Abstract
Tuberculosis (TB) is a complex disease, and both genetic and environmental factors contribute to disease progression. A previous genome-wide linkage study in Thailand determined that chromosome 20p13-12.3 may contain risk factors for young-onset disease. The present study aimed to identify novel susceptibility genes for young-onset TB within a 1-Mbp target region adjacent to the top-ranking risk marker in Chr.20p13-12.3. We performed next-generation sequencing (NGS) of the region in 13 young patients from multi-case families in Thailand. We then selected the functionally interesting single-nucleotide polymorphisms as candidates for subsequent analyses. The detected candidates rs13830 and rs1127354 in ITPA showed an association with young (<45 years old) TB patients. However, there was no association in old (⩾45 years old) patients. These findings confirm that stratifying patients based on age of TB onset can be important for identifying genetic risk factors for TB susceptibility. In addition, in silico expression quantitative trait loci analyses indicated that ITPA expression was associated with rs13830 genotype. This is the first study to use NGS resequencing to gain insight into host genetic factors associated with TB and to report a significant association for ITPA with host susceptibility in young-onset TB. The study also demonstrated the effectiveness of NGS in identifying susceptibility genes in common diseases.
Introduction
Tuberculosis (TB) is one of the three major world-wide infectious diseases in addition to AIDS and malaria. According to the Global Tuberculosis Report 2014 produced by the World Health Organization, there was an estimated 9.0 million incident cases of TB (126 per 100,000 people) reported globally in 2013. Approximately, 1.5 million people were estimated to have died from TB that year.1 TB is a challenging world-health issue, especially because of co-infection with HIV and the existence of multidrug-resistant TB and extensively drug-resistant TB.1 It is estimated that approximately one-third of the world’s population is infected with the TB pathogen Mycobacterium tuberculosis. However, only 5–10% of infected patients will progress and develop the clinical disease.2
Several studies have been conducted to identify the genetic factors involved in TB susceptibility. An early twin study showed that monozygotic twins had a 2.5-fold higher concordance rate for TB when compared with dizygotic twins.3 A recent re-analysis of the twin study data concluded that environmental factors are more important than genetic factors to TB susceptibility. However, genetic factors have a determinate role in patient immune response to infection by M. tuberculosis.4 There are multiple putative TB-associated genes and loci that have been identified via a variety of methods. These methods include linkage studies, candidate gene studies and genome-wide association studies (GWAS).5 A GWAS conducted with an Indonesian cohort identified several potential TB susceptibility loci. However, none of the loci reached genome-wide significance.6 We previously conducted a single-nucleotide polymorphisms (SNP)-based genome-wide linkage study using a group of Thai patients and affected sibling-pair samples.7 The study identified a region on chromosome (Chr.) 20p that had a significant linkage with earlier TB onset based on an ordered subset analysis conducted using minimum age at TB onset. The maximum logarithm of odds score was 3.33.7 A GWAS was also conducted with a cohort of Thai and Japanese individuals. The study identified a risk locus on Chr.20q12 associated with young-onset (<45 years old) TB.8 The difficulty in identifying obvious and reproducible susceptibility loci in genetic studies suggests the presence of additional unknown genetic factors.
The current study attempted to identify new TB susceptibility gene(s) and/or variants within a candidate region on Chr. 20p in a cohort of young Thai individuals. We used next-generation sequencing (NGS) technology to resequence the candidate region. The candidate variants were then selected from the sequencing data for subsequent case–control association analyses.
Materials and methods
Study samples
There were 13 cases used for NGS. These cases were selected from Thai multiplex TB families that were used in our previous genome-wide linkage study.7 The target region for this resequencing study was Chr. 20p13-12.3 and this region shows significant linkage with early-onset TB. We define early-onset TB as disease occurrence in patients ranging from 12 to 23 years of age.7 The youngest affected individual <25 years old in each multiplex family was selected for sequencing.
There were two sample sets used for the association analyses. The first set of samples consisted of 665 TB patients and 777 healthy controls that were studied in the previous Thai GWAS.8 These individuals were recruited from the Chiang Rai, Lampang and Bangkok provinces. Microscopic identification and mycobacterial culture were used to confirm TB diagnosis in 98% of the TB cases. The second set of samples consisted of 545 TB patients and 407 controls recruited from Chiang Rai, Bangkok and Northern Thailand. These patients were also used in the association analysis. Individual and familial histories of TB and TB-associated diseases such as diabetes mellitus (DM) were evaluated. DM status was monitored via fasting blood sugar, HbA1c, rapid testing of capillary blood and history of DM treatment. The numbers of samples in each set are summarized in Supplementary Table S1. The study was approved by the Ethics Review Committees of the Ministry of Public Health in Thailand and the Faculty of Medicine, The University of Tokyo.
