Abstract
The objective of this study was to investigate the genotype–phenotype correlation of the PAX6 gene in aniridia. We clinically examined 5 families and 16 sporadic patients with aniridia. We performed chromosomal analysis and PCR analysis of the PAX6 gene using patient genomic DNA. Chromosomal analysis demonstrated deletions at 11p13 in one allele in four sporadic patients. Seven nonsense mutations, two frameshifts (two insertions), four splice junction errors and two missense mutations were found, and all were heterozygous. The iris phenotype ranged from total to normal in each patient, and the characteristic phenotypes, including cataract, glaucoma or optic nerve hypoplasia, varied widely even among members of the same family. Foveal hypoplasia was detected in all patients except for one. No obvious genotype–phenotype correlation was identified; however, the aniridia phenotype between the two eyes in each patient was quite similar in all patients. Because PAX6 regulates numerous downstream genes and its expression is regulated by several factors during eye development, the aniridia phenotype may be complex even in family members. However, because PAX6 regulation, resulting from both paternal and maternal alleles associated with PAX6, is considered to be roughly similar in both eyes of each patient, the aniridia phenotype may be similar in both eyes of each patient.
Introduction
It is well known that aniridia is caused by mutation of the PAX6 gene.1,2 The PAX6 gene codes a transcriptional regulator and is a master control gene in the development of ocular tissues in invertebrates.3 This is evident from the ectopic formation of eyes on the wings, legs and antennae in Drosophila upon targeted expression of the eyeless gene, a homolog of the human PAX6.4 Furthermore, an expression pattern of the Pax6 gene also indicates the multiple functions of the gene in mammals.5 In situ hybridization for murine Pax6 demonstrated its expression in the developing central nervous system as well as in ocular tissues derived from the ectoderm and neuroectoderm, that is, the corneal epithelium, lens and retina.6 These findings can explain various phenotypic manifestations in the entire eye of aniridia patients, including corneal opacity, absence of the iris, glaucoma, cataract and hypoplasia of the fovea and optic nerve. These findings also present the possibility that mutations of the PAX6 gene cause a variety of developmental anomalies of ocular tissues. In addition to aniridia, PAX6 mutations have been reported in Peters' anomaly,7 corneal dystrophy8 and foveal hypoplasia.9
The PAX6 gene was isolated as a candidate aniridia gene by positional cloning from an overlapping region of chromosomal deletions at 11p13, which are observed in some aniridia patients, especially in those with Wilms' tumor, genitourinary abnormalities and mental retardation (WAGR syndrome).1 In addition to large deletions encompassing the whole gene, more than 330 mutations have been detected in autosomal dominant and sporadic aniridia patients.10 Because most of these mutations are nonsense, frameshifts or splicing errors that result in premature translational termination on one of the alleles,11 haploinsufficiency of the gene,12 or the presence of one amorphic allele, has been suggested to cause the aniridia phenotype.13 We recently identified five additional PAX6 missense mutations in four pedigrees.14 These findings indicate that hypomorphic alleles that significantly disturb an important motif also cause the aniridia phenotype. While different genotypic mutations cause different phenotypes of developmental anomalies, the same PAX6 mutation sometimes resulted in a variety of aniridia phenotypes that ranged from normal-sized irises to the complete absence of the iris in one pedigree,15 which indicated that one genotype produces different phenotypes of aniridia. Thus, the genotype–phenotype correlation of PAX6 mutations in aniridia is still controversial. In the present study, we investigated the correlation between the genotype of amorphic or hypomorphic PAX6 mutations and the phenotype of aniridia.
