Abstract
Activation-induced deaminase (AID) functions by deaminating cytosines and causing U:G mismatches, a rate-limiting step of Ab gene diversification. However, precise mechanisms regulating AID deamination frequency remain incompletely understood. Moreover, it is not known whether different sequence contexts influence the preferential access of mismatch repair or uracil glycosylase (UNG) to AID-initiated U:G mismatches. In this study, we employed two knock-in models to directly compare the mutability of core Sμ and VDJ exon sequences and their ability to regulate AID deamination and subsequent repair process. We find that the switch (S) region is a much more efficient AID deamination target than the V region. Igh locus AID-initiated lesions are processed by error-free and error-prone repair. S region U:G mismatches are preferentially accessed by UNG, leading to more UNG-dependent deletions, enhanced by mismatch repair deficiency. V region mutation hotspots are largely determined by AID deamination. Recurrent and conserved S region motifs potentially function as spacers between AID deamination hotspots. We conclude that the pattern of mutation hotspots and DNA break generation is influenced by sequence-intrinsic properties, which regulate AID deamination and affect the preferential access of downstream repair. Our studies reveal an evolutionarily conserved role for substrate sequences in regulating Ab gene diversity and AID targeting specificity.
Introduction
Secondary Ab gene diversification is required for generating Ag-specific high-affinity isotype-switched Abs in B lymphocytes (1). In mammalian B cells, this secondary diversification process includes somatic hypermutation (SHM) and class switch recombination (CSR). SHM introduces point mutations into the assembled V region exons and immediate downstream intronic J region, whereas deletions or insertions occur infrequently during SHM (2). The resultant point mutations in V regions increase DNA sequence diversity, thus allowing the selection of B cell clones with higher affinity for Ag (3). CSR is a region-specific DNA recombination process that occurs between highly repetitive and evolutionarily conserved sequences termed switch (S) regions (4). S regions are located 5′ of each set of C region (CH) exons except Cδ (4) and undergo double-stranded break (DSB) generation during CSR (5). The broken upstream donor Sμ region rejoins to one of the downstream acceptor S regions, which leads to the switching of the C regions of the Igh locus. CSR renders B cells to acquire different effector functions without affecting Ag specificity because V region exons remain unchanged during CSR. SHM and CSR each require activation-induced deaminase (AID). AID deaminates cytosine in ssDNA and converts it into uracil, resulting in U:G mismatch lesions (3, 6). However, it remains unclear how AID-initiated lesions are preferentially converted into point mutations during SHM versus DSBs during CSR.
AID-initiated U:G mismatches can be subsequently recognized and processed by several competing pathways: 1) the general replication machinery can interpret the U as if it were a T; one of the daughter cells will acquire a C→T transition mutation; 2) uracil glycosylase (UNG) can remove the U, leaving behind an abasic site; error-prone polymerases such as Rev1 can incorporate any nucleotide in place of the U, leading to transitions or transversions at C:G base pairs; 3) MSH2/MSH6 (mutS homolog 2/6), components of the mismatch repair (MMR) pathway, can recognize the U:G mismatches. The strand containing uracil is excised, and error-prone polymerases are recruited to fill the gap at loci that undergo SHM, leading to transition or transversion mutations at A:T base pairs (7). Thus, the mutations in the V region are not directly the result of AID deamination, but rather depend on the UNG and MMR recognition and processing of the AID-induced mismatches. In the absence of MSH2 and UNG, AID-initiated U:G mismatches cannot be recognized by either pathway and are converted to C→T or G→A mutations during replication. Thus, in MSH2−/−UNG−/− mice, almost all the mutations are either C→T or G→A transitions that represent the footprint of AID deamination (8, 9). Although AID deamination is the rate-limiting step of SHM and CSR, the precise molecular mechanisms that regulate the frequency of AID deamination remain to be fully elucidated.
The removal of Us by UNG results in abasic sites that have been suggested to be converted into single-stranded breaks by apurinic/apyrimidinic endonucleases 1 and 2 (10, 11). When single-stranded breaks are near each other on opposite strands, they can generate staggered DSBs; however, when they are distal from each other, MMR appears to be required to generate DSBs (12). Both UNG−/− and MSH2−/− mice exhibit impaired CSR levels in cytokine-activated B cells (8, 13). Ung deficiency leads to more substantial inhibition of CSR, strongly indicating that DNA recombination normally proceeds with a pathway requiring U excision (8, 13). The highly repetitive S regions appear to be the optimal targets of AID during CSR (4). The endogenous Sμ region displays a distinct mutational spectrum with a strong bias toward C:G base pair mutations (8, 9, 14), suggesting a role of UNG in inducing these mutations. The deletion of the core Sμ region significantly reduces CSR level but does not completely ablate CSR (15). However, when the core Sμ deleted allele is crossed into Msh2 deficiency, CSR is almost completely aboragated (16). These data suggest that the residual DSBs occurring in the nonrepetitive part of Sμ regions are mediated by MSH2. Thus, we propose that different sequence contexts of U:G mismatches may preferentially promote distinct usage of DNA repair pathway. The critical roles of MMR and UNG have been well established in SHM and CSR (3, 17). However, after the induction of AID-initiated U:G mismatches, it is unknown whether a given repair pathway preferentially accesses the U:G mismatches present in different sequence contexts. Such a question is of great importance because different repair pathways lead to distinct mutational outcomes.
It remains a longstanding question whether target DNA sequences play a critical role in regulating SHM/CSR. A correlation between hotspot motif positions and mutations has been long suggested (18, 19). Previous studies employing transgenic approaches reached controversial conclusions (20–23), which were also limited, to some extent, by intrinsic complications associated with the transgenic approach (24). Recently, we demonstrate that AID’s mutagenic activity depends on its target sequence at a non-Ig locus (25). However, the role of target DNA sequence in regulating AID activity has not been addressed in the most physiologically relevant locus, the endogenous Igh locus. Furthermore, point mutation versus DSB generation is confounded by a complex interplay between AID deamination and the processing of AID-initiated lesions. To specifically dissect out the role of target DNA sequences in regulating AID deamination and subsequent repair pathway choice, we employed two knock-in (KI) models in which a portion of core Sμ (cSμ) region or a rearranged VDJ exon (VB1–8) was placed into the endogenous V region locus via gene targeting, termed V-cSμ or VB1–8 KI, respectively. Both of these two sequences were inserted into the exactly same genomic location and driven by the same VH186.2 promoter. Thus, our experimental system allows a direct comparison between the mutability of cSμ and VDJ exon sequences and their ability to regulate AID targeting. In the present study, we compare the mutation frequency and pattern of the V-cSμ sequence with the VB1–8 exon sequence in repair factor–sufficient and –deficient backgrounds. Our data reveal a complex interplay between target DNA sequences and repair pathways in determining the outcomes of AID-initiated lesions, namely, point mutations versus DSBs.
Materials and Methods
Embryonic stem cell targeting and generation of V-cSμ KI mice
The targeting construct was employed previously to generate the VB1–8 KI mice, which contained the homologous arms for the JH1–4 locus, the VH186.2 promoter region, leader sequence, and VB1–8 exon sequence (26). An ∼760-bp cSμ region (BbvCI-BbvCI fragment) from the 3-kb endogenous cSμ region was subcloned into the targeting construct, which replaced most of the VB1–8 exon with 21 bp of the V region exon and a 19-bp JH2 exon left flanking the cSμ region. Thus, the cSμ region is under the control of the exactly same VH186.2 promoter as the VB1–8 exon sequence. Additionally, we introduced a stop codon (*) into the leader sequence of VH186.2 preceding the targeted cSμ region. This cSμ region comprises a highly repetitive sequence and contains no open reading frames. The gene targeting was performed as described previously (27). Correctly targeted clones were detected by Southern blot (EcoRI digest) with two probes that hybridized upstream of the 5′ homology arm (DQ52 probe) or downstream of the 3′ homology arm (JH4 probe) (26). For deletion of the neor cassette through two flanking loxP sites, targeted embryonic stem (ES) cell clones were infected with recombinant adenovirus that expressed Cre recombinase. The targeted ES cells were injected into blastocysts to obtain germline transmission in 129 mice, and germline-transmitting mice were termed as V-cSμ KI mice. The KI allele was detected by PCR using the following primers: forward primer (in the leader sequence of VB1–8 exon), 5′-GGTGTTCATCTAATATGTATCCTGCTC-3′; reverse primer (in the inserted cSμ region), 5′-CTCAGCTCAGCCATGCTTTT-3′. Animal work was approved by the Institutional Animal Care and Use Committee of University of Colorado Anschutz Medical Campus (Aurora, CO) and National Jewish Health (Denver, CO).
