Abstract
The 4 dengue virus serotypes (DENV-1–4) cause the most prevalent mosquito-borne viral disease of humans worldwide. DENV-2 Asian 1 (A1) genotype viruses replaced the Asian-American (AA) genotype in Vietnam and Cambodia, after which A1 viruses containing Q or M at envelope (E) residue 160 became more prevalent than those with residue 160K in both countries (2008–2011). We investigated whether these substitutions conferred a fitness advantage by measuring neutralizing antibody titer against reporter virus particles (RVPs) representing AA, A1-160K, A1-160Q, and A1-160M, using patient sera from Vietnam and a well-characterized Nicaraguan cohort. Surprisingly, we found that A1-160Q and A1-160M RVPs were better neutralized by heterologous antisera than A1-160K. Despite this, Vietnamese patients infected with A1-160Q or A1-160M viruses had higher viremia levels than those infected with A1-160K. We thus found that independent lineages in Vietnam and Cambodia acquired a substitution in E that significantly increased polyclonal neutralization but nonetheless were successful in disseminating and infecting human hosts.
Keywords: dengue virus, neutralizing antibodies, envelope protein, fitness, evolution, genotype, lineage, Vietnam, Cambodia
The 4 dengue virus serotypes (DENV-1–4) cause the most medically important arthropod-borne viral disease of humans worldwide. Up to 96 million dengue cases occur annually, including 500 000 hospitalizations due to severe disease [1]. Within each DENV serotype, multiple genetically distinct lineages have evolved in geographically separated regions [2]. Historically in Asia and increasingly in Latin America, all 4 DENV serotypes and often multiple genotypes and/or clades of each serotype circulate simultaneously for extended periods [3]. Antigenic differences between lineages are thought to contribute to disease severity, epidemic cycling, and viral evolution [4–7].
Particular DENV genotypes generally have synchronized epidemic cycles and cause the majority of disease for 3–5 years, and then they become scarce as the prevalence of another DENV serotype increases [8]. During periods of serotype dominance, diversification is often observed for all lineages of that serotype, but as serotype incidence wanes, one lineage disappears while another persists [5–7, 9–12]. Some lineage replacements result from competing clades and genotypes that evolve in close proximity for years, while others are due to introduction of a foreign lineage [6, 13].
One hypothesis is that lineage replacement is stochastic, driven largely by mosquito and virus population bottlenecks, such as annual dry periods with low transmission [9, 12, 14–16]. Another hypothesis is that lineage replacement occurs through natural selection, as replacement events occur over multiple years, suggesting a gradual effect by selective pressure [6]. Furthermore, some clade replacement events coincide with dynamic changes in the prevalence of cocirculating serotypes, suggestive of interaction/competition between antigenically distinct viruses [5].
Two general mechanisms can explain why one virus population may gain a selective advantage over another. First, the dominant clade may have an intrinsic fitness advantage, such as greater viral replication and dissemination in mosquitoes or humans [7, 17]. Alternatively, the dominant clade may have an extrinsic fitness advantage conferred by superior transmission in the presence of host immunity [4, 18]. As for all antigenically variable pathogens, a clade that evades prior host immunity would be able to infect more hosts and replicate better than one constrained by host immunity. However, unique to DENV, a clade that takes advantage of prior host immunity via antibody-dependent enhancement of infection may also have a selective advantage in the face of population immunity [6, 19, 20].
DENV is composed of 90 homo-dimers of envelope (E) glycoprotein, the main antigenic target for neutralizing antibodies. E is composed of 3 domains (EDI, EDII, and EDIII) [21]. EDI is the central structural domain, while EDII contains the highly conserved fusion loop that allows for virus-host membrane fusion. Finally, EDIII contains an immunoglobulin-like fold likely involved in receptor binding [22]. Strongly neutralizing human antibodies target EDIII and the hinge region between EDI and EDII [23–28]. The fusion loop region is targeted by both strongly and weakly neutralizing cross-reactive antibodies [29, 30]. Substitution of just a few amino acid positions can substantially alter DENV type–specific neutralization [31], and antigenic differences between genotypes have been described using monoclonal antibodies [32], as well as antisera from experimentally inoculated animals, human vaccine recipients, and naturally infected humans [4, 7, 33, 34]. However, the specific amino acid substitutions/epitopes that determine lineage differences as recognized by polyclonal sera have not been identified.
Between 2004 and 2008, the DENV-2 Asian 1 (A1) genotype began replacing the resident Asian-American DENV-2 (AA) genotype in Vietnam and Cambodia. In both countries, the genotype replacement was followed by years of DENV-1 circulation, with DENV-2 again emerging as the dominant serotype a few years later. A study found that A1-infected patients had higher viremia levels than AA-infected patients, although no significant difference in replication in mosquito cells or mosquito infectivity was noted [13]. We sought to investigate the possible role of preexisting immunity in driving the genotype replacement and subsequent evolution of the A1 lineage.
Here, we describe the unusual observation of a naturally occurring substitution in the E protein of successful DENV-2 A1 lineages that appears to confer an antigenic fitness cost. During the replacement of the AA by the A1 genotype in Vietnam and Cambodia, 3 separate lineages arose almost simultaneously with substitutions at E position 160 (2 with K160M and 1 with K160Q). We explored the possible role of preexisting immunity on selection of these viruses by testing polyclonal sera against RVPs representing the previously circulating AA, as well as the 3 A1 E-160 variants. Surprisingly, A1-160Q and A1-160M RVPs were better neutralized by polyclonal sera in vitro than A1-160K and AA RVPs. Despite this apparent fitness disadvantage, we found that lineages with 160Q and 160M increased in prevalence in both countries over time and achieved a higher viremia level in patients than lineages with A1-160K.
