Abstract
Th17 cells contribute to the development of some autoimmune and allergic diseases by driving tissue inflammation. However, the function of Th17 cells during cancer progression remains controversial. Here, we show that human memory CD25high Th17 cells suppress T cell immunity in breast cancer. Ectonucleotidase-expressing Th17 cells accumulated in breast cancer tumors and suppressed CD4+ and CD8+ T cell activation. These cells expressed both Rorγt and Foxp3 genes and secreted Th17 related cytokines. We further found that CD39 ectonucleotisase expression on tumor-infiltrating Th17 cells was driven by TGF-βand IL-6. Finally, immunohistochemical analysis of localized breast cancer revealed that high-tumor infiltration by IL-17+ cells was associated with a poor clinical outcome and impeded the favorable effect of high CD8+ infiltration. Altogether, these findings suggest that intratumoral Th17 cells compromise anticancer immune responses in breast cancer patients.
Keywords: breast cancer, Th17, prognosis, ectonucleotidase
Introduction
Th17 cells are a new population of CD4+ T cells involved in cancer immunomodulation.1 These cells are characterized by their capacity to produce IL-17A and IL-17F, but also IL-10, IL-21, IL-22 and TNFα and require the transcription factors RORγt and RORα for their development.2 In humans, Th17 cells are characterized by the expression of the chemokine receptor CCR6.3 While Th17 cells were initially identified in the context of autoimmune responses, we and others have demonstrated that these cells were present in mouse and human cancers.4-7 However, their role in cancer still remains controversial. Adoptive transfer of tumor specific Th17 cells in mouse models demonstrated their potent anticancer activity.8 In human ovarian cancer high expression of IL-17 is also associated with a favorable clinical outcome.9 In contrast, other reports have shown that Th17-derived IL-17A could favor cancer progression because of its capacity to promote vascular endothelial growth factor (VEGF) production and tumor angiogenesis.10,11 We have also previously reported the immunosuppressive functions and the protumorigenic role of murine Th17 lymphocytes.4 We found that TGF-β and IL-6 drove the differentiation of naive mouse CD4+ T cells into Th17 cells that expressed the ectonucleotidases CD39 and CD73.4 These enzymes degrade extracellular ATP into the immunosuppressive molecule adenosine. Accordingly, we found that Th17 could suppress T cell immune responses and promote tumor growth through adenosine. Whether this immunosuppressive pathway is also relevant in human cancer has however remained elusive.
In the present study, we focused our interest on human localized breast cancer (LBC). We observed a specific accumulation of ectonucleotidase-expressing Th17 cells in breast tumors. These cells were found to suppress T cell activation and are associated with poorer prognosis.
Results
Foxp3+ Th17 cells accumulate in locally advanced breast cancer
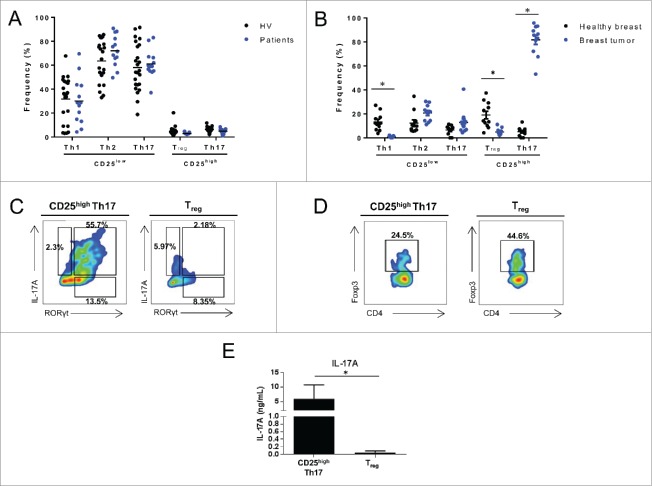
We analyzed the frequency of helper memory CD4+ T cells that were isolated from periheral blood mononuclear cells (PBMCs) of patients with breast cancers and healthy individuals (Table S1 summarizes the clinical characteristic of the patients). Frequency of memory T cell subpopulations was determined using their chemokine receptor expression12 (Fig. S1). In the CD25high population, we observed CCR6 negative and positive populations that respectively shared Treg and Th17 profiles based on expression of transcription factor and cytokine production (Fig. S2). In CD25low population, we isolated CCR6− CXCR3+, CCR6− CXCR3− CCR4+ and CCR6+ CXCR3− populations that respectively shared Th1, Th2 and Th17 profiles (Fig. S2). We did not detect frequency variation of memory helper CD4+ T cell population in PBMCs (Fig. 1A). We then analyzed the frequency of T helper cell populations in 12 samples of normal breast tissue and 13 breast tumors (Table S1). We observed an enrichment of CD4+ T cell infiltration in tumor compared to non-tumor breast tissue. In addition, we observed an accumulation of CD25high expressing Th17 cells in the tumor tissues in comparison to normal breast tissue (Fig. 1B) or blood (Fig. S3). We confirmed that these CD25high CCR6+ tumor-infiltrating cells are Th17 cells, as they express RORγt and IL-17A (Fig. 1C). These cells also co-expressed Foxp3, and are hereafter referred to as CD25high Th17 cells (Fig. 1D). After restimulation CD25high Th17 cells but not Treg produce IL-17A (Fig. 1E). We confirmed that CD25high Th17 cells from PBMCs produce and express other Th17 related cytokines such as IL-17F, IL-21, IL-22, IL-10 and TNFα (Fig. S4). Together, these data demonstrate a specific accumulation of CD25high Th17 cells in the tumor bed of human breast cancer.
Figure 1.
