Abstract
A recently discovered rhodopsin ion pump (DeNaR, also known as KR2) in the marine bacterium Dokdonia eikasta uses light to pump protons or sodium ions from the cell depending on the ionic composition of the medium. In cells suspended in a KCl solution, DeNaR functions as a light-driven proton pump, whereas in a NaCl solution, DeNaR conducts light-driven sodium ion pumping, a novel activity within the rhodopsin family. These two distinct functions raise the questions of whether the conformations of the protein differ in the presence of K+ or Na+ and whether the helical movements that result in the canonical E → C conformational change in other microbial rhodopsins are conserved in DeNaR. Visible absorption maxima of DeNaR in its unphotolyzed (dark) state show an 8 nm difference between Na+ and K+ in decyl maltopyranoside micelles, indicating an influence of the cations on the retinylidene photoactive site. In addition, electronic paramagnetic resonance (EPR) spectra of the dark states reveal repositioning of helices F and G when K+ is replaced with Na+. Furthermore, the conformational changes assessed by EPR spin–spin dipolar coupling show that the light-induced transmembrane helix movements are very similar to those found in bacteriorhodopsin but are altered by the presence of Na+, resulting in a new feature, the clockwise rotation of helix F. The results establish the first observation of a cation switch controlling the conformations of a microbial rhodopsin and indicate specific interactions of Na+ with the half-channels of DeNaR to open an appropriate path for ion translocation.
The microbial rhodopsin family of membrane proteins conducts light energy conversion and light-induced signal transduction in bacteria, archaea, and unicellular eukaryotes.1–3 All rhodopsins share a conserved topology consisting of seven helical transmembrane segments (A–G) that form a pocket for the chromophore retinal, attached in a protonated Schiff base linkage to a lysine residue in the protein’s core. Light-driven isomerization of the retinal chromophore causes a cyclic series of structural rearrangements in the protein scaffold that result in ion transport or sensory signaling.
The best-characterized microbial rhodopsin with regard to the conformational changes occurring during its photocycle is the light-driven proton pump bacteriorhodopsin (BR) from the archaeon Halobacterium salinarum.4–11 The unphotolyzed (dark) conformation of BR exhibits a half-channel to the external medium and a protonated Schiff base poised for the release of a proton to the exterior-facing channel. This dark form is the E conformer (Figure 1, purple). Light induces release of the proton followed by conversion to the C conformer (Figure 1, yellow), in which (i) a half-channel forms from the retinal chromophore’s Schiff base to the cytoplasm and (ii) the Schiff base switches its connection (i.e., accessibility) to the cytoplasmic side for reprotonation. The alternate access of the Schiff base in the E and C conformers combined with appropriate timing of pKa changes that control Schiff base proton release and uptake makes the proton path through the protein vectorial.
Figure 1.
Cytoplasmic view showing the overlap of the E conformer (Protein Data Bank entry 1C3W, purple) and the C conformer (Protein Data Bank entry 1FBK, yellow) of BR demonstrating the helical movements observed in the photochemical reaction cycle of BR. The retinylidene is shown as red sticks. During the E → C conversion, helix F undergoes an outward tilt with counterclockwise rotation and helix G undergoes a lateral outward movement.
The E → C conformational change in BR entails helix displacements that have been characterized by cryoelectron crystallography, X-ray crystallography, and EPR. The largest change observed is a lateral outward tilt of the cytoplasmic half of helix F.6,12,13 The helix F tilt is accompanied by smaller rearrangements of helices C, E, and G on the cytoplasmic side. The movement of helices F and G with respect to each other and to helix B would be expected to expand the structure on the cytoplasmic side, thereby opening a proton-conducting half-channel to the retinylidene Schiff base.
A novel group of microbial rhodopsins with the ability to perform light-driven Na+ transport have been recently identified.14–16 The Na+ pump function is correlated with changes in the canonical proton donor/acceptor Asp-Thr-Asp (DTD) motif from BR. Three proteins containing an Asn-Asp-Gln (NDQ) motif instead of a DTD motif have been tested and shown to perform Na+ transport.14–18 The first of the reported microbial rhodopsin Na+ pumps, DeNaR (also known as KR2) from the bacterium Dokdonia eikasta (formerly Krokinobacter eikastus), uses light to pump protons or sodium ions from the cell depending on the ionic composition of the medium.14 DeNaR is of great interest for two reasons. First, its dual function provides an opportunity to unravel the mechanistic changes that evolved to alter the ion specificity of rhodopsin primary pumps; DeNaR is the only member of the three tested NDQ Na+ transporters reported to exhibit dual functionality by also driving H+ transport under certain conditions.14 Second, microbial rhodopsins are extensively used as tools for optogenetic control of membrane potential in excitable cells such as neurons19,20 and cardiomyocytes.21,22 Na+ is a crucial ion for membrane potential control in these cells, and therefore, the mechanism of conduction of Na+ by microbial rhodopsins is of practical importance.
DeNaR was demonstrated to also effectively pump Li+, but in the presence of mono- or divalent cations with masses larger than that of Na+, DeNaR becomes a pump for only H+.14 There are two possible explanations for the ion selectivity of DeNaR. (i) The DeNaR protein undergoes only one set of structural changes in its photocycle and opens conduction channels of sufficient diameter for passage of Na+, but not larger cations. Therefore, in the presence of cations larger than Na+, only H+ would be transported. (ii) “Cation tuning” occurs, and a novel set of structural rearrangements during the photochemical reaction cycle yields structurally distinct H+ pump and Na+ pump forms of DeNaR. In this report, we demonstrate difierent effects of Na+ and K+ on the structure of DeNaR in both unphotolyzed and photostationary states. Our findings indicate two key results. (i) Under certain conditions, K+ and Na+ induce small but consistent hypsochromic and bathochromic shifts, respectively, in the absorption spectrum of DeNaR, which indicates that the pigment binds these cations in the unphotolyzed state. Further evidence of cation-specific effects on the dark structure of DeNaR are evident in the line broadening–sharpening patterns in EPR spectra in the dark. (ii) Upon illumination, helix F undergoes an outward displacement similar to that of BR in the presence of both Na+ and K+, but with an opposite rotation in the presence of Na+ that is not observed in samples containing K+.
MATERIALS AND METHODS
Overexpression and Purification of Recombinant DeNaR
The gene encoding wild-type D. eikasta sodium-pumping rhodopsin (DeNaR) was synthesized by Genewiz, Inc. (Research Triangle Park, NC). Expression vector pET-24b(+) and BL21(DE3) chemically competent Escherichia coli were from Novagen (San Diego, CA). The XL-10 Gold chemically competent E. coli and Quik-Change XL site-directed muta-genesis kit were from Agilent (Santa Clara, CA), and primers, imidazole, kanamycin sulfate, all trans-retinal, n-methyl-glucamine (NMG+), tetraphenylphosphonium chloride (TPP+), carbonyl cyanide m-chlorophenyl hydrazone, MgCl2, MOPS, and Bis-Tris propane were from Sigma-Aldrich (St. Louis, MO). Enzymes were from New England Biolabs (Ipswich, MA). Amicon YM-10 centrifugal filters were from EMD Millipore (Billerica, MA). The plasmid preparation kit was from Qiagen (Valencia, CA). Ni-NTA agarose, glycerol, Tris-HCl, acetonitrile, Luria-Bertani broth, Slide-a-Lyzer 10000 MWCO dialysis cassettes, NaCl, and KCl were from Fisher (Swanee, GA). n-Decyl β-d-maltopyranoside (DM) and n-dodecyl β-d-maltopyranoside (DDM) were from Anatrace (Maumee, OH), and 10-DG desalting columns were from Bio-Rad (Hercules, CA). S-(1-Oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanethiosulfonate (MTSSL) was from Santa Cruz Biotechnology (Dallas, TX).