METHODS
Cloning of RVPs Structural Plasmids
RVPs were produced as previously reported [35]. One plasmid encodes the West Nile virus (WNV) nonstructural proteins (NS1-NS5) with a green fluorescent protein (GFP) reporter protein substituting the structural proteins (provided by T.C. Pierson, National Institutes of Health), and the second plasmid encodes the DENV structural C-prM-E proteins. Structural genes of the DENV-2 reference strain 16681 were first cloned into a pcDNA3.1/V5-His-TOPO vector (provided by T.C. Pierson) and served as a cloning intermediate. Substitutions were then made in the 16681 C-prM-E plasmid using the QuikChange Site-Directed Mutagenesis kit (Stratagene) to construct the AA genotype and A1 genotype variants A1-160K, A1-160Q, and A1-160M (Table 1).
Table 1.
Amino Acid Positions of Dengue Virus Serotype 2 (DENV-2) E Protein at Which DENV-2 Prototype Strain (16681) and Representative Vietnamese (VN) Asian 1 and Asian/American Genotypes Differ
| Position | 6 | 83 | 120 | 129 | 141 | 160 | 164 | 203 | 226 | 228 | 308 | 346 | 461 | 478 | 484 | 485 | 491 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prototype 16681 | M | N | R | V | I | K | I | N | T | G | V | H | V | T | I | V | V |
| VN Asian1 | I | K | T | I | V | Q/K/M | V | N | K | E | V | Y | V | S | I | V | V |
| VN Asian/American | I | N | T | I | I | K | I | D | T | G | I | H | A | S | V | I | A |
Generation of RVPs
To produce RVPs, 4 × 105 cells were plated per well of a 6-well plate and incubated overnight at 37°C in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco) and penicillin/streptomycin solution (Gibco). The WNV reporter replicon (1 µg), the DENV C-prM-E plasmid (3 µg), and MIRUS TransIT-LT1 Transfection Reagent (12 µL) were transfected into 293 T cells according to the manufacturer's protocol and incubated for 4 hours at 37°C. The medium was then changed to low-glucose DMEM supplemented with 10% FBS and penicillin/streptomycin and incubated for 20 hours at 37°C and 48 hours at 28°C. The RVP-containing supernatant was harvested and frozen at −80°C.
Serum Samples
The Nicaraguan Pediatric Dengue Cohort Study [36] was approved by the institutional review boards of the University of California, Berkeley, and the Nicaraguan Ministry of Health; 18 serum samples collected following primary DENV-1 infection, 10 collected after primary DENV-2 infection, 40 collected after secondary DENV-2 infection, and 20 collected after primary DENV-3 infection were used in neutralization assays. Vietnamese plasma samples used for neutralization tests were from patients with tetanus admitted to the Hospital for Tropical Diseases in 1997–1998 (prior to the AA-A1 genotype replacement event; n = 25) or in 2006–2007 (after the replacement event; n = 27). The scientific and ethics committee of the Hospital for Tropical Diseases in Ho Chi Minh City approved the use of these anonymized, precollected plasma samples for infectious diseases research. Deidentified plasma viremia samples were obtained from Vietnamese patients hospitalized in the Hospital for Tropical Diseases with dengue in 2011 [37].
RVP Titration and Quality Control
Raji–DC-SIGN cells (kindly provided by B. Doranz, Integral Molecular), a human B-cell lymphoma line with the DENV attachment factor DC-SIGN, were used for the RVP neutralization assay as described (Supplementary Figure 1A) [38]. For each RVP lot, the optimal working dilution was determined by titration [38]. To ensure that the antibody-RVP interaction only depended on the neutralization capacity of the antibodies in the serum and was not influenced by the amount of RVPs used, consistent with the law of mass action [39], we performed neutralization assays with a polyclonal DENV-positive control (20 pooled sera from Nicaraguan National Blood Center donors) with concentrations of RVPs above and below the optimal dilution determined from the titration of each lot. The neutralization curves, all with similar 50% neutralization titers (NT50) for the 3 dilutions, are shown in Supplementary Figure 1B [39].
Neutralization Assay
RVPs were diluted in Roswell Park Memorial Institute 1640 complete medium (pH 8.0). RVP neutralization assays were performed as previously described, using serial 3-fold dilutions of sera/plasma [38]. Infection of cells was quantified after 48 hours by measuring GFP-positive cells via flow cytometry, followed by analysis with FlowJo software. Raw data were graphed as the percentage of infected cells versus the log of the reciprocal serum dilution, and a sigmoidal dose response curve with a variable slope was generated using GraphPad Prism 5.0 to determine the NT50, defined as the antibody dilution at which a 50% reduction in infection was observed, compared with the no-antibody control [38, 40]. Stringent quality control rules, including ensuring that viral particles were neutralized according to the law of mass action, the absolute sum of squares was <0.2, and the coefficient of determination (R2) of the nonlinear regression was >0.9, were used to ensure the reproducibility of results. Monoclonal antibodies (MAbs) were obtained as follows: E76, E87, E60, E28, and E18 (kindly provided by M. S. Diamond, Washington University, St. Louis, Missouri) [41]; 87.1 and 82.11 (kindly provided by Federica Sallusto and Antonio Lanzavecchia, Institute for Research in Biomedicine) [23]; and 4G2 (ATCC).
