Abstract
Biogenesis of the 12-subunit RNA polymerase II (Pol II) transcription complex requires so-called GPN-loop GTPases, but the function of these enzymes is unknown. Here we report the first crystal structure of a eukaryotic GPN-loop GTPase, the Saccharomyces cerevisiae enzyme Npa3 (a homolog of human GPN1, also called RPAP4, XAB1, and MBDin), and analyze its catalytic mechanism. The enzyme was trapped in a GDP-bound closed conformation and in a novel GTP analog-bound open conformation displaying a conserved hydrophobic pocket distant from the active site. We show that Npa3 has chaperone activity and interacts with hydrophobic peptide regions of Pol II subunits that form interfaces in the assembled Pol II complex. Biochemical results are consistent with a model that the hydrophobic pocket binds peptides and that this can allosterically stimulate GTPase activity and subsequent peptide release. These results suggest that GPN-loop GTPases are assembly chaperones for Pol II and other protein complexes.
INTRODUCTION
Protein folding was studied mainly for individual proteins (1), but most eukaryotic proteins form complexes, and their assembly is often poorly understood (2). A prominent eukaryotic protein complex is RNA polymerase II (Pol II), a 12-subunit, 520-kDa enzyme that carries out transcription of protein-encoding genes. The structure and function of Pol II have been studied extensively (3), but little is known about its biogenesis. Assembly of Pol II from its 12 subunits apparently occurs in the cytoplasm because the depletion of any Pol II subunit leads to cytoplasmic accumulation of the largest subunit, Rpb1 (4). Assembly of the 10-subunit Pol II core likely involves three subcomplexes, one containing Rpb1 and its associated subunits Rpb5, Rpb6, and Rpb8; one containing the second-largest subunit, Rpb2, and its associated subunit Rpb9; and one containing Rpb3 and its associated subunits Rpb10, Rpb11, and Rpb12 (5). Assembled Pol II binds the import factor Iwr1 between Rpb1 and Rpb2 and is imported to the nucleus (6).
Thus far, Pol II could not be reconstituted in vitro, indicating that its assembly in vivo depends on the help of several factors. Indeed, recent studies identified putative Pol II assembly factors, including the chaperone Hsp90, the RPAP3/R2TP/Prefoldin-like complex, and a recently discovered subfamily of GTPases, the so-called GPN-loop GTPases (4, 7, 8). Eukaryotic cells contain three paralogs of these essential GTPases. In human, these are called GPN1 (also RPAP4, XAB1, or MBDin), GPN2, and GPN3 (also Parcs). Archaea contain a single GPN-loop GTPase, and prokaryotes lack homologs. In the yeast Saccharomyces cerevisiae, deletion of the GPN1 homolog Npa3 or its paralogs GPN2 (YOR262W) and GPN3 (YLR243W) is lethal (9), indicating essential, nonredundant functions of these enzymes. Homo- and heterodimerization of GPN1 and its paralogs were reported previously (10–12). Npa3 contains a nuclear export sequence (NES) (residues 286 to 295) (13), consistent with the predominant cytoplasmic localization of Npa3 in yeast (14, 15) and GPN1 in human cells (7, 13, 16, 17).
The family of GPN-loop GTPases is characterized by a highly conserved motif consisting of the amino acids Gly-Pro-Asn (single-letter code, GPN). Structural studies of the archaeal GPN-loop GTPase Pab0955 suggested that the GPN motif functions in GTP hydrolysis (18). Nucleotide binding and hydrolysis of all GTPases involve five so-called G motifs (G1 to G5) (19). Mutation in the nucleotide-binding site or GPN motif in human GPN1 leads to cytoplasmic accumulation or decreased nuclear levels of Rpb1 (7, 11). Depletion of human GPN3 (20) or mutation of yeast GPN2 or GPN3 (12) also leads to the cytoplasmic accumulation of Rpb1, indicating a general role of all three GPN-loop GTPases in Pol II biogenesis. Depletion of the GPN1 homolog Npa3 from the yeast Saccharomyces cerevisiae leads to the cytoplasmic accumulation of Rpb1 and Rpb3 (21). Rpb1 accumulation is also observed when Npa3 is mutated in its nucleotide-binding site or GPN motif (7, 21). The association of yeast Npa3 with Rpb1 is regulated by GTP binding in whole-cell extracts (21), and a direct interaction of human GPN1 and GPN3 with the recombinant Pol II subunits Rpb4 and Rpb7 and the C-terminal repeat domain (CTD) of Rpb1 has been reported (11).
To gain insights into the structure and function of GPN-loop GTPases and their role in Pol II biogenesis, we studied the yeast GPN1 enzyme Npa3 with a combination of X-ray crystallography, site-directed mutagenesis, enzymatic activity assays, chaperone assays, and a systemic peptide interaction screen. Our results indicate that Npa3 functions as an assembly chaperone during Pol II biogenesis and binds hydrophobic regions in Pol II subunits that are released upon GTP hydrolysis to form interfaces in the mature polymerase complex.
MATERIALS AND METHODS
Npa3 expression and purification.
Wild-type Npa3 from Saccharomyces cerevisiae was amplified from genomic DNA and subcloned into the pOPINI vector (22) (provided by Oxford Protein Production Facility—United Kingdom [OPPF-UK]) containing an N-terminal hexahistidine tag. Mutations were introduced by overlap extension PCR from wild-type Npa3 plasmid DNA, and mutants were subcloned into the pOPINE vector (22) (provided by OPPF-UK) containing a C-terminal hexahistidine tag. Variants of Npa3 were expressed in Escherichia coli Rosetta(DE3) cells (Novagen). The culture was grown in LB medium at 37°C until an absorbance at 600 nm of 0.6 was reached, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the culture was grown for a further 20 h at 20°C. Cells were harvested by centrifugation and frozen at −20°C. Cells were lysed by sonication in buffer A (50 mM Tris [pH 7.5], 300 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol [DTT] supplemented with 5 mM imidazole, 0.2% [vol/vol] Tween 20, and 1× protease inhibitors [100× stock containing 1.42 mg leupeptin, 6.85 mg pepstatin A, 850 mg phenylmethylsulfonyl fluoride {PMSF}, and 1.685 mg benzamidine in 50 ml ethanol]). After centrifugation at 24,000 × g for 30 min, the cleared lysate was loaded onto a 2-ml Ni-nitrilotriacetic acid (Ni-NTA) column (Qiagen) preequilibrated with buffer A containing 10 mM imidazole. The column was washed with 10 column volumes of buffer A containing 10 mM imidazole before elution of the bound protein with buffer A containing 200 mM imidazole. The conductivity of the eluate was adjusted to match that of buffer B (50 mM Tris [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 2 mM DTT), and the eluate was applied to a MonoQ 10/100 GL column (Amersham) equilibrated in buffer B. The protein was eluted with a linear gradient from 100 mM to 1 M NaCl. After concentration, the sample was applied to a HiLoad 16/600 Superdex 200-pg column (GE Healthcare) equilibrated with buffer C (10 mM HEPES [pH 7.5], 5 mM MgCl2, 10 mM DTT) containing either 100 mM NaCl for wild-type Npa3 and full-length Npa3 mutants or 200 mM NaCl for Npa3ΔCΔLoop. Peak fractions were pooled and concentrated as desired.
Crystallization and X-ray structure analysis of Npa3ΔCΔLoop.