Th17 cells accumulate in locally invasive breast cancer. (A) PBMCs from HV or breast cancer patients (patients) were stained with anti-CD4+, anti-CD45RA, anti-CCR6, anti-CCR4, anti-CXCR3, anti-CD25 antibodies and analyzed by flow cytometry. The frequency of memory CD4+ (CD45RA−, CD4+) Th1 (CD25low, CCR6−, CXCR3+), Th2 (CD25low, CCR6−, CXCR3−, CCR4+), Th17 (CD25low, CCR6+, CXCR3−), CD25high Th17 (CD25high, CCR6+, CXCR3−) and Treg (CD25high, CCR6−, CXCR3−) cells is depicted. The data presented represent the analyses performed on 23 HVs and 13 breast cancer patients. (B) CD4+ lymphocytes from breast tumors (n =13) or normal breast tissue (n = 12) were analyzed as in (A). The frequency of memory CD4+ T cells is depicted. (C) Tumor infiltrating CD25high Th17 cells and Treg cells were analyzed for IL-17 and RORγt expression by intracellular staining. Numbers beside outlined areas indicate percent cells in gate. (D) Tumor infiltrating CD25high Th17 cells and Treg cells were analyzed for Foxp3 expression by intracellular staining. Numbers beside outlined areas indicate percent cells in gate. (E) Tumor infiltrating CD25high Th17 and Treg cells sorted from breast tumors were restimulated with anti-CD3 and anti-CD28 antibodies and IL-17A secretion was assessed by ELISA after 3 d. Representative data from one of at least three independent experiments are shown (C–F). *p < 0.05
CD25+ Th17 cells express ectonucleotidases
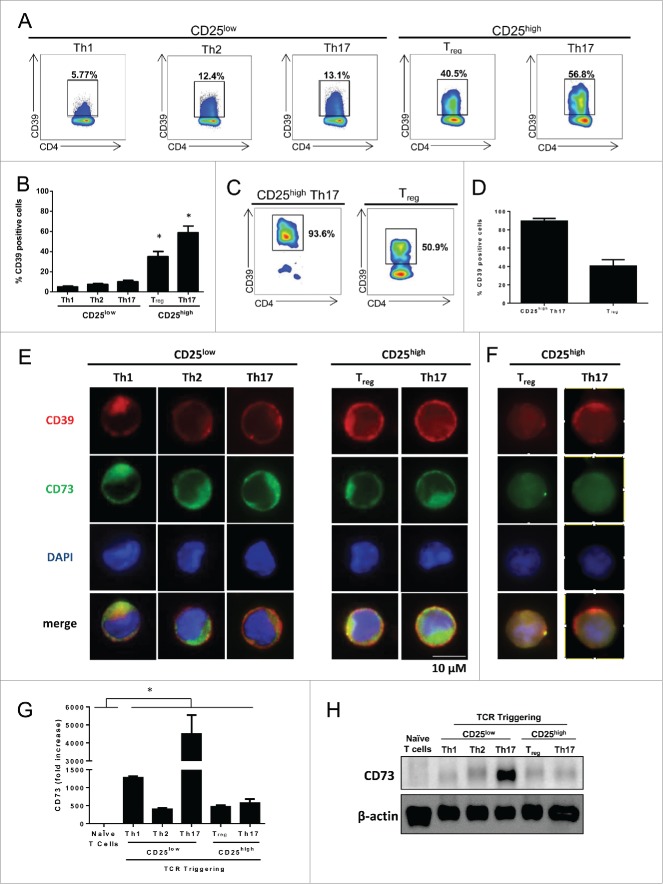
We have tested the expression of ectonucleotidases in CD4+ memory subsets in PBMC from healthy volunteers (HV). We observed that more than 50% of Foxp3+ Treg and CD25high Th17 cells but less than 15% of Th1, Th2 and CD25low Th17 cells expressed CD39 (Figs. 2A, B). We also observed that tumor-infiltrating Th17 cells expressed high levels of CD39 (Figs. 2C,D). CD73 expression could not be detected on Treg and Th17 cells using flow cytometry, however immunofluorescence revealed its submembrane location on both cell types13,14 (Fig. 2E). Moreover, we confirmed that tumor-infiltrating CD25high Th17 cells express ectonucleotidases (Fig. 2F). We confirmed CD73 expression on all activated CD4+ T cell subsets using q-PCR and Western Blotting (Figs. 2G, H). Together these data indicate that human blood and tumor infiltrating Th17 cells express CD39.
Figure 2 (See previous page).
Human CD25high Th17 cells express ectonucleotidases.Memory blood-derived (A, B) or breast-tumor infiltrating (C, D) Th1, Th2, Th17 as well as CD25high Th17 and Tregs were analyzed for CD39 expression using flow cytometry (representative dot plot (A,C) and mean ± SD percentage of cells of 3 independent experiments (B, D). (E) Memory blood-derived Th1, Th2, Th17 as well as CD25high Th17 and Tregs were stained, permeabilized and analyzed for CD39 and CD73 expression using immunofluorescence. Memory blood-derived Th1, Th2, Th17 as well as CD25high Th17 and Tregs were analyzed for CD73 expression using. (F) Breast tumor infiltrating Treg or Th17 CD25high lymphocytes were sorted out and restimulated with anti-CD3 and anti-CD28 antibodies. After 3 d, Entpd1 and Nt5e expression were analyzed using immunofluorescence. (G) q-PCR (mean ± SD percentage of cells of three independent experiments) and (H). Western blotting (One representative of three independent experiments) after 24 and 72 h of in vitro stimulation respectively.
Human Th17 cells exert adenosine dependent suppression
The expression of CD39 and CD73 ectonucleotidases catalyzes the transformation of extracellular ATP into adenosine, which dampens T cell responses.15 Th17 cells had a nucleoside triphosphate diphosphohydrolase activity comparable to Treg cells (Fig. 3A). CD39 mAb blunted adenosine production by both Th17 and Treg subsets (Fig. S5A). Adenosine requires expression of its receptor on the target cell to mediate its effect. We observed that human CD8+ T cells and Th1 CD4+ T cells express selectively the A2A receptor (Fig. 3B). We observed that CD25high Th17 cells decrease the ability of Th1 and CD8+ T cells to produce IFNγ or TNFα in a dose dependent manner (Figs. 3C, D). These cells exert comparable immunosuppressive functions to Treg cells. However, we showed that CD25low Th17 cells do not suppress IFNγ secretion (Fig. S5B). Importantly, we observed that the immunosuppressive effect of Th17 cells is decreased by the addition of CD39 blocking antibody or A2A receptor inhibitor (Fig. 3E). We have tested additional doses of the inhibitor A2A receptor inhibitor. While we noted a dose-dependent effect, doses higher than 10µM fail to be more efficient to revert the effect of CD25high Th17 cells (Fig. S5C).
Figure 3.
