Abstract
Cellular oscillators in the uterus play critical roles in the gestation processes of mammals through entraining of the clock proteins to numerous downstream genes, including growth/differentiation factor (Gdf)10 and Gdf15. The expression of Gdf10 and Gdf15 is significantly increased in the uterus during decidualization, but the mechanism underlying the regulation of Gdf gene expression in the uterus is poorly understood. Here, we focused on the function of the cellular oscillators in the expression of Gdf family by using uterine endometrial stromal cells (UESCs) isolated from pregnant Per2‐dLuc transgenic rats. A significant decline of Per2‐dLuc bioluminescence activity was induced in in vitro decidualized UESCs, and concomitantly the expression of canonical clock genes was downregulated. Conversely, the expression of Gdf10 and Gdf15 of the Gdf was upregulated. In UESCs transfected with Bmal1‐specific siRNA, in which Rev‐erbα expression was downregulated, Gdf10 and Gdf15 were upregulated. However, Gdf5, Gdf7, and Gdf11 were not significantly affected by Bmal1 silencing. The expression of Gdf10 and Gdf15 was enhanced after treatment with a REV‐ERB α antagonist in the presence or absence of progesterone. Chromatin immunoprecipitation‐PCR analysis revealed the inhibitory effect of REV‐ERB α on the expression of Gdf10 and Gdf15 in UESCs by recognizing their gene promoters. Collectively, our findings indicate that the attenuation of REV‐ERB α leads to an upregulation of Gdf10 and Gdf15 in decidual cells, in which cellular oscillators are impaired. Our results provide novel evidence regarding the functions of cellular oscillators regulating the expression of downstream genes during the differentiation of UESCs.
Keywords: Circadian clock, decidualization, growth/differentiation factors, REV‐ERBα
Introduction
Growth/differentiation factors (GDFs) are members of the transforming growth factor‐β (TGF‐β) superfamily, and they are involved in a variety of cellular functions and biological processes such as cell proliferation, differentiation, and remodeling (Lee 1991; McPherron et al. 1997; Whitman 1998). The expression of GDFs is involved in embryonic development and the development of female reproductive tissues. The GDFs such as GDF1, GDF3, GDF10, and GDF15 are spatiotemporally expressed in the embryo and uterus. For example, GDF10 is highly expressed in the uterus during the menstrual cycle and pregnancy (Zhao et al. 1999) and plays a role in head formation (Hino et al. 2004). GDF15, which is also known as macrophage inhibitory cytokine‐1 (MIC‐1), is highly expressed in human placenta and is thought to have a predictive aspect for pregnancy outcomes (Lawton et al. 1997; Fairlie et al. 1999; Tong et al. 2004). GDF15 is principally expressed in villous cytotrophoblast cells, extravillous trophoblasts, decidual stromal cells, and placenta (Lawton et al. 1997; Fairlie et al. 1999; Tong et al. 2004). A study using extravillous trophoblast cells demonstrated an inhibitory effect of GDF15 on cell viability by apoptosis and growth inhibition (Morrish et al. 2001). GDF15 also functions as a potent regulator of matrix metalloproteinases, which controls the degradation of the decidual matrix and thus affects the invasion of trophoblast cells (Marjono et al. 2003). However, the mechanism underlying the regulation of Gdf gene expression in the uterus remains poorly understood.
There are many E‐box and ROR/REV‐ERB response elements (ROREs), which are the circadian clock‐controlled cis‐regulatory elements, in the promoter regions of the Gdf genes such as Gdf10 and Gdf15 (NCBI Reference Sequence: NC_005115.4). Numerous peripheral circadian clocks are partially self‐operative and independent in their responses to external and internal stimuli other than the stimuli originating from the suprachiasmatic nucleus, known as the central circadian clock (Hara et al. 2001; Vollmers et al. 2009; Tahara et al. 2012; Wu et al. 2012). The molecular mechanism of the mammalian circadian clock involves a primary conservative interlocked transcriptional‐translational feedback loop (Ko and Takahashi 2006). This loop is comprised of a core group of clock genes and their protein products, which are mostly the transcription factors. The transcriptional activators BMAL1 and CLOCK form a heterodimer, which drives the expression of the Per1‐3 and Cry1‐2 genes by recognizing E‐box cis‐elements in their promoters (Gekakis et al. 1998; Hogenesch et al. 1998; Ueda et al. 2005). The CLOCK‐BMAL1 heterodimer also induces expression of the nuclear receptor, REV‐ERBα, resulting in the repression of the transcription of Bmal1 through direct binding to the RORE located in the Bmal1 promoter (Albrecht and Eichele 2003; Brown et al. 2005). In addition to regulating each other to sustain oscillations, REV‐ERBα also controls the expression of numerous downstream genes through binding to ROREs at their promoters.
BMAL1, a critical component of clock proteins, is indispensable in maintaining the integrity of the circadian feedback loop and the homeostasis of numerous behaviors and physiological processes (Kondratov et al. 2006; Alvarez et al. 2008; Grechez‐Cassiau et al. 2008; Ratajczak et al. 2009). Several studies provided evidence demonstrating that the physiologic significance of BMAL1 is related to mammalian reproductive functions (Ratajczak et al. 2009; Boden et al. 2010; Liu et al. 2014). REV‐ERBα usually functions as a transcriptional repressor for the lack of activation function (AF‐2) domain present at the C‐terminal of the ligand‐binding domain (Yin and Lazar 2005; Phelan et al. 2010; Crumbley and Burris 2011). REV‐ERBα recruits the endogenous nuclear receptor corepressor (N‐CoR)/histone deacetylase3 complex to repress its target gene transcription, thereby regulating a diverse array of cellular processes (Yin and Lazar 2005; Yin et al. 2006). REV‐ERBα was originally regarded as an orphan nuclear receptor (Miyajima et al. 1989), and thereafter heme was identified as its natural ligand (Yin et al. 2007; Meng et al. 2008; Grant et al. 2010; Kojetin et al. 2011). GSK4112 is synthesized as a chemical agonist of REV‐ERBα, and it represses REV‐ERBα target genes (Grant et al. 2010; Chen et al. 2012; Gibbs et al. 2012; Chini et al. 2013). A chemical REV‐ERBα antagonist, SR8278, was reported (Kojetin et al. 2011), and it increases the transcription of REV‐ERBα target genes (Kojetin et al. 2011; Isayama et al. 2015; Tasaki et al. 2015).
Rev‐erbα has a key role in several physiological actions such as adipocyte differentiation, glucose metabolism, and thermogenesis (Chawla and Lazar 1993; Cho et al. 2012; Gerhart‐Hines et al. 2013). The circadian system consisting of clock genes is also disrupted in differentiating cells of rat ovaries and uteri (Alvarez and Sehgal 2005; He et al. 2007a). Several studies have demonstrated that circadian clock genes are rhythmically expressed in the uterus (Dolatshad et al. 2006; He et al. 2007a; Hirata et al. 2009; Akiyama et al. 2010). In rodents and humans, the uterus endometrial stromal cells (UESCs) undergo proliferation and differentiation into decidual cells in response to ovarian steroids and blastocyst implantation at the early stage of pregnancy (Clarke and Sutherland 1990; Zhang et al. 1994; Dey et al. 2004). Decidualization is critical to the establishment of fetal‐maternal communication and the progression of implantation and this process ultimately results in the formation of the placenta. We observed that the Per2 expression in UESCs is downregulated during decidualization, which influences the expression of the vascular endothelial growth factor gene (Uchikawa et al. 2011). Deregulation of the circadian clock may attenuate or disrupt the expression of clock‐controlled genes (CCGs) and can have a profound influence on organ functions. We have shown that the Bmp genes of the Tgf‐β superfamily are regulated by the attenuation of the cellular circadian clock through binding to the RORE regions of their promoters (Tasaki et al. 2015).
In light of these recent reports, we considered the possibility of cellular circadian oscillators in the regulation of the expression of Gdf10 and Gdf15 genes. In this study, we extended our recent investigations to examine the possible involvement of cellular circadian oscillators in the regulation of Gdf gene expression in stromal cells during decidualization. To do so, we used Bmal1‐specific small interfering (siRNA) and the REV‐ERBα antagonist SR8278, which attenuates or disrupts the expression of cellular circadian oscillators and can have a profound influence on the expression of CCGs. The results presented here demonstrate the regulation of REV‐ERBα in the transcription of Gdf10 and Gdf15 due to REV‐ERBα's recognition of the RORE sites of their promoters.
Materials and Methods
Animals
All the experiments were performed under the control of the guidelines for Animal Experiments in the Faculty of Medicine, Kyushu University, and Law No. 105 and Notification No. 6 of the Government of Japan. All procedures were reviewed and approved by the Committee on the Ethics of Animal Experiments of the Kyushu University (Permit No.: A24‐054‐2). Per2‐dLuc transgenic rats were obtained from our breeding colony. In this transgenic rat, the mouse Per2 promoter region, which is sufficient for circadian oscillation, was fused to a dLuc reporter gene (Ueda et al. 2005). The transgenic rats were maintained under a 12:12‐h light–dark cycle (zeitgeber time, ZT0: 0800 light on; ZT12: 2000 light off) and ad libitum feeding throughout all experiments.