The Npa3ΔCΔLoop protein was concentrated to ∼3.7 mg/ml and incubated at 8°C overnight with either 10 mM GDP (Sigma-Aldrich), 5 mM GDP plus 100 mM NaF–10 mM AlCl3, or 10 mM GMPPCP (Jena Bioscience). Npa3ΔCΔLoop-GDP crystals were grown at 20°C by sitting-drop vapor diffusion after 6 to 15 days using a solution containing 9 mM HEPES (pH 7), 45 mM NaCl, 4.5 mM MgCl2, and 5% (vol/vol) Jeffamine M-600 as the reservoir solution. Npa3ΔCΔLoop-GDP-AlFx was crystallized at 8°C by hanging-drop vapor diffusion with buffer C containing 200 mM NaCl, 100 mM NaF, and 10 mM AlCl3 as the reservoir solution. Npa3ΔCΔLoop-GMPPCP crystals were grown overnight at 8°C in a 1.5-ml Eppendorf tube in buffer C containing 200 mM NaCl. Crystals grew to a maximum size of ∼0.25 by 0.25 by 0.1 mm under all conditions. Cryoprotection was carried out by stepwise transfer to mother solution containing 35% (vol/vol) glycerol before flash-cooling in liquid nitrogen. A single anomalous diffraction experiment from intrinsic sulfur atoms (S-SAD) was performed on Npa3ΔCΔLoop-GDP-AlFx crystals at beamline X06DA of the Swiss Light Source in Villigen, Switzerland (Table 1). Diffraction data were collected at 100 K and a wavelength of 2.066 Å at three different χ angles (0°, 10°, and 20°), as described recently (23). A high-resolution native data set of Npa3ΔCΔLoop-GDP-AlFx was collected from the same crystal at the same beamline. Diffraction data for GDP- and GMPPCP-bound Npa3ΔCΔLoop were collected at 100 K at beamline MX1 at the European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron (EMBL/DESY) in Hamburg, Germany (Table 1). Raw data were processed with XDS (24). The programs SHELXC, SHELXD, and SHELXE from the SHELX suite (25) were used for the detection of 17 sulfur atoms and for SAD phasing. The GDP-bound structure was solved by molecular replacement using PHASER (26) with the Npa3ΔCΔLoop-GDP-AlFx core structure as the search model. The GMPPCP-bound structure was refined by using the Npa3ΔCΔLoop-GDP-AlFx structure with phenix.refine (27). Models were iteratively built with COOT (28) and refined with phenix.refine. Figures were prepared with PyMOL (DeLano Scientific).
TABLE 1.
X-ray diffraction data collection and refinement statistics
| Parameter | Value for data setd |
|||
|---|---|---|---|---|
| Native |
S-SAD GDP-AlFxb,c | |||
| GDPa | GMPPCPa | GDP-AlFxb | ||
| Data collection statistics | ||||
| Space group | C2221 | P41212 | P41212 | P41212 |
| Cell dimensions | ||||
| a, b, c (Å) | 108.0, 119.2, 347.5 | 116.2, 116.2, 56.8 | 116.2, 116.2, 55.9 | 116.2, 116.2, 55.9 |
| Wavelength (Å) | 0.99988 | 0.99888 | 1.0001 | 2.066 |
| Resolution (Å) | 50–2.3 (2.36–2.3) | 50–2.2 (2.26–2.2) | 80–1.85 (1.90–1.85) | 50–2.15 (2.21–2.15)c |
| Rsym (%) | 5.7 (126.9) | 4.5 (155) | 3.8 (145.6) | 5.2 (89.8) |
| I/σI | 20.16 (1.50) | 28.50 (2.07) | 38.32 (2.16) | 66.37 (2.00) |
| Completeness (%) | 99.7 (99.8) | 99.5 (98.0) | 100 (100) | 98.2 (79.9) |
| Redundancy | 7.15 (6.91) | 14.11 (13.12) | 14.39 (14.42) | 63.07 (7.31) |
| CC1/2e (%) | 100 (69.0) | 100 (83.2) | 100 (72.0) | 100 (62.3) |
| Refinement statistics | ||||
| Resolution (Å) | 45.3–2.3 | 36.8–2.2 | 46.2–1.85 | |
| No. of reflections | 99,343 | 20,172 | 33,217 | |
| Rwork/Rfree (%) | 23.8/27.6 | 21.4/22.7 | 20.1/23.9 | |
| No. of atoms | ||||
| Protein | 12,065 | 1,998 | 2,036 | |
| Nucleotide | 168 | 32 | 28 | |
| AlFx | 4 | |||
| Mg2+ | 6 | 1 | 1 | |
| Lauric acid | 14 | 14 | ||
| Glycerol | 24 | 12 | 12 | |
| Water | 121 | 5 | 108 | |
| B factors (Å2) | ||||
| Protein | 83.5 | 121.2 | 59.4 | |
| Nucleotide | 69.6 | 123.6 | 57.6 | |
| AlFx | 107.1 | |||
| Mg2+ | 64.2 | 166.6 | 104.7 | |
| Lauric acid | 108.0 | 49.7 | ||
| Glycerol | 107.7 | 145.2 | 63.8 | |
| Water | 60.1 | 110.7 | 53.5 | |
| RMS deviationsf | ||||
| Bond length (Å) | 0.011 | 0.007 | 0.008 | |
| Bond angle (°) | 1.29 | 1.15 | 1.18 | |
Diffraction data were collected at beamline MX1 at EMBL/DESY, Hamburg, Germany, and processed with XDS (24).
Diffraction data were collected at beamline X06DA, Swiss Light Source, Switzerland, and processed with XDS (24).
Data are merged from three data sets, measured at different χ angles (0°, 10°, and 20°) from a single crystal.
Numbers in parentheses refer to the highest-resolution shell.
CC1/2 is the percentage of correlation between intensities from random half-data sets (53).
RMS, root mean square.
GTPase activity assay.
GTPase activity was measured in 96-well plates by using the Malachite Green phosphate assay kit (BioAssay Systems, Hayward, CA) as recommended by the manufacturer. A total of 0.15 μM Npa3 variants was incubated with 100 μM GTP (Fermentas, St. Leon Rot, Germany) at 37°C in buffer C containing 200 mM NaCl. GTPase activity in the presence of partially unfolded citrate synthase (CS) was measured at 43°C in buffer C containing 100 mM NaCl, pH 7.5 (43°C). Orthophosphate concentrations from three independent experiments were determined at the indicated time points by measuring the absorption at 620 nm using an Infinite M1000 plate reader (Tecan, Männedorf, Switzerland). Control experiments were performed to determine orthophosphate contaminations in individual solutions, and data were corrected for these values.
Analysis of chaperone activity.
Thermal aggregation of CS was essentially carried out as described previously (29), with modifications. A total of 0.15 μM CS (monomer) was incubated in either preheated (43°C) incubation buffer IB (40 mM HEPES-KOH [pH 7.5] [43°C], 5 mM MgCl2) in the absence of nucleotides or preheated (43°C) buffer C containing 100 mM NaCl (pH 7.5) (43°C) in the presence of 1 mM GMPPCP, GTP, or GDP, and various amounts of Npa3 were added. For experiments where GMPPCP or GDP was added, Npa3 samples were incubated with the respective nucleotide overnight at 4°C for nucleotide exchange. CS aggregation was measured by light scattering at 360 nm using a Fluoromax 3 fluorometer (Horiba, Unterhaching, Germany). Data represent the averages of data from three independent experiments. Control experiments were performed to show that Npa3 does not aggregate under the assay conditions (not shown).
Pol II peptide-binding assay.