The gene encoding DeNaR was cloned into the pET-24b(+) plasmid and confirmed by sequencing (Genewiz, Inc.). The DeNaR construct was transformed into BL21(DE3) competent cells, plated onto LB-agar containing 50 μg/mL kanamycin, and incubated at 37 °C overnight. Starter cultures were made by picking single colonies and inoculating 5 mL of LB broth containing 50 μg/mL kanamycin with rotary shaking at 200 rpm and 37 °C overnight. Starter cultures were used to inoculate 100 mL of LB broth cultures containing 50 μg/mL kanamycin and shaking at 200 rpm and 37 °C. One liter LB broth cultures containing 50 μg/mL kanamycin were inoculated using 20 mL of the overnight 100 mL cultures and propagated to an OD600 of 0.4. IPTG was added to a final concentration of 1.0 mM and all trans-retinal (ATR) to a final concentration of 5.0 μM, and the cultures were incubated at 37 °C and 200 rpm for 2.0 h.
The bacteria containing recombinant DeNaR were pelleted at 4000g and 4 °C and resuspended in 50.0 mM Tris-HCl containing 15% glycerol (pH 8.0). Cells were broken using a microfluidizer, and membranes were collected by centrifugation at 100000g and 4 °C for 1.0 h. Each liter of harvested cells was resuspended in 10 mL of 50.0 mM Tris containing 1.5% (w/v) DM or DDM, 15% glycerol, and 500 mM NaCl (pH 8.0), and the mixture was incubated overnight at 4 °C for extraction. The protein extract was centrifuged at 120000g and 4 °C for 1.0 h and the supernatant collected. Ni-NTA resin (1.0 mL/L of culture) equilibrated with 50 mM Tris containing 0.2% (w/v) DM, 15% glycerol, and 500 mM NaCl (pH 8.0) was added to the protein extract and the mixture incubated for 2.0 h at 4 °C. The resin containing bound protein was washed with 5 volumes of 50 mM Tris containing 0.2% (w/v) DM, 15% glycerol, and 500 mM NaCl (pH 8.0) and then 10 volumes of 50 mM Tris containing 0.2% (w/v) DM, 50 mM imidazole, 15% glycerol, and 100 mM NaCl (pH 8.0). The protein was eluted with 5 volumes of 50 mM Bis-Tris containing 0.2% (w/v) DM, 200 mM imidazole, 15% glycerol, and 100 mM NaCl (pH 8.0) and concentrated using Amicon YM-10 centrifugal filters, and imidazole was removed using a 10-DG desalting column. Expression and purification were monitored using 12.5% SDS–PAGE. A typical yield was 3 mg of highly purified DeNaR protein per liter of culture. For samples in DDM, the purification procedures were identical except 0.1% (w/v) DDM was used after extraction.
Preparation of Cysteine Mutants
Positions on helices B, F, and G of DeNaR were selected using a combination of primary sequence alignment and in silico structural prediction using the Robetta server software.23 Site-directed mutagenesis was performed with the QuikChange kit using the wild-type DeNaR construct as a template. DNA sequencing was used to check the mutants, followed by expression and purification as described above. UV–Vis spectroscopy and flash photolysis were used to test the properties of each single- and double-Cys mutant after labeling with MTSSL (see below), and only mutants with photochemical and proton translocation properties similar to those of wild-type DeNaR were used for EPR studies.
Proton Flux Measurements
Freshly harvested E. coli from 1.0 L cultures expressing either wild-type DeNaR or the cysteine mutants were washed with 100 mM NaCl or 100 mM KCl three times by centrifugation and resuspended in 15 mL of 100 mM NaCl, 10 mM MgSO4, 0.1 mM CaCl2, and either 100 mM NaCl or KCl. The OD600 for proton flux measurements was 10. The light source for the illumination of the samples was a 21 V, 150 W tungsten–halogen illuminator (Cole Parmer, Vernon Hills, IL). The light was passed through a heat filter and one long pass >520 nm colored glass filter and focused onto the sample in a cuvette with vigorous magnetic stirring. pH changes were measured by a Ross glass combination micro pH electrode and an Orion Star A211 pH meter (Thermo, Chelmsford, MA) connected to a computer.
Preparation of Cation-Free Samples
For cation-free EPR and UV–Vis sample preparation, sodium was removed from samples by extensive dialysis against a 1000-fold excess of 50 mM Bis-Tris containing 0.2% (w/v) DM and 15% glycerol (pH 6.0), with a total of five buffer changes for 24 h at 4 °C. For samples in DDM buffers, the removal of sodium was identical except 0.1% (w/v) DDM was used.
UV–Vis Spectroscopy and Flash Photolysis
UV–Vis spectra were recorded using a Varian CARY 4000 spectrophotometer equipped with an integrating sphere. Changes in the UV–Vis spectrum of DeNaR upon addition of K+, Na+, NMG+, or Mg2+ to cation-free samples were monitored from 300 to 850 nm. Flash-induced absorption changes in samples containing spin-labeled single- and double-cysteine mutant DeNaR to be used for the EPR measurements were acquired with a laboratory-constructed cross-beam laser flash photolysis system as described previously24 but with a GaGe Octopus digitizer board (model CS8327, DynamicSignals LLC, Lockport, IL) at a sampling rate of up to 40 ns/point. All comparative UV–Vis and flash photolysis data were collected from the same initial batch of salt-free protein with cations added prior to data collection to avoid sample variation. Data analysis was performed with OriginPro version 9.1 (OriginLab, Northampton, MA).
Site-Directed Spin Labeling EPR Measurements
Samples for EPR were prepared by the addition of a 20-fold molar excess of MTSSL to each protein sample [MTSSL dissolved in acetonitrile to a final concentration of 200.0 mM and then diluted in 50.0 mM Bis-Tris containing 0.2% (w/v) DM, 15% glycerol, and 100.0 mM NaCl (pH 6.6) to a final concentration of 5.0 mM prior to addition to protein samples]. The spin-labeling reaction was allowed to proceed for 16 h at 4 °C while the sample was being gently rotated. Excess MTSSL was removed by extensive dialysis as described above. Spin-labeled samples were tested using UV–Vis and flash photolysis to ensure behavior similar to that of wild-type DeNaR.