Immunization and cell sorting
VB1–8/wild-type (wt), Ung−/−, Msh2−/−, and Ung−/−Msh2−/− (DKO) (8) mice were immunized with (4-hydroxy-3-nitrophenyl)-acetyl (NP)–keyhole limpet hemocyanin (KLH) Ag (Sigma-Aldrich) because the VB1–8 exon is NP responsive (28). NP-KLH (20 μg/ml) was dissolved in 1× PBS and mixed with aluminum hydroxide (Thermo Fisher Scientific, catalog number 77161) (1:1 ratio for column), and 200 μl Ag/Alum mixture was injected into each mouse i.p. V-cSμ/wt, Ung−/−, Msh2−/−, and DKO mice were immunized with SRBC Ag, and the immunization protocol was described previously (25). VB1–8/V-cSμ DKO mice were immunized with NP-KLH Ag. Eight or 10 d after immunization, spleens were harvested, splenocytes were stained, and cell sorting was performed as described previously (27).
PCR, mutational analysis, and semiquantitative RT-PCR
Genomic DNA was isolated from splenic B220+PNAhigh germinal center (GC) B cells and employed for PCR. iProof high-fidelity DNA Polymerase (Bio-Rad, Hercules, CA) was used to amplify the VB1–8 or V-cSμ allele, respectively, using two sets of primers (Supplemental Fig. 1B). PCR products were subsequently cloned into the pGEM easy vector (Promega), and miniprep clones were sequenced. Sequences were analyzed with DNASTAR/SeqMan software and were aligned with the corresponding genomic sequences. A Student t test (two samples with equal variance and two tailed) or a Fisher exact test (2 × 2 table, two sided) for statistical significance was applied to compare mutation frequency between different regions or backgrounds. Semiquantitative RT-PCR was performed as described previously (27, 29). Unstimulated B cells were obtained from naive unimmunized mice that carry either the VB1–8 or V-cSμ KI allele or both. To avoid the survival or selection issue conferred by Ag, naive splenic B cells were stimulated with anti-CD40/IL-4 for 4 d as described previously (29), which induces B cell proliferation and survival independent of BCR engagement. Total RNA was purified with TriPure (Roche) and used for reverse transcription reaction according to the manufacturer’s instructions (Promega). Primers were as follows: RT-PCR forward primer for actin, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′, reverse primer for actin, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′; RT-PCR forward primer for V-Cμ transcripts (VH186.2 leader), 5′-CATGGGATGGAGCTGACTCA-3′, reverse primer for V-Cμ transcripts (Cμ exon3), 5′-GTGAGTCACAGTACACACAAATTC-3′. PCR reaction conditions (V-Cμ) were 94°C for 3 min, 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, 34 cycles, 72°C for 10 min.
Results
Generation of V-cSμ KI mice
To test how target DNA sequences influence the mutation frequency and spectrum in the endogenous V region locus, we generated a novel KI mouse model that harbored a 5′ portion of endogenous core Sμ region (5′cSμ) knocked into the Igh V region locus, referred to as the V-cSμ allele (Fig. 1A, Supplemental Fig. 1A, 1B). We employed a similar gene-targeting strategy previously used to generate VB1–8 KI mice (30), which harbor a preassembled productive VDJ allele that contains a VH186.2-DFL16.1-JH2 rearrangement derived from an NP-binding Ab, B1–8 (31). Southern blot analysis showed that the targeted ES cells carry both wt and targeted V-cSμ alleles (Fig. 1B). The KI V-cSμ is a passenger allele that consists of the VH186.2 promoter, a leader sequence containing a translation termination codon, and a 5′cSμ sequence that has no open reading frames, and thus it cannot encode proteins. Hence, this system provides a homogeneous population of B cells carrying a single productive V(D)J rearrangement, VB1–8, that facilitates SHM studies by avoiding the complexity of diverse physiological V(D)J rearrangements, and a passenger allele whose SHM pattern is not influenced by Ag selection. The transcription of the VB1–8 and V-cSμ sequence is driven by the exactly same VH186.2 promoter (Fig. 1C). Because both sequences are placed into the exactly same genomic location and flanked by identical transcription control elements, we predicted that the transcription of the two KI alleles would be similar. Indeed, we employed semiquantitative PCR to assess the transcript levels of two KI alleles, which exhibited no significant difference in both unstimulated and stimulated B cells (Fig. 1D, Supplemental Fig. 1C, 1D). Therefore, such a model system allows us to directly compare the mutability of the cSμ versus VDJ exon sequence.
FIGURE 1.
Gene targeting of 5′cSμ region into endogenous IgH V region locus. (A) Targeting strategy of 5′cSμ region. Restriction endonuclease map of the endogenous Igh locus is shown. The closed circle represents the Igh intronic enhancer (iEμ), and the closed boxes represent DQ52 and JH1–4 elements. Targeting construct was used to introduce the modified 5′cSμ allele (V-5′cSμ) into wt 129 ES cells. The XhoI–EcoRI fragment containing the DQ52 and JH1–4 elements was replaced by the V-5′cSμ cassette and the floxed neomycin gene. Closed triangles represent the loxP sites. Arrow indicates the VH186.2 promoter. An asterisk indicates the stop codon in the leader sequence. Striped box indicates 5′cSμ region. (B) Southern blot analysis of targeted ES cells. Left panel, EcoRI-digested ES cell DNA was hybridized with 3′ probe (JH probe). Right panel, HindIII-digested DNA before and after deletion of the neomycin gene was hybridized with 5′ probe A (DQ52 probe). Germline (GL) and targeted bands (in kb) are indicated by arrows. (C) Schematic of VB1–8 productive and V-cSμ passenger alleles. Top panel: Configuration of VB1–8 allele. The pattern-filled box indicates the VB1–8 exon sequence. The open box (L) indicates the leader of the VB1–8 exon. The V pro indicates the VH186.2 promoter. The oval box indicates Eμ. Bottom panel, Configuration of V-5cSμ allele. A 760-bp of the cSμ region replaced a large portion of the VB1–8 exon sequence with 21 bp of the V region exon and 19-bp JH2 exon left flanking the 5′cSμ region, and a stop codon (*) was introduced into the leader sequence (L). (D) Semiquantitative RT-PCR analysis of V-Cμ transcripts in unstimulated or stimulated B cells from VB1–8 or V-cSμ KI mice. The cDNA samples were prepared from unstimulated or stimulated B cells as described in Materials and Methods, and diluted in 1:5 serials for actin (1:5, 1:25, and 1:125) or 1:3 serials for V-Cμ transcripts (no dilution, 1:3 and 1:9). Representative data are shown from three independent experiments.
The S region is a better target of AID deamination than is the V region
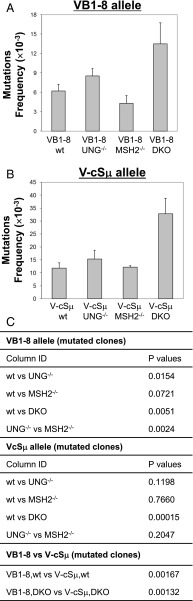
To test the mutability of cSμ and VDJ exon sequences, we induce SHM by immunizing VB1–8/wt mice with NP-KLH and V-cSμ/wt KI mice with SRBC Ag for 8 or 10 d. Of note, this short-term immunization protocol activates GC formation in the absence of appreciable Ag-specific B cell selection because it does not activate affinity maturation (28). Thus, under our short-term immunization conditions, SHM patterns of VB1–8 productive and V-cSμ passenger alleles are not biased by Ag selection (see Discussion). Following immunization, splenic B220+PNAhigh GC B cells were sorted, and genomic DNA was isolated and amplified by PCR using two sets of primers (Supplemental Fig. 1B). Amplified PCR products were subcloned and sequenced. We analyzed similar numbers of sequences for both VB1–8 and V-cSμ alleles and found a similar percentage of clones harboring mutations (Supplemental Tables I, II). Both point mutations and deletions/insertions (indels) were counted toward mutation frequency, albeit the frequency of point mutations was dramatically higher than that of indels (Supplemental Tables I, II). Thus, the mutation frequency largely reflects the level of point mutations. Our data revealed that the V-cSμ sequence is a significantly better SHM target than the VB1–8 exon sequence (Fig. 2, p = 0.00167).
FIGURE 2.
Mutation frequency of VB1–8 and V-cSμ alleles in wt or repair factor–deficient backgrounds. (A) Mutation frequency of VB1–8 allele in wt (n = 4), Ung−/− (n = 5), Msh2−/− (n = 3), and DKO (n = 4) mice (see details in Supplemental Table I). (B) Mutation frequency of the V-cSμ allele in wt (n = 5), Ung−/− (n = 3), Msh2−/− (n = 3), and DKO (n = 4) mice (see details in Supplemental Table II). (C) Statistical significance was calculated with a Student t test between different genetic backgrounds (two tailed, two samples with equal variance).
