Abstract
Background. Results from the Costa Rica Vaccine Trial (CVT) demonstrated partial cross-protection by the bivalent human papillomavirus (HPV) vaccine, which targets HPV-16 and HPV-18, against HPV-31, -33, and -45 infection and an increased incidence of HPV-51 infection.
Methods. A study nested within the CVT intention-to-treat cohort was designed to assess high-risk HPV variant lineage–specific vaccine efficacy (VE). The 2 main end points were (1) long-term incident infections persisting for ≥2 years and/or progression to high-grade squamous intraepithelial lesions (ie, cervical intraepithelial neoplasia grade 2/3 [CIN 2/3]) and (2) incident transient infections lasting for <2 years. For efficiency, incident infections due to HPV-16, -18, -31, -33, -35, -45, and -51 resulting in persistent infection and/or CIN 2/3 were matched (ratio, 1:2) to the more-frequent transient viral infections, by HPV type. Variant lineages were determined by sequencing the upstream regulatory region and/or E6 region.
Results. VEs against persistent or transient infections with HPV-16, -18, -33, -35, -45, and -51 did not differ significantly by variant lineage. As the possible exception, VEs against persistent infection and/or CIN 2/3 due to HPV-31 A/B and HPV-31C variants were −7.1% (95% confidence interval [CI], −33.9% to 0%) and 86.4% (95% CI, 65.1%–97.1%), respectively (P = .02 for test of equal VE). No difference in VE was observed by variant among transient HPV-31 infections (P = .68).
Conclusions. Overall, sequence variation at the variant level does not appear to explain partial cross-protection by the bivalent HPV vaccine.
Keywords: HPV vaccine, cross-protection, HPV variants
Persistent infection with high-risk human papillomaviruses (HPVs) causes cervical cancer, a very common female malignancy worldwide [1]. Although >200 HPV genotypes have been characterized, cervical cancer is associated with a limited set of 12 related types from 5 Alphapapillomavirus species (alpha-5, -6, -7, -9, and -11) [2–4]. HPV-16 and HPV-18 cause approximately 70% of cervical cancers [5] and are targeted by the virus-like particle–based prophylactic HPV vaccines Gardasil and Gardasil9 (Merck, Whitehouse Station, New Jersey) and Cervarix (GlaxoSmithKline Biologicals, Rixensart, Belgium). All are highly effective against HPV-16/18 persistence [6] and cervical intraepithelial neoplasia grade 2/3 (CIN 2/3) [7–9]. Development of neutralizing antibodies against HPV-16/18 is correlated with reported cross-protection against the phylogenetically HPV-16–related types 31 and 33 and the HPV-18–related type 45 [10, 11]. Partial cross-protection has been demonstrated in studies using either the bivalent or quadrivalent vaccine [12], while variable and/or increased rates of infections have been observed for HPV-51, -52, and -58 [12–14].
Variant lineages are classified as viral genomes of a known HPV type with <10% sequence variability [15]. Specific variant lineages show epidemiologic patterns in their distribution and associations with CIN 2/3 and cancer [15, 16]. In addition, HPV variants are associated with hosts of common geographic ancestry and are even evident with population movements; thus, not all geographic regions harbor similar variants (or at similar frequencies) [15, 17–19]. Whether viral genomic polymorphisms associated with variant lineages alter infection rates upon vaccination has not been thoroughly evaluated, but this is of interest because variants exhibit differences in natural history and disease risks [15, 17, 20–22]. Although vaccine protection against HPV-16/18 is uniformly high, cross-protection against genetically related high-risk HPV types is variable and could be due to differential protection against variant lineages [15].
This study was designed to assess the impact of HPV-16/18 vaccination on incident infection(s) at the viral variant level. This study evaluated the relative vaccine efficacy (VE) by variant lineage in samples obtained from young women participating in the Costa Rica Vaccine Trial (CVT), a National Cancer Institute (NCI)–sponsored community-based, double-blind, randomized clinical trial of Cervarix in Guanacaste, Costa Rica. The present study examined the hypothesis that viral variants circulating in Guanacaste were differentially affected by HPV vaccination.
MATERIALS AND METHODS
The study of viral variants was a nested analysis within the previously reported CVT (clinical trials registration NCT00128661). Written informed consent was obtained from all participants in CVT. Institutional review board approval was obtained for the informed consent forms at both the NCI and in Costa Rica.
CVT Study Design
The study design and main outcomes have been published previously [6, 23, 24]. Briefly, the study involved female residents living in Guanacaste and Puntarenas, Costa Rica, with community-based census recruitment occurring during 2004–2005. A total of 7466 women aged 18–25 years were randomly assigned (ratio, 1:1) to receive the HPV-16/18 vaccine, Cervarix, or control (hepatitis A vaccine; Havrix, GSK Biologicals).
Prior to vaccination, all women completed a study survey and had a clinical visit; specimens collected prior to randomization were classified as having been collected at enrollment. Vaccination with 3 doses occurred over 6 months in the majority of women in the CVT; 71.5% of participants received doses within the prespecified vaccination windows at 0 months (enrollment visit) and 1 month and 6 months after enrollment. A total of 3736 women received Havrix, and 3726 women received Cervarix [6]. When the last vaccine dose was administered, at 6 months, women who were sexually active provided a self-collected specimen for HPV testing [23].
Follow-up included annual collection of cervical samples for HPV and cytological testing. Abnormal cytological findings (defined by the Bethesda system) of low-grade squamous intraepithelial lesions (LSIL) or atypical squamous cells of undetermined significance (ASC-US) necessitating triage for HPV testing [25] shifted clinical visits to a 6-month schedule; if 3 consecutive negative results of cytological tests were obtained, annual visits resumed. Participants were referred to undergo colposcopy for a cytological diagnosis of high-grade squamous intraepithelial lesions (HSIL), atypical squamous cells (cannot exclude HSIL), or glandular abnormalities and for repeat cytological testing of LSIL or HPV-positive ASC-US [6, 23].