An array of 1,139 15-mer peptides covering the complete sequence of RNA polymerase II with an overlap of 11 amino acids was synthesized, and triplicates were N-terminally immobilized on a glass surface via a Ttds (1,13-diamino-4,7,10-trioxatridecan-succinic acid) linker by JPT Peptide Technologies, Berlin, Germany. Purified N-terminally hexahistidine-tagged wild-type Npa3 was preincubated with either 10 mM GMPPCP or 10 mM GDP overnight at 4°C in buffer C containing 100 mM NaCl. Incubation with 10 mM GTP was started 5 min prior to the experiment. Microarray incubations and data analysis were carried out by JPT Peptide Technologies. Microarrays were blocked with blocking buffer (Superblock TBS T20; Pierce International) for 60 min, washed with Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBS-T), and incubated with 1 mg/ml of the respective Npa3 sample for 60 min at 4°C. After an additional TBS-T washing step, 0.4 μg/ml penta-His Alexa Fluor 647-labeled antibody (Qiagen) was incubated for 45 min, followed by washing with 3 mM SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer (JPT Peptide Technologies, Berlin, Germany). The microarrays were dried, and the fluorescence signal was analyzed using a GenePix 4200AL scanner (Molecular Devices) and GenePix spot recognition software by JPT Peptide Technologies. In the case of false-positive binding, neighboring overlapping peptides containing partially the same sequence were also not taken into consideration.
Protein structure accession numbers.
Coordinates of the Npa3-GDP and Npa3-GMPPCP complex structures have been deposited with the Protein Data Bank (PDB) under accession numbers 5HCI and 5HCN, respectively.
RESULTS
Domain organization and structure determination.
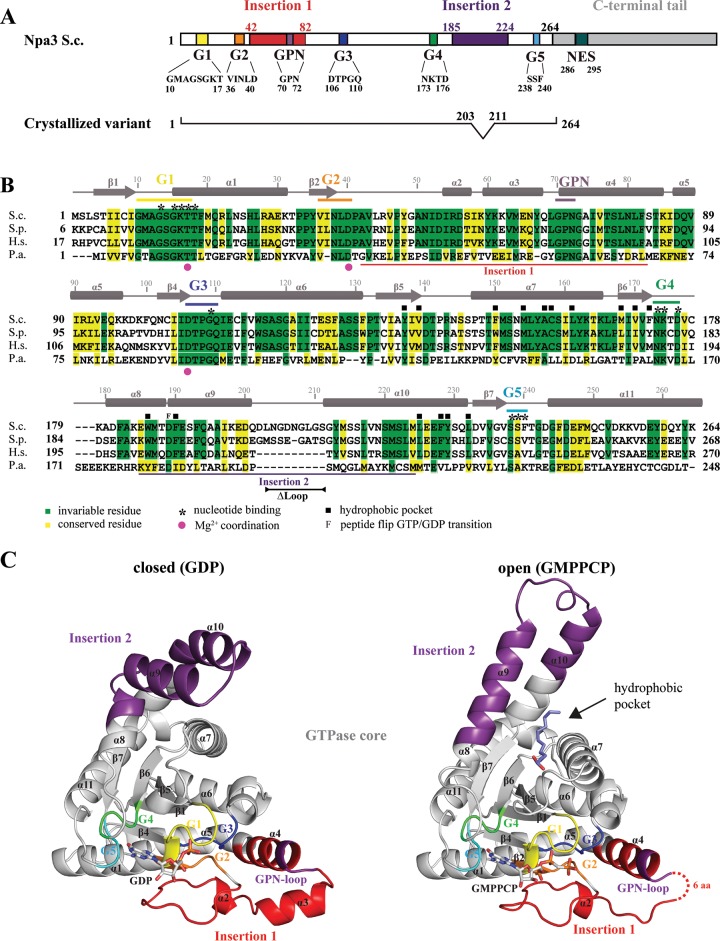
Npa3 contains 385 amino acid residues and consists of a central GTPase core (residues 1 to 41, 83 to 184, and 225 to 263), two protein insertions (“insertion 1” and “insertion 2,” containing residues 42 to 82 and 185 to 224, respectively), and a C-terminal tail (residues 264 to 385) (Fig. 1A). The GTPase core harbors motifs G1 to G5 that are required for GTP binding and hydrolysis (19). The C-terminal tail is poorly conserved among eukaryotes and is absent in archaea. Because efforts to crystallize full-length Npa3 failed, we removed a nonconserved C-terminal tail and part of a loop (residues 203 to 211 in insertion 2) that is predicted to be disordered and absent in most eukaryotic homologs (variant Npa3ΔCΔLoop, comprising residues 1 to 202 and 212 to 264). We cocrystallized Npa3ΔCΔLoop (Fig. 1A) with the nonhydrolyzable GTP analog GMPPCP but also with GDP and with GDP and AlFx (“GDP-AlFx”).
FIG 1.
Crystal structures of Npa3 in GDP-bound (closed) and GMPPCP-bound (open) forms. (A) Schematic representation of yeast (S. cerevisiae [S.c.]) Npa3 domain organization. The color code is used throughout the figures. (B) Amino acid sequence alignment of Npa3 from S. cerevisiae, with eukaryotic homologs from Schizosaccharomyces pombe (S.p.) and GPN1 from Homo sapiens (H.s.) and the archaeal homolog Pab0955 from Pyrococcus abyssi (P.a.). Secondary structure elements are indicated above the sequence (cylinders, α-helices; arrows, β-strands). Amino acid numbering above the sequence corresponds to Npa3 from S. cerevisiae. Motifs G1 to G5 and insertions 1 and 2 are marked with bars. (C) Ribbon representations of the closed (GDP-bound) (left) and open (GMPPCP-bound) (right) conformations. The G motifs and insertion regions are colored as described above for panel A. A fatty acid bound to the hydrophobic pocket that is opened in the Npa3-GMPPCP structure is shown as slate blue sticks. Missing residues are indicated with dashed lines.
The structure with GDP-AlFx was solved by using single anomalous diffraction from intrinsic sulfur atoms, and a protein model was built and refined to a free R factor of 23.9% at a 1.85-Å resolution (Table 1). The model was used to solve the structures containing GMPPCP or GDP, which were refined to free R factors of 22.7% and 27.6% at 2.2-Å and 2.3-Å resolutions, respectively (Table 1). The electron density of the GDP-AlFx complex was not clearly interpretable in the active-site region, where it showed a mixture of different nucleotide configurations. Thus, this structure was discarded, although the protein model was excellent. The structures containing GMPPCP or GDP also showed great stereochemistry and were used for further analysis because they showed very-well-defined densities for bound nucleotides, indicative of defined enzyme states. The structures are relevant for all eukaryotic orthologs because Npa3 is highly conserved, with 50% of the residues in the crystallized variant being identical between S. cerevisiae and human enzymes (Fig. 1B; see also Fig. S1 in the supplemental material).
The Npa3-GDP structure shows eukaryote-specific features.
The Npa3 core consists of a central, six-stranded, parallel β-sheet surrounded by helices (Fig. 1C). The two insertion regions extend the Npa3 core, resulting in an overall L-like shape of the enzyme. The asymmetric unit of the Npa3-GDP crystals contains six enzymes that differ only slightly in regions forming crystal contacts. The Npa3-GDP structure resembles the homologous archaeal structure of Pab0955-MgGDP (PDB accession number 1YRB) (18) but also reveals major differences in insertions 1 and 2 (see Fig. S2 in the supplemental material) and in regions connecting the core to insertion 2. A single residue links strand β7 in the core to the C-terminal end of helix α10, which is bent by ∼50°. In the archaeal structure, strand β7 is shorter and the linker region to α10 comprises 5 residues, allowing for a straight conformation of the helix. Whereas helices α9 and α10 in Npa3-GDP lie side by side, they are tilted in the archaeal structure. Helix α3 in insertion 1 of Npa3-GDP is apparently rotated by ∼40° toward the core, and helix α4 and the GPN loop are shifted by ∼3 Å toward the G3 motif. Taken together, the core of the eukaryotic enzyme generally resembles its archaeal counterpart, whereas the two insertions differ significantly.
The Npa3-GMPPCP structure reveals a novel open conformation.
