Abstract
In the intestine, finger-like villi provide abundant surface area for nutrient absorption. During murine villus development, epithelial Hedgehog (Hh) signals promote aggregation of subepithelial mesenchymal clusters that drive villus emergence. Clusters arise first dorsally and proximally and spread over the entire intestine within 24 h, but the mechanism driving this pattern in the murine intestine is unknown. In chick, the driver of cluster pattern is tensile force from developing smooth muscle, which generates deep longitudinal epithelial folds that locally concentrate the Hh signal, promoting localized expression of cluster genes. By contrast, we show that in mouse, muscle-induced epithelial folding does not occur and artificial deformation of the epithelium does not determine the pattern of clusters or villi. In intestinal explants, modulation of Bmp signaling alters the spatial distribution of clusters and changes the pattern of emerging villi. Increasing Bmp signaling abolishes cluster formation, whereas inhibiting Bmp signaling leads to merged clusters. These dynamic changes in cluster pattern are faithfully simulated by a mathematical model of a Turing field in which an inhibitor of Bmp signaling acts as the Turing activator. In vivo, genetic interruption of Bmp signal reception in either epithelium or mesenchyme reveals that Bmp signaling in Hh-responsive mesenchymal cells controls cluster pattern. Thus, unlike in chick, the murine villus patterning system is independent of muscle-induced epithelial deformation. Rather, a complex cocktail of Bmps and Bmp signal modulators secreted from mesenchymal clusters determines the pattern of villi in a manner that mimics the spread of a self-organizing Turing field.
KEY WORDS: Villus formation, Epithelial-mesenchymal crosstalk, Mathematical model, Activator-inhibitor patterning model, Turing field, Intestinal development, Morphogenesis
Highlighted article: Intestinal villus pattern in the fetal mouse is controlled by mesenchymal BMP signaling, not mechanical forces, and behaves in accordance with a reaction-diffusion Turing mechanism.
INTRODUCTION
The effective absorption of nutrients by the small intestine requires an enormous mucosal surface area. One adaptive mechanism for surface area amplification is the convolution of mucosal surface into a regular array of finger-like projections called villi. Villi arise at embryonic day (E) 14.5 in mice (week 8-10 in humans), when the intestine undergoes a remarkable morphogenic process to convert a pseudostratified epithelial tube surrounded by loose mesenchyme into a field of villi with mesenchymal cores, covered by columnar epithelium. Although the histological changes that accompany villus formation have been studied extensively (Dekaney et al., 1997; Dunn, 1967; Lacroix et al., 1984; Mathan et al., 1976; Matsumoto et al., 2002; Nakamura and Komuro, 1983; Sbarbati, 1982; Trahair and Robinson, 1986), the cellular and molecular drivers of this process are still incompletely understood, especially in mammals.
Recent progress has been made in deciphering the mode of villus emergence in the chick intestine. In that model, the initially flat epithelium gives way to longitudinal ridges, which evolve to regular zigzags and finally to villi, and these sequential morphological stages correlate with maturation of three smooth muscle layers (Coulombre and Coulombre, 1958; Shyer et al., 2013). The deep and regular folding of the epithelium, imposed by mechanical forces from the developing muscles, creates periodic maxima of epithelially secreted Hedgehog (Hh) protein beneath the folded epithelium that direct villus emergence in a regular manner (Shyer et al., 2015).
By contrast, in many mammals, including mouse (Sbarbati, 1982), rat (Dunn, 1967), sheep (Trahair and Robinson, 1986), pig (Dekaney et al., 1997) and human (Lacroix et al., 1984; Matsumoto et al., 2002), ridges and zigzags never form and in several of these systems (including mouse, as shown herein) villus formation is not temporally coordinated with smooth muscle development (de Castro, 2001; Fekete et al., 1996; Georgieva and Gerov, 1975; Kedinger et al., 1990; Keibel, 1910). Thus, localization of the Hh signal via smooth muscle-dependent epithelial deformation cannot explain the establishment of a uniform field of villi in many species; additional patterning forces are required.
In the mouse, villi arise as domes over condensed clusters of mesenchymal cells that express Pdgfra (Karlsson et al., 2000). We previously established that, prior to villus formation, scattered mesenchymal cells express Gli1, Ptc1 (Ptch1) and Pdgfra (Karlsson et al., 2000; Walton et al., 2012). Hh ligands from the epithelium cause agglutination of these cells into clusters beginning at E14.5 (Walton et al., 2012). In mice overexpressing the pan-Hh inhibitor Hhip, clusters and villi fail to form (Madison et al., 2005). Similarly, in cultured intestinal explants inhibition of Hh signaling by cyclopamine or anti-Hh antibody (5E1) abolishes cluster formation and villus emergence, without altering smooth muscle (Fig. S1A,B,D,E) (Walton et al., 2012). By contrast, increasing the strength of the Hh signal increases cluster size, again without changing smooth muscle (Fig. S1C,F) (Walton et al., 2012), confirming that epithelial Hh controls a signaling cascade that drives mesenchymal cluster formation independently of alterations in the surrounding smooth muscle layers.
Forming mesenchymal clusters are regularly positioned in the murine intestine, with an average cluster-to-cluster spacing of ∼60-70 µm (Walton et al., 2012), suggesting that specific signals are required to generate such a well-patterned field. Although Hh signals control cluster agglutination and size, Shh and Ihh ligands are uniformly expressed by the pre-villus epithelium (Kolterud et al., 2009; Shyer et al., 2015); thus, it is unclear how epithelial Hh could establish the regular pattern of clusters, especially in species (e.g. mouse) in which there is no epithelial folding to create local Hh maxima. Since Bmp signaling is known to control a variety of patterning fields in other contexts (Hogan, 1996) and mesenchymal clusters in emerged villi express Bmp2 and Bmp4 (Karlsson et al., 2000), we examined the potential role of this pathway in cluster patterning.
Here, we show that newly established clusters express multiple Bmp ligands and Bmp signal modifiers. Moreover, abolishing Bmp signal reception changes cluster pattern, causing rows of mesenchymal cluster ‘spots’ to merge into ‘stripes’. Merged clusters can also be generated by conditional deletion of Bmpr1a in Hh-responsive cells of the mesenchyme, whereas epithelial Bmpr1a deletion has no effect. This ‘spots to stripes' pattern change is fully consistent with the mathematical predictions of a classical reaction-diffusion model, in which the overlapping activity domains of an activator and an inhibitor create a patterned field, as initially described by Turing (Maini, 2004; Meinhardt, 2012; Turing, 1952). In such a model, progressive saturation of the Turing activator (in this case posited to be a Bmp inhibitor) causes spots to become stripes. Together, our data establish that in the mouse model Bmp signaling controls cluster distribution. The patterning of clusters and emergence of villi in this model are not dependent upon muscular forces or epithelial deformation, but behave in accordance with a Bmp-dependent self-organizing Turing field.