Exfoliated cervical cells were obtained using a Cervex-Brush (Rovers Medical Devices, Oss, the Netherlands) directly applied to the cervix and eluted into PreservCyt medium (Cytyc, Marlborough, Massachusetts) for cytological and HPV testing as previously described [6].
HPV Variant Study Design
Infecting HPV types were selected based upon phylogenetic relatedness to vaccine types and/or previous evidence for partial protection or observed enhanced susceptibility to infection after immunization [6, 13, 26–31]. A total of 7 high-risk HPV types were evaluated: HPV-16 and HPV-18; HPV-31, -33, and -35 (3 types related to HPV-16 in the alpha-9 species); HPV-45 (a type in the alpha-7 species related to HPV-18); and HPV-51 (a type from the alpha-5 species) [3].
Incident infection(s) were eligible for selection, defined as an HPV infection that was detected after vaccination and not present at either enrollment (time of first vaccine dose) or the 6-month visit. Infection duration was estimated as the interval between the initial HPV-positive test result and the time of the last positive HPV test. Long-term/persistent infections were defined as incident infection(s) that lasted for >2 years worth of visits (ie, >660 days) and/or resulted in a CIN 2/3 diagnosis. Short-term/transient infections were defined as those lasting <2 years and not associated with CIN 2/3. For efficiency, all incident persistent infections/CIN 2/3 were selected, whereas a random sampling of the more abundant incident transient infections matched by HPV type were selected in a ratio of 2 short-term infections for each long-term infection. Selection for testing was masked to vaccine arm and number of doses administered.
Laboratory Characterization of HPV Type–Specific Variant Lineages
The laboratory was blinded to all data except HPV type. Briefly, DNA was isolated from exfoliated cells, and HPV genomes were assigned to variant lineages by DNA sequence analyses as previously described [17, 32]. Polymerase chain reaction (PCR) analyses targeted the upstream regulatory region (URR) and/or E6 open reading frame (ORF), because these regions allow classification of HPV variant genome lineages that are also correlated with the major polymorphisms in the L1 and L2 ORFs [15, 33–35]. Samples for which the URR assay did not yield amplification underwent repeat analysis using the E6 assay. PCR primers and conditions are provided in Supplementary Table 1 [17, 33, 36].
To classify HPV-31 lineages for the extended study, an additional PCR assay (L1_FG) amplifying a smaller fragment was developed, based on lineage-specific sequence differences in a hypervariable region of the L1 encoding the FG surface loop [33]. Classification of HPV-31 variants by the L1_FG assay was validated using samples previously characterized by the URR/E6 assays (κ = 0.95; Supplementary Table 2). Samples characterized by at least 1 HPV-31 variant assay were included in the HPV-31 extended analysis; samples with discordant variant results were excluded (n = 9). Thus, there was a difference in the number of HPV-31 infections classified as persistent infections/CIN 2/3 in the main and extended studies.
Statistical Analysis
The lineage-specific VE was calculated for each variant group. All incident persistent infections/CIN 2/3 for the described types and a 2:1 match by type for incident transient infections were included in the analysis. Since the total numbers of women at risk for the end points in the 2 vaccine arms (Cervarix and Havrix) were practically the same for each analysis, we used event counts instead of attack rates. The estimated HPV VE against viral variant lineages for each HPV type was calculated as the ratio of the difference in numbers of variant lineage end points observed between the Havrix arm and the Cervarix arm, divided by the number of events in the Havrix arm.
The expected number of events in the Cervarix arm is a binomial variable, and the sample size is equal to the total number of events in the two arms. Noting that the VE is the complement of the ratio of attack rates in the two arms, we transformed the estimates of the proportions and the confidence bounds to obtain a corresponding confidence interval (CI) for the VE, using the Clopper–Pearson method [37].
Additionally, we tested whether the efficacy of Cervarix varied by variant lineages within the same viral type, separately for each end point (ie, persistent infections/CIN 2/3 and transient infections). The ratio of the attack rates was evaluated by a Poisson test for rates, using the rateratio.test package in R [38]. The null hypothesis of the tests is equal VEs for each viral lineage within a given HPV type.
HPV-31 Extended Analysis: Sample Selection and Variant Detection
To follow-up on a suggestive initial finding in the main analysis, all HPV-31 infections were tested or retested; infections detected within 180 days of enrollment were considered prevalent and used for exploratory analyses.
RESULTS
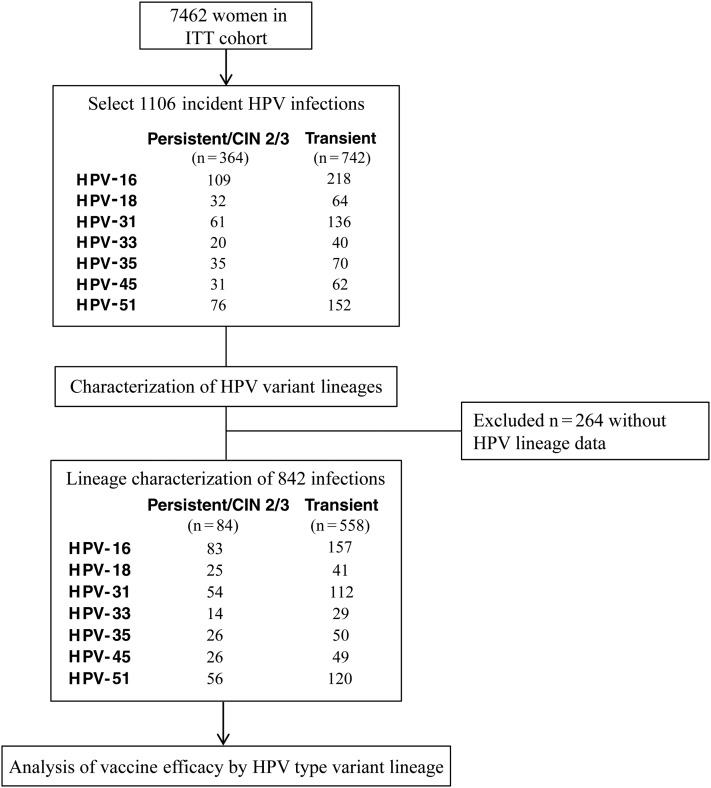
The study design is shown in Figure 1. A total of 1106 incident infections were selected for lineage characterization of high-risk HPV types 16, 18, 31, 33, 35, 45, and 51 (Supplementary Table 3). Of the 1106 incident HPV infections selected, 842 (76.1%) were successfully characterized for variant lineage classification; 264 infections (23.9%) were excluded either because the sample was unavailable (n = 47) or the PCR reactions failed (n = 217). Of the 842 infections characterized, 284 (33.7%) were persistent/CIN 2/3, and 558 (66.3%) were transient.
