that depend on metal presence. Indeed, both micronutrient deficiencies and excesses have adverse effects on humans. Iron deficiency anemia, hypothyroidism, goiter, endemic cretinism, and acrodermatitis enteropathica are examples of easily recognized clinical entities that are associated with deficiencies of iron, iodine, and zinc (Diplock 1987). On the contrary, hemochromatosis is caused by the excessive intestinal absorption of dietary iron. The disruption in trace element homeostasis has also been associated to development impairments (Kambe et al. 2008), immunity disorders (Rink and Haase 2007), and diseases such as diabetes (Wiernsperger and Rapin 2010) or cancer (Margalioth et al. 1983; Díez et al. 1989; Costello and Franklin 2006). At the same time, differences in micronutrient homeostasis between populations can be adaptive. In sub-Saharan Africa, a nonsynonymous variant in the intestinal zinc transporter ZIP4 modifies cellular zinc uptake and shows a genomic signature of recent positive selection (Engelken et al. 2014). In Europe, dietary iron deficiency may have been counteracted by the C282Y mutation in HFE (rs1800562), although it causes hemochromatosis (iron overload) in homozygotes (Bulaj et al. 1996; Bamshad and Wooding 2003; Toomajian et al. 2003). Moreover, a different variant in HFE seems to be under recent positive selection in Asians (Ye et al. 2015). Signatures of recent genetic adaptation have also been described in several selenoprotein genes and related regulatory genes in populations living in selenium deficient regions of Asia (White et al. 2015). At the opposite extreme, the strong genetic differentiation around the AS3MT gene between the Argentinean Andes and the Peruvian populations has been suggested to result from adaptation to tolerate the arsenic-rich environment of the northern Argentinean Andes (Schlebusch et al. 2015).
Genetic variation in humans contributes to interindividual differences in gene expression and to differential relative abundances of proteins and metabolites in cells, tissues, and organs. For instance, several genome-wide association studies (GWAS) have revealed several loci and single nucleotide polymorphism (SNPs) directly related to iron status (McLaren et al. 2011, 2012; van der Harst et al. 2012). At the same time, human trace element homeostasis has been shown to result not only from regulation at gene expression level but at protein trafficking and post-translational levels (Malinouski et al. 2014). In recent years, expression quantitative trait loci studies (eQTL) have successfully linked a variety of SNPs in humans and model organisms to variation in gene expression levels (Nica and Dermitzakis 2013). In parallel, new advances in mass spectrometry-based proteomics have provided consistent quantification measurements of subsets of proteins across many biological samples, thus enabling the analysis of the impact of genetic variation at the protein level (protein quantitative trait loci [pQTL]). Selected reaction monitoring (SRM) (Lange et al. 2008) is one of the main strategies for reproducible quantification of specific peptides and proteins in large sample cohorts and different experimental conditions with a wide dynamic range with high sensitivity and accuracy (Ebhardt et al. 2012; Sabidó et al. 2013). Although the use of these approaches to evaluate genome-wide SNP–protein associations (trans-effects) is still limited due to required number of samples, a few studies have successfully performed pQTL experiments for a restricted number of SNP–protein associations (Lourdusamy et al. 2012; Johansson et al. 2013; Wu et al. 2013, 2014; Battle et al. 2015; Liu et al. 2015). Indeed, by restricting the number of proteins and polymorphisms to the SNPs located in specific genomic regions, or to the proteins involved in a particular metabolic process, the number of associations is reduced and it is possible to identify protein cis-regulation elements by reducing the number of potential false positives. Although one may expect a correlation between mRNA and protein levels, several studies find generally modest correlation, and point to nontranscriptional regulatory mechanisms influencing protein levels (Foss et al. 2011; Ghazalpour et al. 2011; Schwanhausser et al. 2011; Wu et al. 2013, 2014).
Here, we have taken advantage of recent mass spectrometry-based targeted methods to map pQTL for cis-regulatory variants influencing the abundance of proteins involved in trace element homeostasis in human liver samples. In addition, we have sought to describe the diversity of the regulation of trace element concentrations by analyzing four different layers of variation in the same samples: 1) SNPs in genes involved in the regulation of some trace elements; 2) the mRNA expression of those genes; 3) the relative amounts of the proteins coded by those genes; and 4) the concentration of the trace elements themselves. In particular, up to 47 proteins were successfully quantified by targeted proteomics over a large dynamic concentration range; 40 of them are directly related to 6 essential elements (selenium, magnesium, manganese, iron, copper, and zinc). In parallel, and for the same set of 150 liver tissue samples, we characterized the expression levels of about 90 different transcripts of genes also related to metal homeostasis, and quantified up to 18 trace elements. Next, we explored 165 SNPs located around the regions encoding those transcripts (cis-SNPs) for association to the variation in mRNA expression and in protein and trace element concentration. The joint analysis of these four levels reveals individual differences in trace element regulation, and expands our understanding of the interplay between genetic variants, gene expression, and the proteome in the maintenance of micronutrient homeostasis. Besides identifying protein, expression, and micronutrient QTLs (nutriQTL), we explore and discuss the existing evidence for signals of recent selection around them.
Results
Initially, we compiled 150 liver samples originally collected from autopsies of human individuals of European ancestry from the UK Human Tissue Bank (UKHTB). The average age of the subjects was 49.2 ± 14.6 years; 56% of the subjects were males. Next, we evaluated the abundance levels of 18 trace elements as well as of 90 transcripts and 40 proteins related to metal homeostasis including several membrane transporters, storage proteins, regulatory factors, and metal-binding proteins (table 1).
Table 1.
Summary of Metals, Proteins, mRNA Expression Quantified, and SNPs Genotyped.
| Metal | Gene | Protein | SRM Protein Detection | mRNA Expression | SNPs Genotyped |
|---|---|---|---|---|---|
| Cu | ATOX1 | ATOX1 (O00244) | Quantified | ||
| ATP7B | ATP7B (P35670) | Quantified | |||
| COMMD1 | COMD1 (Q8N668) | Quantified | |||
| CP | CERU (P00450) | Quantified | Quantified | ||
| SLC31A1 | COPT1 (O15431) | Quantified | rs3934969a, rs10759636b | ||
| SLC31A2 | COPT2 (O15432) | Quantified | |||
| SOD1 | SODC (P00441) | Quantified | Quantified | rs8132736c, rs9974808a | |
| Fe | ACO1 | ACOC (P21399) | Quantified | Quantified | |
| BDH2 | BDH2 (Q9BUT1) | Quantified | Quantified | ||
| CYBRD1 | CYBR1 (Q53TN4) | Targeted | Quantified | rs2356709c, rs960748a, rs950163a, rs6752812c, rs884409d | |
| EPAS1 | EPAS1 (Q99814) | Quantified | rs4953354e | ||
| FECH | HEMH (P22830) | Quantified | Quantified | ||
| FTH1 | FRIH (P02794) | Quantified | Quantified | ||
| FTL | FRIL (P02792) | Quantified | Quantified | ||
| HAMP | HEPC (P81172) | Quantified | Quantified | ||
| HEPH | HEPH (Q9BQS7) | Quantified | |||
| HFE | HFE (Q30201) | Targeted | Quantified | rs12346a, rs4324798a, rs1800562f | |
| HFE2 | RGMC (Q6ZVN8) | Quantified | |||
| HIF1A | HIF1A (Q16665) | Quantified | |||
| LTF | TRFL (P02788) | Quantified | Quantified | ||
| RHOA | RHOA (P61586) | Quantified | Quantified | ||
| SLC11A22 | NRAM2 (P49281)2 | Targeted | Quantified | rs6580783g2, rs6580784c2 | |
| SLC17A1 | NPT1 (Q14916) | rs17342717h | |||
| SLC40A1 | S40A1 (Q9NP59) | Quantified | rs1437883c, rs2067416a | ||
| STEAP33 | STEA3 (Q658P3)3 | Quantified | Quantified | ||
| TF | TRFE (P02787) | Quantified | Quantified | rs1525889a, rs1799852f, rs3811647f, rs2280673f | |
| TFR2 | TFR2 (Q9UP52) | Targeted | Quantified | rs2075672i | |
| TFRC | TFR1 (P02786) | Targeted | Quantified | ||
| TMPRSS6 | TMPS6 (Q8IU80) | Targeted | Quantified | rs855791i, rs4820268f | |
| ISCU | ISCU (Q9H1K1) | Quantified | |||
| LCN2 | NGAL (P80188) | Quantified | |||
| rs2698530j, rs7787204j, rs987710j | |||||
| Mg | SLC41A1 | S41A1 (Q8IVJ1) | Quantified | rs12743401a, rs823154k, rs960603k, rs6661355c | |
| Mn | SOD2 | SODM (P04179) | Quantified | Quantified | rs2842992a, rs2842980k |
