Abstract.
We propose an active learning (AL) approach for prostate segmentation from magnetic resonance images. Our label query strategy is inspired from the principles of visual saliency that have similar considerations for choosing the most salient region. These similarities are encoded in a graph using classification maps and low-level features. Random walks are used to identify the most informative node, which is equivalent to the label query sample in AL. To reduce computation time, a volume of interest (VOI) is identified and all subsequent analysis, such as probability map generation using semisupervised random forest classifiers and label query, is restricted to this VOI. The negative log-likelihood of the probability maps serves as the penalty cost in a second-order Markov random field cost function, which is optimized using graph cuts for prostate segmentation. Experimental results on the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2012 prostate segmentation challenge show the superior performance of our approach to conventional methods using fully supervised learning.
Keywords: prostate segmentation, visual saliency, graph cuts, random forests, MRI, random walks, active learning, semisupervised learning
1. Introduction
According to the American Cancer Society, prostate cancer is the second leading cause of cancer death in American men, and early diagnosis can potentially increase the survival rate among patients.1 Among all imaging modalities, magnetic resonance imaging (MRI) is the most popular for treatment planning due to its high spatial resolution, soft-tissue contrast, and absence of ionizing radiations. Accurate quantification of prostate volume and location of the prostate relative to adjacent organs are also essential parts of image-guided radiation therapy. Hence, there is a need to develop accurate and robust (semi)automatic algorithms for prostate segmentation.
Manual segmentation of the prostate in MRI is time consuming, and prone to inter- and intraexpert variability. Software algorithms should be able to overcome challenges like (1) variability of prostate size and shape between subjects; (2) variable image appearance and intensity ranges from different MR scanning protocols; and (3) lack of clear prostate boundaries due to similar intensity profiles of surrounding tissues. Due to poor contrast, segmentation methods using specific image features find it hard to distinguish between prostate and nonprostate regions. Machine learning (ML) methods present a formal way to learn and identify features that are highly discriminative for classification and segmentation purposes. A robust algorithm requires many example cases to learn from a wide range of image appearances. However, obtaining sufficient manual annotations is very expensive, time consuming, and requires personnel with high expertise. In this paper, we propose a ML-based segmentation method that requires significantly fewer labeled samples, yet achieves higher segmentation accuracy than conventional ML methods. Our algorithm uses (1) an active learning (AL) strategy that incorporates principles from visual saliency to query labels of informative samples; and (2) semisupervised learning (SSL) that uses a few labeled samples and many unlabeled samples to construct a highly accurate classifier.
1.1. Related Work on Magnetic Resonance Imaging Prostate Segmentation
Many recent studies have proposed automatic segmentation algorithms for prostate MRI, such as multiatlas matching,2,3 active shape models,4 active appearance models,5 and patient-specific deformable models.6 ML methods7,8 use appearance features from training images to train classifiers that calculate probability maps of test images. The final segmentation is then obtained using level sets7 or graph cuts.8 Success of ML methods depends on the discriminative power of handcrafted features. To overcome this shortcoming, Liao et al.9 propose a deep-learning framework using independent subspace analysis to automatically learn the most discriminative features.
The growing importance of prostate MRI segmentation led to the prostate segmentation challenge in MICCAI 2012. Different approaches in the challenge include marginal space learning,10 multiatlas segmentation using local appearance-specific atlases,11 and classification maps with level sets.7 A detailed overview of the different methods and their performance is also given.12
1.2. Our Contribution
Recent work has shown that expert feedback can greatly improve the accuracy of medical image-segmentation algorithms.13,14 This motivated us to explore the efficacy of incorporating expert feedback for improved prostate segmentation. In this paper, we combine AL and SSL to segment the prostate from MR images. The important contribution of this paper is a visual saliency-based approach to select the most informative samples for AL. We show that many of the principles of salient region detection are applicable to query selection in active learning tasks. Hence, selecting the most informative region in MRI becomes a problem of salient region detection by defining an appropriate measure of a region’s importance.
An earlier version of our work segments Crohn’s disease regions in MRI.15 However, this paper has the following characteristics: (1) detailed investigation of the effect of different components of the query strategy; (2) analysis on savings in time and effort by our method; and (3) comparison with popular AL query strategies. Sections 2–5 describe different parts of our method. We describe our dataset and results in Sec. 6 and conclude with discussions in Sec. 7.
2. Overview of Segmentation Method
Training of the random forest- (RF-) based SSL classifier (RF-SSL) starts by taking the labeled samples of the first training patient (denoted as set ). The samples from the remaining training patients are assumed to be unlabeled (denoted as set ). Note that set consists only of the samples that have been annotated by the experts, and does not comprise all voxels of the image. Although including labeled samples in the “unlabeled” set seems contradictory, knowledge of the true labels is essential to quantify the performance of our method. This will help us to calculate the percentage of total samples required to train a robust system and the quantitative advantage over fully supervised methods. The AL method selects the most informative sample in in which the actual label is obtained from the stored label set. This sample is added to set and the classifier is updated using online learning. The query continues until the AL algorithm cannot identify an informative sample, i.e., a sample for which its characteristics have not been added to the training set .
Since analysis of each voxel is time consuming, we use an automatically detected volume of interest (VOI) for each image obtained using our previous method.8 All analysis is restricted to the VOI voxels. A fivefold cross validation is adopted to tune our classifier. The trained RF-SSL classifier is used to segment the test volumes using the VOI probability maps and graph cuts. An overview of our method is given in Algorithm 1.