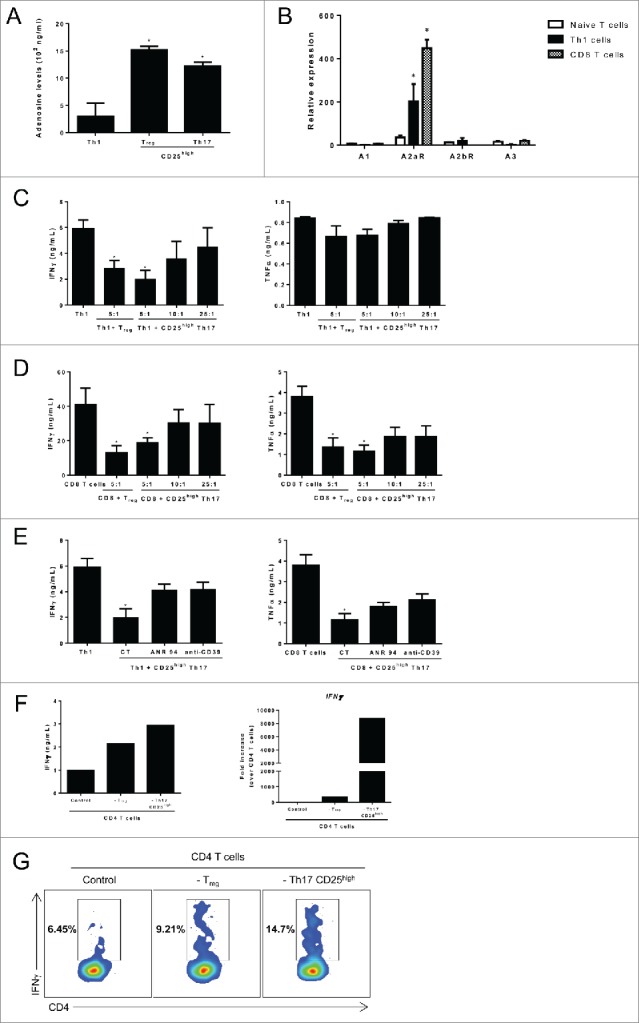
CD25high Th17 cells exert adenosine dependent suppressive functions. (A) Blood-derived memory Th1, CD25high Th17 cells and Tregs were cocultured for 72 h with 2 µM ATP. The concentration of adenosine in the supernatant was determined by enzymatic assay.25 (B) Blood-derived memory Th1 cells and CD8+ T cells were cell sorted using flow cytometry, reactivated using anti-CD3 and anti-CD28. A1, A2AR, A2BR and A3 mRNA expression level was assessed after 72 h by RT-qPCR analysis. CD25high Th17 cells or Tregs were cocultured with (C) CD4+ or (D) CD8+ T cells at different ratios (1,00,000 Th1 or CD8+ T cells to 20,000 (5:1), 10,000 (10:1) or 2,500 (25:1) suppressive cells) for 3 d. IFNγ and TNFα secretion was assessed using ELISA. (E) Same as in (C) and (D) using neutralizing anti-CD39 antibody or an adenosine A2A receptor inhibitor ANR 94. (F) Breast tumor infiltrating lymphocytes (TILs) were sorted out and either total CD4+ TILs or TILs depleted from immunosuppressive populations (Treg or Th17 CD25high cells) were restimulated with anti-CD3 and anti-CD28 antibodies for 3 d. IFNγ secretion was assessed by ELISA and IFNγ expression was analyzed by RT-qPCR (G) same as in (F) but IFNγ secretion was assessed by intracellular staining 3. Data are mean ±SD of three independent experiments. *p < 0.05.
Together these data demonstrate that Th17 cells suppressed both CD8+ and Th1 cell ability to produce IFNγ in an adenosine and A2A receptor dependent manner.
Additionally we observed that CD4+ TILs depleted from CD25high Th17 are able to produce more IFNγ than total CD4+ TILs (Figs. 3F, G), confirming that tumor-infiltrating CD25high Th17 cells suppress T cell responses.
Th17 cell accumulation in breast cancer is due to both proliferation and recruitment
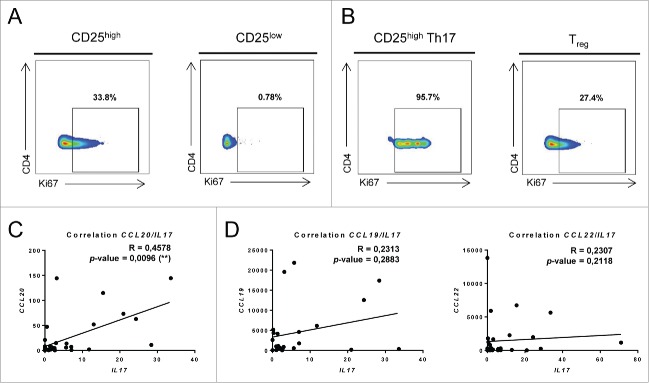
Local accumulation of Th17 cells in breast tumor could be due to their proliferation in situ or to their recruitment. We measured Ki67 expression in CD25high and CD25low tumor infiltrating memory CD4+ T cells. Ki67 was shown to be expressed only during the first 24h after TCR triggering.16 We observed a specific proliferation of CD25high memory CD4+ T cells in tumor bed compared to CD25low tumor infiltrating memory T cells and PBMCs memory T cells (Fig. 4A). Interestingly this proliferation is specifically observed in the CD25high Th17 cells subset and not in Treg (Fig. 4B).
Figure 4.
Th17 cell accumulation in breast cancer is due to both proliferation and recruitment. (A) Ki67 expression in CD25high and CD25low tumor-infiltrating memory CD4+ T cells was determined by flow cytometry using intracellular staining. (B) Ki67 expression was determined as in (A) in CD25high Th17 and Tregs. Representative data from one of three experiments are shown. mRNA was extracted from 36 breast cancer tumor samples and the expression of IL17, CCL19, CCL20 and CCL22 was determined using RT-qPCR. The correlation between (C) IL17 and CCL20 as well as (D) IL17 and CCL19 or IL17 and CCL22 expression was determined using RT-qPCR.
Th17 cells are characterized by the specific expression of CCR6,17 a receptor for the chemokine CCL20.18 CCL20 expression at the tumor site could be involved in the accumulation of Th17 cells. We obtained mRNA from 36 locally advanced breast cancers (Table S2) and observed a strong correlation between mRNA CCL20 expression in tumor bed and IL17 mRNA expression (Fig. 4C). As a control, CCL22 expression which is known to recruit Treg cells and CCL19, which is known to recruit CCR7+ T cells are not associated with IL17 mRNA and protein expression (Fig. 4D). Together these data suggest that CD25high Th17 cells accumulate in breast tumor because of both recruitment via CCL20/CCR6 pathway and in situ proliferation.
Ectonucleotidase expression on Th17 cells is driven by tumor-derived TGF-β and IL-6
We previously reported in mouse models that ENTPD1 and NT5E (encoding CD39 and CD73 respectively) expression in Th17 cells is due to the stimulation by TGF-β and IL-6, which are frequently expressed in breast tumor tissue. We thus speculated that after recruitment in the tumor bed via the CCR6/CCL20 axis, Th17 cells could be locally stimulated to acquire ectonucleotidase immunosuppressive functions.