However, as discussed above, the mutation frequency depends not only on AID deamination but also on error-prone repair. To directly compare the frequency of AID deamination in the V versus S region and exclude the effects of downstream repair, we crossed the V-cSμ or VB1–8 KI allele into Ung−/−Msh2−/− (DKO) mice. In the absence of MSH2 and UNG, AID-initiated deamination events are converted to either C→T or G→A mutations by replication machinery (8, 9); thus, these signature mutations represent the footprint of AID deamination. The resultant VB1–8 DKO or V-cSμ DKO mice were immunized with NP-KLH or SRBC Ag, respectively, and similar approaches were employed to analyze the mutation frequency of both alleles in GC B cells as described above. We found that the mutation frequency of V-cSμ allele was much higher than that of the VB1–8 sequence (Supplemental Table I for VB1–8 and Supplemental Table II for V-cSμ). To minimize the variation caused by different immunizations or Ags, we crossed both V-cSμ and VB1–8 KI alleles into DKO mice (termed V-cSμ/VB1–8 DKO) that were immunized with NP-KLH Ag. Consistent with the data obtained from the DKO mice carrying either allele, we found that the mutation frequency of the V-cSμ allele was significantly higher than that of the VB1–8 allele in the compound mutant V-cSμ/VB1–8 DKO GC B cells (Supplemental Table I for VB1–8 and Supplemental Table II for Vc-Sμ); therefore, all of the data from DKO mice were pooled together and presented as a whole (Fig. 2A, 2B, p = 0.00132 between VB1–8 versus V-cSμ allele in DKO mice). Taken together, our data definitively demonstrate that the cSμ region sequence is more frequently targeted by AID than is the VB1–8 exon sequence, and that different sequence contexts affect the frequency of AID-initiated deamination.
Moreover, our data showed that the DKO GC B cells harbor a higher level of mutations in both VB1–8 and V-cSμ alleles as compared with wt GC B cells (Fig. 2A, 2B). Thus, these data demonstrate that a large fraction of AID-initiated U:G mismatch lesions is in fact corrected by error-free repair pathway under a physiological condition, thereby leading to a lower mutation frequency in the wt group.
Differential effects of different repair factors on mutation frequency
To test whether different sequence contexts influence the processing manner of AID-initiated U:G mismatches, we crossed the V-cSμ or VB1–8 KI allele, respectively, into Ung−/− or Msh2−/− mice. Wt, Ung−/−, and Msh2−/− mice carrying the VB1–8 allele were immunized with NP-KLH Ag whereas the V-cSμ KI mice of various genotypes were immunized with SRBC Ag, and genomic DNA isolated from splenic GC B cells was analyzed as described above. We found that UNG deficiency led to a significant increase in the mutation frequency of the VB1–8 sequence as compared with Msh2−/− mice or, to a lesser extent, wt mice (Fig. 2A). In contrast, Msh2 deficiency reduced the mutation frequency of the VB1–8 sequence, albeit the reduction was not statistically significant compared with wt controls (Fig. 2A). Thus, we conclude that deficiency of MSH2 or UNG affects the SHM of VB1–8 differentially. Contrary to our findings of the VB1–8 sequence, the mutation frequency of the V-cSμ region is comparable among wt, Ung−/−, or Msh2−/− mice (Fig. 2B), demonstrating that deficiency of either repair factor has no obvious effects on the mutability of this sequence.
UNG-dependent deletions were observed more frequently in the V-cSμ region
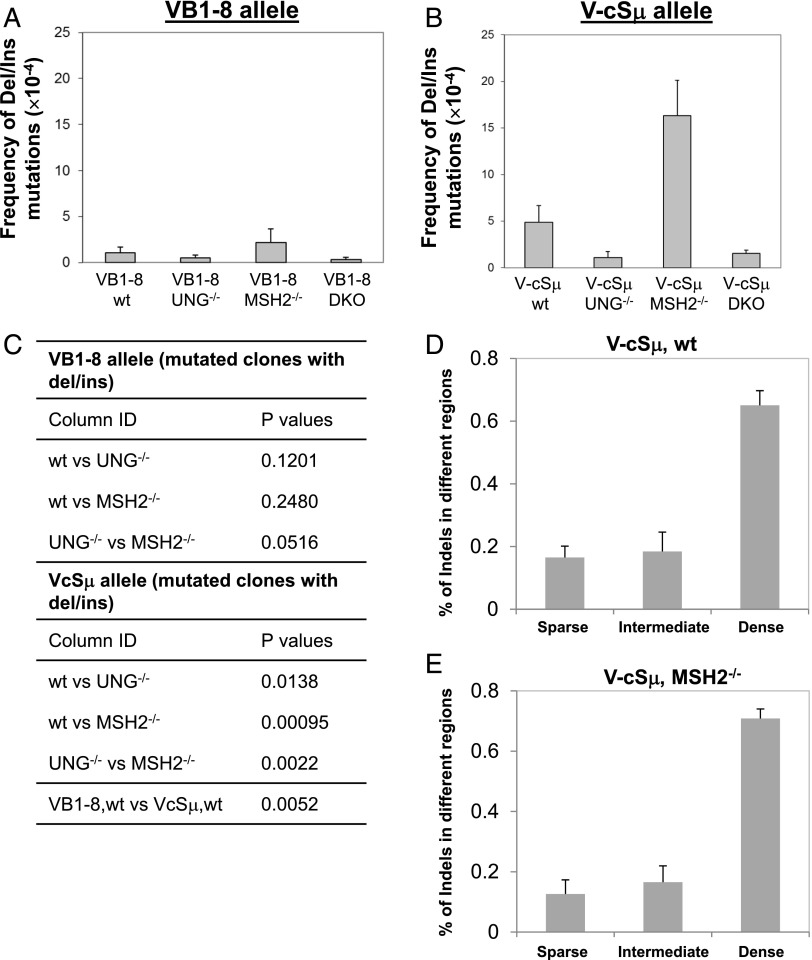
The endogenous V region locus is usually targeted for point mutations whereas the endogenous Sμ region is prone to form DSBs (32–34). To investigate how different sequence contexts affect the susceptibility of U:G mismatches being converted into DSBs, we examined both VB1–8 and V-cSμ alleles for the frequency of deletions and insertions (indels), an indicator of DSB formation. Notably, we found that the targeted cSμ sequence harbored a much higher level of indels than did the VB1–8 sequence (Fig. 3A–C, Supplemental Tables I, II). Among the analyzed indels, most events are deletional mutations whereas insertions occurred much less frequently (82% deletion versus 18% insertion). The deletions range in size from a few base pairs up to 457 bp. Furthermore, we found that these indel events mostly occurred in the AGCT dense region of the KI cSμ sequence (Fig. 3D). Thus, our data reveal that intrinsic features of the cSμ sequence influence the mutational outcome of AID activity in a position-independent manner.
FIGURE 3.
Frequency of deletions and insertions (indels) of VB1–8 and V-cSμ alleles. (A) Frequency of indels in VB1–8 allele in wt (n = 4), Ung−/− (n = 5), Msh2−/− (n = 3), and DKO (n = 4) mice (see details in Supplemental Table I). (B) The frequency of indels in the V-cSμ allele in wt (n = 5), Ung−/− (n = 3), Msh2−/− (n = 3), and DKO (n = 4) mice (see details in Supplemental Table II). (C) Statistical significance was calculated with a Student t test between different genetic backgrounds (two tailed, two samples with equal variance). (D) The percentage of indels that occurred in different regions of the KI cSμ sequence in V-cSμ/wt samples (n = 5). Data represent means ± SEM. (E) The percentage of indels that occurred in different regions of the KI cSμ sequence in V-cSμ/Msh2−/− samples (n = 3). Data represent means ± SEM. Nucleotide position in KI cSμ region: AGCT sparse region, 99–599 bp; intermediate region, 600–717 bp; dense region, 718–1065 bp.