Algorithm 1.
Semisupervised and active learning for prostate segmentation.
| Input Image with voxels. |
| Output Segmented prostate. |
| Sequence of Steps: |
| 1. Bias correction and intensity normalization. |
| 2. Train RF–SSL classifier with labeled samples of the first training patient (set ) and unlabeled samples of remaining training patients set . |
| 3. Determine the most informative sample from and query its label. |
| 4. Add labeled sample to and update RF–SSL using online learning |
| 5. Continue label query until no further queries are required. This completes training of RF–SSL. |
| 6. For test patient, generate classification map for the VOI patches using the final RF–SSL. |
| 7. Segment prostate using final classification map and graph cuts [Eqs. (16) and (17)]. |
3. Image Features
We use intensity statistics, texture and curvature entropy, and spatial context features for voxel classification, as they have been found to be very effective in distinguishing prostate from background 8 and also in identifying disease activity.16 The features are extracted from a patch around each voxel. Intensity, texture, and curvature features combined give an 85 dimensional feature vector, while context features give an additional 96 features. Since our emphasis in this paper is the combination of SSL and AL techniques, we give a brief description of each feature and refer the reader to16 for details.
3.1. Intensity Statistics
MR images commonly contain regions that do not form distinct spatial patterns but differ in their higher-order statistics.17 Therefore, in addition to the features processed by the human visual system, i.e., mean and variance, we extract skewness and kurtosis values from each voxel’s neighborhood.
3.2. Texture Entropy
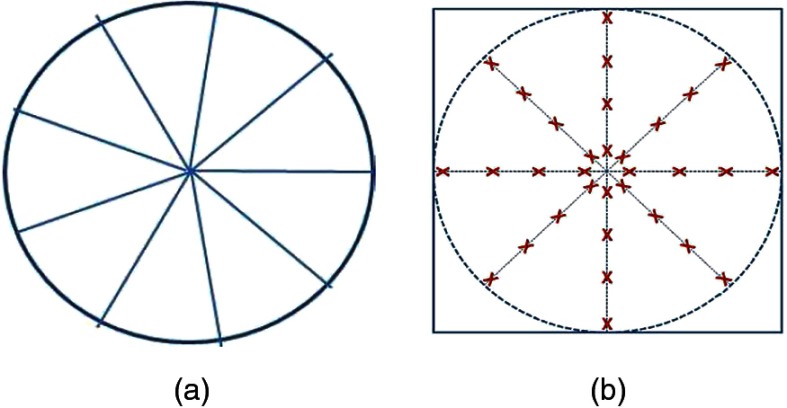
Texture maps are obtained from two-dimensional (2-D) Gabor filter banks for each slice (at orientations 0 deg, 45 deg, 90 deg, 135 deg, and scale 0.5, (1). Each image patch is partitioned into nine equal parts corresponding to nine sectors of a circle. Figure 1(a) shows the template for partitioning a patch into sectors and extracting entropy features. For voxels within each sector, we calculate the texture entropy given by
| (1) |
where denotes the probability distribution of texture values in sector . This procedure is repeated for all the eight texture maps over four orientations and two scales to extract a 72 dimensional feature vector.
Fig. 1.
(a) partitioning of patch for calculating anisotropy features and (b) template for calculating context features.
3.3. Curvature Entropy
Different tissues have different curvature distributions, and we exploit this characteristic for accurate prostate identification. Curvature maps are obtained from the gradient maps of tangent along the three-dimensional (3-D) surface. The second fundamental form () of these curvature maps is identical with the Weingarten mapping and the trace of the matrix gives the mean curvature. This mean curvature map is used for calculating curvature entropy. Details on curvature calculation is given in Ref. 18. Similar to texture, curvature entropy is calculated from nine sectors of a patch and is given by
| (2) |
where denotes the probability distribution of curvature values in sector and denotes the curvature values.
We use 2-D texture and curvature maps as the 3-D maps do not provide consistent features because of low resolution in the direction. Hence the extracted 3-D features are unreliable.
3.4. Spatial Context Features
Context information is particularly important for medical images because of the the regular arrangement of human organs. Figure 1(b) shows the template for context information where the circle center is the current voxel and the sampled points are identified by a red ‘X’. At each voxel corresponding to an ‘X’ we calculate the mean intensity, texture, and curvature values from a patch. The texture values were derived from the texture maps at 90-deg orientation and scale 1. The ‘X’s are located at distances of 1, 3, 5, and 8 voxels from the center, and the angle between consecutive rays is 45 deg. The values from the 32 regions are concatenated into a 96 dimensional feature vector and the final feature vector has values.
4. Random Forest Classifiers
RFs19 have become increasingly popular in classification tasks because of their computational efficiency for large training data, ability to handle multiclass classification, and facility to quantify importance measures of features. A RF is an ensemble of decision trees, where each tree is typically trained with a different subset of the training data (“bagging”), thereby improving the generalization ability of the classifier. Forest training is equivalent to determining the set of tests that best separate the data into the different training classes using a measure of class impurity like class entropy. The tuning parameters are maximal tree depth and number of trees.
4.1. Semisupervised Learning With Random Forests
Semisupervised learning techniques20 train classifiers with a few labeled samples and many unlabeled samples. They leverage the hidden structural information in many unlabeled samples and map them to the labeled data. This is a typical scenario in many medical applications where it is difficult to find qualified experts to label the large number of medical images. A “single-shot” RF method for SSL without the need for iterative retraining was introduced in Ref. 21. Note that this method is exclusively used for semisupervised learning, whereas the active learning part of our approach explains the selection of informative samples for label queries. We use this method for SSL as it is shown to outperform other approaches such as support vector machines.21 We give a brief explanation of this method later.