Phylogenetic Analyses
The sequence set for phylogenetic analyses consisted of all full-length DENV-2 E genes labeled as isolates from Vietnam and Cambodia that were available in GenBank as of June 2015 (n = 261). For the phylogenetic tree, a set that represented DENV-2 genetic diversity was also included (n = 13). Phylogenetic relationships were inferred with the maximum likelihood (ML) method (version 3.0; available at: http://www.atgc-montpellier.fr/phyml/), using the general time reversible (GTR) nucleotide substitution model with 4 discrete Γ categories of among-site rate variation, allowing for invariant sites (GTR + Γ4 + I model). The ML tree topology was estimated using nearest neighbor interchange and subtree pruning and regrafting branch swapping. Trees were unrooted but drawn with American genotype DENV-2 as the outgroup.
RESULTS
Greater Neutralization of A1-160Q RVP Than AA RVP by Vietnamese Serum Samples From 2 Different Periods
We hypothesized that the A1 genotype may have succeeded in replacing the AA genotype by acquiring substitutions that allowed it to better escape population immunity. We first generated RVPs representing AA and A1 to analyze their neutralization profiles with population-level patient sera collected in Vietnam before and after the AA/A1 genotype replacement. Starting with A1 DENV-2 reference strain 16681, we used site-directed mutagenesis to introduce amino acid substitutions to generate the consensus of either the A1 or AA genotype, which differ at 13 amino acids in E (Table 1). We initially constructed an A1 RVP with Q at position 160 (A1-160Q), as it was a major variant in 2006 when AA was almost fully replaced by A1 in Vietnam. We infected human Raji–DC-SIGN cells with A1-160Q and AA RVPs in the presence of 2 sets of Vietnamese plasma samples: 25 samples collected in 1997–1998, prior to the A1/AA lineage replacement, and 27 samples collected in 2006–2007, after the lineage replacement. Both sets were from patients with tetanus with an unknown prior DENV immune history.
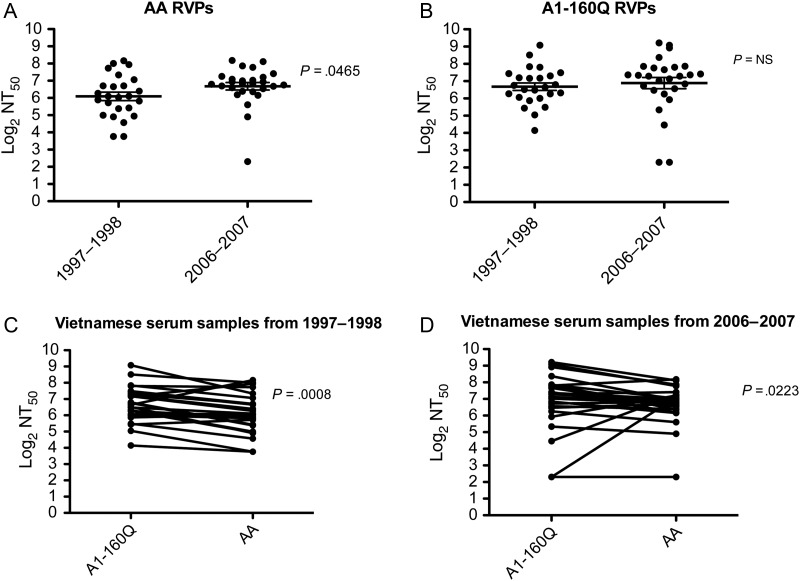
DENV neutralization assays can vary from laboratory to laboratory [42]; here, we used a flow cytometry–based system with human cells, implemented with stringent quality control measures to ensure reproducibility [38]. As expected, AA RVPs were better neutralized by Vietnamese sera collected after the major DENV-2 epidemic (2006–2007), rather than before the epidemic (1997–1998), suggesting that years of intense AA transmission increased the magnitude of the neutralizing antibody response against the AA genotype (Figure 1A). No significant difference in NT50 values was observed when A1-160Q RVPs were tested with sera obtained from Vietnamese patients in 2006–2007, compared with sera obtained in 1997–1998 (Figure 1B). However, when we compared the neutralization titers of the A1-160Q relative to the AA RVPs, we found that A1-160Q RVPs were better neutralized by sera obtained from Vietnamese patients before (Figure 1C) and after (Figure 1D) the DENV-2 epidemic. This observation raised the unusual possibility of a virus with an apparent fitness disadvantage arising naturally in an DENV-endemic setting.
Figure 1.
Neutralization titers to A1-160Q reporter virus particles (RVPs) and Asian-American (AA) RVPs in Vietnamese serum samples from 2 different periods, 1997–1998 and 2006–2007. RVPs were incubated with 3-fold serial dilutions of serum from Vietnamese samples. The RVP-serum mixture was then used to infect Raji–DC-SIGN cells, and after 48 hours, green fluorescent protein expression was recorded by flow cytometry and used to calculate the percentage of infection. The 50% neutralization titer (NT50) was calculated, and the log2 of the NT50 values were plotted on the y-axis. Prism GraphPad was used to compare the NT50 between: AA RVPs, serum samples obtained from Vietnamese subjects during 2 different periods, 1997–1998 and 2006–2007 (A); A1-160Q RVPs, serum samples obtained from Vietnamese subjects during 2 different periods, 1997–1998 and 2006–2007 (B); A1-160Q RVPs and AA RVPs, using serum samples obtained from Vietnamese subjects during 1997–1998 (C); and A1-160Q RVPs and AA RVPs using serum samples obtained from Vietnamese subjects during 2006–2007 (D). Abbreviation: NS, not significant.