RESULTS
Villus patterning in the mouse is not driven by tensile forces
Villus emergence correlates with smooth muscle development in chick (Coulombre and Coulombre, 1958; Shyer et al., 2013); thus, we examined whether this is also true in mouse. Such an analysis requires that the same region of the intestine (here, jejunum) be compared at all time points, since villus emergence propagates from proximal to distal intestine over a 24 h period (Walton et al., 2012). At E13.0, 48 h before villus morphogenesis initiates in the jejunum (Walton et al., 2012), a well-developed inner circular muscle (ICM) is already prominent (Fig. 1A) along the entire length of the intestine. A mature outer longitudinal muscle (OLM) is not seen until E16 (Fig. 1E), 24 h after the first villi emerge in the jejunum. Although a few scattered cells that are weakly positive for alpha smooth muscle actin (αSMA), a marker of mature smooth muscle, can be discerned at E15.0 (Fig. 1D), an organized OLM layer is absent at this time. Finally, the muscularis mucosa, which is immature at E16.5 (desmin positive, but αSMA negative; Fig. 1F), remains discontinuous at E18.5 (Fig. 1G). Thus, in the mouse, villus emergence is not temporally synchronized with the maturation of any of the three muscle layers.
Fig. 1.

Smooth muscle development does not correlate with mouse villus emergence; tensile forces are not required for cluster patterning. αSMA immunostaining (red) marks mature muscle cells in the developing muscle layers at E13 (A), E14 (B), E14.5 (C), E15 (D), E16 (E), E16.5 (F) and E18.5 (G). The ICM is already mature at E13 (A), but a mature OLM is not present until E16 (outer circle, arrow in E). Scattered αSMA+ cells at E14.5 (arrows, C) and E15 (arrows, D) are not organized into a continuous muscle. (F,G) Muscularis mucosa is not present at E16.5 (F) and is still incomplete at E18.5 (G, arrows). Desmin (green) marks smooth muscle precursor cells (F, arrows). DAPI (blue) stains nuclei. (H-K) An E13 intestine from a PdgfraEGFP/+ mouse was opened longitudinally and cultured for 2 days. No clusters were present initially (H). Clusters began to develop by 20 h (I, near arrow). By 38 h, well-patterned clusters were visible (J) and small villi were emerging (magnified in K). The dashed line (K) indicates cut edges of the intestine, where it has rolled back after cutting. Clusters and villi were observed to form in intestines cut open prior to cluster formation and grown in culture for 2 days in more than 25 independent samples from at least eight separate experiments. Scale bars: 50 µm in A-G; 100 µm in H-K.
Since formation of the ICM precedes villus development, we further examined whether confinement forces mediated by this muscle might play a role in cluster formation or villus emergence. E13.5 intestines (prior to cluster formation) were opened longitudinally, interrupting the circularity of the ICM (Fig. 1H). Although the ICM might still impose some force on the overlying tissue, radial confinement is abolished; indeed, opened intestines tend to invert. After 20 h in culture, clusters begin to form at the anterior end of the intestinal segment (Fig. 1I). By 38 h, clear, well-patterned mesenchymal clusters and rudimentary villi are visible (Fig. 1J,K). Villus and cluster size is uniform, even at the cut edges of the intestine, where residual strain is predicted to be lower (Fig. 1K, dashed line). Thus, in mouse, radial confinement from the ICM is not required for cluster formation, cluster patterning or initial villus emergence. We cannot, however, rule out the possibility that confinement from the ICM might facilitate the progression of villus outgrowth after initiation.
Epithelial deformation does not determine cluster pattern in the mouse intestine
In the chick, epithelial bending is an upstream driver of cluster formation and patterning. Thus, we examined the relationship between clusters and epithelial deformation in the mouse intestine. Thick (80-100 µm) vibratome sections of E14.5 mouse intestine were stained for α-tubulin, a marker that reveals epithelial cell shape and is also enriched in clusters, and confocal z-stacks were collected (Movie 1). Individual sections of the top, middle and bottom of a representative stack (Fig. 2A-C) reveal that the apical surface of the epithelium is still flat when the first clusters form basally beneath the epithelium. Thus, deep folds of the entire epithelium, such as those observed in the chick, are not seen in the mouse. On the basal side, however, epithelial cells directly above nascent clusters are shorter, creating shallow but obvious epithelial deformations (Fig. 2A-C, arrows). No basal deformations are detected in regions lacking a mesenchymal cluster or after treatment with cyclopamine (Fig. S1B) (Walton et al., 2012), which abolishes clusters, consistent with the idea that basal shortening is driven by signals from these clusters.
Fig. 2.
Epithelial deformation does not determine cluster pattern in the mouse intestine. (A-C) Single slices from top (A), middle (B) and bottom (C) of a 50 µm thick z-section of an E14.5 intestine immunostained for α-tubulin (red) to outline epithelial cells and mark clusters (outer ring marks the ICM); DAPI (blue) marks nuclei. The arrows indicate the position of two clusters. Note basal epithelial deformation with no apical deformation. Movie 1 steps through the entire z-stack. (D-F) Mesh screens force deformation of the epithelium in E13.5 PdgfraEGFP/+ intestines that were cut open lengthwise and cultured under a mesh screen. (D) Individual villi with single mesenchymal clusters develop under a screen with a 55 µm aperture. (E) Multiple villi with a single cluster per villus develop under a mesh screen with a 75 µm aperture. (F) Note that cluster formation and villus development are delayed in tissue under a mesh screen (anterior, left side) as compared with the posterior side (right side) that was not under the screen. n>12 in four separate experiments for each of the mesh screen cultures. Scale bars: 30 µm in A-C; 50 µm in D-F.
Although the analysis above suggests that cluster formation is upstream rather than downstream of basal epithelial deformation in the mouse, we tested directly whether artificially imposed epithelial deformation can drive cluster and villus pattern in this model. Shyer et al. (2015) deformed the chick intestinal epithelium by placing a grid on the opened epithelial surface and observed precocious induction of villus cluster genes and emergence of single villi through the holes of the grid. To take this analysis one step further, we utilized grids of different mesh sizes, placing them on the opened epithelial surface of the E13.5 mouse intestine, prior to cluster or villus formation. Intestinal explants were observed daily for 4 days. We reasoned that, if epithelial deformation is a critical determinant of cluster and villus pattern in the mouse, grids of increasingly wider mesh size should produce increasingly wider clusters and villi. However, this was not seen (Fig. 2D-F). Mesh sizes that approximate cluster size (55 µm aperture) allowed single villi to grow into the mesh spaces (Fig. 2D), as seen previously (Shyer et al., 2015). However, in grids with larger mesh sizes (75 µm aperture and larger), multiple clusters (and villi), rather than larger clusters and villi, were observed (Fig. 2E). Additionally, analysis of the boundary of the grid revealed that grid placement slowed rather than accelerated cluster formation and villus emergence (Fig. 2F). Together, these experiments demonstrate that, for the mouse intestine, epithelial deformation is not sufficient to impart patterning cues to the field of clusters and emerging villi. Rather, the presence and patterning of mesenchymal clusters determines the presence and pattern of the emerging villi. We therefore sought to identify the signal(s) downstream from Hh-mediated cluster aggregation that is responsible for cluster patterning.
