Abstract
OBJECTIVE
This study aims to analyze the role of artificial neural networks (ANNs) in cytopathology. More specifically, it aims to highlight the importance of employing ANNs in existing and future applications and in identifying unexplored or poorly explored research topics.
STUDY DESIGN
A systematic search was conducted in scientific databases for articles related to cytopathology and ANNs with respect to anatomical places of the human body where cytopathology is performed. For each anatomic system/organ, the major outcomes described in the scientific literature are presented and the most important aspects are highlighted.
RESULTS
The vast majority of ANN applications are related to cervical cytopathology, specifically for the ANN-based, semiautomated commercial diagnostic system PAPNET. For cervical cytopathology, there is a plethora of studies relevant to the diagnostic accuracy; in addition, there are also efforts evaluating cost-effectiveness and applications on primary, secondary, or hybrid screening. For the rest of the anatomical sites, such as the gastrointestinal system, thyroid gland, urinary tract, and breast, there are significantly less efforts relevant to the application of ANNs. Additionally, there are still anatomical systems for which ANNs have never been applied on their cytological material.
CONCLUSIONS
Cytopathology is an ideal discipline to apply ANNs. In general, diagnosis is performed by experts via the light microscope. However, this approach introduces subjectivity, because this is not a universal and objective measurement process. This has resulted in the existence of a gray zone between normal and pathological cases. From the analysis of related articles, it is obvious that there is a need to perform more thorough analyses, using extensive number of cases and particularly for the nonexplored organs. Efforts to apply such systems within the laboratory test environment are required for their future uptake.
Keywords: artificial neural networks, neural networks, artificial intelligence, cytopathology, cytology, review, automation, computer-assisted diagnosis, decision support
Introduction
Computer science and artificial intelligence have enabled the development of computer-aided systems to support clinical diagnosis or therapeutic and treatment decisions. Many machine learning methodologies such as neural networks,1–7 discriminant analysis,1,8,9 classification and regression trees,10,11 genetic algorithms,12 and recently, deep learning13–15 have been successfully used in medicine, whereas other techniques are in the center of current research studies.
Cytopathology is a relatively new medical discipline, which in most countries is considered to be a branch of pathology. In cytopathology, diseases are studied and diagnosed at a cellular level (free cells or small tissue fragments traditionally examined via the microscope). This discipline was founded by Papanicolaou in 192816 and became popular when he proposed the worldwide known Pap test. This test is used as a screening tool for detecting precancerous cervical lesions and thus preventing cervical cancer (CxCa).17,18 However, CxCa is not the sole disease that cytopathology deals with. Even in its early days,19–21 cytopathology was commonly used to investigate thyroid lesions, fluids in body cavities (peritoneal, pleural, pericardial, and cerebrospinal), and in almost the total range of body sites. In addition, cell study is not only used for cancer diagnosis, but it can also be employed for the diagnosis of infectious diseases and inflammatory conditions (eg, viruses, fungi, and bacteria). One of the major advantages of cytopathology practice is its noninvasive or minimally invasive nature, ie, the biological material is extracted from the patients in a painless manner (eg, cells are extracted using a brush, spatula, or fine needle), in contrast to histopathology that examines tissues and thus biopsy, and sometimes anesthesia, is required.
Cytopathology and pathology22 seem to be highly suitable medical sectors for the application of artificial neural networks (ANNs), because diagnosis is usually performed by specialized cytopathologists and involves criteria justified via microscopic examination.23 This procedure rarely involves quantification steps, can be prone to errors, and is relatively subjective. On the other hand, measurements of the examined cells and their formations are possible with the use of digital cameras and image analysis software; thus, it is technologically feasible to introduce a high degree of objectivity through measurements. However, processing a multitude of measurements can also be a barrier, especially because diagnosis should be performed in a timely and objective manner so as to be helpful in supporting decision-making in cytopathology.
In this review, research efforts related to the application of ANNs for decision support in cytopathology diagnosis are presented. From our bibliographic research, there were no other publications summarizing the applications of ANNs in cytopathology. The analysis is performed on the basis of human systems or organs. The structure of this review is as follows: (a) a short introduction of ANNs is provided; (b) the cytopathology subdisciplines are presented, highlighting the human systems in which ANNs can support the diagnostic process; (c) the applications of ANNs in each biological system are presented and the results are analyzed; and finally (d) in conclusion, future applications and directions are presented, barriers for the application of ANNs in the everyday practice are highlighted, gaps in various cytopathology research areas are spotted, and potential applications of ANNs in the cytopathology laboratory of the future are introduced.
Artificial Neural Networks
ANNs are complex computational models inspired by the human nervous system, which are capable of machine learning and pattern recognition.24–26 These models have the ability to learn from the past experience in order to provide outcomes for new data. This capability of learning from a certain data set makes ANNs suitable for classification and prediction tasks in practical situations. Furthermore, ANNs are inherently non-linear and nonconvex, which makes them more suitable for processing complex data patterns, in contrast to many traditional methods based on linear techniques.
Usually, ANNs are constructed by interconnected neurons (Fig. 1). Each neuron sums the weighted inputs (sum of the product of each input i multiplied by the corresponding weight ω), and the summation result is passed from a nonlinear, nonconvex, and nonconcave module (transfer function or activation function). The output of this function is the artificial neuron output as well.
Figure 1.
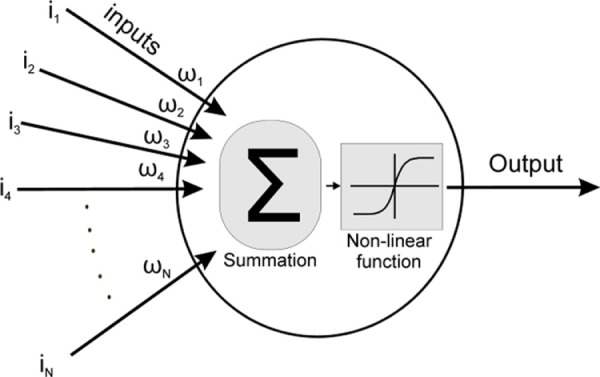
Schematic diagram of an artificial neuron. From left to right: inputs (i1–iN) are multiplied with synaptic weights (ω1–ωN), the products are added (Σ), and the result is passed from a nonlinear function to produce the neuron output.
A network of interconnected artificial neurons forms the ANN; neurons are interconnected, in a way that the output of one is the input of one or more other neurons. In general, the model of interconnections does require a specific structure; however, for algorithm development and simplicity, the most popular interconnection models have a layered approach. More specifically, there is an input layer of neurons, which receives the input data, one or more hidden layers, and finally an output layer providing the output of the network. Therefore, the structure of such ANNs is denoted in the form of X-Y-Z, where X, Y, Z are the number of input, hidden, and output neurons, respectively. This typical structure of a multilayer feed-forward ANN appears in Figure 2. This is the structure of the most widely used neural network: the multilayer perceptron (MLP) network.25,26 As depicted in Figure 2, each layer of the MLP network includes one or more neurons directionally linked with the neurons of the previous and the next layers.
Figure 2.
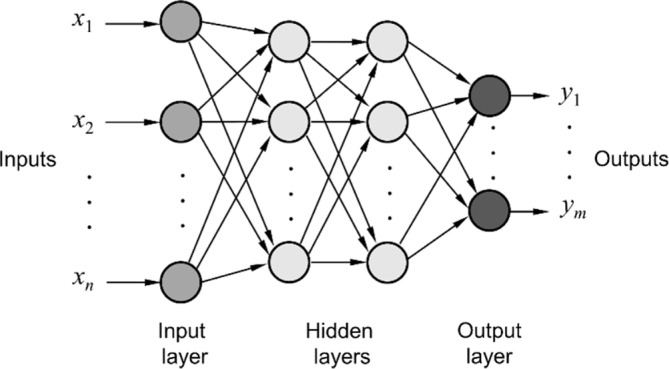
Typical structure of a feed-forward multilayer neural network.
