Abstract
Systematic description and the unambiguous communication of findings and models remain among the unresolved fundamental challenges in systems neuroscience. No common descriptive frameworks exist to describe systematically the connective architecture of the nervous system, even at the grossest level of observation. Furthermore, the accelerating volume of novel data generated on neural connectivity outpaces the rate at which this data is curated into neuroinformatics databases to synthesize digitally systems-level insights from disjointed reports and observations. To help address these challenges, we propose the Neural Systems Language (NSyL). NSyL is a modeling language to be used by investigators to encode and communicate systematically reports of neural connectivity from neuroanatomy and brain imaging. NSyL engenders systematic description and communication of connectivity irrespective of the animal taxon described, experimental or observational technique implemented, or nomenclature referenced. As a language, NSyL is internally consistent, concise, and comprehensible to both humans and computers. NSyL is a promising development for systematizing the representation of neural architecture, effectively managing the increasing volume of data on neural connectivity and streamlining systems neuroscience research. Here we present similar precedent systems, how NSyL extends existing frameworks, and the reasoning behind NSyL’s development. We explore NSyL’s potential for balancing robustness and consistency in representation by encoding previously reported assertions of connectivity from the literature as examples. Finally, we propose and discuss the implications of a framework for how NSyL will be digitally implemented in the future to streamline curation of experimental results and bridge the gaps among anatomists, imagers, and neuroinformatics databases.
INDEXING TERMS: systems neuroscience, neuroanatomy, connectomics, informatics, motivated behavior
Effective scientific communication—especially about a physical object as complex as the nervous system—requires the use of clearly defined terms for parts and clearly defined relationships between the terms. Without a defined nomenclature, it will not be possible to understand in a systematic way the arrangement of nervous system circuitry and its organizing principles. Other arenas of life science research have had systematic, defined nomenclatures for many years. For example, the flood of reports of novel plants and animals from the New World in the middle of the Seventeenth Century inspired Linnaeus to develop a revolutionary binomial nomenclature system with a set of rules for biological classification that is still used today in a highly evolved form (Melville, 1995; International Commission on Zoological Nomenclature 1999). However, as Swanson (2000) and Bota et al. (2003) have discussed, there is still no widely accepted system of terms and relationships between terms for the nervous system’s parts and their interconnections, despite ongoing analysis with ever-increasing sophistication since antiquity.
Pressure to standardize neuroanatomical nomenclature and description has recently come from two modern sources: connectomics and neuroinformatics. According to Sporns et al. (2005), in its purest form, a connectome is the complete set of neural connections between defined parts of a nervous system, and connectomics is the subfield of systems neuroscience concerned with acquiring, analyzing, and applying information about nervous system connectivity. Neuroinformatics, the application of principles and techniques from information science to neuroscience, provides support in the form of methods and tools to manage and analyze neuroscientific information in digital knowledge-management systems. As Bota and Swanson (2007a) observed, automated inference engines used to manipulate massive data sets automatically and build systems-level understanding of the structure and function of the nervous system within and between different species require the use of controlled vocabularies of nervous system parts with internally consistent sets of terms.
Over the last several decades, our group has used experimental pathway tracing methods that are several orders of magnitude more sensitive than earlier approaches to help define the structural organization of mammalian nervous system networks that control survival behaviors. This has led to a flood of new data, over 5,000 newly reported axonal connections between gray matter regions and neuron types; the development of systematic traditional and interactive digital atlases of the adult and developing rat brain with accompanying hierarchically ordered, complete and nonoverlapping tables of nervous system parts; the implementation of an online Brain Architecture Knowledge Management System (BAMS); and the creation of a Foundational Model of Structural Connectivity (FMC) for the nervous system in general (Alvarez-Bolado and Swanson, 1996; Swanson, 2001, 2003; Bota et al., 2003, 2012; Swanson and Bota, 2010). A critically missing component is a modeling language and notation system (easily readable by humans and machines) for systematically representing neural circuitry and aiding in automating “biocuration,” the practice of acquiring biological data from the corpus of existing literature or the experimental workbench for input into knowledge management systems.
A significant unsolved challenge in systems neuroscience concerns exactly how connectomic and neuroanatomical information is curated and organized for analysis in neuroinformatics databases. The current method of biocuration requires expert neuroscientists to read individual published manuscripts and manually enter data and metadata. This presents two challenges. First, the rate at which novel connectivity data is generated drastically outpaces the rate of biocuration. If this is to change, more efficient methods of biocuration must be developed in which curation takes place in a decentralized fashion, “closer” to the sources of data generation (experimentalists). Furthermore, manual biocuration requires a significant consumption of time, money, and labor that could be better utilized conducting experiments or developing new data analysis methods to further our knowledge of the nervous system. Natural language processing—digital systems that parse and build meaningful representations of human-generated language—has yet to achieve robust machine reading capable of reliable automated curation of data and metadata from the nuanced language found in the scientific literature. The generation of semiautomated and automated systems for biocuration that do not rely on natural language processing would allow a more efficient allocation of time and energy in the laboratory in a more immediate, technologically tractable manner. As we explore here, the challenge of systematically representing and communicating nuanced, domain-specific information using modeling languages in a manner that is comprehensible to both humans and computers has, to different levels of success, already been addressed in closely related fields. We believe the approaches employed in other fields may be transferable to addressing the aforementioned challenges faced in systems neuroscience.
Modeling languages, artificial languages for representing nuanced, domain-specific information, were developed to represent real-world problems in forms that machines could manipulate and communicate with one another with minimal human involvement. Historically, these languages have been used to help solve problems and streamline operations and communication in engineering, chemistry, computer science, and electronics (Weiner, 1948; Boute, 1986; Weininger, 1988; Claessen and Sands, 1999; Gakh and Burnett, 2001). These new languages were methods to take advantage of newly available computational power, to systematically represent information, and to bridge the communication gap between human and machine. More recently, modeling languages have been developed by Le Novère et al. (2009) in biochemistry and Raikov et al. (2011) in computational neuroscience for increasing systematicity and efficiency of representation and communication. For example, the Systems Biology Graphical Notation (SBGN) is a graphical modeling language for systematically representing metabolic, cell signaling, and genetic interactions. The Notation’s symbol semantics allow it to communicate nuanced biochemistry in an intuitive, unambiguous manner. NineML, a computational neuroscience modeling language, interfaces between different neural network simulation software systems to promote greater interoperability of artificial neural network model data. Both of these modeling languages decrease ambiguity in nuanced domains of knowledge and promote effective communication through their use of standardized representations. While neither SBGN nor NineML is the sole solution to the problem of systematic representation and communication in their fields, and have not been universally adopted, they serve as examples of how these challenges can be addressed.