Preparation of total RNA from uterine tissues
Uterine tissues were collected at 2000 h (ZT12) on days 4.5 (D4.5) and 6.5 of gestation. Uterine tissues from day 6.5 rats were cut into pieces carefully at interimplantation site and implantation site. Samples from the implantation site contained the whole uterine components and embryos. Pieces of uterine tissues (ca. 20 mg) were homogenized in 1 mL of lysis buffer (Qiagen, Hilden, Germany) with a disposable homogenizer (BioMasher, Funakoshi, Tokyo). During homogenization, the perimetric tissues were removed. Total RNA was isolated using an RNeasy Mini kit (Qiagen) and cDNAs were generated by RT with Oligo (dT)15 and Random Primers according to the manufacture's protocol. The RNA concentration was determined using 260/280‐nm spectrophotometry (Pharmacia Biotech, Buckinghamshire, UK), and RNA integrity was checked by agarose gel electrophoresis.
Preparation and culture of UESCs
The UESCs were isolated from Per2‐dLuc transgenic rats on day 4.50 (D4.50) of gestation as reported previously (He et al. 2007a; Oozono et al. 2008; Matsumoto et al. 2009). The uterine lumens were filled with PBS containing 0.1% collagenase (Invitrogen, Carlsbad, CA) and incubated at 37°C for 1 h in a shaking water bath. The harvested cells were washed thrice with fresh DMEM/F12 (Invitrogen), and seeded onto 35 mm collagen‐coated dishes (Iwaki, Tokyo) at the density of 2 × 105 cells/dish with 2 mL of culture medium [phenol red‐free DMEM/F12 supplemented with 10% charcoal‐treated FBS (Invitrogen) and 1× PS (Nacalai Tesque, Kyoto)]. The culture medium was replaced at 15 min after cell seeding to remove epithelial cells. Cells were cultured in a humidified atmosphere of 5% CO2 at 37°C for 2 days. Then, cells were cultured in serum‐free medium supplemented with 1× antibiotic–antimycotic (AA; Nacalai Tesque), 1× Insulin‐Transferrin‐Selenium (ITS, Life Technologies, Grand Island, NY), 0.1% bovine serum albumin (BSA, Sigma‐Aldrich, St Lousis, MO), and 100 nmol/L progesterone (P4, Sigma‐Aldrich) for additional 2 days prior to other treatments.
In vitro decidualization
Confluent UESCs were further cultured for 5 days in DMEM/F12 supplemented with 0.1 mmol/L medroxyprogesterone acetate (MPA, Sigma‐Aldrich), 0.5 mmol/L 2‐O‐dibutyryl cAMP (db‐cAMP, Sigma‐Aldrich), 1× AA, 1× ITS, 0.1% BSA, as previously described (Matsumoto et al. 2009). The differentiating status was revealed by the expression of the Prl8a2 gene (Jabbour and Critchley 2001).
Real‐time monitoring of Per2‐dLuc oscillations
The cultured UESCs were synchronized with 100 nmol/L dexamethasone (DXM, Sigma‐Aldrich) for 2 h in the serum‐free medium containing 1× AA. Then, cells were given the serum‐free medium DMEM/F12 supplemented with 0.1 mmol/L luciferin (Wako, Tokyo), 0.1% BSA, 1× AA, and 1× ITS, and subjected to luminescence determination. Luciferase activity was monitored at 37°C with a Kronos Dio AB‐2550 luminometer (ATTO, Tokyo) interfaced to a computer for continuous data acquisition (He et al. 2007b; Hirata et al. 2009). In some experiments, confluent UESCs were synchronized with DXM, and then monitoring was performed in the presence of 10 μmol/L SR8278 (Sigma‐Aldrich) or 0.1% DMSO (vehicle control). The data are presented as photon counts per min. Bioluminescence data were detrended by subtracting the 24‐h running average from the raw data. Detrended datasets were smoothed by taking 2‐h running averages. The amplitude and period of Per2‐dLuc oscillations were documented by the single Cosinor method using Timing Series Single 6.3 (Expert Soft Tech., Richelieu, France).
Microarray analysis
RNA samples isolated from cultured UESCs at 30, 36, 42, and 48 h after synchronization with DXM were used for microarray analysis using the Whole Rat Genome Microarray 4x44K Ver3.0 (Agilent Technologies, Santa Clara, CA) representing 30,367 probe sets. Bioinformatics analysis was performed using Agilent Future Extraction software (Agilent Technologies). The data were filtered for signal intensity values (P < 0.05, detectable), which allowed removing very low signal values (Tasaki et al. 2013). The ratio of signal intensity values was calculated for the Gdf gene family.
Bmal1 siRNA transfection
RNA oligos targeting the Bmal1 mRNA (F: GAAUGUCACAGGCAAGUUUdTdT; R: AAACUUGCCUGUGUGACAUUCdTdT) and nonsilencing RNA (mission_SIG‐001) were purchased from Sigma‐Aldrich. Cultured UESCs were first plated in 35‐mm collagen‐coated dishes with 2 mL DMEM/F12 supplemented with 1× AA, 1× ITS, 0.1% BSA. The medium was removed after 24 h, and the cells were transfected with the RNA oligos diluted in Opti‐MEM using Lipofectamine® RNAiMAX reagent (Life Technologies) according to the manufacturer's protocol (Chen et al. 2013a). Both the Bmal1‐specific siRNA and nonsilencing RNA were used at a final concentration of 25 nmol/L. The cells were maintained with transfection medium for duration of 12 h. Then the medium was replaced with DMEM/F12 supplemented with 1× AA, 1× ITS, 0.1% BSA, and 100 nmol/L P4. After 48 h in culture, cells were synchronized with DXM for monitoring of luciferase activity.
RNA extraction and RT‐PCR
Uterus tissues were collected from pregnant rats on D4.50 (ZT4) of gestation. The pieces of uterine tissues (0.1 g) were homogenized in 1 ml of Sepasol‐RNA I Super (Nacalai Tesque) for 2 min (Uchikawa et al. 2011), and total RNA was isolated using an RNeasy Mini kit (Qiagen) according to the manufacturer's protocol. RNA samples were treated with RNase‐free DNase (Qiagen). The cDNAs were generated by RT with Oligo (dT)15 and Random Primers using a GoTaq® 2‐Step RT‐qPCR System (Promega). The PCR reaction was performed in 10 μL of 1× PCR buffer, 0.2 mmol/L each of dNTP, 0.25 U AmpliTaq Gold (Applied Biosystems), 0.2 μmol/L each of the synthetic primer sets (Table 1), and 20 ng cDNA. The amplification was performed in 42 cycles and resulting PCR products were analyzed by electrophoresis on 1.8% agarose gels.
Table 1.
Primer sequences for the targeted genes in real‐time qPCR and RT‐PCR
| Gene | Accession No. | Sequence 5′–3′ | Amplicon (bp) |
|---|---|---|---|
| Bmal1 | NC_005100 | F: CCGTGGACCAAGGAAGTAGA | 97 |
| R: CTGTGAGCTGTGGGAAGGTT | |||
| Rev‐erbα | NM_031134 | F: ACAGCTGACACCACCCAGATC | 102 |
| R: CATGGGCATAGGTGAAGATTTCT | |||
| Per2 | NM_031678 | F: GACGGGTCGAGCAAAGGA | 90 |
| R: CCCTTTTCAGGTGTATAGGTAAGT | |||
| Dbp | NM_012543 | F: GCAAGGAAAGTCCAGGTGCCCG | 95 |
| R: GCGTCTCTCGACCTCTTGGCT | |||
| Gdf5 | XM_001066344 | F: ATCTTTAGGCCAGGGGGTCA | 143 |
| R: GGTCCTGGCTTGGTTTCAGA | |||
| Gdf7 | NM_001170350 | F: TCACAGACCAAGCAACTGAAG | 98 |
| R: ATTCACCACCTCGTGGGAG | |||
| Gdf10 | NM_024375 | F: CCTACTACTGTGCTGGAGCC | 75 |
| R: TCTGGATGGTGGCATGGTTG | |||
| Gdf11 | XM_343148 | F: GGGCAAGAGGGCTAACACAT | 102 |
| R: TCTGAACTGCTTCCGTGAAC | |||
| Gdf15 | NM_019216 | F: CCAGCTGTCCGGATACTCAG | 106 |
| R: GGTAGGCTTCGGGGAGACC | |||
| Prl8a2 | NM_017008 | F: ATCCAGCGAGCTGAAGTCAT | 178 |
| R: CATGAAGTGGTGGGTTTGTG | |||
| Gapdh | NM_017008 | F: ATGGCCTTCCGTGTTCCTAC | 122 |
| R: CTTTACAAAGTTGTCGTTGA |
RT‐qPCR
Cultured cells were harvested at indicated time points. Total RNA was isolated using an RNeasy Mini kit (Qiagen) and cDNAs were generated by RT with Oligo (dT)15 and Random Primers as described above. RT‐qPCR was performed in a 50‐μL volume containing a 20‐ng cDNA sample in GoTaq® qPCR Master Mix and 250 nmol/L specific primers listed in Table 1, with the Mx3000P Real‐time qPCR System (Agilent Technologies, Santa Clara, CA) using the parameters as described in our previous report (Chen et al. 2013b). All reactions were performed in triplicate and displayed amplification efficiency between 80% and 120%. Relative quantification of each mRNA was performed using the comparative quantity (copies) method creating standard curves. The quantity for each sample was normalized to Gapdh.