For labeled samples the information gain over data splits at each node is maximized and encourages separation of the labeled data.19,21 However, for SSL, the objective function encourages separation of the labeled training data and simultaneously separates different high-density regions. It is achieved via the following mixed information gain:
| (3) |
where is the information gain from the labeled data; is the entropy of training points, and and are the subsets going to the left and right children of node , and is a user defined weight. depends on both labeled and unlabeled data and is defined using differential entropies over continuous parameters as
| (4) |
where is the covariance matrix of the assumed multivariate distributions at each node. Further details are given in Ref. 21.
5. Semisupervised Learning-Active Learning-Based Segmentation From Magnetic Resonance Images
Initial Preprocessing: the given images are first bias-corrected22 to remove intensity inhomogeneities due to the magnetic field of MR machines. The intensities are then normalized to [0, 1]. Pixels with the lowest 5% intensities usually indicate noise, and are all set to zero. Similarly, higher intensity tails in the intensity histogram indicate artifacts and outliers. Hence, the mean of the 5% brightest intensity values is taken to be the maximum image intensity. All intensity values are divided by this threshold and ratios exceeding one are set to one. Normalization also increases the image contrast.
5.1. Active Learning Query Strategy
Learning starts with the labeled samples (voxels) of the first training dataset (set ). The features of and “unlabeled” voxels (set from the remaining labeled training datasets) are used as inputs to an RF-based SSL classifier (denoted as RF–SSL), which predicts the class labels and probabilities of the unlabeled patches. The label of the most informative voxel is obtained from the stored label set, added to set , and the RF classifier is updated using online learning.23 Figure 2 shows results of different stages of our method. Although set has labeled data, we still refer to it as the unlabeled set in order to stick to conventional terminology and avoid confusion.
Fig. 2.
(a) Original image divided into patches (green). Manual ground truth segmentation is in red; (b) prostate probability map after first patch selection; (c) second set of selected patches; (d) prostate probability map after second patch selection; (e) final prostate probability map after 4 patch selections; (f) segmented prostate by our method in green and manual segmentation in red; (g) probability maps through FSL; and (h) final segmentation by FSL.
Our query strategy selects a sample with the following characteristics: (1) high classification uncertainty to obtain information from each labeling instance; (2) situated in a dense region such that it is representative of many other samples; and (3) minimal overlap of influence with previously labeled samples to minimize redundancy in labeling effort. Choosing the query sample is similar to selecting salient regions of an image, which have the following characteristics: (1) their feature values are significantly different from the surroundings (high local contrast); and (2) contrast magnitude is higher than other regions and hence it stands out visually. Visual saliency models selecting the most salient region in an image are based on, among others, local contrast,24 entropy,25 and graph-based random walks.26,27
High-contrast regions have maximum information and hence higher entropy.25 Regions with high classification uncertainty also have high entropy, indicating a correspondence between information content of salient regions and classification uncertainty. Salient regions are located on regular objects (or dense regions of the sample space) and different salient regions are far away from each other, i.e., their influence areas have minimum overlap. Thus, we see that the properties of salient regions have similarities with the desired characteristics of query samples. Hence, saliency models can be adapted for active learning tasks using appropriate similarity metrics.
Image patches are represented as nodes of a graph and connected by set of edges . Based on the similarity between any two nodes and , a weight is assigned to edge connecting and . Random walks on the graph determine the most frequently visited node, which is also the most salient node. The weights between two nodes and are given by
| (5) |
where is the feature vector of node , also termed its informativeness ; , and denotes norm. Informativeness of node (or voxel ) is given by
| (6) |
where is the classification uncertainty of given by the label entropy as
| (7) |
where indicates all possible labels (in this case two) for , and is calculated by RF-SSL. High entropy indicates greater uncertainty. incorporates contextual information, and is the collection of intensity, texture, and curvature differences defined as
| (8) |
where is the sum of the exponential of intensity differences between node and all unlabeled nodes in (a neighborhood of ), and . For similar nodes, takes higher values. and are the corresponding texture (from the oriented map at 90 deg) and curvature differences. Note that we do not average the feature differences over the neighborhood. In a high-density region, is calculated by summing over more voxels than in a sparsely populated region. Since is not divided by a normalization constant, its value is higher in the high-density region.
Context Information for Informativeness: medical images have inherent context information because the relative arrangement of organs in the human body is constant, and similar tissues are clustered together. An unlabeled sample close to a labeled sample is assigned lower importance because it has a higher probability of having the same label than a sample far away. If the radiologist were to annotate samples close to an already labeled sample, it “does not” generally lead to significant information gain. Thus, incorporates context information and is equal to ‘s distance from the nearest labeled sample
| (9) |
where denotes all the labeled samples (or nodes) and denotes the Euclidean distance based on voxel coordinates.
Thus, we observe that Eq. (7) encodes the classification uncertainty of a node (or image voxel), Eq. (8) quantifies the patch’s density and Eq. (9) determines the influence of a labeled sample on a test voxel. Since the features for all voxels are precomputed, we normalize them by subtracting the mean of each feature dimension from the corresponding element and dividing by the standard deviation. Thus, all the features are normalized to the same range, which improves the accuracy of detecting salient nodes.