EPR spectra were recorded on a Bruker EMX X-band spectrometer using a modulation amplitude of 2.0 G, a modulation frequency of 100 kHz, and a microwave power of 16 mW for room-temperature measurements and 0.1 mW for frozen samples. Room-temperature spectra were recorded in a 50 μL capillary tube placed at the center of a 5.0 mm outer diameter quartz EPR tube (KIMAX-51, Gerreshimer, Quretaro, Mexico). Illumination of the sample was achieved with an illuminator (FOI-250, Titan Tool Supply Inc., Buffalo, NY) with a 250 W tungsten–halogen lamp equipped with a light guide. The light beam was passed through a heat filter and a long-pass filter transmitting >520 nm light through the front window of the 4119HS resonator. For liquid N2 EPR experiments, the sample was illuminated for 5 min inside a 5.0 mm tube using the combination of light source and filters described above. The EPR tube containing the sample was kept in a 15 mL conical tube lined with foil to create a reffective surface, and actinic light was applied from above. After illumination, ethanol from an ethanol/dry ice bath was poured around the sample tube to quickly freeze the sample while under continuous illumination and then quickly transferred to a liquid N2 Dewar prior to placement in the instrument. All EPR data were collected from the same initial batch of salt-free protein with cations added prior to data collection to avoid sample variation. All data acquisition and analyses were conducted using WinEPR.
RESULTS
Light-Induced pH Changes in E. coli Suspensions
Following the protocol and criteria of Inoue et al.,14 we confirmed the Na+ and H+ pumping activities of wild-type DeNaR expressed in E. coli cells (Figure S1 of the Supporting Information). The results confirmed the function of DeNaR as an outward H+ pump in the presence of K+, becoming a Na+ pump when Na+ is substituted for K+ as previously reported.14 The key criteria used to establish light-induced Na+ pumping based on pH measurements of cell suspensions are (i) a light-induced pH increase corresponding to flux of protons into the cells, (ii) CCCP enhancement of the proton influx showing that proton movement is passive, (iii) TPP+ elimination of the passive proton flux, showing that the flux was derived from electrical hyperpolarization of the membrane, (iv) dependence of the CCCP-enhanced proton influx on Na+, and (v) independence of the nature of the anion in suspension. The criteria for light-induced H+ pumping are (i) light-induced outward proton flux and (ii) eliminated by CCCP.
These criteria were tested to confirm that our recombinant system expresses DeNaR with transport activity as reported by Inoue et al.,14 and in later experiments, where mutations were introduced, as a benchmark for proper wild-type DeNaR behavior. We also confirmed the previously reported pumping behavior of DeNaR in the presence of alkali metals14 (data not shown). DeNaR behaves as a H+ pump in the presence of all group I cations, except for Li+ and Na+. We also confirmed that in our recombinant system DeNaR is also a H+ pump in the presence of all group II cations, which further supports the dual function of DeNaR as a H+ pump and a Na+ pump. For purposes of discussion, DeNaR in the presence of K+ or Na+ will be termed the H+ pump form or Na+ pump form, respectively.
Effects of Cations on the Unphotolyzed (dark) State of DeNaR
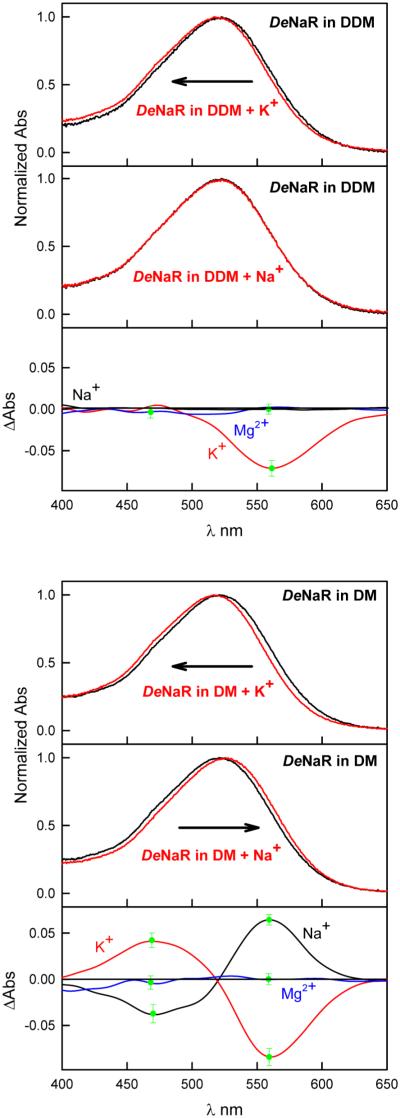
In purified recombinant wild-type DeNaR in detergent micelles, we observed a red shift in the visible absorption maximum (λmax) caused by replacement of K+ with Na+, shifting the λmax from 517 nm in the presence of 100 mM KCl to 525 nm in DeNaR samples containing 100 mM Na+ (Figure 2 and Figure S2 of the Supporting Information). We considered the possibility that the 8 nm red shift caused by the substitution of K+ with Na+ was due to an ionic strength effect. To determine whether this was the case, we substituted NMG+, a large cation that is not pumped by DeNaR, for Na+ and K+. NMG+ did not change the UV–Vis spectrum from that of the salt-free condition (Figure S2 of the Supporting Information). Further confirmation that the effects of K+ and Na+ on the UV–Vis spectrum of DeNaR were not due to a salt effect was obtained by adding Mg2+ to a cation-free sample of the pigment. The absorption spectrum of DeNaR in the presence of Mg2+ is indistinguishable from the cation-free sample (Figure 2).
Figure 2.
Comparison of the effect of the cation on the UV–Vis spectrum of DeNaR in DM and DDM micelles. In each case, the absorption spectra are the average of wavelength scans from four independent measurements normalized to their absorption at their λmax. The difierence spectra are from the spectra above fast Fourier-transform-smoothed for the sake of clarity. The green bars at peaks in the difierence spectra indicate the standard error of the mean at that wavelength.
Inoue et al. reported that spectral shifts were not observed in their DeNaR samples.14 A primary difierence between our measurements and those reported by Inoue and co-workers was their use of DDM instead of DM for extraction and subsequent experimental procedures. We repeated our measurements using DDM for extraction and confirmed that the DeNaR visible spectrum in DDM detergent micelles is not sensitive to Na+ or Mg2+ (Figure 2) as reported by Inoue et al.14 We observed a similar blue shift in the optical spectrum of DeNaR in the presence of K+ (Figure 2) in both DM- and DDM-extracted pigment. The difierences in absorption spectra do not answer the question of whether there is a cation-induced structural difierence in the dark-state protein, which we investigated by SDSL EPR measurements (next section).
Expression and Characterization of Cys Mutants for SDSL
Site-directed spin labeling and EPR have been used to map helix rearrangements during the photocycle of several microbial rhodopsins, including the prokaryotic pigments BR,4,5,7,25 sensory rhodopsins I and II,26,27 and more recently extended to an algal channelrhodopsin.28,29 The approach entails nitroxide spin labeling of endogenous or introduced Cys residues within the scaffold of the protein, using interactions of Cys-specific spin-labels (MTSSL) to triangulate the direction of helix movements. In BR and SRII, light-induced helix movements are nearly identical and are most evident by comparison of sites on helices B, F, and G in pairwise combinations. Because the prokaryotic DeNaR sequence is most similar to that of other prokaryotic opsins, we selected the helix positions for substitution of Cys on the hypothesis that DeNaR undergoes light-induced helix movements in its H+ pump form similar to those in BR and SRII. As in BR and SRII, there are no endogenous Cys residues in DeNaR.