Besides MLP network, there are many other types of neural networks, including probabilistic neural networks (PNNs), general regression neural networks, radial basis function (RBF) networks, cascade networks, Kohonen networks or self-organizing maps (SOMs), learning vector quantization (LVQ), Hebb networks, recurrent neural networks (RNNs), and hybrid networks.25,26 These networks may have different structures compared with MLP network, some of them may be more complex than others and generally they are based on different learning algorithms. The selection of the appropriate type of neural network depends on the machine learning problem at hand, as each ANN type has been designed to address specific issues. However, the learning and the predictive abilities of these networks are not only problem dependent but are also determined by several other factors, such as the parameters of the specific type, the network’s architecture (topology), the learning algorithm chosen for training, and the characteristics of the data provided to the network.
Depending on the type of machine learning problems that they are called to solve, ANNs can be grouped under three main categories, namely, clustering, classification, and regression. As far as the learning algorithms are concerned, they can also be categorized into supervised, unsupervised, and semisupervised learning.
In unsupervised learning (clustering problem),24 there is neither an explicit teacher nor training samples. This technique is applied when the classes of the samples of a data set are known in advance. The classification of the feature vectors is accomplished by examining the similarity among them based on the criteria defining the desired properties of groups. The SOMs, also known as Kohonen maps, are the most popular ANNs for clustering problems.
In supervised learning (classification and regression problems),24 a set of training examples are presented to the model in order to train it. More specifically, for each training sample, a feature vector characterizing the sample along with its correct class is provided to the ANN. Based on these training samples, the classifier learns how to assign a new feature vector to a correct class; thus, it obtains the ability to learn from past experience in order to provide outcomes for new data. In developing intelligent clinical decision support systems, supervised classification methods are more commonly used than unsupervised methods since the task requires outcome prediction, eg, prediction of disease status. The most common ANNs used for supervised classification are the MLP network, the PNN, and the RBF network, whereas in cases of regression problems, the most widely used are the general regression neural network and the MLP network.
Essentially, the construction of an ANN involves the development of algorithms addressing a specific problem. The difference between an ANN and a conventional computational system is the learning through training process, which resembles the learning capability of the brain. Via training/learning, the ANN self-adapts and changes its structural characteristics. This is based on the information that flows through the network neurons.
The pipeline for creating a system with intelligence powered by an ANN and designed to be used for a production system has several, but typical, steps (Fig. 3).
Figure 3.
Typical cycle for ANN system development.
The first step requires the collection of the data. The data set size is closely related to the problem’s nature, and several times, this data size is required to be augmented after evaluation of the ANN system’s results. Feature extraction and feature selection algorithms have to be applied in this step in order to create an appropriate processing by the ANN input dataset.
The second step is data preprocessing. More specifically, data are processed so that any inconsistent values are removed. Then, mathematical transformations are applied in order to map alphanumeric data to numbers suitable for the specific ANN model employed. Additionally, the available data are separated into three sets as follows: the training set, which is used to train the ANN (ie, to add the knowledge to the system); the validation set, which is used as a test platform for fine-tuning the model’s parameters and selecting the best-performing ANN; and the test set, which is used to assess the performance of the developed model on data that have not been used in any previous step of the designing process. More advanced performance assessment methods such as cross-validation27 are also very popular. By cross-validation, data are divided many times in different training and test sets. Subsequently, many models are created and tested; finally, the model performance is assessed on the basis of the outcomes. Cross-validation is an important technique when the size of the available data is small and/or when the number of parameters in the model is large. Via cross-validation, overfitting and model robustness can be assessed.
The third step involves the selection of the appropriate ANN model and the definition of its parameters and characteristics. These characteristics are related to the architecture of the ANN. For example, determining the right architecture of an MLP network involves selecting the optimal parameters for the network, such as the number of hidden layers, the number of neurons of each hidden layer, the transfer functions of the neurons, and the learning rate and momentum of the learning algorithm. In general, according to the ANN type, there are individual characteristics that should be adjusted in advance, or, sometimes, defined during the development of the system, via trial and error method.
Next, the ANN training takes place. More specifically, the training data set is presented to the network layers, and the fine-tuning of the system parameters is performed by testing it on the validation set. During this step, the ANN’s learning algorithm is applied. The role of this algorithm is to minimize the total errors of the system. Toward this purpose, a large number of cases with a known outcome are provided to the network. The backpropagation (BP) algorithm25,26 is the most common learning technique for training a typical MLP network. During training with the BP algorithm, information about the errors of the network on known data is propagated backward from the output layer to the input layer and is used to adjust the connections between the layers and their neurons (the weights and biases of the network) in order to minimize the error and thus improve performance.
In the final step of the pipeline, the performance of the system is evaluated using the test set, which contains samples that have not been used previously in the designing and training process (unseen data). Satisfactory results of the final step indicate that the system has an acceptable performance and, therefore, can be used in a production environment for routine usage. In the opposite case, all the steps should be reexamined from the beginning. The system trainer should go one or more steps backward, and after the appropriate changes are made, the process should be repeated until satisfactory performance is achieved. Acceptable performance means that the ANN can predict the outcome of cases that have never been presented to the system before.
Cytopathology is a medical specialty that is heavily based on images; therefore, the application of ANNs is based on image features. The typical pipeline to create ANNs (Fig. 4) includes: step a) image acquisition, usually via a digital camera attached to a microscope; step b) according to the type of ANNs: b1) nuclei isolation (segmentation) and subsequent measurements or b2) transformation of the images, for example, via Fourier transform or b3) no processing (when the image itself is the direct input to the ANN). The outcome of step b is a data set characterizing the nuclei. This data set (step c) may be augmented by additional data, for example, clinical details (results from other examinations), demographic information (such as patient age), or quantified descriptive characteristics of the specimen under examination. In step d, the data are separated into training and test sets, and as mentioned earlier (Fig. 3), an additional validation set is created. These three sets are used for ANN model training, testing, and validation (step e). Typical nuclei morphometry characteristics include (but are not limited to) geometric characteristics such as nucleus area, major and minor axis length, perimeter, or elongation and pixel-based characteristics such as mean optical density for each color component, standard deviation of color histograms, or more complex features related to texture (eg, gray-level cooccurrence matrix characteristics or fractal properties).
Figure 4.
Typical application of ANNs in the field of cytopathology.
Cytopathology Areas of Interest
Nowadays, cytopathology is applied to the majority of human organs and systems. Specifically, its subdisciplines can be considered as follows:
gynecologic cytopathology, applied in the female reproductive tract such as the cervix, the endometrium, or the ovaries;
cytopathology of the urinary tract, for the examination of ureters, the bladder, and urethra;
effusion cytopathology, involving body fluids, specifically from the peritoneum, pleura, and pericardium;
breast cytopathology, evaluating cells from the breast;
thyroid cytopathology, for the thyroid gland via fine-needle aspiration (FNA);
lymph node cytopathology, concerning the lymph nodes;
cytopathology of the respiratory system, relevant to the lungs and airways;
cytopathology of the gastrointestinal system, relevant to the alimentary tract, but in a broader definition to the complete system from mouth to anus;
cytopathology of soft tissues, bones, and skin;
cytopathology for material obtained from the kidney and adrenal;
liver and pancreas cytopathology;
central nervous system cytopathology, mainly for the examination of cerebrospinal fluid;
cytopathology of the eye; and
cytopathology of the salivary gland.
Subsequently, the identified efforts to apply ANNs in the cytopathology subdisciplines are presented.