5.2. Random Walks and Most Salient Node
The weights [Eq. (5)] are used to define the affinity matrix as
| (10) |
The degree of a node , is defined as the sum of all weights connected to node and the degree matrix is defined as the diagonal matrix with the degrees of the node in its main diagonal, i.e., and . The transition matrix for the random walk on the fully connected graph is, .
A Markov chain with states is completely specified by the transition matrix and an initial probability distribution on the states. is the probability of moving from state to state . An ergodic Markov chain is one in which it is possible to go from any state to any other state, not necessarily in a single step. A Markov chain is regular if some power of its transition matrix has only positive elements. A random walk starting at any given state of an ergodic Markov chain reaches equilibrium after a certain amount of time. This equilibrium condition is characterized by the equilibrium probability distribution of the states, , which satisfies the relation
| (11) |
where is the row vector of equilibrium probability distributions of states in the Markov chain. The matrix is the matrix obtained by times stacking of row vector . The fundamental matrix of an ergodic Markov chain is defined as
| (12) |
The following three quantities are defined: —the expected number of steps to return to state for the first time if the Markov chain is started in state at time ; —the expected number of steps to reach state for the first time if the Markov chain is started in state at time . It is also known as the hitting time; —the expected number of steps to reach state for the first time if the Markov chain is started in the equilibrium distribution at time . These three quantities can be derived from and as
| (13) |
where is the th element of ; , , and are the respective elements of . A detailed proof of the above derivations is given in Ref. 28. Since we have to compute the globally most “salient” node, the global saliency of node is given by the sum of hitting times from all other nodes to node on a complete graph, i.e.,
| (14) |
and the most salient node is given by the maximum as .
Stopping Criteria: irrespective of the number of labeled samples, there will always be one unlabeled sample with maximum informativeness. In order to ensure that the label query does not continue indefinitely, we determine the probability values of the two classes for the most informative sample. If the probability value for any one class is (or for the other class) we do not query the label for that sample. If we encounter such samples for five consecutive iterations then we stop the label query because this indicates that the classifier has obtained sufficient samples to have high confidence on its classification output.
5.3. Graph Cut Segmentation
A spatially smooth solution is obtained by formulating the segmentation as a labeling problem within a second-order Markov random field (MRF) cost function. The labels are obtained for each voxel and not the individual patches by optimizing the cost function using graph cuts.29 A second-order MRF energy function is given by
| (15) |
where denotes the set of pixels and is the set of neighboring pixels for pixel . is a weight that determines the relative contribution of penalty cost () and smoothness cost (). is given by
| (16) |
where is the likelihood (from probability maps) previously obtained using RF classifiers and is a very small value to ensure that the cost is a real number. ensures a spatially smooth solution by penalizing discontinuities and is defined as
| (17) |
denotes the intensity. Smoothness cost is determined over an eight-neighborhood system. The labels are obtained by optimizing Eq. (15) using alpha-expansion and graph cuts.29
6. Experiments and Results
The MICCAI 2012 prostate MR image segmentation (PROMISE) prostate segmentation challenge 30 has 50 training and 30 test datasets of transversal T2-weighted MR images. The datasets are acquired under different clinical settings. They are multicenter and multivendor, and have different acquisition protocols (e.g., differences in slice thickness, with/without endorectal coil). The volume dimensions and voxel resolutions are different for different images. Each slice of the different volumes is of size (voxel resolution of ) or (resolution ). The set is selected such that there is a spread in prostate sizes and appearance. Reference segmentations are available only for the training dataset. We employ a 10-fold cross validation on the training data for RF–SSL training and testing. Average results are reported for training and test data. Each patient was segmented by our algorithm only once. Our whole pipeline was implemented in MATLAB on a 2.66-GHz quad core CPU running Windows 7. The quality of our segmentation results with respect to manual segmentations was evaluated using two measures by the organizers: (1) dice metric (DM) and (2) 95% Hausdorff distance (HD).
The results between two different methods were compared using a paired -test that determines whether the two sets of results are statistically different or not. The paired -test was performed using MATLAB’s ttest2 function. The function performs a -test of the null hypothesis that the set of DM values from any two methods are independent random samples from normal distributions with equal means and equal but unknown variances, against the alternative that the means are not equal. The test is conducted at the 5% significance level and the result is returned as the -value. indicates that the difference in results is statistically significant. Otherwise, the performance of the two methods being compared is similar. There are many standard software packages that perform the tests on a set of observations, such as MATLAB’s Statistical Toolbox and the R software package. We use the MATLAB function because it integrates better into our workflow. The -values obtained by the method so described are denoted as . Another set of values are obtained using Bonferroni correction as available in the “multcompare” function in MATLAB. The second set of values are referred to as .
6.1. Markov Random Field Regularization Strength [Eq. (15)]
To fix we choose a separate group of eight patient volumes and perform segmentation using our method but with taking different values from 10 to 0.001. The maximum average DM was obtained for , and this value was chosen for the subsequent experiments. Note that these eight datasets were “not part” of the test or training dataset used for evaluating our algorithm.
6.2. Influence of Number of Trees
We also examine the effect of varying number of trees () in the RF classifier on the average DM value. With increasing , DM increases along with the time taken for training. Table 1 shows the training time () for different as a multiple of the training time for . For , there is no significant increase in DM (), but the training time increases significantly. The best trade-off between and DM is achieved for 50 trees and is the reason we have 50 trees for our RF ensemble.
Table 1.