Chromatin immunoprecipitation (ChIP assay)
Confluent UESCs were synchronized with DXM and then harvested at 48 h and 3.0 × 106 cells were used per one IP reaction. ChIP assay was performed by using SimpleChIP Plus enzymatic chromatin IP kit (Cell Signaling Technology, Beverly, MA), as instructed by the manufacturer's protocol. Briefly, cells were fixed with 1.0% formaldehyde for 10 min to cross‐link proteins to DNA, and the cross‐linking was stopped with glycine. The extracted genomic DNA was digested with 0.5 μL micrococcal nuclease for 20 min. After centrifugation at 16,500 g for 1 min, the nuclear pellet was suspended in the ChIP buffer containing protease inhibitors and lysed with sonication (3 pulses, 20 sec). After determining DNA concentration, the cross‐linked chromatin (2 μg) was incubated overnight at 4°C with an REV‐ERBα antibody (5 μg/500 μL; Cell Signaling Technology) as the positive control and normal rabbit IgG (1 μg/500 μL) as the negative control and incubated with Protein G agarose beads for additional 2 h. The chromatin was eluted from the agarose beads by incubating for 30 min at 65°C and the cross‐links were reversed by incubating with 5 mol/L NaCl (6 μL/150 μL) and proteinase K (2 μL/150 μL) for 2 h at 65°C. Purified DNA was amplified by PCR with specific primer sets as follows: Gdf10: forward 5′‐GCTTGCACAGATTGCTTCTTGT‐3′ (‐2702/‐2681) and reverse 5′‐GCCTCATTTTACTGCCGAACAG‐3′ (‐2526/‐2505): (NC_005115.4); Gdf15: forward 5′‐TCTACAGGAGGAGGGGGACTA‐3′ (‐1163/‐1143) and reverse 5′‐GCCAGGTAGGTGCATGGTAAG‐3′ (‐1002/‐982) (NC_005115.4). The amplification was performed in 35 cycles and PCR products were analyzed by electrophoresis on 1.8% agarose gels.
Statistical analyses
All data are expressed as means ± S.E.M. of at least three separate experiments, each performed with duplicate samples. The amplitude of Per2‐dLuc was determined by the single Cosinor method using Timing Series Single 6.3 (Expert Soft Tech.). The statistical differences in examined values were determined by one‐way or two‐way ANOVA followed by a Bonferroni's post hoc test using SigmaPlot software (Ver. 11.2; Systat Software, San Jose, CA). Differences were considered significant at P < 0.05 or less.
Results
Expression levels of the Gdf genes in cultured UESCs by microassay
To gain insight into the cellular clocks of UESCs and the expression of the Gdf genes, we analyzed the global expression of the Gdf genes by performing DNA microarrays (Tasaki et al. 2013). The expression levels of Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15, designated as core Gdf genes, were relatively high compared to those of Gdf2, Gdf3, Gdf6, and Gdf9 (Fig. 1A). Of the core Gdf genes, the alterations of Gdf5, Gdf10, Gdf11, and Gdf15 transcript levels were significant (P < 0.05). We also confirmed the detection of the transcripts of core Gdf genes in UESCs by RT‐PCR (Fig. 1B). The core Gdf genes were analyzed in the following experiments.
Figure 1.
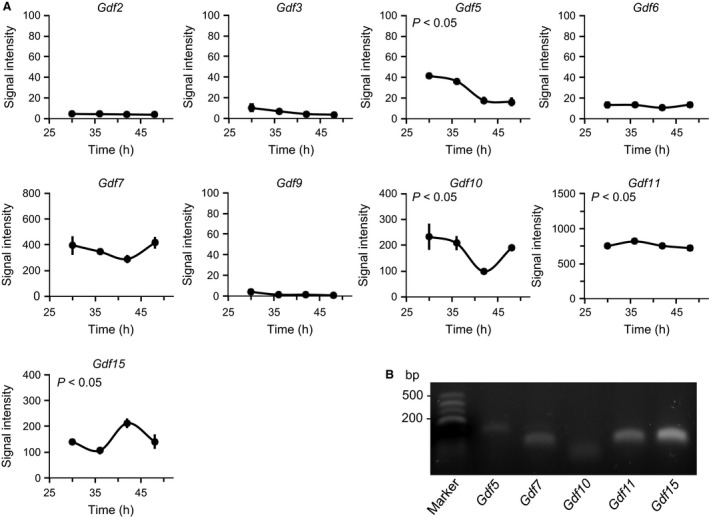
Expression profiles of the Gdf genes in UESCs isolated from pregnant rats as determined by DNA microarray and RT‐PCR analyses of core Gdf genes. (A) According to the indicated protocols, RNA samples isolated from cultured UESCs and subjected to microarray analysis. Each value represents the means ± S.E. (n = 3) of signal intensity from the microarray results. (B) Total RNA was isolated from UESCs and subjected to RT‐PCR for core Gdf genes (Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15) with specific primer sets.
Expression levels of core Gdf genes in whole‐uterus tissues during decidualization
To further analyze the expression of core Gdf genes in the early pregnancy period, we measured the expression levels of Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15 in the whole uterus of pregnant rats on D4.5, defined as the implantation period, by RT‐PCR. As shown in Figure 2A, the transcripts of all core Gdf genes were detected. To investigate the temporal changes in Gdf genes during the implantation and decidualization periods, we analyzed the transcript levels of Gdf genes in the rat uterus on D4.5, at interimplantation sites (D6.5), and at implantation sites on day 6.5 (D6.5E). As shown in Figure 2B, the transcript level of Gdf10 was significantly increased on D6.5 compared to that on D4.5 (P < 0.05). The transcript levels of Gdf10 and Gdf15 were significantly higher at implantation sites on D6.5E than those on D4.5 (P < 0.05). Conversely, the expression levels of Gdf5, Gdf7, and Gdf11 in the pregnant rat uteri were not significantly altered during the implantation or decidualization periods (Fig. 2B).
Figure 2.
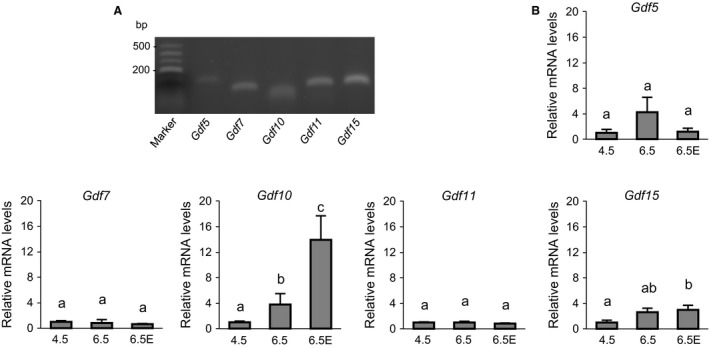
Expression levels of core Gdf genes in pregnant rat uterus during implantation and decidualization. (A) Total RNA samples were collected from the uterus of pregnant rats on pregnant day 4.5 and subjected to RT‐PCR for the expression levels of core Gdf genes. (B) Total RNA samples were collected from the uterus of pregnant rats on pregnant day 4.5 (implantation) or ZT12 (D4.5) and D6.5 (decidualization) or ZT12 (D6.5) and subjected to RT‐qPCR. The D6.5 samples were divided into implantation sites (D6.5E) and inter‐implantation sites (D6.5). The transcript levels were calculated and normalized to each value given by the D4.5 sample. Values with different letters are significantly different (P < 0.05).
Downregulation of Per2‐dLuc oscillation and canonical clock gene expression in decidual cells
To further investigate whether decidualization induces changes in the cellular clockwork, we used MPA and db‐cAMP to induce the in vitro decidualization of UESCs prepared from pregnant rats on D4.5. After the control and decidualized UESCs were synchronized with 100 nmol/L DXM for 2 h, luciferase activity was chronologically monitored. Both cell groups generated several Per2‐dLuc oscillations, but a significant attenuation of Per2‐dLuc bioluminescence was observed in the decidual cells (P = 0.0054, vs. CONT) (Fig. 3A). A significant increase in the transcript level of Prl8a2, a decidualization marker, was observed in the treated cells (two‐way ANOVA, P < 0.001). Circadian rhythms of Bmal1, Per2, Rev‐erbα, and Dbp were not detected in the decidual cells, in which the transcript levels of these genes were significantly reduced compared to the levels in the control cells (two‐way ANOVA, P < 0.01). Thus, the expression of canonical clock genes was attenuated in decidual cells.