| Se | AKAP6 | AKAP6 (Q13023) | rs8013938l | ||
| DIO1 | IOD1 (P49895) | Quantified | Quantified | rs2235544m | |
| EEFSEC | SELB (P57772) | Quantified | Quantified | ||
| EIF4A3 | IF4A3 (P38919) | Quantified | Quantified | ||
| FABP1 | FABPL (P07148) | Quantified | Quantified | rs1545223a | |
| GPX1 | GPX1 (P07203) | Quantified | Quantified | rs3172494n, rs3774800o, rs9873994o, rs1865741o, rs1050450p, rs3448q, rs6797765o, rs6779524o, rs1568661c, rs4625n | |
| GPX2 | GPX2 (P18283) | Quantified | Quantified | rs10133290c | |
| GPX3 | GPX3 (P22352) | Quantified | Quantified | rs6773575ç*, rs4958427q, rs1478392c, rs8177409s, rs3828599t, rs3792796t, rs4958434t | |
| GPX4 | GPX4 (P36969) | Quantified | Quantified | rs1046040a, rs713041u | |
| KCNMA14 | KCMA1 (Q12791)4 | rs2619641l4 | |||
| PRKG1 | KGP1 (Q13976) | rs10508958l | |||
| SARS | SYSC (P49591) | Quantified | Quantified | ||
| SCLY | SCLY (Q96I15) | Quantified | rs2264132a | ||
| SECISBP2 | SEBP2 (Q96T21) | Quantified | rs3002340c, rs10908903v | ||
| SELENBP1 | SBP1 (Q13228) | Quantified | Quantified | rs11806477c, rs17564336a | |
| SELH | SELH (Q8IZQ5) | Quantified | |||
| SELK | SELK (Q9Y6D0) | Targeted | Quantified | rs13317328c | |
| SELM | SELM (Q8WWX9) | Quantified | rs8135353c | ||
| SELO | SELO (Q9BVL4) | Quantified | Quantified | rs4838862c | |
| VIMP | SELS (Q9BQE4) | Quantified | Quantified | rs34713741x | |
| SELT | SELT (P62341) | Targeted | Quantified | rs7639294k | |
| SEP15 | SEP15 (O60613) | Targeted | Quantified | rs17129522c, rs5845y | |
| SEPHS1 | SPS1 (P49903) | Quantified | rs3802587c, rs10737068a, rs7475361a | ||
| SEPHS2 | SPS2 (Q99611) | Quantified | Quantified | ||
| SEPP1 | SEPP1 (P49908) | Targeted | Quantified | rs4292454q, rs28551699a, rs6873545a, rs10941583o, rs12521192o, rs7579p, rs13168440o, rs1862629o, rs13154178o, rs11746790o, rs13177679o, rs6451656o, rs305229p* | |
| SEPSECS | SPCS (Q9HD40) | Quantified | Quantified | ||
| SEPW1 | SELW (P63302) | Quantified | Quantified | rs17304884c | |
| SGCD | SGCD (Q92629) | rs32076l | |||
| TXN | THIO (P10599) | rs2986669a | |||
| TXNRD1 | TRXR1 (Q16881) | Quantified | Quantified | ||
| TXNRD2 | TRXR2 (Q9NNW7) | Quantified | rs6518585a, rs1139793q, rs8141610a | ||
| RPL30 | RL30 (P62888) | Quantified | |||
| Zn | GPR39 | GPR39 (O43194) | Quantified | ||
| IL6 | IL6 (P05231) | Quantified | rs1800795q, rs2069832t, rs2069840t | ||
| IL6R | IL6RA (P08887) | rs2228145q, ñ | |||
| MT1A5 | MT1A (P04731)5 | Quantified | TagSNPs MT regionr5 | ||
| MT1E5 | MT1E (P04732)5 | Quantified | TagSNPs MT regionr5 | ||
| MT1F5 | MT1F (P04733)5 | Targeted | TagSNPs MT regionr5 | ||
| MT1G5 | MT1G (P13640)5 | Quantified | TagSNPs MT regionr5 | ||
| MT1H5 | MT1H (P80294)5 | Quantified | rs904777k5, rs1001362k5, rs4784706k5, rs11076164a5, rs4784718k5, rs11932508n5*, rs12917875b5, TagSNPs MT regionr5 | ||
| MT2A5 | MT2 (P02795)5 | Quantified | rs12918376v5, rs10636c5, TagSNPs MT regionr5 | ||
| MT45 | MT4 (P47944)5 | Targeted | TagSNPs MT regionr5 | ||
| MTF15 | MTF1 (Q14872)5 | Targeted | Quantified | rs4634868c | |
| MTF25 | MTF2 (Q9Y483)5 | Quantified | rs4484937a5 | ||
| SLC11A16 | NRAM1 (P49279)6 | rs2695343k6 | |||
| SLC30A17 | ZNT1 (Q9Y6M5)7 | Quantified | Quantified | rs2166104c7, rs7530862a7 | |
| SLC30A108 | ZNT10 (Q6XR72)8 | Quantified | rs2275707b8, rs2647462a8, rs34672561c8 | ||
| SLC30A2 | ZNT2 (Q9BRI3) | Targeted | Quantified | rs6681452c | |
| SLC30A3 | ZNT3 (Q99726) | Quantified | |||
| SLC30A4 | ZNT4 (O14863) | Targeted | Quantified | rs8029246c | |
| SLC30A59 | ZNT5 (Q8TAD4)9 | Quantified | rs35243v9, rs10471766c9 | ||
| SLC30A6 | ZNT6 (Q6NXT4) | Quantified | rs177083c | ||
| SLC30A7 | ZNT7 (Q8NEW0) | Targeted | Quantified | rs12568338c | |
| SLC30A8 | ZNT8 (Q8IWU4) | Quantified | |||
| SLC30A9 | ZNT9 (Q6PML9) | Targeted | Quantified | rs7684181v, rs3804186c, rs2660341w, rs2660346z, rs9291209o, rs17838947o, rs10433709n, rs6447123o, rs7659700o, rs7677472o, rs17446061a, rs2880666n, rs6447133o | |
| SLC39A1 | S39A1 (Q9NY26) | Quantified | Quantified | rs4971100b | |
| SLC39A10 | S39AA (Q9ULF5) | Quantified | rs6733196a, rs4850376k, rs1489802a | ||
| SLC39A11 | S39AB (Q8N1S5) | Quantified | rs1126967a, rs17248364c | ||
| SLC39A12 | S39AC (Q504Y0) | Quantified | rs7085542a | ||
| SLC39A13 | S39AD (Q96H72) | Quantified | rs749067c, rs1113480v, rs3942852v | ||
| SLC39A14 | S39AE (Q15043) | Quantified | Quantified | rs1051708a | |
| SLC39A2 | S39A2 (Q9NP94) | Quantified | |||
| SLC39A3 | S39A3 (Q9BRY0) | Quantified | rs4806874c | ||
| SLC39A4 | S39A4 (Q6P5W5) | Quantified | rs11992877c, rs10876864a* | ||
| SLC39A5 | S39A5 (Q6ZMH5) | Quantified | rs11171856c | ||
| SLC39A6 | S39A6 (Q13433) | Quantified | rs3737473c | ||
| SLC39A7 | S39A7 (Q92504) | Quantified | |||
| SLC39A810 | S39A8 (Q9C0K1)10 | Quantified | rs151413b10, rs9331k10, rs13107325a10, rs10031415c10 | ||
| SLC39A9 | S39A9 (Q9NUM3) | Quantified | rs8017816v, rs12050204c | ||
| STAT3 | STAT3 (P40763) | Quantified | Quantified | ||
| B1 | SLC4A11 | S4A11 (Q8NBS3) | Targeted | Quantified | rs6051629c |
| As | AS3MT | AS3MT (Q9HBK9) | Quantified | ||
| Mo | MFSD5 | MFSD5 (Q6N075) | Quantified | ||
| Na1/I1 | SLC5A5 | SC5A5 (Q92911) | Targeted | ||
| Ca1 | TRPM2 | CLUS (P10909) | Quantified |
Note.—Protein codes refer to the Uniprot database entry names without the suffix _HUMAN indicating the UniProtKB identifier in brackets. 1Not quantified; 2Also tested with Mn, Co, Cd, Ni, V, and Pb; 3Also tested with Cu; 4Also tested with Mg; 5Also tested with Cd, Cu, Se, Hg, Rb, and As; 6Also tested with Fe, Mn, and Cd; 7Also tested with Cd; 8Also tested with Mn; 9Also tested with Co; 10Also tested with Cd; aGreenawalt et al. (2011); bSchadt et al. (2008); cInnocenti et al. (2011); çQiu et al. (2011); dConstantine et al. (2009); eHanaoka et al. (2012); fBenyamin et al. (2009), gVeyrieras et al. (2008); hPichler et al. (2011); ivan der Harst et al. (2012); jMcLaren et al. (2011); kZeller et al. (2010); lSavas et al. (2010); mPanicker et al. (2008); nDixon et al. (2007); oTagSNP; pHesketh (2008); qFST; rTagSNPs MT region: rs12444489, rs8052106, rs1811322, rs11642055, rs904773, rs7189840, rs1875232, rs7197489, rs7194895, rs1587479, rs1599933, rs4784714, and rs8064100; sVoetsch et al. (2007); tVerlaan et al. (2009); uGautrey et al. (2011); vMontgomery et al. (2010); wGe et al. (2009); xMéplan et al. (2010); ySutherland et al. (2010); zBell et al. (2011); ñReich et al. (2007); *TransQTLs.
SNP Genotyping and Metal, Protein, and Transcript Quantification
A total of 169 SNPs that were related to the transcripts and/or proteins of genes involved in metal homeostasis, as well as 13 additional SNPs, were genotyped in the same samples (table 1 and supplementary tables S1 and S2, Supplementary Material online). Most of the selected SNPs were chosen for their potential regulatory role based on prior knowledge from different association (Panicker et al. 2008; Benyamin et al. 2009; Constantine et al. 2009; Méplan et al. 2010; Savas et al. 2010; Sutherland et al. 2010; Pichler et al. 2011; van der Harst et al. 2012; McLaren et al. 2012) and eQTL studies (Dixon et al. 2007; Voetsch et al. 2007; Schadt et al. 2008; Veyrieras et al. 2008, 2012; Ge et al. 2009; Verlaan et al. 2009; Montgomery et al. 2010; Zeller et al. 2010; Greenawalt et al. 2011; Innocenti et al. 2011; Qiu et al. 2011; Rye et al. 2013). However, we also included 7 SNPs with other functional roles (Reich et al. 2007; Voetsch et al. 2007; Hesketh 2008; Ge et al. 2009; Bell et al. 2011; Gautrey et al. 2011; Hanaoka et al. 2012), 6 SNPs for their known high population differentiation (FST), 13 tagSNPs around the metallothionein (MT) cluster of chromosome 16 and 19 additional SNPs that could be used as tagSNPs for those genomic regions around the studied genes that presented other previously known signatures of recent positive selection (GPX1, SEPP1, SLC30A9). Four of the SNPs were transQTLs; the remaining 165 SNPs were located at a median distance of 13 kb of their related closest gene. A principal component analysis (PCA) on SNP genotypes (supplementary fig. S1, Supplementary Material online) showed that our samples were indistinguishable from the European populations of the 1000 Genomes Project (Altshuler et al. 2012) while clearly different from Asians and Africans. Indeed, when the allele frequencies were compared with those in the GBR (British in England and Scotland) population of the 1000 Genomes Project by means of the Fisher’s exact test (supplementary table S2, Supplementary Material online) and P values were pooled with Fisher’s combined probability test, the result was not statistically significant (P = 0.230), implying that the set of allele frequencies is not statistically different between GBR and our sample.