Sera Obtained After Primary DENV-2 AA Infection Neutralize A1-160Q and AA Equally, but Sera Obtained After Secondary DENV-2 AA Infection Neutralize A1-160Q Better Than AA
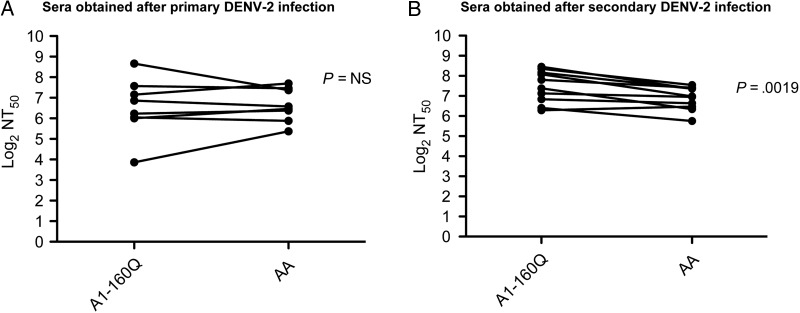
We further probed the difference between A1 and AA DENV-2 genotypes by titrating the RVPs against a panel of sera from the Nicaraguan Pediatric Dengue Cohort Study, which has been ongoing for 12 years and provides continuous monitoring of all DENV infections that occur in the cohort [43, 44]. This allows for identification of the infecting DENV serotype in primary and secondary infections [38]. Coincidentally, all DENV-2 viruses circulating in the Nicaraguan cohort were AA genotype, enabling us to test the role of primary AA and secondary AA immunity against the A1 and AA lineages. With sera obtained after primary DENV-2 infection, we did not observe significant differences between neutralization of genotypes A1-160Q and AA (Figure 2A). However, sera obtained after secondary DENV-2 infection better neutralized A1-160Q, compared with AA RVPs (Figure 2B). This unexpected observation suggested that A1-160Q viruses may be better neutralized by serotype cross-reactive antibodies.
Figure 2.
Neutralization titers (NTs) to A1-160Q reporter virus particles (RVPs) and Asian-American (AA) RVPs in serum samples from the Nicaraguan Pediatric Dengue Cohort Study. Neutralization assays were performed as described in the legend for Figure 1. NTs were compared between A1-160Q RVPs and AA RVPs, using sera obtained from Nicaraguan subjects after primary dengue virus serotype 2 (DENV-2) infection (A) and after secondary DENV-2 infection (B). Abbreviation: NS, not significant.
Changes in Prevalence of A1-160K, A1-160Q, and A1-160M Following AA/A1 Genotype Replacement
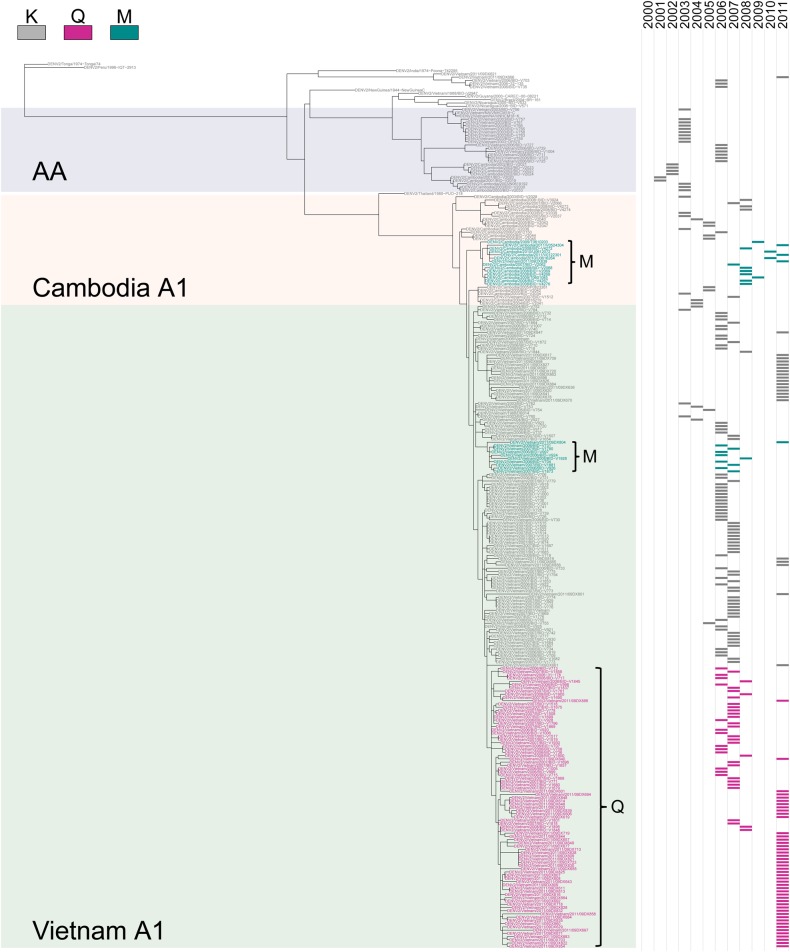
We estimated the phylogenetic and temporal relationships among all DENV-2 E genes listed as from Vietnam and Cambodia in GenBank and identified 2 distinct lineages with variation at E-160 that arose in Vietnam in 2006: one with A1-160Q and another with A1-160M (Figure 3). The proportion of A1 isolates with 160Q or M increased until 2008, at which point DENV-1 genotype I viruses were the dominant serotype (2007–2010) and no DENV-2 was sequenced from clinical cases, although DENV-2 did circulate at low levels during this period. When DENV-2 reemerged as the dominant serotype in 2011, the majority (65%) of A1 sequenced in Vietnam contained 160Q, with a smaller number of A1-160M and K viruses still in circulation (Table 2).