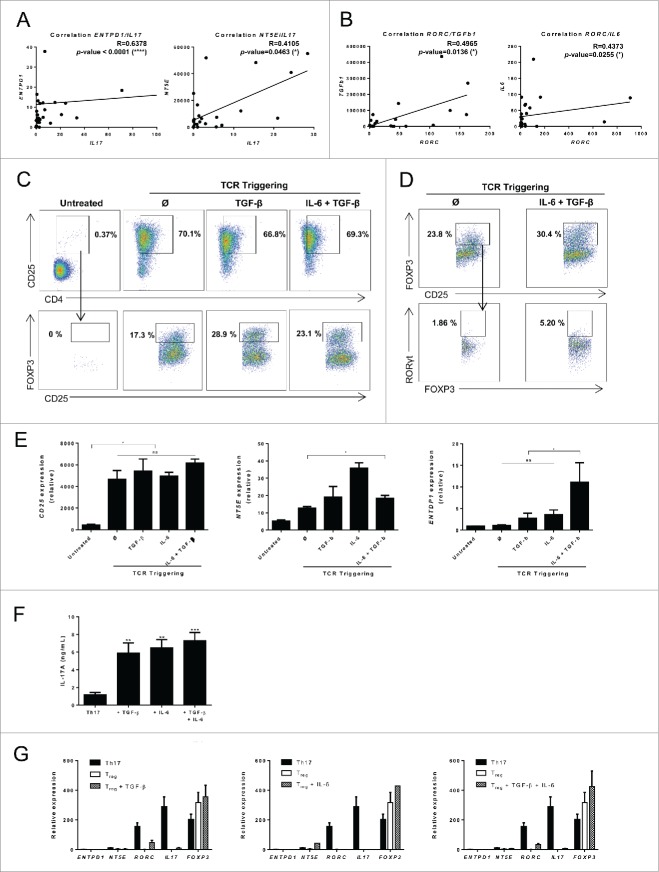
Using a series of 36 locally advanced breast cancers we observed a correlation between IL17 mRNA expression in tumor and ENTPD1 and NT5E mRNA expression (Fig. 5A). In addition, we noted a correlation between RORC and IL6 as well as RORC and TGFb1 expression (Fig. 5B), thus suggesting that these two cytokines could are involved in accumulation of immunosuppressive Th17 cells. In contrast, neither IL6 nor ENTPD1 and NT5E were correlated with FOXP3 expression (Fig. S6). We cell sorted CD25low Th17 from healthy donor PBMCs and stimulated these cells with TCR triggering and TGF-β or IL-6 or the combination of both cytokines. Using flow cytometry we found that TCR triggering is sufficient to induce CD25 expression and that TGF-β or the combination of TGF-β and IL-6 induced Foxp3 expression in Th17 cells (Fig. 5C). As a control, Treg cells could not be differentiated into CD25high Th17 cells by TGF-β and IL-6 (Fig. 5D). TCR triggering induced CD25 and NT5E but not ENTPD1 mRNA expression. NT5E expression is boosted by cytokine stimulation. In contrast, only cytokine stimulation with TGF-β and IL-6 enhanced ENTPD1 mRNA expression (Fig. 5E). We observed that IL-17 secretion was maintained in these conditions (Fig. 5F). As a control, TGF-β or IL-6 or the combination of both cytokines did not enhance ectonucleotidase expression in Treg cells (Fig. 5G). These data suggest that tumor production of TGF-β and IL-6 could promote differentiation of conventional Th17 cells into CD25high Th17 cells that expressed CD39 and CD73.
Figure 5 (See previous page).
Th17 cell ectonucleotidase expression is driven by tumor-derived TGF-β and IL-6. (A) mRNA was extracted from 36 breast cancer tumor samples and the expression of IL17, ENTPD1 and NT5E was determined using RT-qPCR. The correlation between IL17 and ENTPD1 as well as IL17 and NT5E expression was determined using RT-qPCR. (B) mRNA was extracted from 36 breast cancer tumor samples and the expression of RORC, TGFb1 and IL6 was determined using RT-qPCR. The correlation between RORC and TGFb1 as well as RORC and IL6 expression was determined using RT-qPCR. (C) CD25low memory Th17 cells were cell sorted from PBMCs and restimulated with anti-CD3 and anti-CD28 antibodies for 3 d in the presence of TGF-β, IL-6 or TGF-β and IL-6. After 3 d, the expression of CD25 and FOXP3 were determined by flow cytometry. Representative data from one of three independent experiments are shown. (D) Tregs were cell sorted from PBMCs and restimulated with anti-CD3 and anti-CD28 antibodies for 3 d in the presence of TGF-β and IL-6. After 3 d, the expression of CD25, FOXP3 and RORC were determined by flow cytometry. Representative data from one of three independent experiments are shown. (E) CD25high memory Th17 cells were cell sorted from PBMCs and restimulated with anti-CD3 and anti-CD28 antibodies for 3 d in the presence of TGF-β, IL-6 or TGF-β and IL-6. After 3 d, mRNA was extracted and the expression of CD25 (left panel), NT5E (middle panel) and ENTPD1 (right panel), was determined. (F) CD25low memory Th17 cells were cell sorted from PBMCs and were restimulated with anti-CD3 and anti-CD28 antibodies for 3 d in the presence of TGF-β, IL-6 or TGF-β and IL-6. After 3 d, IL-17A secretion was assessed by ELISA. ***p <0.001.(G) Treg were cell sorted from PBMCs and restimulated with anti-CD3 and anti-CD28 antibodies for 3 d in the presence of TGF-β (left panel), IL-6 (middle panel) or TGF-β and IL-6(right panel). After 3 d, mRNA was extracted and the expression of ENTPD1, NT5E, RORC, IL17 and FOXP3 was determined. Th17 cells were used as control. Data are mean ± SD of two independent experiments (E, F). * p < 0.05 (unless otherwise indicated).
IL-17-infiltrating cells drive immunosuppression in human breast cancer
We evaluated the functional characteristics of immune cell infiltrates using low array immune gene expression in our series of 36 patients with local breast cancer. In this population we performed IL-17 immunohistochemistry and observed that 15 tumors are highly infiltrated by IL-17+ cells, and 21 were poorly infiltrated by IL-17+ cells. Upon gene expression we observed that gene related to the Th17 cluster (IL17, IL21, RORC, TNFα) and CD39 were significantly overexpressed among IL-17Hi tumors (Fig. 6A), thus demonstrated that IL-17 labeling is a valuable surrogate marker of Th17 infiltration.
Figure 6.