ANNs in Gynecological Cytopathology, the PAPNET System
ANNs are most commonly applied in cytopathology of the female reproductive system and particularly in the cytopathology of the cervix. A few hundreds of articles can be found in the literature related to ANNs for CxCa, whereas semiautomated commercial products based on ANNs are available. Based on flow cytometry (FCM), the development of these techniques has been constantly evolving from the 1950s up to present.28
One of the most well-known applications of ANNs in CxCa detection is the automated cytological screening system for cervical smears, known as the PAPNET system.29 According to the authors’ knowledge, PAPNET was the most successful commercial ANN-based product for CxCa detection. PAPNET has been mainly used in various comparative studies to test within the subcategories of cervical cytology reporting, in comparison with the light microscope examination (aka human observers).30–45 A meta-analysis study by Abulafia and Sherer46 concluded that compared with manual screening, PAPNET identified 20% more abnormal cases, it has 50% less false-negative cases, and it can classify as abnormal 33% of manually screened false-negative slides in rescreening. Additionally, better performance is reported on high-grade squamous intraepithelial lesion (HSIL) cases47 and in most of the cytological categories used for CxCa reporting. Especially for cases of squamous cell carcinoma (SCC), which were difficult to identify by the light microscope, PAPNET was able to identify all of them.39,48 As seen in the literature, this system has been extensively used in quality control and assurance,42,49,50 as well as, with regard to quality control stan-dards36 and for inter- and intraobserver variability studies.40 From these papers, it is concluded that PAPNET reduced the number of false-negative cases.51
The cost-effectiveness of PAPNET as a commercial ANN-based system has been assessed,52–55 and the results are contradicting. Some reports indicated that the cost per woman increased,56 taking into consideration that the cost-effectiveness depends heavily on the setting and varies from country to country. Other studies concluded that the cost of rescreening all negative slides with the PAPNET system, despite the fact that it provided increased sensitivity, could not be justified.57,58 Finally, there were cases in which the system was found to provide an economical solution.59
There are two major settings proposed for the use of PAPNET, either in primary CxCa screening or for rescreening purposes. For primary screening, the results showed that ANNs (or screeners) had better60–62 or similar41 performance with cytopathologists, while the screening process in one study is reported as a faster method to obtain diagnosis.43 For rescreening purposes,63–67 PAPNET achieved the desirable target to reduce the number of false-negative cases; however, in one paper, the cost-effectiveness of the system was considered to be questionable.68
Moreover, a third setting for PAPNET application is hybrid screening,69 in which case slides diagnosed by the ANN system are also reviewed by light microscope. The outcome of these research efforts suggests that pure ANN provides optimized prediction of HSIL or negative cases, but for the detection of low-grade squamous intraepithelial lesions (LSILs), the light microscope examination presents additional value.
The major drawback of the majority of comparative studies that are not targeting cytological categories of HSIL or cancer is that there is no evidence of comparisons with histological outcomes,70 ie, most of the research papers compare light microscope examination vs. the ANN system on cases without histological confirmation. In our opinion, it is extremely difficult to have histological confirmation on a large number of cases, because it is neither ethical nor legal to perform biopsies for cytological categories with normal, LSIL, and atypical squamous cells of undetermined significance. Additionally, comparative studies incorporate tens of thousands of cases; thus, it is not possible to have biopsies for all of them.
Finally, PAPNET was used for comparative studies involving human papillomavirus-related molecular tests31 with colposcopy techniques71 and has been compared with other non-ANN-based semiautomated screening systems, such as the Imager® by Hologic®72 or the AutoPap® Guided System.52
Cervix is not the sole organ of the female reproductive system. Uterus, ovaries, and the fallopian tubes are part of the complete system, and these organs are also of interest for pathology and cytopathology. Uterus, and more specifically endometrium, is a relatively easily accessible organ to extract cells, thus ideal for cytological diagnosis. Despite the applications of ANNs for the classification of images obtained via hysteroscopy,73 the literature related to cytological images is rather poor. In 2002, the potential of the LVQ ANN for the classification of cells from endometrial lesions in postmenopausal women was reported.74 The study involved a relatively small number of cases: 12 cases of atrophic endometrium (normal), 48 cases of hyperplasia without cytologic atypia, 12 cases of hyperplasia with cytologic atypia, and 48 cases of adenocarcinoma. The study applied morphometry in ~100 nuclei from each slide, which were subsequently used to train and test the LVQ ANN. LVQ appeared to be a good classifier at a cellular level and a useful tool for classification on the individual patient level. Specifically, the LVQ assigned 86% of atrophy nuclei, 75% of hyperplasias, and 90% of neoplasias correctly. Further discrimination between simple and complex hyperplasias and complex atypical hyperplasia was not possible, while discrimination of individual patients was not reported.
In the case of ovarian cancer, a great number of reports regarding the role of ANNs for the discrimination between malignant and benign tumors75 (ie, in histological material) can be found. However, in cytological material and despite the fact that it is feasible to obtain cells via image-guided FNA, no reports involving the application of ANNs in the cytopathology of ovaries are available.
ANNs in the Cytopathology of Gastrointestinal System
The gastrointestinal system is responsible for consuming and digesting food, absorption of nutrients, and expelling of wastes. In a broad definition, this system includes all organs and structures between the mouth and anus; esophagus, stomach and duodenum, small and large intestines, salivary glands, and pancreas are included in this system and, therefore, are of particular interest to cytopathology.
The application of ANNs to gastrointestinal cytopathology was first attempted in 1993.76 More specifically, Molnar et al analyzed 21 normal, 15 dysplastic, and 23 malignant, gastric imprint smears stained by Feulgen technique. The mean DNA content, the 2c deviation index (2cDI), 5c exceeding rate (5cER), G1, S, and G2 phase fraction ratios, cell nucleus area, and form factor were measured in this study. They applied discriminant analysis, yielding a diagnostic accuracy of 96% for the malignant cases, 87% for dysplasias, and 81% for normal cases. Subsequently, the BP ANN was applied, and all the normal and malignant cases (100%) and all but one of the dysplasias (98%) were correctly classified. In 1996, a second article77 investigated nuclear morphometry and the BP ANN for the discrimination of benign and malignant gastric lesions. The study group had 23 cancer, 19 gastritis, and 58 ulcer cases. In contrast to the method applied by Molnar et al,76 image morphometry on cell nuclei was employed in order to measure geometric and textural characteristics. In that particular study, the description of two different architectures of the BP ANN can be found. Ultimately, 97.6% of benign nuclei and 95% of malignant nuclei were correctly classified. These preliminary results indicated that image morphometry and ANNs may effectively discriminate malignant from benign gastric cell nuclei; however, no discrimination at single patient level was applied. One year later, the comparison of BP and LVQ was presented;78 the two ANNs gave comparable results, with an overall accuracy of ~97% for benign and malignant nuclei classification. After this paper, a study79 compared three variations of the LVQ that had similar performance. In 2000, a cascaded RNN to investigate cell nuclei from 19 cases of cancer, 19 cases of gastritis, and 56 cases of ulcer as a test set was applied.80 The application of the RNN gave correct classification in 96% of benign nuclei and 89% of malignant nuclei. In these papers, different ANN architectures were tested giving encouraging results in the classification of cell nuclei from the stomach, but without using the patient as end point.
Despite the fact that this PAPNET was originally designed for cervical cytopathology, in 1998 Koss et al81 used this system to examine cells from 138 esophageal smears obtained from an equal number of patients. PAPNET identified potentially malignant images, which were subsequently examined by medical experts. Specifically, 128 cell images are classified in each case. In this study, all 35 patients with cancer—esophageal, gastric, oral, or metastatic—were correctly identified; in 11 cases, effects of radiotherapy were detected; in 8 patients, SCC was recognized; 6 patients were found with adenocarcinoma; and 3 additional cases revealed carcinoma not further specified. Interestingly, PAPNET enabled the detection of one case of esophageal carcinoma in situ that was not previously recognized on a smear or biopsy specimens, as well as one case of gastric adenocarcinoma; thus, ANN-based systems succeeded with greater accuracy than human experts in an organ that the system was not originally designed to be applied.
In 1998, PAPNET was used to screen oral cytological material. In this setting, PAPNET proved to have a performance of 61% in detecting oral SCC,82 with false-negative cases being attributed to bad preparation. PAPNET was also used to investigate sputum cytological samples.83 The authors used 122 single slides of sputum specimens. According to the results from the 31 smears with cancer cells, 30 were appropriately identified by PAPNET (sensitivity: 97.1%), while PAPNET missed only one case of small cell carcinoma that contained just one cluster of neoplastic cells. PAPNET also resulted in reviewing 20 of the negative cases, which on reevaluation were identified as bronchial cells with squamous metaplasia and altered benign squamous cells of inflammatory type. According to the authors, before applying PAPNET for screening of sputum samples, a prospective study is needed in order to establish the clinical value of the method.