Effect of number of trees in RF classifiers () on segmentation accuracy and training time.
| 5 | 7 | 10 | 20 | 50 | 70 | 100 | 150 | |
| DM | 77.6 | 82.1 | 84.0 | 85.6 | 90.2 | 90.9 | 91.0 | 91.2 |
| 0.1T | 0.2T | 0.3T | 0.6T | T | 1.6T | 2.3T | 3.2T |
6.3. Results on Training Data
Since we have access to the manual segmentations of the training data, we can perform different analysis to determine the effect of different components of our method. We present comparative results for
-
1.
SSL–AL: our proposed segmentation algorithm using RF, SSL, and AL. A fivefold cross-validation setting is used where labels are queried “only for training datasets”.
-
2.
FSL: our automatic fully supervised learning-based method.8 The classifier is trained on all the manually labeled samples given by experts and does not use active learning. A fivefold cross-validation approach is used to tune the classifier.
-
3.
: SSL–AL without incorporating context information through .
-
4.
FSL–AL: fully supervised segmentation using active learning. The difference from SSL–AL is that instead of a semisupervised classifier we use a fully supervised classifier trained only on the labeled data. Online learning is used for updating the classifier.
Since other methods have been applied on the PROMISE12 segmentation challenge, we refer the reader to 12 for details on their performance. Figure 3 shows the box plots of the average DM and HD measures for all 50 training datasets, and Fig. 4 shows segmentation results on patient 15 using the above methods. SSL–AL gives the best segmentation results, and significantly improves over FSL‘s performance with and . Note that the values were obtained by comparing the DM values of SSL–AL and FSL. SSL–AL achieves higher segmentation accuracy using fewer labeled samples, which translates to less labeling time. gives lower segmentation accuracies than due to absence of context information. This clearly indicates that inclusion of leads to more accurate classifiers, and hence better segmentations. FSL–AL has a similar performance as FSL, but is significantly worse than SSL–AL. Although FSL–AL uses active learning, it does not incorporate SSL. This also highlights the importance of SSL in our method. Figure 5 shows segmentation results for SSL–AL and FSL on the more challenging regions of the prostate apex and base. As is the case for previous examples, SSL–AL outperforms FSL by a considerable degree. Table 2 summarizes the sensitivity, specificity, and accuracy using the above four approaches, and SSL–AL is superior to the other methods by a considerable degree.
Fig. 3.
Box plots of: (a) DM and (b) HD values for all 50 training patients.
Fig. 4.

Segmentation results for Patient 15. The manual annotations are shown in red with the algorithm segmentations in green: SSL–AL (column 1), FSL (column 2), (column 3) and FSL–AL (column 4). Each row shows results for different slices of the same volume.
Fig. 5.

Segmentation results for the base and apex of two patients in the training data. The manual annotations are shown in red with the algorithm segmentations in green: (a) SSL–AL results for prostate base; (b) FSL results for prostate base; (c) SSL–AL results for prostate apex; and (d) FSL results for prostate apex. The results are for Patient 20 (row 1) and Patient 32 (row 2).
Table 2.
Quantitative measures for segmentation accuracy on training data. Acc, overall accuracy of identifying prostate and background samples; Sen, sensitivity (percent of correctly classified prostate samples); Spe, specificity; DM, Dice metric; HD, 95% Hausdorff distance; and , , results of a -test in comparison with the results of SSL–AL. Dice metric scores are considered for this comparison.
| SSL–AL | FSL | FSL–AL | ||
|---|---|---|---|---|
| Sen | ||||
| Spe | ||||
| Acc | ||||
| DM | ||||
| — | 0.0031 | 0.00094 | 0.0019 | |
| — | 0.0113 | 0.0081 | 0.0225 | |
| HD |
Computation Time: recall that once the training of different classifiers (e.g., SSL–AL, FSL, and ) are complete, they are used to generate probability maps of the VOIs of test images. These VOIs are the same for all classifiers. Since we also use the same feature set for all methods, the computation times for segmenting the test image are almost identical for the different methods: 11.8 min for SSL–AL, 11.9 min for FSL, 11.8 min for , and 11.7 min for FSL–AL.
6.4. Results on MICCAI PROMISE12 Online Challenge Dataset
To apply our method on the online challenge dataset, we train our SSL–AL classifier on all 50 volumes of the training data. Then, we use the trained classifier to segment the test images of the PROMISE MICCAI 2012 prostate segmentation challenge12 and submit the results online. The test data (also called the online challenge data) have 30 patient volumes, although its manual segmentations are not available to the participants. The results are submitted online and the user gets feedback on the algorithm’s performance with evaluation metric values for all 30 cases and a ranking compared to other methods on the same dataset. The rankings are based on an overall score that combines the scores of individual metrics and is explained in Ref. 12. The rankings and performance on individual volumes is publicly available on the challenge website at Ref. 31. Our proposed SSL–AL method is ranked third among all the methods, while our previous FSL method 8 is ranked 14th. This is a significant improvement due to the use of SSL and AL algorithms for training the classifier.
For SSL–AL, the average of different evaluation metrics are , mm, and . The corresponding values for FSL are , , and boundary distance . A plot of DM and HD values for our method on individual datasets is shown in Fig. 6. For SSL–AL, we had two cases with , while 24 cases had including nine cases having . This clearly indicates a significant improvement in segmentation accuracy for SSL–AL over FSL despite not having access to labels of the target scans.
Fig. 6.

Individual values for SSL–AL on 30 patients of the MICCAI 2012 online challenge dataset: (a) DM and (b) 95% HD.