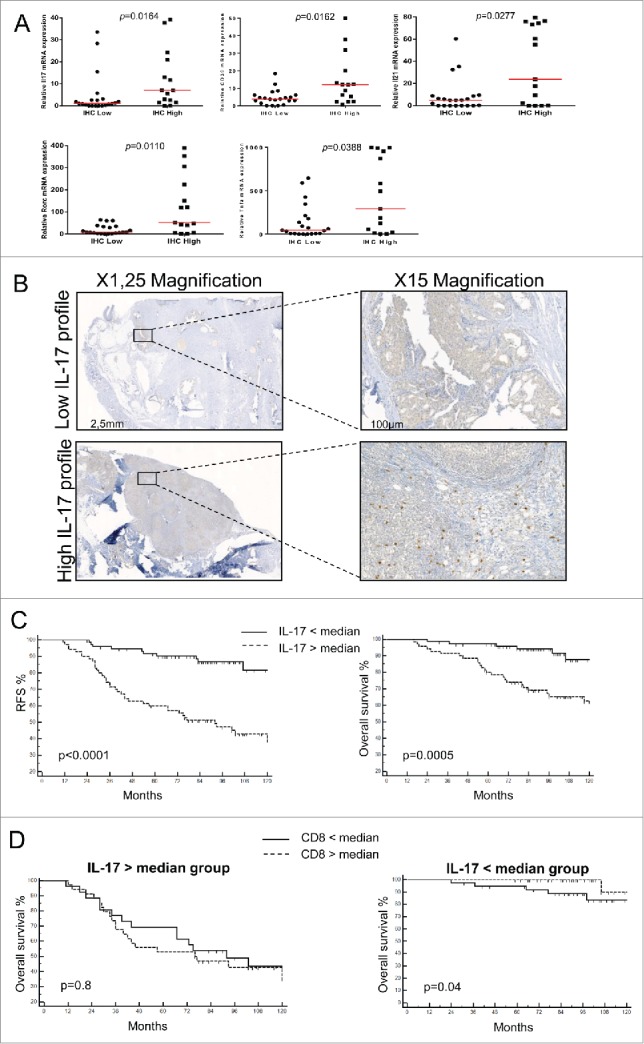
IL-17-infiltrating cells drive immunosuppression in human breast cancer/IL-17-infiltrating cells negatively affect human breast cancer prognosis. (A) IL-17 expression in 36 breast cancer tumor samples was determined by immunohistochemistry (IHC). Samples were then classified as IL-17high and IL-17low based on IHC results and the expression of IL17, ENTPD1, IL21, RORC and TNFA was determined using RT-qPCR. (B) Representative picture of IL-17+ staining of breast tumor using immunhistochemistry. We represent a tumor with a low (upper panel) and high (lower panel) IL-17 infiltrates. N = 145 patients. (C) Kaplan–Meier curves of relapse free survival (left panel) and overall survival (right panel) for patients with local breast cancer according to the presence of a high or low density of IL-17+ cells in the tumor stroma. N =145 patients. (D) Kaplan–Meier curves of overall survival according to the presence of a high or low density of CD8+ cells in the tumor stroma in the group of a high (left panel) or low (right panel) density of IL-17+ cells infiltrate. N = 70.
To test the global immune response, we constructed a correlation matrix for that we submitted to hierarchical clustering. We determined two major clusters of genes displaying similar correlation profile. One cluster contained genes coregulated with CD3e (black rectangle) and associated with T cell density. The other one contained genes coregulated with CD33 (red rectangle) and associated with myeloid derived suppressor cells (MDSC) infiltration and immune suppression (Fig. S7A). We performed analysis using ClueGO software to determine interconnection between each gene clusters. We identified a network of intercorrelated genes associated with a coordinated CD4+ and CD8+ T cell response associated with a Th17 immune response (Fig. S7B). Together these data underscore that in breast cancer a coordinated immune response is present with a T cell immune response drift toward Th17 polarization in association with a myeloid immunosuppressive pathway.
In a retrospective series of 145 patients with node positive breast cancer homogenously treated by surgery followed by anthracycline-based adjuvant chemotherapy (Table S3), we quantified by immunohistochemistry stromal and tumor nest CD8+ and IL-17+ cells. We then separated patients into two groups (high vs. low), using median as a cut-off for each marker (Fig. 6B). We observed that only high stromal IL-17+ cells were significantly associated with poor relapse-free survival (RFS) and overall survival (OS) in both univariate and multivariate models (Fig. 6C and Tables S4 and S5). Upon testing the prognostic role of CD8+ cells in the two IL-17 groups we observed no imbalance between prognosis factors (Table S6). High infiltration in CD8+ cells is associated with a better survival only in the group of low IL-17+ infiltrates (Fig. 6D). Taken together, these data suggest that Th17 presence in human breast cancer is associated with ectonucleotidase expression, and with poorer outcome.
Discussion
Tumor-infiltrating lymphocytes (TILs) have been shown to profoundly affect breast cancer prognosis. However, the nature and functions of TILs in breast cancer microenvironment remains incompletely defined. Here, we found that CD25high Th17 cells accumulated in human breast cancer. These cells express CD39 and CD73 and suppress CD4+ and CD8+ T cell activation through adenosine release. Breast cancer patients featuring high IL-17+ cell infiltration had a poor clinical prognosis; even in case of a high CD8+ T cell infiltration. These novel human data underscore the clinical relevance of this immunosuppressive pathway in breast cancer.
In contrast to the well-defined contributions of the effector T cells in breast cancer, the role of Th17 cells in this disease remains elusive. Tumor infiltration by IL-17-producing cells has been proposed to be a negative prognostic factor,19 suggesting that Th17 cells may be detrimental in this context. However, the underlying biological mechanisms have not yet been reported. In breast cancer, CCR6+ Foxp3+ cells were proposed to dampen CD8+ T cell activation, leading to poorer clinical prognosis.20 However, whether these cells presented Th17 cell features was unclear, and the immune suppression mechanism was not documented. Our findings here revealed that in the breast tumor microenvironment, CD25high Th17 cells that expressed both Foxp3 and RORc are Th17 cells characterized by their ability to express Th17 cytokines, but are also immunosuppressive cells that contribute to tumor escape.
We had previously reported that mouse Th17 cells could harbor immunosuppressive functions that relied primarily on their expression of ectonucleotidases.4 The expression of the ectonucleotidases CD39 and CD73 allow the conversion of ATP into adenosine, which dampens T cell activation through binding to A2A receptors. Unlike effector T cells, Treg express high levels of CD39, which contributes to immune suppression. Our findings here report for the first time in breast cancer the existence of ectonucleotidase-expressing CD25high Th17 cells. These cells expressed CD39 and CD73 ectonucleotidases at comparable levels to Treg cells. Interestingly, in ulcerative colitis, CD39 expression was similarly found comparable between immunosuppressive Foxp3+ Th17 cells and bona fide Treg from patients, confirming the ability of CD25high Th17 cells to suppress immune activation in various contexts.21 However, as effector Th1 cells or CD8+ T cells expressed CD73 (Fig. 2E and data not shown) we cannot exclude a possible cooperation between CD25high Th17 cells and effector cells to induce adenosine production.