Candidate region selection
The region on Chr. 20p13-12.3 was previously demonstrated to be significantly linked with early-onset TB based on an ordered subset analysis.7 The SNP marker rs750702 located in this region shows a peak logarithm of odd score of 3.33. Thus, a 1-Mbp region centered on rs750702 was selected for the current study because it covers the linkage peak (Supplementary Figure S1).
Sequencing of TB cases
The samples were subjected to candidate region capture using the Ion TargetSeq Custom Enrichment Kit 500 kb-2 Mb (Thermo Fischer Scientific, Waltham, MA, USA). The kit was designed and customized by providing chromosomal physical position information on the target region to the manufacturer (Supplementary Information; Supplementary Figure S1).
The samples were prepared and sequenced on the Ion Torrent Personal Genome Machine (PGM; Thermo Fischer Scientific) according to manufacturer’s protocols. The samples’ genomic DNA (gDNA) were sheared into 200-bp fragments with the Ion Xpress Plus Fragment Library Kit (Thermo Fischer Scientific). The fragmented gDNA were ligated to sequencing adapters and individually barcoded using the Ion Xpress Barcode Adapters 1–16 Kit (Thermo Fischer Scientific). The prepared gDNA were then amplified. The Invitrogen SOLID Library Size Selection 2% E-gel (Thermo Fischer Scientific) and the E-Gel Safe Image Real-Time Transilluminator (Thermo Fischer Scientific) were used to select optimal-sized fragments with attached adapters and barcodes of ~330 bases in length. The fragments were then extracted from the gel before being purified and pooled to make sets of libraries consisting of ~500 ng of gDNA.
The Ion TargetSeq Custom Enrichment Kit was used to hybridize and capture the target region for each library set. The molarity of each hybridized library was measured with the Agilent 2100 Bioanalyzer High Sensitivity Kit (Agilent Technologies, Santa Clara, CA, USA). The libraries were then combined into three sequencing sets. The sequencing sets consisted of seven, six and five pooled-sample libraries. Sequencing templates were prepared from the pooled libraries by emulsion PCR with the Ion OneTouch (Thermo Fischer Scientific) and Ion OneTouch ES system (Thermo Fischer Scientific) using reagents from the Ion OneTouch 200 Template Kit v2 DL (Thermo Fischer Scientific).
The sequencing was conducted on the Ion Torrent PGM using the Ion PGM 200 Sequencing Kit and Ion 318 Chips (Thermo Fischer Scientific) according to manufacturer’s protocol.
Variant detection and selection of candidate variants
All sequencing data were analyzed with two software programs; Ion Variant Caller plugin (Thermo Fischer Scientific) and CLC Genomics Workbench v5.5 (Qiagen, Venlo, Limburg, Netherlands). The sequence data from the Ion Torrent PGM were subjected to the Ion Variant Caller plugin variant detection workflow. A separate analysis and variant detection workflow was created for the use on CLC Genomics Workbench v5.5 (Supplementary Information). The sequencing data were aligned to human genome build hg19 in both programs. The detected variants were required to have 20× read depth coverage in both software programs to be included for further analysis. These requirements helped to improve accuracy of detected variants.
Information on each variant was obtained from the UCSC Genome Browser (GRCh37/hg19 assembly) (http://genome.ucsc.edu/)9 and the HapMap10 database for the population of Han Chinese in Beijing, China (CHB). The detected variants observed in only one sequenced sample were excluded to increase stringency. Variants that were already studied in the Thai GWAS8 and the proxy SNPs (r 2⩾0.8 in CHB and Japanese in Tokyo, Japan (JPT) populations) were also excluded. The proxy SNPs were identified using the SNAP website Version 2.2,11 and the 1000 Genomes Project12 Pilot 1 data were used as a reference. The variants with minor allele frequency<0.05 in the HapMap database and/or 1000 Genome Project CHB population were also excluded. All remaining non-synonymous variants reported in exon regions were selected as candidates. A second filtering and selection step was conducted on the remaining variants with minor allele frequency⩾0.05. Candidate variants located in the 3ʹ-untranslated region (UTR) of genes were screened with two databases; microRNA.org—Targets and Expression (http://www.microrna.org/microrna/home.do)13,14 and miRDB (http://mirdb.org/miRDB/).15,16 This screening was used to determine whether the variants were located in predicted microRNA-binding sites. A prediction score >80 was set as the threshold for this selection. The variants located within predicted microRNA-binding sites were also listed as candidates. In addition, variants were also examined to determine whether they were located within DNaseI hypersensivity sites or gene regulatory sites (e.g., promoter regions) using HaploReg v2 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)17 and to prioritize functionally interesting variants. Variants located in a DNaseI hypersensivity site with motif change or regulatory regions were also included as candidates. The site of each candidate was examined with the UCSC Genome Browser9 and variants within regions with high histone H3 lysine 27 acetylation (H3K27Ac) markers were selected as candidates.