Materials and methods
Patients and clinical data
The study included five families (autosomal dominant trait) and 16 sporadic patients with aniridia. Visual acuity was examined by Landordt's ring procedure, and examination of ocular anterior and posterior segments was performed with a slit-lamp biomicroscope and a binocular indirect ophthalmoscope. Glaucoma was diagnosed by measurement of intraocular pressure, examination of the iridocorneal angle, a slit-lamp biomicroscope, and visual field measurement if available. Full-field electroretinographies (ERGs) were recorded in six patients (Patient no. 12, 19 and 21–24) according to the International Society for Clinical Electrophysiology of Vision (ISCEV) standards. Some examinations in infants were performed under general anesthesia when necessary.
Optic nerve hypoplasia was diagnosed when the ratio of DM (the distance from disc to macular) to DD (the mean disc diameter) was greater than 3.2 and the disc showed a typical double ring sign, simultaneously. The pediatrician also evaluated systemic and neurological findings, and screening for the presence of Wilms' tumor was performed by echography.
Chromosomal and genetic analysis
These studies were conducted in accordance with the Declaration of Helsinki. Our use of human subjects was conducted with patients' informed consent, and approved by the Ethics Committee of National Center for Child Health and Development (Approval no. 518). Blood samples of the patients and family members were collected from the peripheral veins. High-resolution G-banded chromosomes were obtained from phytohemagglutinin -synchronized blood lymphocyte culture after 72 h. Twenty metaphases were analyzed. Genomic DNA was prepared from isolated leukocytes using a standard procedure.14 For mutation analysis, the extracted DNA was amplified using PCR primer sets (Supplementary Table S1) for 13 exons of PAX6 (NCBI; M93650; http://www.ncbi.nlm.nih.gov/nuccore/189632/) under the PCR conditions previously described.16 The PCR product was purified and directly sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI 3100 autosequencer (Applied Biosystems, MA, USA), as previously described.17 The sequences were compared with the reference in the database at the National Center for Biotechnology Information (NCBI; NC_000011.9; http://www.ncbi.nlm.nih.gov/nuccore/NC_000011.9).
Results
Clinical findings
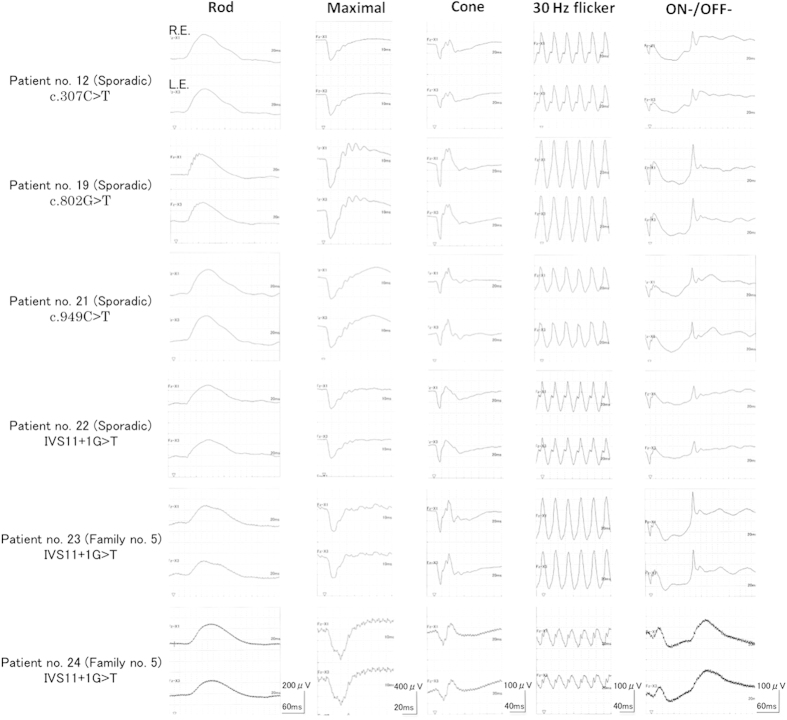
The lists of clinical phenotypes associated with mutations and chromosomal defects are summarized in Tables 1 and 2, respectively. Regarding the aniridia phenotype, the iris was totally absent in 50 eyes (89%), partially absent in 4 eyes (7%) and had an almost normal appearance in 2 eyes (4%). Corneal opacity was observed in 10 eyes (18%), congenital or developmental cataract in 50 eyes (89%), glaucoma in 12 eyes (21%), foveal hypoplasia in 54 eyes (96%) and optic nerve hypoplasia in 2 eyes (4%). Visual acuity ranged from 0.06 to 0.6 but was not greater than 0.3 in any of the eyes except for both eyes of Patient no. 11 