Our findings indicating the predominance of CD25high Th17 cells over Treg in the breast cancer tumor microenvironment suggest that these cells primarily drive immune suppression at the tumor site. In addition our observations indicate that CD25high Th17 cells are able to proliferate at the tumor site, and are recruited via the CCR6/CCL20 axis. In addition, we identified here that TGF-β and IL-6 favor the conversion of conventional CCR6+ peripheral Th17 into CD25high immunosuppressive Th17 cells. Together these results support the hypothesis that CD25low CCR6+ peripheral Th17 cells could be recruited in the tumor bed by CCL20, and then activated by local antigen stimulation, thus suggesting local priming and a coordinated Th17 derived immune response. While the ability of Treg to differentiate into Th17 cells in proinflammatory environments has been thoroughly studied,22 the conversion of conventional Th17 cells into suppressive Th17 has not been yet explored, and the influence of the tumor microenvironment on Th17 cells remains incompletely understood.
The prognostic value of CD8+ and Treg tumor infiltration has been established in large cohorts of breast cancer patients.23,24 Our retrospective analysis of CD8+ and IL17+ breast cancer infiltration by immunohistochemistry indicates that the combined analysis of these two cell populations refines breast cancer patient's prognosis. The observation that high CD8+ T cell infiltration is associated with favorable outcome only in the presence of low associated-IL-17+ cell infiltration further suggests an important immunosuppressive role of CD39-expressing Th17 cells on CD8+ dependent breast anticancer immunity. These results also support the use of therapies designed to neutralize ectonucleotidases in breast cancer.
Materials and Methods
Patients and healthy donors
Between January, 2013 and December 2013, we collected PBMCs and tumor samples from patients with breast cancer in the Centre Georges François Leclerc. The study group (n = 14) comprised patients who underwent surgery for an invasive ductal non-metastatic breast carcinoma. The control group (n = 23) consisted of sex and age matched HV. Tumor stage was determined according to the 2002 International Union against Cancer TNM classification system. All patients gave informed consent approved by the local Ethics Committee. Review of pathology reports confirmed the diagnosis. Information regarding clinical pathological characters of patients is presented in Table S1.
Cohort 1: We retrospectively studied frozen cancer-tissue specimens from 36 consecutive patients who underwent surgery for an invasive ductal non-metastatic breast carcinoma at the Georges Francois Leclerc Cancer Centre, Dijon France from January 2010 to June 2010.
A second cohort of 145 consecutive patients, treated by surgery and adjuvant therapy by anthracycline-based chemotherapy for HER2-negative invasive ductal non-metastatic breast carcinoma between 2002 and 2004 at the Georges Francois Leclerc Cancer Centre. The study was approved by the ethical local committee, and patients gave written informed consent for the use of samples from their tumors for future investigations at the time of the diagnosis. Histoprognostic grade was defined according to the modified Bloom and Richardson method. The steroid hormone receptor status was determined using enzyme immunoassays (Abbott Diagnostics, Rungis, France).
T cell isolation from tumors
Tumors were removed aseptically and minced with scissors into 1–2 mm3 pieces. The minced tumors were then stirred in 40 mL complete RPMI 1640 containing 40 mg collagenase, type IV (Sigma), 4 mg deoxyribonuclease (Sigma) for 3 h at room temperature. The tumor cell suspension was filtered through a nylon-mesh screen with pores of 50 μm to remove cell clumps, and the filtrate was then centrifuged (250×g, 10 min). The cell pellet was washed twice with serum-free RPMI 1,640 and resuspended in complete RPMI 1,640.
T cell culture
Memory CD4+ T cells (CD4+ CD45RA−) were first enriched from blood PBMCs using the Human CD4+ T Cell Enrichment Cocktail (Stemcell, 50 µl/mL) and then cell-sorted. Isolated CD4+ T cells were stimulated with plate-bound antibodies against CD3 (OKT3, 2 µg/mL, BioXcell) and CD28 (CD28.2, 2 µ g/mL, BioXcell) with or without cytokines. Human IL-6 (20 ng/mL) and TGF-β (5 ng/mL) were all purchased from MiltenyiBiotec. Cells were classically harvested on day three (unless otherwise specified) for detection of cytokines by ELISA and real-time quantitative PCR analysis.
Suppression assays
To test CD4+ CD25high Th17 cell suppressive activity, 105 Th1 or CD8+ T cells were activated with 2 μg/mL anti-CD3 (BioXcell) and anti-CD28 (BioXcell) as effector cells and cocultured with or without Treg or CD4+ CD25high Th17 cells at ratios of 5:1, 10:1 and 25:1 for 72 h in complete medium (XVivo 15, gibco). IFNα and TNFα secretion was assessed subsequently after 3 d. In some experiments, an anti-CD39 antibody (A1, 10 µg/mL, AbD serotec) or an adenosine A2A receptor inhibitor ANR94 (10 µmol/L) was added.
Quantification of adenosine levels
Adenosine levels were assessed as previously described.25 Briefly, 30 µL of conditioned medium after reactivation of different CD4+ T cell subsets were transferred into separate wells in 96-well microplates containing combinations of the following enzymes in a final volume of 170 µL buffer: 0.3 U/mL adenosine deaminase (type IX from bovine spleen), 0.25 U/mL bacterial purine nucleoside phosphorylase and 0.15 U/mL microbial xanthine oxidase (all from Sigma). After 20 min at room temperature, 30 µL of H2O2-detecting mixture containing HRP (1 U/mL, Invitrogen, Molecular Probes) and Amplex Red reagent (60 µM, Invitrogen, Molecular Probes) was added to the microwells, followed by measurement of the fluorescence intensity at the emission and excitation wavelengths of 545 and 590 nm, respectively. Two different enzymatic cocktails were used: mix A containing the complete enzymatic cascade and mix B containing the same mixture without adenosine deaminase. For adenosine sensing, the background fluorescence determined in the absence of adenosine deaminase (mix B) has been substracted from the fluorescence in well containing the complete adenosine-converting cascade.
Flow cytometry
Antibodies anti-CXCR3 (G025H7), anti-CCR6 (G034E3), anti-CCR4 (TG6), anti-CD25 (BC96), anti CD45RA (HI100) and anti-CD4+ (OKT4) were purchased from Biolegend. All events were acquired by a BD LSR-II cytometer equipped with BD FACSDiva software (BD Biosciences) and data were analyzed using FlowJo software (Tree Star, Ashland, Oregon).
Measurement of cytokines
Cell culture supernatants were assayed by ELISA for human IFNγ, IL-4, IL-10, IL-17A, IL-17F, IL-21, IL-22 and TNF-α (Biolegend, St Quentin, France) according to manufacturer's protocol.
For intracellular cytokine staining, cells were stimulated for 4 h at 37˚C in culture medium containing PMA (50 ng/mL; Sigma-Aldrich), ionomycin (1 µg/mL; Sigma-Aldrich) and monensin (GolgiStop; 1 µl/mL; BD Biosciences). After staining for surface markers and 7-Amino-Actinomycin D (7-AAD) to exclude dead cells, cells were fixed and permeabilized according to the manufacturer's instructions (Cytofix/Cytoperm kit, BD Biosciences), then stained for intracellular products. Antibodies used for intracellular staining were as follows: Allophycocyanin-conjugated anti-RORγt (REA278, Miltenyi), phycoerythrin (PE)-conjugated anti-Foxp3 (PCH101, eBioscience) or PE-conjugated anti-RORγt (Miltenyi) or Brilliant Violet 421-conjugated anti-IL-17A (eBio64DEC17, eBioscience).