Mesenchymal clusters express multiple Bmp signaling molecules
Bmps are secreted ligands responsible for patterning in many developmental contexts (Hogan, 1996), and several Bmp ligands are known targets of Hh signaling (Roberts et al., 1995). To better assess the potential involvement of Bmp signaling during cluster formation and patterning of nascent clusters, we examined the localization of RNA transcripts for several Bmp ligands and modifiers during the initial round of cluster formation as well as in clusters associated with emerged villi (Fig. 3). Prior to cluster formation, Bmp4, Bmp5 and Bmp7 are expressed in many cells of the subepithelial mesenchyme, whereas Bmp2 is primarily epithelial (Fig. 3A-D). As clusters form (E14.5), Bmp2 expression is initiated in clustered cells (Fig. 3A, inset). As villi emerge (E15.5), all Bmp genes except Bmp7 are expressed robustly in mesenchymal clusters (Fig. 3I-L); Bmp2 continues to be the most specific cluster marker (Fig. 3I). The expression pattern of the Bmp modifier twisted gastrulation 1 (Twsg1) (Fig. 3E,M) is similar to that of Bmp5, while the tolloid-like molecule Bmp1 is weakly expressed at E14.5 (Fig. 3F) but is high in mesenchymal clusters at E15.5 (Fig. 3N). The Bmp inhibitor noggin (Nog) is expressed in mesenchymal clusters only after emergence (Fig. 3G,O), whereas follistatin-like 1 (Fstl1) is highly expressed throughout the mesenchyme at both stages (Fig. 3H,P). Thus, multiple Bmp ligands and Bmp signaling modifiers are dynamically expressed both in unclustered mesenchyme and in nascent and mature mesenchymal clusters.
Fig. 3.

RNA in situ hybridization of Bmp pathway ligands and modifiers. Analysis was performed at E14.5 just prior to cluster formation and villus emergence (A-H), and at E15.5 once villi have begun to emerge (I-P). The inset (A) shows cluster-specific expression of Bmp2 in nascent clusters at a slightly later stage, when expression is switching from epithelial to mesenchymal. Scale bars: 100 µm.
Modulation of Bmp signaling affects cluster formation and pattern
To determine whether Bmp ligands or signaling modifiers could modulate cluster pattern, intestines from E13.5 PtclacZ/+ embryos (prior to cluster formation) were cultured on transwell filters in the presence of agarose beads soaked in bovine serum albumin (BSA, control) or recombinant Bmp2, Bmp4, Bmp5, Bmp7, or heterodimerized Bmp2/7 or Bmp4/7. After 2 days, clusters formed in the expected pattern near control beads (Fig. 4A,D,G,J). However, all Bmp-soaked beads inhibited mesenchymal cluster formation and subsequent villus emergence in the region surrounding the bead, but not on the opposite side of the intestine (Fig. 4B,C,E,F,H,I,K, Fig. S2). Lack of cluster formation near Bmp beads was confirmed by immunostaining for Pdgfrα, a marker of mesenchymal clusters (Karlsson et al., 2000) (Fig. 4K). Bmp2 was the most potent ligand for inhibiting cluster formation near the beads, strongly inhibiting cluster formation at 125 ng/ml and above (Fig. 4L, Fig. S2E).
Fig. 4.

Bmp ligand-soaked agarose beads inhibit cluster formation and villus emergence. (A-I) E13.5 PtclacZ/+ intestines were harvested prior to cluster formation and cultured for 2 days with agarose beads soaked in BSA or recombinant protein, as indicated. X-gal staining shows the pattern of PtclacZ/+ clusters that form in whole (A-F) or transverse sectioned (G-I) intestines. Hatched ovals (B,C,E,F) outline the clearance of PtclacZ/+ clusters around the Bmp-soaked bead; faint clusters that appear within the hatched oval are on the transwell/opposite side of the intestine, as seen in the transverse sections in H and I. (J,K) Pdgfrα immunostaining (green) marks clusters formed near a BSA-soaked bead (J), which are absent around the Bmp-soaked bead (K). Inhibition of cluster formation near Bmp-soaked beads was observed in at least 100 beads placed on more than 25 intestines. (L) Scatterplot of the mean of the distances from the edge of Bmp2-soaked beads or Bmp4-soaked beads to the nearest clusters measured in a sampling from those experiments (at least six beads placed on at least three different intestines). Error bars are s.d. See also Fig. S2 for measurements of clearing around other Bmp ligand-soaked beads. Scale bars: 50 μm.
Similarly, we tested the effects of several of the Bmp signaling modulators that are expressed in clusters (Fig. 5). The cluster patterning perturbations were more subtle, consisting of larger or merged clusters rather than a clearing of clusters (Fig. 5A-C). Pattern changes for Twsg1 were most obvious (Fig. 5C), although cluster sizes for intestines treated with Bmp1, Nog or Twsg1 in the medium (10 ng/ml) were all statistically different from clusters treated with BSA (Fig. 5E). Clusters closest to Twsg1-soaked beads (250 ng/ml) placed on top of the intestine were significantly larger than clusters more distant from the bead (Fig. 5D). Clusters in BSA-treated intestines did not differ in size from clusters distant from Twsg1 beads (Fig. 5E).
Fig. 5.
Bmp modifiers alter cluster pattern and villus shape. E13.5 PtclacZ/+ intestines were harvested prior to cluster formation and cultured for 2 days with 10 ng/ml recombinant protein as indicated (A-C) or 250 ng/ml Twsg1 on agarose beads (D, hatched circle). (A-D) X-gal staining shows the pattern of PtclacZ/+ clusters that form in whole intestines. Note that the merging of clusters is most dramatic in Twsg1-treated intestines in C and near the Twsg1 beads in D. (E) Box and whisker plot of cluster areas showing the largest, smallest, median (center line) and mean (+) for each treatment. Cluster areas for Bmp1-treated and Nog-treated intestines were significantly different from those of BSA-treated and Twsg1-treated intestines (*P<0.0001), but were not different from each other. Twsg1-treated intestines were significantly different from BSA-treated intestines (*P<0.0001). Clusters immediately surrounding Twsg1-soaked beads were significantly different in size from clusters at a distance from the beads (**P<0.0001), but not from clusters in BSA-treated intestines (P=0.689). Scale bars: 250 µm in A,B; 100 µm in C,D.
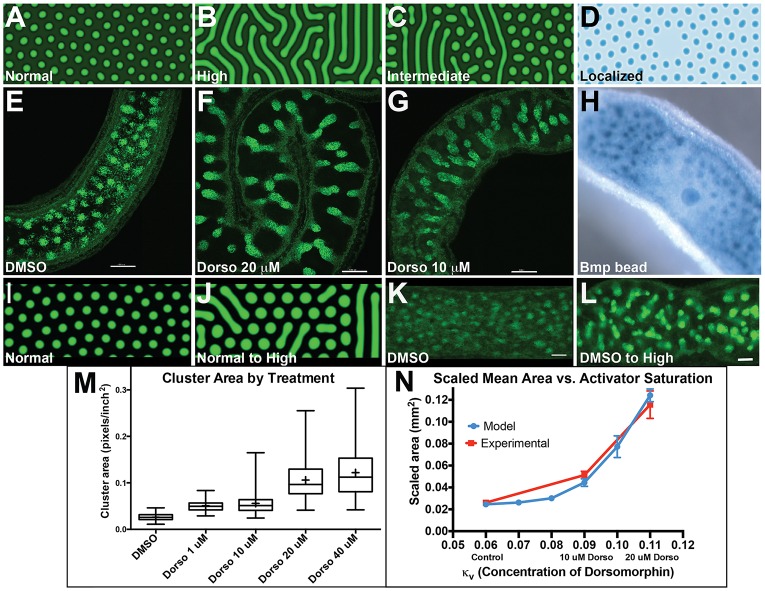
Next, we tested the effect of complete Bmp signal inhibition on cluster formation and villus emergence. Intestines were harvested prior to cluster formation and cultured for 2 days with a small-molecule inhibitor of Bmp signaling, dorsomorphin (Fig. 6). Inhibition of Bmp signaling in this manner alters cluster patterning dramatically. Clusters are two to three times larger and often connected, so that the ‘spot-like' distribution of clusters in control intestines (Fig. 6A-C) becomes ‘striped' in the presence of dorsomorphin (Fig. 6D-F). In transverse sections of the intestines, the larger merged clusters are seen to alter the shapes of associated villi (compare Fig. 6G-J). These changes occur without altering smooth muscle (Fig. 6H-L), again confirming that cluster pattern, not muscle tension, determines the pattern of villus emergence in the mouse.