Figure 3.
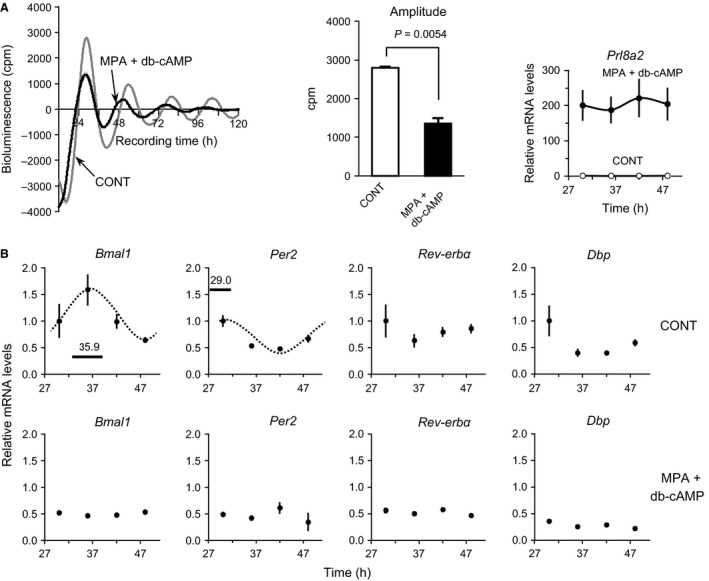
Per2 oscillation profiles and expression levels of core clock genes in UESCs during in vitro decidualization. (A) UESCs were isolated from the uterine horns on day 4.5 of pregnancy and cultured for 3 days prior to any treatment. Decidual cells were obtained by treating confluent UESCs for 4.5 days with 0.1 mmol/L MPA plus 0.5 mmol/L db‐cAMP. Both types of cells were synchronized with DXM for 2 h prior to monitoring. The amplitude of Per2‐dLuc oscillations was determined by the single Cosinor method. The mRNA level of Prl8a2, a marker for UESCs decidualization, was analyzed in the control and decidual cells by RT‐qPCR. (B) According to the first Per2‐dLuc phase in panel A, total RNA samples were collected from control and decidual cells at the indicated times after synchronization and RT‐qPCR analyses of transcript levels were performed. The transcript levels were calculated and normalized to each value given by the control sample at 30 h. The Cosinor analysis method was used to determine the rhythmic expression of examined genes. Statistical rhythmicity (dotted curves, P < 0.05) and peak time with 95% confidential intervals (line length) are shown in each panel.
Expression levels of core Gdf genes in decidual cells
We analyzed the expression of Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15 in decidual cells induced by MPA and db‐cAMP. The transcripts of Gdf10 and Gdf15 were increased in the decidual cells by more than 30‐fold and fourfold, respectively (two‐way ANOVA, P < 0.001, Fig. 4). Conversely, the transcript levels of Gdf5, Gdf7, and Gdf11 were not significantly altered in the decidual cells.
Figure 4.
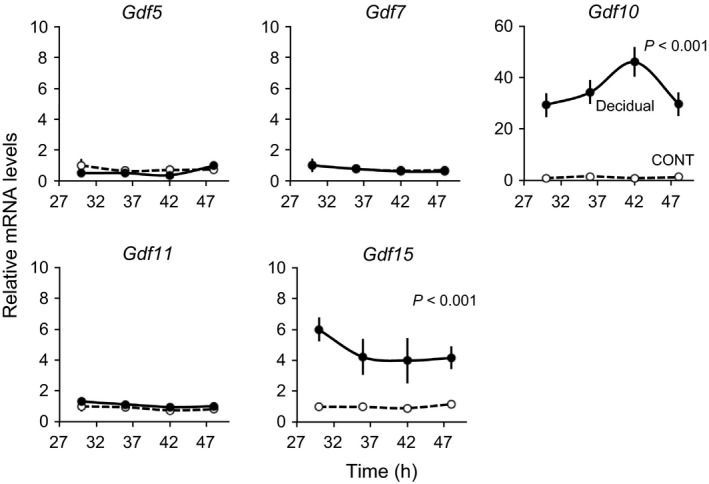
Expression profiles of core Gdf genes in decidual cells. According to the first Per2‐dLuc phase shown in Figure 3A, total RNA samples were collected from control (dot line) and decidual (solid line) cells at the indicated times after synchronization. RT‐qPCR analyses of transcript levels were performed using their specific primers for Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15. The transcript levels were calculated and normalized to each value given by the control sample at 30 h.
Effects of Bmal1 siRNA treatment on the expression of canonical clock genes and Gdf genes in UESCs
To analyze whether the cellular circadian clock in UESCs contributes to the expression of Gdf genes, we transfected Bmal1‐specific siRNA (siRNA) into UESCs. Given the critical role of Bmal1 in the sustaining of the cellular circadian rhythm, we investigated Per2‐dLuc oscillations using UESCs transfected with siRNA or nonsilencing RNA (CONT). Both cell groups generated several Per2‐dLuc oscillations (Fig. 5A). However, the amplitude of Per2‐dLuc bioluminescence was significantly reduced in the siRNA‐treated cells (P = 0.0022). The circadian rhythm of Bmal1 was not detected after the siRNA transfection (Fig. 5B). Concomitantly, the circadian rhythms at the transcript levels of Per2, Rev‐erbα, and Dbp, which are downstream genes of Bmal1, were attenuated in the siRNA‐treated cells. The transcripts of Gdf10 and Gdf15 were significantly increased in the siRNA‐treated cells (two‐way ANOVA, P < 0.001 and P < 0.05, respectively) (Fig. 6). No significant alterations in Gdf5, Gdf7, and Gdf11 transcript levels were observed in the siRNA‐treated cells. Thus, the expression of Gdf10 and Gdf15, but not those of Gdf5, Gdf7, and Gdf11, was upregulated by attenuation of the cellular circadian clocks.
Figure 5.
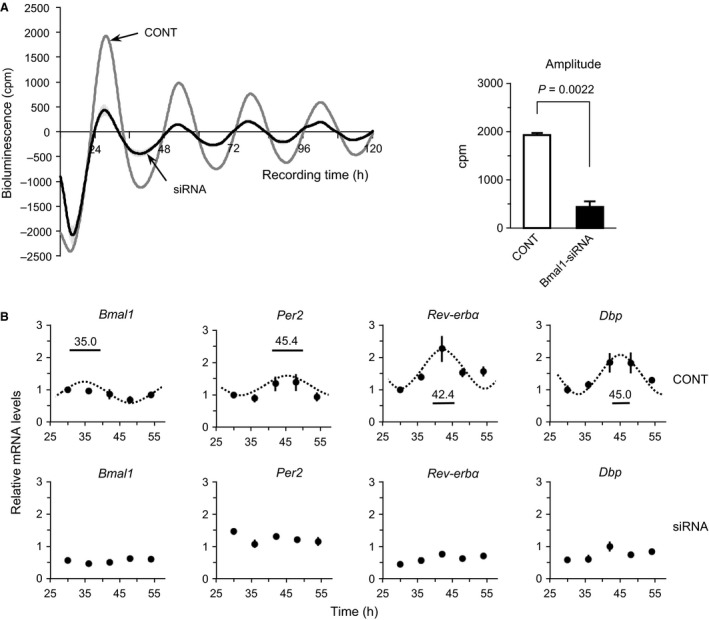
Effect of Bmal1‐specific siRNA treatment on Per2‐dLuc oscillations in UESCs. (A) Representative records of bioluminescence showing the effect of Bmal1 interference on the rhythmic expression of Per2‐dLuc oscillations in UESCs. UESCs were treated with Bmal1‐specific siRNA (Bmal1‐siRNA) or nonsilencing RNA (CONT) according to the indicated protocols. The cells were then synchronized with DXM for bioluminescence determination (time: 0 h). Amplitude of Per2‐dLuc oscillation in UESCs was estimated with or without Bmal1 siRNA treatment. (B) According to the first Per2‐dLuc phase (solid line) in panel A, total RNA samples were collected from control and siRNA‐treated cells at the indicated times after synchronization. RT‐qPCR analyses of transcript levels were performed using their specific primers. The transcript levels were calculated and normalized to each value given by the control sample at 30 h. The Cosinor analysis method was used to determine the rhythmic expression of examined genes. Statistical rhythmicity (dotted curves, P < 0.05) and peak time with 95% confidential intervals (line length) are shown in each panel.
Figure 6.