In the same sample set, the concentration of 18 trace elements was determined using inductively coupled plasma mass spectrometry (ICP-MS). The quantified trace elements included As, Cd, Co, Cr, Cu, Fe, Hg, Li, Mg, Mn, Mo, Ni, Pb, Rb, Se, Sn, V, and Zn (table 2 and supplementary table S3, Supplementary Material online). Lithium and nickel are the trace elements that displayed most interindividual variability, whereas magnesium and selenium appeared as the most stable micronutrients. Micronutrient concentrations were significantly different between sexes for Fe (P = 0.042), Cd (P < 0.001), Mo (P = 0.035), and Sn (P = 0.015). In parallel, we measured the expression levels of 90 genes related to 6 essential elements (copper, iron, magnesium, manganese, selenium, and zinc) and 16 additional genes using quantitative real-time polymerase chain reaction (qRT-PCR; fig. 1A and supplementary tables S1 and S4, Supplementary Material online). After Bonferroni correction, none of the 90 mRNAs quantified showed significant differences between sexes.
Table 2.
Element Raw Concentrations Measured in Human Liver Samples (mg/kg dry weight).
| Element | Mean | 95% CI | CV |
|---|---|---|---|
| As | 0.017 | 0.014–0.020 | 0.917 |
| Cd | 1.66 | 1.299–2.021 | 0.921 |
| Co | 0.090 | 0.081–0.098 | 0.534 |
| Cr | 0.298 | 0.169–0.427 | 1.747 |
| Cu | 11.69 | 10.57–12.81 | 0.528 |
| Fe | 336.5 | 272.2–400.9 | 0.846 |
| Hg | 0.109 | 0.070–0.149 | 1.336 |
| Li | 0.006 | −0.022 to 0.035 | 7.456 |
| Mg | 497.2 | 461.9–532.5 | 0.411 |
| Mn | 3.213 | 2.959–3.467 | 0.450 |
| Mo | 1.474 | 1.304–1.643 | 0.596 |
| Ni | 0.085 | −0.079 to 0.248 | 5.174 |
| Pb | 0.206 | 0.157–0.256 | 1.095 |
| Rb | 6.783 | 6.160–7.407 | 0.507 |
| Se | 0.768 | 0.711–0.825 | 0.423 |
| Sn | 0.248 | 0.184–0.312 | 1.070 |
| V | 0.038 | 0.026–0.049 | 1.482 |
| Zn | 141.6 | 127.8–155.4 | 0.538 |
Note.—Mean = geometric mean (ppm); CI = confidence interval; CV = coefficient of variation.
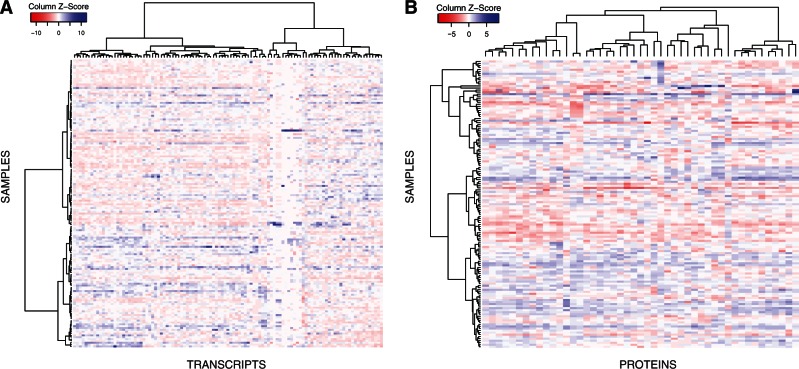
Fig. 1.
Quantitative data matrix corresponding to the transcriptomics (A) and proteomics (B) assays represented as heatmaps. Rows and columns in each heatmap have been sorted according to a hierarchical clustering (distance: correlation; cluster method: ward), which is shown on the left and on the top in the form of dendrograms.
These measurements were subsequently complemented with protein quantification by SRM. SRM is a targeted mass spectrometry method, usually performed on triple quadrupole instruments, in which specific peptides and their fragments are quantified. Each peptide-fragment pair is called a transition, and multiple transitions are measured to conclusively identify and quantify the peptides of interest in complex samples. A subset of 40 targeted proteins involved in the homeostasis of the same 6 essential elements (copper, iron, magnesium, manganese, selenium, and zinc) together with 7 additional proteins were consistently quantified in all samples using one or two peptides per protein (supplementary tables S1 and S5, Supplementary Material online). Each peptide was quantified with two or three light–heavy transition pairs free of interferences (supplementary tables S6 and S7, Supplementary Material online). Transition groups corresponding to the targeted peptides were evaluated with MultiQuant (v. 2.1.1; AB Sciex) based on the following: 1) coelution of the transition traces associated with a targeted peptide, both in its light and heavy form; 2) presence coeluting transition traces for a given peptide exceeding a signal‐to‐noise ratio of three; 3) rank correlation between the light SRM relative intensities and the heavy counterparts; 4) rank correlation between the SRM relative intensities and the intensities obtained in the MS2 spectra during the SRM assay development; and 5) consistency among replicates. These measurements generated a complete protein quantification matrix with few missing values, comparable with those obtained in transcriptomics (fig. 1B). After Bonferroni correction, only ferritin light chain (FTL) had significantly different abundances between sexes (P = 8 × 10−4). Overall, in 41 instances we quantified the mRNA and protein levels of the same gene (table 1).
Genetic Influence on Protein Abundances and Transcript Expression Levels
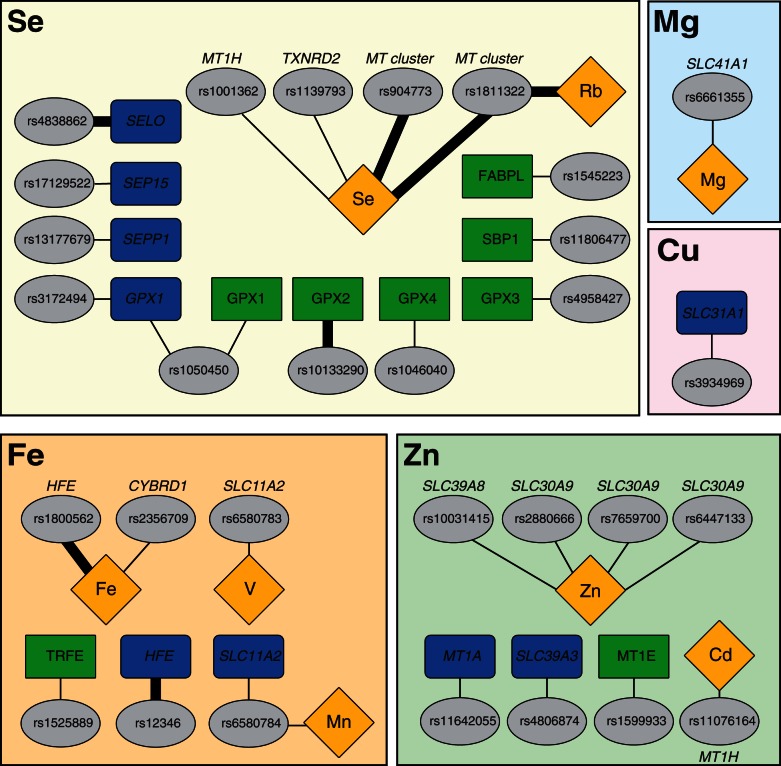
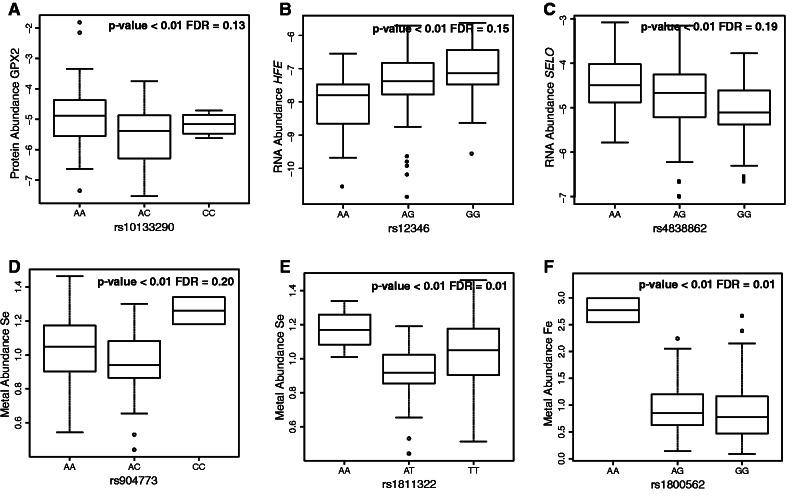
pQTL analysis enabled us to correlate protein abundance with genetic variation in the studied human liver samples (supplementary table S8, Supplementary Material online). We identified eight cis-associations with P < 0.05, which involved proteins GPX1 (rs1050450), GPX2 (rs10133290), GPX3 (rs4958427), GPX4 (rs1046040), FABPL (rs1545223), SBP1 (rs11806477), MT1E (rs1599933), and TRFE (rs1525889) (fig. 2 and supplementary table S9 and fig. S2, Supplementary Material online). However, only the pQTL influencing GPX2 protein levels remained significant after false discovery rate (FDR)-based P-value adjustment (FDR ≤ 0.20; supplementary fig. S2, Supplementary Material online, and fig. 3A). Except for the serotransferrin (TRFE) and the MT1E cases, the remaining identified pQTL involved proteins related to selenium (fig. 2). Five of these cis-associated SNPs had been previously described as eQTLs in liver, either in the study of Greenawalt et al. (2011) or in that of Innocenti et al. (2011) (table 1). Besides having been previously identified as an eQTL, rs1525889 is in almost complete linkage disequilibrium (r2 = 0.98) with rs3811647 (supplementary fig. S3, Supplementary Material online), which had been associated with serum transferrin levels in a GWAS (Benyamin et al. 2009). We found that both the T allele (P = 0.034) and the TT genotype (P = 0.005) at rs1525889 were associated with lower TRFE levels (supplementary table S9, Supplementary Material online), essentially replicating and extending to liver the finding by Benyamin et al. (2009). Along the same lines, rs1050450 in GPX1 had also been previously associated with altered glutathione peroxidase 1 (GPX1) function (Hamanishi et al. 2004). In particular, in vitro functional analyses suggested that the derived allele of rs1050450 (which involves a Pro (C) to Leu (T) substitution) implied lower GPX1 activity (Hamanishi et al. 2004). In accordance with these functional analyses, we found that genotypes CC and CT displayed higher GPX1 protein abundances in liver than the TT homozygotes at this nonsynonymous SNP (P = 0.036; supplementary fig. S2, Supplementary Material online).