The candidate variants were validated by direct Sanger sequencing. Sequencing primers (Supplementary Table S2) were designed using Primer3 v.0.4.0 or Primer3web version 4.0.0.18,19 The amplification of sequencing amplicons was conducted on a GeneAmp PCR System 9700 (Thermo Fischer Scientific) or a TGradient Thermocycler (Biometra, Göttingen, Germany) using FastStart Taq DNA Polymerase, dNTPs, and 10× PCR Buffer with MgCl2 (Roche, Basel, Switzerland). The sequencing was conducted using an ABI PRISM 3130xl Genetic Analyzer (Thermo Fischer Scientific). The sequencing results were viewed with Sequence Scanner v1.0 (Thermo Fischer Scientific). Detailed protocols are described in the Supplementary Information.
TagSNPs were designed to capture the ITPA gene in detail for association analysis and was performed using Haploview 4.2 (Supplementary Information). The following SNPs were selected to cover the gene region: rs11087570, rs8362, and rs6139034.
Genotyping and association analysis
The candidate variants were genotyped by TaqMan assays in the first sample set. The variants rs1127354 and rs13830 were further genotyped in the additional sample set. The results from all sample sets were also analyzed together to improve statistical power. All genotyping probes were ordered from and manufactured by Applied Biosystems (Thermo Fischer Scientific) (Supplementary Table S3). The genotyping was conducted with KAPA PROBE FAST qPCR Master Mix (KAPA Biosystems, Wilmington, MA, USA). The Invader assay (Hologic, Bedford, MA, USA) was used to genotype rs13830 in the second sample set.
The Hardy–Weinberg equilibrium test was performed, and the χ 2-test was applied to evaluate differences between allele and genotype frequencies between all cases and controls. These tests were also performed for the subgroup analyses. In this study, allelic and genotypic models were used in addition to the dominant and recessive models tested. A subgroup analysis was also conducted. The cases were divided into two groups; a ‘young’ subgroup (<45 years old) and an ‘old’ subgroup (⩾45 years old). The subgroups were then compared with the controls. The 45-year-old threshold was also applied empirically in the previous GWAS and was based on the age of onset distribution for TB patients in the studied countries.8 A Fisher’s exact test was applied to improve statistical accuracy when subgroups had small sample numbers.
Linkage disequilibrium and haplotype analysis
Linkage disequilibrium and haplotype analyses were conducted using Haploview 4.2,20 and a 10,000-time permutation P value was calculated for each haplotype.
In silico SNP–gene expression quantitative trait loci analysis
An in silico expression quantitative trait loci (eQTL) analysis was conducted with Genevar software to assess the association between the SNP and gene expression.21 SNP–gene association and eQTL–gene analyses were conducted with expression profile data obtained from lymphoblastoid cell lines from 80 CHB subjects in the HapMap3 project. The NCBI36/Ensembl 50 database was used as the reference. A Spearman’s rank correlation coefficient (rho) with 10,000-time permutation was used as an analysis parameter.
Results
NGS coverage, candidate variant detection, and selection
The Ion TargetSeq Probe Kit coverage for the candidate region was reportedly 87.6% (Supplementary Figure S1). The mean raw accuracy and average sequencing target statistics are summarized in Supplementary Table S4. The numbers of detected variants in each sequenced sample are summarized in Supplementary Table S5. After between-sample duplicates were removed, there were 1,878 variants. There were seven variants selected as candidates for the association analysis after filtering all detected variants. The SNP rs13830 was not observed in any microRNA-binding site or DNaseI hypersensivity site. However, it was detected in the 3′-UTR of ITPA in more than one sample by NGS. This SNP (rs13830) was reported22 to have a high linkage disequilibrium with a detected non-synonymous variant rs1127354 in a Japanese population. The SNP rs13830 was included as a candidate and was examined for functional effects. Furthermore, there were more variants in ITPA than other genes. Thus, we examined the gene using tagSNPs. There were a total of 11 variants tested for association (Table 1).
Table 1. Candidate variants selected for genotyping.
| SNP | Gene | Site | Description |
|---|---|---|---|
| rs1127354 | ITPA | Exon | Non-synonymous variants detected by NGS |
| rs2280090 | ADAM33 | Exon | |
| rs17857295 | MAVS | Exon | |
| rs6115814 | ITPA | DHS | Functional variants detected variants by NGS |
| rs6116080 | PANK2 | DHS | |
| rs6084506 | PANK2 | DHS | |
| rs1132922 | MAVS | 3′-UTR | |
| rs11087570 | ITPA | Intron | TagSNPs covering ITPA gene |
| rs8362 | ITPA | Exon | |
| rs6139034 | ITPA | Intron | |
| rs13830 | ITPA | 3′-UTR | Additional variant |
Abbreviations: DHS, DNaseI hypersensitivity site; NGS, next-generation sequencing; SNP, single-nucleotide polymorphism; UTR, untranslated region.