To further elucidate the mechanisms that induce indels, we analyzed the GC B cells from VcSμ/Ung−/−, VcSμ/Msh2−/−, and VcSμ/DKO mice for the frequency of indels. Our data showed that UNG deficiency significantly reduced the frequency of indels as compared with wt controls (Fig. 3B, Supplemental Table II), thereby demonstrating an essential role of UNG in promoting deletional/insertional events in the V-cSμ sequence. In contrast, Msh2 deficiency remarkably increased the frequency of such indels compared with wt control or Ung−/− samples (Fig. 3B, Supplemental Table II). Consistent with our findings of wt samples, most of the indels are deletional mutations in VcSμ/Msh2−/− samples whereas insertions occurred much less frequently (91% deletion versus 8.7% insertion). Additionally, we observed that most of these indels occurred in the AGCT dense region of the KI cSμ sequence (Fig. 3E). We conclude that the MMR pathway normally suppresses the formation of such indels and that its deficiency significantly promotes the generation of such events in the V-cSμ sequence. In the absence of UNG and MSH2, the frequency of indels remains at an extremely low level, similar to that observed in V-cSμ/Ung−/− samples (Fig. 3B, Supplemental Table II). The VB1–8 sequence does not generate indels frequently, as shown by all the samples of various genotypes (Fig. 3A–C, Supplemental Table I). Thus, we conclude that the V-cSμ sequence is prone to form indels, an indicator of DSBs, which depends on UNG.
Differential effects of MSH2 and UNG on mutation spectrum
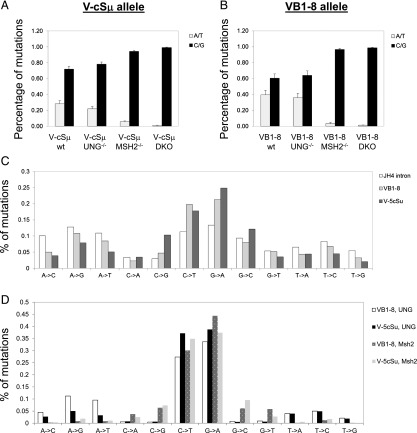
Recognition of AID-initiated lesions by UNG results in mutations at C/G pairs, whereas the MMR pathway essentially causes mutations solely at A/T pairs (17). The compiled database from V gene, non–V gene, and V gene–flanking mutations shows that mutations occur at C/G or A/T pairs with a roughly equal frequency (C/G, 47%; A/T, 53%) (35), indicating that a U:G lesion could be recognized by either pathway equivalently. However, we predict that an S region sequence will behave differently from other sequences even when it is placed at the V region locus. Indeed, we found that the percentage of C:G base pair mutations was significantly increased as compared with that of A:T base pair mutations in the V-cSμ region (Fig. 4A, 71% C:G versus 29% A:T). In contrast, the percentage of C:G and A:T base pair mutations were relatively comparable in the VB1–8 sequence (Fig. 4B). Of note, the base composition of the two KI sequences is rather comparable (Supplemental Fig. 1E). A more detailed analysis revealed that, among the increased C:G base pair mutations, the highest percentage of increase was C→G transversions; to a lesser extent, G→C mutations were also increased in the V-cSμ KI allele (Fig. 4C). Thus, we conclude that the V-cSμ sequence exhibits a strong bias toward C:G base pair mutations, suggesting that the initial U:G lesions in the KI cSμ region is preferentially recognized and processed by the UNG pathway.
FIGURE 4.
Mutation spectrum in different repair factor–deficient backgrounds. (A) Percentage of A/T and C/G mutations in VB1–8 allele. (B) Percentage of A/T and C/G mutations in VcSμ allele. (C) Mutation spectrum of JH4 intron, VB1–8 and VcSμ alleles in wt mice. (D) Mutation spectrum of VB1–8 and VcSμ alleles in UNG or MSH2-deficient mice.
To investigate how the repair pathway affects the mutation spectrum in both alleles, we analyzed the GC B cells from Ung−/−, Msh2−/−, or DKO mice carrying either or both KI alleles. In the absence of Ung, the overall percentage of A:T or C:G mutations was not significantly altered in both KI alleles compared with wt samples (Fig. 4A, 4B). However, Ung deficiency drastically affected the spectrum of C:G base pair mutations in both KI alleles: a vast majority of C:G base pair mutations were C→T or G→A transitions presumably generated by replication machinery, whereas transversions at C:G base pairs were almost absent (Supplemental Fig. 2). Consistent with previous studies, we found that the mutations at A:T base pairs were largely dependent on Msh2 for both KI alleles (Fig. 4A, 4B). Notably, C→T and G→A mutations are the most abundant type of mutations observed in both VB1–8 and V-cSμ alleles as compared with the JH4 intronic sequence (Fig. 4C); furthermore, such phenotypes become more prominent in the absence of MSH2 or UNG (Fig. 4D).
In the absence of Ung, U:G mismatches can be processed by MSH2 or replication machinery, and presumably MSH2 might gain a better access to the V-cSμ region due to the lack of UNG’s competition. However, our data showed that the percentage of A base pair mutations in the VB1–8 sequence was much higher than that in the V-cSμ sequence even in the absence of Ung (Fig. 4D). Thus, these results suggest that MSH2 prefers to access the VB1–8 instead of the V-cSμ region. In the absence of UNG and MSH2, almost all mutations were C→T or G→A transitions in both KI alleles (Supplemental Fig. 2), which represent the footprints of AID deamination. Overall, we conclude that target DNA sequences influence the processing manner of the AID-initiated lesions and function together with repair factors to generate a different mutation spectrum.
Distributions of hotspots in the VB1–8 allele
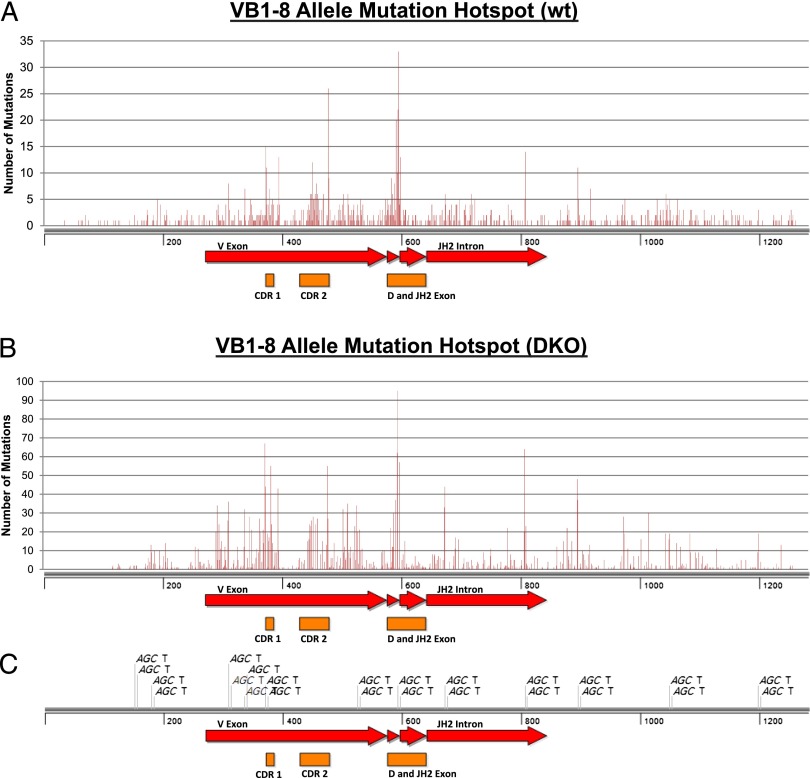
To further elucidate the molecular mechanism of AID targeting, we analyzed the distribution pattern of frequently targeted hotspots in the VB1–8 KI allele from wt samples. Our analysis reveals that the hotspots in the productive VB1–8 sequence cluster within a few nucleotides whereas most base pairs have a relatively low frequency of mutations (Fig. 5A). Such a distribution pattern strongly implicates a specific targeting mechanism to these hotspots. In this regard, the hotspots in the VB1–8 sequence clearly coincide with the CDRs (Fig. 5A), consistent with previous studies suggesting that CDRs are highly evolved to target a high level of SHM (36, 37). In particular, nucleotide 596 in CDR3 was the most frequently targeted hotspot (Table I), and CDR1 and CDR2 also contained multiple highly targeted hotspots (Fig. 5A). Additionally, we found that the hotspots outside of CDRs colocalized with AGCT motifs (Fig. 5A, 5C, Table I). Interestingly, most of the hotspot mutations (>90%) are C:G base pair mutations (Table I). Overall, we conclude that most of the hotspots in the productive VB1–8 sequences are predominantly associated with CDRs, and AGCT motifs serve as a secondary determinant for certain hotspots.
FIGURE 5.