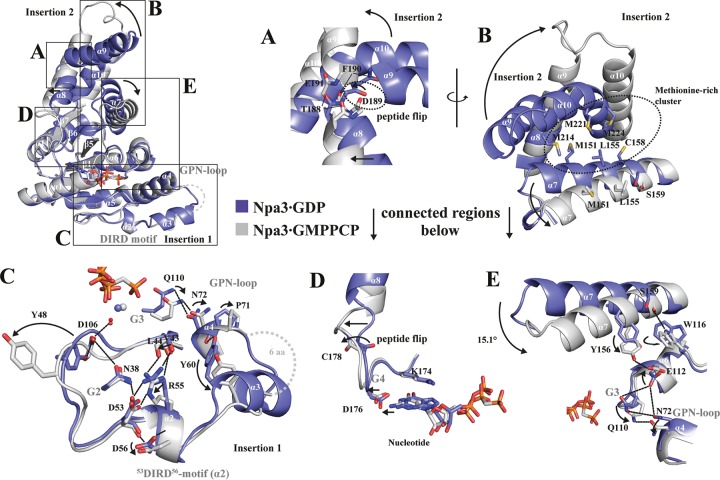
Our Npa3-GMPPCP structure reveals a novel open conformation that differs significantly from the Npa3-GDP structure (Fig. 1C and 2). Both insertions are rearranged, and the position of helix α7 is changed, opening an extended hydrophobic pocket. Insertion 2 adopts a different fold and remotely resembles the helical bundle of the N domain of the prokaryotic GTPase Ffh from Thermus aquaticus (PDB accession number 1LS1) (30). In the GMPPCP-bound state, residue D189 of insertion 2 and residue C179 show peptide bond flips that enable a straight conformation of helices α8 and α9 and the formation of a single helix (Fig. 2A and D). Helix α10, which is bent in the Npa3-GDP structure, adopts a straight conformation, and helix α7 is rotated by ∼15°. This disrupts a methionine-rich hydrophobic core formed between helices α7, α8, and α9 that kept the pocket closed in the Npa3-GDP structure (Fig. 2B and E). Rotation of helix α7 enables it to hydrogen bond to residue E112, which no longer interacts with Q110 in motif G3 and N72 in the GPN loop, as observed in the closed Npa3-GDP structure (Fig. 2E). Mutation of any one of these residues (E112, Q110, or N72) is lethal in yeast (7, 10, 21).
FIG 2.
Superposition of closed Npa3-GDP and open Npa3-GMPPCP structures. Magnesium ions are shown as blue (Npa3-GDP) or gray (Npa3-GMPPCP) spheres, water molecules are shown as red spheres, and hydrogen bonds are shown as dashed lines. (A) Peptide flip of D189 enables pocket opening and formation of a single helix from helices α8 and α9. (B) Conformational changes in insertion 2 and helix α7 facilitate the opening of an extended, hydrophobic pocket. (C) A set of residues in insertion 1 rearranges, including the GPN loop and a DIRD motif, leading to increased flexibility of this region in the GMPPCP-bound state. (D) Pocket opening allosterically alters the active site via the G4 motif. (E) Conformational changes in helix α7 are linked to the G3 motif and the GPN loop.
In addition, insertion 1 is changed in the Npa3-GMPPCP structure, including motif G3, which binds the GPN loop (Fig. 2C). Insertion 1 is more flexible than in the GDP-bound structure. Its hydrogen bonds with Y48 to D106 (G3) and with Y60 to the carbonyl of P71 (GPN loop) are lost, helix α3 is unfolded, and residues 64 to 69 are mobile. The positioning of insertion 1 apparently involves a novel “DIRD” motif that comprises the invariant residues D53, I54, R55, and D56 (Fig. 2C). The DIRD motif is, along with the GPN motif, the most highly conserved region within the sequence of the yeast Npa3 paralogs GPN2 and GPN3 (see Fig. S3 in the supplemental material). Residue N38 in motif G2 bridges between D106 in motif G3 and D53 in the DIRD motif to keep helix α2 and thus insertion 1 in close proximity to the core. The side chain of D56 in the DIRD motif is flipped in the Npa3-GMPPCP structure and hydrogen bonds to D53, possibly stabilizing the changed position of insertion 1. Taken together, Npa3 can adopt two very different conformations, a closed conformation observed with bound GDP and a novel open conformation observed when the GTP analogon GMPPCP is bound.
Nucleotide binding and conformational states.
The two structures reveal atomic details of how Npa3 binds GMPPCP or GDP using its G1 to G5 motifs (Fig. 3). In the Npa3-GMPPCP structure, strong electron density corresponding to the γ-phosphate of the nucleotide is observed (Fig. 3A). The octahedral coordination of the magnesium ion by oxygen atoms of the β- and γ-phosphates of GMPPCP and Thr17 of the G1 motif (P loop [10-GMAGSGKT-17]) is completed by three water molecules, hydrogen bonded by D40 in the G2 motif (36-VINLD-40) and D106 and T107 in the G3 motif (106-DTPGQ-110) (Fig. 3B). After GTP hydrolysis, an additional water molecule apparently replaces the coordination sphere previously occupied by the γ-phosphate oxygen of GMPPCP (Fig. 3C). Residues in the G1 motif stabilize the negative charge of the phosphate ions. Guanine specificity is conferred by hydrogen bonds of the Watson-Crick edge of GMPPCP to motifs G4 (173-NKTD-176) and G5 (238-SSF-240). F240 in motif G5 stacks against the guanine base. Thus, our structures explain how Npa3 specifically binds GDP and the GTP analog.
FIG 3.
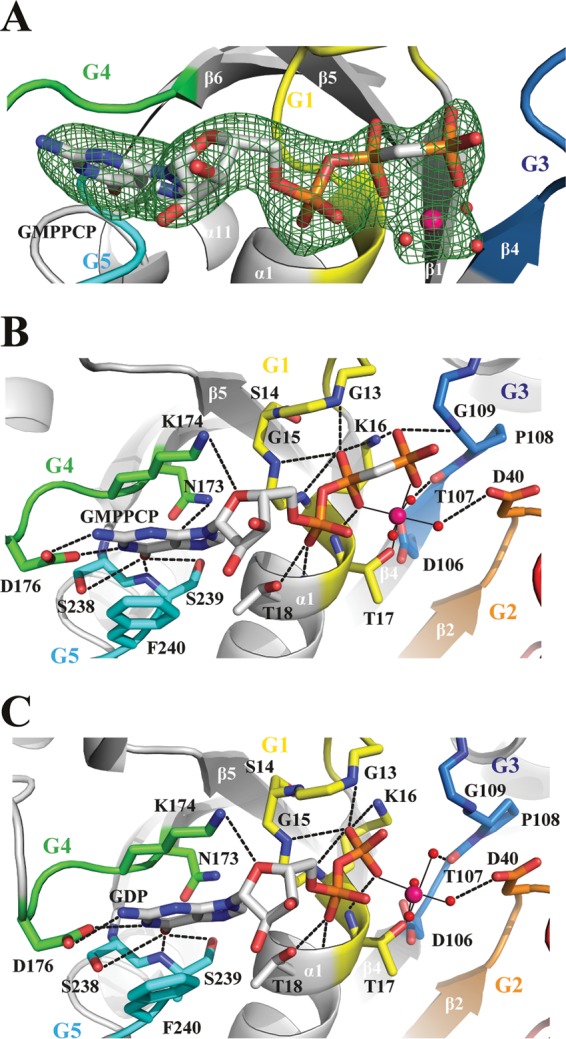
Nucleotide-binding pocket and active site. (A) Initial unbiased Fo-Fc difference electron density of GMPPCP and the magnesium ion, including coordinated water molecules contoured at 3σ (green mesh). The final Npa3-GMPPCP model is superimposed. Motifs G1 to G5 are color-coded as described in the legend to Fig. 1A. Water molecules are shown as red spheres, magnesium ions are shown as pink spheres, and hydrogen bonds are shown as dashed lines. (B and C) Nucleotide interaction network of Npa3 with GMPPCP (B) and GDP (C). Metal ion-ligand interactions are shown as solid black lines.