Association, linkage disequilibrium, and haplotype analyses
Allelic, genotypic, dominant, and recessive models were used to analyze the genotyped variants. The subgroup analyses were performed by age and then compared with controls. There were no variants that showed deviation from the expected genotype counts in the Hardy–Weinberg equilibrium test. There were marginal associations observed for rs1127354 in the allelic and recessive models (P=0.015; odds ratio (OR)=1.41; 95% confidence interval (CI)=1.07–1.87 and P=0.013; OR=1.50; 95% CI=1.09–2.06, respectively) in the young subgroup for the non-synonymous variants detected by NGS. There were no significant associations for any other non-synonymous variants (Supplementary Table S6). The variant rs13830 showed marginal association in the allelic and recessive models (P=4.4E–03, OR=1.50, 95% CI=1.13–1.99 and P=3.7E–03, OR=1.61, 95% CI=1.16–2.22, respectively) in the young subgroup analysis (Supplementary Table S6). In addition, marginal associations were observed for rs6139034 in the recessive model in the old subgroup analysis (P=0.017, OR=1.37, 95% CI=1.06–1.79) and for rs8362 in the recessive model in the young subgroup analysis (P=0.024, OR=1.56, 95% CI=1.06–2.30; Supplementary Table S6).
No tested variants showed a stronger association than rs1127354 or rs13830. These two variants were selected for genotyping in the second sample set. The data from both sample sets were analyzed to increase statistical power. Both rs1127354 and rs13830 passed the Hardy–Weinberg equilibrium test and showed lower P values in the young subgroup for the allelic (P=1.3E–03, OR=1.39, 95% CI=1.14–1.70 and P=5.1E–05, OR=1.52, 95% CI=1.24–1.86, respectively) and recessive (P=1.1E–03, OR=1.47, 95% CI=1.17–1.85 and P=4.5E–05, OR=1.62, 95% CI=1.28–2.04, respectively) models (Table 2).
Table 2. Genotyping results for rs1127354 and rs13830 with first and second sample sets.
| Subgroup | Variant (alleles 1/2) | Sample set | Risk allele |
Risk allele freq.
|
Genotype counts (proportion %)
|
Allelic model
|
Recessive model
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control |
Case
|
Control
|
P value | OR (95% CI) | P value | OR (95% CI) | ||||||||
| 1/1 | 1/2 | 2/2 | 1/1 | 1/2 | 2/2 | ||||||||||
| All | rs1127354 (C/A) | 1st | C | 0.822 | 0.796 | 452 (68.1) | 188 (28.3) | 24 (3.6) | 488 (63.1) | 255 (33.0) | 30 (3.9) | 0.077 | 1.18 (0.98–1.43) | 0.050 | 1.25 (1.00–1.55) |
| 2nd | C | 0.846 | 0.813 | 389 (71.5) | 142 (26.1) | 13 (2.4) | 265 (66.3) | 120 (30.0) | 15 (3.8) | 0.058 | 1.26 (0.99–1.61) | 0.084 | 1.28 (0.97–1.69) | ||
| 1st + 2nd | C | 0.833 | 0.802 | 841 (69.6) | 330 (27.3) | 37 (3.1) | 753 (64.2) | 375 (32.0) | 45 (3.8) | 5.60E–03 | 1.23 (1.06–1.43) | 4.90E-03 | 1.28 (1.08–1.52) | ||
| rs13830 (G/A) | 1st | G | 0.814 | 0.788 | 447 (67.4) | 186 (28.1) | 30 (4.5) | 483 (62.2) | 258 (33.2) | 36 (4.6) | 0.073 | 1.18 (0.98–1.42) | 0.038 | 1.26 (1.01–1.57) | |