In 2008, a group of researchers published a report84 for preoperative prediction of tumor staging by ANNs using clinicopathological datasets and genetic variables. The study involved 121 patients with primary gastric cancer; the accuracy of the method used was 81.8%. These researchers discovered important factors associated with tumor staging, specifically age and polymorphisms of a number of specific genes. It was concluded that the proposed method had the potential to serve as a prognostic system for staging and for individualized patient care.
In summary, with regard to the application of ANNs to the organs of the gastrointestinal system, the investigation resulted in <10 articles with rather small number of cases. However, the results for all cases can be considered as encouraging and have the potential to trigger more extensive efforts in the everyday laboratory setting. In Table 1, the technical and performance characteristics of the applied ANNs in the gastrointestinal system organs are summarized.
Table 1.
Summary of technical characteristics of ANNs applied in cytopathology of the gastrointestinal system.
| REFERENCE NUMBER | DESCRIPTION OF CASES | VARIABLES | METHOD | RESULTS | DISCRIMINATION LEVEL |
|---|---|---|---|---|---|
| 76 | 21 normal, 15 dysplastic and 23 malignant cases | M-DNA, 2cDI, 5cER, G1, S, G2, nucleus aarea, nucleus form factor | Back propagation | OA = 100% on malignant cases (complete data set) OA = 98% on dysplasias (complete data set) |
Classification of patients |
| 77 | 23 cases of cancer, 19 of gastritis and 58 of ulcer | Nucleus morphometry variables (2,500 nuclei in the training set, 8524 nuclei in the test set) | Two variants of back propagation | OA = 97.6% of benign nuclei OA = 95% of malignant nuclei OA = 95.8% and 97.3% on the test set |
Classification of nuclei as benign or malignant |
| 78 | 30 cases of cancer, 26 of gastritis and 64 of ulcer | Nucleus morphometry variables (3,000 nuclei in the training set, 10,300 nuclei in the test set) | Comparison of back propagation and LVQ ANNs | Back propagation O.A = 98–99% (training set) O.A = 95.8% and 97.3% (test set) LVQ1 OA = 96.79 (complete set) LVQ2.1 OA = 96.73 (complete set) LVQ3 OA = 97.51 (complete set) |
Classification of nuclei as benign or malignant |
| 79 | 23 cases of cancer, 19 of gastritis and 58 of ulcer | Nucleus morphometry variables | LVQ ANN | O.A in four data splits ranged from 98.69%–98.82% sensitivity: 94.40%–95.00% specificity: 99.68%–99.71% (data set not specified) | Classification of nuclei as benign or malignant |
| 80 | 19 cases of gastritis and 56 cases of ulcer | Nucleus morphometry variables (20% of nuclei in the training set and 80% in the test set) | Cascaded recurrent neural network | 96% of benign nuclei and 88% of malignant nuclei (data set not specified) | Classification of nuclei as benign or malignant |
| 81 | 138 esophageal smears | Cytological images evaluated by PAPNET | PAPNET | All malignant cases were correctly identified, two cases not identified in previous examinations were identified (test set only) | Classification of patients according to the esophageal disease |
| 82 | 62 oral smears from 27 patients | Cytological images evaluated by PAPNET | PAPNET | 100% correlation between human and PAPNET examination. 61% of the patients were confirmed by biopsy (there is no training set) | Identification of patients with oral cancer |
| 83 | 122 sputum slides: 6 inadequate, 81 negative, 3 atypical, 1 suspect, 31 positive | Evaluation by PAPNET | PAPNET | Sensitivity: 97.1% (missed 1 small cell carcinoma) (there is no training set) | Identification of patients with disease |
| 84 | 121 patients with primary gastric cancer | Clinical data and pathological findings were collected and genetic polymorphisms of candidate genes were evaluated | Quick propagation | O.A. 81.82% in predicting tumor staging, and identification of important gene polymorphisms | Prediction of tumor staging |
ANNs in the Cytopathology of Thyroid Gland
Thyroid examination is one of the success stories of cytopathology because it is based on FNA; the method is relatively painless; cells extracted via this method are either directly placed on the glass slide or immersed in a solution and subsequently placed on a single cell layer. Thyroid FNA is performed with the help of ultrasonography (US-guided FNA) so that specific thyroid nodules can be accessed. According to the cytological examination findings, a decision is made whether a surgical operation of the thyroid gland is required and also the extent of the operation.
ANNs appeared in thyroid disease evaluation in 1993.85 However, applications in cytopathology appeared in 1996.86 The BP ANN was tested on the discrimination of benign from malignant thyroid nuclei, according to nuclear morphometry. More specifically, nuclei from 51 patients with thyroid diseases were processed. A correct classification of 90.6% of nuclei was achieved with the application of the BP NN. Subsequently, classification at a single patient level had an overall accuracy of 98% of the cases. In 1999,87 four variations of the LVQ classifier (LVQ1, LVQ2.1, LVQ3, and OLVQ1) were tested on a larger set: 100 cases of goiter and follicular adenomas (FAs), 11 cases of follicular carcinoma, 35 cases of papillary carcinoma, 24 cases of oncocytic adenoma, 8 cases of oncocytic carcinoma, and 20 cases of Hashimoto thyroiditis. In this paper, the mean value and standard deviation of nucleus morphometry features were used; thus, patients were represented from cumulative nuclei measurements. LVQ variations enabled 97.7% classification of benign vs. malignant patients, but no important results in the classification of other histological subgroups were reported. In 2006, Cochand-Priollet et al88 performed nuclear morphometry followed by statistical preselection of significant features (only four features were found to be important). In this study, four classifiers were subjected to comparison, namely, (a) a linear classifier, (b) a two-layer feed-forward ANN (2L-FNN), (c) combined 2L-FNN generated by the AdaBoost, and (d) the k-Nearest Neighbors classifier. The results of the classifiers were in the range of 83%–94%; however, the linear classifier had the worst performance (65%) in patient discrimination. This proved that ANNs can be more accurate than traditional methods due to their nonlinear nature.
In 2007, Shapiro et al89 analyzed 197 thyroid follicular tumors (adenomas and carcinomas), and the research group tested several types of ANNs with different designs. They used nuclear morphometric parameters (area, perimeter, and shape factor) and density features of chromatin texture (mean value and standard deviation of gray levels). The ANNs were based on the means of the cytological features’ characteristics in order to represent the outcome for a patient instead of single nuclei. The method increased the accuracy of diagnosis for follicular tumors to 97% (75 out of 78) of cases and ANNs distinguished adenomas from carcinomas by 87% (73 out of 84). In conclusion, the authors reported that ANNs raised the sensitivity of cytological diagnosis of follicular tumors to 90%, compared with the usual cytologic method (sensitivity of 56%).
In 2004, a research team studied the problem of thyroid cases with not determinate cytological diagnosis.90 The study had 453 patients and used a feed-forward ANN fed with cytological and clinical data. The classification aim was to categorize patients from high to low risk. The researchers discovered that only the cytological parameters contributed to this classification. Comparing the receiver-operating characteristic (ROC) curves, the ANN model discriminated with higher sensitivity and specificity between benign and malignant nodules compared with standard cytologic criteria and there was no degradation between the training and test set; thus, the method was confirmed to be robust and valuable in the gray zone of cytopathology of the thyroid, ie, in cases of undetermined significance.
In 2006, the use of a complex of quantitative cytological characteristics to differentiate between follicular cancer (FC) and FA (84 patients in the test set) with an overall accuracy of 96% on the test set was reported,91,92 and an effort to discriminate FAs from FCs was reported. This was an extremely interesting article, regarding the manner in which the ANN was applied. A two-layer ANN was employed. The first layer had one input assigned to each training image and the classification was based on image characteristics extracted via Fourier frequency bands; thus, no morphometry was applied. Also, the second interesting characteristic of this approach was that the system discriminated each case as FC, FA, or unknown. In this way, the research team not only applied an ANN directly on images but also additionally handled cases not possible to be otherwise discriminated with acceptable accuracy.