Figure 1.
Study design to determine the relative efficacy of bivalent human papillomavirus (HPV) vaccine against HPV variant lineages in the Costa Rica HPV Vaccine Trial (CVT). Design of sample selection for analysis of incident HPV infections in a subset of women in the intention-to-treat (ITT) cohort of the CVT. This study was designed to assess whether the HPV vaccine relative efficacy differed for incident type-specific variant infections, using 2 different outcomes: (1) transient infection and (2) persistent infection and/or cervical intraepithelial neoplasia grade 2/3 (CIN 2/3)–associated infection (see “Materials and Methods” section).
To assess whether differences in type-specific VE resulted from differences in HPV variant lineage infections, the genomes were partially sequenced to classify variant lineages, categorized by infection outcome and vaccine arm, and compared with respect to lineage-specific VE. For vaccine types HPV-16 and HPV-18, there was strong protection upon vaccination, irrespective of variant lineage or infection duration (Table 1). We next assessed vaccine variant efficacies for HPV-31, -33, -35, -45, and -51 (Table 1). Insufficient variant heterogeneity for HPV-33 and HPV-35 limited estimation of VE by lineage. Although HPV-45 infections showed cross-protection in the CVT [6], no difference was observed in comparison of lineages A (n = 35) and B (n = 40) for VE among transient infections (P = .52) or persistent infections/CIN 2/3 (P = .99; Table 1).
Table 1.
Relative Vaccine Efficacy (VE) Against Incident Infection With High-Risk Human Papillomavirus (HPV) Variant Lineages
| Species, Lineage, Arm | Transient Infection |
Persistent Infection and/or CIN 2/3 Progression |
Total Evaluated, No. | ||||
|---|---|---|---|---|---|---|---|
| No. (%) | Lineage-Specific VE, (95% CI) | P Valuea | No. (%) | Lineage-Specific VE, (95% CI) | P Valuea | ||
| HPV-16 (alpha-9) | .97 | .99 | |||||
| Lineage A | |||||||
| Cervarix | 16 (51.6) | 88.1 (81.5–93.1) | 15 (48.4) | 76.2 (63.8–86.0) | 31 | ||
| Havrix | 135 (68.2) | … | 63 (31.8) | … | 198 | ||
| Lineage D | |||||||
| Cervarix | 1 (50.0) | 80.0 (28.4–99.5) | 1 (50.0) | 75.0 (19.4–99.4) | 2 | ||
| Havrix | 5 (55.6) | … | 4 (44.4) | … | 9 | ||
| HPV-18 (alpha-7) | .99 | .93 | |||||
| Lineage A | |||||||
| Cervarix | 2 (40.0) | 92.0 (74.0–99.0) | 3 (60.0) | 66.7 (29.9–92.5) | 5 | ||
| Havrix | 25 (73.5) | … | 9 (26.5) | … | 34 | ||
| Lineage B | |||||||
| Cervarix | 1 (33.3) | 92.3 (64.0–99.8) | 2 (66.7) | 81.8 (48.2–97.7) | 3 | ||
| Havrix | 13 (54.2) | … | 11 (45.8) | … | 24 | ||
| HPV-31 (alpha-9) | .68 | .02 | |||||
| Lineages A/B | |||||||
| Cervarix | 19 (55.9) | 50.0 (33.4–66.6) | 15 (44.1) | −7.1 (−33.9 to 0) | 34 | ||
| Havrix | 38 (73.1) | … | 14 (26.9) | … | 52 | ||
| Lineage C | |||||||
| Cervarix | 15 (83.3) | 62.5 (45.8–77.3) | 3 (16.7) | 86.4 (65.1–97.1) | 18 | ||
| Havrix | 40 (64.5) | … | 22 (35.5) | … | 62 | ||
| HPV-33 (alpha-9) | |||||||
| Lineage A | |||||||
| Cervarix | 10 (66.7) | 44.4 (21.5–69.2) | 5 (33.3) | 37.5 (8.52–75.5) | 15 | ||
| Havrix | 18 (69.2) | … | 8 (30.8) | … | 26 | ||
| Lineage B | |||||||
| Cervarix | 0 (0.0) | …c | 0 (0.0) | …c | 0 | ||
| Havrix | 1 (50.0) | … | 1 (50.0) | … | 2 | ||
| HPV-35 (alpha-9) | .34 | ||||||
| Lineage A | |||||||
| Cervarix | 18 (58.1) | 25.0 (9.77–46.7) | 13 (41.9) | 0.0 (.0–24.7) | 31 | ||
| Havrix | 24 (64.9) | … | 13 (35.1) | … | 37 | ||
| Sublineage A2 | |||||||
| Cervarix | 1 (100.0) | 85.7 (42.1–99.6) | 0 (0.0) | …c | 1 | ||
| Havrix | 7 (100.0) | … | 0 (0.0) | … | 7 | ||
| HPV-45 (alpha-7) | .52 | .99 | |||||
| Lineage A | |||||||
| Cervarix | 4 (44.4) | 78.9 (54.4–93.9) | 5 (55.6) | 28.6 (3.67–71.0) | 9 | ||
| Havrix | 19 (73.1) | … | 7 (26.9) | … | 26 | ||
| Lineage B | |||||||
| Cervarix | 8 (61.5) | 55.6 (30.8–78.5) | 5 (38.5) | 44.4 (13.7–78.8) | 13 | ||
| Havrix | 18 (66.7) | … | 9 (33.3) | … | 27 | ||
| HPV-51 (alpha-5) | .46 | .80 | |||||
| Lineage A | |||||||
| Cervarix | 51 (61.4) | 0.0 (.0–6.98) | 32 (38.6) | −100.0 (−79.4 to −1.0) | 83 | ||
| Havrix | 51 (76.1) | … | 16 (23.9) | … | 67 | ||
| Lineage B | |||||||
| Cervarix | 12 (75.0) | −100.0 (−54.1 to −100.0) | 4 (25.0) | 0.0 (0.0–60.2) | 16 | ||
| Havrix | 6 (60.0) | … | 4 (40.0) | … | 10 | ||
Abbreviations: CI, confidence interval; CIN 2/3, cervical intraepithelial neoplasia grade 2/3.