| 2nd | G | 0.847 | 0.805 | 390 (71.7) | 141 (25.9) | 13 (2.4) | 263 (64.6) | 129 (31.7) | 15 (3.7) | 0.017 | 1.34 (1.05–1.70) | 0.020 | 1.39 (1.05–1.83) | ||
| 1st + 2nd | G | 0.829 | 0.793 | 837 (69.3) | 327 (27.1) | 43 (3.6) | 746 (63.0) | 387 (32.7) | 51 (4.3) | 1.70E-03 | 1.26 (1.09–1.46) | 1.10E-03 | 1.33 (1.12–1.57) | ||
| Younga | rs1127354 (C/A) | 1st | C | 0.847 | 0.796 | 169 (71.9) | 60 (25.5) | 6 (2.6) | 488 (63.1) | 255 (33.0) | 30 (3.9) | 0.015 | 1.41 (1.07–1.87) | 0.013 | 1.50 (1.09–2.06) |
| 2nd | C | 0.851 | 0.813 | 189 (73.0) | 63 (24.3) | 7 (2.7) | 265 (66.3) | 120 (30.0) | 15 (3.8) | 0.068 | 1.32 (0.98–1.78) | 0.069 | 1.38 (0.98–1.94) | ||
| 1st + 2nd | C | 0.849 | 0.802 | 358 (72.5) | 123 (24.9) | 13 (2.6) | 753 (64.2) | 375 (32.0) | 45 (3.8) | 1.30E–03 | 1.39 (1.14–1.70) | 1.10E-03 | 1.47 (1.17–1.85) | ||
| rs13830 (G/A) | 1st | G | 0.848 | 0.788 | 169 (72.5) | 57 (24.5) | 7 (3.0) | 483 (62.2) | 258 (33.2) | 36 (4.6) | 4.40E–03 | 1.50 (1.13–1.99) | 3.70E-03 | 1.61 (1.16–2.22) | |
| 2nd | G | 0.859 | 0.805 | 192 (74.1) | 61 (23.6) | 6 (2.3) | 263 (64.6) | 129 (31.7) | 15 (3.7) | 0.011 | 1.48 (1.09–2.00) | 0.010 | 1.57 (1.11–2.21) | ||
| 1st + 2nd | G | 0.854 | 0.793 | 361 (73.4) | 118 (24.0) | 13 (2.6) | 746 (63.0) | 387 (32.7) | 51 (4.3) | 5.10E–05 | 1.52 (1.24–1.86) | 4.50E-05 | 1.62 (1.28–2.04) | ||
| Olda | rs1127354 (C/A) | 1st | C | 0.809 | 0.796 | 283 (66.0) | 128 (29.8) | 18 (4.2) | 488 (63.1) | 255 (33.0) | 30 (3.9) | 0.458 | 1.08 (0.88–1.34) | 0.326 | 1.13 (0.88–1.45) |
| 2nd | C | 0.840 | 0.813 | 200 (70.2) | 79 (27.7) | 6 (2.1) | 265 (66.3) | 120 (30.0) | 15 (3.8) | 0.182 | 1.21 (0.91–1.62) | 0.278 | 1.20 (0.86–1.66) | ||
| 1st + 2nd | C | 0.821 | 0.802 | 483 (67.6) | 207 (29.0) | 24 (3.4) | 753 (64.2) | 375 (32.0) | 45 (3.8) | 0.136 | 1.14 (0.96–1.35) | 0.126 | 1.17 (0.96–1.42) | ||
| rs13830 (G/A) | 1st | G | 0.797 | 0.788 | 278 (64.7) | 129 (30.0) | 23 (5.3) | 483 (62.2) | 258 (33.2) | 36 (4.6) | 0.608 | 1.06 (0.86–1.30) | 0.391 | 1.11 (0.87–1.42) | |
| 2nd | G | 0.835 | 0.805 | 198 (69.5) | 80 (28.1) | 7 (2.5) | 263 (64.6) | 129 (31.7) | 15 (3.7) | 0.150 | 1.23 (0.93–1.63) | 0.183 | 1.25 (0.90–1.72) | ||
| 1st + 2nd | G | 0.812 | 0.793 | 476 (66.6) | 209 (29.2) | 30 (4.2) | 746 (63.0) | 387 (32.7) | 51 (4.3) | 0.169 | 1.12 (0.95–1.33) | 0.116 | 1.17 (0.96–1.42) | ||
Abbreviations: CI, confidence interval; freq., frequency; OR, odds ratio.
Allelic model: difference in allele frequencies between cases and controls (1 vs 2).
Recessive model: homozygous risk allele versus heterozygous and homozygous non-risk alleles (1/1 vs. 1/2+2/2).
‘Young’ cases were those <45 years old and ‘old’ cases were those ⩾45 years old.
The non-synonymous variant rs1127354 showed a strong linkage disequilibrium with rs13830 (r 2=0.88; Figure 1). There were no tested haplotypes that reached significant permutated P values and none of the haplotypes had P values lower than the SNPs rs13830 and rs1127354 (Supplementary Tables S7 and S8).
Figure 1.