The method by Vincent et al.32 is ranked first, while the method by Birkbeck et al.10 is ranked second. Their performance is summarized as follows: (1) Vincent et al.32—, , and ; and (2) Birkbeck et al.10—, , and . For DM, SSL–AL ranked fourth with 33 having , ranked third. For all other metrics, our method was ranked third. Importantly, our method’s DM and HD values are very close to the two methods ranked higher than ours. The overall score used to determine the rankings was as follows: (1) Vincent et al.32 ; (2) Birkbeck et al.10; (3) ; (4) Malmberg et al.33 ; and (5) . The significant improvement in performance of SSL–AL over FSL clearly indicates the advantage of using SSL and AL in training a classifier. The difference in their performance is also significant . The comparative performance is summarized in Table 3.
Table 3.
Quantitative measures for segmentation accuracy for the online challenge dataset. DM, dice metric in %; HD-95%, Hausdorff distance in mm, and BD, boundary distance in mm.
6.5. Importance of Context Parameter () in [Eq. (6)]
Recall that in Eq. (6) is the Euclidean distance of an unlabeled sample to the nearest labeled sample while other query selection strategies use a user defined parameter.34 We run our segmentation algorithm by varying the value of from 0 to 10 in steps of 0.1 (denoted by in Table 4) and results of cross validation showed that best segmentation results was obtained for . We use the same eight datasets as used to set the value of . The DM value is lower than our adaptive because a constant discards contextual information from the nearest labeled sample.
Table 4.
Segmentation performance for constant .
|
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.5 | 1 | 1.5 | 2 | 2.5 | 3 | 5 | 7 | 9 | 10 | |
| DM | 80.6 | 81.5 | 83.6 | 84.6 | 86.6 | 88.2 | 88.0 | 87.3 | 87.0 | 86.5 | 86.3 | 86.2 |
6.6. Comparison with Other Query Strategies
The effectiveness of our query strategy is compared with other widely used query strategies such as uncertainty sampling,35 query-by-committee (QBC) 36 and density weighting.34 The first two methods select samples with high classification uncertainty obtained from the label entropy. QBC employs a committee of models to test different hypotheses. The most informative query is one which leads to maximum disagreement between the different models. We get the following DM values with these methods: QBC-85.9%, and IDW-86.2%. The corresponding comparisons with SSL–AL by -tests gave , in all cases.
For QBC, we generate two different hypotheses: Eq. (6) is calculated with determined from (1) fully supervised RFs; and (2) RF-SSL (as proposed in our method). The sample which gives maximum disagreement (or maximum distance according to Eq. (5) is chosen as the query sample). For uncertainty sampling, we use only the value in Eq. (7) as a measure of a sample’s informativeness. For IDW the informativeness of a sample is given by
| (18) |
Further details on parameter selection are given in Ref. 34.
Our query strategy’s superior performance is due to use of random walk-based global graph optimization that selects the most informative sample, and also due to context information . Other query strategies do not analyze the global information from graphs and hence perform poorly.
7. Discussion and Conclusion
We have developed a query strategy for active learning-based prostate segmentation. The problem of query sample selection is similar to detecting visually salient regions on a graph. Our method combines semisupervised classification and active learning to achieve higher segmentation accuracy than fully supervised methods, but with fewer labeled samples.
Savings in Labeling Effort and Time: one of the primary objectives of our method is to reduce training time. By querying labels of most informative patches, we also expect to reduce the redundancy of labels such that new labels provide truly information to the classifier. Also, savings in labeling effort can be estimated by the number of queried voxels during active learning and compared with the number of manually labeled voxels.
In fivefold cross validation FSL uses of manual labels for training while SSL–AL requires 42% of manual annotations and still achieves higher segmentation accuracy. Thus, SSL–AL performs better than FSL with nearly half of its labeled data. The figure of 42% was determined by counting the total number of voxels labeled for SSL–AL during query over all the 50 training patients, and dividing by the total number of manually annotated voxels.
Interestingly, although FSL has access to more training samples, it performs poorly compared to SSL–AL. This quite clearly indicates that more training samples do not necessarily translate to better performance. More samples could introduce noise, especially if the annotations are not accurate enough. In such a scenario, it is beneficial to ask experts to label only the most-informative samples. This leads to savings in time, effort, and also improves algorithm’s performance.
Online learning leads to significant time savings. Updating the classifiers with every new label requires about . On the other hand, lots of time is required to retrain the entire classifier using the entire training set, from 0.3 s (with four labeled samples) to 43 s (with 676 labeled samples). This gives an estimate of the time saved by using our method.
The total “training time” for the 50 training patients is 38 min using FSL and 20 min using SSL–AL. The lower training time for SSL–AL is due to the fact that less number of samples are required for each class. Fewer labeled voxels lead to time savings of during training. Although labels are queried for both prostate and background voxels, the number of queried prostate samples (62%) is higher compared to the background samples (38%). This is due to the prostate samples having different characteristics in different patients. The average segmentation time for FSL, SSL–AL, , and FSL–AL is approximately the same, i.e., 12 min. After training is complete the RF-based classifiers take the same time to classify each voxel, generate probability maps and obtain the final segmentation using graph cuts.