To guide the selection of positions on the protein that would be suitable for mutations, we used the Rosetta structural prediction software30 in the Robetta Full-Chain Protein Structure Prediction Server, to identify the rhodopsin template structure that most closely matches DeNaR and predict structural models of DeNaR. These models were used to select positions for the introduction of single- and double-Cys mutations. Robetta predicted a DeNaR structure based on xanthorhodopsin31 (Protein Data Bank entry 3DDL) by homology modeling (Figure S3 of the Supporting Information). According to the phylogenetic analysis by Inoue et al.,14 DeNaR is most closely related to xanthorhodopsin than to other microbial rhodopsin pumps. Using a combination of sequence alignment and our in silico structural model, we chose 16 difierent positions on helices B (F57, S60, N61, I62, L63, and V67), F (I203, L204, S205, N206, I207, and W208), and G (L262, G263, N264, and L265) for single-point mutations. These single-Cys mutants were expressed in E. coli and tested for transport activity by light-induced proton flux measurements. Of the 16, only V67C showed poor expression under our conditions. Expression levels of the mutants were used as a screening factor, and poor expression (i.e., <80% compared to that of wild-type DeNaR) was considered as an indication of improper folding.
We next identified the single Cys mutations that do not perturb the pumping activity and show normal photochemistry by flash photolysis (Figures S4 and S5 of the Supporting Information) and purified the single-mutant DeNaR proteins for SDSL experiments. Room-temperature CW EPR was used to screen which single-Cys mutants showed good immobilization of the MTSSL spin-label identified as considerable line broadening when compared to free spin-label (data not shown). Many of the single-Cys mutants (F57C, N61C, I62C, L63C, L204C, S205C, N206C, W208C, L262C, and G263C), although functional in ion transport and expressed at levels similar to that of wild-type DeNaR, exhibited EPR spectra with high mobility of the spin-label very similar to that of free spin-label and were excluded.
The mutants with wild-type activity and good MTSSL immobilization based on signal broadening observed in the EPR spectra were used as templates for mutations on a second helical position. We made several pairs of mutants to generate a library of double mutants to cover B/F, B/G, and F/G helical pairs. The double mutants were tested for expression level, ion pumping, and flash photolysis (Figures S4 and S6 of the Supporting Information). Finally the double-mutant pairs B60/F203, B60/F207, B60/G264, B60/F265, F203/G264, F203/G265, F207/G264, and F207/G265 (Figure 3; bold letters indicate helix) were selected as the final candidates for EPR studies because their function most closely resembled that of the wild-type protein and their spin labeling resulted in samples with suitably immobile probes. The room-temperature EPR spectra of these Cys mutant pairs all showed significant signal broadening consistent with good immobilization of the spin-label via ligation with the protein, in combination with line broadening effects caused by dipolar coupling of the nitroxide spin-labels at each position.32 To eliminate the possibility that the broadening and sharpening patterns in the room-temperature EPR experiments had significant contributions from the rotational motion of the spin-label, we selected four mutants with the highest signal intensity for liquid N2 EPR experiments (Figure S7 of the Supporting Information). Our results show that the broadening and sharpening patterns were consistent upon comparison of the signals at room temperature to those collected at liquid N2 temperatures. Moreover, the mobility of the spin-label headgroup is not a significant contributor to our room-temperature spectra because the rotational correlation time is mostly dictated by the protein motion. Because our samples are detergent-solubilized, the apparent molecular weight is much larger than that predicted by a DeNaR monomer. The rotational correlation time estimated for a 2.5 kDa molecule yields approximately 1 ns based on the Stokes–Einstein relationship.33 We routinely use spin columns with a molecular weight cutoff of 100 kDa without protein loss, which suggests that in detergent micelles the effective molecular weight of the protein is large enough so that the major contribution to the line broadening is expected to be from the dipolar interaction of the MTSSL labels. Moreover, the outermost hyperfine line separation in the room-temperature EPR spectra of all singly labeled mutants ranged from 66 to 68 G, near the value of Az, suggesting that the mobility of the label is not a concern in our measurements.
Figure 3.
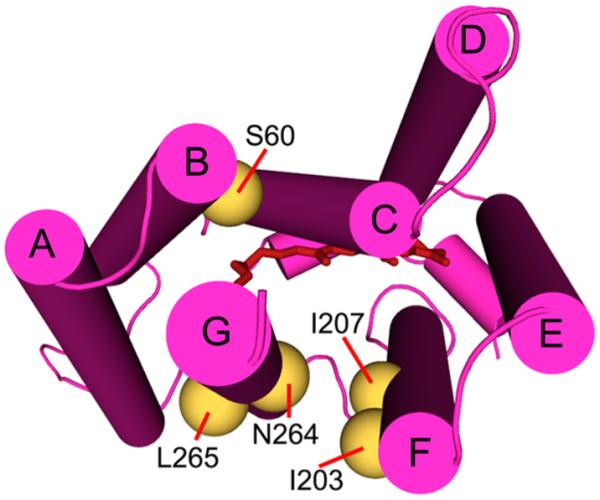
Cytoplasmic view of the predicted structure of DeNaR with the residues chosen for Cys mutations used in SDSL experiments shown as gold spheres.
Cation-Specific Helical Arrangement in the Dark State
Line shape broadening and sharpening in CW EPR measurements are used as fingerprints for the interaction of two magnetically coupled spin-labels. The degree of broadness of the line shape of an EPR spectrum is a function of solvent accessibility and distance between two labels. As the distance between two spin-labels decreases, the dipolar coupling (HD) between two nitroxide spins follows:
where μe is the electron magnetic dipole moment, θ is the angle between the external magnetic field and the vector connecting the two interacting spins, and r is the distance between them. Thus, the EPR signal broadening due to dipolar interaction of a specific spin pair is a very sensitive function of distance, inversely proportional to the third power.
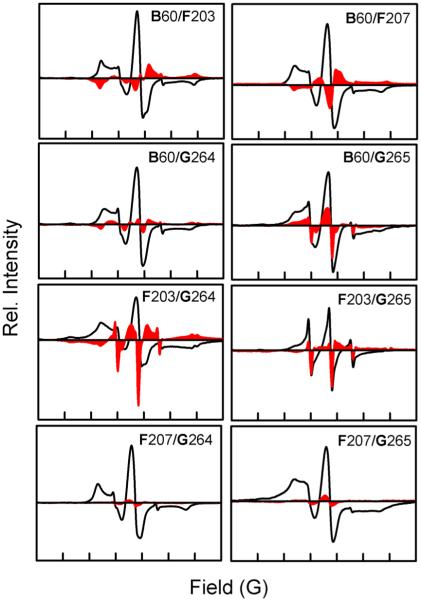
Considering the dual function observed in DeNaR, we used spin labeling to test whether the H+ pump and Na+ pump forms of the protein have a structurally difierent initial dark state. Difierence EPR spectra between the dark states of H+ and Na+ pump forms of DeNaR reveal that DeNaR undergoes a helical rearrangement caused by the substitution of Na+ for K+ in the dark. First, helix F tilts inward toward helix B when Na+ substitutes for K+. This conclusion was based on line broadening (i.e., the difierence signal intensity is in the opposite direction of the original signal) observed in the K+ minus Na+ difierence dark EPR spectra (Figure 4) for in the B60/F203 and B60/F207 mutants. Because both of these mutants show a signal broadening when Na+ substitutes for K+, we can also conclude that there is no significant rotational motion between helices B and F.
Figure 4.
Effect of cation on the initial helical organization of DeNaR during the dark state. The solid black lines are the dark-state EPR spectra of DeNaR in the presence of K+; the red areas represent the difierence spectra of the EPR spectra of DeNaR in the presence of K+ minus DeNaR in the presence of Na+. Bold letters indicate helices. The width of the abscissa in each of the panels represents 150 G. All first-derivative EPR spectra were normalized to a constant spin concentration prior to subtraction.