a Data denote no. of the specified type of infection and/or disease state (% of all infections per specified lineage and vaccine arm).
b For comparison of the equality of vaccine efficacy for each HPV variant lineage by infection duration.
c Not determined, because of insufficient sample size.
Incident HPV-51 infections were previously found to be increased in the Cervarix arm, compared with the Havrix arm, of the CVT [6]. Infection with this relatively common type was due to lineage A variants in 85.2% of cases (n = 150) and due to B variants in 14.8% (n = 26); no statistically significant differences for VE were observed for either persistent infections/CIN 2/3 (P = .46) or transient infections (P = .80) by lineage. Lineage A infections were more common in persistent infections/CIN 2/3 after vaccination, whereas lineage B infections were more common in transient infections after vaccination; the interaction was not statistically significant (Table 1).
HPV-31 infections were dichotomized into variant lineages A/B or C, representing the divergence of the lineages from a most recent common ancestor [33]. In women who received Cervarix and developed HPV-31 persistent infection/CIN 2/3, VE against HPV-31 variant lineage C infections was 86.4% (95% CI, 65.1%–97.1%), compared with −7.1% (95% CI, −33.9% to 0%) for HPV-31 A/B variant lineage infections (P = .02; Table 1). No significant difference was observed for VE against transient HPV-31 infection by variant lineage.
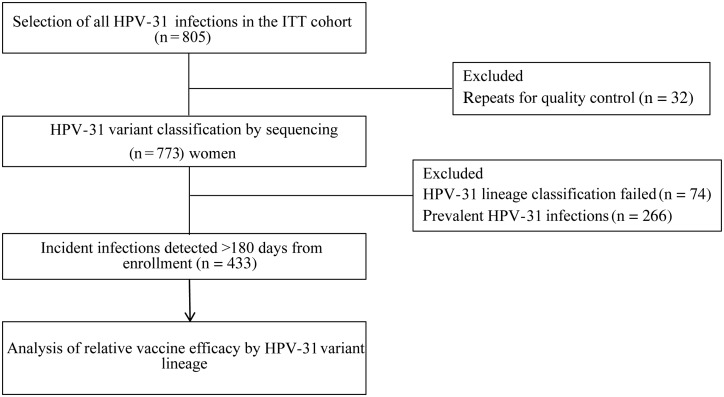
Because of the intriguing difference between persistent infections/CIN 2/3 and transient infections due to HPV-31, a post hoc study was performed to test all incident HPV-31 infections (Figure 2). An additional PCR assay was developed to characterize HPV-31 variant lineages by amplifying and sequencing a small fragment within the L1 FG loop (see “Materials and Methods” section). Seventy-four HPV-31 infections (9.2%) that were unamplified by all 3 HPV-31 variant assays were excluded.
Figure 2.
Expanded study design to assess the relative efficacy of the bivalent human papillomavirus (HPV) vaccine against HPV-31 variant infections. Samples in which HPV-31 was previously detected [6] were selected for variant lineage classification, and incident infections were evaluated for lineage-specific vaccine efficacy. A total of 433 incident HPV-31 infections, classified into variant lineages, were evaluated for relative vaccine efficacy, based on infection duration ([1] transient infection or [2] persistent infection and/or cervical intraepithelial neoplasia grade 2/3). Prevalent infections (n = 266), defined as HPV-31 detected within 180 days of study enrollment, were excluded in this analysis. Repeats for QC were an additional sample selected from a different time point in 32 women. Abbreviation: ITT, intention to treat.
To determine whether differences in infection duration modify the effect of vaccination on HPV-31 variants, the number of HPV-31 positive tests during follow-up was used to categorize infection periods: transient single infections, transient multiple infections, or persistent infections/CIN 2/3 (Table 2). The terms “single” and “multiple” refer to the number of HPV-31–positive tests detected within the <2-year period. The end point of ≥2 years for persistent infection was the same as that used in the main study, although the numbers are slightly different, owing to use of an additional assay.
Table 2.