In 2008, Daskalakis et al93 reported on the use of a multiclassifier system for distinguishing benign and malignant thyroid nodules taken via FNA. The researchers constructed a multiclassifier system (ensemble of classifiers) using several combinations of rules and mixtures of the different classifiers involved. The study was based on nuclear morphological and textural features. The results indicated that the best ensemble of classifiers had better accuracy (95.7%) compared with the best single classifier (89.6%). The involved classifiers were linear least-squares minimum distance, statistical quadratic Bayesian, k-NN, support vector machine (SVM), PNN, and, of course, ensembles of these. This approach highlights a new combinatorial methodology for cytopathology that has the potential to obtain better accuracy.
In 2011, a new study94 investigated the potential of the LVQ on nuclei measured from monolayer (ThinPrep) smears. The study involved 335 patients and nuclear morphometry describing size, shape, and texture of ~100 nuclei per case. LVQ discriminated individual cells as benign or malignant, and a cascaded second LVQ ANN subsequently discriminated the patients. The application of the proposed combined ANNs had an overall accuracy of 94%. The study concluded that the diagnostic accuracy of thyroid FNA can be improved by the use of ANNs. More interesting results were for follicular neoplasms suspicious for malignancy and in Hürthle cell tumors. This is the second combinatorial application of ANNs, although it is using a sequential approach rather than the ensemble proposed in the study by Daskalakis et al.93
The most recent research in thyroid cytopathology was published in 2014.95 Ozolek et al described a method to distinguish between follicular lesions of the thyroid. They utilized the optimal transport-based linear embedding for segmented nuclei96 together with an adaptation of existing classification methods. The classification results were nearly perfect as they correlated well with the clinical diagnosis. The data set came from 94 patients, and despite that it was based on histological sections, the classification was based on isolated nuclei using a supervised method.97
The diverging techniques employed in these research efforts and the nonstandardized method of presenting the results do not allow comparison of the techniques. However, the main characteristics in each paper, ie, the cases involved in each study, the variables/features used, the ANN model, and methodology, as well as the major results of the ANNs applied in thyroid cytopathology, are summarized in Table 2.
Table 2.
Summary of technical characteristics of ANNs applied in thyroid cytopathology and performance metrics.
| REFERENCE NUMBER | DESCRIPTION OF CASES | VARIABLES | METHOD | RESULTS | DISCRIMINATION LEVEL |
|---|---|---|---|---|---|
| 86 | 25 cases of goiter and follicular adenomas, 1 case of follicular carcinoma, 12 cases of papillary carcinoma, 6 cases of oncocytic adenoma, 3 cases of oncocytic carcinoma and 4 cases of Hashimoto thyroiditis (13850 feature vectors) | Nucleus morphometry variables (training set: 2,770 nuclei, test set: 11,080 nuclei) | Back propagation used to classify nuclei and subsequently majority logic classifier for patients | Nuclei classification OA: 97.29% (training set) OA: 90.61% (test set) Patient classification OA: 98% of the patients classified correctly |
Nuclei and subsequently patients |
| 87 | 100 cases of goiter and follicular adenomas, 11 case of follicular carcinoma, 35 cases of papillary carcinoma, 24 cases of oncocytic adenoma, 8 cases of oncocytic carcinoma and 20 cases of Hashimoto thyroiditis | Nucleus morphometry features for each patient (training set: measurements from 59 patients, test set: measurements from 139 patients) | LVQ ANN | OA = 97.7% on benign from malignant patients (sensitivity: 94.9%, specificity: 98.9%) No important results in the classification of disease subgroups |
Classification of patients by the ANN on the basis of nuclei features |
| 88 | 157 cases from thyroid FNA | Nucleus morphometry, applied statistical pre-selection of features that revealed only four features as important | Comparison of a) Linear classifier b) 2 Layer Feed Forward Classifier 2L-FNN, c) combined 2L-FNN generated by the Ada-Boost d) k-NN classifier | OA = 83–94% except of the linear classifier (65%) Cumulative results in the training and test set |
Classification of individual nuclei and subsequently classification of patients via a threshold |
| 89 | 197 thyroid follicular tumors (100 adenomas and 97 carcinomas) | Nuclear morphometry and morphological features evaluated by experts And image features based on Fourier transform | Several types of back propagation ANN | Increased accuracy to 97% in follicular tumors, the use of color microscopic images enabled correct classification of adenomas from carcinomas in 87% of the cases (results in the test set) | Discriminating patients with adenoma form patients with carcinoma |
| 90 | 453 cases of indeterminate cytological thyroid results | Cytological and clinical data (371 cases in the training set and 82 cases in the test set) | Back propagation ANN | Increased accuracy comparing to cytology on the basis of ROC curves Only cytological data contributed to the classification Training and test set performance had no difference |
Classification of patients |
| 91, 92 | 55 cases of follicular adenoma and 49 cases of follicular carcinoma | Images and their Fourier trans form band characteristics (10 cases from each category were used in the training set and the remaining cases in the test set) | Two layer MLP ANN | Depending on threshold 82% of follicular adenomas and 59% of follicular carcinomas | Images and the related patients (3 images per patient) |
| 93 | 115 cases of thyroid FNA (53 benign and 62 malignant) | Nucleus morphological and textural features after the application of segmentation algorithm | Multiple classifiers: linear least squares minimum distance, statistical quadratic Bayesian, k-NN, SVM, PNN and ensembles of classifiers | OA = 95.7% for the best ensemble compared to OA = 89.6% for the best single classifier Sensitivity of the best ensemble classifier = 96.8% and specificity = 94.3% |
Classification of patient as benign nor malignant on the basis of one image per patient |
| 94 | 335 cases of thyroid FNAs spanning all cytological and histological categories | Nuclear morphometry (32,887 measured nuclei separation 50% in the training set and 50% in the test set) | Cascaded LVQ classifiers for nuclei and subsequently patient classification | Nuclei classification OA = 95.98% at the cellular level (training set) and 92.25 at the test set Patient classification All patients classified correctly (in the training and test set) |
Nuclei classification and subsequently patient classification as benign or malignant |
| 95 | 94 cases of thyroid FNAs | Linear optimal transportation of nucleus pictures (automated nucleus identification was applied) | k-NN | 100% in almost all follicular variants | Classification of patients on the basis of isolated nucleus pictures |
ANNs in the Cytopathology of the Breast
ANNs have been intensively used in breast cancer on the basis of imaging methods. A search in PubMed using the terms “breast” and “neural network” results in >50 publications, most of them related to ANNs for the evaluation of imaging material such as X-rays, US images, and thermographs. This is probably due to the fact that the standard screening method for breast cancer is based on imaging modalities. However, there have been efforts to evaluate cytological material as well.
In 1990, Wolberg and Mangasarian98 created three systems on the basis of cytological descriptive parameters using 369 cases as the training set and 70 cases as the test set (57 benign and 13 malignant). The first system generated a multisurface pattern separation and misclassified one malignant test sample; the ANN system misclassified two benign test samples and a classification and regression tree-based system misclassified three of the benign test samples. The results were promising as no malignant case was missed by the ANN. The following year (1991), Dawson et al used the CAS-100 (Cell Analysis Systems) system to measure and analyze nuclear morphometric and texture features of cytological preparations from 35 breast carcinomas (well, moderate, and poorly differentiated) as well as benign lesions.99 The extracted features were morphometric parameters and Markovian texture feature data from nuclei of different grades. Two methods were tested, namely, multivariate Bayesian analysis and an ANN. The researchers tried to perform deep classification (ie, to identify the nuclear grade of individual nuclei). Both classification systems were able to assign a correct grade to low-grade lesions (~70% correct) more often than to high-grade tumors (~20%). The researchers reported that difficulties in correct assignment of high-grade tumors could be explained by the presence of nuclear heterogeneity in these tumors, ie, high-grade nuclei frequently did not predominate in high-grade smears. This research highlighted the problem of completely automated systems, where the landscape of the cytological slide is not dominated by abnormal cells; thus, such systems should have the capability to assist diagnosis based on a few cells or clusters.
In a 1992 study,100 an ANN was used successfully to predict the clinical outcome of node-positive breast cancer patients. The aim was to predict the clinical outcome for 1008 patients. Input information involved clinical laboratory data, specifically tumor hormone receptor status, DNA index and S-phase measured by FCM, tumor size, number of axillary lymph nodes involved with tumor, and age of the patient, as well as the length of clinical follow-up, relapse status, and time of relapse. The ANN was tested on a separate set of 960 cases. The ANN was as powerful as Cox regression modeling in identifying breast cancer patients at high and low risk for relapse.