Fig. 2.
Summary of significant QTL identified for genes related to trace element homeostasis. pQTLs (P ≤ 0.05) are indicated in green, eQTLs (P ≤ 0.05) in blue, and nutriQTLs (P ≤ 0.025) in yellow. Those QTLs surpassing an FDR cutoff of 0.20 are highlighted with a thick line.
Fig. 3.
Box plots representing significant quantitative trait loci (FDR ≤ 0.20) for GPX2 protein abundance (A), HFE and SELO transcript expression levels (B, C), and selenium and iron content (D, F). Protein abundances are represented as the log2 ratio of the endogenous abundance and the reference isotopically labeled internal standard; mRNA abundances are represented as ΔΔCT; and trace element abundances refer to the corrected concentrations in mg/kg (ppm).
At the transcript level, we found two significant eQTLs surpassing an FDR cutoff of 0.20 (fig. 3B and C): rs12346, affecting the expression levels of HFE (FDR = 0.15), and rs4838862, influencing the transcription of SELO (FDR = 0.19). The two eQTLs had been previously identified by Greenawalt et al. (2011) and Innocenti et al. (2011), respectively. In both instances we detected significant additive effects (with P < 0.001 and P = 0.001, respectively). Although not surviving multiple testing correction (i.e., P ≤ 0.05 but not surpassing the FDR cutoff of 0.20), we found suggestive evidence for eight additional eQTLs in genes related to selenium (SEP15, SEPP1, and GPX1), iron (SLC11A2), zinc (SLC39A3 and MT1A), and copper (SLC31A1) (supplementary tables S10 and S11 and fig. S4, Supplementary Material online). Except for those eQTLs detected in MT1A and SEPP1, which were genotyped for representing tagSNPs of their corresponding genomic regions, the remaining additional eQTLs detected had already been identified as eQTLs, although not always in liver tissue (Dixon et al. 2007; Greenawalt et al. 2011; Innocenti et al. 2011). Moreover, the eQTLs described here in SLC39A3 and MT1A were confirmed in the liver GTEX data (Ardlie et al. 2015; Mele et al. 2015), which comprises RNAseq data for 32 liver donor samples. The two eQTLs affecting GPX1 transcript levels (rs3172494 and rs1050450) were not in linkage disequilibrium (supplementary fig. S3, Supplementary Material online) and thus may represent two independent association signals (supplementary fig. S4, Supplementary Material online). As with the differences in GPX1 protein levels, genotypes CC and CT at rs1050450 displayed higher GPX1 transcript abundances than the TT homozygotes (P = 0.035; supplementary fig. S4, Supplementary Material online).
Genetic Influence on Trace Element Homeostasis
Next, we explored associations between SNP genotypes and trace element abundances, and in doing so replicated the known association between rs1800562 and iron concentration in liver (FDR = 0.01) and identified up to three new nutriQTLs influencing the liver concentrations of rubidium (rs1811322, FDR = 0.01) and selenium (rs1811322 with FDR = 0.01 and rs904773 at an FDR = 0.20) along the MT cluster in chromosome 16, which among other genes comprises MT1A, MT1E, MT1F, MTH1, MT1G, MT1H, MT2A, and MT4 (fig. 3D–F and supplementary fig. S5, Supplementary Material online). However, given the moderate linkage disequilibrium between rs1811322 and rs904773 (r2 = 0.43), they may not represent independent signals. The SNP rs1800562 corresponds to the C282Y nonsynonymous amino acid change in the HFE gene (table 3), which when homozygous causes hereditary hemochromatosis, and has been repeatedly recognized to be associated with iron content (Bulaj et al. 1996; Benyamin et al. 2014). Considering all significant nutriQTLs (P ≤ 0.025 even if not all exhibited an FDR < 0.20), we detected up to five nutriQTLs around the MT genes (supplementary tables S12, S13, and fig. S5, Supplementary Material online). They were related to three different metals (selenium, cadmium, and rubidium), which is in agreement with the distinct metal-binding capacities of MTs as well as with the MTs role in the homeostasis and transport of physiological metals and heavy-metal detoxification. Similarly, we detected two different nutriQTLs around the SLC11A2 gene influencing the liver content of vanadium and manganese (P ≤ 0.025 but FDR > 0.20 in both instances); the protein product of this gene is involved in iron absorption but also transports other divalent metals. Finally, the three nutriQTLs related to zinc detected within SLC30A9, which presented P-values ≤ 0.025 but did not survive the FDR correction, were in moderate linkage disequilibrium (supplementary fig. S3, Supplementary Material online) and thus may be also reporting the same association signal. Given that, as stated above, iron and cadmium have significantly different concentrations between the sexes, we tested the effects of their nutriQTLs specifically by sex. SNP rs1800562 had practically the same effect on the iron concentration found in males (P = 0.001) and females (P = 0.013), while rs2356709 (supplementary tables S12 and S13, Supplementary Material online) affected only males (P = 0.001; in females, P = 0.664). A sex-specific effect was also observed for rs1107614 (supplementary tables S12 and S13, Supplementary Material online), which was detected as nutriQTL for cadmium in females (P = 0.018) but not in males (P = 0.270).
Table 3.
Genomic Features and Derived Allele Frequencies of Significant QTLs (FDR ≤ 0.20).
| SNP | Alleles | Gene | Genomic Location | CEUb | CHBb | JPTb | YRIb | GBRb | This study |
|---|---|---|---|---|---|---|---|---|---|
| rs10133290 | C|A | GPX2 | 3′ downstream | 0.771 | 0.866 | 0.865 | 0.540 | 0.831 | 0.799 |
| rs12346 | C|T | HFE | 3′ downstream | 0.371 | 0.124 | 0.096 | 0.011 | 0.253 | 0.333 |
| rs4838862 | G|A | SELO | Intronic | 0.506 | 0.510 | 0.466 | 0.676 | 0.506 | 0.534 |
| rs1800562 | G|A | HFE | Nonsynonymous (Cys > Tyr)a | 0.053 | 0.000 | 0.000 | 0.000 | 0.056 | 0.092 |
| rs1811322 | T|A | MT region | 5′ upstream (MT2A) | 0.194 | 0.325 | 0.343 | 0.114 | 0.180 | 0.160 |
| rs904773 | A|C | MT region | Intronic (MT2A) | 0.212 | 0.273 | 0.331 | 0.193 | 0.202 | 0.174 |
Note.—JPT = Japanese in Tokyo, Japan.
aPredicted as “Damaging” by SIFT and as “Probably damaging” by Polyphen.
bData from the 1000 Genomes Project (http://www.1000genomes.org, last accessed September 10, 2015).
Dependence among Micronutrient, Protein, and RNA Abundances
Pairwise correlations between RNA, protein, and trace element concentrations yielded 24 significant correlations (FDR ≤ 0.05; supplementary table S14, Supplementary Material online). Among those significant cases, five attained high correlation values (R > 0.50): Namely, iron content in liver correlated with both ferritin heavy chain (FTH1) and light chain (FTL), whereas zinc correlated with both the abundance of the MT1E, and also with the transcription levels of two additional MT genes, MT1G and MTH1 (supplementary fig. S6, Supplementary Material online). Because both the abundances of FTL and iron were different between males and females, we tested their correlation separately by sex and found that it is present and similar in both sexes (R = 0.715 in females and R = 0.768 in males). As expected, the correlation between FTH1 and iron followed a similar pattern (R = 0.694 in females and R = 0.642 in males) because FTL and FTH1 are different constituent chains of the same protein, ferritin.
Signatures of Genetic Adaptation in Detected QTLs
Among the complete set of eight pQTLs (P ≤ 0.05), 10 eQTLs (P ≤ 0.05), and 15 nutriQTLs (P ≤ 0.025) detected (supplementary tables S9, S11, and S13, Supplementary Material online), we recognized some genes for which evidences of positive selection have been previously shown (Toomajian and Kreitman 2002; Toomajian et al. 2003; Foster et al. 2006; Zhang et al. 2015). This led us to further investigate the patterns of variation around the 20 gene regions in which we detected any QTL at significant P values (even if they did not reach FDR correction) through use of publicly available genetic data from the 1000 Genomes project (Altshuler et al. 2012). Specifically, we explored for signatures of natural selection with statistics for site frequency spectrum, population differentiation, and linkage disequilibrium structure as well as with the hierarchical boosting scores for different types of selective sweeps as implemented in the 1000 Genomes Selection Browser (Pybus et al. 2014) for three continental populations: Yoruba in Ibadan, Nigeria (YRI), Han Chinese in Beijing, China (CHB), and Utah residents with ancestry from northern and western Europe (CEU) (supplementary note S1, Supplementary Material online). Moreover, we also tested whether the detected QTLs were SNPs unusually differentiated between the different pairs of the same HapMap populations because these populations live in geographical areas with potential micronutrient content differences in soil (supplementary table S15, Supplementary Material online). Notably, we found significant evidence for positive selection in several genes related to zinc and selenium homeostasis: SLC30A9, SLC39A8, and GPX1 showed clear signatures of recent positive selection in Asia; while GPX3 and SELENBP1 displayed signals for selective sweeps in Europe (supplementary note S1, Supplementary Material online). In all cases, the signatures of selection identified are plausibly linked to the QTLs described. Moreover, except for GPX1, all QTLs detected in this subset of genes exhibited significant population differentiation (FST values above the 97.5% percentile of the corresponding genome-wide FST distribution).