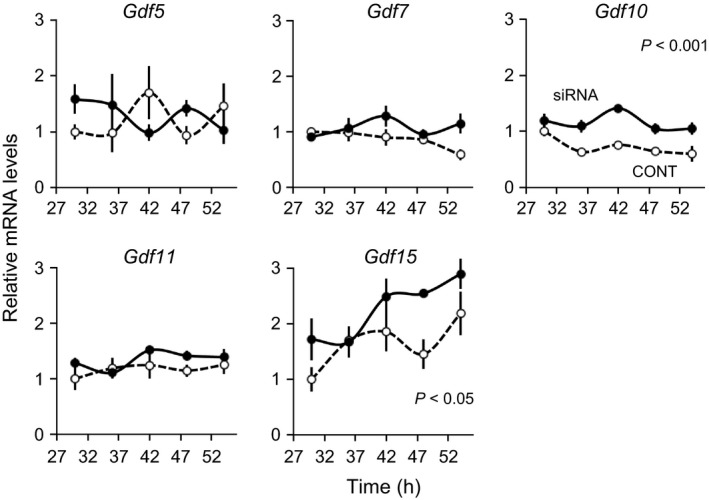
Expression profiles of core Gdf genes in UESCs transfected with Bmal1‐specific siRNA or nonsilencing RNA. According to the first Per2‐dLuc phase shown in Figure 5A, total RNA samples were collected from control (dot line) and decidual (solid line) cells at the indicated times after synchronization. RT‐qPCR analyses of transcript levels were performed using their specific primers for Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15. The transcript levels were calculated and normalized to each value given by the control sample at 30 h.
Effect of SR8278 treatment on the expression of Gdf10 and Gdf15 in UESCs
To further understand the physiological function of the UESC circadian clock and to detect whether Gdf10 and Gdf15 are controlled under the regulation of REV‐ERBα, we treated UESCs with the REV‐ERBα antagonist SR8278. A significant decline of Per2‐dLuc bioluminescence oscillation amplitude was observed in the SR8278‐treated cells (P = 0.0194, vs. CONT) (Fig. 7A). However, the circadian rhythms of the canonical clock genes were not significantly altered by the SR8278 treatment (Fig. 7B). We observed that the transcripts of Gdf10 and Gdf15 were significantly increased in the SR8278‐treated cells (P < 0.001, vs. CONT) (Fig. 8). Conversely, the transcript levels of Gdf5, Gdf7, and Gdf11 were not upregulated by SR8278 treatment. Regardless of the presence or absence of P4, the SR8278 treatment caused significant increases in the expression levels of Gdf10 and Gdf15.
Figure 7.
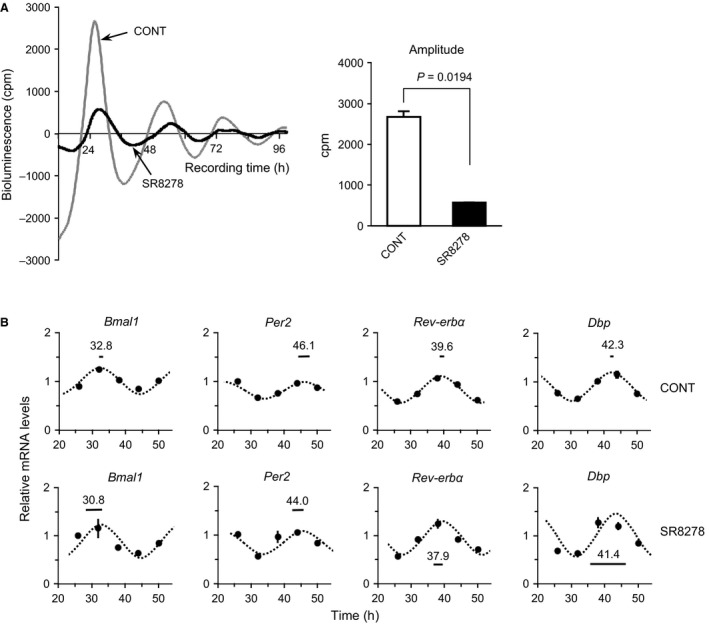
Effect of the REV‐ERB α antagonist SR8278 treatment on Per2‐dLuc oscillations and clock genes expression in UESCs. (A) Representative records of bioluminescence showing the effect of SR8278 on the rhythmic expression of Per2‐dLuc oscillations in UESCs. Cultured UESCs were washed twice with serum‐free medium and subjected to Per2‐dLuc bioluminescence determination with 10 μmol/L SR8278 or 0.1% DMSO (vehicle control) (time: 0 h). Amplitude of Per2‐dLuc oscillation was estimated in the cells with or without SR8278 treatment. (B) According to the first Per2‐dLuc phase, total RNA samples were collected from control and SR8278‐treated cells at the indicated times after synchronization. RT‐qPCR analyses of transcript levels were performed using their specific primers. The transcript levels were calculated and normalized to each value given by the control sample at 26 h. The Cosinor analysis method was used to determine the rhythmic expression of examined genes. Statistical rhythmicity (dotted curves, P < 0.05) and peak time with 95% confidential intervals (line length) are shown in each panel.
Figure 8.
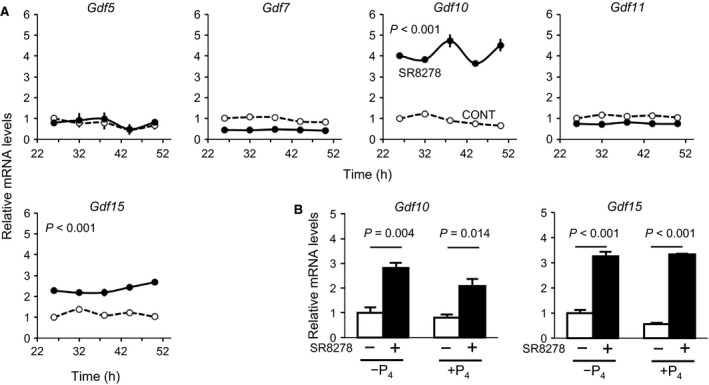
Expression profiles of core Gdf genes in UESCs treated with the REV‐ERB α antagonist SR8278. (A) According to the first Per2‐dLuc phase shown in Figure 7A, total RNA samples were collected from control and SR8278‐treated cells at the indicated times after synchronization. RT‐qPCR analyses of transcript levels were performed using their specific primers. Effect of SR8278 treatment on the expression of Gdf10 and Gdf15 was also performed in the presence or absence of P4. The transcript levels were calculated and normalized to each value given by the control sample at 26 h.
REV‐ERBα binding to the putative RORE sites in the upstream regions from transcriptional start sites of Gdf10 and Gdf15
To further investigate whether REV‐ERBα exerts an inhibitory effect on the expression of Gdf10 and Gdf15 through direct binding to the RORE sites, we performed a ChIP‐PCR analysis. We searched putative RORE sites within the 3000‐bp upstream regions from the transcriptional start sites of Gdf10 and Gdf15. Many putative RORE sites are located in these regions. Of these putative RORE sites, we selected several sites with a mass and short distance (less than 200 bp) at distal or proximal regions (Fig. 9A). We prepared the protein/DNA cross‐linked chromatin and immunoprecipitated it with an anti‐REV‐ERBα antibody and normal rabbit IgG (negative control). The results of this ChIP‐PCR analysis revealed specific bands detected in both the Gdf10 and Gdf15 samples precipitated with the antibody (Fig. 9B). These results indicated that REV‐ERBα directly bound to at least GTGACC−2634/−2629, CCCAGT−2631/−2626, and TCCAGT−2553/−2548 in the Gdf10 promoter and GGGTCA−1078/−1073 and TCCAGT−1052/−1047 in the Gdf15 promoter.
Figure 9.
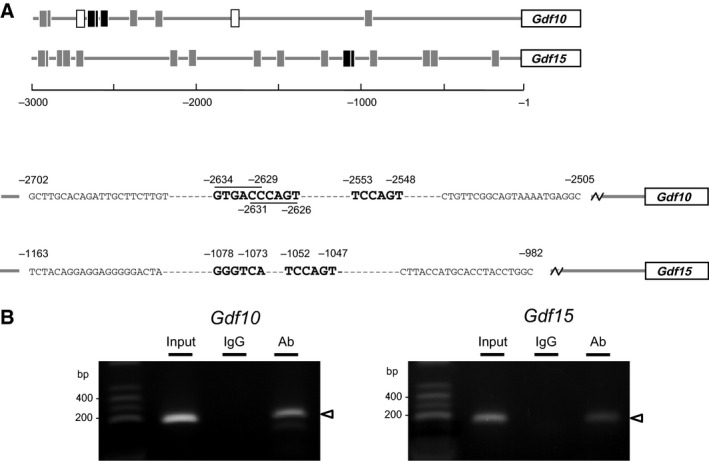
ChIP‐PCR analysis of the putative RORE sites in upstream regions from transcriptional start sites of Gdf10 and Gdf15. (A) The putative RORE (black and gray squares) and E‐box sites (open squares) located in upstream regions from transcriptional start sites of Gdf10 and Gdf15 are listed, and distal and proximal sites were analyzed, respectively (black squares). Three and two putative RORE sites (gothic letters), GTGACC −2634/−2629, CCCAGT −2631/−2626, and TCCAGT −2553/−2548 for Gdf10 and GGGTCA −1078/−1073 and TCCAGT −1052/−1047 for Gdf15, located in upstream regions from transcriptional start sites of Gdf10 and Gdf15 were targeted, respectively. Purified DNA was amplified by PCR with specific primer sequences (small letters) (B) Confluent UESCs (3.8 × 106 cells) were synchronized with DXM, and then harvested at 48 h and fixed with formaldehyde to cross‐link proteins to DNA as described in Materials and Methods. The extracted genome DNA was digested with Micrococcal Nuclease into fragments. After DNA purification, the cross‐linked chromatin (2 μg) was used for immunoprecipitation using REV‐ERB α antibody and normal rabbit IgG (negative control). Purified DNA was amplified with specific primer sets for Gdf10 and Gdf15. Input, input sample; IgG, negative control; Ab, anti‐REV‐ERB α antibody.