In 1997, two studies investigated the role of ANNs in breast cytological material. In the first study,101 the BP ANN was tested for the distinction of benign from malignant breast cell nuclei. The study was carried out on 68 carcinomas and 32 benign breast lumps. A set of image nuclear features was investigated in the discrimination at the nucleus level (achieving an overall accuracy of 87%), and subsequently at the patient level, correct diagnosis was achieved in 98% of cases. On the same data set, the LVQ ANN102 was applied that correctly classified 87.4% of the cells; subsequently, a threshold-based method achieved the same accuracy on individual patients. Both studies indicated that ANNs and image morphometry have the potential to assist the diagnosis of breast cytological material.
One year later (1998), Ohno-Machado and Bialek103 compared the selection of variables for building two classification models: ANN and logistic regression. The researchers used a set of 460 cases to build the models that would classify cell samples obtained by FNA as malignant or benign, depending on nine pathology features. Variables selected by a step-down logistic regression model were compared with those selected by a measure of relevance derived from ANN weights. Despite that both types of models resulted in similar predictive accuracy, the same variables were not identified as important for classification. Thus, variable relevance based on weights for neural network models does not seem to be a consistent index of the importance of that variable for multivariate models such as logistic regression. This study highlighted the use of ANNs in the evaluation of discrimination power of individual features.
The same year, Reigosa et al104 used 46 cases of intraductal breast carcinoma aiming to classify them into high- or low-grade nuclear grade on the basis of histological images and a BP ANN. Despite the fact that the authors used histological material, the results seem to be reproducible in cytological material because all the studied characteristics were based on nuclear measurements. At least 200 nuclei per case were measured using various morphometric characteristics. However, the variance of the measurements was used as an input to the ANN in order to approximate the complete picture. The ANN system had an accuracy of 97.5% as compared with the consensus of at least two of the three expert pathologists who decided on the grading. Note that consensus between any two pathologists was no higher than 85%, while consensus of the BP ANN with any of the three pathologists was no smaller than 87.5%. This work demonstrated that ANNs could act as decision support tools in the definitive classification of intraductal carcinomas.
Einstein et al,105 in 1998, explored the use of fractal analysis in the numerical description of chromatin appearance in breast cytology. They used images of nuclei from FNA of the breast characterized in terms of their Minkowski and spectral fractal dimensions. The study involved 19 patients with benign epithelial cell lesions and 22 with invasive ductal carcinomas. The authors proved that chromatin appearance in breast epithelial cell nuclear images could be considered as fractal. Not only statistically significant differences between the fractal properties of benign and malignant nuclei were found but it was also suggested that it is possible to assign human understandable texture description to fractal properties. The research team applied logistic regression, which correctly diagnosed 95.1% of the cases and ANNs that classified all cases correctly.
The most recent papers of ANNs in breast cytological material appeared in 2013 and 2014. In the first article, Dey et al106 used histology-proven breast lesions consisting of 20 fibroadenomas, 28 infiltrating ductal carcinomas (IDCs), and 16 infiltrating lobular carcinomas (ILCs). The morphometric analysis was performed on hematoxylin and eosin (H&E)-stained smears. The novelty of this work was the introduction of some objective morphological information produced by human experts such as sample cellularity and nucleoli characteristics, among others. The researchers applied a BP ANN, which, in the test set, classified all the benign and ILC cases and six of the seven IDC cases. Thus, the authors implemented an ANN able to differentiate IDC from ILC on FNA material and introduced contextual features evaluated by cytopathology experts. In 2014,107 the application of a BP ANN model constructed on the basis of cytomorphological data, morphometry, nuclear densitometry data, and gray-level cooccurrence matrix information was reported. The researchers used 52 cases of fibroadenomas and 60 cases of IDC on H&E-stained smears. The BP model identified all cases of fibroadenomas and infiltrating carcinomas in the test set.
Table 3 summarizes the results of the efforts on breast cytopathology, technical details of the applied systems, as well as the performance metrics.
Table 3.
Summary of technical characteristics of ANNs applied in breast cytopathology and performance metrics.
| REFERENCE NUMBER | DESCRIPTION OF CASES | VARIABLES | METHOD | RESULTS | DISCRIMINATION LEVEL |
|---|---|---|---|---|---|
| 98 | 369 cases of breast FNAs as training set and tested on 70 cases (57 benign and 13 malignant) | Cytological descriptive features | Multisurface pattern separation, MLP and CART | The expert system misclassified one malignant test sample, the ANN misclassified two benign test samples and the CART misclassified three benign test samples | Classification of patients as benign or malignant |
| 99 | 35 breast carcinomas (well, moderate, and poorly differentiated), testing was on 31 unknown cases | Morphometric and Markovian texture feature data from breast cancer nuclei | Multivariate analysis and MLP | OA = 70% in low-grade lesions (both systems had similar performance) OA = 20% in high-grade tumors |
Assigning grading to nuclei |
| 100 | 1008 patients were used for training and internal test and 960 cases were used for validation | Clinical and laboratory data, including DNA index and S-phase measured by flow cytometry | MLP and Cox regression model | Similar results of ANN and Cox regression model | Aim was to predict recurrence of disease |
| 101 | 68 carcinomas and 32 benign breast lesions | Nucleus morphological and textural features (3000 nuclei in training set, about 6000 nuclei in the test set) | BP at the nucleus level and 50% threshold to discriminate patients | OA = 87% at the nucleus level and 98% at the patient level (training and test set together) | Classification of nuclei by the ANN, subsequently patient classification |
| 102 | 68 carcinomas and 32 benign breast lesions | Nucleus morphological and textural features (3000 nuclei in training set, about 6000 nuclei in the test set) | LVQ ANN classified individual nuclei and subsequently patients were classified via a threshold | OA = 87.41% at the nucleus level and OA = 98% at the patient level | Classification of nuclei by the ANN, subsequently patient classification |
| 103 | 687 cases of breast cytology (450 malignant and 237 benign) | 9 morphology variables describing cytological features (460 cases for the training set and 227 cases for the test set) | ANN and logistic regression to identify important variables | Similar classification results (ROC AUC = 98%) however variables selected by the ANN were not the same as in logistic regression for ANN | Patient classification by morphology variables of cytological specimens |
| 104 | 46 intraductal breast carcinoma cases to be classified into high or low grade nuclear | Nucleus morphometry features (images from 6 cases were used in the training set and 40 in the test set) | Back propagation ANN | OA = 97.5% in the test set | Classification of lesions as high or low grade |
| 105 | 19 patients with benign epithelial cell lesions and 22 patients with invasive ductal carcinomas | Fractal dimensions of nuclei (not training and test set separation) | MLP and logistic regression model | OA = 100% for the ANN AO = 95.1% for the logistic regression (leave one out validation) | Classification of patients as benign or malignant |
| 106 | 64 cases of histology proven breast lesions: 20 fibroadenomas, 28 infiltrating ductal carcinomas (IDC), and 16 infiltrating lobular carcinomas (ILC) | Cytomorphological and morphometric features of the specimens | Back propagation (training set: 40 cases, validation set: 8 cases, test set: 16 cases) | In the test set all benign cases and IDC were classified correctly and 6 out of 7 IDC cases | Discrimination of patients into three groups: benign, IDC and ILC |
| 107 | 52 cases of fibroadenomas and 60 cases of infiltrating ductal carcinoma | Cytomorphological information, and morphometry | Back propagation (training set: 71 cases, validation set: 9 cases, test set: 22 cases) | The BP model identified all cases of fibroadenomas and infiltrating carcinomas in the test set and missed one malignant case in the validation set | Differentiation of patients with fibroadenomas from patients with infiltrating ductal carcinomas |
ANNs in the Cytopathology of the Urinary System
The majority of efforts related to urological cancer and ANNs are based on clinical data,108,109 specifically for prostate cancer prognosis, bladder cancer diagnosis, grading and prognosis, testicular and renal cancer staging on the basis of tissue specimens, and additionally for image interpretation and planning of treatment. Cytology of the urinary system is based on urine collection and, therefore, is a completely painless and relatively easy process to apply for the patient, in contrast to the discomfort caused when a tissue sample has to be acquired. Thus, it is a very popular discipline.