Incorporation of contextual information into the active learning framework and using random walks to query the most informative sample leads to improved accuracy. Additionally, a combination of context information, and classification uncertainty leads to a query strategy that outperforms FSL methods. Experimental results on real patient prostate MR volumes from the public MICCAI 2012 Prostate segmentation challenge dataset show our method is ranked third amongst 16 methods and its performance is quite close to the two methods ranked above ours. This clearly demonstrates the improvement in segmentation accuracy obtained using SSL and AL even without knowledge of labels of the test image.
Intensity normalization and inhomogeneity correction in medical images is important for subsequent analysis. These steps pose challenges in MR images due to the lack of pulse sequence dependent standardized intensity scale like the Hounsfield units in computed tomography. This affects the post processing of the acquired images. Different intensity normalization techniques include registration of joint histograms,37 landmarking matching between histograms,38 histogram estimation using Gaussian mixtures,39 and multiplicative correction field.40 Other methods41,42 that perform simultaneous normalization and image segmentation are equally effective as preprocessing tools. Histogram model-based approaches for intensity normalization were limited by the fact that the images were taken from different clinical settings. Hence, we chose to use a simple method that achieves equally effective results. If we normalize our images using the methods in Refs. 41 and 42, the final DM values were not significantly different compared to our method as shown by . Hence, we may conclude that our simple intensity normalization strategy was effective in meeting its desired objective.
Our method’s use in actual clinical scenarios is limited by the relatively high computation time of 12 min. Since our framework is based on MATLAB, it is possible to improve the computation speed by using other software packages or exploring faster ways for semisupervised classification and active learning.
Although our algorithm results in good segmentation results with high DM and low HD values, its primary limiting factor is the quality of the labels provided by the annotator. If the annotator can correctly identify prostate and background the resulting classifier and final segmentation output is accurate, and better than fully supervised methods. However, if the labels are inaccurate then the final segmentation output is also poor. Our algorithm can also be used for other relevant applications where some initial annotations are available. It may require some changes to feature extraction, but the principles of SSL and AL can be used without major changes.
Biography
Biographies for the authors are not available.
References
- 1.American Cancer Society, “Cancer facts and figures 2014,” American Cancer Society, Atlanta, http://www.cancer.org (2014). [Google Scholar]
- 2.Klein S., et al. , “Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information,” Med. Phys. 35(4), 1407–1417 (2008). 10.1118/1.2842076 [DOI] [PubMed] [Google Scholar]
- 3.Martin S., Daanen V., Troccaz J., “Automated segmentation of the prostate 3D MR images using a probabilistic atlas and a spatially constrained deformable model,” Med. Phys. 37(4), 1579–1590 (2010). 10.1118/1.3315367 [DOI] [PubMed] [Google Scholar]
- 4.Pasquier D., et al. , “Automatic segmentation of pelvic structures from magnetic resonance images for prostate cancer radiotherapy,” Int. J. Radiat. Oncol. Biol. Phys. 68(2), 592–600 (2007). 10.1016/j.ijrobp.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Toth R., Madabhushi A., “Multifeature landmark-free active appearance models: application to prostate MRI segmentation,” IEEE Trans. Med. Imaging 31(8), 1638–1650 (2012). 10.1109/TMI.2012.2201498 [DOI] [PubMed] [Google Scholar]
- 6.Chandra S., et al. , “Patient specific prostate segmentation in 3D magnetic resonance images,” IEEE Trans. Med. Imaging 31(10), 1955–1964 (2012). 10.1109/TMI.2012.2211377 [DOI] [PubMed] [Google Scholar]
- 7.Ghose S., et al. , “A random forest based classification approach to prostate segmentation in MRI,” in Proc. MICCAI Prostate Segmentation Challenge, pp. 20–27 (2012). [Google Scholar]
- 8.Mahapatra D., Buhmann J., “Prostate MRI segmentation using learned semantic knowledge and graph cuts,” IEEE Trans. Biomed. Eng. 61(3), 756–764 (2014). 10.1109/TBME.2013.2289306 [DOI] [PubMed] [Google Scholar]
- 9.Liao S., et al. , “Representation learning: a unified deep learning framework for automatic prostate MR segmentation,” in Proc. Medical Image Computing and Computer-Assisted Intervention (MICCAI 2013), Part-II, pp. 254–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birkbeck N., Zhang J., Zhou S., “Region-specific hierarchical segmentation of mr prostate using discriminative learning,” in Proc. MICCAI Prostate Segmentation Challenge, pp. 4–11 (2012). [Google Scholar]
- 11.Gao Q., Rueckert D., Edwards P., “An automatic multi-atlas based prostate segmentation using local appearance-specific atlases and patch-based voxel weighting,” in Proc. MICCAI Prostate Segmentation Challenge, pp. 12–19 (2012). [Google Scholar]
- 12.Litjens G., et al. , “Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge,” Med. Image Anal. 18(2), 359–373 (2014). 10.1016/j.media.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahapatra D., et al. , “Semi-supervised and active learning for automatic segmentation of crohn’s disease,” in Proc. Medical Image Computing and Computer-Assisted Intervention (MICCAI 2013), Part 2, pp. 214–221 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Iglesias J., et al. , “Combining generative and discriminative models for semantic segmentation of ct scans via active learning,” in Information Processing in Medical Imaging, pp. 25–36 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Mahapatra D., et al. , “Active learning based segmentation of crohn’s disease using principles of visual saliency,” in Proc. IEEE 11th Int. Symp. on Biomedical Imaging (ISBI 2014), pp. 226–229 (2014). 10.1109/ISBI.2014.6867850 [DOI] [Google Scholar]
- 16.Mahapatra D., et al. , “Automatic detection and segmentation of crohn’s disease tissues from abdominal MRI,” IEEE Trans. Med. Imaging 32(12), 2332–2348 (2013). 10.1109/TMI.2013.2282124 [DOI] [PubMed] [Google Scholar]
- 17.Petrou M., Kovalev V., Reichenbach J., “Three-dimensional nonlinear invisible boundary detection,” IEEE Trans. Image Process. 15(10), 3020–3032 (2006). 10.1109/TIP.2006.877516 [DOI] [PubMed] [Google Scholar]
- 18.Arridge S., “Geometry of images,” 2015, http://www.cs.ucl.ac.uk/staff/S.Arridge/teaching/ndsp/ (13 July 2015).