without foveal hypoplasia. Wilms' tumor, hepatoblastoma and polydactylia were observed in one patient each, and mental retardation was observed in three patients. A series of ERGs was obtained from six patients (Patient no. 12, 20–24) (Figure 1). Mild to moderate abnormal ERGs were detected in five patients, but ERGs were within the normal limit in Patient no. 19. Patient no. 12, who had a c.307C>T mutation in the paired domain (PD), exhibited abnormal maximum and cone ERG with a mild reduction of amplitude. Patient no. 19, who had a c.802G>T mutation in the proline/serine/threonine-rich region (PST), exhibited a normal ERG response. Patient no. 21, who had a c.949C>T mutation in PST, exhibited subnormal cone ERGs with a mild reduction of cone and 30 Hz flicker amplitude. Patient no. 22, who had an IVS11+1G>T mutation, exhibited nearly negative ERG in maximum response, accompanied by a mild reduction in cone ERGs. Patient no. 23, who was a familial case (family no. 5) and had the same mutation as Patient no. 22, only exhibited a reduced b-wave of maximum response with subnormal Rod ERG. Patient no. 24, the father of Patient no. 23, exhibited a relatively severe phenotype with delay and reduced amplitude throughout the ERGs, except for a subnormal Rod response.
Table 1. Mutations and phenotype of each patient.
| Patient no. | Family no. | Inheritance | Exon/Intron | Nucleotide change | Mutation | Amino acid change | Pax6 domain | Likely NMD | Age | Sex | Aniridia (R/L) | CO | Cataract | Glaucoma | FH | ONH | BCVA | WT | MR | Other remarks |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Sporadic | Exon5 | c.50A>G | Missense | p.Asn17Ser | PD | No | 9 | F | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 2 | Sporadic | Exon5 | c.131G>A | Missense | p.Arg44Gln | PD | No | 21 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 3 | Sporadic | Exon5 | c.79C>T | Nonsense | p.Gln27X | PD | Yes | 9 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.2/0.2 | − | − | − | |
| Intron6 | IVS6+1G>A | Splice error | NA | PD | Yes | |||||||||||||||
| 4 | 1 | Familial | Exon5 | c.109_110insG | Frameshift | p.Ala37fs | PD | Yes | 31 | F | Total/total | +/+ | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − |
| 5 | 1 | Exon5 | c.109_110insG | Frameshift | p.Ala37fs | PD | Yes | 10 | M | Total/total | +/+ | +/+ | +/+ | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 6 | 1 | Exon5 | c.109_110insG | Frameshift | p.Ala37fs | PD | Yes | 2 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | ND | − | − | − | |
| 7 | 2 | Familial | Intron5 | IVS5+1G>T | Splice error | NA | PD | Yes | 36 | F | Total/total | −/− | +/+ | +/+ | +/+ | −/− | 0.1/0.1 | − | − | Hepatoblastoma |
| 8 | 2 | Intron5 | IVS5+1G>T | Splice error | NA | PD | Yes | 1 | M | Total/total | +/+ | +/+ | +/+ | +/+ | −/− | ND | − | + | − | |
| 9 | 3 | Familial | Intron5 | IVS5+2T>G | Splice error | NA | PD | Yes | 64 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − |
| 10 | 3 | Intron5 | IVS5+2T>G | Splice error | NA | PD | Yes | 38 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 11 | 3 | Intron5 | IVS5+2T>G | Splice error | NA | PD | Yes | 30 | M | Partial/partial | −/− | −/− | −/− | −/− | −/− | 0.6/0.6 | − | − | − | |
| 12 | Sporadic | Exon6 | c.307C>T | Nonsense | p.Arg103X | PD | Yes | 13 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.15/0.15 | − | − | − | |
| 13 | Sporadic | Exon8 | c.551_552insG | Frameshift | p.Glu184fs | LNK | Yes | 4 | F | Total/total | −/− | +/+ | −/− | +/+ | −/− | ND | − | − | − | |
| 14 | Sporadic | Exon8 | c.607C>T | Nonsense | p.Arg203X | LNK | Yes | 11 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0. 1/ 0.1 | − | − | − | |
| 15 | Sporadic | Exon8 | c.607C>T | Nonsense | p.Arg203X | LNK | Yes | 9 | F | Partial/partial | −/− | +/+ | −/− | +/+ | −/− | ND | − | − | − | |
| 16 | Sporadic | Exon9 | c.718C>T | Nonsense | p.Arg240X | HD | Yes | 27 | F | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.3/0.3 | − | − | − | |
| 17 | 4 | Familial | Exon9 | c.718C>T | Nonsense | p.Arg240X | HD | Yes | 29 | M | Total/total | −/− | +/+ | −/− | +/+ | +/+ | 0.08/0.06 | − | − | − |
| 18 | 4 | Exon9 | c.718C>T | Nonsense | p.Arg240X | HD | Yes | 5 | F | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 19 | Sporadic | Exon10 | c.802G>T | Nonsense | p.Glu268X | PST | Yes | 6 | F | Total/total | −/− | −/− | −/− | +/+ | −/− | 0.2/0.15 | − | − | − | |
| 20 | Sporadic | Exon10 | c.829C>T | Nonsense | p.Gln277X | PST | Yes | 13 | F | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.1/0.1 | − | − | − | |
| 21 | Sporadic | Exon11 | c.949C>T | Nonsense | p.Arg317X | PST | Yes | 10 | M | Total/total | −/− | −/− | +/+ | +/+ | −/− | 0.03/0.1 | − | − | − | |
| 22 | Sporadic | Intron11 | IVS11+1G>T | Splice error | NA | PST | Yes | 4 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.15/ 0.15 | − | − | − | |
| 23 | 5 | Familial | Intron11 | IVS11+1G>T | Splice error | NA | PST | Yes | 7 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.2/0.3 | − | − | − |
| 24 | 5 | Intron11 | IVS11+1G>T | Splice error | NA | PST | Yes | 36 | M | Normal/normal | −/− | +/+ | −/− | +/+ | −/− | 0.2/0.3 | − | − | − |
Abbreviations: BCVA, best corrected visual acuity; CO, Corneal opacity; F, female; FH, foveal hypoplasia; HD, homeodomain; L, left; LNK, linker region; M, male; MR, mental retardation; N/A, not applicable; ND, not detected; NMD, nonsense-mediated decay; ONH, optic nerve hypoplasia; PD, paired domain; PST, proline/serine/threonine-rich region; R, right; WT, Wilms' tumor.
Patients list shows correlation between genotype and phenotype.
Family no. 2 and no. 3 are supposed to have splice error at intron 5, while no obvious similarity of clinical phenotype is identified. Sporadic cases of no.14 and no.15 have c. 607C>T mutation, but exhibit different iris phenotype, partial and normal iris. Sporadic case no. 16 and family no. 4 exhibit same genotype, c. 718C>T, while visual acuity shows variety. In addition, only case no. 17 exhibits optic nerve hypoplasia. Meanwhile, phenotype of two eyes of the same patient is almost completely similar.