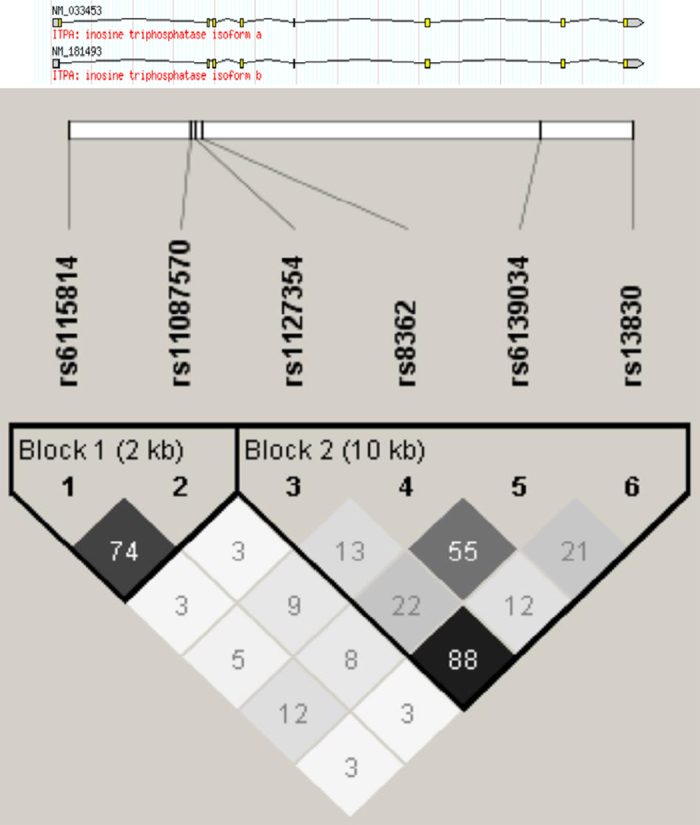
LD structure of variants on ITPA. LD structure of variants on the ITPA gene in the genotyped ‘young’ (<45 years old) Thai population. The numbers in each box show the r 2 value between variants. The variants rs1127354 and rs13830 showed strong LD (r 2=0.88). LD, linkage disequilibrium.
In silico eQTL analysis
The SNP–gene association analysis for ITPA and rs13830 showed a significant correlation between expression level of ITPA rs13830 genotype (P perm=0.0048). Lower expression was associated with the ‘G’ allele (Figure 2).
Figure 2.
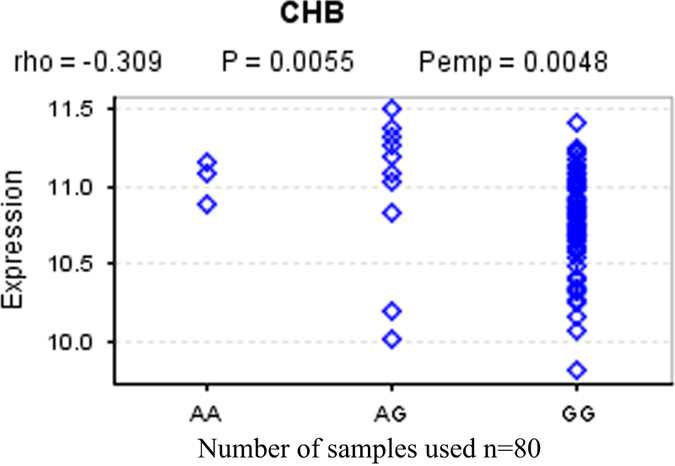
Correlation between rs13830 genotype and ITPA expression. Differential expression levels of ITPA gene for the different genotypes. The risk ‘G’ allele is observed to have significantly lower expression compared with the ‘A’ allele.
Discussion
There are no previously published studies using NGS to search for human genetic factors that affect TB susceptibility. NGS has largely been used for genetic studies of the pathogen M. tuberculosis.23,24 The current study is the first attempt to use NGS to gain insight into host genetic factors associated with TB. However, analyses of data obtained from NGS have always posed a challenge.25,26 In the current study, the number of variants detected by NGS in each sequenced sample for the 1-Mbp region was large. As a result, the variants were filtered and selected using the described methods (Supplementary Information).
The variants rs1127354 (missense) and rs13830 (3′-UTR) located in the ITPA gene showed moderate association with TB susceptibility. The variant rs13830 showed the strongest association. These findings were observed primarily in the ‘young’ subgroup analysis. The association was not as prominent when all age groups were included in the analysis. This finding supports the results from previous studies that show stratification by TB age of onset effectively identifies genetic factors associated with TB susceptibility.7,8,27
ITPA encodes the enzyme inosine triphosphate pyrophosphatase, which functions to catalyze the hydrolysis of inosine triphosphate to inosine monophosphate and pyrophosphate.28,29 The role of inosine triphosphate pyrophosphatase in humans is not well defined. However, it is important for the maintenance of genomic stability by preventing DNA damage and mutagenesis in human cells.30,31 A previous study reported that there was a relationship between low ITPA activity and adverse effects of the immunosuppressive drug azathioprine.32 In addition, it has been speculated that ITPA has a role in immunity.33 The variant rs13830 is located in the 3′-UTR of ITPA and polymorphisms in the 3′-UTR of a gene may affect regulation of messenger RNA transcripts and gene expression.34–36 The eQTL analysis showed that the expression level of ITPA in lymphocyte cell lines differed according to rs13830 genotype, with higher expression in the minor ‘A’ allele. The understanding of TB disease progression has largely focused on the host immune response and the roles of T cells37 and B cells in response to M. tuberculosis infection.38,39 The results observed in our association and in silico eQTL analyses indicate that expression of ITPA may be affected in immune cells and is associated with rs13830 genotype. The association with young-onset TB observed with the ‘G’ allele suggests that ITPA could have a role in host immune response and subsequent progression of TB. Furthermore, investigations in the UCSC genome browser (http://genome.ucsc.edu/) Gene Sorter database40 revealed that expression of ITPA is high in peripheral blood CD4+ T cells and in the lungs. Thus, it may be reasonable to speculate that the risk ‘G’ allele of rs13830 may affect translation and/or regulation of ITPA and reduce expression. The decreased expression impairs the host immune response to M. tuberculosis infection in young-onset cases. It has been suggested that genetic effects that affect susceptibility to infectious diseases may be stronger in younger patients than in older patients.41 The results of the association analysis for rs13830 may reflect the genetic effect of rs13830 for TB susceptibility. These results suggest that it has a more profound role for young-onset TB cases than in older patients. Older patients may be susceptible due to other factors such as a compromised immune reaction to M. tuberculosis infection due to age or secondary infection.