Fig. 6.

Inhibition of Bmp signaling alters cluster pattern and villus size. PtclacZ/+ intestinal pieces were harvested at E13.5 and cultured with DMSO (A-C,G,H,K,M), 20 μM dorsomorphin (D-F,I,J,L,N) or 40 μM dorsomorphin (O). Treatments were performed in at least 25 control and 35 dorsomorphin-treated intestines; representative images of duodenum (A,D), jejunum (B,E,G-O) and ileum (C,F) are shown. After 2 days, intestines were fixed and X-gal stained to show the pattern of PtclacZ/+ clusters (blue) in whole intestines (A-F) and 100 µm sections (G,J). Arrows in D,E mark areas where clusters merge to stripes with dorsomorphin treatment. Arrowheads in J mark large fused clusters and large villi. (H,I,K,L) Dorsomorphin treatment does not alter smooth muscle (αSMA, red). Sections are shown with (K,L) and without (H,I) DAPI staining. (M-O) Epithelial cells (outlined by E-cadherin immunostaining, green) remain pseudostratified in intervillus regions (red arrowheads) when Bmp signaling is inhibited (N,O), similar to controls (M). Scale bars: 100 µm in A-F; 50 µm in G-O.
Studies in other systems have shown that inhibition of Bmp signaling in the epithelium causes cells to shorten and become columnar or cuboidal (Eom et al., 2011; Gibson and Perrimon, 2005; Rajagopal et al., 2009; Shen and Dahmann, 2005). However, dorsomorphin treatment does not cause widespread columnar conversion; epithelium above the merged clusters is columnar, while epithelium between these clusters remains pseudostratified (Fig. 6M-O). Doubling the dose of dorsomorphin generates larger clusters and larger villi, but does not convert all epithelial cells to a columnar shape (Fig. 6O). Thus, inhibition of Bmp signaling throughout the intestine affects cluster pattern without altering cell shape in the epithelium.
We previously showed that increased Hh signaling also produces larger clusters (Walton et al., 2012), although a striped pattern was not seen. Here, dorsomorphin treatment does not appear to dramatically affect the expression levels of the Hh target gene Ptc1 in these Ptc1lacZ/+ intestines. Lack of a significant effect of dorsomorphin on the Hh signaling pathway was further confirmed by qRT-PCR (Fig. S3).
Genetic loss of Bmpr1a in mesenchymal clusters impacts cluster size and villus morphology
Although the experiments above reveal the importance of Bmp signaling in modulating cluster pattern, they do not establish which compartment (epithelial, mesenchymal or both) must transduce Bmp signals to control this process. Since Bmp signaling activity is detected in both the epithelium and mesenchyme (Fig. S4), we used a conditional Bmpr1af/f mouse model (Mishina et al., 2002), in combination with tissue-specific Cre drivers, to genetically delete Bmpr1a signaling in either the mesenchyme (using Gli1CreERT2/+) (Bai et al., 2002) or the epithelium (with ShhCre) (Harfe et al., 2004) prior to cluster formation. Demonstration of reporter expression and qRT-PCR showing the efficacy of deletion are shown in Fig. S5.
Loss of Bmpr1a in the epithelium (ShhCre; activated by E9.5) (Harfe et al., 2004) does not affect muscle development, cluster size, cluster distribution or epithelial morphology (Fig. 7A,B). However, loss of Bmpr1a in Hh-responsive mesenchymal cells results in larger clusters and wide villi (Fig. 7C,D), a phenotype closely resembling that seen in intestines treated with dorsomorphin. Scanning electron microscopy (SEM) confirms larger/merged villi in the Bmpr1af/f; Gli1CreER/+ mutants as compared with control littermates (Fig. S6). Indeed proliferation was increased with loss of Bmpr1a in the mesenchyme; however, neither the amount (Fig. S7) nor the pattern of proliferation (Fig. S8) was altered in the epithelium. Together, these data demonstrate that the patterning of mesenchymal clusters is determined by the level of Bmp signal transduction in the Hh-responsive mesenchymal compartment.
Fig. 7.
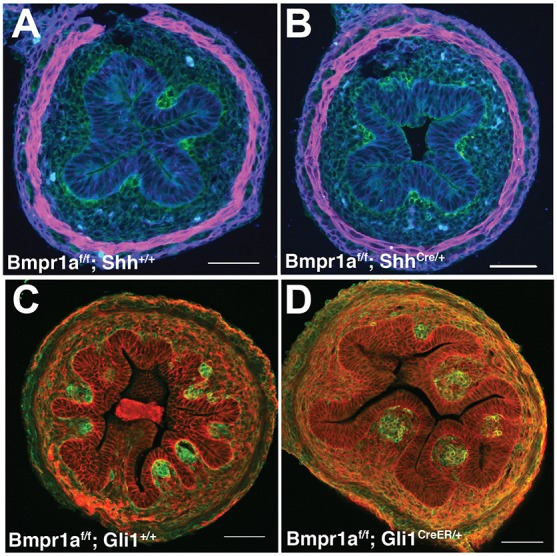
Conditional loss of Bmp signaling in Hh-responsive mesenchymal cells, but not in epithelial cells, results in fused clusters and wide villi. (A,B) Bmpr1af/f mice were mated to ShhCre mice. ShhCre is activated by E10 in the intestinal epithelium (Kolterud et al., 2009). No changes in epithelial (E-cadherin, blue) or mesenchymal pattern (Pdgfrα, green) were noted; muscle was unchanged (αSMA, magenta). (C,D) Bmpr1af/f mice were mated with Gli1CreERT2/+ mice. Recombination was induced with three doses of tamoxifen, beginning at E12.5 prior to cluster formation, and tissues were harvested at E15.5. Large clusters (Pdgfrα, green) and wide villi (E-cadherin, red) are seen in Bmpr1af/f; Gli1CreER mutant intestines (D). Eight mutant and eight control littermate intestines from three separate litters were analyzed for Bmpr1af/f; ShhCre. Twenty mutant and 21 control littermate intestines from six separate litters were analyzed for Bmpr1af/f; Gli1CreER. Scale bars: 50 µm.