Catalytic mechanism.
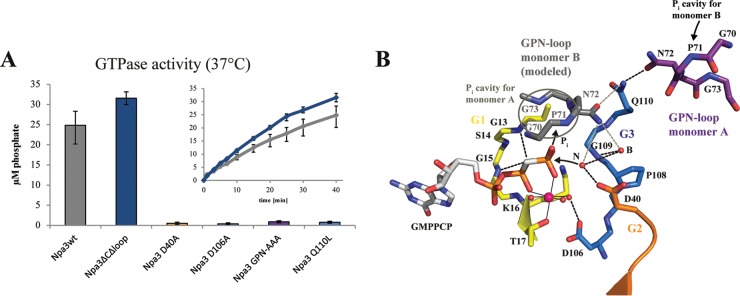
The catalytic mechanism for the hydrolysis of GTP to GDP and inorganic phosphate by GPN-loop GTPases was previously suggested for the archaeal enzyme (18). To probe the catalytic mechanism in the eukaryotic system, we prepared Npa3 variants with point mutations and tested their ability to hydrolyze GTP (Fig. 4). Whereas residue D106 in motif G3 stabilizes the Mg2+ ion, residue D40 in motif G2 positions the nucleophilic water molecule (N), as observed for the bacterial GTPase FtsY (31). Consistent with this, the D106A and D40A variants of Npa3 were both inactive (Fig. 4A).
FIG 4.
Catalytic mechanism. (A) GTPase activity of Npa3 mutants. Bars represent free orthophosphate concentrations after 40 min at 37°C. Kinetics are shown for wild-type Npa3 (Npa3wt) (gray) and the crystallized variant Npa3ΔCΔLoop (blue). (B) Schematic of the mechanism of GTP hydrolysis. The active site of Npa3-GMPPCP is shown. The GPN loop of monomer B is modeled by superpositioning of two Npa3-GMPPCP enzymes on the archaeal Pab0955 dimer (PDB accession number 1YRB) (18). The nucleophilic water (N) that attacks the γ-phosphate and the buttressing water (B) are superimposed from the Npa3-GDP structure. Hydrogen bonds are shown as dashed black lines, potential hydrogen bonds derived from dimer modeling are shown as dashed gray lines, water molecules are shown as red spheres, the magnesium ion is shown as a pink sphere, and metal ion-ligand interactions are shown as solid black lines.
The catalytic mechanism was suggested to also involve the GPN loop. In the archaeal enzyme dimer, the GPN loop of one monomer protrudes into the active site of the other monomer, where it binds the hydrolyzed GTP γ-phosphate (18). Indeed, Npa3 variants with a mutation of the GPN loop (70GPN72 changed to 70AAA72) lacked GTPase activity, supporting the suggested mechanism (Fig. 4). In addition, a single Q110L mutation, predicted to disrupt buttressing of the GPN loop by residue Q110 in motif G3, was also inactive. All mutated residues shown here to be involved in catalysis in vitro are essential in vivo (7, 21). Furthermore, purified Npa3 formed a dimer, and in the Npa3-GDP crystals, two symmetry-related complexes formed a dimer that contained the GPN loop of one monomer in the active site of the other monomer, as observed for the archaeal structure.
From these results, the catalytic mechanism of GPN-loop GTPases emerges. The nucleophilic water molecule is positioned in-line of the scissile phosphodiester bond at the γ-phosphate of GTP by residue D40 and a buttressing water molecule bound to the backbone of residue G109 of motif G3, which stabilizes the GPN loop via residue Q110 and positions it in the active site of monomer B. The negative charge in the transition state of the SN2 reaction is partially neutralized by a magnesium ion that is also positioned by D106 in motif G3. Modeling of the Npa3 dimer, based on the archaeal dimer structure, shows that the GPN loop of one monomer protrudes into the active site of the other monomer even in the open enzyme conformation and could bind the hydrolyzed orthophosphate.
Putative peptide-binding pocket.
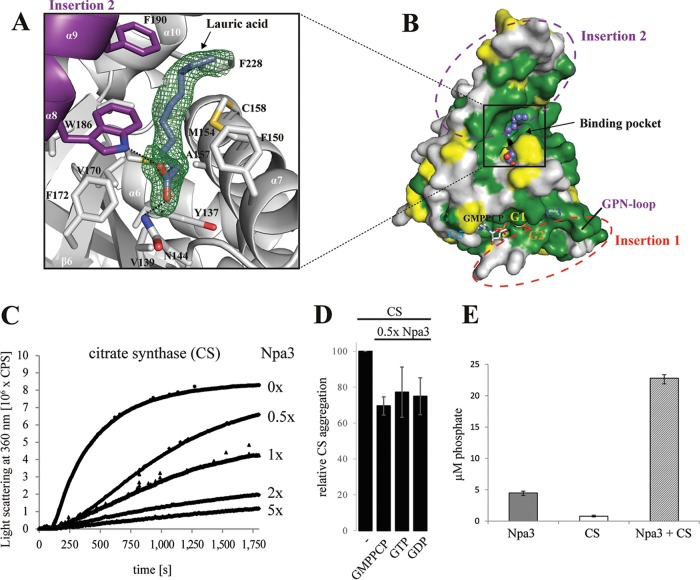
In the Npa3-GMPPCP structure, we observed a strong, extended electron density in the hydrophobic pocket that is created upon the transition from the closed to the open state of the enzyme (Fig. 5A). This density could be explained by a molecule of lauric acid, a C12 fatty acid that may have been derived from the hydrolysis of Tween 20, a lauric acid ester that was present in the lysis buffer (Fig. 5A). The fatty acid carboxyl group binds residues N144 and W186, whereas the acid tail is close (within 5 Å) to the hydrophobic residues Y137, V139, F150, M154, A157, C158, L161, M168, V170, F172, W186, F190, L225, F228, Y229, and L232. The surface of the pocket is highly conserved between species (Fig. 5B).
FIG 5.
Putative peptide-binding pocket and GTPase-stimulating chaperone activity. (A) Initial unbiased Fo-Fc difference electron density for lauric acid (slate blue sticks) bound to the putative peptide-binding pocket of Npa3, contoured at 2σ (green mesh). (B) Highly conserved surface of the putative peptide-binding pocket. Invariant residues are in green, conserved residues are in yellow, and variable residues are in gray. Insertion regions and G motifs are depicted. Lauric acid is shown as blue spheres. (C) Npa3 has chaperone-like activity in vitro. Npa3 suppresses thermally induced (43°C) aggregation of the general nonnative chaperone substrate protein citrate synthase (CS). Different amounts of wild-type Npa3 were added, as indicated on the right. (D) Chaperone-like activity of Npa3 is independent of added nucleotides under limiting conditions. A total of 1 mM GMPPCP, GTP, or GDP was added, and relative CS aggregation in the presence of 0.5× molar amounts of Npa3 was determined after 30 min. (E) GTPase activity of Npa3 is stimulated >4-fold in the presence of the nonnative chaperone-substrate protein citrate synthase. Free orthophosphate concentrations were determined after 40 min at 43°C (see Materials and Methods).
GPN1 is known to interact with assembling Pol II subunits (4), which expose hydrophobic peptide regions buried in subunit interfaces of the Pol II assembly (32). Therefore, it is likely that the newly observed hydrophobic pocket of Npa3 binds hydrophobic peptides in the cellular context. We therefore considered that the hydrophobic pocket naturally binds to hydrophobic protein regions as described for molecular chaperones that prevent misassembly and aggregation of multisubunit complexes (1, 2). Indeed, modeling showed that hydrophobic peptides may be accommodated in the pocket in an extended conformation.
Npa3 has GTPase-stimulating chaperone activity.