Hardware description languages, such as Verilog, are electrical engineering modeling languages used in the design, simulation, and construction of electronic circuitry (Thomas and Moorby, 1996). A hardware description language systematically encodes electronic circuit component specifications (such as resistance or capacitance) and a circuit’s wiring diagram as specially encoded text for digital storage, communication, and analysis. Despite superficial differences, these systematic encodings of electronic circuit structure are pertinent to systems neuroscience. Theory, philosophy, and clinical and basic research support a model of the nervous system as a biological computing network composed of highly interconnected neural circuitry observable at multiple levels of granularity (Weiner, 1948; Haugeland, 1981; Swanson and Bota, 2010; Thompson and Swanson, 2010; Delanda, 2011; Swanson, 2012). At its core, systems neuroscience is the practice of reverse engineering the structure and function of this computing network (Swanson, 2012). In light of the utility of hardware description languages in describing the network structure of manmade circuitry and the absence of database-independent modeling languages for systematically describing nervous system connective architecture, we propose a novel Neural Systems Language (NSyL).
MATERIALS AND METHODS
The NSyL is a modeling language to be used by investigators to encode and communicate results or models of neural connectivity systematically. NSyL’s rigid syntax and flexible semantics allow it to be consistently used (without linguistic modification) irrespective of the animal taxon described, experimental or observational technique implemented, anatomical nomenclature referenced, or neuroinformatics system used for data storage or analysis. NSyL is internally consistent, concise, and comprehensible to both humans and computer algorithms.
The emerging application of network science and graph theory to systems neuroscience has given the field a toolkit for analyzing neural network topology (the connective relationships among different brain regions, neuron types, and individual neurons) in terms of networks of abstracted “nodes” and “edges.” NSyL encodes neural network architecture by representing connectivity as graph theoretical abstractions. These types of abstractions allow for analysis that has recently helped to elucidate both the commonalities between different forms of networks and the relationships between network structure and network function. For example, graph theoretical abstractions of neural connectivity performed by Bassett et al. (2008) suggest that brain networks both exhibit structural “motifs” found in other types of networks (for example, gene expression relationships and the internet) and that certain parameters describing these “motifs” may correlate with disease states such as schizophrenia. These findings support a model of the nervous system as a biological information-processing network in which network function (behavior) is emergent from, and constrained by, network structure. Furthermore, these findings demonstrate the value that network science and graph theory bring to connectomics and systems neuroscience. While “black-box” abstractions of networks in terms of nodes and edges may obfuscate fine-grained biological detail, graph theory’s associated analytic techniques offer a unique toolkit of approaches for exploring the relationship between the nervous system’s network structure and its function in generating behavior. It is with these novel insights and techniques in mind that we formulate NSyL from the perspective of graph theoretical approaches to describing network topology and propose NSyL as an extension of Bota and Swanson’s (2010) foundational model of connectivity (FMC).
The FMC describes the nervous system through established axioms, domain-specific assumptions, interrelated principles, and a tightly controlled vocabulary of parts and relationships that work together to allow systematic discourse about the nervous system, a necessity when analyzing data about connectivity in neuroinformatics databases and with automated inference engines. To develop NSyL consistent with the FMC, NSyL’s overall domain of discourse is that of the nervous system described as a network of “nodes” and “connection” relationships between nodes at three nested levels of observation. We assert that the fundamental structural information-processing unit of the nervous system is the neuron, denoted in the FMC as a “micronode,” at the finest level of granularity. As outlined by Bota and Swanson (2007b), a unique set of neurons characterized by its location in the nervous system, expressed molecules, cellular morphology, and population input–output (axonal) connectivity pattern is known as a “neuron type” and is denoted in the FMC as a “mesonode” (Bohland et al., 2009a). At the grossest level of granularity, a gray matter region is characterized by parameters including but not limited to its topography, gross cytoarchitectural properties, unique set of neuron types, intraregional connectivity, and input–output interregional connectivity and is known as a “macronode.” We propose an update of the FMC formally to incorporate these three granular levels of nervous system description into the model’s controlled vocabulary of parts and relationships. We denote the levels here as the “macrolevel,” the “mesolevel,” and the “microlevel.” Each level describes the scope of observation at which description of its nodes and connections is valid. These three levels provide a nested, hierarchical framework for describing the architecture of the nervous system, with the node as a recurring basic unit (Fig. 1) at three nested levels of granularity.
Figure 1.
Three levels of granularity in description of the nervous system. As specified in the Foundational Model of Connectivity, the neuronal connective architecture of the nervous system may be modeled as a complex network of “nodes” and connections at three distinct levels of granularity. The least granular, the macrolevel, describes gray matter regions (macronodes) and macroconnections as inputs and outputs of a brain region treated as a black box. Descriptions at the mesolevel describe unique neuron types (mesonodes) and their associated mesoconnectivity. The finest level of granularity, the microlevel, describes individual neurons (micronodes) and microconnectivity. Although the general terms “node” and “connection” are elements of description at each level, the details required to describe a node or connection accurately at a given level vary and are a function of that level. As displayed graphically, a macronode is composed of a unique set of mesonodes, each of which is composed of numerous micronodes.
Just as we establish the node as the basic unit of the nervous system at any level of observation, we describe the “connection” (the information-conducting relationship between nodes) as the basic connectomic relationship between nodes. A connection may be experimentally asserted (either as “structurally observed” axonal connections or as “correlationally inferred” connections) or theoretical for modeling purposes. Any connection is described at one of three levels of granularity: macroconnectivity, mesoconnectivity, or microconnectivity. A macroconnection is a connection between two macronodes (gray matter regions), a mesoconnection is a connection between two mesonodes (neuron types), and a microconnection is a connection between two micronodes (individual neurons).
A structurally observed connection is an anatomical linkage between two nodes in the nervous system that is asserted from direct observations of biological neural network structure (Swanson, 2012). Structurally observed connections may be asserted only from experimental or normal anatomical techniques, including but not limited to pathway tracing and electron microscopic serial section analysis, respectively. The physical course taken by any structurally observed connection through gray matter regions and white matter tracts is called its “route,” and route information is part of the description of an observed structural connection and should ideally be included in the description of a structurally observed connection when possible. Unlike an inferred correlational connection, the assertion of a structurally observed connection between two nodes implies but does not ensure, as observed by Issac et al. (1995), a statistical correlation between the neural activity levels of two nodes.
A correlationally inferred connection is a relationship of activation or influence between two nodes in the nervous system that is inferred to be a connection between nodes. Correlationally inferred connections are inferred from experiments showing a statistical correlation (or dependence) between the direct or indirect neural activity levels (for example, membrane electrical potential or local blood oxygen concentration, respectively) of two nodes in the nervous system (Bullmore and Sporns, 2009). A correlationally inferred connection may be asserted from methods such as MR diffusion imaging and tractography; resting state, task-specific, and volumetric fMRI; invasive and noninvasive electrophysiology; and immediate early gene activation patterns (Fox et al., 1997; Brovelli et al., 2004; Bassett et al., 2008; Iturria-Medina et al., 2008; Behrens and Sporns, 2011; Sporns, 2011). Because of limits in spatial and temporal resolution inherent to many of the techniques used to assert correlationally inferred connections, the assertion of a correlationally inferred connection between two nodes does not imply the existence of an observable structural (that is, neuron-to-neuron synaptic) connection between two nodes (Sporns, 2011). A correlationally inferred connection may be directional (if the technique used establishes causal relationships between node activity levels) but is more commonly directionless (which is distinct from “bidirectional”), in which the connection source and target are not distinguishable by the technique used (Brovelli et al., 2004).