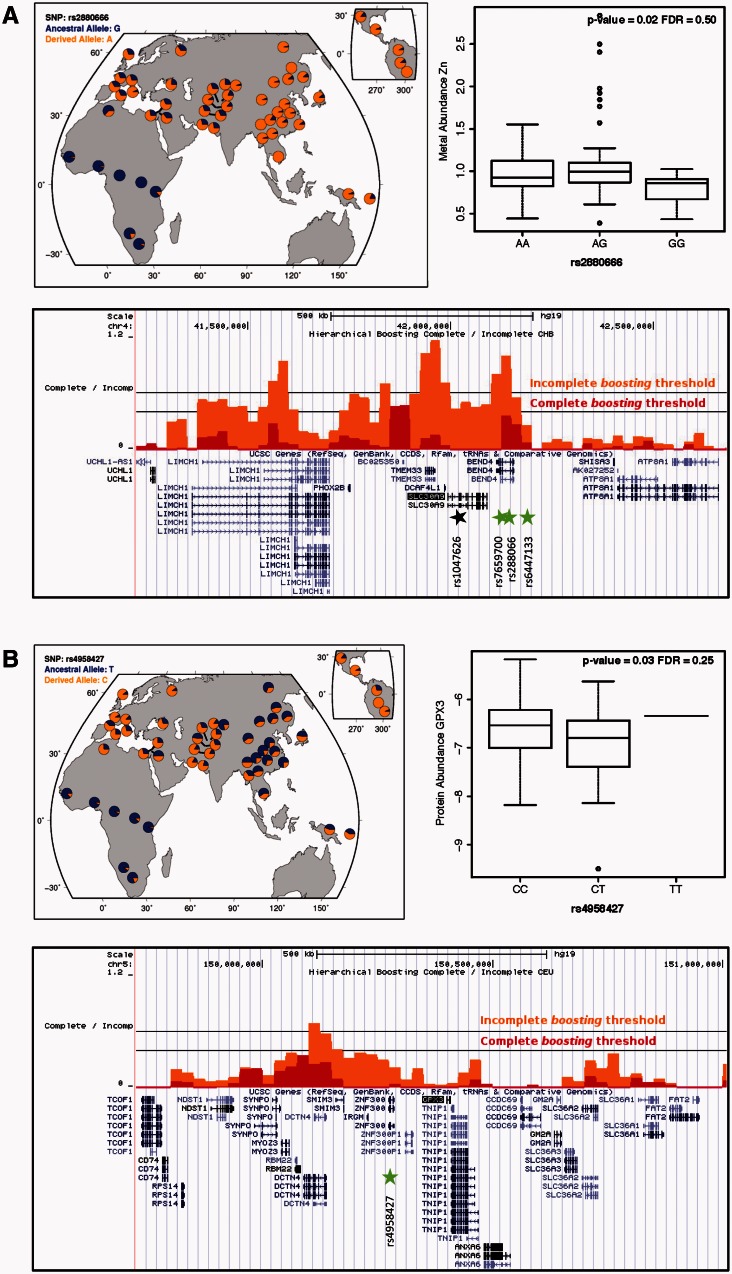
Among all genomic regions analyzed, the strongest evidence for a selective sweep, which extended along 500 kb, was found around SLC30A9 in the CHB population (fig. 4A). In fact, the same region had already been reported to exhibit one of the most significant signatures of adaptation found in Asians in several genome-wide scans of selection (Carlson et al. 2005; Sabeti et al. 2007; Williamson et al. 2007; Pickrell et al. 2009). Recently, Zhang et al. (2015) not only reported positive selection at SLC30A9 in East Asians but also in Africans and attributed these adaptive signals to a nonsynonymous SNP (rs1047626), whose allelic variants are almost fixed in opposite directions in Africa and Asia, respectively. In particular, the ancestral A allele, which encodes a methionine at amino acid position 50 of SLC30A9, has a frequency of 92% in the Yoruba population, while the derived G allele, which encodes valine, is found at a frequency of 96.4% in the Han Chinese. However, no direct functional evidence was demonstrated for this highly differentiated nonsynonymous SNP and in silico prediction in PolyPhen predicted the substitution as benign (Zhang et al. 2015). Interestingly, when comparing the allele frequencies found in CEU, CHB, and YRI at the three nutriQTLs detected in the 3′ flanking region of SLC30A9 with those of rs1047626, we found that the pattern of variation at rs2880666 closely resembles that of the nonsynonymous substitution (fig. 4A and supplementary note S1, Supplementary Material online). Furthermore, the three nutriQTLs displayed significant high differentiation values between different HapMap population pairs (FST values above the 95% percentile of the corresponding genome-wide distributions; supplementary table S15, Supplementary Material online) and presented moderate linkage disequilibrium with rs1047626 in the CEU population (with r2 values between 0.57 and 0.59). Although we cannot determine which is the true adaptive variant (or if both contribute), our results point to the possible existence of differences in liver zinc content between the two main haplotypes described worldwide (Zhang et al. 2015) with the major SLC30A9 haplotype in Asia possibly being associated with higher liver zinc concentrations than the African one.
Fig. 4.
Signatures of natural selection around the nutriQTL and pQTL detected for SLC30A9 (A) and GPX3 (B), respectively. In each section, the upper left panel shows the worldwide distribution of allele frequencies as provided by the HGPD Selection Browser (http://hgdp.uchicago.edu, last accessed May 12, 2015) (Pickrell et al. 2009); the upper right panel is a box plot of metal (A) or protein abundance (B) by SNP genotype; and the bottom panel contains the selective sweep tracks in Han Chinese (CHB) or Europeans (CEU) as provided by the 1000 Genomes Selection Browser 1.0 (http://hsb.upf.edu, last accessed May 19, 2015) (Pybus et al. 2014). Each track has its own significance threshold (at a 1% false positive rate) indicating when a signal can be considered significant for selective sweeps predicted to be complete (red) versus incomplete (orange).
Signatures of positive natural selection have also been previously described at GPX1 (Foster et al. 2006; White et al. 2015) and SLC39A8 (Zhang et al. 2015). Foster et al. (2006) reported the first evidence for a selective sweep at the GPX1 locus in Asian populations linked to the high frequencies of the Pro allele at the rs1050450 in Asia. More recently, White et al. (2015) described multiple allele frequency shifts in several selenoprotein genes and related regulatory genes in East Asia in response to the low dietary selenium levels found across much of this geographical region. Interestingly, the nonsynonymous substitution at rs1050450 and an additional functional variant at the promoter region of GPX1 (rs3811699), in strong linkage disequilibrium with rs1050450 and known to affect GPX1 transcription (Hamanishi et al. 2004), were identified in the same study among those SNPs contributing most to the signatures of positive selection detected in East Asia (White et al. 2015). Besides confirming the known geographical distribution and functional differences at rs1050450, we find that the ancestral G allele at the new eQTL that we describe for GPX1 (rs3172494) is almost fixed in Asia and is associated with higher expression (P = 0.021). This pattern is along the same direction as that of the two previously known functional variants at GPX1, for which alleles in East Asia confer higher enzymatic (rs1050450) and transcriptional activity (rs3811699), respectively (Hamanishi et al. 2004). As for SLC39A8, Zhang et al. (2015) reported four highly differentiated eQTL SNPs across populations and significant signals of positive selection in southern Han Chinese when using the iHS test. Here, we have detected a significant peak for a selective sweep and significant P values in the XP-EHH test, specifically in the CHB population and within an∼161 kb region between two hotspots of recombination, which comprises part of the SLC39A8 gene as well as the detected putative nutriQTL in its 5′ region (rs10031415). Moreover, in this case the allele fixed in East Asia is associated with lower liver zinc content (P = 0.018; supplementary note S1, Supplementary Material online).
The CEU population displayed significant signatures for incomplete selective sweeps at the 5′ regions of GPX3 and SELENBP1, which coincided with the genomic location of the pQTLs detected for GPX3 and SBP1: rs4958427 and rs11806477, respectively (fig. 4B and supplementary note S1, Supplementary Material online). Moreover, in both cases the derived allele was found at high frequencies in Europe and showed a tendency toward higher protein abundances than the ancestral allele. In particular, although we only detected significant differences among the three genotypes at rs11806477 (P = 0.048), at rs4958427 we detected clear significant differences between genotypes (P = 0.010, when comparing the CC and the CT genotypes) as well as between alleles (P = 0.030). Even though the derived allele at rs11806477 was nearly fixed in East Asia, CHB did not display clear departures from neutrality in our analysis (supplementary note S1, Supplementary Material online). However, at this SNP we detected high population differentiation not only between the HapMap populations CEU and CHB, but also between YRI and CHB (FST values above the 97.5th and the 95th percentile of the corresponding FST genome-wide distributions, respectively). On the contrary, rs4958427 showed extreme population differentiation between CEU and the two non-European populations (FST values above the 99th percentile of the two corresponding genome-wide distributions). Thus, only for GPX3 can we convincingly link the geographical distribution of the genetic variation at the pQTL with the signatures of positive selection identified.
Discussion
In this study, we used a candidate-based approach to explore the impact of genetic variation on trace element homeostasis at three different levels of phenotypic variation: Trace element content, expression level, and protein abundance. All three molecular phenotypes were interrogated in the same set of tissue samples, that is, about 150 liver samples from individuals of European ancestry. Among many other vital functions, the liver stores several essential trace elements such as iron, cooper, and magnesium, and it regulates an important number of metal transport proteins (Heuberger 2007). Hence, even though only a fraction of eQTLs are reproducible across different tissues (Schadt et al. 2008; Greenawalt et al. 2011), we have interrogated a physiologically relevant tissue to identify the effects of genetic variation on trace element homeostasis. Genotyped SNPs mostly included previously described eQTLs, although not always described in liver, and other known functional SNPs on the same targeted genes (table 1). As a result, we confirmed known results but also identified new QTLs for several micronutrients as well as for the expression levels and protein abundances of different genes and proteins related to metal homeostasis. In particular, of the 522 potential cis-associations tested and considering an FDR correction of 0.20, we found the following results: 1) one pQTL affecting the selenium-dependent GPX2 (rs10133290, FDR = 0.13); 2) two previously described eQTLs affecting the expression of HFE (rs12346, FDR = 0.15; Innocenti et al. 2011) and SELO (rs4838862, FDR = 0.19; Greenawalt et al. 2011); and 3) four nutriQTLs including (the already known C282Y mutation at HFE regulating iron status (rs1800562, FDR = 0.01; Bulaj et al. 1996; Toomajian and Kreitman 2002), and three different SNPs among the clustered MT genes on chromosome 16, for which genotypes were associated to differential liver concentrations of rubidium (rs1811322, FDR = 0.01) and selenium (rs1811322 with an FDR = 0.01 and rs904773 at an FDR = 0.20).