The existence of a putative hydrophobic peptide-binding pocket prompted us to ask whether Npa3 has chaperone activity. We tested for chaperone-like activity of Npa3 in vitro with a standard assay that uses citrate synthase (CS) as a general chaperone substrate protein. In this assay, the temperature-induced aggregation of CS is suppressed when a chaperone is added (29). Indeed, Npa3 was able to suppress temperature-induced aggregation of CS, revealing that this GPN-loop GTPase has chaperone activity (Fig. 5C). We next investigated whether the chaperone activity was nucleotide dependent. Because no GTPase activity was observed in the standard chaperone assay buffer, we changed the buffer to buffer C containing 100 mM NaCl (see Materials and Methods) and adjusted the pH value to 7.5 at 43°C. This buffer supported GTP hydrolysis albeit at ∼5-fold-reduced levels, apparently due to the increased temperature of 43°C (see Materials and Methods). Chaperone-like activity was readily observed in the presence of nucleotides, but no significant differences occurred after the addition of GMPPCP, GTP, or GDP (Fig. 5D). These results indicate that the open Npa3 conformation is not induced by GTP binding but rather by the binding of hydrophobic unfolded protein peptide regions. Pocket opening in Npa3 goes along with a conformational change in motifs G3 (Fig. 2C and E) and G4 (Fig. 2D) that widens the nucleotide-binding pocket. Indeed, a similar conformational change in Ffh from T. aquaticus was proposed previously to allosterically alter the active site via the G4 motif (33). Thus, we speculated that peptide binding would allosterically widen the active center, promote GDP displacement, and facilitate GTP binding and thus may lead to an increased GTP turnover rate.
To investigate this, we analyzed the GTPase activity of Npa3 in both the presence and the absence of partially unfolded CS at 43°C. Indeed, the presence of CS stimulated GTPase activity >4-fold (Fig. 5E), providing a link between chaperone and GTPase functions and indicating that the structural changes inferred by crystallography occur in solution. The GTP hydrolysis-driven release of CS and subsequent rebinding and pocket opening could thus explain the similar chaperone activities observed with our assay upon GTP addition. Taken together, our results show that Npa3 exhibits chaperone-like activity and indicate that it can bind unfolded protein peptide regions without GTP addition. These results also suggested that peptide binding triggers the opening of the pocket, promotes GDP displacement, and thus facilitates GTP rebinding, whereas peptide release is regulated by GTP hydrolysis.
Npa3 binds peptides derived from Pol II subunit interfaces.
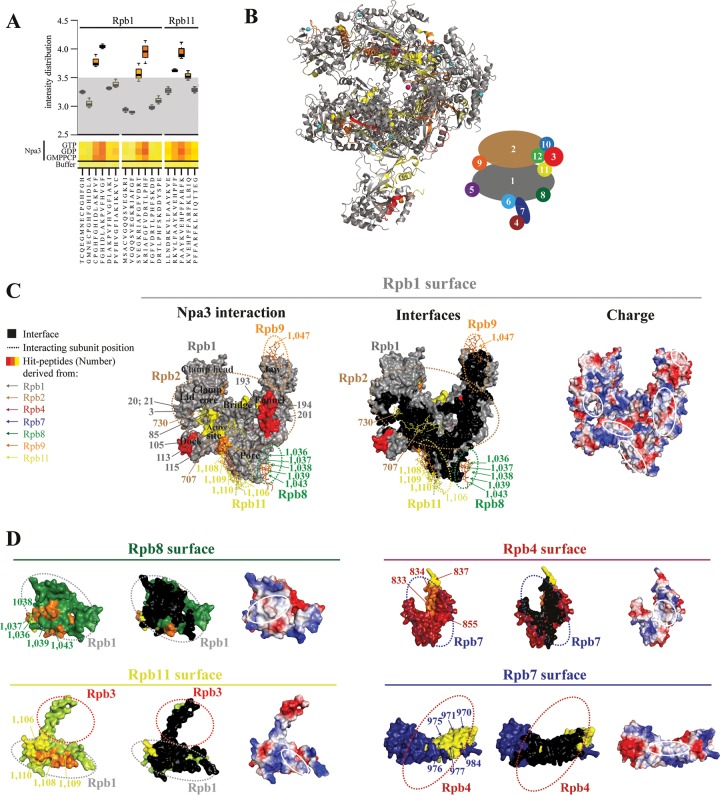
Because Npa3 was implicated in Pol II biogenesis and exhibits general chaperone activity, as observed for other assembly chaperones (34, 35), we investigated whether Npa3 may function in the assembly of Pol II. Unfortunately, this could not be tested directly, because Pol II is not available in a recombinant form and has never been assembled from its subunits in vitro, and endogenous Pol II is not amenable to chaperone assays. However, we asked whether Npa3 would be able to bind peptides derived from Pol II subunits that may be exposed during Pol II assembly. To address this in an unbiased fashion, we used peptide arrays to screen a total of 1,139 Pol II-derived 15-residue-long peptides (overlap of 11 residues, respectively), covering all regions of all 12 Pol II subunits, for binding Npa3 in the presence of GMPPCP, GTP, or GDP (Fig. 6A; see also Table S1 and Fig. S4 in the supplemental material). In this assay, 55 peptides bound Npa3 significantly (signal intensity of >3.5) (see Table S2 in the supplemental material). The binding efficiency generally did not depend on nucleotides, consistent with the model that opening of the hydrophobic pocket is triggered by peptide binding. Npa3 did not interact with assembled, mature Pol II (not shown), consistent with the model that Npa3 binds Pol II assembly intermediates.
FIG 6.
Npa3 binds Pol II-derived peptides located at subunit interfaces. (A) Box plot representation of a representative portion of the heat map describing the Npa3 peptide-binding landscape. Tested were a total of 1,139 15-mer Pol II-derived peptides, covering the complete sequence of the 12-subunit Pol II in the presence of GMPPCP, GTP, and GDP. Control experiments were performed without Npa3 and nucleotides to test the cross-reactivity of the anti-His antibody. Intensity distribution is shown on a logarithmic scale. Peptides with a signal intensity of <3.5 were defined as unbound peptides (gray area). (B) Location of Npa3-binding peptides in the assembled Pol II complex (PDB accession number 1WCM) (32). Npa3 binding to Pol II peptides is depicted in yellow (signal intensity of 3.5 to 3.75), orange (3.75 to 4), and red (>4), whereas unbound regions are in gray (<3.5). A schematic representation of the 12 Pol II subunits Rpb1 to Rpb12 in the folded Pol II complex is shown on the right. (C and D) Surface representations of Pol II subunits showing that Npa3 interacts with hydrophobic Pol II-derived peptides located at subunit interfaces (left). Peptides in Pol II subunit surfaces involved in interactions with other subunits are colored as described above for panel B. Numbers correspond to the peptide numbers from the array (see Table S1 in the supplemental material for a list of all peptides, Table S2 in the supplemental material for a list of all peptides that interact with Npa3, and Fig. S4 in the supplemental material for original peptide interaction analysis) and are color-coded according to the subunit from which they are derived, as shown on the left and in the schematic of Pol II subunit organization in panel B. Npa3-bound peptides from other subunits that interact with Rpb1 are shown as sticks. Residues involved in subunit interfaces are shown in black (middle). White solid lines highlight hydrophobic interface regions, bound by Npa3 (right). Dashed lines indicate where other Pol II subunits are positioned in the assembled complex.