Cluster patterning resembles a spreading Turing field
We previously showed that mesenchymal clusters are established in an anterior to posterior and dorsal to ventral wave, eventually generating a regular pattern along the length of the intestine (Walton et al., 2012). Turing first postulated that similar patterns can be generated by the reactions of diffusive chemicals; small perturbations in initially uniformly distributed chemicals can increase in amplitude over time, leading to a regular spatial pattern. The Turing system employs an activator and an inhibitor, both expressed by the same cell or group of cells. The inhibitor spreads further than the activator, and patterned spots become stripes when the activator is saturating (Turing, 1952).
Cluster patterning in vivo shares several features with the Turing system. (1) An initially homogeneous state, lacking clusters and villi, evolves quickly (within 12 h) to a patterned state that then spreads (Walton et al., 2012). (2) The pattern can be altered by Bmp ligands, which inhibit the formation of clusters where Bmp signaling is high (Fig. 4, Fig. S2); thus, one or more Bmp ligands may act as Turing inhibitors. (3) Dorsomorphin treatment changes the cluster pattern from spots to stripes (Fig. 6) and beads soaked with several Bmp inhibitors/modulators cause cluster fusion into short stripes (Fig. 5), suggesting that an inhibitor of Bmp signaling could be the Turing activator. (4) Multiple Bmp ligands and signaling modulators are expressed by cluster cells (Fig. 3), in accordance with the requirement for expression of activator and inhibitor by the same cells. (5) The genetic experiments above show that patterning requires the reception of Bmp signals in the mesenchymal compartment, including the cluster cells themselves (Fig. 7).
To test the degree to which a mathematical model of a Turing system resembles the experimentally derived data, we constructed a Turing system comprising two modulators of Bmp signaling following a ‘pure activator-inhibitor' system (Dillon and Othmer, 1993) and incorporating cell density (see the supplementary Materials and Methods for details). We propose that the Turing activator is a Bmp inhibitor and that the Turing inhibitor consists of one or more Bmp ligands. The evolution of the resulting pattern, represented as cell density, in the absence or presence of saturating activator (Bmp inhibition/dorsomorphin) is displayed in Fig. 8A,B. Light-green regions represent high concentrations of Bmp inhibitor (Turing activator), while the dark regions represent low concentrations. In our model, mesenchymal cluster cells are assumed to produce these proteins, whose interactions give rise to these gradients, so that regions of high Bmp inhibitor correspond to the location of mesenchymal cells where clustering occurs. As predicted by the Turing system, spots are seen when the activator concentration is well below saturation, whereas saturation of the activator produces larger clusters and areas with a stripy pattern. These patterns closely resemble those observed experimentally, including the 2- to 3-fold increase in cluster size with saturating activator (Bmp inhibitor) (plotted in Fig. 8M,N).
Fig. 8.
A Turing field model of cluster cell patterning recapitulates the experimental data. (A-D) Still images of the patterns predicted by the simulations (further details are provided in the supplementary Materials and Methods). A regularly spaced pattern of clusters (spots) is predicted in control (A) Turing simulations. The pattern is altered to stripes (B) when the Turing activator (Bmp inhibitor) is saturated. An intermediate level of activator results in shorter stripes (C, lower dose of dorsomorphin), while a localized increase in Turing inhibitor (excess Bmp ligand at center) prevents cluster formation near the source (D). (E-H) Representative images of experimental results obtained under treatment conditions that match the Turing simulations. Treatments were as follows: control (E), 20 μM dorsomorphin (F), 10 μM dorsomorphin (G) and Bmp-soaked bead (250 ng/µl) (H). (I-L) Dynamic changes in Bmp inhibitor concentration change patterns in simulations and experimental tests. A developed spot pattern (I) evolved toward stripes (J) when the inhibitor concentration was computationally increased. Experimentally, established cluster spots (K) merge to form short stripes (L) when dorsomorphin (20 µM) is added (n=18 intestines). Scale bars: 100 µm. (M) Box and whisker plot showing the largest, smallest, median (middle line) and mean (+) cluster area for intestines treated for 2 days with vehicle (DMSO) or increasing doses of dorsomorphin. n=5 fields for at least five different intestines for DMSO, 10 µM, 20 µM, 40 µM dorsomorphin; n=5 fields for three different intestines for 1 µM dorsomorphin. (N) Scatter plot comparing the average cluster area at increasing concentrations of dorsomorphin (experimental) verses the simulated model. Error bars are s.e. The s.d. increases with increasing concentrations of Bmp inhibitor due to striped patterns and boundary conditions (n=73, 75, 74, 34, 15, 3 for the increasing concentrations of dorsomorphin).
We then used the model to predict the patterns that would evolve with intermediate levels of activator. At such intermediate levels, spots and stripes were mixed and stripes were predicted to be shorter and less frequent (Fig. 8C). This was indeed borne out experimentally, after exposure of intestinal explants to an intermediate dose of dorsomorphin (Fig. 8G). We also modeled a localized source of high concentration of Turing inhibitor (Bmp ligand) and found that the resulting simulation (Fig. 8D) mirrored our experimental results of intestines cultured with Bmp ligand-soaked agarose beads (Fig. 4, Fig. 8H, Fig. S2E). Finally, we examined a scenario in which a spot-like pattern was allowed to evolve for 24 h in the absence of saturated activator, and then activator concentration was computationally increased in a stepwise fashion. In this simulation, the initial spot-like pattern evolved to become more stripe-like (Fig. 8I,J, Movie 2). Experimentally, we allowed E13.5 intestines to develop for 48 h on transwells, until the spot-like pattern of clusters became apparent (Fig. 8K) and then added dorsomorphin to the culture for an additional 48 h. The initial spot pattern filled in to become stripy, in a manner closely resembling the computed simulation (Fig. 8L). Overall, these results provide strong evidence that cluster patterning and subsequent villus emergence are controlled, at least in part, by Bmp signaling and that the patterning field evolves in a manner consistent with a self-organizing Turing field within the mesenchyme.
DISCUSSION
Optimal absorptive function by the small intestine depends upon the generation of a tightly packed and well-organized field of villi, a process that begins in fetal life. Substantial evidence over the past several decades has emphasized the role of complex epithelial-mesenchymal crosstalk in the process of villus formation [for a recent review, see Wells and Spence (2014)]. Indeed, we previously established that one of the earliest steps in villus development takes place at E14.5 in the mouse, when Hh signals, expressed uniformly from the epithelium (Kolterud et al., 2009), act on evenly distributed Ptc1+ Gli1+ Pdgfra+ subepithelial mesenchymal cells, causing their aggregation into mesenchymal clusters (Walton et al., 2012). Here, we provide evidence that, downstream from this cluster-forming epithelial Hh signal, a Bmp signaling network that operates entirely within the mesenchyme is responsible for establishment of cluster spacing and pattern. We show that cluster pattern can be dynamically altered simply by modifying the concentration of Bmp ligands or Bmp signaling modifiers and that the pattern evolves in a manner consistent with a Turing activator/inhibitor field.