A second difierence is that in the Na+ pump form of DeNaR the distance between helices B and G is larger than in the H+ pump form. The line sharpening effect (i.e., signal intensity is in the same direction of the original spectrum) is observed in the B60/G264 and B60/G265 mutants (Figure 4), again indicating helix tilting. Note that because of the proximity of residues 264 and 265 on helix G, it is dificult to detect rotational changes in the position of that helix, and further studies will be needed to determine if such a change exists. In our attempts to produce spin-labeled DeNaR mutants that were both functionally similar to the wild-type protein and showed good immobilization of the MTSSL nitroxide label, residues 264 and 265 were the only positions on helix G in the selected region that met our criteria.
Third, the substitution of K+ with Na+ also causes an effect on the relative positions of helices F and G. The dark-state spectrum of the F203/G264 mutant shows a slight broadening effect that is reffected on a shifted spectrum, whereas in the F203/G265 mutant, sharpening is observed. Line sharpening is observed also in both the F207/G264 and F207/G265 mutants. This pattern of broadening and sharpening indicates that helix F moves toward helix G with a slight clockwise rotation because the broadening observed for the F203/G264 mutant does not result in a full flip of the difierence EPR signal and is demonstrated more as a shift, and the intensities of the signal sharpening observed for the F207/G264 and F207/G265 mutants are quite small in comparison.
Cation-Specific Light-Induced Conformational Changes
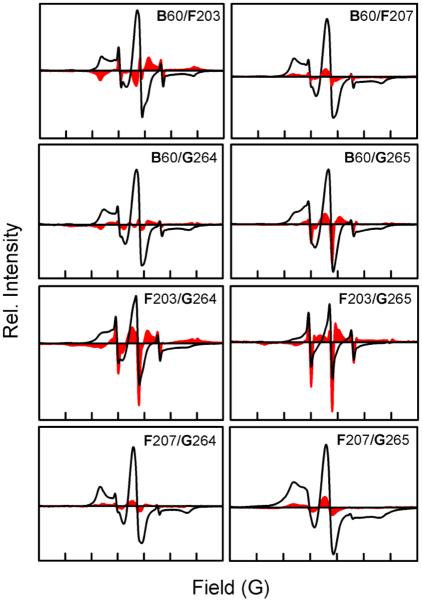
The results mentioned above demonstrate that the H+ pump and Na+ pump forms of DeNaR exhibit a cation-specific structural rearrangement between their unphotolyzed (dark) states. This observation suggested that further difierences might exist in the set of light-induced helical movements for each form of DeNaR. Confirming this possibility, we observed a cation-specific modulation of light-induced helix motion in DeNaR, evident in the movement of helix F with respect to helix B in the illuminated photostationary state. Analysis of the light-minus-dark difierence EPR spectra of the H+ pump form of DeNaR shows signal sharpening in the B60/F203 mutant (Figure 5). When Na+ substitutes for K+, helix F undergoes clockwise rotation as demonstrated by the line broadening observed in the B60/F203 mutant and the contrasting sharpening observed in the B60/F207 mutant, as evident in the K+-minus-Na+ difierence spectra (Figure 6). An outward displacement of helix G is also observed in the EPR spectra for both forms of the protein, as demonstrated by the sharpening of the EPR spectra upon illumination of the B60/G264 and B60/G265 mutants. The difierence spectra of K+ minus Na+ for both B/G helical pairs also show sharpening, which indicates that helix G undergoes a light-induced outward movement with respect to helix B in the Na+-pumping form of DeNaR.
Figure 5.
Room-temperature CW EPR spectra of helix B/G, B/F, and F/G spin-labeled mutants in the presence of Na+ or K+. The solid black lines are the dark spectra, and the red areas are the light-minus-dark difierence spectra. The width of the abscissa axis in each of the panels represents 150 G.
Figure 6.
Effect of cation on the helical movement of DeNaR during the photostationary illuminated state. The solid black lines are the dark-state EPR spectra of DeNaR in the presence of K+; the red areas represent the difierence spectra of the illuminated EPR spectra of DeNaR in the presence of K+ minus DeNaR in the presence of Na+. Bold letters indicate helices. The width of the abscissa axis in each of the panels represents 150 G. All first-derivative EPR spectra were normalized to a constant spin concentration prior to subtraction.
With mixing of the anisotropic g and A tensors into the X-band EPR spectrum and the compounding mobility of the nitroxide probe and the labeled protein, the room-temperature EPR spectrum of the spin-label is intrinsically complex.34 To confirm our findings from the room-temperature experiments, we performed duplicate EPR measurements under cryogenic conditions for four doubly labeled mutants showing the largest signals using frozen powdered samples by fitting to the resolved broadening function contributed by dipolar interaction only, to quantify only the protein conformational changes induced by the specific cations (Figure S7 of the Supporting Information). We can establish an approximation of the effect of cations on the helical movements in DeNaR by analyzing the broadening and sharpening effects of the EPR line shapes in the presence of K+ or Na+. The helical movements can be summarized as modulation of helices F and G by the presence of K+ versus Na+. Our interpretation is that DeNaR organizes itself into difierent conformations for transport of H+ or Na+ in the dark state (Figure 4), as shown above, and that the two conformations exhibit difierences in helix motions during illumination (Figures 5 and 6). Overall, both forms exhibit helix movements similar to that of the canonical rhodopsin H+ pump BR, and the difierences observed can be described as cation-modified E → C conformer conversions.3
DISCUSSION
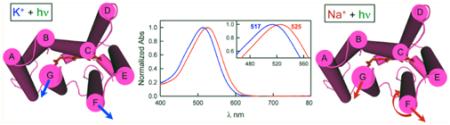
The first indications of cation binding of DeNaR that we observed were the small shifts in the visible spectrum in the presence of Na+ (λmax = 525 nm) relative to the spectra in the presence of K+ (λmax = 517 nm) and a salt-free solution of the protein (λmax = 521 nm) (Figure 2 and Figure S2 of the Supporting Information). These shifts indicate either (i) a direct effect of cation binding near the retinal Schiff base (RSB) chromophore altering its charge environment or (ii) an allosteric effect on the chromophore, possibly caused by the cation-specific helical repositioning that we observed in the unphotolyzed state. The spectral shifts are not an effect of ionic strength changes because the UV–Vis spectrum of DeNaR in a salt-free solution is indistinguishable from that of DeNaR in the presence of NMG+ and Mg2+ (Figure 2 and Figure S2 of the Supporting Information), which we interpret as a lack of accessibility of those large cations to binding sites accessible to Na+ and K+. Our findings therefore show a cation-specific effect on the optical spectrum of DeNaR. Binding near the protonated Schiff base (PSB) is an appealing possibility because retinal photoisomerization-induced PSB proton transfer or proton repositioning1–3,8 is a central part of the mechanism for transport of other ions by microbial rhodopsins. If the spectral shifts were due to a direct effect on the chromophore, the data would imply size-limited specific interaction site(s) near the PSB.