There is a relatively small number of reports on cytological material. The idea of computer-based discrimination of urothelial cells according to our search first appeared in 1975.110 Approximately 15 years later, Moallemi111 presented an ANN-based system that could serve as the front end of any image analysis application targeting urothelial cells. More specifically, he implemented an MLP network-based algorithm that operates using automated image measurements on cytological images. The MLP network decides if the objects under investigation are cells that require further processing or should be rejected as not important. The proposed methodology achieved an accuracy of 93.4% in detecting objects that require subsequent processing. Despite the fact that the ANN does not perform the classification step into medical diagnoses, it introduces the concept of selecting objects in the cytological field of view for subsequent analysis via the use of ANNs. This introduces the idea of the use of ANNs for fully automated systems.
In 1998, Pantazopoulos et al112,113 applied the BP and LVQ ANN on 50 cases of lithiasis, 61 of inflammation, 99 of benign prostatic hyperplasia, 5 of carcinoma in situ, 71 of grade I, and 184 of grade II or III transitional cell carcinoma of the bladder. The approach was based on image morphometry and subsequently on ANN application on nuclei measurements. At the patient level, the methods gave correct classification of 100% of benign patients and 94.51% of malignant patients with 96.96% overall accuracy. The comparison of BP and LVQ114 indicated that both ANNs had similar performance at the patient level. The same year Van Biesen et al115 reported on the use of unsupervised ANNs, specifically on the use of 10 × 10 neuron Kohonen’s SOM on 41 mainly clinical and a few cytological parameters from 75 patients aiming to discriminate patients with glomerular from tubular disease. Their results showed that SOMs have higher sensitivity and predictive value than the nephrologists’ diagnoses and that the best classification was based on a hybrid system. Specifically, sensitivity and predictive value for the diagnosis glomerular were 100% and 88% for the SOM, 90% and 83% for nephrologists, and 95% and 96% for the hybrid system, respectively. The authors concluded that ANNs may give a new insight into the diagnostic approach of renal disease and that the approach uncovered misconceptions in the problem-solving strategies of the clinicians. According to the literature search, this was the sole article that reports the application of a nonsupervised ANN on cytological material.
Two years later (2000), Vriesema et al116 performed a pilot study of the PAPNET system. The researchers used PAPNET to detect urothelial cell carcinoma of the bladder. They employed 85 bladder wash samples and compared the results obtained with a commercial image analysis system QUANTICYT (BioProcon) and human examination using the microscope. They provided risk estimation as low, intermediate, and high. The sensitivity of ANNs for diagnosing was 92%, whereas microscope and QUANTICYT showed sensitivities of 50% and 69%, respectively. For the prediction of a positive cystoscopy, the highest area under the curve (AUC) for ANNs was found to be 0.71, while it was 0.58 for the microscope. For predicting tumor recurrence after a negative cystoscopy, QUANTICYT had the highest AUC value (0.62), whereas ANN had 0.50.
ANNs in the Cytopathology of Effusions
Effusion cytology is related to fluid collections, usually from the peritoneum, pleura, and pericardium. Such fluids are collected with the use of a needle; the fluid is centrifuged to obtain cells and subsequently create slides. Despite that effusion cytology is extensively applied, according to the search results, there are only two reports of ANNs’ applications.
In 1995, Truong et al117 applied BP ANNs on image morphometry data for 112 Papanicolaou smears of lymphocyte-rich effusions. The aim was to differentiate reactive lymphocytosis from malignant lymphoma. An architecture of 7-10-1 neurons with sigmoid transfer function, using five morphometry and two densitometry features as inputs, enabled the correct classification of 89.3% of the test set, with sensitivity of 76.9% and specificity of 93.0%. The same BP architecture with a step transfer function gave an overall accuracy of 95.3%, sensitivity of 85.7%, and specificity of 97.6%. Obviously, the transfer function had an important effect on performance. Additionally, the results on the nucleus level classification were encouraging. This paper highlighted the role of the ANNs’ parameterization in the final accuracy.
The second report was in 2012; Barwad et al118 built a 25-2-1 BP ANN for differentiating carcinomas from benign cases in effusion cytology. The researchers used 114 cases (57 benign and 57 malignant). Image morphometric, densitometric, and chromatin texture data were used as inputs to the system. Simultaneously, a logistic regression analysis was performed for direct comparison with the ANN: both the ANN and the logistic regression identified correctly all the benign and malignant cases. Both reports provided consistent results in cellular and patient level, indicating the potential of ANNs in effusion cytopathology, considering that larger data sets are needed and extensive validation is required in the clinical practice.
Discussion and Conclusions
In general, ANNs and artificial intelligence have been used in clinical decision support since the early days of computing. Even from the 1990s,5 the concept of introducing ANNs in cytopathology tasks was presented, aiming mainly to create and operate prototype ANNs designed and developed to solve simple pattern recognition problems. In addition, ANNs have been proposed for database analysis and machine vision.5 There are several advantages in choosing such an approach: ANNs have the ability to tolerate ambiguous, noisy, and spurious data, and since they can be trained, they can handle unfamiliar cases. ANNs were proposed to be used alongside other traditional algorithmic processing techniques for the development of systems useful in quantitative pathology.
The analysis of the literature proved that the vast majority of ANN applications are for cervical cytopathology. Additionally, there are commercialized ANN-based systems and the literature relates not only to diagnostic performance but also to the application of ANNs in quality control and cost-effectiveness studies. This is an expected result as the first widespread application of cytopathology is related to CxCa. Other cytopathology disciplines have exploited ANN technologies to a much smaller degree. These are in agreement with the popularity of cytological examinations according to the biological systems/organs. The majority of the proposed ANN applications are related to breast and thyroid cytopathology as well as cytopathology of the urinary tract. ANNs have been applied to a smaller degree on cytopathology of the gastrointestinal system and to a lesser degree in effusion cytopathology. Yet, there are cytopathology subdisciplines that have not yet used ANNs, especially, cytopathology of the lymph node, respiratory system, soft tissues, bone and skin, liver and pancreas, central nervous system, and the eye, among others. This is probably due to the fact that there are significantly few patients who benefit from these cytology examinations. Additionally, the biological material is difficult to be collected, and the impact is smaller because it affects few patients. In summary, application of ANNs on such types of biological material is still an open research topic today.
Excluding cervical cytopathology, in the vast majority of articles, results of the application of ANNs on cell nuclei measurements were presented. This is to our opinion a disadvantage, because, despite that cytological diagnosis is based mainly on nuclear characteristics, the contextual characteristics should not be neglected (ie, structures formed by nuclei are important diagnostic criteria). Only a few articles have taken into account the contextual characteristics, although in a subjective manner, because there is no measurement process but a human-based context characterization. In some papers, the standard deviation of nuclei measures was used to approximate such conditions. Thus, in the majority of the efforts, contextual characteristics were missing or not objectively evaluated. Moreover, other important findings, for example colloid presence in thyroid cytopathology, were not considered at all. The use of contextual parameters to our opinion is very important to achieve accurate diagnosis; thus, efforts toward the inclusion of scene context in an objective (ie, measurable) manner are required. In fact, commercialized systems take into account such characteristics as the first slide evaluation is in low magnification.
The isolation and selection of nuclei in most of the cases were performed by human experts. This is due to technical difficulties in automated segmentation of the images for automatic nuclei detection. In fully automated systems, nuclei detection, separation from artifacts, identification and separation of touching, or overlapping nuclei are crucial. Techniques creating slides with single-cell layer (monolayer) come to facilitate this procedure; however, image-processing algorithms with acceptable performance both in speed and detection accuracy for nuclei isolation in a routine setting are missing. Such algorithms are heavily dependent on both staining and cell characteristics of each tissue type and anatomical site; note that the human body has more than 200 different cell types.