A theoretical connection is neither observed nor inferred but nonetheless proposed for model-building purposes. Theoretical connections can be used, for example, when describing experimental hypotheses or when building models that lack complete experimental data.
In addition to describing nodes and connections, NSyL is equipped to represent the relationship between a node and an effector (a muscle or gland) as well as influences among the nervous system, the effectors, and the environment. The addition of these nonneuronal components and relationships allows for a more complete description of the nervous system’s relationship with the rest of an animal and its environment for modeling purposes.
Language implementation
The NSyL, as we present it here, is currently developed to describe macroconnectivity at the macrolevel (as connections between brain regions). Macrolevel NSyL is one of three languages that we propose be potentially developed (conceptually, the remaining two would describe mesoconnectivity and microconnectivity). As already mentioned, the conserved elements of nodes and connections may grant continuity in NSyL among the three granularities and languages. The decision to constrain our discourse currently to the macrolevel stems from the following.
There exists a body of literature with which we may validate the effectiveness of NSyL at describing macroconnectivity (Swanson, 2003).
The current state of the art in human connectomics is limited mostly to describing macroconnectivity (Bullmore and Sporns, 2009).
Greater consensus must be reached with regard to what parameters of mesoconnectivity and microconnectivity are necessary for modeling and encoding using NSyL (Bohland et al., 2009b).
NSyL syntax
Macrolevel NSyL is written and communicated as specially encoded sentences of text called “NSyL expressions.” NSyL expressions have stereotyped syntax that allows for reports of connectivity asserted from experiments using different experimental or normal techniques, animal taxa, and nomenclatures to be described while still maintaining consistent expression form, a requirement for computer algorithms to decode and manipulate NSyL expressions to help automate the process of biocuration and extracting assertions of connectivity from the literature. Expressions follow a set of “formation rules” (see below) ensuring that connectivity is systematically represented.
In summary, a NSyL expression has two parts: “experimental parameters,” with corresponding values, and “connection assertions.” These two parts are separated by a delineating “;” character, giving a NSyL expression the following generic morphology.
{Experimental Parameters; Connection Assertions}
Connection assertions describe the actual connections reported in a particular expression, and experimental parameters describe a minimum amount of information about the nature of the experiment to provide metadata context for the reported connectivity. At a minimum, metadata about three specific experimental parameters are required—nomenclature, taxon, and technique—and these experimental parameters are paired with their corresponding values by the “:” character. For example, a valid set of experimental parameters may be:
‘Taxon’ : ‘rat’, ‘Nomenclature’ : ‘Swanson 2003’, ‘Technique’ : ‘Anterograde PHAL’
Other experimental parameters might possibly include route information, sex of the animal, and age of the animal. With regard to experimental parameters, taxon (for example, species) and method (for example, fMRI or Fluorogold retrograde tracer) are relatively straightforward entities. As discussed in the introductory paragraphs, nomenclature is foundationally important but is often confused and misunderstood. Therefore, it is critical to have a clear definition for the purposes of macrolevel NSyL:
A robust nomenclature is a single complete and nonoverlapping set of defined terms for nervous system gray matter regions and white matter tracts, and the boundary relationships between them, as presented in a systematic and documented 2D or 3D atlas and, preferably, as a digitally encoded database or knowledge-management system ontology (see Bota et al., 2003; Bota and Swanson, 2008, 2010; Swanson and Bota, 2010). Any set of terms used to describe neural circuitry that is less stringent yields proportionally less accurate descriptions of the circuitry and less accurate inferences from knowledge-management systems.
Another potential (but not required) experimental parameter is connection strength. NSyL can, in principle, accommodate any quantitative or qualitative measure of connection strength. The importance and desirability of accurately quantified descriptions of connection strength cannot be ignored. While current techniques exhibit limited consistency in observed or inferred connection strength, it may ultimately be fruitful and desirable to possess quantitative descriptions of connectivity strength. As techniques improve and the consistency and accuracy of quantified measures of connection strength improve, systems such as macrolevel NSyL ideally should be structured to encode these improved quantitative measures of connectivity strength systematically. As presented here, macrolevel NSyL is equipped to describe observed structural or inferred correlational macroconnection strength as binary values (a connection is present but strength could not be determined), qualitative descriptors along an ordinal scale, or quantitative measures of connection strength. Regardless of the descriptive metric that an investigator choses to use when reporting his or her findings, NSyL is designed to encode the reported strength effectively so long as the investigator specify, in the metadata of an expression, which of the aforementioned measures was used when describing their findings and the appropriate experimental parameters associated with that technique. This flexibility allows NSyL to be used to systematically encode observed connectivity regardless of how the investigator quantifies or qualifies the strength of a connection.
Connection assertions may be grouped into “experimental sets” if the assertions share a complete set of identical values for experimental parameters. Experimental sets may be nested (i.e., sets may contain other sets). Any connection assertions “contained” by one or more nested sets “inherit” their experimental parameters from those sets. Once the value of an experimental parameter is specified in an experimental set, it may not be specified again until that experimental set is “closed.” For example:
- {‘Taxon’ : ‘Rat’,
- {‘Technique’ : ‘Anterograde PHAL’;
- (Connection Assertions)
- }
- {‘Technique’ : ‘Retrograde Flurogold’;
- (Connection Assertions)
- }
}
In the above expression, the specification of ‘rat’ as the described ‘taxon’ applies to all contained experimental sets and connection assertions. The parameter ‘technique’ may be specified with different values between sets that share a “parent” set so long as those sets do not contain one another. The nesting of experimental sets allows for more concise phrasing of experimental parameters by eliminating redundancy between sets that repeat identical parameters.
Different values of experimental parameter metadata (such as technique, species, nomenclature, and so on) listed at the beginning of an experimental set not only describe the nature of the experiment, but may determine whether or not additional information is required to accurately describe asserted connectivity (such as “route” in certain anterograde pathway tracing methods used for observing structural connections).
Reflecting our decision to design macrolevel NSyL with a graph-theoretical approach, NSyL expresses connectivity data as connections between “source” components and a series of output “target” components as edge partners between nodes in a (directed) graph. All node input can be described as output from a different node (or series of nodes). As such, describing only output prevents redundant “double encoding” that would result in the same connection being described twice (once as an input to a node, once as an output from a node), creating unnecessarily verbose expressions and ambiguity.