Mutations in DeNaR characterized by H+ flux measurements, optical and vibrational spectroscopy, and laser flash photolysis led to the conclusion that unphotolyzed DeNaR contains a Na+ binding site located on the extracellular side of the protein but that binding of Na+ to DeNaR does not alter its visible absorption spectrum.14,35 The difierences between our finding of a Na+-induced absorption spectrum shift and these findings appear to be largely due to difierences in the detergent used. The measurements we reported above used DeNaR samples prepared in DM micelles, whereas the previously published work14,35 used DDM. Accordingly, we repeated the UV–Vis experiments using DDM-extracted protein. We confirm that as previously reported,14 Na+ does not affect the UV–Vis spectrum of DeNaR in DDM micelles (Figure 2). In our measurements, K+ induced a slight blue shift in the optical spectrum of DeNaR in both DM and DDM micelles. Addition of Mg2+ did not affect the optical spectra of DeNaR under our conditions. Our results indicate the following. (i) In DM micelles, a Na+ binding site is accessible in DeNaR in the unphotolyzed state, whereas in DDM, this site is not accessible or the bathochromic shift is prevented by a difierence in the structure of the protein in this detergent. (ii) K+ binds to DeNaR in both DM and DDM micelles. (iii) Na+ and K+ induce bathochromic and hypsochromic shifts, respectively, in the optical spectrum of DeNaR that are not due to a salt/ionic strength effect, as evidenced by the lack of spectral shifts in the presence of NMG+ and Mg2+. The Na+ binding we observed has important mechanistic implications, namely that DeNaR has evolved to selectively recognize and pump Na+. Binding of Na+ near the RSB was recently proposed on the basis of a high-affinity site for Na+ forming transiently during the photochemical reaction cycle (photocycle) of the homologous light-driven Na+ pump from Gillisia limnaea (GLR, in our nomenclature GlNaR).16
In BR, there is an outward tilting and counterclockwise rotation of helix F and outward movement of helix G that take place in late stages of the photocycle after deprotonation of the Schiff base (Figure 1).3 These movements lead to the conversion of the extracellularly open E conformer to the cytoplasmically open C conformer. SRII exhibits the same canonical helix motions indicating an E → C conformer conversion25–27 despite being a signaling protein rather than a transporter, a striking similarity that is understood in terms of the shared features of the respective mechanisms of BR and SRII.36 Light-induced displacement/rotation of helix F in the photocycle of the SRI–HtrI attractant signaling is in the direction that is the opposite of that in BR and SRII, which agrees with other data showing that SRI undergoes a C → E conversion in the SRI–HtrI molecular complex.24,26,37
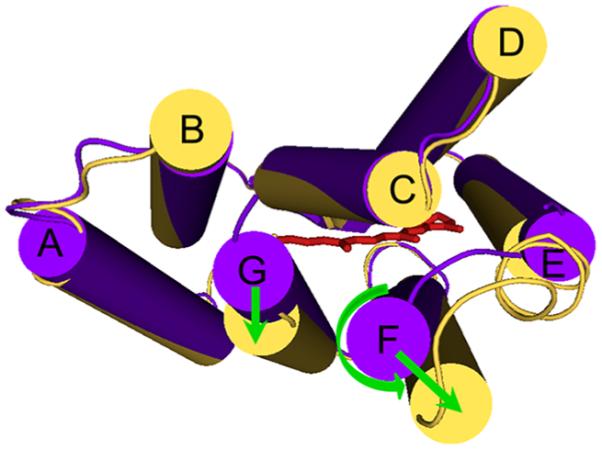
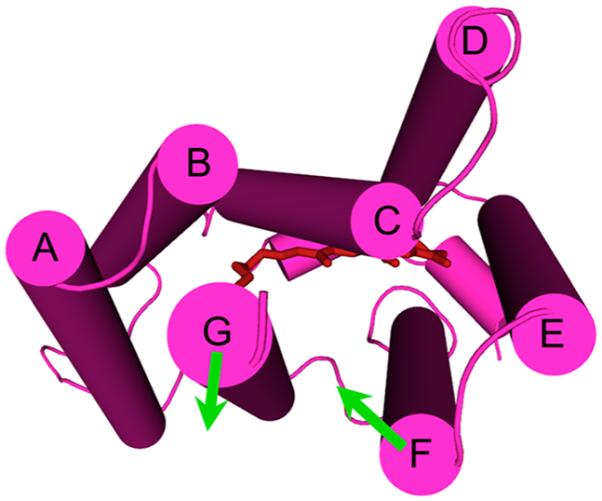
A priori, from its H+ pumping BR-like functionality in the absence of Na+, one would expect that DeNaR in K+ undergoes an E → C conformational change similar to that of BR. Comparing the light-induced helix movements of the proton pump form of DeNaR (Figure 5) with those of BR confirms their expected similarity in that the principal movements in BR, lateral tilts of helices F and G away from the protein’s center that open the cytoplasmic half-channel (Figure 1), occur in DeNaR to similar extents. Difierences appear in the presence of Na+ both in the positions of these two functionally crucial helices in the dark state and in their movements during the light-induced conformational change. In the dark state, the Na+ pump form is altered by helix F tilting inward toward helices G and B and helix G tilting away from helix B toward helix F (Figure 7). As in the H+ pumps, both helices F and G move laterally outward during the photocycle. However, in contrast to the counterclockwise rotation of helix F exhibited by BR (and SRII), helix F in the DeNaR Na+ pump form undergoes a clockwise rotation (Figure 8). A possible consequence of Na+ tuning of the H+ transport mechanism on cytoplasmic-side helix positions is a rearrangement of helices F and G in the dark, preparing the molecule for an altered light-induced channel opening accommodating transport of the larger Na+ ion. In this model, the difierence in the helix positions in the dark state and their altered motion would be responsible for the opening of half-channels for selective H+ or Na+ conduction.
Figure 7.
Structural summary of the helical rearrangement in DeNaR caused by substitution of Na+ for K+ in the dark (cytoplasmic view). The difierence EPR spectra in the unphotolyzed state show that Na+ causes a movement of helix F toward the center of the protein, with a movement of helix G away from helix B and toward helix F.
Figure 8.
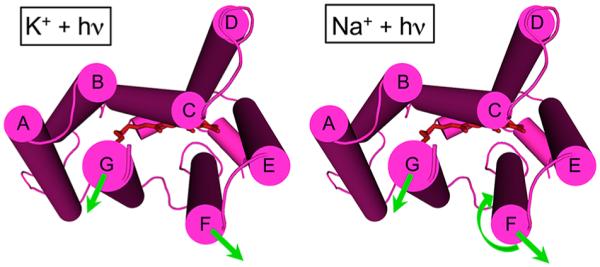
Structural summary of the light-induced helical movements observed in DeNaR. In the presence of K+ (left), we observed an outward lateral tilt of helix F away from both helices B and G and an outward lateral tilt of helix G (cytoplasmic view). In the presence of Na+ (right), we observed an outward movement of helix F, an outward movement of helix G, and a clockwise rotation of helix F relative to helix G. In the presence of K+, DeNaR undergoes a BR-like rearrangement that is consistent with the H+ pumping activity observed under those conditions. In the presence of Na+, helix F undergoes a rotation that is the opposite of that in BR.