Table 2. Chromosomal abnormalities and phenotype of each patient.
| Patient no. | Inheritance | Chromosomal anomaly | Age | Sex | Aniridia (R/L) | Corneal opacity | Cataract | Glaucoma | FH | ONH | BCVA | Wilms' tumor | MR | Other remarks |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | Sporadic | del(11) (p12p14.2) | 1 | F | Total/total | −/− | +/+ (cyst) | −/− | +/+ | −/− | ND | − | ND | − |
| 26 | Sporadic | del(11) (p13p14) | 3 | M | Total/total | −/− | +/+ | −/− | +/+ | −/− | 0.06/0.08 | − | − | − |
| 27 | Sporadic | del(11) (p11.2p14.2) | 3 | F | Total/total | +/+ | +/+ | +/+ | +/+ | −/− | ND | − | + | Polydactylia |
| 28 | Sporadic | del(11) (p12p14) | 8 | F | Total/total | +/+ | +/+ | +/+ | +/+ | −/− | 0.1/0.1 | + | + | − |
Abbreviations: BCVA, best corrected visual acuity; F, female; FH, foveal hypoplasia; L, left; M, male; MR, mental retardation; ONH, optic nerve hypoplasia; R, right. A chromosomal deletion including 11p13 was detected in four sporadic patients. Almost all eyes exhibit completely hypoplastic iris and fovea, and have cataracts. Mental retardation is observed in patients with cloudy cornea and glaucoma. Wilms' tumor and polydactylia is identified in one patient each. Those two patients are clinically diagnosed as WAGR syndrome.
Figure 1.
Electroretinographies of the patients (Patient no. 12, 20–24). Patient no. 12 exhibited a mild reduction of maximum and cone ERG. Patient no. 19 exhibited almost normal ERGs. Patient no. 21 exhibited subnormal cone ERGs. Patient no. 22 exhibited a reduced maximum response with amplitude reduction of b-wave, and ON-/OFF ERG were abnormally reduced. Patient no. 23 exhibited a mild reduction of maximum response. Patient no. 24 exhibited a relatively severe abnormality in maximum, cone, 30 Hz flicker and ON-/OFF ERG with both reduction of amplitude and delayed response. R.E., right eye; L.E., left eye. Calibrations are presented in the bottom of the respective ERGs. ERGs, electroretinographies.
PAX6 mutations
Seven nonsense mutations located in exons 5–11 of the PAX6 gene, including two frameshifts (two insertions), four splicing errors and two missense mutations, were found in five families and 12 sporadic patients. Among those mutations, eight mutations were associated with PD, two with the linking region, two with the homeodomain and four with PST. Of these nonsense mutations, frameshifts and splicing errors, the transcripts of mRNA were likely degraded by nonsense-mediated decay.18 Patient no. 3 had two mutations: a nonsense mutation (p.Q27X) in exon 5 and a splicing error in intron 6. These mutations were not found in his parents or siblings (data not shown). Novel mutations, including c.79C>T (p.Gln27X), c.109_110insG (p.Ala2GlyfsX19), IVS5+1G>T, IVS5+2T>G, c.551_552insG (p.Glu9ArgfsX15), c.802G>T (p.Glu268X) and IVS11+1G>T, were identified in the current study. As previously reported, the two missense mutations c.131G>A (p.Arg44Gln) and c.50A>G (p.Asn17Ser) were found in two sporadic patients.
Genotype–phenotype correlation
Although PAX6 mutations were found throughout the region from exon 5 to 11, where the PD to PST was coded, no clear correlation was identified between phenotype and genotype. Moreover, even in members of the same family who had the same mutation, the phenotype of aniridia was partially different: the appearances of corneal opacity and glaucoma were variable in family no. 1; the appearance of corneal opacity was variable in family no. 2; the appearances of iris remnant, cataract and foveal hypoplasia were variable in family no. 3; and the appearance of optic nerve hypoplasia was variable in family no. 4. ERG also showed variation even in Patient no. 22–24 who had the same IVS11+1G>T mutation. However, all ocular findings were almost completely symmetrical in both eyes of each patient.