Although the Chr. 20q12 locus was identified as a risk locus in the previous Thai GWAS,8 our analysis of the association results in the region showed no significant associations. The variants studied here and their proxies were not included in the Thai GWAS. This finding indicates that the filtering and selection method employed here can successfully identify novel candidate variants.
There are several limitations in the current study. The P values of rs13830 and rs1127354 did not reach genome-wide significance, which might be due to the limited sample size of the ‘young’ subgroup. The associations should be confirmed by increasing the sample numbers and/or conducting replication studies in other Asian populations that are genetically close to the Thai population. The possibility that rs13830 is not the causative variant cannot be excluded. Thus, further investigations of rs13830 proxies are warranted.
In conclusion, this study has demonstrated that the current NGS method can be coupled with rigorous filtering and selection processes. Our approach can successfully identify novel genetic susceptibility loci and contribute to the elucidation of genetic factors with roles in TB disease progression. In addition, targeted resequencing methods may also reveal unidentified susceptibility loci for other common diseases. This is the first report of a potential association of ITPA with young age-at-onset TB in the Thai population. The findings may improve our understanding of TB pathogenesis and may be useful for studies that attempt to uncover effective drug targets.
Acknowledgments
This work was partly supported by a Grant in Aid for Scientific Research (B), KAKENHI Grant Number 24406010 and 15H05271, and SATREPS from Japan Agency for Medical Research and Development and Japan International Cooperative Agency. We also used the remaining samples from the previous studies collected by the TB/HIV research project in Thailand, which is a collaborative research project under the Research Institute of Tuberculosis, Japan Anti-tuberculosis Association and is supported by the Ministry of Health, Labor, and Welfare of Japan and the Department of Medical Sciences, Ministry of Public Health of Thailand.
The authors declare no conflict of interest.
Footnotes
Supplementary Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv)
References
- World Health Organization. Global Tuberculosis Report 2014. Available at http://www.who.int/tb/publications/global_report/gtbr14_main_text.pdf?ua=1.
- Vynnycky E , Fine PE . Lifetime risks, incubation period, and serial interval of tuberculosis. Am J Epidemiol 2000; 152: 247–263. [DOI] [PubMed] [Google Scholar]
- Comstock GW . Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis 1978; 117: 621–624. [DOI] [PubMed] [Google Scholar]
- van der Eijk EA , van de Vosse E , Vandenbroucke JP , van Dissel JT . Heredity versus environment in tuberculosis in twins: the 1950s United Kingdom Prophit Survey Simonds and Comstock revisited. Am J Respir Crit Care Med 2007; 176: 1281–1288. [DOI] [PubMed] [Google Scholar]
- Qu HQ , Fisher-Hoch SP , McCormick JB . Knowledge gaining by human genetic studies on tuberculosis susceptibility. J Hum Genet 2011; 56: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png E , Alisjahbana B , Sahiratmadja E , Marzuki S , Nelwan R , Balabanova Y et al. A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Med Genet 2012; 13: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahasirimongkol S , Yanai H , Nishida N , Ridruechai C , Matsushita I , Ohashi J et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun 2009; 10: 77–83. [DOI] [PubMed] [Google Scholar]
- Mahasirimongkol S , Yanai H , Mushiroda T , Promphittayarat W , Wattanapokayakit S , Phromjai J et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet 2012; 57: 363–367. [DOI] [PubMed] [Google Scholar]
- Kent WJ , Sugnet CW , Furey TS , Roskin KM , Pringle TH , Zahler AM et al. The human genome browser at UCSC. Genome Res 2002; 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. The International HapMap Project. Nature 2003; 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Johnson AD , Handsaker RE , Pulit SL , Nizzari MM , O'Donnell CJ , de Bakker PI . SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis GR , Auton A , Brooks LD , DePristo MA , Durbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B , Enright AJ , Aravin A , Tuschl T , Sander C , Marks DS . Human MicroRNA targets. PLoS Biol 2004; 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D , Wilson M , Gabow A , Marks DS , Sander C . The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X . miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 2008; 14: 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X , El Naqa IM . Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 2008; 24: 325–332. [DOI] [PubMed] [Google Scholar]