Human Papillomavirus (HPV) Lineage–Specific Vaccine Efficacy (VE) for All Incident HPV-31 Infections in the Costa Rica HPV Vaccine Trial
| Lineage, Vaccine Arm | Transient Infection (Single)a |
Transient Infection (Multiple)b |
All Transient Infections |
Persistent Infection and/or CIN 2/3 Progression |
Total Evaluated, No.c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%d) | Lineage-Specific VE, % (95% CI) | P Valuee | No. (%d) | Lineage-Specific VE, % (95% CI) | P Valuee | No. (%d) | Lineage-Specific VE, % (95% CI) | P Valuee | No. (%d) | Lineage-Specific VE, % (95% CI) | P Valuee | ||
| HPV-31 A/B | .90 | .76 | .90 | .017 | |||||||||
| Cervarix | 39 (52.0) | 58.1 (47.4–68.2) | 19 (25.3) | 58.7 (43.2–73.0) | 58 (77.3) | 58.3 (49.6–66.6) | 17 (22.7) | −6.3 (−.15 to 30.2) | 75 | ||||
| Havrix | 93 (60.0) | … | 46 (29.7) | … | 139 (89.7) | … | 16 (10.3) | … | 155 | ||||
| HPV-31 C | |||||||||||||
| Cervarix | 32 (60.4) | 56.8 (44.7–68.0) | 17 (32.1) | 66.7 (52.1–79.2) | 49 (92.5) | 60.8 (51.7–69.4) | 4 (7.5) | 84.0 (63.9–95.5) | 53 | ||||
| Havrix | 74 (49.3) | … | 51 (34.0) | … | 125 (83.3) | … | 25 (16.7) | … | 150 | ||||
Abbreviations: CI, confidence interval; CIN 2/3, cervical intraepithelial neoplasia grade 2/3.
a Data are for infections with 1 positive HPV-31 test result.
b Data are for infections with >1 positive HPV-31 test result within 2 years of the previous positive test.
c Data are combined number of all transient infections and persistent infections and/or CIN 2/3 progression, by lineage and vaccine arm.
d Data denote no. of the specified type of infection and/or disease state (% of all infections per specified lineage and vaccine arm).
e For comparison of the equality of lineage-specific VE per variant lineage and infection duration.
The majority of HPV-31 infections (any lineage) were transient, and, among these, no significant difference was observed in lineage-specific VE, whether for single or multiple transient infections (Table 2). As previously observed, persistent infections/CIN 2/3 due to HPV-31 displayed significant differences by variant lineage. Incidence of persistent infections/CIN 2/3 due to HPV-31 A/B was similar by vaccine arm (17 infections in the Cervarix arm and 16 in the Havrix arm), resulting in a VE of −6.3% (95% CI, −.15% to 30.2%), whereas the VE against HPV-31C (4 infections in the Cervarix arm and 25 in the Havrix arm) was 84.0% (95% CI, 63.9%–95.5%; P = .017).
To determine whether the vaccine influenced the natural history of existing HPV-31 infections, we tested 266 infections that were prevalent at the time of vaccination. No lineage-specific effect on infection duration was observed among the prevalent HPV-31 infections (data not shown). To evaluate an HPV-naive group that should have maximum protection from Cervarix, we limited the analyses to women who received 3 vaccine doses and were negative for HPV by PCR and HPV-16/18 serologic analysis at baseline. HPV-31 results were similar as reported above (13 transient HPV-31 A/B infections in the Cervarix arm and 52 in the Havrix group, with a VE of 75%; 9 and 47 transient HPV-31 C infections, respectively, with a VE of 81%; 3 and 4 persistent infections/CIN 2/3 due to HPV-31 A/B, respectively, with a VE of 25%; and 0 and 5 persistent infections/CIN 2/3 due to HPV-31 C, respectively, with a VE of 100%).
DISCUSSION
This study evaluated whether the partial protection against or increased susceptibility to some high-risk HPV types after administration of the HPV-16/18 AS04-adjuvanted vaccine could be due to viral type genetic heterogeneity. The evolution of HPV genomes has resulted in the emergence of stable viral variant lineages with highly correlated nucleotide changes across the viral genome [15, 35]. To determine whether synthetic selective pressure created by the recent introduction of HPV vaccines limits the dispersion of specific HPV variants, we used specimens and data from the ITT cohort of the CVT. We selected HPV types previously shown to be significantly protected, partially protected, or increased after HPV vaccination in either the CVT [6, 24] or other studies [12]. We chose our end point based on the most important predictors of cancer risk following infection, viral persistence and/or CIN 2/3. We observed moderate-to-high VE against variants of HPV-31 lineage C for both transient infections (VE, 61%) and persistent infections/CIN 2/3 (VE, 84%), whereas, lineages A/B showed only partial protection for infections that were transient (VE, 58%) but not for those that led to persistent infection/CIN 2/3 (VE, −6%). If corroborated, this finding could be of clinical importance, since HPV-31 lineages A/B have been observed to more likely lead to development of CIN 3, compared with HPV-31 lineage C [17, 22]. We evaluated whether the difference in VE of HPV-31 A/B lineages for persistent infections/CIN 2/3 might be related to changes in duration of infection, but we did not observe a difference by vaccine arm (data not shown). Thus, it is unlikely that the vaccine acts subsequent to acquisition of an HPV-31A/B variant infection to change the natural history and/or duration of these infections [39].
In women with no prior evidence of HPV exposure, vaccination is highly efficacious against autologous HPV type acquisition and persistence [6, 13, 27, 39]. As reported in the CVT, the bivalent vaccine displayed 91% VE (95% CI, 82%–96%) against 1-year persistent HPV-16/18 and 90% (95% CI, 40%–100%) against related CIN 2/3 in the ATP cohort [24], as well as a VE of 49% (95% CI, 38%–58%) in the ITT group [6]. A more stringent end point of ≥2 years for persistence used in the current variant analysis confirmed a high efficacy against vaccine-types (HPV-16 and HPV-18) irrespective of variant lineage, maintaining high-type specific protection.
As previously reported in the CVT, VE against the HPV-16/18–related types HPV-31, -33, and -45 was 44% (95% CI, 18%–63%) in the ATP and 16% (95% CI, −5% to 32%) in the ITT group for protection against 1-year persistence [6]. Analysis by HPV genotype in the ATP revealed that only the closest phylogenetically HPV-16/18–related types, HPV-31 and HPV-45, were significantly protected [6]. Vaccine cross-protective efficacy against 6-month and 1-year persistence in the ATP arm of the CVT was significant for HPV-31, with a 1-year persistence VE of 46% (95% CI, 8%–69%). For HPV-45, only 6-month persistence VE of 73% (95% CI, 45 to 88) was significant [6]. We observed no significant differences in VE for HPV-45 lineages A and B (Table 1).