Chromosomal anomalies
A chromosomal deletion including 11p13 was detected in four sporadic patients. In all of the affected eyes, one of which also had posterior lenticular cyst, the iris and fovea were totally absent, and cataracts were observed. Patients with severe phenotype accompanied with cloudy cornea and glaucoma showed mental retardation, and one of the patients was affected by Wilms' tumor, while the other had polydactylia. Those two patients were clinically considered to have WAGR syndrome before chromosomal analysis.
Discussion
The present study demonstrated some novel genotypes of the PAX6 gene in aniridia. Patient no. 3 had two mutations, a nonsense mutation (p.Q27X) in exon 5 and a splicing error in exon 6. It is well known that PAX6 mutations’ effects are dose-dependent,19 and the abnormalities depend on the severity of the mutation of PAX6. In mutants of the small eye (Sey) locus, where the murine homolog of the PAX6 gene is located,20 the inactivation of both alleles causes the eyes not to develop and results in severe craniofacial disorders,21 whereas inactivation of one allele results in the development of small eyes and mild central nervous system disorders, the latter of which are detectable only at the embryonic stage. In humans, compound heterozygotes are usually embryonic lethal, but two cases have survived with no or small eyes and central nervous system disorders.19,22 In the present study, Patient no. 3 exhibited only aniridia; thus, the two mutations may be present in one allele and the former one (p.Q27X) first terminates the PAX6 protein.
Three chromosomal abnormalities of 11p13 and seven nonsense mutations, two frameshifts and four splicing errors of the PAX6 gene were found in our patients. Although some mutations in the PST region are reported to cause run-on translation into the 3’UTR, which results in a dominant negative mutation and a severe aniridia phenotype,23,24 all mutations in the current study were limited in location from exon 5 to exon 11 and likely induced haploinsufficiency caused by nonsense mediated decay. The other two missense mutations in the N-terminal subdomain of the paired domain had already been identified in Japanese patients,14 which confirmed that significant disturbance of an important motif by the hypomorphic allele also causes the aniridia phenotype.
In our aniridia patients, an obvious genotype–phenotype correlation was not observed. Although haploinsufficiency of the PAX6 gene causes aniridia, which is associated with characteristic phenotypes including iris hypoplasia, foveal hypoplasia, cloudy cornea, glaucoma and cataract, the variety of the phenotype was not totally correlated with the genotype. In the current study, even among the same family members, the aniridia phenotype varied widely. Notably, in family no. 5, the father (Patient no. 24) exhibited less severe iris hypoplasia than his son (Patient no. 23); however, the ERGs demonstrated a more severely abnormal phenotype than the son, which also suggested that the phenotype was variable even in patients carrying the same mutation. In a spatio-temporal manner, Pax6 regulates various downstream targets, including Ngn2, Lews X, Wnt7b and δ-crystallin,25 and Pax6 is regulated by at least six enhancer regions, three promoters, including P0, P1 and Pα,26 and sometimes by Pax6 itself. In addition, co-activators, such as Sox2 and Maf,27 and co-repressors, such as Smad328 and Nkx6.1,29 also have vital roles in the expression of downstream targets of Pax6. This complex gene expression associated with PAX6, which is regulated by both paternal and maternal alleles, may result in phenotypic variation even in cases with the same PAX6 genotype. Simultaneously, both of the eyes of each patient, which have the same PAX6 genotype and associated regulatory mechanisms, may exhibit almost identical phenotypes.
The authors declare no conflict of interest.
Footnotes
Supplemental Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv).