Mutation hotspots in VB1–8 allele. (A) Point mutations from all VB1–8/wt samples were compiled and plotted against base pair position. Mutation hotspots show a strong correlation with the CDR1, CDR2, and CDR3 regions (see details in Table I). Additional mutation hotspots correlated with the position of AGCT motifs on the VB1–8 allele [displayed in (C)]. (B) Point mutations from all VB1–8/DKO samples were compiled and plotted against base pair position. The overall mutation frequency of VB1–8/DKO samples was higher than that of the VB1–8/wt samples (A). The correlation between mutation hotspots and CDR regions or AGCT motifs was observed. (C) Position of AGCT motifs along the VB1–8 KI allele (1–1284 bp).
Table I. Highly mutated nucleotides in the VB1–8 allele.
| Base Pair Location | No. of Mutations | Base Pair | Location | AGCT |
|---|---|---|---|---|
| VB1–8/wt highly mutated nucleotides | ||||
| 596 | 33 | C | D/J junction and CDR 3 | Yes |
| 478 | 26 | G | CDR 2 | No |
| 595 | 22 | G | D/J junction and CDR 3 | Yes |
| 592 | 20 | G | D and CDR 3 | No |
| 373 | 15 | G | CDR 1 | Yes |
| 809 | 14 | C | JH2 intron | Yes |
| 395 | 13 | G | V exon | No |
| 599 | 13 | C | JH2 exon and CDR 3 | No |
| 451 | 12 | G | CDR 2 | No |
| 374 | 11 | G | CDR 1 | Yes |
| 897 | 11 | G | After JH2 intron | Yes |
| 594 | 10 | A | D/J junction | Yes |
| 479 | 9 | C | CDR 2 | No |
| 584 | 9 | C | D and CDR 3 | No |
| 311 | 8 | G | V exon | Yes |
| 458 | 8 | G | CDR 2 | No |
| 589 | 8 | G | D and CDR 3 | No |
| 338 | 7 | G | V exon | Yes |
| 379 | 7 | G | CDR 1 | No |
| 918 | 7 | G | After JH2 intron | No |
| VB1–8/DKO highly mutated nucleotides | ||||
| 596 | 95 | C | D/J junction and CDR 3 | Yes |
| 373 | 67 | G | CDR 1 | Yes |
| 809 | 64 | C | JH2 intron | Yes |
| 595 | 62 | G | D/J junction and CDR 3 | Yes |
| 599 | 57 | C | JH2 and CDR 3 | No |
| 383 | 55 | G | CDR 1 | No |
| 478 | 55 | G | CDR 2 | No |
| 897 | 48 | G | After JH2 intron | Yes |
| 374 | 44 | G | CDR 1 | Yes |
| 675 | 44 | C | JH2 intron | Yes |
| 395 | 43 | G | V exon | No |
| 592 | 37 | G | D and CDR 3 | No |
| 898 | 37 | C | After JH2 intron | Yes |
| 312 | 36 | C | V exon | Yes |
| 512 | 35 | C | V exon | No |
| 293 | 34 | G | V exon | No |
| 527 | 34 | G | V exon | Yes |
| 674 | 33 | G | JH2 intron | Yes |
| 339 | 32 | C | V exon | Yes |
| 504 | 32 | C | V exon | No |
Next, we compared wt and DKO samples to investigate whether AID deamination and the DNA repair pathway differentially influence the frequency and distribution of mutations in the VB1–8 sequence. As described above, we found that the frequency of mutations in the VB1–8 allele was much higher in DKO samples (Fig. 2A), although a similar number of sequences was mutated in wt versus DKO samples (155 versus 171, Supplemental Table I). Consistently, the number of mutations was also much higher in the VB1–8/DKO samples for individual hotspots (Fig. 5B). These results demonstrate that a large portion of AID-initiated lesions were actually repaired in an error-free manner in wt samples, thereby leading to a lower mutation frequency. We found that the hotspot distribution in the VB1–8 allele was not significantly altered in the absence of MSH2 and UNG (Fig. 5B). For instance, the most frequently targeted hotspot remained exactly the same between wt and DKO samples, and their association with CDRs and AGCT motifs was also largely maintained (Fig. 5, Table I). However, we did notice that the hotspot association with CDR2 was reduced because the number of mutations within CDR2 was relatively decreased as compared with that in CDR1 or CDR3 (Fig. 5B). Additionally, the association with AGCT motifs was enhanced in the absence of MSH2 and UNG because there were a few more hotspots identified outside of CDRs located at 5′ of CDR1, 3′ of CDR2, or within the JH2 intronic region that colocalized with AGCT motifs (Fig. 5B, 5C). Notably, there were a few hotspots that did not associate with either CDRs or AGCT motifs (Fig. 5B), suggesting that the sequence context surrounding these hotspots might promote the generation of mutations. Whereas the hotspot distribution pattern can be attributed to both AID deamination and repair pathway in wt samples, it should solely reflect the contribution of AID deaminase activity in the absence of UNG and MSH2. Taken together, our data demonstrate a predominant association between hotspots and CDRs in wt and DKO samples, suggesting sequence-intrinsic mechanisms targeting these hotspots.
Hotspot distribution in the V-cSμ allele provides mechanistic insights into AID targeting
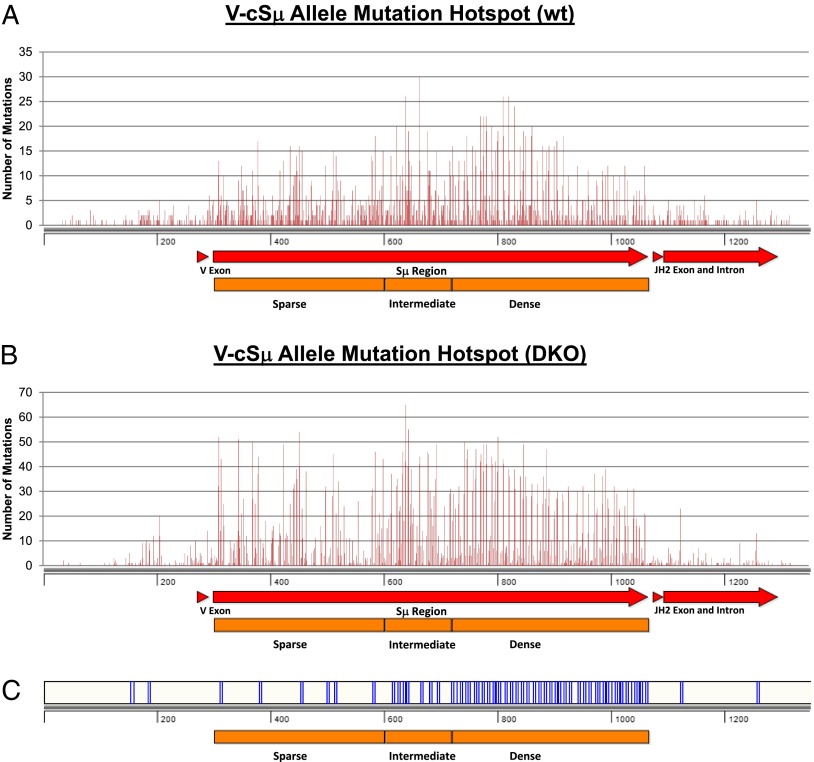
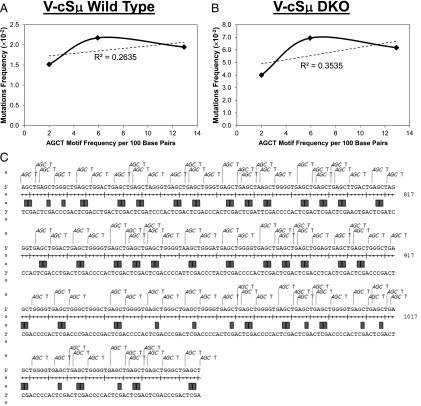
In contrast to the highly clustered hotspots in the VB1–8 allele, the hotspots in the V-cSμ allele exhibit a more evenly distributed pattern (Fig. 6A). Previous studies proposed that the density of AGCT motifs may influence the efficiency of AID targeting. Thus, we performed correlative analysis between the density of AGCT motifs and the frequency of mutations in the KI cSμ sequence. We divided the targeted cSμ sequence into three distinct regions: 1) 299–599 bp as the AGCT sparse region; 2) 600–717 bp as the AGCT intermediate region; and 3) 718–1065 bp as the AGCT dense region (Fig. 6C). Our results showed that the mutation hotspots in V-cSμ did not appear to correlate significantly with the density of AGCT motifs in a linear fashion (Fig. 7A, 7B). In particular, the most frequently targeted nucleotide was not located in the AGCT dense region; instead, it was at the boundary of the sparse and dense AGCT regions, namely, the intermediate region (Fig. 6A, Table II). Moreover, we did not detect a directly proportional increase of mutations to the density of AGCT motifs (Fig. 7A, 7B), and the AGCT sparse region displayed a quite high level of mutations (Fig. 6A). Thus, our data suggest that AID targeting efficiency is not correlated to the density of AGCT motifs in a linear fashion. On the contrary, we propose that AID targeting can be induced efficiently once the density of AGCT motifs reaches a threshold.