The report of a connection between a source and at least one target is known as a “connection assertion.” A new connection assertion is started with the “@” character for clarity in long NSyL expressions, and the nature of the connection (for example, denoted with the “>+” for “unidirectional and excitatory”) separates the connection source from its target(s). NSyL expressions can be formed in which a connection assertion describes a single source that may connect to more than one target:
{‘Experimental Parameter’ : ‘Value’; @Source1 > + Target1A, Target1B}
In this generic example, a connection assertion is described with a single source (“Source1”) that has unidirectional, excitatory connections (“>+”) to two comma-delimited target nodes (“Target1A” and “Target1B”). NSyL expressions may also be formed with multiple connection assertions that describe connectivity between multiple distinct sources and their corresponding target nodes:
{‘Experimental Parameter’ : ‘Value’; @Source1 > + Target1A, Target1B, @Source2 > −Target2A}
In this example, the target nodes of “Source1” (“Target1A” and “Target1B”) are comma-delimited. The source node “Source1” is separated from its target nodes by two characters describing the nature of the connection (“>+”). A second connection assertion (asserting a unidirectional, inhibitory connection between “Source2” as a source and “Target2A” as its target) is delimited from the first assertion by the “@” symbol (which symbolizes the beginning of a new assertion.)
Formation rules
A syntactically sound NSyL expression abides by a set of “formation rules” that detail the constraints of its formation (Figs. 2, 3). Although the semantic value of a NSyL expression (i.e., the neural architecture that it encodes) is a function of the species-dependent nomenclature used, all macrolevel NSyL expressions share a common structure because of the rigid nature of the formation rules. Adherence to these rules ensures that connectivity is described systematically and is the basis upon which NSyL may be interpreted by computers for automated biocuration.
Figure 2.
Macrolevel NSyL textual formation rules. To ensure systematicity in encoding novel assertions of neural connectivity, we propose that NSyL expressions follow a series of syntax-defining formation rules. These rules specify how connections are encoded irrespective of the animal studied, technique used, or nomenclature referenced.
Figure 3.
Macrolevel NSyL textual formation rules. Continued from Figure 2.
Graphical notation
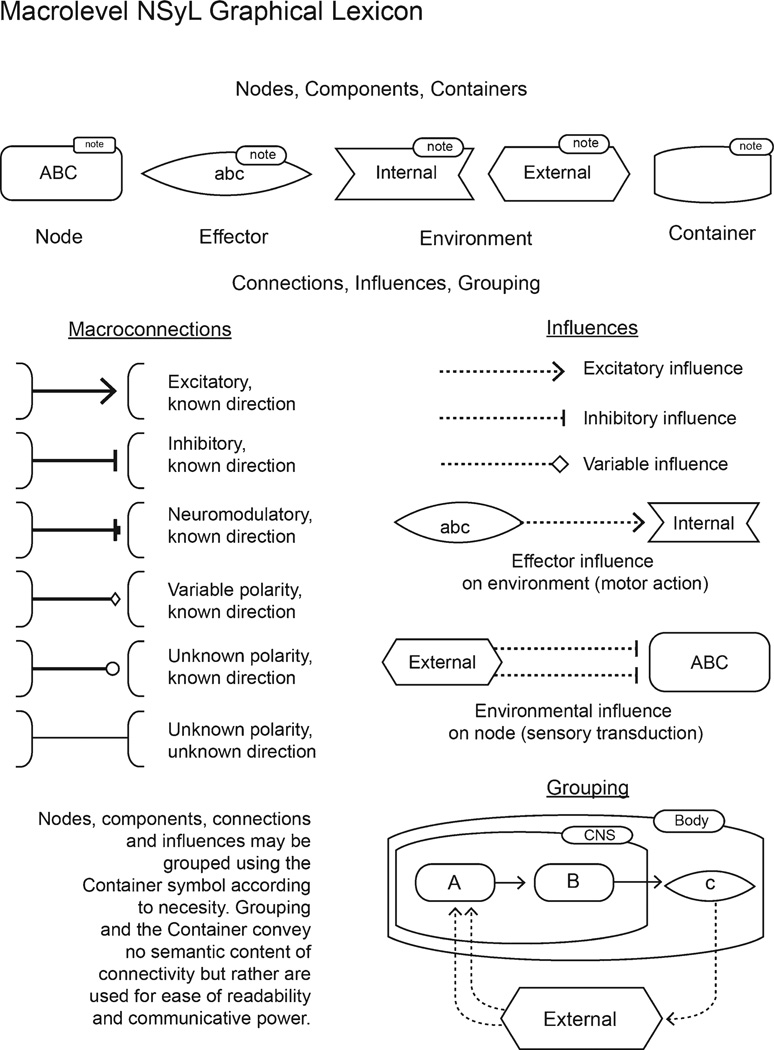
In addition to outlining a textual framework for systematically encoding neural connectivity, macrolevel NSyL outlines a specification for visually conveying connectivity in a systematic way. For graphical macrolevel NSyL, we propose a standard set of symbols that may be used to display visually macrolevel neural connectivity in schematics or wiring diagrams (Fig. 4). Following the example of work by Le Novère et al. (2009) on the systems biology graphical notation (SBGN), graphical NSyL specifies only the definitions of symbols and visual elements while allowing for flexible arrangement, formatting, and adornment in schematics as needed. These symbols represent components (nodes, effectors, and the environment), relationships (connectivity and influence), and techniques (“containers” and “labels”) for visually grouping and labeling components as necessary. Containers are visual boundaries that can be used for grouping nodes or components to make a diagram more easily understood. Labels, small fragments of text, can adorn nodes or containers to help describe a diagram beyond its anatomical abbreviations. Neither containers nor labels are encoded in textual NSyL and are regarded as containing no information about the connective architecture of the network described.
Figure 4.
Macrolevel NSyL graphical lexicon. Nodes and components (effectors and the environment) may be related to one another through macroconnections and influences. Container symbols may be used to group nodes or components as needed in a schematic but do not convey any information about connective topology; rather, they may be used for organizing the layout of schematic elements in more understandable ways.
Graphical formation rules
Graphical macrolevel NSyL, as proposed here, offers a way to display systematically neural connectivity in wiring diagrams and schematics through the use of standardized symbols with clearly defined symbol meanings. We propose a minimum set of simple graphical formation rules that must be followed for systematic schematic formation (Fig. 5).
Figure 5.
Macrolevel NSyL graphical formation rules. Although the absolute placement, adornment, grouping, and labeling of a graphical macrolevel NSyL schematic is at the discretion of the schematic’s author, we propose a minimum set of formation rules for the systematic spatial arrangement of elements, components, connections, and influences in a schematic.