Out of the 30 SNPs involved in the complete set of significant QTLs (including those that do not reach FDR correction), we found 12 instances with exceptionally high levels of population differentiation, which were mainly related to zinc, iron, and selenium (supplementary table S15, Supplementary Material online). Thus, we provide evidence for potential differences among different human populations in the homeostasis of those micronutrients. These putative functional differences between populations are probably relevant because micronutrient status is known to influence the immune response (Rink and Haase 2007) and diseases such as diabetes (Wiernsperger and Rapin 2010) or cancer (Margalioth et al. 1983; Díez et al. 1989; Costello and Franklin 2006), which indeed present differential prevalence among populations. Moreover, in addition to these patterns of extreme differentiation, we detected signatures of positive selection around the QTLs of at least five genes related to the homeostasis of zinc (SLC30A9, SLC39A8) and selenium (GPX1, GPX3, and SELENBP1). However, even if the QTLs identified appear linked to signals of positive selection, we cannot directly infer whether they were the driving force of such adaptive signatures or whether they hitchiked in a selective sweep triggered by other functional neighboring genetic variants. Additional hints, such as the knowledge of important geographical micronutrient differences in soil and the lack of additional functional variants or alternative putative selective phenotypes associated to the surrounding genomic regions, should be carefully considered in each case. Notably, different in-depth studies that focused on zinc transporter genes (Zhang et al. 2015) as well as on selenoproteins and genes regulating selenium and the amino acid selenocysteine (Foster et al. 2006; White et al. 2015) reported signatures of positive selection for several of these genes. These studies also proposed local micronutrient differences, especially deficiencies, as the potential selective pressures behind such cases, even if in most cases no putatively functional variants were identified. Here, we focused first on localizing regulatory functional variation for genes related to trace element homeostasis and then we explored for potential signatures of adaptation. In this way, the functional basis for adaptation for a number of genes related to micronutrient homeostasis might be identified, for which previously only the genomic footprints of selection had been detected. Because several examples of immune response changes related to iron, manganese, zinc, and copper have been reported (Hood and Skaar 2012), other selective forces, besides local and dietary micronutrient availability, could also explain the different signatures of selection identified. Indeed, we recently reported a case of positive selection for a nonsynonymous substitution in the human intestinal zinc uptake transporter ZIP4 (SLC39A4) and suggested that the derived allele may have a selective advantage in sub-Saharan Africa by starving pathogens of zinc (Engelken et al. 2014).
Besides exploring statistics for site frequency spectrum, population differentiation, and linkage disequilibrium structure, we made use of the hierarchical boosting scores implemented in the 1000 Genomes Selection Browser for detecting selective sweeps (Pybus et al. 2014; 2015). This method not only uses several neutrality tests to uncover different selective sweep properties but also allows classifying selective sweeps according to their completeness and age. Thus, even if departures from neutrality are not significant among the individual neutrality statistics explored, significant signals for different selective sweep types might emerge. Indeed, different composite strategies that combine signatures from many neutrality tests have been shown to increase power and resolution when detecting positive selection around putative adaptive variants (Zeng et al. 2007; Grossman et al. 2010, 2013). This strategy, when applied to genome-wide sequencing data provided by the 1000 Genomes project, has allowed us here to detect adaptive signatures in the flanking regulatory regions of genes that displayed no deviations from neutrality when previously interrogated. For example, Foster et al. (2006) already analyzed sequencing data on the coding and untranslated regions of six selenoprotein genes (including GPX1, GPX2, GPX3, GPX4, SEPP1, and TXNRD1) and reported strong evidence for a selective sweep only on GPX1, while we have been able to capture a selective sweep also on the 5′ region of GPX3. At the same time, particular adaptive events around the detected QTLs may have eluded our detection, either because of the nature of the signal they are associated with or because we did not interrogate the appropriate populations. For instance, we did not detect significant selective sweeps at either the HFE gene or at SLC39A3. However, the haplotype structure and frequency of the C282Y substitution at HFE (rs1800562) have been suggested to be consistent with a selective advantage for this nutriQTL, even if significant departures from neutrality were not detected with several neutrality statistics (Toomajian and Kreitman 2002; Bamshad and Wooding 2003; Toomajian et al. 2003). Also, significant signals with the iHS test were previously reported for the zinc transporter SLC39A3 in populations with European and Native American ancestries (Zhang et al. 2015).
Additionally, other forms of adaptation such as polygenic selection (Berg and Coop 2014) may also operate on the regulation of micronutrients. These could be further interrogated in conjunction with the QTLs identified through compilation of additional genetic population data and information on the corresponding micronutrient soil availability each population encounters. Certainly, data on micronutrient soil availability (Sillanpää 1982) would be more meaningful for studies of prehistoric human adaptation to micronutrient deficiencies than studies on modern day stunting or dietary intake (Wessells and Brown 2012). Indeed, by exploring sequencing data on different worldwide human populations, White et al. (2015) recently demonstrated multiple allele frequency shifts on several selenoproteins and genes involved in the regulation of selenium and selenocysteine, especially for East Asian populations living in selenium deficient areas. Interestingly, among those top genes contributing to the signatures of local adaptation in East and Central South Asia, they identified GPX1 as described above together with SEPP1 and SEP15, among others. Albeit not statistically significant, we detected patterns of variation suggestive of positive selection around the QTLs detected for SEP15 and SEPP1, in both Europe and Asia, as well as for GPX2 and SLC39A3, in Europe (supplementary note S1, Supplementary Material online). Moreover, with the exception of GPX2, these QTLs also exhibited extremely high population differentiation among different HapMap population pairs with FST values above the 97.5 and 99th percentiles of the corresponding genome-wide distributions (supplementary table S15, Supplementary Material online). Thus, even though the classical genetic signals for positive selection in these cases were rather diffuse, they might reflect polygenic adaptation, because the micronutrient status in the individual is influenced by many different genes involved in absorption, transport, storage, and excretion.
In this study, we explored for the first time three different molecular phenotypes (trace element concentrations, RNA expression levels, and protein abundance) in human liver samples of European origin and identified a number of QTLs influencing trace element homeostasis. Given the paramount importance of essential trace elements in health and disease, this work may represent a pilot study that motivates future research integrating data on genomics, proteomics, and trace elements (ionomics). The integration of trace element quantification from different tissues would be valuable in GWAS studies, which are already common in plant research (Atwell et al. 2010). Moreover, ionomics could easily be incorporated to the existing layers (genomics, proteomics, transcriptomics, and clinical data) in the current international cancer genomics projects (Hudson et al. 2010; Chang et al. 2013). In multitissue projects such as the GTEX study (Ardlie et al. 2015; Mele et al. 2015), the integration of ionomics would open the door toward unraveling the expression levels and hierarchies of trace element–associated genes and proteins, and these could be many, taking into account the hundreds of zinc finger transcription factors alone. In this study, a substantial fraction of the detected QTLs displayed allele frequencies that were highly differentiated among human populations, thus implying possible worldwide differences in trace element homeostasis with potential relevance for health and disease. Besides population differentiation, in several instances, we found additional signals of positive selection linked to the described QTLs, which probably reflect both classical sweeps as well as polygenic adaptation. Moreover, we provide a functional basis for selection as we searched for adaptive events only in QTLs related to trace element homeostasis. Overall, these results illustrate the importance of regulatory variants in explaining phenotypic differences among populations as well as in the human adaptive response to environmental pressures.
Materials and Methods
Samples
Liver samples originally collected from 150 autopsies of human individuals of western European origin were obtained from the UKHTB under appropriate ethical approval. The project obtained the ethics approval from UKHTB and the Institutional Review Board of the local institution (Comitè Ètic d'Investigació Clínica - Institut Municipal d'Assistència Sanitària) in Barcelona, Spain. All analyses were performed on anonymized samples. None of the known causes of death implied directly liver disease (the information was not available for two samples). Moreover, the median time between death and sample extraction and freezing was 17.09 h, but this information was only available for 98 samples.
RNA, DNA, and Protein Extraction
Coextraction of RNA, DNA, and proteins from each tissue sample was performed using the AllPrep DNA/RNA/Protein Mini Kit (QIAGEN) following the standard manufacturer’s protocol. RNA and DNA concentrations were quantified using a NanoDrop spectrophotometer and the RNA quality was further analyzed using an Agilent 2100 Bioanalyser. The RNA integrity number ranged from 7 to 9.8. Precipitated proteins were dissolved in freshly prepared digestion buffer (6 M urea, 0.2 M NH4HCO3), sonicated for 15 min and quantified using BCA Protein Quantification Kit (Thermo Fisher Scientific). A total of 50 µg of protein extract at a concentration of 0.33 µg/µl was reduced with dithiothreitol (150 nmol, 1 h, 37 °C) and alkylated in the dark with iodoacetamide (300 nmols, 30 min, 25 °C). The resulting protein mix was diluted one-third with 200 mM NH4HCO3 and digested with 5 µg LysC (Wako) overnight at 37 °C, then diluted one-half with 200 mM NH4HCO3 and digested with 5 µg of trypsin (Promega) for 8 h at 37 °C. Finally, the peptide mix was acidified with formic acid and desalted with a home-made Empore C18 column (3 M Inc.) prior to Liquid Chromatography Tandem Mass Spectrometry analysis (Rappsilber et al. 2007).
SRM Assay Development
A total of 67 proteins, mainly involved in the homeostasis of metal elements, were selected for quantification using SRM (table 1 and supplementary table S1, Supplementary Material online). SRM assays for the selected proteins were developed following the general high‐throughput strategy previously reported (Picotti et al. 2010). Initially, 2–4 unique tryptic peptides ranging from 6 to 20 amino acids in length were chosen for each protein (supplementary tables S6 and S7, Supplementary Material online). Unique peptides previously observed in discovery MS experiments (Martens et al. 2005; Desiere et al. 2006) were prioritized during the peptide selection process. All peptides containing amino acids prone to undergo unspecific reactions (e.g., Met, Trp, Asn, and Gln) were generally avoided and only selected when no other options were available (Lange et al. 2008). The selected peptides were chemically synthesized using C-terminal 13C6,15N2-Lysine or 13C6,15N4-Arginine (JPT, Berlin, Germany) and used for SRM assay development. Fragment ion spectra were collected for each peptide using a shotgun data-dependent acquisition method in an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a nano-flow HPLC (EasyLC Proxeon, Odense, Denmark). The obtained MS2 data were analyzed with Proteome Discoverer software suite (v1.3.0.339) and the Mascot search engine (v2.3, Matrix Science; Perkins et al. 1999). Search parameters were set to 7 ppm for the precursor mass tolerance, 0.5 Da for the fragment mass tolerance, and fully tryptic peptides and no missed cleavages were selected. An identification FDR of 1% based on decoy assignments was used (Elias and Gygi 2007). Oxidation of methionine and protein acetylation at the N-terminal were defined as variable modification, whereas carbamidomethylation on cysteines, and 13C6,15N2-Lysine and 13C6,15N4-Arginine were set as fixed modifications.