Discussion
Circadian oscillators are expressed in various peripheral tissues including the ovary and uterus and may play critical roles in the regulation of reproductive physiological processes (Dolatshad et al. 2006; He et al. 2007a; Hirata et al. 2009; Akiyama et al. 2010; Uchikawa et al. 2011). To address these possible roles, we focused on the function of the circadian oscillators in the expression of Gdf family members by using rat UESCs. The results indicate that a few Gdf family members are repressed by the binding of REV‐ERBα, a canonical clock component, to the RORE sites located at Gdf putative promoters. We found that the expression levels of Gdf10 and Gdf15 were significantly increased in the embryo implantation sites of the pregnant D6.5 rat uterus compared to that at pregnant D4.5. We and other research group also observed that the canonical clock genes Bmal1, Rev‐erbα, and Per2 were downregulated in UESCs during the in vitro decidualization (Isayama et al. 2015; Muter et al. 2015; Tasaki et al. 2015).
These results are consistent with our previous observations of Per2 protein levels using whole‐uterus tissues (Uchikawa et al. 2011). The expression levels of Gdf10 and Gdf15 were significantly increased in the decidual cells, and the knockdown of Bmal1‐specific mRNA induced significant increases in the expression levels of Gdf10 and Gdf15. We also found that Gdf10 and Gdf15 were upregulated by the inhibition of REV‐ERBα function by using its antagonist SR8278. However, the expression of Gdf10 and Gdf15 was not altered regardless of the presence or absence of P4, suggesting that REV‐ERBα does not coordinate with P4 in the expression of these genes. After validating the RORE sites, we assessed the inhibitory effect of REV‐ERBα on the expression of Gdf10 and Gdf15 in the uterus prior to embryo implantation.
Previous reports implicated the Gdf genes in the regulation of prenatal uterine development (Lawton et al. 1997; Zhao et al. 1999; Tong et al. 2004), and Gdf family members were demonstrated to play critical roles in the regulation of oocyte development (Gui and Joyce 2005). Analyses using DNA microarrays, RT‐qPCR, and RT‐PCR revealed the expression of Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15 in the UESCs of pregnant D4.5 rats (Figs. 1, 2). Of these core Gdf genes, it is noted that in this study the transcripts of Gdf10 and Gdf15 were also increased in the uterus on D6.5, in particular at the implantation site (D6.5E), compared to D4.5, suggesting roles of Gdf10 and Gdf15 in embryo implantation (Fig. 2). These results may support the contention that Gdf10 and Gdf15 are critical for the formation of placenta (Zhao et al.1999; Moore et al. 2000; Segerer et al. 2012).
In rodents and humans, uterine receptivity implies a dialog between UESCs and free‐floating blastocysts. The UESCs undergo proliferation and differentiation into decidual cells, and the placenta is ultimately formed. In this study, the decidual cells induced by the in vitro treatment with MPA and db‐cAMP displayed attenuation of the rhythmic expression of canonical clock genes as well as the generation of Per2‐dLuc oscillations (Fig. 3). Consistently, our results demonstrated that the canonical clock genes were significantly downregulated in the decidual cells and lost rhythmic expression. The underlying mechanism of attenuated clockwork in decidual cells is not yet clear. One possibility is that regular interaction of clock proteins and chromosomal DNA is disrupted or impaired at the level of gene transcription due to the requirement of differentiation‐specific gene transcription. Especially, BMAL1 is indispensable in maintaining the integrity of the circadian feedback loop and the homeostasis of numerous physiological states (Rudic et al. 2004; Shimba et al. 2005). Recent our studies also indicate the downregulation of Bmal1 in in vivo and in vitro experiments (Isayama et al. 2015; Tasaki et al. 2015). However, differentiation‐inducing factor(s) in UESCs are not identified yet.
However, the relationship between the circadian clockwork and the Gdf gene expression during the differentiation of the UESCs into the decidual cells remains largely undefined. We suggested that some of the Gdf genes are under the direct regulation of uterine circadian oscillators. To investigate this hypothesis, we examined highly expressed Gdf genes such as Gdf5, Gdf7, Gdf10, Gdf11, and Gdf15 from the DNA microarray (Fig. 1) and measured the transcript levels of these core Gdf genes in the UESCs of pregnant D4.5 rats. The results indicate that the Gdf genes are differentially regulated in the UESCs.
It is well established that Bmal1 is indispensable in sustaining the transcriptional‐translational feedback loop. In this study using Bmal1‐specific siRNA, the expression of the core clock genes Per2, Rev‐erbα, and Dbp was significantly reduced, as was the amplitude of Per2‐dLuc oscillations (Fig. 5). These results are similar to those of an earlier investigation in which the levels of Per1, Per2, Rev‐erbα, and Dbp mRNA were low in Bmal1 null mice (Bunger et al. 2000; Boden et al. 2010). Our findings indicate that the expression of clock genes is impaired in Bmal1 siRNA‐treated UESCs, providing evidence for further studies of CCGs in the uterus.
Interestingly, the Bmal1 silencing induced significant increases in Gdf10 and Gdf15 mRNA levels, but it did not affect the expression of Gdf5, Gdf7, or Gdf11 (Fig. 6). Because of the presence of clock‐controlled cis‐regulatory elements within their promoters and their rhythmic expression, the present results suggest that these genes are directly under the regulation of canonical clock genes.
REV‐ERBα is an important nuclear hormone receptor (NHR) in the regulation of cell physiology. Substantial evidence has shown that REV‐ERBα is a circadian clock component in the maintenance of circadian rhythms (Preitner et al. 2002). SR8278, a synthetic antagonist of REV‐ERBα, can inhibit the activity of REV‐ERBα to increase the expression of its target genes (Kojetin et al. 2011; De et al. 2015). In this study, we examined the expression of Gdf10 and Gdf15 in cultured UESCs using the antagonist SR8278. The addition of SR8278 increased the Gdf10 and Gdf15 transcript levels due to the decreased activity of REV‐ERBα (Fig. 8). REV‐ERBα is recruited to the target gene promoters by its binding to the agonist heme (Raghuram et al. 2007). In the presence of SR8278, this increase in Gdf10 and Gdf15 transcript levels may be the result of a competitive inhibition of REV‐ERBα binding to heme. Intriguingly, we observed that the increased transcriptions of Gdf10 and Gdf15 in cultured UESCs treated with SR8278 were not affected in the presence of P4 (Fig. 6). Thus, P4 is not related to the expression of Gdf10 or Gdf15 in UESCs. Interestingly, Gdf5, Gdf7, and Gdf11 did not display upregulation after SR8278 treatment, similar to those observed in the pregnant uteri during the implantation or decidualization periods. These results conclude that the expression of Gdf5, Gdf7, and Gdf11 is independent of circadian clockwork in UESCs.
The present results might indicate that REV‐ERBα has an inhibitory effect on the regulation of Gdf10 and Gdf15 expression in UESCs of pregnant D4.5 rats. It is interesting to note that a mass of RORE [5′‐(A/G)GGTCA‐3′ or 5′‐TGACC(C/T)‐3′] sites exist at the 5′‐upstream region of the Gdf10 and Gdf15 genes (−2939/−2215 for Gdf10, and −1240/−922 and −2959/−2707 for Gdf15), whereas there are few or no canonical E‐box (5′‐CACGTG‐3′ or 5′‐CACGTT‐3′) sites. In light of our observation of highly expressed Gdf10 and Gdf15 genes in cultured UESCs treated with the REV‐ERBα antagonist, we further analyzed the binding of REV‐ERBα to their upstream regions from their transcriptional start sites. The ChIP assay results indicated that REV‐ERBα could bind to the RORE sites on the putative promoters of Gdf10 and Gdf15 (Fig. 9). We therefore suggest that the expression of Gdf10 and Gdf15 genes may be directly under the regulation of REV‐ERBα as a repressor by recognizing the RORE sites of the Gdf10 and Gdf15 promoters. Both Gdf10 and Gdf15 displayed low expression levels in the UESCs of the pregnant D4.5 rats.
Conversely, the expression levels of Gdf10 and Gdf15 were significantly increased in the decidual cells. Taken together with these findings, we propose that the clock oscillator silencing may contribute to the upregulation of the Gdf gene expression in decidual cells. Thus, REV‐ERBα is recruited to the Gdf gene promoters in the cultured UESCs and is directly bound to the RORE sites in the Gdf gene promoters. Consequently, REV‐ERBα acts as a transcriptional silencer to repress the expression of Gdf10 and Gdf15 in UESCs.