The obtained Npa3-binding peptides stemmed from all Pol II subunits except Rpb3, Rpb6, and Rpb12. When we mapped the Npa3-binding peptides onto the Pol II structure (Fig. 6B), we found that 42 of the 55 peptides were at least partially located in interfaces between Pol II subunits and were enriched in hydrophobic residues (Fig. 6C and D; see also Table S2 and Fig. S4 in the supplemental material). We therefore mapped residues in subunit interfaces in the mature Pol II complex using CoCoMaps (https://www.molnac.unisa.it/BioTools/cocomaps) and compared them with peptides that were bound by Npa3 (Fig. 6C and D). We identified numerous Npa3-binding peptides within the interface between the two largest Pol II subunits, Rpb1 and Rpb2. In particular, many peptides mapped to the extended Rpb1-Rpb2 interface within the clamp domain of Pol II (Rpb1 peptides 3, 20, 21, 24, 57, and 85 and Rpb2 peptide 730) (Fig. 6C). We also detected three Npa3-binding peptides (peptides 193, 194, and 201) in the funnel domain of Rpb1.
Npa3 also interacted with peptides from the subunits Rpb8, Rpb9, and Rpb11 that are located in critical interfaces with Rpb1 (Fig. 6D). Three overlapping Npa3-binding peptides (peptides 1037 to 1039) contained a hydrophobic region in Rpb8 (117-SFGGLLMR-124) that forms an interface with Rpb1. In Rpb11, 18 of a total of 25 residues that form the interface with Rpb1 were found in overlapping Npa3-binding peptides (peptides 1106 and 1108 to 1110). Peptide 1109 covers most interface residues and shows the strongest Npa3 binding among Rpb11-derived peptides. We also identified interactions of Npa3 with 10 peptides in the Rpb4-Rpb7 interface (Rpb4 peptides 833, 834, 837, and 855 and Rpb7 peptides 970, 971, 975, 976, 977, and 984) (Fig. 6D), consistent with a previously reported interaction of human GPN1 with Rpb4 and Rpb7 (11). Npa3 did not interact with a purified yeast Rpb4-Rpb7 complex (not shown), supporting the model that Npa3 binds peptide regions located at the subunit interface that are accessible only during subcomplex assembly. Unfortunately, attempts to use the identified peptides for interaction analysis in solution and for cocrystallization trials with Npa3 failed due to the insolubility of such synthesized peptides. Nevertheless, our data show that Npa3 binds numerous Pol II-derived hydrophobic peptides that are located at subunit interfaces, consistent with a function of Npa3 in Pol II assembly.
DISCUSSION
Many macromolecular complexes have been shown to require assembly chaperones for their biogenesis (2), including the nucleosome (36, 37), RuBisCO (38), the proteasome (39), spliceosomal snRNPs (40), and the ribosome (41). In contrast, biogenesis of the 12-subunit Pol II complex remains poorly understood. It was shown previously that Pol II biogenesis requires the R2TP/Prefoldin-like complex and the ATPase Hsp90 (4), a chaperone that is involved in the assembly of several protein complexes (42), but additional factors are likely involved in Pol II assembly.
Here we study the structure and function of the new family of GPN-loop GTPases and provide evidence that these enzymes assist in Pol II assembly. We present the first structure of a eukaryotic GPN-loop GTPase, the GPN1 homolog Npa3 from yeast. We show that Npa3 can adopt an open state with a hydrophobic pocket, that it can bind peptides derived from Pol II subunit interfaces, that it has general chaperone activity, and that a chaperone substrate protein can stimulate its GTPase activity. The latter observation is reminiscent of the previously reported stimulation of the ATPase activity of the chaperones Hsp70 (43–45) and Hsp90 (46) by substrate binding. GTPases were also shown previously to play a key role during ribosome assembly in bacteria (47) and in eukaryotes (48).
Together with previously reported data, our results suggest a model for Pol II biogenesis (Fig. 7). In this model, GPN-loop GTPases are involved in the correct assembly of Pol II in the cytoplasm. The assembled Pol II would then be recognized by Iwr1 and imported into the nucleus (6). Several lines of evidence argue for a role of GPN-loop GTPases in Pol II assembly. First, GPN1, GPN2, and GPN3 interact with Pol II assembly intermediates (4). Second, GPN1 interacts with the CCT complex (7), a chaperone complex with various functions (49) that interacts with Pol II subunits (50). Third, in patients with myofibrillar myopathies, a neuromuscular disorder characterized by protein aggregates, human GPN1 shows increased expression and accumulates with Rpb1 in the cytoplasm of muscle cells (51), consistent with a chaperone function of GPN1.
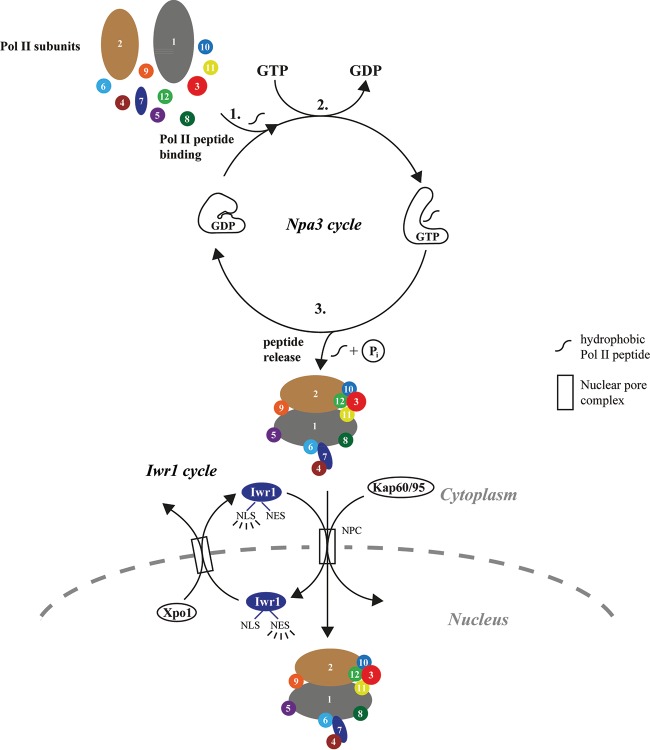
FIG 7.
Model for Pol II biogenesis. Whereas the “Npa3 cycle” drives cytoplasmic assembly of Pol II, the “Iwr1 cycle” drives Pol II nuclear import. In the Npa3 cycle, pocket opening of Npa3·GDP is induced by binding of hydrophobic regions of Pol II subunits that form interfaces in the assembled Pol II complex, thereby preventing misassembly (step 1). Pocket opening allosterically communicates with the active site, stimulates GDP displacement, and thereby facilitates GTP rebinding (step 2). GTP hydrolysis leads to the release of Pol II peptides, facilitating the formation of Pol II subunit interfaces and the assembly of Pol II in the cytoplasm (step 3). In the Iwr1 cycle, assembled Pol II is recognized by Iwr1, which provides an import adaptor for nuclear import via its nuclear localization sequence (NLS). Iwr1 is recycled with the use of its nuclear export signal (NES).
In our model, an exposed hydrophobic peptide region in a newly synthesized Pol II subunit would trigger the opening of the Npa3 pocket in its GDP-bound state. Npa3 would then trap exposed hydrophobic regions of Pol II subunits, preventing their misassembly and opening a time window for association with the cognate Pol II subunit (Fig. 7). Peptide binding would then alter the active site allosterically, decreasing its affinity for GDP, provoking GDP displacement, and increasing its affinity for GTP, to facilitate GTP rebinding. Subsequent GTP hydrolysis would then lead to the release of the bound Pol II subunit to enable association with cognate subunits and Pol II assembly. The assembled Pol II would then be recognized by Iwr1, which binds between the two largest polymerase subunits and serves as an adaptor for the import of Pol II into the nucleus (6).