SRM measurements were performed on a 5500 Q-Trap mass spectrometer (AB Sciex, Framingham, MA) coupled to a nanoLC Ultra-1DPlus (AB Sciex). Briefly, peptide mixtures were loaded onto a C18 Acclaim PepMap precolumn (Thermo Fisher Scientific, cat # 164564) and were separated by reversed-phase chromatography using a 12-cm column with an inner diameter of 75 μm, and packed with 5 μm C18 particles (Nikkyo Technos Co, Japan). The chromatographic gradient started at 98% buffer A (0.1% formic acid in water) and 2% buffer B (0.1% formic acid in acetonitrile) with a flow rate of 300 nl/min for 5 min and gradually increased to 60% buffer A and 40% buffer B in 35 min. After each analysis, both the precolumn and the column were washed for 10 min with 2% buffer A and 98% buffer B. Digested beta-galactosidase was analyzed between the SRM measurements of biological samples to avoid sample carryover and to assure stability of the instrument. Measurements were done in scheduled SRM mode, using a retention time window of 5 min. Around 450 transitions were packed per method with a maximum total cycle time of 2 s, to ensure dwell times over 10 ms per transition. Raw data have been deposited in the PASSEL repository with the data set identifier PASS00678. Relative concentrations for the total set of 47 proteins successfully quantified are presented in supplementary table S5, Supplementary Material online, as the log2 ratio between the endogenous peptide abundances in each sample and their corresponding reference.
Gene Expression Analysis
RNA was normalized to a concentration of 200 ng/µl and subsequently reverse transcribed to cDNA using the High-Capacity cDNA reverse transcription kit (Life Technologies) following the standard manufacturer’s protocol. Two sets of 56 different GEx assays were designed and subsequently analyzed with the OpenArray Real Time PCR System (Life Technologies). We included assays for 90 genes related to metal homeostasis as well as for 5 genes related to other micronutrients (vitamin D and thiamine) and several technical assays (table 1 and supplementary table S1, Supplementary Material online). Three different assays for the housekeeping GAPDH gene were used as internal controls and all samples were processed in duplicate. The OpenArray Real-Time qPCR Analysis and the DataAssist software packages from LifeTechnologies were used for subsequent gene expression data analysis. Relative expression quantifications as measured by the 2−ΔΔCT values are presented in supplementary table S4, Supplementary Material online. Effect sizes and P values obtained for all SNPs tested in the eQTL analysis (supplementary table S10, Supplementary Material online) were extracted from the GTEx portal (http://www.gtexportal.org/home/testyourown, last accessed September 14, 2015) (Ardlie et al. 2015; Mele et al. 2015), which includes results from 32 RNAseq samples.
Genotyping
Genomic DNA was quantified using PicoGreen. Up to 182 SNPs (table 1 and supplementary tables S1 and S2, Supplementary Material online) were genotyped at the Spanish “Centro Nacional de Genotipado” (CEGEN-ISCIII)” (www.cegen.org, last accessed December 2, 2015) by using the VeraCode GoldenGate GT Assay Technology (format 192-plex; Illumina Inc., San Diego, CA) and the GenomeStudio Software (2011.1). Genotypes and additional sample information have been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/, last accessed June 19, 2015), under accession number EGAS00001001292.
Ionomics
Snap-frozen tissues were dissected with a ceramic knife while frozen and subsequently heat dried at 60°C for 24 h. Acid digestion (with HNO3 and H2O2) of 40–160 mg of tissue sample was performed in Teflon closed reactors at 90 °C. After their corresponding dilution, determination of As, Cd, Co, Cr, Cu, Fe, Hg, Li, Mg, Mn, Mo, Ni, Pb, Rb, Se, Sn, V, and Zn was performed in three technical replicates per sample by ICP-MS in a 7500 CE Agilent instrument at the Serveis Cientifico-Tècnics at the University of Barcelona. Raw element concentration in mg/kg (ppm) as indicated in supplementary table S3, Supplementary Material online, was that obtained from the ICP-MS analysis after considering the corresponding correction for dilution factor and the dry weight of each sample. Subsequently, the raw element concentration of each sample was normalized by the mean of all samples. Finally, the element corrected concentration, which is the value that was used in all subsequent analyses, corresponds to the normalized element concentration divided by the mean of the normalized value obtained for Mg, Mn, Fe, Cu, Se, and Mo. This correction was necessary to adjust for the effect of variable fat content and subsequent dilution effects in the different samples.
Statistical Analysis
PCA was performed on individual SNP genotypes with the SmartPCA software package (Patterson et al. 2006). We estimated haplotypes using the PHASE software as implemented within SNPator (Morcillo-Suarez et al. 2008). Linkage disequilibrium analysis and r2 calculation was performed with the Haploview program (Barrett et al. 2005). Logarithmic transformation of the 2−ΔΔCT values for gene expression, protein log2 ratios for protein abundance, and normalized metal abundances were used as the response variable for QTL analyses. Based on the allele composition of the selected genotypes, each SNP constituted the single independent variable, and associations between genotyped cis-SNPs and 1) protein abundances, 2) essential metal content, and 3) transcript levels were evaluated using linear regression. A total of nine SNPs (5.3%) not in Hardy–Weinberg equilibrium (P < 0.005; tested within SNPator) were excluded from association analysis. Similarly, only one SNP was used for quantitative trait analysis among sets of SNPs with minimum r2 > 0.8 (supplementary fig. S3, Supplementary Material online). Unless otherwise stated, multiple testing was addressed by means of the Benjamini–Hochberg false discovery correction (Benjamini and Hochberg 1995).
For all pQTL (P ≤ 0.05), eQTL (P ≤ 0.05), and nutriQTLs (P ≤ 0.025) detected (even if not surpassing FDR correction), we performed a further detailed comparison of micronutrient content and RNA and protein abundances, between alleles and genotypes at cis-SNPs, through analysis of variance within the SPSS Statistics software package (version 19). Correlation analysis was performed between RNA and protein abundances as well as between micronutrient content and protein or RNA abundances. We have estimated the statistical power to detect significant associations of our study design by drawing samples from independent normal distributions with sizes equal to 2Np and 2N(1 − p), where N is the size of the study (i.e., N = 150) and p is the minor allele frequency of each SNP we genotyped. We found that the average difference that can be detected in our study equals 0.409 standard deviations, assuming an equal variance scenario. Only significant correlations (FDR ≤ 0.05) with a minimum absolute R value of 0.50 were considered. Signatures of natural selection were explored for all pQTL (P ≤ 0.05), eQTL (P ≤ 0.05), and nutriQTLs (P ≤ 0.025) detected (even if not surpassing FDR correction) with statistics for site frequency spectrum (Tajima’s D, Fay and Wu H, Composite Likelihood Ratio Test), population differentiation (FST, cross-population composite likelihood ratio test), and linkage disequilibrium structure (iHS, cross-population extended haplo-type homozygosity) as well as with the hierarchical boosting scores for different types of selective sweeps as implemented in the 1000 Genomes Selection Browser (Pybus et al. 2014) for three HapMap populations (YRI, CHB, and YRI) Global genome-wide FST pairwise scores between CEU–YRI, CHB–YRI, and CEU–CHB were extracted from the 1000 Genomes Selection Browser (Pybus et al. 2014) and subsequently used to compute the percentiles of each corresponding FST pairwise score of all detected QTL occupy.
Supplementary Material
Supplementary notes S1–S60, figures S1–S6, and tables S1–S15 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This article is dedicated to the memory of our friend and colleague Johannes Engelken, who passed away unexpectedly on October 23, 2015. We would like to thank Marc Pybus, Juan Antonio Rodríguez, and Elena Carnero for their help and insightful comments on the manuscript. This work was supported by Ministerio de Ciencia e Innovación, Spain (grants BFU2008-01046 and SAF2011-29239) and by Direcció General de Recerca, Generalitat de Catalunya (2009SGR-1101 and 2014SGR-866). The CRG/UPF Proteomics Unit is part of the “Plataforma de Recursos Biomoleculares y Bioinformáticos (ProteoRed)” supported by grant PT13/0001 of the Instituto de Salud Carlos III (ISCIII). J.E. was supported by a Postdoctoral scholarship from the Volkswagenstiftung (Az: I/85 198).