In conclusion, our experiments demonstrated that the cellular circadian oscillators of rat UESCs regulate the transcription of Gdf10 and Gdf15 genes. REV‐ERBα can repress the expression of Gdf10 and Gdf15 genes by recognizing the RORE sites of the target gene promoters. The attenuation of REV‐ERBα may lead to an upregulation of Gdf genes in decidual cells, in which cellular oscillators are impaired. This study revealed novel evidence regarding the physiology functions of circadian oscillators regulating the expression of downstream genes during the differentiation of UESCs.
Conflict of Interest
No conflicts of interest, financial or otherwise, are declared by the authors.
Zhao L., Isayama K., Chen H., Yamauchi N., Shigeyoshi Y., Hashimoto S., Hattori M.‐A.. The nuclear receptor REV‐ERB α represses the transcription of growth/differentiation factor 10 and 15 genes in rat endometrium stromal cells. Physiol Rep, 4 (2), 2016, e12663, doi: 10.14814/phy2.12663
Funding Information
This work was supported in part by a Grant‐in‐Aid for Scientific Research (B) from the Japan Society for the Promotion of Sciences (JSPS No. 22380152, 24658246) (to M‐A Hattori). Lijia Zhao (Grant No. 201308050010) and Huatao Chen (Grant No. 2010630069) are sponsored by the China Scholarship Council. Keishiro Isayama was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (Grant No. 6117).
References
- Akiyama, S. , Ohta H., Watanabe S., Moriya T., Hariu A., Nakahata N., et al. 2010. The uterus sustains stable biological clock during pregnancy. Tohoku J. Exp. Med. 221:287–298. [DOI] [PubMed] [Google Scholar]
- Albrecht, U. , and Eichele G.. 2003. The mammalian circadian clock. Curr. Opin. Genet. Dev. 13:271–277. [DOI] [PubMed] [Google Scholar]
- Alvarez, J. D. , and Sehgal A.. 2005. The thymus is similar to the testis in its pattern of circadian clock gene expression. J. Biol. Rhythms 20:111–121. [DOI] [PubMed] [Google Scholar]
- Alvarez, J. D. , Hansen A., Ord T., Bebas P., Chappell P. E., Giebultowicz J. M., et al. 2008. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms 23:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden, M. J. , Varcoe T. J., Voultsios A., and Kennaway D. J.. 2010. Reproductive biology of female Bmal1 null mice. Reproduction 139:1077–1090. [DOI] [PubMed] [Google Scholar]
- Brown, S. A. , Ripperger J., Kadener S., Fleury‐Olela F., Vilbois F., Rosbash M., et al. 2005. PERIOD1‐associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308:693–696. [DOI] [PubMed] [Google Scholar]
- Bunger, M. K. , Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., et al. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, A. , and Lazar M. A.. 1993. Induction of Rev‐erbα, an orphan receptor encoded on the opposite strand of the α thyroid hormone receptor gene, during adipocyte differentiation. J. Biol. Chem. 268:16265–16269. [PubMed] [Google Scholar]
- Chen, H. T. , Chu G. Y., Zhao L. J., Yamauchi N., Shigeyoshi Y., Hashimoto S., et al. 2012. Rev‐erbα regulates circadian rhythms and StAR expression in rat granulosa cells as identified by the agonist GSK4112. Biochem. Biophys. Res. Commun. 420:374–379. [DOI] [PubMed] [Google Scholar]
- Chen, H. T. , Zhao L. J., Chu G. Y., Kito G., Yamauchi N., Shigeyoshi Y., et al. 2013a. FSH induces the development of circadian clockwork in rat granulosa cells via a gap junction protein Cx43‐dependent pathway. Am. J. Physiol. Endocrinol. Metab. 304:E566–E575. [DOI] [PubMed] [Google Scholar]
- Chen, H. T. , Zhao L. J., Kumazawa M., Yamauchi N., Shigeyoshi Y., Hashimoto S., et al. 2013b. Downregulation of core clock gene Bmal1 attenuates expression of progesterone and prostaglandin biosynthesis‐related genes in rat luteinizing granulosa cells. Am. J. Physiol. Cell Physiol. 304:C1131–C1140. [DOI] [PubMed] [Google Scholar]
- Chini, C. C. S. , Escande C., Nin V., and Chini E. N.. 2013. DBC1 (Deleted in Breast Cancer 1) modulates the stability and function of the nuclear receptor Rev‐erbα . Biochem. J. 451:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., et al. 2012. Regulation of circadian behaviour and metabolism by REV‐ERB‐α and REV‐ERB‐β . Nature 485:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, C. L. , and Sutherland R. L.. 1990. Progestin regulation of cellular proliferation. Endocr. Rev. 11:266–301. [DOI] [PubMed] [Google Scholar]
- Crumbley, C. , and Burris T. P.. 2011. Direct regulation of CLOCK expression by REV‐ERB. PLoS ONE 6:e17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De, M. C. , Ercolani L., Parodi C., Veronesi M., Vecchio C. L., Bottegoni G., et al. 2015. Dual inhibition of Rev‐erbβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 34:2597–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, S. K. , Lim H., Das S. K., Reese J., Paria B. C., Daikoku T., et al. 2004. Molecular cues to implantation. Endocr. Rev. 25:341–373. [DOI] [PubMed] [Google Scholar]
- Dolatshad, H. , Campbell E. A., O'Hara L., Maywood E. S., Hastings M. H., and Johnson M. H.. 2006. Developmental and reproductive performance in circadian mutant mice. Hum. Reprod. 21:68–79. [DOI] [PubMed] [Google Scholar]
- Fairlie, W. D. , Moore A. G., Bauskin A. R., Russell P. K., Zhang H. P., and Breit S. N.. 1999. MIC‐1 is a novel TGF‐β superfamily cytokine associated with macrophage activation. J. Leukoc. Biol. 65:2–5. [DOI] [PubMed] [Google Scholar]
- Gekakis, N. , Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., et al. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564–1569. [DOI] [PubMed] [Google Scholar]
- Gerhart‐Hines, Z. , Feng D., Emmett M. J., Everett L. J., Loro E., Briggs E. R., et al. 2013. The nuclear receptor Rev‐erbα controls circadian thermogenic plasticity. Nature 503:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, J. E. , Blaikley J., Beesley S., Matthews L., Simpson K. D., Boyce S. H., et al. 2012. The nuclear receptor REV‐ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. U. S. A. 109:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, D. , Yin L., Collins J. L., Parks D. J., Orband‐Miller L. A., Wisely G. B., et al. 2010. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev‐erbα . ACS Chem. Biol. 5:925–932. [DOI] [PubMed] [Google Scholar]
- Grechez‐Cassiau, A. , Rayet B., Guillaumond F., Teboul M., and Delaunay F.. 2008. The circadian clock component BMAL1 is a critical regulator of p21 WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283:4535–4542. [DOI] [PubMed] [Google Scholar]
- Gui, L. M. , and Joyce I. M.. 2005. RNA interference evidence that growth differentiation factor‐9 mediates oocyte regulation of cumulus expansion in mice. Biol. Reprod. 72:195–199. [DOI] [PubMed] [Google Scholar]
- Hara, R. , Wan K. K., Wakamatsu H., Aida R., Moriya T., Akiyama M., et al. 2001. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells 6:269–278. [DOI] [PubMed] [Google Scholar]
- He, P. J. , Hirata M., Yamauchi N., Hashimoto S., and Hattori M.‐A.. 2007a. The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real‐time monitoring system. J. Endocrinol. 193:413–420. [DOI] [PubMed] [Google Scholar]
- He, P. J. , Hirata M., Yamauchi N., Hashimoto S., and Hattori M.‐A.. 2007b. Gonadotropic regulation of circadian clockwork in rat granulosa cells. Mol. Cell. Biochem. 302:111–118. [DOI] [PubMed] [Google Scholar]
- Hino, J. , Kangawa K., Matsuo H., Nohno T., and Nishimatsu S.. 2004. Bone morphogenetic protein‐3 family members and their biological functions. Front Biosci. 9:1520–1529. [DOI] [PubMed] [Google Scholar]
- Hirata, M. , He P. J., Shibuya N., Uchikawa M., Yamauchi N., Hashimoto S., et al. 2009. Progesterone, but not estradiol, synchronizes circadian oscillator in the uterus endometrial stromal cells. Mol. Cell. Biochem. 324:31–38. [DOI] [PubMed] [Google Scholar]
- Hogenesch, J. B. , Gu Y. Z., Jain S. J., and Bradfield C. A.. 1998. The basic‐helix‐loop‐helix‐PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. U. S. A. 95:5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isayama, K. , Zhao L. J., Chen H. T., Yamauchi N., Shigeyoshi Y., Hashimoto S., et al. 2015. Removal of Rev‐erbα inhibition contributes to the prostaglandin G/H synthase 2 expression in rat endometrial stromal cells. Am. J. Physiol. Endocrinol. Metab. 308:E650–E661. [DOI] [PubMed] [Google Scholar]
- Jabbour, H. N. , and Critchley H. O. D.. 2001. Potential roles of decidual prolactin in early pregnancy. Reproduction 121:197–205. [DOI] [PubMed] [Google Scholar]