Real-time quantitative PCR
Total RNA from T cells was extracted with TriReagent (Ambion), reverse transcribed using M-MLV Reverse Transcriptase (Invitrogen) and was analyzed by real-time quantitative PCR (RT-qPCR) with the Sybr Green method according to the manufacturer's instructions using the 7,500 Fast Real Time PCR system (Applied Biosystems). Expression was normalized to the expression of human ACTB. Primers designed to assess gene expression are as reported in Table S1.
Western blotting
Purified naive T cells were differentiated for 72 h into Th1, Th2, Th17, Treg or CD25high Th17 cells, then collected and pelleted by centrifugation (5 min, 1,500 × g). Cells were lysed in boiling buffer [1% SDS, 1 mmoL/L sodium orthovanadate, and 10 mmoL/L Tris (pH 7.4)] containing protease inhibitor cocktail for 20 min at 4°C. Cell lysates were subjected to sonication (10 s at 10%) and protein concentrations were assessed using a Bio-Rad DC Protein Assay Kit. Proteins were then denaturated, loaded, and separated by SDS-PAGE and transferred onto nitrocellulose membranes (Schleicher & Schuell). After blockade of nonspecific binding with 5% bovine serum albumin (BSA) in Tris-buffered saline containing 0.1% tween 20 (TBST), membranes were incubated overnight with primary antibody diluted in TBST containing 1% BSA, washed and incubated for 1 h with secondary antibody diluted in TBST–1% BSA. After additional washes, membranes were incubated with Luminol reagent (Santa Cruz Biotechnology) and exposed to X-ray films. The following human monoclonal antibody was used: anti-CD73 (NBP1-85740, Novus).
Immunohistochemical labeling
Immunohistochemistry used monoclonal antibodies against the T-cell marker CD8+ (Dako, Trappes, France), and IL-17 (Santa Cruz Biotechnology, Santa Cruz, USA). Antigen retrieval was carried out by heating slides for 15 min at 95°C in 1 mmoL/l EDTA. Labeling was detected using the Dako Envision system (Dako). The stained arrays were counterstained with haematoxylin and mounted in Aquamount (Dako). Positive and negative staining controls were carried out with paraffin tonsil sections using IL-17 and CD8+ monoclonal antibody and an isotype-matched negative control antibody.
Levels of lymphocytic infiltration were evaluated by two independent physicians (FG and SL). All samples were previously anonymized and blinded to the clinicopathological data. On the slide, the number of stained cells was analyzed by enumeration of positive cells in three high-power fields (40×) in the adjacent stromal area and in the tumor bed. The mean count of the three fields was used for statistical analysis. The results of the analyses conducted by each independent pathologist were subsequently compared. Discrepancies between the two observers were reviewed jointly to reach a consensus when the mean count differ from more than 1%. The kappa coefficient of correlation between the observers was 95%.
Immunofluorescence microscopy
Double immunofluorescence staining was performed for CD39 and CD73 markers. The following primary antibodies were used: mouse anti-CD39 (Abcam) and rabbit anti-CD73 (Novus). Secondary antibodies were a goat anti-rabbit Alexa Fluor 488 (A11034, Invitrogen) and a donkey anti-mouse Alexa Fluor 564(A10037, Invitrogen). Images were acquired using a Z-stac acquisition (x63) (AxioImager M2, Zeiss, Germany).
Cluego analysis
To further understand the biological relevance of the hub genes and their regulators in breast tumor, we performed functional enrichment analysis using ClueGO.26 ClueGO facilitates the visualization of functionally related genes displayed as a clustered network and chart. The statistical test used for the enrichment was based on right-sided hypergeometric option with a Benjamini–Hochberg correction and kappa score of 0.3.
Correlation matrix analysis
Correlations were performed using the Spearman test. Correlation matrix was represented using Treeview viewer. Unsupervised hierarchical clustering of samples was performed by using Gene Cluster 3.0 software. The hierarchical clustering was performed using Euclidean distance measure and complete linkage analysis. Positive correlation appears in red and negative correlation in green.
Statistical analyses
Statistical analysis was performed using Prism software (Graph Pad software, La Jolla, CA, USA) for biological studies. For the analysis of experimental data, comparison of continuous data was achieved by the Mann–Whitney U test and comparison of categorical data by Fisher's exact test, as appropriate. All p values are two tailed. P values < 0.05 were considered significant. Data are represented as mean ± SD.
Regarding the analysis of clinical data, patient or disease characteristics were examined using the χ2 test or Fisher's exact test for qualitative variables, and the Student t or Mann–Whitney tests for continuous variables, as appropriate. All patients were followed up until death or the end of data recording (September 1st, 2014). OS was calculated from the date of diagnosis until the date of death (all causes). Alive patients were censored at the last follow-up. RFS was calculated from the date of diagnosis until the date of relapse (local or metastatic). Alive or dead patients without relapse were censored at the last follow-up. Follow-up was calculated using the reverse Kaplan–Meier method. OS and RFS probabilities were estimated using the Kaplan–Meier method and were compared by the log-rank test. Hazards ratios (HRs) with a 95% confidence interval (CI) were calculated using univariate Cox proportional hazards regression modeling. All variables with a univariate Cox p value ≤ 0.20 were eligible for multivariate analyses. Correlations between co-variables were firstly tested for eligible variables. To prevent collinearity, when two variables were significantly correlated, one variable was retained according to its clinical relevance or to the value of the likelihood ratio. Finally, multivariate Cox proportional hazards regression modeling was applied to assess the independent prognosis effect for OS and RFS. Analyses were performed using MedCalc Software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
The authors are supported by grants from the Ligue Nationale contre le Cancer (F.Gh., F.V.), the Fondation de France (L.A.), the Institut National du Cancer (F.Gh.), the Association pour la recherche sur le cancer (F.Gh., G.M.), the Conseil Régional Bourgogne/INSERM (M.T., H.B., L.A.), the CNRS, FEDER, Le Studium, Orléans and Fondation pour la Recherche Médicale (F.Gh., B.R.), the French National Research Agency [ANR-13-JSV3-0001] (L.A.) and [ANR-11-LABX-0021], the Ligue Régionale contre le cancer Comité Grand-Est (L.A., S.L), the Cancéropole Grand Est (Emergence) (L.A.), and the European Community (Marie Curie Fellowship PCIG10-GA-2011-303719) (L.A.). We thank members from the Dijon flow cytometry facility for cell sorting.