- 19.Breiman L., “Random forests,” Mach. Learn. 45(1), 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 20.Chapelle O., Scholkopf B., Zien A., Semi-Supervised Learning, MIT Press, Cambridge, MA: (2006). [Google Scholar]
- 21.Criminisi A., Shotton J., Decision Forests for Computer Vision and Medical Image Analysis, Springer, New York City: (2013). [Google Scholar]
- 22.Cohen M., Dubois R., Zeineh M., “Rapid and effective correction of RF inhomogeneity for high field magnetic resonance imaging,” Human Brain Mapp. 10(4), 204–211 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffari A., et al. , “On-line random forests,” in IEEE 12th Int. Conf. on Computer Vision Workshops (ICCV Workshops 2009), pp. 1393–1400 (2009). 10.1109/ICCVW.2009.5457447 [DOI] [Google Scholar]
- 24.Itti L., Koch C., Niebur E., “A model of saliency based visual attention for rapid scene analysis,” IEEE Trans. Pattern Anal. Mach. Intell. 20(11), 1254–1259 (1998). 10.1109/34.730558 [DOI] [Google Scholar]
- 25.Kadir T., Brady M., “Saliency, scale and image description,” Int. J. Comput. Vision 45(2), 83–105 (2001). 10.1023/A:1012460413855 [DOI] [Google Scholar]
- 26.Gopalakrishnan V., Hu Y., Rajan D., “Random walks on graphs to model saliency in images,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR 2009), pp. 1698–1705 (2009). 10.1109/CVPR.2009.5206767 [DOI] [Google Scholar]
- 27.Harel J., Koch C., Perona P., “Graph based visual saliency,” in Neural Information Processing systems (NIPS 2006), pp. 545–552 (2006). [Google Scholar]
- 28.Aldous D., Fill J., “Reversible markov chains and random walks on graphs,” 2002, http://www.stat.berkeley.edu/~aldous/RWG/book.html, (2014).
- 29.Boykov Y., Veksler O., “Fast approximate energy minimization via graph cuts,” IEEE Trans. Pattern Anal. Mach. Intell. 23, 1222–1239 (2001). 10.1109/34.969114 [DOI] [Google Scholar]
- 30.Litjens G., et al. , “The PROMISE12 segmentation challenge,” 2012 http://promise12.grand-challenge.org/.
- 31.Litjens G., et al. , “The PROMISE12 segmentation challenge,” 2012 http://promise12.grand-challenge.org/Results/Overview.
- 32.Vincent G., Guillard G., Bowes M., “Fully automatic segmentation of the prostate using active appearance models,” in Proc. MICCAI Prostate Segmentation Challenge, pp. 75–81 (2012). [Google Scholar]
- 33.Malmberg F., et al. , “Smart paint a new interactive segmentation method applied to MR prostate segmentation,” in Proc. MICCAI Prostate Segmentation Challenge, pp. 4–11 (2012). [Google Scholar]
- 34.Settles B., Craven M., “An analysis of active learning strategies for sequence labeling tasks,” in Empirical Methods in Natural Language Processing, pp. 1070–1079 (2008). [Google Scholar]
- 35.Lewis D., Catlett J., “Heterogenous uncertainty sampling for supervised learning,” in Proc. of the Eleventh Int. Conf. on Machine Learning, pp. 148–156 (1994). [Google Scholar]
- 36.Freund Y., et al. , “Selective sampling using the query by committee algorithm,” Mach. Learn. 28(2), 133–168 (1997). 10.1023/A:1007330508534 [DOI] [Google Scholar]
- 37.Jager F., et al. , A New Method for MRI Intensity Standardization with Application to Lesion Detection in the Brain: Vision Modeling and Visualization, Aka GmBH, Augsburg, Germany: (2006). [Google Scholar]
- 38.Nyúl L., Udupa J., “On standardizing the MR image intensity scale,” Magn. Reson. Med. 42(6), 1072–1081 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Hellier P., “Consistent intensity correction of MR images,” in Proc. 2003 Int. Conf. on Image Processing (ICIP 2003), pp. 1109–1112 (2003). [Google Scholar]
- 40.Weisenfeld N., Warfield S., “Normalization of joint image-intensity statistics in MRI using the kullback-leibler divergence,” in Proc IEEE Int. Symp. on Biomedical Imaging: Nano to Macro (ISBI 2004), pp. 101–104 (2004). [Google Scholar]
- 41.Li C., et al. , “A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI,” IEEE Trans. Imag. Proc. 20(7), 2007–2016 (2011). 10.1109/TIP.2011.2146190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee S., Acton S., “Region based segmentation in the presence of intensity inhomogeneity using Legendre polynomials,” IEEE Signal Process Lett. 22(3), 298–302 (2015). 10.1109/LSP.2014.2346538 [DOI] [Google Scholar]