NSyL semantics
NSyL’s ability to describe connectivity independent of one fixed nomenclature requires flexible definitions of anatomical abbreviations and the explicit declaration of what nomenclature an investigator used. The semantic content of a node abbreviation, the nervous system part denoted by that abbreviation, is a function of the nomenclature specified in the experimental parameters of a NSyL expression. To draw a historical example for the need to specify a nomenclature when describing neural architecture, in the rat the abbreviations “ILA” and “IL” both denote the “infralimbic area” macronode in Swanson’s (1998) and Krettek and Price’s (1977) nomenclatures, respectively. In this case, ambiguity arises because one structure is referenced by two terms. Additionally, the term “IL” may denote either the “infralimbic area” or “pituitary gland, intermediate lobe” macronodes in Krettek and Price (1977) and Swanson (1998), respectively. Here, one term may ambiguously denote two structures. As seen in these two examples, knowing the specified nomenclature is needed to determine the semantic content (corresponding nervous system part) represented by an abbreviation. NSyL resolves potential semantic ambiguity and introduces the systematicity needed by neuroinformatics systems and inference engines by requiring a nomenclature to be specified in each NSyL expression.
Obviously, direct comparisons between NSyL expressions can be made only when the same nomenclature is used. As knowledge-management systems fill with connectional data, the need for wide adoption of a particular nomenclature, or different nomenclatures with clearly defined mappings between them, will become increasingly obvious (see Swanson and Bota, 2010).
Species independence
Macrolevel NSyL is developed to describe connectivity found in any animal species (in which macrolevel observations can be made) with no changes to the formation rules of the language needed to describe different species. The same formation rules apply when (for example) describing macrolevel neural network structure in any species (for which an appropriate macrolevel anatomical nomenclature has been released). Although the rules governing expression formation do not change between species, the exact definition of a given macronode abbreviation does change in accordance with the species-specific anatomical nomenclature declared in the expression metadata. This “rigid form–flexible function” approach allows NSyL to act as a general descriptive framework regardless of the species described. As such, there is no such thing as a “species-specific version of NSyL.” However, as mentioned in the preceding section, direct comparison of NSyL expressions for circuitry within and between species requires use of a single nomenclature. The choice of that nomenclature is up to individual researchers, and any comparisons made are performed not by NSyL itself but rather in the context of online knowledge-management systems.
Comparisons within and between species
Challenges associated with comparing assertions of neural connectivity between species and between different anatomical parcellations and their associated nomenclatures within a species share a common challenge: to what extent are neuroanatomical findings generalizable? It is well known that multiple nomenclatures, and thus multiple interpretations of how to parcel gray matter regions and white matter tracts, exist for any given species. In the end, the choice of a nomenclature within a species should be based on a critical analysis of the available evidence and not on authority or popularity. Unfortunately, for many (but certainly not all) nervous system parts, there is not enough evidence to choose between nomenclatures. Nomenclature comparisons between species are considerably more problematic and rest on considerations of common evolutionary and developmental origins (Bota and Arbib, 2004) and on comparisons of morphological features and functional measures (from molecular to cognitive). As mentioned above, the choice of one nomenclature over another is up to individual researchers, and any comparisons made are performed not by NSyL itself but rather in the context of online knowledge-management systems.
Digital framework implementation
Aside from formalizing a notation for nervous system connective architecture, NSyL holds the potential to help address challenges that systems neuroscience faces in biocuration and how data is imported into neuroinformatics databases. For formal modeling languages NSyL could serve as a “lingua franca” between experimentalists and as a “middle ground” between natural language in manuscripts and connectivity data stored in appropriate formats in neuroinformatics databases. Currently, natural language in manuscripts still remains indecipherable to computers. As such, the task of biocuration is left to human experts. In contrast, modeling languages such as NSyL are machine-readable, making NSyL expressions amenable to automated extraction from the literature and digital curation and storage without explicitly requiring human effort. The implementation and adoption of a formal modeling language, such as NSyL presented here, for encoding connectomic results could accelerate biocuration.
NSyL is designed to aid investigators and neuroinformaticians in systematically encoding and communicating novel, previously unreported assertions of macroconnectivity. Macrolevel NSyL as we describe it here, and tools we propose be developed to implement it, hold the potential to help further disperse results from novel manuscripts and investigations farther and faster than possible by manual biocuration into neuroinformatics databases.
Although NSyL can be used to encode connectivity described in the earlier literature, so-called legacy data, its ideal use (as proposed here) is as a tool for investigators to report novel macroconnections (in a systematic way amenable to automated biocuration) that have not yet been either asserted in the literature or incorporated into the manuscript submission process. Investigators’ use of NSyL from the outset would ensure that their findings could be promptly distributed, and cited, more efficiently than would be the case if the encoding of their findings were left to arbitrary manual biocuration.
We are committed to the development of a suite of web-based tools that will allow an investigator to easily create NSyL expressions for incorporation into a manuscript. These user-friendly, visually driven programs will help experimentalists and modelers encode connectivity without having to learn NSyL’s formation rules. Incorporating best practices in user-interface design and feedback, the tools will require a user to verify the fidelity of the connectivity asserted in a newly formed expression before releasing a final form of the expression ready for publication. This verification will help ensure that connectivity data later extracted from an expression accurately reflects the investigator’s observations or theories.
To streamline the use of NSyL and the process of automated biocuration, we propose the option that textual NSyL expressions be encoded as two-dimensional barcodes and entered as figures in manuscripts. These barcodes would consume a minimum amount of space in an article while adequately encoding a NSyL expression. Two-dimensional barcodes, such as the commonly used “QR codes,” are easily generated and deconstructed to-and-from text and are a convenient vehicle for NSyL expressions. With even 177 pixels square, a single QR Code can store over 4200 characters. This level of information density allows for relatively large amounts of connectivity to be systematically encoded (in both print and digital medium) using very little space even at standard print resolution (International Standards Organization, 2000). In the event an expression is larger than the informational capacity of a standard barcode (as may occur if one were to encode the entire connectome of a species or a large connectivity adjacency matrix), we propose that NSyL QR barcodes could instead contain an encoded identification number and decryption key that, as a pair, specify an identification number and access key to the full expression stored in an online expression repository. Such a system would allow authors securely to create an expression encoding up to thousands of macroconnections that would remain private until a published article is available to the community in public manuscript-indexing databases such as PubMed.