There is mounting evidence for the validity of Turing-based models to explain pattern evolution in several diverse biological systems, including feather bud arrangements (Baker et al., 2009), hair follicle spacing (Maini et al., 2006; Sick et al., 2006), palatal rugae distribution (Economou et al., 2012), tongue papilla patterning (Zhou et al., 2006), digit patterning (Raspopovic et al., 2014) and zebrafish mesodermal pigmentation (Eom et al., 2012; Kondo and Miura, 2010), and it is interesting that Bmp ligands appear to act as Turing inhibitors in several of these systems (Garfinkel et al., 2004; Harris et al., 2005; Mou et al., 2011). In the intestine, we have seen that several different Bmp ligands are expressed by clusters (Bmp2, 4, 5, 7) and functional assays indicate that high concentrations of all of these Bmps act to inhibit the formation of clusters (Fig. 4, Fig. S2). Similarly, multiple Bmp signaling modifiers are expressed by clusters (Nog, Twsg1, Bmp1, Fstl1) and several of these cause pattern perturbations in the explant agarose bead assay (Fig. 5). Thus, although we show that pattern formation in the intestine is faithfully modeled by a computational framework that embodies a two-component system, as originally described by Turing (1952), it is highly likely that pattern establishment and maintenance in vivo are actually a product of a much more complex combination of Bmp pathway components. In fact, in the introduction to his classical paper describing such patterning fields, Turing himself stated that his model is an idealization and simplification of reality (Turing, 1952).
In addition to their role in patterning, our data suggest that signals from mesenchymal clusters are responsible for the epithelial cell shape changes that initiate villus emergence. Epithelial cells begin to shorten apicobasally as clusters first form (Fig. 2). If clusters do not form [for example, after inhibition of Hh signaling (Madison et al., 2005; Walton et al., 2012) or in the vicinity of a Bmp-soaked bead], the epithelium remains pseudostratified. By contrast, induction of larger clusters [e.g. smoothened agonist (SAG; a synthetic Hh pathway agonist that binds smoothened) or dorsomorphin treatment] results in larger villi, over which more epithelial cells take on a columnar shape (Walton et al., 2012). Since, in the fly wing disc, clonal loss of the Bmp receptor thickveins causes cells to become columnar (Gibson and Perrimon, 2005), we predicted that epithelial Bmpr1a deletion or addition of dorsomorphin to cultured intestines would cause widespread conversion to epithelial columnar morphology. This was not observed. Thus, our studies suggest that Bmp signaling alone does not mediate the epithelial cell shape changes that occur over villus clusters. Determining the pathway(s) responsible for this morphogenic process, which is likely to provide an important part of the driving mechanism for villus outgrowth, remains an important future goal.
It is interesting that, although the use of intestinal villi to expand intestinal surface area is a well-conserved attribute in multiple species, divergent strategies for patterning of the villi have emerged during evolution. In the chick intestine, recent studies have shown that tensile forces from developing smooth muscles progressively deform the epithelium to create localized peaks of Hh protein underneath sharply bent epithelial alcoves; these Hh maxima seem to determine the location of the villi (Shyer et al., 2015). However, our data show that a different epistatic relationship between cluster formation, muscular forces and epithelial deformation portends in the mouse. Maturation of the various smooth muscle layers does not correspond temporally with the process of villus emergence in the murine intestine. Additionally, the epithelium is not remodeled into ridges or zigzags prior to villus formation; rather, villi arise as discrete domes directly from a flat epithelial surface. Although we did note soft basal epithelial deformations above nascent clusters, previous modeling studies suggest that such minimal bending is unlikely to create a substantial concentration of Hh signals (Shyer et al., 2015). Moreover, we never observed these soft basal deformations in the absence of a cluster; indeed, our evidence suggests that the deformations are a consequence of unknown signals from the underlying clusters. We provide extensive evidence that mesenchymal clusters and Bmp signaling in cluster cells control villus pattern in the mouse. Directly perturbing mesenchymal cluster pattern by altering Bmp signaling does not affect smooth muscle development (Figs 6 and 7) but does alter the pattern of clusters, thereby producing predictable changes in the pattern of the villi (Fig. 8). In fact, dramatic changes in Bmp concentration can even alter established patterns (Fig. 8I-L). Strikingly, however, despite the different modes of villification in the chick and mouse, conserved signaling pathways are involved: Hh signals from the epithelium play central roles in the induction of cluster genes, such as Bmp, in both species.
As in mouse, the intestines of human, pig and rat lack the zigzag-like structures of chick intestine (Dekaney et al., 1997; Lacroix et al., 1984; Matsumoto et al., 2002; Nakamura and Komuro, 1983). Although the proximal human intestine may contain short ridge-like structures that are later broken up into individual villi (Johnson, 1910), the human distal small intestine develops villi directly, as in the mouse (Johnson, 1910). Additionally, only the ICM, which may play a confinement role, aiding but not initiating villus emergence, is formed prior to villus emergence, and maturation of the remaining smooth muscle layers in the human (Fekete et al., 1996; Keibel, 1910), pig (de Castro, 2001; Georgieva and Gerov, 1975) and rat (Kedinger et al., 1990) occurs well after the initiation of villus formation, as in mouse. It is therefore likely that these mammalian species also rely on a villification patterning process that is controlled by gradient fields of signaling proteins rather than employing the avian model of muscle-directed epithelial deformation. It is also important to note that several rounds of villus formation have been demonstrated in the mouse (Walton et al., 2012) and are likely to occur in all species. Once the initial pattern is set, a Turing-like patterning mechanism in a growing domain could act to establish the arrangement of subsequent mesenchymal clusters, thereby generating a field of uniformly patterned villi in the intestine of all these species, including chick.
MATERIALS AND METHODS
Mice
Mice were handled humanely according to UCUCA guidelines. The following lines were used: C57BL/6 and CD1 (Charles River); RosamT/mG (Muzumdar et al., 2007), PdgfraEGFP/+ (Hamilton et al., 2003), ShhEGFPCre/+ (Harfe et al., 2004), PtclacZ (Goodrich et al., 1997), Gli1CreERT2 (Bai et al., 2002) (all Jackson Labs); and Bmpr1af/f (Mishina et al., 2002) (Dr Yuji Mishina).
Tamoxifen induction of recombination
Pregnant females were gavage fed daily from E12.5-14.5 with 250 µl 20 mg/ml tamoxifen dissolved in corn oil. Embryos were collected at E15.5.
Cultures, recombinant proteins and inhibitors
Embryonic intestines were harvested at E12.5 or E13.5 and grown in culture with protein-soaked agarose beads or dorsomorphin as described (Walton and Kolterud, 2014). Media were changed twice daily. Bmp2, Bmp4, Bmp5, Bmp7, Nog, Bmp1, Twsg1, and heterodimerized Bmp2/7 and 4/7 recombinant proteins were obtained from R&D Systems.
Cluster area measurements
Using ImageJ software (NIH), ellipses were drawn around clusters to measure area. All statistical tests were performed in Excel (Microsoft) or Prism (GraphPad). Unless otherwise noted, t-tests were two-tailed and non-parametric.
Mesh screen cultures
E13.5 intestines were harvested from PdgfraEGFP/+ embryos, cut open lengthwise and placed on a transwell membrane to expose the luminal surface. Mesh screens (55 or 75 µm, the Mesh Company, #300 or #230) were cut to size and placed on top of the intestines to culture for 1 week, with images acquired daily.