Light-induced outward displacement of helix F and to a lesser extent helix G occurs in the cytoplasmic half-channel of BR as the largest changes during the E → C conformer conversion11,38 (Figure 1). SRI and SRII have been shown to undergo a similar movement of helix F in their light-induced conversions between E and C conformers.26,27 A two-conformer model of BR is supported well by the library of structures obtained from structures of spectral intermediates in the BR photocycle.39 The structures of the dark-state conformation and the early intermediates K and L are approximated well by the E conformer, while the structures of late intermediates M–O are approximated well by the C conformer. The conformational change is believed to occur during the lifetime of the M intermediate in a spectrally silent M1 → M2 conversion.38 We observe that in both K+- and Na+-bound forms of DeNaR, the outward movements of helices F and G match those of the E → C conformer conversion of BR. This result provides confidence that we are observing the analogous conformational change in the light-minus-dark EPR difierence spectra. In the Na+-bound form but not in the K+-bound form, we see a rotation of helix F that is the opposite of that observed in EPR measurements with spin-labels at the corresponding positions in BR.
The rotation difierence between the K+- and Na+-bound forms may be due to this helix movement not occurring in the K+-bound form. However, we cannot exclude the possibility that the known difierent photocycle kinetics in the Na+- and K+-bound forms14 lead to accumulation of difierent C conformer substates in the light because the conversion from the E to C conformer may not be concerted but may involve a sequence of steps. The difierence in the dark structures we observed suggests that the light-induced conformational changes may also exhibit difierences, but we cannot distinguish between an absolute difierence and a difierence in conformational substate accumulation in photostationary-state measurements.
The cation tuning of structure exhibited by DeNaR is a unique observation among microbial rhodopsins; however, there are other transmembrane proteins that exhibit an analogous phenomenon. The well-characterized potassium channel from Streptomyces lividans (KcsA)40 binds both K+ and Na+, each of which induces conformational changes in the protein that alter its conductive state.41 Na+ ions increase the helical content of KcsA with respect to that in K+, rendering it nonconductive.40 The cation selectivity of KcsA involves not only Asp and Glu residues and backbone carbonyl groups in the pore but also a selectivity filter at the mouth of the channel.42–44 The selectivity filter of KcsA entails the protein assuming a difierent conformation upon Na+ binding (i.e., cation tuning analogous to that of DeNaR) that prevents K+ transport. Another example is the cation-driven rabbit isoform of the Na+/glucose cotransporter SGLT1.45 In SGLT1, the protein’s transport activity is modulated by H+, Li+, and Na+;46 however, it is only in the presence of Na+ that full activation of the transporter is achieved. DeNaR also shows a similar promiscuity, as it is able to transport Li+ and Na+ and becomes a H+ pump in the presence of K+.14
While this work was under review, two simultaneous reports of crystal structures of dark-state DeNaR were published.47,48 Two aspects of the structures are of direct relevance to the interpretation of the work presented here. First, the predicted structure we used as a basis for the selection of mutants for the SDSL EPR measurements matches closely the published structures, with the helix positions and the residues used for spin labeling overlapping almost perfectly (Figure S8 of the Supporting Information). Hence, the crystal structures validate our predicted dark structure, confirming the conformation of DeNaR to be nearly identical to that of the E conformer of BR, and validate our interpretation of the relative positions of the residues used for spin labeling. Second, neither study reported internally bound Na+, but the acidic pH of the mother liquor solutions used for crystallization may have precluded sodium binding. Also it is challenging to distinguish water from Na+ even at the high resolutions obtained by both groups. Gushchin and co-workers47 identified a well-defined sodium binding site at the DeNaR oligomerization interface in their crystals of DeNaR pentamers. The ion is bound to the Tyr25 side chain and Thr83 and Phe86 backbone oxygens of one protomer and to the Asp102 side chain of the other protomer. Cross protomer interaction (hydrogen-bonding rather than ion-mediated) has been observed in crystals of proteorhodopsin oligomers (pentamers and hexamers) and shown to have a functional role in ion (proton) transport in that protein.49 The question of whether the Na+ binding observed in the crystal structures is responsible for the Na+-induced absorption shift we report here remains open. In the former case, the absorption effect would be expected to be indirect because it is not near the retinylidene chromophore.
Our observations indicate that DeNaR has evolved a unique set of helical rearrangements to support its dual function as a H+ and Na+ pump. Further investigation is needed to identify the specific structural constraints in DeNaR that impart this cation tuning and to identify the path of Na+ through the protein, including the initial cation binding/filter site or region. Our results demonstrate that microbial rhodopsins show cation tuning as seen in transporter proteins from higher organisms, and that variation exists in the light-induced structural rearrangement of the canonical E → C conformer conversion in microbial rhodopsins.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Elizabeth Lane for her expert technical assistance and Dr. Oleg Sineshchekov for extensive discussions.
Funding
The work was supported by National Institutes of Health Grants R01GM027750 (J.L.S.), R21MH098288 (J.L.S.), and HL095820 (A.-L.T.) and the Hermann Eye Fund and Endowed Chair AU-0009 from the Robert A. Welch Foundation to J.L.S.
ABBREVIATIONS
- DeNaR
D. eikasta sodium-pumping rhodopsin (also known as KR2)
- BR
bacteriorhodopsin
- SRI
sensory rhodopsin I
- SRII
sensory rhodopsin II
- EPR
electron paramagnetic resonance
- NMG
n-methyl-glucamine
- TPP
tetraphenylphosphonium chloride
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- MOPS
3(N)-morpholinopropanesulfonic acid
- DM
n-decyl β-d-maltopyranoside
- DDM
n-dodecyl β-d-maltopyranoside
- MTSSL
S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanethiosulfonate
- LB
Luria-Bertani
- IPTG
isopropyl β-d-thiogalactopyranoside
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- RSB
retinylidene Schiff base
- PSB
protonated Schiff base
- ATR
all trans-retinal
- λmax
wavelength of maximal absorption
Footnotes
Supporting Information
Control measurements for light-induced H+ fluxes of wild-type DeNaR and cysteine mutants, laser flash spectroscopy data for singly and doubly spin-labeled mutants, optical spectra of DeNaR in solutions containing Na+, K+, and NMG+, room-temperature EPR spectra used to calculate the difierence spectra provided in the text, liquid nitrogen EPR control experiments, and the DeNaR structural model compared to crystal structures. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/bi501386d.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- (2).Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Spudich JL, Sineshchekov OA, Govorunova EG. Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta. 2014;1837:546–552. doi: 10.1016/j.bbabio.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- (5).Thorgeirsson TE, Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Transient channel-opening in bacteriorhodopsin: An EPR study. J. Mol. Biol. 1997;273:951–957. doi: 10.1006/jmbi.1997.1362. [DOI] [PubMed] [Google Scholar]