Figure 3.
Phylogenetic tree of Vietnamese and Cambodian dengue virus serotype 2 (DENV-2) isolates. Maximum likelihood phylogenetic tree of the ancestral relationships among full-length DENV-2 E gene sequences from Vietnam and Cambodia and 13 DENV-2 reference sequences. Strain names are colored by amino acid at position 160. The background of the tree is shaded to indicate genotype and country for relevant lineages. A time series (right) shows the year of virus isolation. Abbreviation: AA, Asian-American.
Table 2.
Increasing Percentage Over Time of A1-160Q Virus in Vietnam and A1-160M Virus in Cambodia
| Year | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2011 |
|---|---|---|---|---|---|---|---|---|---|
| Vietnam | |||||||||
| 160K, no. | NA | NA | 11 | 3 | 2 | 44 | 34 | 1 | 23 |
| 160Q, no. | NA | NA | 0 | 0 | 0 | 12 | 22 | 5 | 47 |
| 160M, no. | NA | NA | 0 | 0 | 0 | 5 | 3 | 1 | 2 |
| Q, % | NA | NA | 0 | 0 | 0 | 20 | 37 | 71 | 65 |
| Cambodia | |||||||||
| 160K, no. | 2 | 5 | 9 | 3 | 5 | 0 | 5 | 6 | 2 |
| 160M, no. | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 18 | 2 |
| M, % | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 75 | 50 |
Analysis was based on sequence analysis of E protein amino acid 160, using all sequences listed as from Vietnam and Cambodia in GenBank.
Abbreviation: NA, not available.
While the A1-160Q/M substitution arose in Vietnam, a distinct lineage of A1-160M simultaneously emerged in Cambodia. Although fewer sequences were available on GenBank from Cambodia, all isolates of A1 in Cambodia before 2005 contained 160K. However, in 2007, a lineage of A1 emerged that contained 160M, and by 2008, when DENV-1 genotype I also dominated in Cambodia, A1-160M was more commonly isolated than A1-160K (Table 2). Although few isolates are available on GenBank after 2008, virological data from Cambodia indicates that A1-160M continued to be isolated in 2010 and 2011 (Philippe Buchy, personal communication). Thus, based on analyses of the available sequences in GenBank, it appears that as DENV-1 genotype I became dominant in both Vietnam and Cambodia, A1 DENV-2 viruses evolved 2 different amino acids substitutions at E position 160 in 3 independent lineages.
Sera Obtained After Secondary DENV-2 Infection and After Primary DENV-1 and DENV-3 Infection Neutralize A1-160Q and AI-160M Better Than A1-160K
The physiochemical properties of the variants at position 160 differ; the initial lysine (K) is positively charged, while glutamine (Q) is polar uncharged and methionine (M) is hydrophobic. Position 160, located in a valley on the surface of EDI (Supplementary Figure 2A), is a contact residue of 2 potently neutralizing type-specific DENV-1 human antibodies, 1F4 [45] and HM14c10 [46], and is adjacent to a site shown to substantially alter DENV-1 type-specific immunity [31]. In DENV-3 viruses, position 160 is adjacent to the site of amino acid deletions (E157 and E158 in DENV-1, -2, and -4 are absent in DENV-3).
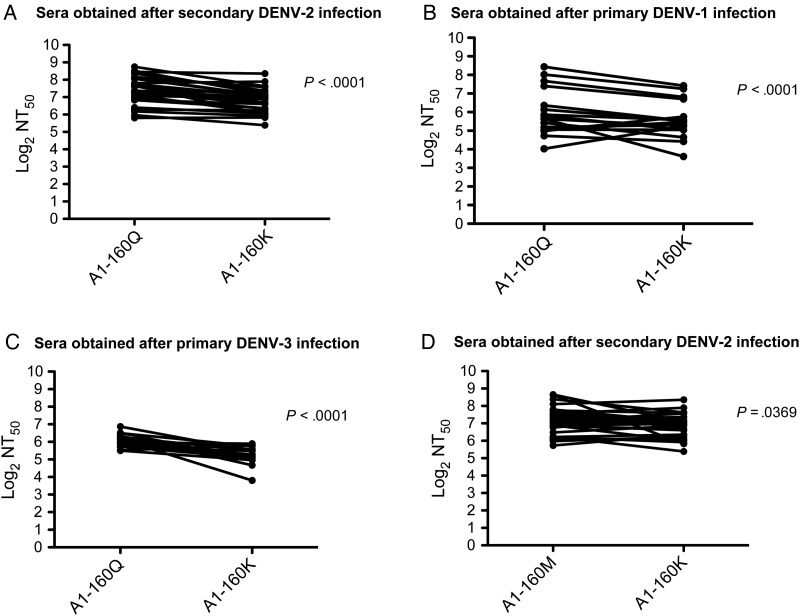
To investigate whether the difference in serotype cross-reactive neutralization by human sera was due to amino acid variation at E-160, we generated A1-160K and A1-160M RVPs and compared their neutralization by Nicaraguan sera obtained after primary DENV-1 and DENV-3 infections, as well as after secondary DENV-2 infections. Interestingly, the high neutralization titers to A1-160Q RVPs were significantly reduced when the A1-160K RVPs were tested with Nicaraguan cohort sera obtained after secondary DENV-2 infection (P < .0001; Figure 4A), after primary DENV-1 infection (P < .0001; Figure 4B), and after primary DENV-3 infection (P < .0001; Figure 4C). Like A1-160Q, A1-160M RVPs were also significantly better neutralized than A1-160K RVPs by sera obtained after secondary DENV-2 infection ( P = .0369; Figure 4D). Thus, A1-160Q, and to some extent A1-160M, changed the neutralization profile of polyclonal sera to A1 RVPs, making them significantly better neutralized by serotype-cross-reactive sera.