HPV-51 1-year persistence was significantly increased in the ATP vaccine arm, with a VE of −64% (95% CI, −151% to −8%). Enigmatically, HPV-51 lineage A and HPV-51 lineage B variants were differentially increased in persistent infections/CIN 2/3 and transient infections, respectively, although VE was not statistically significantly different by lineage. In contrast, the PATRICIA trial that also used the bivalent HPV-16/18 vaccine observed weak but significant cross-protection for HPV-51 6-month persistence [13]. The reasons for the differences in VE for HPV-51 variants in this study and the HPV-51 increased incidence between studies could be related to statistical instability due to the limited number of HPV-51 lineage B infections reported here.
In a previous report, HPV-31 partial protection was associated with anti–HPV-16 antibody titers, the ability of sera to neutralize HPV-31 pseudovirions, and antibody avidity [28]. We evaluated the number of vaccine doses received by women with HPV-31 infections. There were no significant differences between the proportion of women receiving 3 versus <3 doses either by HPV-31 variant lineage (ie, A/B vs C) or by outcome, transient infection versus persistent infection/CIN 2/3 (data not shown). Thus, it is unlikely that viral titers are related to HPV-31 variant differences in partial protection for persistent infection/CIN 2/3. Theoretically, differences in protection of HPV-31 variants could result from polymorphisms within the L1 sequence that alter conformational epitopes recognized by vaccine-induced antibodies. Based on our previously reported data on the heterogeneity of HPV-31 variant complete genome sequences (supplementary figure 2A in the article by Chen et al [33]), we noted only 1 consistent L1 amino acid change difference between variants HPV-31 A/B and C lineages, and it was located in the L1 FG loop (HPV-31 amino acids L1-26; Supplementary Table 4). This conformational epitope corresponds to amino acids HPV-31 L1 262–276 and was previously shown to facilitate HPV-31 cross-reactivity and neutralization by HPV-16 [40–43] and HPV-31 [44, 45] antibodies. HPV-31 C lineage contains an alanine at L1–267, similar to that seen in HPV-16, as well as the HPV-16 L1 amino acid sequence used in Cervarix (NCBI accession no. AAC09292.1) [46]. HPV-31 A/B variants contain threonine at this position (Supplementary Table 4). Thus, it is plausible that this genetic variation could contribute to HPV-31C lineage being better protected than the A/B lineages, consistent with the enhanced activity of monoclonal antibody 31.F16 to recognize HPV-31C variant as compared to HPV-31A/B variants [45]. Nevertheless, it is not clear why this differential protection was not seen when using HPV-31 transient infections as the outcome. It is possible that persistence involves both protection and natural history differences of HPV-31 variants, as HPV-31 lineage C appears to persist longer than A/B [17, 47].
The data suggest that VE by lineage for persistent infections/CIN 2/3 was generally lower than that observed for transient infections (eg, for HPV-45 A lineage, VE against transient infection was 79%, and VE against persistent infections/CIN 2/3 was 29%). Since the number of events was limited and because this was not an a priori hypothesis, we have not pursued this analysis in detail. It is possible that lower VE in long-term persistent infections was a result of misclassification of incident infections that were low-level prevalent infections, as discussed by Malagon et al [12].
This study has limitations. The number of infections in each outcome was limited by stratification on variant lineage. The study was well powered to detect clinically significant differences, but it does not rule out variant differences in other populations. Our study did not access rare variations in the viral variant genomes, particularly in the L1 ORF containing the neutralizing epitopes, but evaluated common changes conserved among variant lineages, based on knowledge of the evolution and conservation of HPV genetic variations.
In conclusion, we tested the hypothesis that HPV variant lineages might explain the partial protection or increased susceptibility to the HPV vaccine. Although we did not find a consistent pattern of variants having differing vaccine efficiencies, HPV-31 A/B versus C lineages showed differences in the main outcome, persistent infection/CIN 2/3. We are perplexed by these observations, as transient HPV-31 infections showed no differences. Based on the knowledge that the vaccine induces neutralizing antibodies that should affect both persistent infections/CIN 2/3 and acquisition of transient infections, we have no underlying mechanism that can explain these observations. However, data on HPV-31 variant natural history suggest that lineage C tends to be more persistent, whereas, lineages A/B are more associated with CIN 3. These observations are not consistent with the model derived from HPV-16 variants in which persistence and oncogenicity are related [15]. Complexities in viral-host relationships beyond current understanding might be at play, or there may be random effects of population sampling yielding data that are difficult to interpret. Nevertheless, considering the enhanced oncogenicity of HPV-31 A/B lineages, this study requires confirmation or refutation in additional vaccine cohorts to determine whether HPV-31 variants show differential protection by the bivalent vaccine.
STUDY GROUP MEMBERS
Investigators in the CVT group are as follows: Bernal Cortés (specimen and repository manager), Paula González (long-term follow-up [LTFU] co–principal investigator), Roland Herrero (CVT co–principal investigator), Silvia E. Jiménez (trial coordinator), Carolina Porras (coinvestigator), and Ana Cecilia Rodríguez (coinvestigator), Proyecto Epidemiológico Guanacaste, Fundación INCIENSA (San José, Costa Rica); Allan Hildesheim (co–principal investigator and NCI co–project officer), Aimée R. Kreimer (LTFU coinvestigator and NCI co–project officer), Douglas R. Lowy (HPV virologist), Mark Schiffman (CVT medical monitor and NCI co–project officer), John T. Schiller (HPV virologist), Mark Sherman (CVT quality control pathologist), and Sholom Wacholder (statistician), National Cancer Institute (Bethesda, MD); Ligia A. Pinto and Troy J. Kemp (HPV immunology laboratory), Leidos Biomedical Research, Frederick National Laboratory for Cancer Research, NCI-Frederick (Frederick, MD); Mary Sidawy (CVT histopathologist), Georgetown University (Washington, DC); Wim Quint (virologist, HPV DNA testing), Leen-Jan van Doorn (HPV DNA testing), and Linda Struijk (HPV DNA testing), DDL Diagnostic Laboratory (Rijskijk, Netherlands); Joel M. Palefsky (expert on anal HPV infection and disease diagnosis and management) and Teresa M. Darragh (pathologist and clinical management), University of California (San Francisco); and Mark H. Stoler (QC pathologist), University of Virginia (Charlottesville).