Tissue fixation and immunostaining
Epithelial-mesenchymal separation is described in the supplementary Materials and Methods. Tissues were fixed for 2 h at room temperature or overnight at 4°C in 4% paraformaldehyde. Vibratome, paraffin, and frozen sections were prepared as previously described (Walton and Kolterud, 2014; Walton et al., 2012). Antibodies used were: Pdgfrα (Santa Cruz, sc338; 1:200), E-cadherin (BD Biosciences, 610181; 1:1000), αSMA (Sigma, C6198; 1:500), desmin (Abcam, ab8592; 1:500), ezrin (Sigma, E8897; 1:500), α-tubulin (Sigma, T6199; 1:1000). Additional antibodies used were Ki67 (NovaCastra, L111859; 1:750), BrdU (Accurate, OBT0030G; 1:200) and CD44v6 (eBiosciences, BMS145; 1:1000) followed by tyramide signal amplification with the Molecular Probes T20932 Kit. BrdU immunostaining to determine proliferation index is described in the supplementary Materials and Methods.
Scanning electron microscopy (SEM)
Tissues were prepared for SEM as described in the supplementary Materials and Methods and the luminal structure was imaged on an AMRAY 1910 field emission scanning electron microscope.
RNA in situ hybridization
RNA in situ hybridization was performed on 8 µm frozen sections as described previously (Li et al., 2007).
Quantitative RT-PCR
Purified RNA was reverse transcribed and then subjected to qRT-PCR as described in the supplementary Materials and Methods using the primers listed in Table S1.
Turing field simulations
In our model, the activator-inhibitor interactions assume a saturating source of both the activator species (u) and the inhibitor species (v), the latter of which is altered by pharmacological inhibitors of Bmp signaling such as dorsomorphin (Gierer and Meinhard, 1972). The saturating source of u is also inhibited by the presence of v, according to the classic Turing activator-inhibitor system. We include density-dependent cell proliferation, along with the diffusive and chemotactic movement of the mesenchymal cells. Additional details of the Turing model are provided in the supplementary Materials and Methods.
Acknowledgements
We gratefully acknowledge mouse lines shared by Dr Yuji Mishina and Dr Alexandra Joyner. We thank University of Michigan colleagues for helpful discussions and advice: Drs Yuji Mishina, Doug Engel, Jason Spence, Ben Allen, Deneen Wellik and Linda Samuelson. We are grateful to Dr Cliff Tabin for providing unpublished data on chick villus patterning and for stimulating discussions on villus patterning.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.D.W., M.W., S.K.S., A.K., S.S. and D.L.G. contributed to concepts and approaches; K.D.W., A.K., M.J.C., J.K., N.P., D.C. and A.M.F. performed experiments; K.D.W., M.W., S.K.S., A.K., S.S. and D.L.G. analyzed data; K.D.W. and D.L.G. prepared the manuscript; K.D.W., S.K.S., A.K., S.S. and D.L.G. edited the manuscript.
Funding
Support was provided by National Institutes of Health (NIH) grants [R01 DK065850 and R01 DK089933 to D.L.G.] and an NIH Institutional Research and Academic Career Development Award Fellowship (to M.W.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.130112/-/DC1
References
- Bai C. B., Auerbach W., Lee J. S., Stephen D. and Joyner A. L. (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753-4761. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Schnell S. and Maini P. K. (2009). Waves and patterning in developmental biology: vertebrate segmentation and feather bud formation as case studies. Int. J. Dev. Biol. 53, 783-794. 10.1387/ijdb.072493rb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombre A. J. and Coulombre J. L. (1958). Intestinal development. I. Morphogenesis of the villi and musculature. J. Embryol. Exp. Morphol. 6, 403-411. [PubMed] [Google Scholar]
- de Castro A. M. M. G., Alvares E. P., Zocoller-Seno M. C., Neves M. F. and Passipieri M. (2001). Morphological study of the small intestine of moura pigs (Sus scrofa- Lineaus, 1758) during fetal development. Braz. J. Morphol. Sci. 18, 95-101. [Google Scholar]
- Dekaney C. M., Bazer F. W. and Jaeger L. A. (1997). Mucosal morphogenesis and cytodifferentiation in fetal porcine small intestine. Anat. Rec. 249, 517-523. [DOI] [PubMed] [Google Scholar]
- Dillon R. and Othmer H. G. (1993). Control of gap junction permeability can control pattern formation in limb development. Exp. Theor. Adv. Biol. Pattern Format. 259, 65-81. 10.1007/978-1-4615-2433-5_9 [DOI] [Google Scholar]
- Dunn J. S. (1967). The fine structure of the absorptive epithelial cells of the developing small intestine of the rat. J. Anat. 101, 57-68. [PMC free article] [PubMed] [Google Scholar]
- Economou A. D., Ohazama A., Porntaveetus T., Sharpe P. T., Kondo S., Basson M. A., Gritli-Linde A., Cobourne M. T. and Green J. B. (2012). Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat. Genet. 44, 348-351. 10.1038/ng.1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom D. S., Amarnath S., Fogel J. L. and Agarwala S. (2011). Bone morphogenetic proteins regulate neural tube closure by interacting with the apicobasal polarity pathway. Development 138, 3179-3188. 10.1242/dev.058602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom D. S., Inoue S., Patterson L. B., Gordon T. N., Slingwine R., Kondo S., Watanabe M. and Parichy D. M. (2012). Melanophore migration and survival during zebrafish adult pigment stripe development require the immunoglobulin superfamily adhesion molecule Igsf11. PLoS Genet. 8, e1002899 10.1371/journal.pgen.1002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete E., Benedeczky I., Timmermans J. P., Resch B. A. and Scheuermann D. W. (1996). Sequential pattern of nerve-muscle contacts in the small intestine of developing human fetus. An ultrastructural and immunohistochemical study. Histol. Histopathol. 11, 845-850. [PubMed] [Google Scholar]
- Garfinkel A., Tintut Y., Petrasek D., Bostrom K. and Demer L. L. (2004). Pattern formation by vascular mesenchymal cells. Proc. Natl. Acad. Sci. USA 101, 9247-9250. 10.1073/pnas.0308436101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva R. and Gerov K. (1975). The morphological and functional differentiation of the alimentary canal of the pig during ontogeny. II. Development and differentiation of the jejunum. Anat. Anz. 137, 16-20. [PubMed] [Google Scholar]
- Gibson M. C. and Perrimon N. (2005). Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307, 1785-1789. 10.1126/science.1104751 [DOI] [PubMed] [Google Scholar]
- Gierer A. and Meinhardt H. (1972). Theory of biological pattern formation. Kybernetik 12, 30-39. 10.1007/BF00289234 [DOI] [PubMed] [Google Scholar]
- Goodrich L. V., Milenkovic L., Higgins K. M. and Scott M. P. (1997). Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109-1113. 10.1126/science.277.5329.1109 [DOI] [PubMed] [Google Scholar]