FIGURE 6.
Mutation hotspots in V-cSμ allele. (A) The point mutations from all V-cSμ/wt samples were compiled and plotted against base pair position. Most highly targeted mutation hotspots occur in the AGCT intermediate or dense regions at the position of an AGCT motif (see details in Table II). The AGCT sparse region also contains a relatively high amount of point mutations. (B) The point mutations from all VB1–8/DKO samples were compiled and plotted against base pair position. The position of mutation hotspots also correlated with the position of AGCT motifs [displayed in (C)]. The overall mutation frequency of V-cSμ/DKO samples was higher than that of the V-cSμ/wt samples (A). (C) Position of AGCT motifs along the V-cSμ KI allele (1–1352 bp).
FIGURE 7.
Correlation between AGCT density and mutation frequency. (A and B) Correlation plots for mutation frequency and AGCT density. The mutation frequency of the AGCT sparse, intermediate, and dense regions were correlated with the AGCT motif density (frequency of AGCT motifs per 100 bp) in the V-cSμ/wt (A) or V-cSμ/DKO (B) samples. The frequency of mutations did not correlate with the density of the AGCT motif proportionally. (C) A detailed analysis of the highly targeted hotspots in the AGCT dense region of the V-cSμ KI allele in DKO mice. The highly targeted hotspots (orange rectangles) almost exclusively occur at C/G base pairs within the AGCT motifs, reflecting the footprint of AID deamination in this region. A highly targeted hotspot was defined as any nucleotide with >15 substitutions, which ranks roughly in the 73rd percentile for all mutations.
Table II. Highly mutated nucleotides in the V-cSμ allele.
| Base Pair Position | No. of Mutations | Base Pair | Location | AGCT |
|---|---|---|---|---|
| V-cSμ/wt highly mutated nucleotides | ||||
| 665 | 30 | G | Intermediate | Yes |
| 641 | 26 | C | Intermediate | Yes |
| 813 | 26 | G | Dense | Yes |
| 823 | 26 | G | Dense | Yes |
| 833 | 24 | G | Dense | Yes |
| 773 | 22 | G | Dense | Yes |
| 779 | 22 | C | Dense | Yes |
| 783 | 22 | G | Dense | Yes |
| 625 | 20 | G | Intermediate | Yes |
| 793 | 20 | G | Dense | Yes |
| 864 | 20 | C | Dense | Yes |
| 646 | 19 | C | Intermediate | No |
| 680 | 19 | G | Intermediate | Yes |
| 784 | 19 | C | Dense | Yes |
| 804 | 19 | C | Dense | Yes |
| 849 | 19 | C | Dense | Yes |
| 587 | 18 | C | Sparse | No |
| 749 | 18 | G | Dense | Yes |
| 814 | 18 | C | Dense | Yes |
| 853 | 18 | G | Dense | Yes |
| 863 | 18 | G | Dense | Yes |
| 919 | 18 | C | Dense | Yes |
| V-cSμ/DKO highly mutated nucleotides | ||||
| 641 | 65 | C | Intermediate | Yes |
| 646 | 55 | C | Intermediate | No |
| 453 | 54 | G | Sparse | Yes |
| 311 | 52 | C | Sparse | Yes |
| 804 | 52 | C | Dense | Yes |
| 346 | 51 | C | Sparse | No |
| 370 | 50 | G | Sparse | No |
| 745 | 50 | C | Dense | Yes |
| 425 | 49 | C | Sparse | No |
| 695 | 49 | C | Intermediate | Yes |
| 779 | 49 | C | Dense | Yes |
| 784 | 49 | C | Dense | Yes |
| 849 | 49 | C | Dense | Yes |
| 750 | 47 | C | Dense | Yes |
| 765 | 47 | C | Dense | Yes |
| 889 | 47 | C | Dense | Yes |
| 587 | 46 | G | Sparse | No |
| 636 | 46 | C | Intermediate | Yes |
| 680 | 46 | G | Intermediate | Yes |
| 513 | 45 | C | Sparse | Yes |
| 681 | 45 | C | Intermediate | Yes |
| 774 | 45 | C | Dense | Yes |
In the absence of MSH2 and UNG, the distribution of hotspots was not significantly different from that observed in wt samples (Fig. 6B, Table II), suggesting that the distribution pattern of hotspots was largely determined by AID deaminase activity in the V-cSμ allele. Notably, we found that the number of mutations in the cSμ region was significantly increased in the DKO samples compared with wt controls (Fig. 6A, 6B) (p < 0.001). Thus, we conclude that a large fraction of AID-initiated lesions are processed by error-free repair in this region, thereby resulting in the lower number of mutations in wt controls.
Mechanistic insights into AID targeting were revealed by a more detailed analysis of mutations in the AGCT dense region in the absence of MSH2 and UNG. We found that the hotspots indeed colocalized with AGCT motifs in the AGCT dense region. Remarkably, only the GC base pair within AGCT motifs was frequently targeted by AID (Fig. 7C). Moreover, there were recurrent gaps identified between the highly targeted hotspots in cSμ regions, which constitute a conserved short stretch of sequences such as GGGGTG. These recurrent G-rich stretches were much less frequently targeted by AID compared with the GC hotspots within AGCT motifs, despite the immediate proximity of these two motifs (Fig. 7C). We propose that these recurrent and conserved short stretches might serve as spacers to facilitate conformational changes of DNA sequences during AID targeting.
Discussion
The targeting specificity of AID in V versus S regions remains a central unresolved question in the field of CSR and SHM. In the present study, we compared the mutability of two optimal targets of AID, a VDJ exon sequence versus an S region, in repair factor–sufficient and –deficient backgrounds. Our studies led to several critical and fundamental discoveries: 1) the S region sequence is an intrinsically more efficient AID deamination target than is the V region sequence; 2) the AID-initiated lesions can undergo error-free repair in both V and S regions; 3) the S region harbors more UNG-dependent deletions, an indicator of DSBs, which are significantly enhanced by MMR deficiency; and 4) recurrent and conserved S region motifs were identified that potentially function as spacers between AID deamination hotspots. Overall, we conclude that target DNA sequences directly modulate AID deamination frequency and promote differential accessibility of repair factors (UNG versus MMR) to AID-initiated lesions, thereby leading to distinct outcomes of AID.
Our previous studies showed that target DNA sequences influence their own mutability at a non–Ig gene locus, Bcl6 (25); however, it remains to be addressed whether target DNA sequences at the V region locus, the most physiologically relevant locus, affect AID targeting specificity. To date, it is impossible to directly compare the frequency of AID deamination in V versus S regions because the two sequences are controlled by different cis regulatory elements in their normal endogenous loci. To address this question, we developed a novel KI model in which we targeted a portion of the 5′cSμ sequence (∼760 bp) into the endogenous V region locus. The targeted 5′cSμ sequence possesses all the unique features of the endogenous cSμ region, such as a high density of AGCT motifs, yet it allows for efficient amplification by PCR and mutational analysis by sequencing. Owing to their high repetitiveness and excessive length, mutational analysis of endogenous S regions is extremely difficult, especially for the repetitive cSμ region (38), which has not been achieved.
In our model system, the targeted cSμ sequence and VB1–8 exon share the identical transcription control elements, including the VH186.2 promoter and other cis regulatory elements of the Igh locus; indeed, the transcription of two KI alleles is rather similar, thereby allowing a direct comparison of the mutability between V and S region sequences. We found that mutation frequency of the cSμ sequence is significantly higher than that of the VB1–8 exon sequence in the absence of UNG and MSH2. In MSH2−/−UNG−/− mice, almost all mutations are either C→T or G→A transitions that represent the footprint of AID deamination; therefore, our data demonstrate that the cSμ sequence is indeed a more efficient AID deamination target. One potential caveat of our model is that the difference in SHM of the two sequences might be influenced by Ag selection within GCs. However, under our short-term immunization conditions, SHM patterns of the VB1–8 productive allele are not biased by Ag selection (28). Furthermore, the VB1-8 exon sequence exhibited a similar mutation frequency and pattern, including hotspot distribution in both productive and passenger alleles that shared the identical transcription control elements and essentially identical sequence except that translation termination codons were introduced in the passenger allele (39). Thus, these data demonstrated that the SHM pattern of the VB1–8 productive allele shows no influence of Ag selection (39). Taken together, we conclude that the SHM difference between cSμ and VDJ exon sequence is driven by sequence-intrinsic mechanisms. It remains possible that different sequences might display differential binding affinity for AID, thereby recruiting a different amount of AID. Alternatively, it is also possible that no matter what test sequences are inserted into the V region locus, the amount of AID recruited remains similar. Instead, the deaminase activity of AID on different substrates might differ, especially at certain hotspot sequences. In line with this possibility, the SHM of the VB1–8 sequence often clustered around a few hotspots whereas the rest of the sequence was much less frequently targeted (Fig. 5). These data suggest that the intrinsic property of sequences appear to determine the deaminase activity of AID, thereby resulting in the distinct targeting pattern of SHM in the VB1–8 sequence. Because the cSμ region harbors more hotspot sequences, such sequence-intrinsic mechanisms may operate more efficiently, which leads to increased deamination frequency. Another possibility is that the cSμ sequence might recruit AID cofactors that preferentially bind to AGCT motifs such as 14-3-3 adaptor proteins (40), thereby enhancing AID deamination frequency. It remains to be determined which mechanisms operate to enhance AID deamination in the targeted cSμ sequence or the certain hotspot sequences of VB1–8, which may require additional studies focused on disrupting unusual aspects of the sequence via mutagenesis.