- Ko, C. H. , and Takahashi J. S.. 2006. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15:R271–R277. [DOI] [PubMed] [Google Scholar]
- Kojetin, D. , Wang Y. J., Kamenecka T. M., and Burris T. P.. 2011. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV‐ERB. ACS Chem. Biol. 6:131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov, R. V. , Shamanna R. K., Kondratova A. A., Gorbacheva V. Y., and Antoch M. P.. 2006. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 20:530. [DOI] [PubMed] [Google Scholar]
- Lawton, L. N. , Bonaldo M. D., Jelenc P. C., Qiu L., Baumes S. A., Marcelino R. A., et al. 1997. Identification of a novel member of the TGF‐β superfamily highly expressed in human placenta. Gene 203:17–26. [DOI] [PubMed] [Google Scholar]
- Lee, S. J. 1991. Expression of growth/differentiation factor‐I in the nervous system conservation of a bicistronic structure. Proc. Natl. Acad. Sci. U. S. A. 88:4250–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Johnson B. P., Shen A. L., Wallisser J. A., Krentz K. J., Moran S. M., et al. 2014. Loss of BMAL1 in ovarian steroidogenic cells results in implantation failure in female mice. Proc. Natl. Acad. Sci. U. S. A. 111:14295–14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjono, A. B. , Brown D. A., Horton K. E., Wallace E. M., Breit S. N., and Manuelpillai U.. 2003. Macrophage inhibitory cytokine‐1 in gestational tissues and maternal serum in normal and pre‐eclamptic pregnancy. Placenta 24:100–106. [DOI] [PubMed] [Google Scholar]
- Matsumoto, K. , Yamauchi N., Watanabe R., Oozono S., Kubota K., Nishimura K., et al. 2009. In vitro decidualization of rat endometrial stromal cells. Cell Tissue Res. 335:575–583. [DOI] [PubMed] [Google Scholar]
- McPherron, A. C. , Lawler A. M., and Lee S. J.. 1997. Regulation of skeletal muscle mass in mice by a new TGF‐β superfamily member. Nature 387:83–90. [DOI] [PubMed] [Google Scholar]
- Meng, Q. J. , McMaster A., Beesley S., Lu W. Q., Gibbs J., Parks D., et al. 2008. Ligand modulation of REV‐ERBα function resets the peripheral circadian clock in a phasic manner. J. Cell Sci. 121:3629–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima, N. , Horiuchi R., Shibuya Y., Fukushige S., Matsubara K., Toyoshima K., et al. 1989. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell 57:31–39. [DOI] [PubMed] [Google Scholar]
- Moore, A. G. , Brown D. A., Fairlie W. D., Bauskin A. R., Brown P. K., Munier M. L. C., et al. 2000. The transforming growth factor‐β superfamily cytokine macrophage inhibitory cytokine‐1 is present in high concentrations in the serum of pregnant women. J. Clin. Endocrinol. Metab. 85:4781–4788. [DOI] [PubMed] [Google Scholar]
- Morrish, D. W. , Dakour J., and Li H.. 2001. Life and death in the placenta: new peptides and genes regulating human syncytiotrophoblast and extravillous cytotrophoblast lineage formation and renewal. Curr. Protein Pept. Sci. 2:245–259. [DOI] [PubMed] [Google Scholar]
- Muter, J. , Lucas E. S., Chan Y. W., Brighton P. J., Moore J. D., Quenby S., et al. 2015. The clock protein period 2 synchronizes mitotic expansion and decidual transformation of human endometrial stromal cells. FASEB J. 29:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oozono, S. , Yamauchi N., Nishimura K., Matsumoto K., Watanabe R., Kubota K., et al. 2008. Expression of rat uterine serine proteinases homologous to mouse implantation serine proteinase 2. J. Exp. Zool. B Mol. Dev. Evol. 310B:642–649. [DOI] [PubMed] [Google Scholar]
- Phelan, C. A. , Gampe R. T., Lambert M. H., Parks D. J., Montana V., Bynum J., et al. 2010. Structure of Rev‐erbα bound to N‐CoR reveals a unique mechanism of nuclear receptor‐co‐repressor interaction. Nat. Struct. Mol. Biol. 17:808–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner, N. , Damiola F., Molina L. L., Zakany J., Duboule D., Albrecht U., et al. 2002. The orphan nuclear receptor REV‐ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260. [DOI] [PubMed] [Google Scholar]
- Raghuram, S. , Stayrook K. R., Huang P. X., Rogers P. M., Nosie A. K., McClure D. B., et al. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV‐ERBα and REV‐ERBβ . Nat. Struct. Mol. Biol. 14:1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, C. K. , Boehle K. L., and Muglia L. J.. 2009. Impaired steroidogenesis and implantation failure in Bmal1 −/− mice. Endocrinology 150:1879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic, R. D. , McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., et al. 2004. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2:1893–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerer, S. E. , Rieger L., Kapp M., Dombrowski Y., Muller N., Dietl J., et al. 2012. MIC‐1 (a multifunctional modulator of dendritic cell phenotype and function) is produced by decidual stromal cells and trophoblasts. Hum. Reprod. 27:200–209. [DOI] [PubMed] [Google Scholar]
- Shimba, S. , Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., et al. 2005. Brain and muscle Arnt‐like protein‐1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 102:12071–12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara, Y. , Kuroda H., Saito K., Nakajima Y., Kubo Y., Ohnishi N., et al. 2012. In vivo monitoring of peripheral circadian clocks in the mouse. Curr. Biol. 22:1029–1034. [DOI] [PubMed] [Google Scholar]
- Tasaki, H. , Zhao L. J., Isayama K., Chen H. T., Nobuhiko Y., Yasufumi S., et al. 2013. Profiling of circadian genes expressed in the uterus endometrial stromal cells of pregnant rats as revealed by DNA microarray coupled with RNA interference. Front. Endocrinol. 4:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki, H. , Zhao L. J., Isayama K., Chen H. T., Yamauchi N., Shigeyoshi Y., et al. 2015. Inhibitory role of REV‐ERBα in the expression of bone morphogenetic protein gene family in rat uterus endometrium stromal cells. Am. J. Physiol. Cell Physiol. 308:C528–C538. [DOI] [PubMed] [Google Scholar]
- Tong, S. , Marjono B., Brown D. A., Mulvey S., Breit S. N., Manuelpillai U., et al. 2004. Serum concentrations of macrophage inhibitory cytokine 1 (MIC 1) as a predictor of miscarriage. Lancet 363:129–130. [DOI] [PubMed] [Google Scholar]
- Uchikawa, M. , Kawamura M., Yamauchi N., and Hattori M.‐A.. 2011. Down‐regulation of circadian clock gene period 2 in uterine endometrial stromal cells of pregnant rats during decidualization. Chronobiol. Int. 28:1–9. [DOI] [PubMed] [Google Scholar]
- Ueda, H. R. , Hayashi S., Chen W. B., Sano M., Machida M., Shigeyoshi Y., et al. 2005. System‐level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37:187–192. [DOI] [PubMed] [Google Scholar]
- Vollmers, C. , Gill S., DiTacchio L., Pulivarthy S. R., Le H. D., and Panda S.. 2009. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl Acad. Sci. U. S. A. 106:21453–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman, M. 1998. Smads and early developmental signaling by the TGFβ superfamily. Genes Dev. 12:2445–2462. [DOI] [PubMed] [Google Scholar]
- Wu, T. , Fu O., Yao L., Sun L., ZhuGe F., and Fu Z. W.. 2012. Differential responses of peripheral circadian clocks to a short‐term feeding stimulus. Mol. Biol. Rep. 39:9783–9789. [DOI] [PubMed] [Google Scholar]
- Yin, L. , and Lazar M. A.. 2005. The orphan nuclear receptor Rev‐erbα recruits the N‐CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 19:1452–1459. [DOI] [PubMed] [Google Scholar]
- Yin, L. , Wang J., Klein P. S., and Lazar M. A.. 2006. Nuclear receptor Rev‐erbα is a critical lithium‐sensitive component of the circadian clock. Science 311:1002–1005. [DOI] [PubMed] [Google Scholar]
- Yin, L. , Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., et al. 2007. Rev‐erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318:1786–1789. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Funk C., Glasser S. R., and Mulholland J.. 1994. Progesterone regulation of heparin‐binding epidermal growth factor‐like growth factor gene expression during sensitization and decidualization in the rat uterus: effects of the antiprogestin, ZK 98.299. Endocrinology 135:1256–1263. [DOI] [PubMed] [Google Scholar]
- Zhao, R. B. , Lawler A. M., and Lee S. J.. 1999. Characterization of GDF‐10 expression patterns and null mice. Dev. Biol. 212:68–79. [DOI] [PubMed] [Google Scholar]