Figure 4.
A1-160Q, A1-160M, and A1-160K reporter virus particle (RVP) variants were tested against serum samples obtained from Nicaraguan subjects from the Pediatric Dengue Cohort Study. Neutralization assays were performed as described in the legend to Figure 1. A–C, Neutralization titers (NTs) were compared between A1-160Q RVPs and A1-160K RVPs, using sera obtained from Nicaraguan subjects after secondary dengue virus serotype 2 (DENV-2) infection (A), after primary DENV-1 infection (B), and after primary DENV-3 infection (C). D, NTs were compared between A1-160M RVPs and A1-160K RVPs, using sera obtained from Nicaraguan subjects after secondary DENV-2 infection.
Substitution at E-160 Does Not Result in an Overall Change to Virion Structure
Amino acid substitutions may affect antibody binding by directly modifying the corresponding epitope, but they can also affect distant sites by causing a global change to virion structure or affecting the number of epitopes exposed by the virion through so-called breathing [47]. To test whether the 160Q and 160M substitutions modified cross-reactive antibody binding by inducing a global change to virion structure, we tested the neutralization profiles of A1-160Q and A1-160K RVPs using a panel of MAbs. We tested MAb 87.1 and E76 (which target the EDIII A strand, a cryptic viral epitope only accessible with viral breathing or global changes to virion structure), E87 (which targets the EDIII C-C loop in the lateral ridge), and MAbs E60, 82.11, E28, E18, and 4G2 (which target the EDII fusion loop) against A1-160Q and A1-160K RVPs, but we did not find significant differences in their NT50 values (Supplementary Figure 2B). The target of E76 in the EDIII A strand is a temperature-dependent epitope, but we observed similar E76 neutralization profiles of A1-160Q and A1-160K at 2 different temperatures (4°C and 23°C; Supplementary Figure 2C). These data provide preliminary evidence that the substitution at position 160 does not alter the overall virion structure and suggest that heterologous antibodies sensitive to the 160 substitutions may directly target an epitope that includes position 160.
Individuals Infected With A1-160Q Have a Significantly Higher Viremia Level Than Those Infected With A1-160K
We hypothesized that Q and M at position 160, rather than K, must have some in vivo fitness advantage to explain their evolutionary success, despite apparently being better neutralized by polyclonal sera. We compared plasma viremia data for 70 Vietnamese adults infected with A1 viruses in 2011. Of these, 70% were secondary, 11% primary, and 19% indeterminate DENV infections. Primary and secondary DENV infections were classified as previously described [37]. The majority of specimens (67%) were collected on day 3 of illness, with 24% collected on day 2 and 9% collected on day 4. Adults infected with A1-160Q/M were statistically more likely to have secondary immune responses than those infected with A1-160K (83% vs 43%; P < .002, by the 2-sample test for equality of proportions with continuity correction). The day of viremia level measurement did not differ significantly between groups (0.17 days; P = .23, by the 2-sided t test).
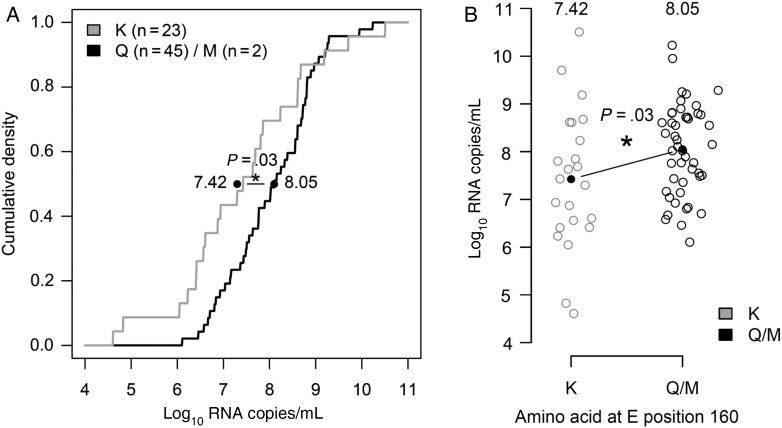
On average, individuals infected with A1-160Q/M had 4-fold higher viral titers than those infected with A1-160K (A1-160Q/M, 8.05 log10 RNA copies/mL; A1-160K, 7.42 log10 RNA copies/mL; difference, 0.63 log10 RNA copies/mL; P = .03, as measured by linear regression). The difference in viremia levels between A1-160Q/M and A1-160K remained significant when controlling for day of illness (difference, 0.59 log10 RNA copies/mL; P < .05) and only modestly decreased when also controlling for immune status (difference, 0.50 log10 RNA copies/mL; P = .12). Figure 5 shows the cumulative distributions of viremia titers for those infected with A1-160K, compared with A1-160Q/M, along with the raw viremia level data. Thus, although A1-160Q and A1-160M were better neutralized by serotype cross-reactive sera in vitro, viruses with these substitutions achieved higher viremia levels in vivo and were more often observed to cause secondary infections, providing a possible explanation for their increasing rate of detection in clinical cases in Vietnam and Cambodia.
Figure 5.