AID deamination leads to U:G mismatches recognized by MMR or UNG pathways. After MMR or UNG recognition, in theory, both error-free and error-prone repair can be recruited to the lesions. It has been suggested that error-prone repair might be preferentially recruited to Ig loci whereas error-free repair functions predominantly in non–Ig loci (41, 42). However, based on our data, we propose that error-free repair is also involved in the processing of AID-initiated lesions at Ig loci. We found that the mutation frequencies of both V and S regions were significantly higher in DKO mice than those in wt mice. Because the mutation frequency in the absence of MSH2 and UNG reflects the frequency of AID deamination, we reason that the reduced mutation frequency in UNG/MSH2-proficient mice is due to the error-free repair, which can correct the U:G mismatches and generate no mutations. These data led us to conclude that AID-initiated lesions at the Igh locus can be processed by error-free repair, similar to the non–Ig loci, and that the mutation level at the Igh locus probably exceeds the capacity of error-free repair, thereby resulting in the recruitment of error-prone repair, which in turn leads to mutations.
We found that the cSμ sequence harbors more C:G base pair mutations, which is consistent with previous findings of the endogenous Sμ region (38). Thus, the unique mutational outcome of an S region appears to be associated with its sequence rather than locus position. The biased C:G base pair mutations might be due to increased Rev1 functionality at the cSμ region. REV1 is a deoxycytidyl transferase that catalyzes the incorporation of deoxycytidines opposite deoxyguanines and abasic sites. In Rev1−/− mice, there is a dramatic reduction of C→G or G→C mutations (43). It would be of interest to further investigate how different error-prone polymerases influence the mutation spectrum. A strong bias of mutations at C:G base pairs suggests a preferential recognition of the UNG pathway (17). Previous studies showed that sequence context influences UNG-initiated error-prone versus error-free repair of AID-induced lesions (44). Thus, we propose that the initial U:G lesion in S regions is located in a sequence context facilitating its recognition by UNG, which in turn leads to more DSBs. Furthermore, we hypothesize that the processing manner of the U:G mismatches can be influenced by their sequence context (1): U:G mismatches can be recognized by UNG. The architecture of the UNG active site suggests that the enzyme must bind U that is extrahelical, or “flipped out,” from the DNA base stack. If actual flipping out of the U base is rate limiting, as suggested by data from the human, Escherichia coli, and HSV-1 enzymes (45–47), then the DNA sequence surrounding the U may influence the cleavage rate of UNG. Namely, the sequence context of U:G mismatches could affect the U base accessibility to the active site of the UNG enzyme, thereby determining UNG’s overall activity (2). U:G mismatches are recognized by MSH2/MSH6, which form a heterodimer and slide along the duplex of DNA to identify mismatches. If a sequence is prone to form higher order structures such as S regions, it is conceivable that U:G mismatches might be less accessible to this repair pathway. Consistent with these notions, our data show that S region–specific indels require UNG, whereas MSH2 deficiency enhances the frequency of such events. These data suggest that MSH2/MSH6 normally suppress the formation of these indels, probably by competing with UNG to access U:G mismatches.
Based on our data, we hypothesize that AID-initiated U:G lesions in S regions prefer UNG recognition, which contributes to more frequent DSBs. Our hypothesis is in line with another long-postulated idea for DSB formation, which suggests that: 1) removal of U by UNG results in abasic sites, 2) these sites could be converted into single-strand nicks by apurinic/apyrimidinic endonucleases 1 and 2, and 3) the adjacent nicks could be converted into staggered DSBs (48, 49). However, an alternative mechanism, which is not mutually exclusive to our hypothesis, is that higher AID deamination frequency in S regions contributes to more frequent DSBs. We indeed found that the mutation frequency of S regions is higher than that of V regions in the absence of MSH2 and UNG, suggesting that this mechanism might also contribute to the frequent DSB formation in S regions. Taken together, we propose that the combination of a higher AID deamination frequency and the preferential recognition of UNG leads to more DSBs in S regions.
It remains possible that the lack of a high frequency of indels in the productive VB1–8 allele might be influenced by selection for survival because indels in the coding V exons could be detrimental to a B cell and selected against, albeit it has been shown that V region exons can harbor indels in the productive allele (50, 51). In this regard, our recent study showed that the nonproductive VB1–8 sequence indeed harbored more indels as compared with its productive counterpart in Peyer’s patch GC B cells (39). However, in the cytokine-activated B cells that are not subject to Ag selection, both the productive and passenger VB1–8 sequences harbor a very low level of indels, whereas the cSμ sequence contains many more indels (39). Taken together, our data demonstrate that the cSμ sequence is intrinsically prone to internal deletions.
Computational and biochemical analysis has predicted certain hotspot motifs such as RGYW and cold-spot motifs such as SYC for AID targeting (18, 52). The density of RGYW/AGCT motifs may influence the efficiency of AID deamination in vitro (53) and correlate with the recombination junctions (54). However, we found that the density of AGCT motifs does not exhibit a proportional correlation to the mutation frequency in the cSμ region, which suggests that a certain threshold of AGCT density is sufficient to induce a high level of mutations. Nevertheless, we indeed found that the deletion/insertion events mostly occurred in the AGCT dense region (Fig. 3D). These data collectively suggest that the high density of AGCT motifs serves as the prone target of DSBs. Additionally, we identified conserved and recurrent S region motifs as G stretches that interspersed between AGCT motifs (Fig. 7C). We propose that such motifs might play a scaffolding or conformational role in facilitating AID targeting. Further analysis of the sequence context of these motifs may help us to better understand the specificity of AID targeting and provide mechanistic insights into how AID interacts with its DNA substrates.
Supplementary Material
Acknowledgments
We thank Drs. Frederick W. Alt for generous support of this study and Janet Stavnezer for MSH2−/−UNG−/− mice. We thank Dr. Yu Zhang for critical reading of the manuscript and thoughtful comments. We apologize to those whose work was not cited due to length restrictions.
This work was supported by University of Colorado School of Medicine and Cancer Center startup funds, a Boettcher Foundation Webb–Waring biomedical research award, an American Society of Hematology scholar award, a fund from the Cancer League of Colorado, and by National Institutes of Health Grants R21AI110777-01A1, R21CA184707-01A1, and R01CA166325-01A1 (to J.H.W.). M.T.E. is supported by National Institutes of Health Grant 3R01CA166325-02S1. X.C. is supported by National Institutes of Health Training Grant T32 AI074491.
The online version of this article contains supplemental material.