- Ward LD , Kellis M . HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A , Cutcutache I , Koressaar T , Ye J , Faircloth BC , Remm M et al. Primer3--new capabilities and interfaces. Nucleic Acids Res 2012; 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T , Remm M . Enhancements and modifications of primer design program Primer3. Bioinformatics 2007; 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Barrett JC , Fry B , Maller J , Daly MJ . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Yang TP , Beazley C , Montgomery SB , Dimas AS , Gutierrez-Arcelus M , Stranger BE et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 2010; 26: 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y , Kurosaki M , Nishida N , Sugiyama M , Matsuura K , Sakamoto N et al. Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic hepatitis C. Hum Mol Genet 2011; 20: 3507–3516. [DOI] [PubMed] [Google Scholar]
- Loman NJ , Pallen MJ . XDR-TB genome sequencing: a glimpse of the microbiology of the future. Future Microbiol 2008; 3: 111–113. [DOI] [PubMed] [Google Scholar]
- Roetzer A , Diel R , Kohl TA , Rückert C , Nübel U , Blom J et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 2013; 10: e1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J , Ji H . Next-generation DNA sequencing. Nat Biotechnol 2008; 26: 1135–1145. [DOI] [PubMed] [Google Scholar]
- Pop M , Salzberg SL . Bioinformatics challenges of new sequencing technology. Trends Genet 2008; 24: 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant AV , El Baghdadi J , Sabri A , El Azbaoui S , Alaoui-Tahiri K , Abderrahmani Rhorfi I et al. Age-dependent association between pulmonary tuberculosis and common TOX variants in the 8q12-13 linkage region. Am J Hum Genet 2013; 92: 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SL , Turner BM , Hirschhorn K . Human inosine triphosphatase: catalytic properties and population studies. Clin Chim Acta 1979; 97: 143–153. [DOI] [PubMed] [Google Scholar]
- Sumi S , Marinaki AM , Arenas M , Fairbanks L , Shobowale-Bakre M , Rees DC et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet 2002; 111: 360–367. [DOI] [PubMed] [Google Scholar]
- Lin S , McLennan AG , Ying K , Wang Z , Gu S , Jin H et al. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene. J Biol Chem 2001; 276: 18695–18701. [DOI] [PubMed] [Google Scholar]
- Menezes MR , Waisertreiger IS , Lopez-Bertoni H , Luo X , Pavlov YI . Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells. PLoS ONE 2012; 7: e32313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipkova M , Franz J , Abe M , Klett C , Wieland E , Andus T . Association between adverse effects under azathioprine therapy and inosine triphosphate pyrophosphatase activity in patients with chronic inflammatory bowel disease. Ther Drug Monit 2011; 33: 321–328. [DOI] [PubMed] [Google Scholar]
- Bakker JA , Bierau J , Drent M . A role for ITPA variants in the clinical course of pulmonary Langerhans' cell histiocytosis? Eur Respir J 2010; 36: 684–686. [DOI] [PubMed] [Google Scholar]
- Song C , Chen LZ , Zhang RH , Yu XJ , Zeng YX . Functional variant in the 3'- untranslated region of Toll-like receptor 4 is associated with nasopharyngeal carcinoma risk. Cancer Biol Ther 2006; 5: 1285–1291. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A , Li L , Gaedigk A , Bradford LD , Benson EA , Flockhart DA et al. In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4α expression. Drug Metab Dispos 2012; 40: 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak E , Gebauer F . Translational control by 3'-UTR-binding proteins. Brief Funct Genomics 2013; 12: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JD . The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012; 12: 581–591. [DOI] [PubMed] [Google Scholar]
- Maglione PJ , Chan J . How B cells shape the immune response against Mycobacterium tuberculosis. Eur J Immunol 2009; 39: 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J , Mehta S , Bharrhan S , Chen Y , Achkar JM , Casadevall A et al. The role of B cells and humoral immunity in Mycobacterium tuberculosis infection. Semin Immunol 2014; 26: 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ , Hsu F , Karolchik D , Kuhn RM , Clawson H , Trumbower H et al. Exploring relationships and mining data with the UCSC Gene Sorter. Genome Res 2005; 15: 737–741, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A , Quintana-Murci L , Thaler DS , Schurr E , Abel L , Casanova JL . Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann NY Acad Sci 2010; 1214: 18–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