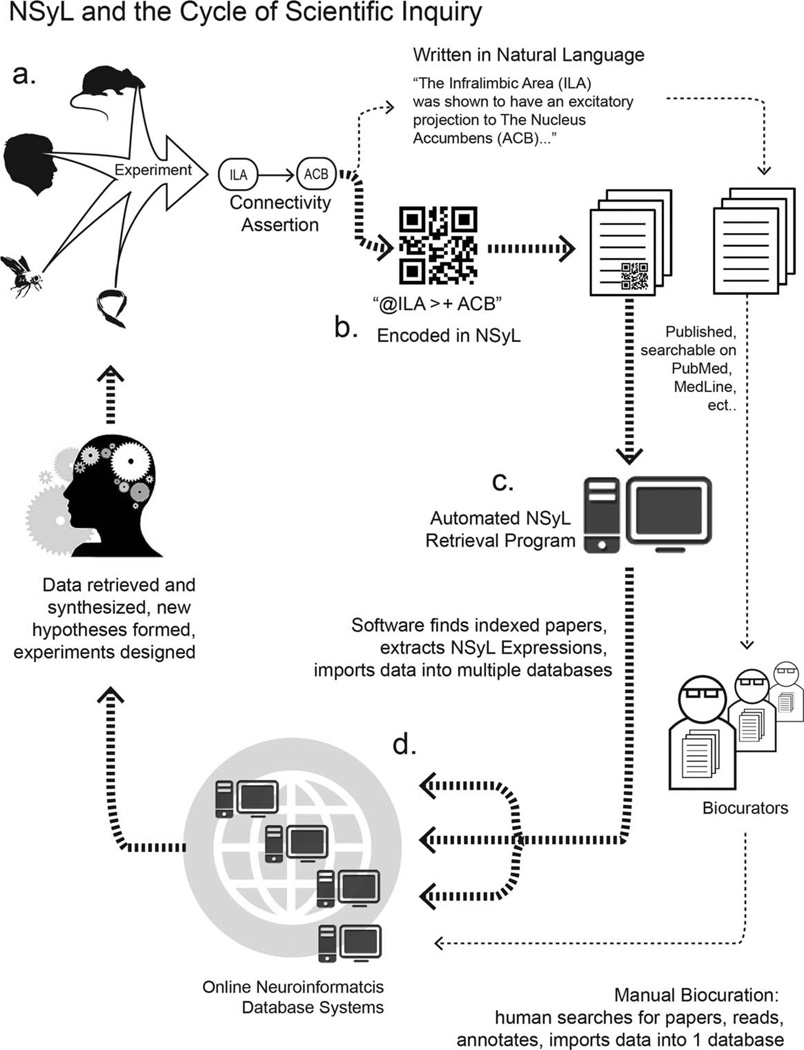
Additionally, we are committed to develop and implement web-based programs for the automated retrieval and extraction of NSyL expressions from published and indexed manuscripts and the automated distribution of this extracted data to neuroinformatics databases. NSyL expressions incorporated into manuscripts as text or figures (for example, using the proposed two-dimensional barcode example; Figs. 6–8) could be automatically retrieved and decoded for their connectivity data with little manual biocurative effort (Fig. 9). We propose that, once decoded, these programs would be able to share the extracted data with existing neuroinformatics databases in a free and open manner using established web protocols and standards (for example, such as by using an XML serialization and web service API). This proposed streamlined curation of connectivity data via automated expression retrieval and distribution would accelerate biocuration as a whole, increase the quantity of data available to any informatics system, and spread novel information faster and farther throughout the systems neuroscience community.
Figure 6.
Observed structural connectivity. A subset of the results of a multitracer double coinjection pathway tracing experiment are shown above. Note that, per NSyL’s formation rules, experimental sets may be nested and a given source node, such as the infralimbic area (ILA), may appear more than once in the entire expression. Adapted with permission from Thompson and Swanson (2010).
Figure 8.
Theoretical connectivity. Although this model applies equally to the general taxon Vertebrates, the nomenclature used necessitates declaring rat as the taxon. Note the closed-loop relationship of reciprocal influence between the organism and the environment, both internal and external. Figure 6 is also an example of the use of grouping container symbols, labels, and multiple arrowheads. Adapted with permission from Swanson (2012).
Figure 9.
Proposed implementation and workflow. a: Systems neuroscience experiments performed generate information (assertions) about neural connectivity. b: Experimentalists who encode their findings in NSyL can place NSyL expressions as text (as Supplemental Information or directly in manuscripts) or as figures (for example, as encoded in standard two-dimensional QR Code barcodes) c: Automated web software can parse publication repositories (such as PubMed) for papers containing NSyL expressions, automatically decoding the expressions. d: Communicating the contained connectivity information into different neuroinformatics databases. Use of our proposed online, user-friendly tools for encoding results in NSyL will effectively “precurate” experimental results for streamlined entry into neuroinformatics workbenches.
RESULTS
To be an effective modeling language, NSyL must be able to encode connectivity of different types (structurally observed, correlationally inferred, or theoretical) in different taxa using different techniques and nomenclatures. Although the goal of NSyL as a framework is not primarily to re-encode connectivity already reported in the literature but rather systematically and efficiently to encode and semiautomatically curate future assertions of connectivity, we can test the effectiveness of NSyL as a system for encoding connectivity by encoding real-world examples from legacy literature as case studies. Here we present legacy experimental assertions (from different species) of observed structural (Fig. 6), inferred correlational (Fig. 7), and theoretical model (Fig. 8) macroconnectivity from the existing literature encoded as macrolevel NSyL expressions, graphical schematics, and two-dimensional barcodes that could be incorporated as figures into published articless to help with automate biocuration via online automated retrieval and encoding. We show that the formation rules (both graphical and textual) for macrolevel NSyL balance systematicity with robustness and are able adequately to encode connectivity flexibly but consistently.
Figure 7.
Inferred correlational connectivity. A subset of an inferred correlational human brain network shown to be atypical in schizophrenia. Per the author’s nomenclature selection, the numbers listed inside nodes correspond to Brodmann cerebral cortical areas in the human. Generated from data used with permission from Bassett et al. (2008).
The example shown in Figure 6 is a subset of observed structural connectivity findings from Thompson and Swanson (2010). In accordance with macrolevel NSyL formation rules, the results are contained in a series of nested experimental sets with varying experimental parameters. Although all connectivity assertions in the expression are valid for the male Sprague-Dawley strain rat using Swanson’s (2003) brain atlas and nomenclature, their experimental techniques vary (between different forms of experimental pathway tracing). As such, four experimental sets (varying in experimental technique) are contained within the larger set describing nomenclature, taxon, and sex.
Figure 7 displays the connectivity results of a human fMRI experiment (Bassett et al. 2008). Note that, whereas Figure 7 represents an experiment with a different technique, species, and nomenclature (as reflected in the experimental parameters), the entire expression strongly resembles that in Figure 6. This consistency highlights NSyL’s ability to encode connectivity systematically regardless of whether it is used to encode assertions in the human or a model system. As specified in macrolevel NSyL’s formation rules, inferred correlational connections in which the functional polarity (excitatory, inhibitory, or neuromodulatory) and directionality (which node is the connection source, which node is the connection target) are not specified use a different set of symbols to communicate connectivity in the NSyL expression and a different line segment to connect nodes in the graphical NSyL schematic. It is worth noting that, although there exist a multitude of different nomenclatures and atlases by which human (or any species) data may be interpreted, the nomenclature listed in the expression is the nomenclature with which the authors interpreted their results as expressed in their article. As a framework for encoding and communicating connectivity, NSyL concerns itself with the effective description of findings and relegates the tasks of nomenclature selection and internomenclature comparisons to expression authors and neuroinformatics databases, respectively. As proposed here, the relationship that a system such as NSyL would have with neuroinformatics databases relegates the task of mapping between different nomenclatures chosen by different authors to the databases.