Supplementary Data
Supplementary materials are available at (http://jid.oxfordjournals.org/). Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the women of Guanacaste and Puntarenas, Costa Rica, who gave of themselves in participating in this effort; the staff in Costa Rica involved in this project, for their tremendous effort and dedication; members of the team from Information Management Services, who were responsible for the development and maintenance of the data system used in the trial and who serve as the data management center for this effort; Jean Cyr, Julie Buckland, and John Schussler; members of the Data and Safety Monitoring Board charged with protecting the safety and interest of participants in our trial (Steve Self, Chair, Adriana Benavides, Luis Diego Calzada, Ruth Karron, Ritu Nayar, and Nancy Roach); and members of the external Scientific HPV Working Group, who have contributed to the success of our efforts over the years (Joanna Cain, Chair, Diane Davey, David DeMets, Francisco Fuster, Ann Gershon, Elizabeth Holly, Silvia Lara, Henriette Raventós, Wasima Rida, Luis Rosero-Bixby, Kristen Suthers, and Sarah Thomas).
S. W. made important contributions to the CVT project in general and to this analysis in particular. His untimely death is a loss to all who knew him.
Disclaimer. The Costa Rica HPV Vaccine Trial is a long-standing collaboration between investigators in Costa Rica and at the National Cancer Institute. The NCI and Costa Rica investigators are responsible for the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation of the manuscript. On efficacy-related manuscripts, GlaxoSmithKline has the right to review and comment.
Financial support. This work was supported by the National Cancer Institute, National Institutes of Health (NIH; awards CA78527, P30 CA13330, and N01-CP-11005); the NIH Office of Research on Women's Health; and GlaxoSmithKline Biologicals (clinical trials agreement FDA BB-IND 7920).
Potential conflicts of interest. J. T. S. and D. R. L. are named inventors on US government–owned HPV vaccine patents that are licensed to GlaxoSmithKline and Merck and for which the National Cancer Institute receives licensing fees. They are entitled to limited royalties, as specified by federal law. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Collaborators: Costa Rica HPV Vaccine Trial (CVT) group, Paula González, Roland Herrero, Silvia E. Jiménez, Carolina Porras, Ana Cecilia Rodríguez, Allan Hildesheim, Aimée R. Kreimer, Douglas R. Lowy, Mark Schiffman, John T. Schiller, Mark Sherman, Sholom Wacholder, Ligia A. Pinto, Troy J. Kemp, Mary Sidawy, Wim Quint, Leen-Jan van Doorn, Linda Struijk, Joel M. Palefsky, Teresa M. Darragh, and Mark H. Stoler
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Bernard HU. Taxonomy and phylogeny of papillomaviruses: An overview and recent developments. Infect Genet Evol 2013; 18:357–61. [DOI] [PubMed] [Google Scholar]
- 3.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010; 401:70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology 2013; 445:2–10. [DOI] [PubMed] [Google Scholar]
- 5.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer 2011; 128:927–35. [DOI] [PubMed] [Google Scholar]
- 6.Herrero R, Wacholder S, Rodriguez AC et al. Prevention of persistent human papillomavirus infection by an HPV16/18 vaccine: a community-based randomized clinical trial in Guanacaste, Costa Rica. Cancer Discov 2011; 1:408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garland SM, Hernandez-Avila M, Wheeler CM et al. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N Engl J Med 2007; 356:1928–43. [DOI] [PubMed] [Google Scholar]
- 8.Paavonen J, Jenkins D, Bosch FX et al. Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: an interim analysis of a phase III double-blind, randomised controlled trial. Lancet 2007; 369:2161–70. [DOI] [PubMed] [Google Scholar]
- 9.Group FIIS, Dillner J, Kjaer SK et al. Four year efficacy of prophylactic human papillomavirus quadrivalent vaccine against low grade cervical, vulvar, and vaginal intraepithelial neoplasia and anogenital warts: randomised controlled trial. BMJ 2010; 341:c3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper E, Bissett SL, Howell-Jones R et al. Neutralization of non-vaccine human papillomavirus pseudoviruses from the A7 and A9 species groups by bivalent HPV vaccine sera. Vaccine 2011; 29:8585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemp TJ, Hildesheim A, Safaeian M et al. HPV16/18 L1 VLP vaccine induces cross-neutralizing antibodies that may mediate cross-protection. Vaccine 2011; 29:2011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malagon T, Drolet M, Boily MC et al. Cross-protective efficacy of two human papillomavirus vaccines: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:781–9. [DOI] [PubMed] [Google Scholar]
- 13.Wheeler CM, Castellsague X, Garland SM et al. Cross-protective efficacy of HPV-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by non-vaccine oncogenic HPV types: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol 2012; 13:100–10. [DOI] [PubMed] [Google Scholar]
- 14.Herrero R. Human papillomavirus (HPV) vaccines: limited cross-protection against additional HPV types. J Infect Dis 2009; 199:919–22. [DOI] [PubMed] [Google Scholar]
- 15.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology 2013; 445:232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xi LF, Schiffman M, Koutsky LA et al. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst 2014; 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffman M, Rodriguez AC, Chen Z et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Research 2010; 70:3159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi LF, Kiviat NB, Hildesheim A et al. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst 2006; 98:1045–52. [DOI] [PubMed] [Google Scholar]