Viremia levels of 70 Vietnamese patients infected with A1-160K, Q, or M in 2011. A, Cumulative density plot of log10 RNA copies/mL for patients infected with A1-160K or M (n = 23 and n = 2, respectively) or A1-160Q (n = 45). Linear regression shows the effect of infecting virus (A1-160Q/M vs K) on viremia levels. The black line corresponds to the difference in viremia levels (a significant difference, P = .03), and average values for those infected with A1-160Q/M and A1-160K are printed adjacent to the line. B, Jitter plot of log10 RNA copies/mL in individuals infected with A1-160K or A1-160Q/M.
DISCUSSION
Here we report a naturally occurring single substitution in E that significantly alters polyclonal neutralization. We observed that A1 DENV-2 lineages circulating in both Cambodia and Vietnam underwent an amino acid substitution at E position 160, making the viruses more susceptible to in vitro polyclonal antibody neutralization, yet more evolutionarily successful. Based on available DENV-2 E gene sequences, 3 separate lineages, 2 with 160M and 1 with 160Q, arose simultaneously in Vietnam and Cambodia in 2006–2007 and, in both countries, increased in relative rate of detection over time. Further, individuals infected with A1-160M and A1-160Q viruses had significantly higher early viremia levels than those infected with A1-160K isolates, which were more likely to occur in secondary infections. Our findings suggest that A1-160Q and A1-160M substitutions confer a fitness advantage that allows them to overcome the fitness cost of being better neutralized by serotype cross-reactive sera. However, the specific causal mechanisms underlying the evolutionary success of the A1-160M and A1-160Q lineages remain elusive.
One scenario is that A1-160Q and, possibly, A1-160M are more successful at replicating in the presence of poorly neutralizing heterotypic antibodies by taking advantage of antibody-dependent enhancement. Globally, DENV-2 is more often isolated from secondary infections [48], and based on our findings, it is plausible that A1-160Q and A1-160M replicate better than A1-160K in DENV-immune individuals. An alternate scenario is that the substitution at position 160 results in an intrinsic fitness advantage in humans and, possibly, mosquitoes, improving viral replication and dissemination independent of preexisting anti-DENV antibodies [7, 17].
It is possible that another mutation in the genome explains the fitness advantage. We searched for other variants in E, but position 160 was the only highly variable position; the next most variant site, position 201, was 96% conserved. Full genomes are not available for the 70 viremic adults we studied. However, for all full-length sequences of Vietnamese DENV-2 Asian 1 viruses in GenBank (n = 127), E position 160 was the only major variant (>20% variation) in the E gene. Two positions in the NS5 gene (one in the methyltransferase and one in the polymerase) were major variants, but distinct amino acids at these positions did not directly correlate with E-160 variants.
In conclusion, the successful emergence and expanded circulation for multiple years in Vietnam and Cambodia of viruses with a substitution in the E protein (K160Q, K160M) that induces increased susceptibility to cross-neutralization calls for an expanded view of the mechanism(s) of selection driving DENV evolution.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank Shayna M. Cave, Molly OhAinle, Claire Quiner, Lisha Wang, and Ignacio Salas, for their contribution to generating the AA and A1-160Q RVP constructs; David Burke, for sharing code for assisting with generation of phylogenetic trees; Philippe Buchy, for providing sequence information about recent Cambodian DENV-2 isolates; Michael Diamond, for MAbs E76, E87, E60, E28, and E18; Federica Sallusto and Antonio Lanzavecchia, for MAbs 87.1 and 82.11; and Angel Balmaseda and the Pediatric Dengue Cohort Study team at the Centro de Salud Sócrates Flores Vivas, the Laboratorio Nacional de Virología of the Centro Nacional de Diagnóstico y Referencia at the Nicaraguan Ministry of Health, and the Sustainable Sciences Institute-Nicaragua, for providing the serum samples from the Nicaraguan cohort study.
E. H. and C. P. S. contributed the samples and clinical data; C. W., L. C. K., and E. H. conceived and designed the experiments; C. W., M. M., and K. D. T. H. performed the experiments; C. W., L. C. K., M. M., and E. H. analyzed the data; and C. W., L. C. K., and E. H. drafted the manuscript; all authors reviewed the manuscript.
Financial support. This work was supported by the Bill and Melinda Gates Foundation and the Instituto Carlos Slim de la Salud (FIRST Program); the Nicaraguan Pediatric Dengue Cohort Study was supported by the Pediatric Dengue Vaccine Initiative (grant VE-1 to E. H.) and the National Institutes of Health (NIH) R01 AI099631 (to Dr Angel Balmaseda); and L. C. K. was supported by a Gates Cambridge Scholarship and the NIH Oxford-Cambridge Scholars Program.