Finally, Figure 8 displays the use of multiple types of connections, influences, components, and grouping containers to encode Swanson’s (2012) Four-Subsystems Model of vertebrate nervous system architecture. Note that the containers used are labeled, and both influences between components and the environment (both internal and external) may cross the boundaries of containers. Consistent formation rules and NSyL expression structure are maintained to describe systematically theoretical models of connectivity in a manner consistent with observed structural and inferred correlational connectivity.
DISCUSSION
NSyL systematically encodes observations and inferences about the connective architecture of the nervous system in different species, using optional nomenclatures and different techniques within a common descriptive framework. This systematicity in description represents an advance in the field’s ability effectively to describe and communicate findings among model systems and has the potential to leverage advances in neuroinformatics and digital standards of communication. These advances will help build systems-level understanding of neural network architecture from otherwise disjointed observations and will streamline the process of biocuration.
The neuroanatomy community has historically struggled with systematicity. Since the time of Aristotle, the community has lacked both controlled vocabularies for terminology and parts as well as methods to map between different nomenclatures or parcellation schemes of the nervous system (Swanson, 2000). NSyL’s compatibility with any chosen internally consistent, hierarchically organized nomenclature does not coerce experimentalists into conforming to one naming system or parcellation. However, to address the classical challenges in ambiguous naming and the description of findings, NSyL requires experimentalists to state explicitly the anatomical nomenclature with which their assertions were made. Theoretically, the most descriptively powerful requirement that could be asked of the author may be requiring both an internally consistent (complete and nonoverlapping), hierarchically organized reference nomenclature with an accompanying carefully constructed anatomical reference neuroanatomical atlas. Inroads are being made to map different nomenclatures to one another and map nomenclatures to reference atlases (as mentioned), but we believe that the most prudent requirement, given the state of the field, is to require an expression author to state explicitly a single reference nomenclature. Requiring experimentalists to reference their nomenclature minimizes ambiguity regarding the circuitry described and allows different assertions to be compared against one another between nomenclatures online in databases, such as through topological mappings as performed in the Brain Architecture Knowledge Management System (Bota et al., 2005). It is only through this standardization in communication and description that systems neuroscience can leverage modern informatics tools to build integrative understandings of the nervous system.
Systems neuroscience and neuroinformatics are at a disadvantage in description and systematicity of communication (that NSyL may ameliorate) compared with other fields of scientific inquiry and engineering. Powerful analytic tools and data management frameworks for understanding the brain have been developed, and numerous experiments have been performed to describe the nervous system, all without any formalized methods for systematically describing, communicating, and curating experimental results into said data frameworks. This stands in stark contrast to other fields. For example, no one could imagine modern electrical engineering or computer science without the systematic conventions those communities employ in the design and development of digital systems or electronic circuitry. However, for neural circuitry, communication and representation of connectivity remain ambiguous, neuroinformatics databases remain sparsely populated compared with the amount of information available in the existing literature, and a significant gap remains between the producers and the consumers of connectivity data.
The current solution to bridge the gap between the two, manual biocuration, remains dauntingly time, expertise, and labor intensive. To optimize the information flow between those who discover or propose novel connections in the nervous system and those who structure this data for mass storage and analysis, a common systematic, machine- and human-readable discourse must be established. As proposed, NSyL could enable those who conduct experiments to describe their results in a manner that, by its controlled formation rules, would engender unambiguous communication between experimentalists within their own community as well as semiautomated communication between experimentalists and informaticians. With careful implementation and adoption, frameworks such as NSyL could improve the systematicity of encoding findings in neuroanatomy and systems neuroscience and increase the rate at which novel data are imported to neuroinformatics knowledge management systems as the field of connectomics grows and more experiments are performed.
NSyL, as we propose it here, is an open and evolving project. As mentioned above, macrolevel NSyL is one of three languages that we believe must be developed. We are committed to developing macrolevel NSyL and are beginning the process of charting out the steps we must take to develop the necessary digital infrastructure for the language. We look forward to creating community portals online in which we would like to engage the systems neuroscience community on extending macrolevel NSyL and determining the vision and implementation of macrolevel NSyL and the proposed remaining languages. As an extension of the Foundational Model of Connectivity (Swanson and Bota, 2010), NSyL is subject to change as the FMC and the fields of neuroanatomy and systems neuroscience continue to grow.
Just as anatomy ought not be dictated by authority but rather by evidence, need, and consensus, so too is NSyL controlled by no central authority but rather may be augmented or modified according to evidence, need, and consensus in harmony with a NSyL Community Guidance Committee to be established. We urge that our colleagues see how formal descriptive systems such as NSyL help to address the classical and contemporary challenges outlined here, become involved in the use and development of NSyL, and adopt NSyL into their practices. The need for NSyL—and automation and systematicity in neuroanatomy and connectomics—has never been more pressing. It is only through engaging anatomists, imagers, modelers, neuroinformaticians, and biocurators in active discussion that NSyL and formal descriptive systems like it will achieve its goals in minimizing ambiguity, systematizing communication, and increasing the impact and efficiency of systems neuroscience research.
ACKNOWLEDGMENTS
Grant sponsor: National Institutes of Health; grant numbers: 5R01NS050792; 5R37NS016686.
We thank Drs. Dani Bassett, Mihail Bota, Ed Bullmore, Gully A.P.C. Burns, Joel Hahn, and Richard H. Thompson and Mr. T. Dalton Combs for their valuable discussions and assistance.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ROLE OF AUTHORS
Drafted the preliminary structure of the language itself and the manuscript: RAB. Contributed equally to the development, refinement, and revision of the language and the manuscript: RAB, LWS.