- AID
- activation-induced deaminase
- cSμ
- core Sμ
- CSR
- class switch recombination
- DKO
- double knockout
- DSB
- double-stranded break
- ES
- embryonic stem
- GC
- germinal center
- KI
- knock-in
- KLH
- keyhole limpet hemocyanin
- MMR
- mismatch repair
- NP
- (4-hydroxy-3-nitrophenyl)-acetyl
- SHM
- somatic hypermutation
- S region
- switch region
- UNG
- uracil glycosylase
- wt
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Kato L., Stanlie A., Begum N. A., Kobayashi M., Aida M., Honjo T. 2012. An evolutionary view of the mechanism for immune and genome diversity. J. Immunol. 188: 3559–3566. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs H., Bross L. 2001. Towards an understanding of somatic hypermutation. Curr. Opin. Immunol. 13: 208–218. [DOI] [PubMed] [Google Scholar]
- 3.Di Noia J. M., Neuberger M. S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76: 1–22. [DOI] [PubMed] [Google Scholar]
- 4.Hackney J. A., Misaghi S., Senger K., Garris C., Sun Y., Lorenzo M. N., Zarrin A. A. 2009. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv. Immunol. 101: 163–189. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri J., Alt F. W. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4: 541–552. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z., Wang J. H. 2014. Generation and repair of AID-initiated DNA lesions in B lymphocytes. Front. Med. 8: 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chahwan R., Edelmann W., Scharff M. D., Roa S. 2012. AIDing antibody diversity by error-prone mismatch repair. Semin. Immunol. 24: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada C., Di Noia J. M., Neuberger M. S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16: 163–171. [DOI] [PubMed] [Google Scholar]
- 9.Xue K., Rada C., Neuberger M. S. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2–/– ung–/– mice. J. Exp. Med. 203: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guikema J. E., Linehan E. K., Tsuchimoto D., Nakabeppu Y., Strauss P. R., Stavnezer J., Schrader C. E. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 204: 3017–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrader C. E., Guikema J. E., Wu X., Stavnezer J. 2009. The roles of APE1, APE2, DNA polymerase β and mismatch repair in creating S region DNA breaks during antibody class switch. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eccleston J., Schrader C. E., Yuan K., Stavnezer J., Selsing E. 2009. Class switch recombination efficiency and junction microhomology patterns in Msh2-, Mlh1-, and Exo1-deficient mice depend on the presence of mu switch region tandem repeats. J. Immunol. 183: 1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada C., Williams G. T., Nilsen H., Barnes D. E., Lindahl T., Neuberger M. S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12: 1748–1755. [DOI] [PubMed] [Google Scholar]
- 14.Rada C., Ehrenstein M. R., Neuberger M. S., Milstein C. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity 9: 135–141. [DOI] [PubMed] [Google Scholar]
- 15.Luby T. M., Schrader C. E., Stavnezer J., Selsing E. 2001. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J. Exp. Med. 193: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min I. M., Schrader C. E., Vardo J., Luby T. M., D’Avirro N., Stavnezer J., Selsing E. 2003. The Sμ tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity 19: 515–524. [DOI] [PubMed] [Google Scholar]
- 17.Neuberger M. S., Rada C. 2007. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J. Exp. Med. 204: 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogozin I. B., Kolchanov N. A. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta 1171: 11–18. [DOI] [PubMed] [Google Scholar]
- 19.Rogozin I. B., Pavlov Y. I., Bebenek K., Matsuda T., Kunkel T. A. 2001. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat. Immunol. 2: 530–536. [DOI] [PubMed] [Google Scholar]
- 20.Klotz E. L., Hackett J., Jr., Storb U. 1998. Somatic hypermutation of an artificial test substrate within an Igκ transgene. J. Immunol. 161: 782–790. [PubMed] [Google Scholar]
- 21.Storb U., Klotz E. L., Hackett J., Jr., Kage K., Bozek G., Martin T. E. 1998. A hypermutable insert in an immunoglobulin transgene contains hotspots of somatic mutation and sequences predicting highly stable structures in the RNA transcript. J. Exp. Med. 188: 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael N., Martin T. E., Nicolae D., Kim N., Padjen K., Zhan P., Nguyen H., Pinkert C., Storb U. 2002. Effects of sequence and structure on the hypermutability of immunoglobulin genes. Immunity 16: 123–134. [DOI] [PubMed] [Google Scholar]
- 23.Yélamos J., Klix N., Goyenechea B., Lozano F., Chui Y. L., González Fernández A., Pannell R., Neuberger M. S., Milstein C. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature 376: 225–229. [DOI] [PubMed] [Google Scholar]
- 24.Jolly C. J., Neuberger M. S. 2001. Somatic hypermutation of immunoglobulin κ transgenes: association of mutability with demethylation. Immunol. Cell Biol. 79: 18–22. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., Viboolsittiseri S. S., O’Connor B. P., Wang J. H. 2012. Target DNA sequence directly regulates the frequency of activation-induced deaminase-dependent mutations. J. Immunol. 189: 3970–3982. [DOI] [PubMed] [Google Scholar]
- 26.Fukita Y., Jacobs H., Rajewsky K. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity 9: 105–114. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z., Ranganath S., Viboolsittiseri S. S., Eder M. D., Chen X., Elos M. T., Yuan S., Hansen E., Wang J. H. 2014. AID-initiated DNA lesions are differentially processed in distinct B cell populations. J. Immunol. 193: 5545–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss U., Zoebelein R., Rajewsky K. 1992. Accumulation of somatic mutants in the B cell compartment after primary immunization with a T cell-dependent antigen. Eur. J. Immunol. 22: 511–517. [DOI] [PubMed] [Google Scholar]
- 29.Chen Z., Getahun A., Chen X., Dollin Y., Cambier J. C., Wang J. H. 2015. Imbalanced PTEN and PI3K signaling impairs class switch recombination. J. Immunol. 195: 5461–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonoda E., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., Rajewsky K. 1997. B cell development under the condition of allelic inclusion. Immunity 6: 225–233. [DOI] [PubMed] [Google Scholar]
- 31.Bothwell A. L., Paskind M., Reth M., Imanishi-Kari T., Rajewsky K., Baltimore D. 1981. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a γ2a variable region. Cell 24: 625–637. [DOI] [PubMed] [Google Scholar]
- 32.Dudley D. D., Manis J. P., Zarrin A. A., Kaylor L., Tian M., Alt F. W. 2002. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA 99: 9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M. C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 37: 235–239. [DOI] [PubMed] [Google Scholar]
- 34.Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R. F., Nussenzweig A., Nussenzweig M. C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197: 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milstein C., Neuberger M. S., Staden R. 1998. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA 95: 8791–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner S. D., Milstein C., Neuberger M. S. 1995. Codon bias targets mutation. Nature 376: 732. [DOI] [PubMed] [Google Scholar]
- 37.Wei L., Chahwan R., Wang S., Wang X., Pham P. T., Goodman M. F., Bergman A., Scharff M. D., MacCarthy T. 2015. Overlapping hotspots in CDRs are critical sites for V region diversification. Proc. Natl. Acad. Sci. USA 112: E728–E737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal D., Maul R. W., Ghosh A., Chakraborty T., Khamlichi A. A., Sen R., Gearhart P. J. 2009. Immunoglobulin switch μ sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J. Exp. Med. 206: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeap L. S., Hwang J. K., Du Z., Meyers R. M., Meng F. L., Jakubauskaitė A., Liu M., Mani V., Neuberg D., Kepler T. B., et al. 2015. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell 163: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z., Fulop Z., Wu G., Pone E. J., Zhang J., Mai T., Thomas L. M., Al-Qahtani A., White C. A., Park S. R., et al. 2010. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 17: 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M., Duke J. L., Richter D. J., Vinuesa C. G., Goodnow C. C., Kleinstein S. H., Schatz D. G. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845. [DOI] [PubMed] [Google Scholar]
- 42.Liu M., Schatz D. G. 2009. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 30: 173–181. [DOI] [PubMed] [Google Scholar]
- 43.Jansen J. G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203: 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Durán P., Belver L., de Yébenes V. G., Delgado P., Pisano D. G., Ramiro A. R. 2012. UNG shapes the specificity of AID-induced somatic hypermutation. J. Exp. Med. 209: 1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verri A., Mazzarello P., Spadari S., Focher F. 1992. Uracil-DNA glycosylases preferentially excise mispaired uracil. Biochem. J. 287: 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett S. E., Sanderson R. J., Mosbaugh D. W. 1995. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry 34: 6109–6119. [DOI] [PubMed] [Google Scholar]
- 47.Parikh S. S., Putnam C. D., Tainer J. A. 2000. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 460: 183–199. [DOI] [PubMed] [Google Scholar]
- 48.Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R. T., Datta A., et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94: 157–214. [DOI] [PubMed] [Google Scholar]
- 49.Stavnezer J. 2011. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 32: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briney B. S., Willis J. R., Crowe J. E., Jr. 2012. Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun. 13: 523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson P. C., de Bouteiller O., Liu Y. J., Potter K., Banchereau J., Capra J. D., Pascual V. 1998. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J. Exp. Med. 187: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham P., Bransteitter R., Petruska J., Goodman M. F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424: 103–107. [DOI] [PubMed] [Google Scholar]
- 53.Chaudhuri J., Khuong C., Alt F. W. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 430: 992–998. [DOI] [PubMed] [Google Scholar]
- 54.Zarrin A. A., Alt F. W., Chaudhuri J., Stokes N., Kaushal D., Du Pasquier L., Tian M. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5: 1275–1281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.