- 19.Chan SY, Bernard HU, Ong CK, Chan SP, Hofmann B, Delius H. Phylogenetic analysis of 48 papillomavirus types and 28 subtypes and variants: a showcase for the molecular evolution of DNA viruses. J Virol 1992; 66:5714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burk RD, Terai M, Gravitt PE et al. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res 2003; 63:7215–20. [PubMed] [Google Scholar]
- 21.Xi LF, Koutsky LA, Galloway DA et al. Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J Natl Cancer Inst 1997; 89:796–802. [DOI] [PubMed] [Google Scholar]
- 22.Xi LF, Schiffman M, Koutsky LA et al. Association of human papillomavirus type 31 variants with risk of cervical intraepithelial neoplasia grades 2–3. Int J Cancer 2012; 131:2300–7. [DOI] [PubMed] [Google Scholar]
- 23.Herrero R, Hildesheim A, Rodriguez AC et al. Rationale and design of a community-based double-blind randomized clinical trial of an HPV 16 and 18 vaccine in Guanacaste, Costa Rica. Vaccine 2008; 26:4795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildesheim A, Wacholder S, Catteau G et al. Efficacy of the HPV-16/18 vaccine: Final according to protocol results from the blinded phase of the randomized Costa Rica HPV-16/18 vaccine trial. Vaccine 2014; 32:5087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon D, Davey D, Kurman R et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA 2002; 287:2114–9. [DOI] [PubMed] [Google Scholar]
- 26.Kreimer AR, Gonzalez P, Katki HA et al. Efficacy of a bivalent HPV 16/18 vaccine against anal HPV 16/18 infection among young women: a nested analysis within the Costa Rica Vaccine Trial. Lancet Oncol 2011; 12:862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paavonen J, Naud P, Salmeron J et al. Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): final analysis of a double-blind, randomised study in young women. Lancet 2009; 374:301–14. [DOI] [PubMed] [Google Scholar]
- 28.Safaeian M, Kemp TJ, Pan DY et al. Cross-protective vaccine efficacy of the bivalent HPV vaccine against HPV31 is associated with humoral immune responses: results from the Costa Rica Vaccine Trial. Hum Vaccin Immunother 2013; 9:1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Einstein MH, Baron M, Levin MJ et al. Comparison of the immunogenicity of the human papillomavirus (HPV)-16/18 vaccine and the HPV-6/11/16/18 vaccine for oncogenic non-vaccine types HPV-31 and HPV-45 in healthy women aged 18–45 years. Hum Vaccin 2011; 7:1359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemp TJ, Safaeian M, Hildesheim A et al. Kinetic and HPV infection effects on cross-type neutralizing antibody and avidity responses induced by Cervarix((R)). Vaccine 2012; 31:165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safaeian M, Porras C, Pan Y et al. Durable antibody responses following one dose of the bivalent human papillomavirus l1 virus-like particle vaccine in the costa rica vaccine trial. Cancer Prev Res (Phila) 2013; 6:1242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, de Freitas LB, Burk RD. Evolution and classification of oncogenic human papillomavirus types and variants associated with cervical cancer. Methods Mol Biol 2015; 1249:3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Schiffman M, Herrero R et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One 2011; 6:e20183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Schiffman M, Herrero R et al. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One 2013; 8:e72565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Terai M, Fu L, Herrero R, DeSalle R, Burk RD. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. J Virol 2005; 79:7014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler CM, Yamada T, Hildesheim A, Jenison SA. Human papillomavirus type 16 sequence variants: identification by E6 and L1 lineage-specific hybridization. J Clin Microbiol 1997; 35:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clopper CJ, Pearson ES. The Use of Confidence or Fiducial Limits Illustrated in the Case of the Binomial. Biometrika 1934; 26:404–13. [Google Scholar]
- 38.Team RC. R: A language and environment for statistical computing. Version 3.2. Vienna, Austria: R Foundation for Statistical Computing, 2015. [Google Scholar]
- 39.Hildesheim A, Herrero R, Wacholder S et al. Effect of human papillomavirus 16/18 L1 viruslike particle vaccine among young women with preexisting infection: a randomized trial. JAMA 2007; 298:743–53. [DOI] [PubMed] [Google Scholar]
- 40.Carpentier GS, Fleury MJ, Touze A et al. Mutations on the FG surface loop of human papillomavirus type 16 major capsid protein affect recognition by both type-specific neutralizing antibodies and cross-reactive antibodies. J Med Virol 2005; 77:558–65. [DOI] [PubMed] [Google Scholar]
- 41.Fleury MJ, Touze A, Alvarez E et al. Identification of type-specific and cross-reactive neutralizing conformational epitopes on the major capsid protein of human papillomavirus type 31. Arch Virol 2006; 151:1511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen ND, Dillner J, Eklund C et al. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 1996; 223:174–84. [DOI] [PubMed] [Google Scholar]
- 43.Guan J, Bywaters SM, Brendle SA et al. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 2015; 483:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleury MJ, Touze A, Maurel MC, Moreau T, Coursaget P. Identification of neutralizing conformational epitopes on the human papillomavirus type 31 major capsid protein and functional implications. Protein Sci 2009; 18:1425–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bissett SL, Godi A, Fleury MJ, Touze A, Cocuzza C, Beddows S. Naturally occurring capsid protein variants of human papillomavirus genotype 31 represent a single L1 serotype. J Virol 2015; 89:7748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deschuyteneer M, Elouahabi A, Plainchamp D et al. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccin 2010; 6:407–19. [DOI] [PubMed] [Google Scholar]
- 47.Xi LF, Schiffman M, Koutsky LA et al. Persistence of newly detected human papillomavirus type 31 infection, stratified by variant lineage. Int J Cancer 2013; 132:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.