LITERATURE CITED
- Alvarez-Bolado G, Swanson LW. Developmental brain maps: structure of the embryonic rat brain. Amsterdam: Elsevier; 1996. [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TEJ, Sporns O. Human connectomics. Curr Opin Neurobiol. 2011;22:1–10. doi: 10.1016/j.conb.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Wu C, Barbas H, Bokil H, Bota M, Breiter HC, Cline HT, Doyle JC, Freed PJ, Greenspan RJ, Haber SN, Hawrylycz M, Herrera DG, Hilgetag CC, Huang ZJ, Jones A, Jones EG, Karten HJ, Kleinfeld D, Kötter, Lester HA, Lin JM, Mensh BD, Mikula S, Panksepp J, Price JL, Safdeih J, Saper CB, Schiff ND, Schmahmann JD, Stillman BW, Svodoba K, Swanson LW, Toga AW, Van Essen DC, Watson JD, Mitra PP. A proposal for coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at the mesoscopic level scale. PLoS Comput Biol. 2009a;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Bokil H, Allen CB, Mitra PP. The brain atlas concordance problem. PLoS One. 2009b;4:e7200. doi: 10.1371/journal.pone.0007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Arbib MA. Integrating databases and expert systems for the analysis of brain structures: connections, similarities, and homologies. Neuroinformatics. 2004;4:19–58. doi: 10.1385/NI:2:1:019. [DOI] [PubMed] [Google Scholar]
- Bota M, Swanson LW. The neuron classification problem. Brain Res Rev. 2007a;5:79–88. doi: 10.1016/j.brainresrev.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Swanson LW. Online workbenches for neural network connections. J Comp Neurol. 2007b;500:807–814. doi: 10.1002/cne.21209. [DOI] [PubMed] [Google Scholar]
- Bota M, Swanson LW. BAMS neuroanatomical ontology: design and implementation. Front Neuroinform. 2008;2:1–8. doi: 10.3389/neuro.11.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Swanson LW. Collating and curating neuroanatomical nomenclatures: principles and use of the Brain Architecture Knowledge Management System (BAMS) Front Neuroinform. 2010;4:1–16. doi: 10.3389/fninf.2010.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Dong H-W, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6:795–799. doi: 10.1038/nn1096. [DOI] [PubMed] [Google Scholar]
- Bota M, Dong H-W, Swanson LW. Brain Architecture Management System. Neuroinformatics. 2005;3:15–48. doi: 10.1385/NI:3:1:015. [DOI] [PubMed] [Google Scholar]
- Bota M, Dong H-W, Swanson LW. Combining collation and annotation efforts with an automated inference engine for completion of the rat and mouse connectomes in BAMS. Front Neuroinform. 2012;6:1–10. doi: 10.3389/fninf.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute RT. System semantics and formal circuit description. IEEE Trans Circuits Syst. 1986;33:1219–1231. [Google Scholar]
- Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressleret SL. Beta oscillations in a large-scale sensorimotor cortical network: directional influences revealed by Granger causality. Proc Nat Acad Sci U S A. 2004;101:9849–9854. doi: 10.1073/pnas.0308538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Claessen K, Sands D. Observable sharing for functional circuit description. Lect Notes Comput Sci. 1999;1742:78. [Google Scholar]
- Delanda M. Philosophy and simulation: the emergence of synthetic reason. New York: Continuum International Publishing Group; 2011. [Google Scholar]
- Fox P, Ingham R, George MS, Mayberg H, Ingham I, Roby J, Martin C, Jerabek P. Imaging human intra-cerebral connectivity by PET during TMS. Neuroreport. 1997;8:2787–2791. doi: 10.1097/00001756-199708180-00027. [DOI] [PubMed] [Google Scholar]
- Gakh A, Burnett M. Modular Chemical Descriptor Language (MCDL): composition, connectivity, and supplementary modules. J Chem Inform Comput Sci. 2001;41:1494–1499. doi: 10.1021/ci000108y. [DOI] [PubMed] [Google Scholar]
- Haugeland J. Mind design. Montgomery, VT: Bradford Books; 1981. [Google Scholar]
- International Commission on Zoological Nomenclature. International Code of Zoological Nomenclature. 4th ed. London: International Trust for Zoological Nomenclature; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Standards Organization. Information technology—Automatic identification and data capture techniques—Bar code symbology—QR code. Geveva, Switzerland: ISO Copyright Office; 2000. [Google Scholar]
- Issac JT, Nicoll RA, Malenka RC. Evidence for silent synapses: implications for the expression of LTP. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodríguez EJ, Alemán-Gómez Y, Melie-García L. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage. 2008;40:1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Krettek JE, Price JL. The cortical projections of the mediodorsal nuclei and adjacent thalamic nuclei in the rat. J Comp Neurol. 1977;171:157–192. doi: 10.1002/cne.901710204. [DOI] [PubMed] [Google Scholar]
- Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, Bergman FT, Gauges R, Ghazal P, Kawaji H, Li L, Matsuoka Y, Villèeger A, Boyd SE, Calzone L, Courtot M, Dogrusoz U, Freeman TC, Funahashi A, Ghosh S, Jouraku A, Kim S, Kolpakov F, Luna A, Sahle S, Schmidt E, Watterson S, Wu G, Goryanin I, Kell DB, Sander C, Sauro H, Snoep JL, Kohn K, Kitano H. The Systems Biology Graphical Notation. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Melville RV. Towards stability in the names of animals. London: International Trust for Zoological Nomenclature; 1995. [Google Scholar]
- Raikov I, Cannon R, Clewley1 R, Cornelis H, Davison A, Schutter ED, Djurfeldt M, Gleeson P, Gorchetchnikov1 A, Plesser HE, Hill S, Hines M, Kriener B, Le Franc Y, Lo CC, Morrison A, Muller E, Ray1 S, Schwabe1 L, Szatmary B. NineML: The Network Interchange for Neuroscience Modeling Language. BMC Neurosci. 2011;12:330. [Google Scholar]
- Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:245–251. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW. Brain maps: structure of the rat brain: a laboratory guide with printed and electronic templates for data, models, and schematics. 2nd ed. Amsterdam: Elsevier; 1998. [Google Scholar]
- Swanson LW. What is the brain? Trends Neurosci. 2000;23:519–527. doi: 10.1016/s0166-2236(00)01639-8. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Interactive brain maps and atlases. In: Arbib MA, Grethe JG, editors. Computing the brain: a guide to neuroinformatics. San Diego: Academic Press; 2001. pp. 167–177. [Google Scholar]
- Swanson LW. Brain maps: structure of the rat brain. 3rd ed. Amsterdam: Elsevier; 2003. [Google Scholar]
- Swanson LW. Brain architecture: understanding the basic plan. 2nd ed. New York: Oxford University Press; 2012. [Google Scholar]
- Swanson LW, Bota M. Foundational Model of Structural Connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc Nat Acad Sci U S A. 2010;107:20610–20617. doi: 10.1073/pnas.1015128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Moorby P. The Verilog hardware description language. Boston: Kluwer Academic Publishers; 1996. [Google Scholar]
- Thompson RH, Swanson LW. Hypothesis-driven structural connectivity analysis supports network over hierarchical model of brain architecture. Proc Nat Acad Sci U S A. 2010;107:15235–15239. doi: 10.1073/pnas.1009112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner N. Cybernetics: or control and communication in the animal and the machine. Cambridge, MA: The Technology Press; 1948. [Google Scholar]
- Weininger D. SMILES: a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inform Comput Sci. 1988;28:31–36. [Google Scholar]