- (6).Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- (7).Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Light-induced rotation of a transmembrane α-helix in bacteriorhodopsin. J. Mol. Biol. 2000;304:715–721. doi: 10.1006/jmbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- (8).Lanyi JK, Luecke H. Bacteriorhodopsin. Curr. Opin. Struct. Biol. 2001;11:415–419. doi: 10.1016/s0959-440x(00)00226-8. [DOI] [PubMed] [Google Scholar]
- (9).Neutze R, Pebay-Peyroula E, Edman K, Royant A, Navarro J, Landau EM. Bacteriorhodopsin: A high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta. 2002;1565:144–167. doi: 10.1016/s0005-2736(02)00566-7. [DOI] [PubMed] [Google Scholar]
- (10).Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- (11).Wickstrand C, Dods R, Royant A, Neutze R. Bacteriorhodopsin: Would the real structural intermediates please stand up? Biochim. Biophys. Acta. 2014;1850:536–553. doi: 10.1016/j.bbagen.2014.05.021. [DOI] [PubMed] [Google Scholar]
- (12).Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Vonck J. A three-dimensional difference map of the N intermediate in the bacteriorhodopsin photocycle: Part of the F helix tilts in the M to N transition. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- (14).Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, Kandori H. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- (15).Kwon SK, Kim BK, Song JY, Kwak MJ, Lee CH, Yoon JH, Oh TK, Kim JF. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol. Evol. 2013;5:187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Balashov SP, Imasheva ES, Dioumaev AK, Wang JM, Jung KH, Lanyi JK. Light-Driven Na+ Pump from Gillisia limnaea: A High-Affinity Na+ Binding Site Is Formed Transiently in the Photocycle. Biochemistry. 2014;53:7549–7561. doi: 10.1021/bi501064n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Riedel T, Gomez-Consarnau L, Tomasch J, Martin M, Jarek M, Gonzalez JM, Spring S, Rohlfs M, Brinkhoff T, Cypionka H, Goker M, Fiebig A, Klein J, Goesmann A, Fuhrman JA, Wagner-Dobler I. Genomics and physiology of a marine flavobacterium encoding a proteorhodopsin and a xanthorhodopsin- like protein. PLoS One. 2013;8:e57487. doi: 10.1371/journal.pone.0057487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Brown LS. Eubacterial rhodopsins: Unique photo- sensors and diverse ion pumps. Biochim. Biophys. Acta. 2014;1837:553–561. doi: 10.1016/j.bbabio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- (19).Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Knollmann BC. Pacing lightly: Optogenetics gets to the heart. Nat. Methods. 2010;7:889–891. doi: 10.1038/nmeth1110-889. [DOI] [PubMed] [Google Scholar]
- (22).Entcheva E. Cardiac optogenetics. Am. J. Physiol. 2013;304:H1179–H1191. doi: 10.1152/ajpheart.00432.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sineshchekov OA, Sasaki J, Phillips BJ, Spudich JL. A Schiff base connectivity switch in sensory rhodopsin signaling. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16159–16164. doi: 10.1073/pnas.0807486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Radzwill N, Gerwert K, Steinhoff HJ. Time- resolved detection of transient movement of helices F and G in doubly spin-labeled bacteriorhodopsin. Biophys. J. 2001;80:2856–2866. doi: 10.1016/S0006-3495(01)76252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sasaki J, Tsai AL, Spudich JL. Opposite displacement of helix F in attractant and repellent signaling by sensory rhodopsin-Htr complexes. J. Biol. Chem. 2011;286:18868–18877. doi: 10.1074/jbc.M110.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wegener AA, Chizhov I, Engelhard M, Steinhoff HJ. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 2000;301:881–891. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- (28).Sattig T, Rickert C, Bamberg E, Steinhoff HJ, Bamann C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew. Chem., Int. Ed. 2013;52:9705–9708. doi: 10.1002/anie.201301698. [DOI] [PubMed] [Google Scholar]
- (29).Krause N, Engelhard C, Heberle J, Schlesinger R, Bittl R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by EPR spectroscopy. FEBS Lett. 2013;587:3309–3313. doi: 10.1016/j.febslet.2013.08.043. [DOI] [PubMed] [Google Scholar]
- (30).Bonneau R, Tsai J, Ruczinski I, Chivian D, Rohl C, Strauss CE, Baker D. Rosetta in CASP4: Progress in ab initio protein structure prediction. Proteins. 2001;(Suppl. 5):119–126. doi: 10.1002/prot.1170. [DOI] [PubMed] [Google Scholar]
- (31).Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- (33).Millhauser GL, Fiori WR, Miick SM. Electron spin labels. Methods Enzymol. 1995;246:589–610. doi: 10.1016/0076-6879(95)46026-8. [DOI] [PubMed] [Google Scholar]
- (34).Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ono H, Inoue K, Abe-Yoshizumi R, Kandori H. FTIR spectroscopy of a light-driven compatible sodium ion-proton pumping rhodopsin at 77 K. J. Phys. Chem. B. 2014;118:4784–4792. doi: 10.1021/jp500756f. [DOI] [PubMed] [Google Scholar]
- (36).Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sineshchekov OA, Sasaki J, Wang J, Spudich JL. Attractant and repellent signaling conformers of sensory rhodopsin-transducer complexes. Biochemistry. 2010;49:6696–6704. doi: 10.1021/bi100798w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lanyi JK. Bacteriorhodopsin. Annu. Rev. Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- (39).Subramaniam S, Lindahl M, Bullough P, Faruqi AR, Tittor J, Oesterhelt D, Brown L, Lanyi J, Henderson R. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 1999;287:145–161. doi: 10.1006/jmbi.1999.2589. [DOI] [PubMed] [Google Scholar]
- (40).Renart ML, Barrera FN, Molina ML, Encinar JA, Poveda JA, Fernandez AM, Gomez J, Gonzalez-Ros JM. Effects of conducting and blocking ions on the structure and stability of the potassium channel KcsA. J. Biol. Chem. 2006;281:29905–29915. doi: 10.1074/jbc.M602636200. [DOI] [PubMed] [Google Scholar]
- (41).Yellen G. Single channel seeks permeant ion for brief but intimate relationship. J. Gen. Physiol. 1997;110:83–85. doi: 10.1085/jgp.110.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhou Y, MacKinnon R. The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 2003;333:965–975. doi: 10.1016/j.jmb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- (43).MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- (44).Zhou Y, MacKinnon R. Ion binding affinity in the cavity of the KcsA potassium channel. Biochemistry. 2004;43:4978–4982. doi: 10.1021/bi049876z. [DOI] [PubMed] [Google Scholar]
- (45).Wright EM, Loo DD, Turk E, Hirayama BA. Sodium cotransporters. Curr. Opin. Cell Biol. 1996;8:468–473. doi: 10.1016/s0955-0674(96)80022-6. [DOI] [PubMed] [Google Scholar]
- (46).Hirayama BA, Loo DD, Wright EM. Cation effects on protein conformation and transport in the Na+/glucose cotransporter. J. Biol. Chem. 1997;272:2110–2115. doi: 10.1074/jbc.272.4.2110. [DOI] [PubMed] [Google Scholar]
- (47).Gushchin I, Shevchenko V, Polovinkin V, Kovalev K, Alekseev A, Round E, Borshchevskiy V, Balandin T, Popov A, Gensch T, Fahlke C, Bamann C, Willbold D, Buldt G, Bamberg E, Gordeliy V. Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 2015;22:390–395. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- (48).Kato HE, Inoue K, Abe-Yoshizumi R, Kato Y, Ono H, Konno M, Hososhima S, Ishizuka T, Hoque MR, Kunitomo H, Ito J, Yoshizawa S, Yamashita K, Takemoto M, Nishizawa T, Taniguchi R, Kogure K, Maturana AD, Iino Y, Yawo H, Ishitani R, Kandori H, Nureki O. Structural basis for Na transport mechanism by a light-driven Na pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- (49).Ran T, Ozorowski G, Gao Y, Sineshchekov OA, Wang W, Spudich JL, Luecke H. Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr. 2013;D69:1965–1980. doi: 10.1107/S0907444913017575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.