Potential conflicts of interests. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bhatt S, Gething PW, Brady OJ et al. The global distribution and burden of dengue. Nature 2013; 496:504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 2003; 3:19–28. [DOI] [PubMed] [Google Scholar]
- 3.Messina JP, Brady OJ, Scott TW et al. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol 2014; 22:138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochel TJ, Watts DM, Halstead SB et al. Effect of dengue-1 antibodies on American dengue-2 viral infection and dengue haemorrhagic fever. Lancet 2002; 360:310–2. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Mammen MP Jr, Chinnawirotpisan P et al. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol 2005; 79:15123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams B, Holmes EC, Zhang C et al. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc Natl Acad Sci U S A 2006; 103:14234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.OhAinle M, Balmaseda A, Macalalad AR et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med 2011; 3:114ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings DA, Irizarry RA, Huang NE et al. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature 2004; 427:344–7. [DOI] [PubMed] [Google Scholar]
- 9.Sittisombut N, Sistayanarain A, Cardosa MJ et al. Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am J Trop Med Hyg 1997; 57:100–8. [DOI] [PubMed] [Google Scholar]
- 10.Wittke V, Robb TE, Thu HM et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 2002; 301:148–56. [DOI] [PubMed] [Google Scholar]
- 11.Klungthong C, Zhang C, Mammen MP Jr, Ubol S, Holmes EC. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 2004; 329:168–79. [DOI] [PubMed] [Google Scholar]
- 12.Thu HM, Lowry K, Myint TT et al. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg Infect Dis 2004; 10:593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ty Hang VT, Holmes EC, Duong V et al. Emergence of the Asian 1 genotype of dengue virus serotype 2 in viet nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl Trop Dis 2010; 4:e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster JE, Bennett SN, Carrington CV, Vaughan H, McMillan WO. Phylogeography and molecular evolution of dengue 2 in the Caribbean basin, 1981–2000. Virology 2004; 324:48–59. [DOI] [PubMed] [Google Scholar]
- 15.A-Nuegoonpipat A, Berlioz-Arthaud A, Chow V et al. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology 2004; 329:505–12. [DOI] [PubMed] [Google Scholar]
- 16.Hay SI, Myers MF, Burke DS et al. Etiology of interepidemic periods of mosquito-borne disease. Proc Natl Acad Sci U S A 2000; 97:9335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quiner CA, Parameswaran P, Ciota AT et al. Increased replicative fitness of a dengue virus 2 clade in native mosquitoes: potential contribution to a clade replacement event in Nicaragua. J Virol 2014; 88:13125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messer WB, Yount B, Hacker KE et al. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl Trop Dis 2012; 6:e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson N, Anderson R, Gupta S. The effect of antibody-dependent enhancement on the transmission dynamics and persistence of multiple-strain pathogens. Proc Natl Acad Sci U S A 1999; 96:790–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings DA, Schwartz IB, Billings L, Shaw LB, Burke DS. Dynamic effects of antibody-dependent enhancement on the fitness of viruses. Proc Natl Acad Sci U S A 2005; 102:15259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn RJ, Zhang W, Rossmann MG et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 2002; 108:717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 1995; 375:291–8. [DOI] [PubMed] [Google Scholar]
- 23.Beltramello M, Williams KL, Simmons CP et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010; 8:271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Alwis R, Smith SA, Olivarez NP et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 2012; 109:7439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fibriansah G, Tan JL, Smith SA et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun 2015; 6:6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fibriansah G, Ibarra KD, Ng TS et al. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 2015; 349:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 2015; 520:109–13. [DOI] [PubMed] [Google Scholar]
- 28.Brien JD, Austin SK, Sukupolvi-Petty S et al. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol 2010; 84:10630–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SA, de Alwis AR, Kose N et al. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio 2013; 4:e00873–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai CY, Williams KL, Wu YC et al. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl Trop Dis 2013; 7:e2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanBlargan LA, Mukherjee S, Dowd KA, Durbin AP, Whitehead SS, Pierson TC. The type-specific neutralizing antibody response elicited by a dengue vaccine candidate is focused on two amino acids of the envelope protein. PLoS Pathog 2013; 9:e1003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrestha B, Brien JD, Sukupolvi-Petty S et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog 2010; 6:e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochel TJ, Watts DM, Gozalo AS, Ewing DF, Porter KR, Russell KL. Cross-serotype neutralization of dengue virus in Aotus nancymae monkeys. J Infect Dis 2005; 191:1000–4. [DOI] [PubMed] [Google Scholar]
- 34.Katzelnick LC, Fonville JM, Gromowski GD et al. Dengue viruses cluster antigenically but not as discrete serotypes. Science 2015; 349:1338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 2008; 381:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuan G, Gordon A, Aviles W et al. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 2009; 170:120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyet MN, Duong TH, Trung VT et al. Host and viral features of human dengue cases shape the population of infected and infectious Aedes aegypti mosquitoes. Proc Natl Acad Sci U S A 2013; 110:9072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montoya M, Gresh L, Mercado JC et al. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 2013; 7:e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson TC, Sanchez MD, Puffer BA et al. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology 2006; 346:53–65. [DOI] [PubMed] [Google Scholar]
- 40.Mattia K, Puffer BA, Williams KL et al. Dengue reporter virus particles for measuring neutralizing antibodies against each of the four dengue serotypes. PLoS One 2011; 6:e27252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukupolvi-Petty S, Austin SK, Engle M et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 2010; 84:9227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas SJ, Nisalak A, Anderson KB et al. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: How alterations in assay conditions impact performance. Am J Trop Med Hyg 2009; 81:825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon A, Kuan G, Mercado JC et al. The Nicaraguan pediatric dengue cohort study: incidence of inapparent and symptomatic dengue virus infections, 2004–2010. PLoS Negl Trop Dis 2013; 7:e2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balmaseda A, Standish K, Mercado JC et al. Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J Infect Dis 2010; 201:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fibriansah G, Ng TS, Kostyuchenko VA et al. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J Virol 2013; 87:7585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teoh EP, Kukkaro P, Teo EW et al. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 2012; 4:139ra83. [DOI] [PubMed] [Google Scholar]
- 47.Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol 2014; 88:11726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.OhAinle M, Harris E. Dengue pathogenesis: viral factors. In: Gubler DJ, Ooi EE, Vasudevan S, Farrar J, eds. Dengue and dengue hemorrhagic fever. 2nd ed. Wallingford, UK: CABI, 2014:231–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.