References
- Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell 1991; 67: 1059–1074. [DOI] [PubMed] [Google Scholar]
- Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A et al. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet 1992; 1: 328–332. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells 1996; 1: 11–15. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 1995; 267: 1788–1792. [DOI] [PubMed] [Google Scholar]
- Kioussi C, O'Connell S, St-Onge L, Treier M, Gleiberman AS, Gruss P et al. Pax6 is essential for establishing ventral-dorsal cell boundaries in pituitary gland development. Proc Natl Acad S USA 1999; 96: 14378–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 1991; 113: 1435–1449. [DOI] [PubMed] [Google Scholar]
- Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ et al. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet 1994; 6: 168–173. [DOI] [PubMed] [Google Scholar]
- Mirzayans F, Pearce WG, MacDonald IM, Walter MA. Mutation of the PAX6 gene in patients with autosomal dominant keratitis. Am J Hum Genet 1995; 57: 539–548. [PMC free article] [PubMed] [Google Scholar]
- Azuma N, Nishina S, Yanagisawa H, Okuyama T, Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet 1996; 13: 141–142. [DOI] [PubMed] [Google Scholar]
- Hingorani M, Hanson I, van Heyningen V. Aniridia. Eur J Hum Genet 2012; 20: 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DO, Howarth RJ, Williamson KA, van Heyningen V, Beal SJ, Crolla JA. Genetic analysis of chromosome 11p13 and the PAX6 gene in a series of 125 cases referred with aniridia. Am J Med Genetics A 2008; 146A: 558–569. [DOI] [PubMed] [Google Scholar]
- Fisher E, Scambler P. Human haploinsufficiency--one for sorrow, two for joy. Nat Genet 1994; 7: 5–7. [DOI] [PubMed] [Google Scholar]
- Vincent MC, Pujo AL, Olivier D, Calvas P. Screening for PAX6 gene mutations is consistent with haploinsufficiency as the main mechanism leading to various ocular defects. Eur J Hum Genet 2003; 11: 163–169. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hotta Y, Tanaka H, Yamada M. Missense mutations in the PAX6 gene in aniridia. Invest Ophthalmol Vis Sci 1998; 39: 2524–2528. [PubMed] [Google Scholar]
- Mintz-Hittner HA, Ferrell RE, Lyons LA, Kretzer FL. Criteria to detect minimal expressivity within families with autosomal dominant aniridia. Am J Opthalmol 1992; 114: 700–707. [DOI] [PubMed] [Google Scholar]
- Azuma N, Yamaguchi Y, Handa H, Hayakawa M, Kanai A, Yamada M. Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. Am J Hum Genet 1999; 65: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K, Ishigami C, Takahashi M, Park DH, Hirami Y, Nakanishi H et al. Two novel mutations in the EYS gene are possible major causes of autosomal recessive retinitis pigmentosa in the Japanese population. PLoS One 2012; 7: e31036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype-phenotype correlations. BMC Genet 2005; 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet 1994; 7: 463–471. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 1991; 354: 522–525. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol 1986; 97: 95–110. [PubMed] [Google Scholar]
- Solomon BD, Pineda-Alvarez DE, Balog JZ, Hadley D, Gropman AL, Nandagopal R et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A 2009; 149A: 2543–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S, Jinda W, Limwongse C, Atchaneeyasakul LO, Phadke SR. Run-on mutation in the PAX6 gene and chorioretinal degeneration in autosomal dominant aniridia. Mol Vis 2011; 17: 1305–1309. [PMC free article] [PubMed] [Google Scholar]
- Singh S, Tang HK, Lee JY, Saunders GF. Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J Biol Chem 1998; 273: 21531–21541. [DOI] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 2008; 26: 1663–1672. [DOI] [PubMed] [Google Scholar]
- Haubst N, Berger J, Radjendirane V, Graw J, Favor J, Saunders GF et al. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development 2004; 131: 6131–6140. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int J Dev Biol 2004; 48: 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocott T, Frost V, Maillard M, Johansen T, Wheeler GN, Dawes LJ et al. The MH1 domain of Smad3 interacts with Pax6 and represses autoregulation of the Pax6 P1 promoter. Nucleic Acids Res 2007; 35: 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier BR, Gosmain Y, Mamin A, Philippe J. The beta-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6. Biochem J 2007; 403: 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.