References
- Altshuler D, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, Flicek P, Gabriel SB, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M, et al. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Wooding SP. 2003. Signatures of natural selection in the human genome. Nat Rev Genet. 4:99–111. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. 2015. Impact of regulatory variation from RNA to protein. Science 347:664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gögele M, Anderson D, Broer L, Podmore C, et al. 2014. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 5:4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, Palotie A, Peltonen L, Martin NG, Montgomery GW, Whitfield JB, et al. 2009. Variants in TF and HFE explain 40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 84:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JJ, Coop G. 2014. A population genetic signal of polygenic adaptation. PLoS Genet. 10:e1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulaj ZJ, Griffen LM, Jorde LB, Edwards CQ, Kushner JP. 1996. Clinical and biochemical abnormalities in people heterozygous for hemochromatosis. N Engl J Med. 335:1799–1805. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA. 2005. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 15:1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, Ally A, Balasundaram M, Birol I, Butterfield YSN, et al. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine CC, Anderson GJ, Vulpe CD, McLaren CE, Bahlo M, Yeap HL, Gertig DM, Osborne NJ, Bertalli NA, Beckman KB, et al. 2009. A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. Br J Haematol. 147:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello LC, Franklin RB. 2006. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Cancer. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. 2006. The PeptideAtlas project. Nucleic Acids Res. 34:D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez M, Arroyo M, Cerdàn FJ, Muñoz M, Martin MA, Balibrea JL. 1989. Serum and tissue trace metal levels in lung cancer. Oncology 46:230–234. [DOI] [PubMed] [Google Scholar]
- Diplock T. 1987. Trace elements in human health with special reference to selenium. Am J Clin Nutr. 45:1313–1322. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KCC, Taylor J, Burnett E, Gut I, Farrall M, et al. 2007. A genome-wide association study of global gene expression. Nat Genet. 39:1202–1207. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Sabidó E, Hüttenhain R, Collins B, Aebersold R. 2012. Range of protein detection by selected/multiple reaction monitoring mass spectrometry in an unfractionated human cell culture lysate. Proteomics 12:1185–1193. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 4:207–214. [DOI] [PubMed] [Google Scholar]
- Engelken J, Carnero-Montoro E, Pybus M, Andrews GK, Lalueza-Fox C, Comas D, Sekler I, de la Rasilla M, Rosas A, Stoneking M, et al. 2014. Extreme population differences in the human zinc transporter ZIP4 (SLC39A4) are explained by positive selection in sub-Saharan Africa. PLoS Genet. 10:e1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss EJ, Radulovic D, Shaffer SA, Goodlett DR, Kruglyak L, Bedalov A. 2011. Genetic variation shapes protein networks mainly through non-transcriptional mechanisms. PLoS Biol. 9:e1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CB, Aswath K, Chanock SJ, McKay HF, Peters U. 2006. Polymorphism analysis of six selenoprotein genes: support for a selective sweep at the glutathione peroxidase 1 locus (3p21) in Asian populations. BMC Genet. 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautrey H, Nicol F, Sneddon AA, Hall J, Hesketh J. 2011. A T/C polymorphism in the GPX4 3′UTR affects the selenoprotein expression pattern and cell viability in transfected Caco-2 cells. Biochim Biophys Acta. 1810:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B Pokholok DK Kwan T Grundberg E Morcos L Verlaan DJ Le J Koka V Lam KCL, Gagné V et al. 2009. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 41:1216–1222. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7:e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts SK, et al. 2011. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Shlyakhter I, Shylakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327:883–886. [DOI] [PubMed] [Google Scholar]
- Hamanishi T, Furuta H, Kato H, Doi A, Tamai M, Shimomura H, Sakagashira S, Nishi M, Sasaki H, Sanke T, et al. 2004. Functional variants in the glutathione peroxidase-1 (GPx-1) gene are associated with increased intima-media thickness of carotid arteries and risk of macrovascular diseases in Japanese type 2 diabetic patients. Diabetes 53:2455–2460. [DOI] [PubMed] [Google Scholar]
- Hanaoka M, Droma Y, Basnyat B, Ito M, Kobayashi N, Katsuyama Y, Kubo K, Ota M. 2012. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PLoS One 7:e50566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh J. 2008. Nutrigenomics and selenium: gene expression patterns, physiological targets, and genetics. Annu Rev Nutr. 28:157–177. [DOI] [PubMed] [Google Scholar]
- Heuberger R. 2007. Liver disease. In: Marian MJ, Williams-Muller P, Muir Bowers J, editors. Integrating therapeutic and complementary nutrition. Boca Raton (FL): CRC Press, Taylor & Francis; p. 323–354. [Google Scholar]
- Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol. 10:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, et al. 2010. International network of cancer genome projects. Nature 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, et al. 2011. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 7:e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson Å, Enroth S, Palmblad M, Deelder AM, Bergquist J, Gyllensten U. 2013. Identification of genetic variants influencing the human plasma proteome. Proc Natl Acad Sci U S A. 110:4673–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T, Weaver BP, Andrews GK. 2008. The genetics of essential metal homeostasis during development. Genesis 46:214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. 2008. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Buil A, Collins BC, Gillet LC, Blum LC, Cheng LY, Vitek O, Mouritsen J, Lachance G, Spector TD, et al. 2015. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 11:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourdusamy A, Newhouse S, Lunnon K, Proitsi P, Powell J, Hodges A, Nelson SK, Stewart A, Williams S, Kloszewska I, et al. 2012. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 21:3719–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinouski M, Hasan NM, Zhang Y, Seravalli J, Lin J, Avanesov A, Lutsenko S, Gladyshev VN. 2014. Genome-wide RNAi ionomics screen reveals new genes and regulation of human trace element metabolism. Nat Commun. 5:3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalioth EJ, Schenker JG, Chevion M. 1983. Copper and zinc levels in normal and malignant tissues. Cancer 52:868–872. [DOI] [PubMed] [Google Scholar]
- Martens L, Van Damme P, Van Damme J, Staes A, Timmerman E, Ghesquière B, Thomas GR, Vandekerckhove J, Gevaert K. 2005. The human platelet proteome mapped by peptide-centric proteomics: a functional protein profile. Proteomics 5:3193–3204. [DOI] [PubMed] [Google Scholar]
- McLaren CE, Garner CP, Constantine CC, McLachlan S, Vulpe CD, Snively BM, Gordeuk VR, Nickerson DA, Cook JD, Leiendecker-Foster C, et al. 2011. Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS One 6:e17390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren CE, McLachlan S, Garner CP, Vulpe CD, Gordeuk VR, Eckfeldt JH, Adams PC, Acton RT, Murray JA, Leiendecker-Foster C, et al. 2012. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS One 7:e38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavatá I, Vrána D, Vodicka P, Hesketh JE. 2010. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis 31:1074–1079. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464:773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo-Suarez C, Alegre J, Sangros R, Gazave E, de Cid R, Milne R, Amigo J, Ferrer-Admetlla A, Moreno-Estrada A, Gardner M, et al. 2008. SNP analysis to results (SNPator): a web-based environment oriented to statistical genomics analyses upon SNP data. Bioinformatics 24:1643–1644. [DOI] [PubMed] [Google Scholar]
- Nica AC, Dermitzakis ET. 2013. Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci. 368:20120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker V, Cluett C, Shields B, Murray A, Parnell KS, Perry JRB, Weedon MN, Singleton A, Hernandez D, Evans J, et al. 2008. A common variation in deiodinase 1 gene DIO1 is associated with the relative levels of free thyroxine and triiodothyronine. J Clin Endocrinol Metab. 93:3075–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, Porcu E, Pattaro C, Busonero F, Zanon A, et al. 2011. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum Mol Genet. 20:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B. 2010. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, Dall’Olio GM, Luisi P, Uzkudun M, Carreño-Torres A, Pavlidis P, Laayouni H, Bertranpetit J, Engelken J. 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42:D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, Luisi P, Dall’Olio GM, Uzkudun M, Laayouni H, Bertranpetit J, Engelken J. 2015. Hierarchical boosting: a machine-learning framework to detect and classify hard selective sweeps in human populations. Bioinformatics first published online August 26, 2015. doi:10.1093/bioinformatics/btv493. [DOI] [PubMed] [Google Scholar]
- Qiu W, Cho MH, Riley JH, Anderson WH, Singh D, Bakke P, Gulsvik A, Litonjua AA, Lomas DA, Crapo JD, et al. 2011. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS One 6:e24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, Choy E, Hu D, Tamraz B, Pawlikowska L, et al. 2007. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet. 80:716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink L, Haase H. 2007. Zinc homeostasis and immunity. Trends Immunol. 28:1–4. [DOI] [PubMed] [Google Scholar]
- Rye MS, Wiertsema SP, Scaman ESH, Thornton R, Francis RW, Vijayasekaran S, Coates HL, Jamieson SE, Blackwell JM. 2013. Genetic and functional evidence for a role for SLC11A1 in susceptibility to otitis media in early childhood in a Western Australian population. Infect Genet Evol. 16:411–418. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabidó E, Wu Y, Bautista L, Porstmann T, Chang CY, Vitek O, Stoffel M, Aebersold R. 2013. Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after a sustained high-fat diet. Mol Syst Biol. 9:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas S, Briollais L, Ibrahim-Zada I, Jarjanazi H, Choi YH, Musquera M, Fleshner N, Venkateswaran V, Ozcelik H. 2010. A whole-genome SNP association study of NCI60 cell line panel indicates a role of Ca2+ signaling in selenium resistance. PLoS One 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, Gattepaille LM, Engstrom K, Vahter M, Jakobsson M, Broberg K. 2015. Human adaptation to arsenic-rich environments. Mol Biol Evol. 32(6):1544–1555. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. [DOI] [PubMed] [Google Scholar]
- Sillanpää M. 1982. Micronutrients and the nutrient status of soils: a global study. FAO Soils Bulletin 48. Rome (Italy): Food and Agriculture Organization of the United States. [Google Scholar]
- Sutherland A, Kim DH, Relton C, Ahn YO, Hesketh J. 2010. Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr. 5:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C, Ajioka RS, Jorde LB, Kushner JP, Kreitman M. 2003. A method for detecting recent selection in the human genome from allele age estimates. Genetics 165:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C, Kreitman M. 2002. Sequence variation and haplotype structure at the human HFE locus. Genetics 161:1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, et al. 2012. Seventy-five genetic loci influencing the human red blood cell. Nature 492:369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KCL, Koka V, Dias J, Gurd S, Martin NW, Mallmin H, et al. 2009. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 19:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Gaffney DJ, Pickrell JK, Gilad Y, Stephens M, Pritchard JK. 2012. Exon-specific QTLs skew the inferred distribution of expression QTLs detected using gene expression array data. PLoS One 7:e30629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4:e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voetsch B, Jin RC, Bierl C, Benke KS, Kenet G, Simioni P, Ottaviano F, Damasceno BP, Annichino-Bizacchi JM, Handy DE, et al. 2007. Promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene: a novel risk factor for arterial ischemic stroke among young adults and children. Stroke 38:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells KR, Brown KH. 2012. Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS One 7:e50568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L, Romagné F, Müller E, Erlebach E, Weihmann A, Parra G, Andrés A, Castellano S. 2015. Genetic adaptation to levels of dietary selenium in recent human history. Mol Biol Evol. 32:1507–1518. [DOI] [PubMed] [Google Scholar]
- Wiernsperger N, Rapin J. 2010. Trace elements in glucometabolic disorders: an update. Diabetol Metab Syndr. 2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. 2013. Variation and genetic control of protein abundance in humans. Nature 499:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. 2014. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158:1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Cao C, Lin X, O’Brien KO, Gu Z. 2015. Natural selection on HFE in Asian populations contributes to enhanced non-heme iron absorption. BMC Genet. 16:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. 2010. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5:e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Shi S, Wu CI. 2007. Compound tests for the detection of hitchhiking under positive selection. Mol Biol Evol. 24:1898–1908. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li J, Tian L, Lu D, Yuan K, Yuan Y, Xu S. 2015. Differential natural selection of human zinc transporter genes between African and Non-African populations. Sci Rep. 5:9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.