References
- 1.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005; 201:233-40; PMID:15657292; http://dx.doi.org/ 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol 2009; 27:485-517; PMID:19132915; http://dx.doi.org/ 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- 3.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol 2007; 8:639-46; PMID:17486092; http://dx.doi.org/ 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- 4.Chalmin F, Mignot G, Bruchard M, Chevriaux A, Vegran F, Hichami A, Ladoire S, Derangère V, Vincent J, Masson D et al.. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppres activity via the regulation of ectonucleotidase expression. Immunity 2012; 36:362-73; PMID:22406269; http://dx.doi.org/doi: 10.1016/j.immuni.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 5.Zhang JP, Yan J, Xu J, Pang XH, Chen MS, Li L, Wu C, Li SP, Zheng L. Increased intratumoral IL-17-producing cells correlate with poor survival in hepatocellular carcinoma patients. J Hepatol 2009; 50:980-9; PMID:19329213; http://dx.doi.org/ 10.1016/j.jhep.2008.12.033 [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Wan J, Liu J, Xie W, Diao X, Xu J, Zhu B, Chen Z. Increased IL-17-producing cells correlate with poor survival and lymphangiogenesis in NSCLC patients. Lung Cancer 2010; 69:348-54; PMID:20022135; http://dx.doi.org/ 10.1016/j.lungcan.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 7.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F et al.. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011; 71:1263-71; PMID:21303976; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2907 [DOI] [PubMed] [Google Scholar]
- 8.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ et al.. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 2011; 35:972-85; PMID:22177921; http://dx.doi.org/ 10.1016/j.immuni.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, Huang E, Finlayson E, Simeone D, Welling TH et al.. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood 2009; 114:1141-9; PMID:19470694; http://dx.doi.org/ 10.1182/blood-2009-03-208249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruchard M, Mignot G, Derangere V, Chalmin F, Chevriaux A, Vegran F, Boireau W, Simon B, Ryffel B, Connat JL et al.. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat Med 2013; 19:57-64; PMID:23202296; http://dx.doi.org/ 10.1038/nm.2999 [DOI] [PubMed] [Google Scholar]
- 11.Chung AS, Wu X, Zhuang G, Ngu H, Kasman I, Zhang J, Vernes JM, Jiang Z, Meng YG, Peale FV et al.. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nat Med 2013; 19:1114-23; PMID:23913124; http://dx.doi.org/ 10.1038/nm.3291 [DOI] [PubMed] [Google Scholar]
- 12.Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med 2007; 27:469-85, v; PMID:17658403; http://dx.doi.org/ 10.1016/j.cll.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saze Z, Schuler PJ, Hong CS, Cheng D, Jackson EK, Whiteside TL. Adenosine production by human B cells and B cell-mediated suppression of activated T cells. Blood 2013; 122:9-18; PMID:23678003; http://dx.doi.org/ 10.1182/blood-2013-02-482406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuler PJ, Saze Z, Hong CS, Muller L, Gillespie DG, Cheng D et al.. Human CD4 CD39 regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73 exosomes or CD73 cells. Clin Exp Immunol 2014; 177(2):531-43; PMID:24749746; http://dx.doi.org/ 10.1111/cei.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Harasymczuk M, Mandapathil M, Lang S, Jackson EK et al.. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 2007; 204:1257-65; PMID:17502665; http://dx.doi.org/ 10.1084/jem.20062512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez F, Belloc F, Lacombe F, Dumain P, Reiffers J, Bernard P, Boisseau MR. Modalities of synthesis of Ki67 antigen during the stimulation of lymphocytes. Cytometry 1991; 12:42-9; PMID:1999122; http://dx.doi.org/ 10.1002/cyto.990120107 [DOI] [PubMed] [Google Scholar]
- 17.Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol 2008; 180:214-21; PMID:18097022; http://dx.doi.org/12948524 10.4049/jimmunol.180.1.214 [DOI] [PubMed] [Google Scholar]
- 18.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Ggrowth Factor Rev 2003; 14:409-26; PMID:12948524; http://dx.doi.org/ 10.1016/S1359-6101(03)00049-2 [DOI] [PubMed] [Google Scholar]
- 19.Chen WC, Lai YH, Chen HY, Guo HR, Su IJ, Chen HH. Interleukin-17-producing cell infiltration in the breast cancer tumour microenvironment is a poor prognostic factor. Histopathology 2013; 63:225-33; PMID:23738752; http://dx.doi.org/ 10.1111/his.12156 [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Xu W, Qiu S, Xiong S. Enrichment of CCR6+Foxp3+ regulatory T cells in the tumor mass correlates with impaired CD8+ T cell function and poor prognosis of breast cancer. Clin Immunol 2010; 135:466-75; PMID:20181533; http://dx.doi.org/ 10.1016/j.clim.2010.01.014 [DOI] [PubMed] [Google Scholar]
- 21.Longhi MS, Moss A, Bai A, Wu Y, Huang H, Cheifetz A, Quintana FJ, Robson SC. Characterization of human CD39+ Th17 cells with suppressor activity and modulation in inflammatory bowel disease. PloS One 2014; 9:e87956; PMID:24505337; http://dx.doi.org/ 10.1371/journal.pone.0087956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valmori D, Raffin C, Raimbaud I, Ayyoub M. Human RORgammat+ TH17 cells preferentially differentiate from naive FOXP3+Treg in the presence of lineage-specific polarizing factors. Proc Natl Acad Sci U S A 2010; 107:19402-7; PMID:20962281; http://dx.doi.org/ 10.1073/pnas.1008247107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladoire S, Arnould L, Apetoh L, Coudert B, Martin F, Chauffert B, Fumoleau P, Ghiringhelli F. Pathologic complete response to neoadjuvant chemotherapy of breast carcinoma is associated with the disappearance of tumor-infiltrating foxp3+ regulatory T cells. Clin Cancer Res 2008; 14:2413-20; PMID:18413832; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-4491 [DOI] [PubMed] [Google Scholar]
- 24.Ladoire S, Mignot G, Dabakuyo S, Arnould L, Apetoh L, Rebe C, Coudert B, Martin F, Bizollon MH, Vanoli A et al.. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. J Pathol 2011; 224:389-400; PMID:21437909; http://dx.doi.org/ 10.1002/path.2866 [DOI] [PubMed] [Google Scholar]
- 25.Helenius M, Jalkanen S, Yegutkin G. Enzyme-coupled assays for simultaneous detection of nanomolar ATP, ADP, AMP, adenosine, inosine and pyrophosphate concentrations in extracellular fluids. Biochim Biophys Acta 2012; 1823:1967-75; PMID:22967714; http://dx.doi.org/ 10.1016/j.bbamcr.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 26.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009; 25:1091-3; PMID:19237447; http://dx.doi.org/ 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.