- Hamilton T. G., Klinghoffer R. A., Corrin P. D. and Soriano P. (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013-4025. 10.1128/MCB.23.11.4013-4025.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B., Scherz P., Nissim S., Tian H., McMahon A. P. and Tabin C. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Harris M. P., Williamson S., Fallon J. F., Meinhardt H. and Prum R. O. (2005). Molecular evidence for an activator-inhibitor mechanism in development of embryonic feather branching. Proc. Natl. Acad. Sci. USA 102, 11734-11739. 10.1073/pnas.0500781102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. L. M. (1996). Bone morphogenetic proteins in development. Curr. Opin. Genet. Dev. 6, 432-438. 10.1016/S0959-437X(96)80064-5 [DOI] [PubMed] [Google Scholar]
- Johnson F. P. (1910). The development of the mucous membrane of the oesophagus, stomach and small intestine in the human embryo. Am. J. Anat. 10, 521-575. 10.1002/aja.1000100116 [DOI] [Google Scholar]
- Karlsson L., Lindahl P., Heath J. K. and Betsholtz C. (2000). Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457-3466. [DOI] [PubMed] [Google Scholar]
- Kedinger M., Simon-Assmann P., Bouziges F., Arnold C., Alexandra E. and Haffen K. (1990). Smooth muscle actin expression during rat gut development and induction in fetal skin fibroblastic cells associated with intestinal embryonic epithelium. Differentiation 43, 87-97. 10.1111/j.1432-0436.1990.tb00434.x [DOI] [PubMed] [Google Scholar]
- Keibel F. and Mall F. P. (1910). Manual of Human Embryology. Philadelphia: J. B. Lippincott Company. [Google Scholar]
- Kolterud A., Grosse A. S., Zacharias W. J., Walton K., Kretovich K. E., Madison B. B., Waghray M., Ferris J. E., Hu C., Merchant J. L. et al. (2009). Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology 137, 618-628. 10.1053/j.gastro.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S. and Miura T. (2010). Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616-1620. 10.1126/science.1179047 [DOI] [PubMed] [Google Scholar]
- Lacroix B., Kedinger M., Simon-Assmann P. and Haffen K. (1984). Early organogenesis of human small intestine: scanning electron microscopy and brush border enzymology. Gut 25, 925-930. 10.1136/gut.25.9.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Madison B. B., Zacharias W., Kolterud A., States D. and Gumucio D. L. (2007). Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol. Genomics 29, 290-301. 10.1152/physiolgenomics.00269.2006 [DOI] [PubMed] [Google Scholar]
- Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T. and Gumucio D. L. (2005). Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279-289. 10.1242/dev.01576 [DOI] [PubMed] [Google Scholar]
- Maini P. K. (2004). The impact of Turing's work on pattern fomation in biology. Math. Today 40, 73-92. [Google Scholar]
- Maini P. K., Baker R. E. and Chuong C.-M. (2006). Developmental biology: the Turing model comes of molecular age. Science 314, 1397-1398. 10.1126/science.1136396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan M., Moxey P. C. and Trier J. S. (1976). Morphogenesis of fetal rat duodenal villi. Am. J. Anat. 146, 73-92. 10.1002/aja.1001460104 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Hashimoto K., Yoshioka T. and Otani H. (2002). Occlusion and subsequent re-canalization in early duodenal development of human embryos: integrated organogenesis and histogenesis through a possible epithelial-mesenchymal interaction. Anat. Embryol. 205, 53-65. 10.1007/s00429-001-0226-5 [DOI] [PubMed] [Google Scholar]
- Meinhardt H. (2012). Turing's theory of morphogenesis of 1952 and the subsequent discovery of the crucial role of local self-enhancement and long-range inhibition. Interface Focus 2, 407-416. 10.1098/rsfs.2011.0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y., Hanks M. C., Miura S., Tallquist M. D. and Behringer R. R. (2002). Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32, 69-72. 10.1002/gene.10038 [DOI] [PubMed] [Google Scholar]
- Mou C., Pitel F., Gourichon D., Vignoles F., Tzika A., Tato P., Yu L., Burt D. W., Bed'hom B., Tixier-Boichard M. et al. (2011). Cryptic patterning of avian skin confers a developmental facility for loss of neck feathering. PLoS Biol. 9, e1001028 10.1371/journal.pbio.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nakamura K. and Komuro T. (1983). A three-dimensional study of the embryonic development and postnatal maturation of rat duodenal villi. J. Electron Microsc. 32, 338-347. [PubMed] [Google Scholar]
- Rajagopal R., Huang J., Dattilo L. K., Kaartinen V., Mishina Y., Deng C.-X., Umans L., Zwijsen A., Roberts A. B. and Beebe D. C. (2009). The type I BMP receptors, Bmpr1a and Acvr1, activate multiple signaling pathways to regulate lens formation. Dev. Biol. 335, 305-316. 10.1016/j.ydbio.2009.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspopovic J., Marcon L., Russo L. and Sharpe J. (2014). Modeling digits. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 345, 566-570. 10.1126/science.1252960 [DOI] [PubMed] [Google Scholar]
- Roberts D. J., Johnson R. L., Burke A. C., Nelson C. E., Morgan B. A. and Tabin C. (1995). Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development 121, 3163-3174. 10.1126/science.1252960 [DOI] [PubMed] [Google Scholar]
- Sbarbati R. (1982). Morphogenesis of the intestinal villi of the mouse embryo: chance and spatial necessity. J. Anat. 135, 477-499. [PMC free article] [PubMed] [Google Scholar]
- Shen J. and Dahmann C. (2005). Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science 307, 1789-1790. 10.1126/science.1104784 [DOI] [PubMed] [Google Scholar]
- Shyer A. E., Tallinen T., Nerurkar N. L., Wei Z., Gil E. S., Kaplan D. L., Tabin C. J. and Mahadevan L. (2013). Villification: how the gut gets its villi. Science 342, 212-218. 10.1126/science.1238842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer A. E., Huycke T. R., Lee C., Mahadevan L. and Tabin C. J. (2015). Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569-580. 10.1016/j.cell.2015.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sick S., Reinker S., Timmer J. and Schlake T. (2006). WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science 314, 1447-1450. 10.1126/science.1130088 [DOI] [PubMed] [Google Scholar]
- Trahair J. and Robinson P. (1986). The development of the ovine small intestine. Anat. Rec. 214, 294-303. 10.1002/ar.1092140309 [DOI] [PubMed] [Google Scholar]
- Turing A. M. (1952). The chemical basis of morphogenesis. Philos. Trans. R. Soc. B Biol. Sci. 237, 37-72. 10.1098/rstb.1952.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D. and Kolterud A. (2014). Mouse fetal whole intestine culture system for ex vivo manipulation of signaling pathways and three-dimensional live imaging of villus development. J. Vis. Exp. 91, e51817 10.3791/51817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. D., Kolterud A., Czerwinski M. J., Bell M. J., Prakash A., Kushwaha J., Grosse A. S., Schnell S. and Gumucio D. L. (2012). Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl. Acad. Sci. USA 109, 15817-15822. 10.1073/pnas.1205669109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M. and Spence J. R. (2014). How to make an intestine. Development 141, 752-760. 10.1242/dev.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liu H.-X. and Mistretta C. M. (2006). Bone morphogenetic proteins and noggin: inhibiting and inducing fungiform taste papilla development. Dev. Biol. 297, 198-213. 10.1016/j.ydbio.2006.05.022 [DOI] [PubMed] [Google Scholar]