Our model does not include any nuclear roles of Npa3, although they may exist, because nucleocytoplasmic shuttling of GPN1/Npa3 was reported previously (7, 13, 21, 52). However, GPN-loop GTPases lack a nuclear localization signal (NLS), and mutations of GPN2 or GPN3 cannot be rescued by the fusion of a NLS to Rpb3 (12), consistent with the main function of these enzymes being cytosolic. In the future, a system to coexpress all 12 Pol II subunits may be derived, which would allow one to test whether Pol II assembly is facilitated in the presence of GPN-loop GTPases. Future research on the GPN-loop family of essential GTPases is guided by our structure-function analysis of Npa3 and its apparent function as a chaperone in the assembly of protein complexes such as Pol II.
Supplementary Material
ACKNOWLEDGMENTS
J.N. carried out experiments, structure determination, and modeling. F.R.W. supported Pol II-Npa3 interaction studies (data not shown). D.K. and W.M. advised on X-ray data analysis. P.C. initiated and supervised research. J.N. and P.C. prepared the manuscript.
We declare that we have no conflict of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01009-15.
REFERENCES
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Ellis RJ. 2013. Assembly chaperones: a perspective. Philos Trans R Soc Lond B Biol Sci 368:20110398. doi: 10.1098/rstb.2011.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 4.Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, Lamond AI, Bertrand E. 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wild T, Cramer P. 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci 37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P. 2011. Iwr1 directs RNA polymerase II nuclear import. Mol Cell 42:261–266. doi: 10.1016/j.molcel.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 7.Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics 9:2827–2839. doi: 10.1074/mcp.M110.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 10.Alonso B, Beraud C, Meguellati S, Chen SW, Pellequer JL, Armengaud J, Godon C. 2013. Eukaryotic GPN-loop GTPases paralogs use a dimeric assembly reminiscent of archeal GPN. Cell Cycle 12:463–472. doi: 10.4161/cc.23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carre C, Shiekhattar R. 2011. Human GTPases associate with RNA polymerase II to mediate its nuclear import. Mol Cell Biol 31:3953–3962. doi: 10.1128/MCB.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minaker SW, Filiatrault MC, Ben-Aroya S, Hieter P, Stirling PC. 2013. Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics 193:853–864. doi: 10.1534/genetics.112.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes-Pardo H, Barbosa-Camacho AA, Perez-Mejia AE, Lara-Chacon B, Salas-Estrada LA, Robledo-Rivera AY, Montero-Moran GM, Lara-Gonzalez S, Calera MR, Sanchez-Olea R. 2012. A nuclear export sequence in GPN-loop GTPase 1, an essential protein for nuclear targeting of RNA polymerase II, is necessary and sufficient for nuclear export. Biochim Biophys Acta 1823:1756–1766. doi: 10.1016/j.bbamcr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. 2004. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol 24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 16.Lembo F, Pero R, Angrisano T, Vitiello C, Iuliano R, Bruni CB, Chiariotti L. 2003. MBDin, a novel MBD2-interacting protein, relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol Cell Biol 23:1656–1665. doi: 10.1128/MCB.23.5.1656-1665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitta M, Saijo M, Kodo N, Matsuda T, Nakatsu Y, Tamai H, Tanaka K. 2000. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res 28:4212–4218. doi: 10.1093/nar/28.21.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gras S, Chaumont V, Fernandez B, Carpentier P, Charrier-Savournin F, Schmitt S, Pineau C, Flament D, Hecker A, Forterre P, Armengaud J, Housset D. 2007. Structural insights into a new homodimeric self-activated GTPase family. EMBO Rep 8:569–575. doi: 10.1038/sj.embor.7400958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne HR, Sanders DA, McCormick F. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 20.Calera MR, Zamora-Ramos C, Araiza-Villanueva MG, Moreno-Aguilar CA, Pena-Gomez SG, Castellanos-Teran F, Robledo-Rivera AY, Sanchez-Olea R. 2011. Parcs/Gpn3 is required for the nuclear accumulation of RNA polymerase II. Biochim Biophys Acta 1813:1708–1716. doi: 10.1016/j.bbamcr.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ. 2011. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem 286:35553–35561. doi: 10.1074/jbc.M111.286161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ. 2007. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinert T, Olieric V, Waltersperger S, Panepucci E, Chen L, Zhang H, Zhou D, Rose J, Ebihara A, Kuramitsu S, Li D, Howe N, Schnapp G, Pautsch A, Bargsten K, Prota AE, Surana P, Kottur J, Nair DT, Basilico F, Cecatiello V, Pasqualato S, Boland A, Weichenrieder O, Wang BC, Steinmetz MO, Caffrey M, Wang M. 2015. Fast native-SAD phasing for routine macromolecular structure determination. Nat Methods 12:131–133. doi: 10.1038/nmeth.3211. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr A 64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 27.Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr 61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Buchner J, Grallert H, Jakob U. 1998. Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods Enzymol 290:323–338. doi: 10.1016/S0076-6879(98)90029-5. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. 2002. Structural basis for mobility in the 1.1 A crystal structure of the NG domain of Thermus aquaticus Ffh. J Mol Biol 320:783–799. doi: 10.1016/S0022-2836(02)00476-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voigts-Hoffmann F, Schmitz N, Shen K, Shan SO, Ataide SF, Ban N. 2013. The structural basis of FtsY recruitment and GTPase activation by SRP RNA. Mol Cell 52:643–654. doi: 10.1016/j.molcel.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armache KJ, Mitterweger S, Meinhart A, Cramer P. 2005. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 33.Freymann DM, Keenan RJ, Stroud RM, Walter P. 1997. Structure of the conserved GTPase domain of the signal recognition particle. Nature 385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 34.Godin KS, Walbott H, Leulliot N, van Tilbeurgh H, Varani G. 2009. The box H/ACA snoRNP assembly factor Shq1p is a chaperone protein homologous to Hsp90 cochaperones that binds to the Cbf5p enzyme. J Mol Biol 390:231–244. doi: 10.1016/j.jmb.2009.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Duan J, Li D, Ma S, Ye K. 2011. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J 30:5010–5020. doi: 10.1038/emboj.2011.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avvakumov N, Nourani A, Cote J. 2011. Histone chaperones: modulators of chromatin marks. Mol Cell 41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.De Koning L, Corpet A, Haber JE, Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, Beckmann R, Hayer-Hartl M. 2010. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463:197–202. doi: 10.1038/nature08651. [DOI] [PubMed] [Google Scholar]
- 39.Murata S, Yashiroda H, Tanaka K. 2009. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 40.Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 41.Karbstein K. 2010. Chaperoning ribosome assembly. J Cell Biol 189:11–12. doi: 10.1083/jcb.201002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makhnevych T, Houry WA. 2012. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta 1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Smock RG, Rivoire O, Russ WP, Swain JF, Leibler S, Ranganathan R, Gierasch LM. 2010. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol Syst Biol 6:414. doi: 10.1038/msb.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. 2007. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell 26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuravleva A, Gierasch LM. 2011. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci U S A 108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaughlin SH, Smith HW, Jackson SE. 2002. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol 315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 47.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 48.Kressler D, Hurt E, Bassler J. 2010. Driving ribosome assembly. Biochim Biophys Acta 1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Leroux MR, Hartl FU. 2000. Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol 10:R260–R264. doi: 10.1016/S0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- 50.Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR. 2008. The interaction network of the chaperonin CCT. EMBO J 27:1827–1839. doi: 10.1038/emboj.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guglielmi V, Marini M, Masson EF, Malatesta M, Forget D, Tomelleri G, Coulombe B, Vattemi G. 17 April 2015. Abnormal expression of RNA polymerase II-associated proteins in muscle of patients with myofibrillar myopathies. Histopathology doi: 10.1111/his.12715. [DOI] [PubMed] [Google Scholar]
- 52.Forget D, Lacombe AA, Cloutier P, Lavallee-Adam M, Blanchette M, Coulombe B. 2013. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res 41:6881–6891. doi: 10.1093/nar/gkt455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karplus PA, Diederichs K. 2012. Linking crystallographic model and data quality. Science 336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.