Understanding the cell nucleus textural content is another important issue, as nucleus chromatin and other organelles are crucial in malignant cases; thus, specific textural measures are required. Most of the texture measurement methods are not performing well, for example in the differentiation of nuclei with a chromatin pattern distributed in the periphery. A link of the nucleus textural morphometry, as this is perceived by human beings, to fractal-based texture measurements was reported in only one article. Methods where ANNs are applied on nuclei images seem to have an advantage in the textural characterization of the nucleus.
The number of nuclei required for the classification of a case is important. However, only one study stated the minimum number of nuclei required. The selection of nuclei that will be measured and subsequently evaluated by ANNs is important. In most cases, there is a manual nuclei selection; thus, a selection process is applied in advance. However, in the field of view, there are coexisting cells presenting malignant characteristics along with benign nuclei. In an automated setting, proper differentiation should be performed.
Accurate reproduction of measurements in different imaging systems is important. For example, microscope light intensity may affect pixel-based measurement and hence some researchers have performed histogram equalization, while others have performed light adjustment in order to achieve specific color values in an empty field.
In terms of the type of ANNs employed, the popular multilayer feed-forward BP architecture was the most used. Only one article exploits the capabilities of unsupervised techniques, such as the SOM and <5 papers using LVQ, RBF, and other architectures and algorithms. There was only one paper in which the effect of the transfer function in the ANN performance was evaluated. Thus, ANN-related studies may consider the effect of the parameters on the performance metrics.
Recently, deep learning algorithms have shown impressive performance in machine vision applications, including medical image processing and histopathology image analysis.13–15,119,120 A deep neural network (DNN) is an ANN with multiple hidden layers of units between the input and output layers.121 DNNs have the exact same structure as classical MLP neural networks, except that the number of hidden layers is greater so that they can be considered deep. Similar to shallow ANNs, DNNs can model complex nonlinear relationships. One of the main advantages of the DNNs compared with classical ANNs is the fact that these models do not need to be provided with predefined features chosen by an engineer, but they can learn features from the data set by themselves, ie, they perform automatic feature extraction without human intervention, unlike most traditional machine learning algorithms. The ability to automatically extract hierarchies of features from images is the reason why DDNs perform so well in image recognition and classification tasks13–15,119–121 and are so promising for cytopathology applications directly on microscopic images.
Modern cytopathology laboratories apply molecular biology techniques as an adjunct to cytological examination. FCM is one of these methods,122 as it provides new quantitative information that complements the morphological examination during microscopic inspection. It is particularly useful for assessing lymphoid lesions, from FNAs, cerebrospinal fluid, or effusions. FCM may quantify a lot of parameters simultaneously on the basis of cell differentiation markers and DNA quantification. In FCM, the number of measured cells is in the order of tens of thousands, and there are various parameters measured for every cell. Thus, the development of ANN-based methods seems to be of interest if applied on FCM data.123 Actually, some applications of ANNs in FCM124–128 have been reported; some authors report that for field studies, the problem of obtaining suitable training data needs, as well as the number of cell categories, is an issue. This is clearly a field that needs further research.125
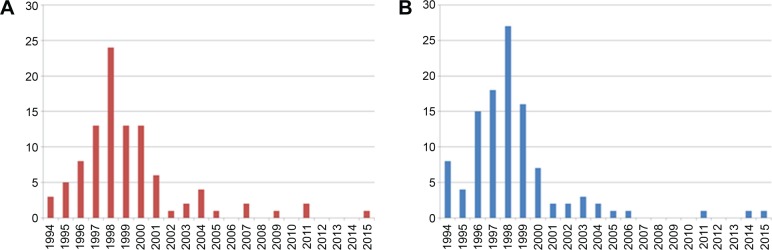
According to search results from SCOPUS and PUBMED bibliographical database for articles relevant to PAPNET, the number of relevant publications peaks in 1997–2000. Since 2000, the literature can be considered rather poor (Fig. 5).
Figure 5.
Historical evolution of publications with the keyword PAPNET according to SCOPUS bibliographic database (A) and PUBMED (B).
Efforts to apply ANNs in cytopathology seem to have stopped around 2000. We can only speculate about this phenomenon as follows: (a) the PAPNET use as a commercial system has declined during these years; therefore, the PAPNET-related publications (dominating this research) seem that followed this trend, (b) a second reason could be overfitting; in some papers, there is no separation of data into training and test sets, and in the majority of articles, the robustness of the ANNs is not evaluated, and (c) obtaining morphometry data is an extremely hard process, because there are no well-established nuclei segmentation algorithms, and the use of ANNs directly on the images is a very new field, after deep architectures have emerged.
Comparison of the performance of the applied techniques is not always feasible. The overall accuracy is not the ideal performance index, as misclassifying a disease case has a different effect compared with misclassifying a nondisease case. Specificity and sensitivity are more representative performance indices; however, these metrics are not provided in all articles. Additionally, the proportion of data included into the training and test set is not standardized, and in some cases, a test set is not there at all. Therefore, comparison of the involved techniques cannot be objective. This fact points toward the requirement of providing the used datasets to the research community and to standardize performance metrics.
It is well known that ANNs have some disadvantages as follows: (1) a large number of variables is required that have to be parameterized during learning, comparing for example to decision trees, (2) they are black box models, thus it is not possible to perform knowledge discovery, as for example in decision trees, where it is possible to extract human understandable rules that lead to the classification result, (3) some architectures can be trapped to local minima, and not lead to the optimal solution, (4) the training process, especially for the BP algorithm, requires a lot of computational power. Hopefully, modern computers will be able to provide such resources, and GPUs (Graphics Processing Units) can be used to decrease training by hundreds of times and training algorithms can be parallelized, (5) overfitting on the training data set is a frequent issue, thus special care should be taken to ensure their robustness, and (6) there are no established algorithms allowing to add new knowledge to ANNs without training them again, ie, when more data become available it is not possible to use only this new data and gain new knowledge from an already-trained ANN. Despite the disadvantages, the literature research showed that ANNs have the capability of learning and may assist cytopathologists in their decision-making. If they are used correctly, ANN models can be superior to standard statistical methods, allowing flexible data interrogation and reliable prediction of disease status. Advances in image analysis and intensive tests on large data collections in the cytopathology environment are the requirements for the widespread implementation of the method. ANNs may become an important asset in the cytopathology laboratory of the future and lead toward better decision support systems.
Acknowledgments
The authors thank the anonymous reviewers for their constructive comments; these comments changed the shape of the paper.
Footnotes
ACADEMIC EDITOR: Kayvan Najarian, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1612 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and Designed the experiments: AP, EK, NM, PB, MH. Analyzed the data: AP, EK, NM. Wrote the first draft of the manuscript: AP, PB, MH. Contributed to the writing of the manuscript: EK, NM, PB, MH, JP, DK, PK. Agree with manuscript results and conclusions: AP, JP, DK, PK. Jointly developed the structure and arguments for the paper: NM, JP, DK, PK. Made critical revisions and approved final version: AP, EK, PB, MH JP, DK, PK. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Marchevsky AM, Tsou JA, Laird-Offringa IA. Classification of individual lung cancer cell lines based on DNA methylation markers: use of linear discriminant analysis and artificial neural networks. J Mol Diagn. 2004;6(1):28–36. doi: 10.1016/S1525-1578(10)60488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramaniam S, Mehrotra M, Gupta D. Support vector machine based classification model for screening Plasmodium falciparum proliferation inhibitors and non-inhibitors. Biomed Eng Comput Biol. 2011;3:13–24. [Google Scholar]
- 3.Karar ME, El-Brawany MA. Automated cardiac drug infusion system using adaptive fuzzy neural networks controller. Biomed Eng Comput Biol. 2011;3:1–11. [Google Scholar]
- 4.Mobasser F, Hashtrudi-Zaad K. A comparative approach to hand force estimation using artificial neural networks. Biomed Eng Comput Biol. 2012;4:1–15. doi: 10.4137/BECB.S9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dytch HE, Wied GL. Artificial neural networks and their use in quantitative pathology. Anal Quant Cytol Histol. 1990;12(6):379–393. [PubMed] [Google Scholar]
- 6.Lisboa PJ, Taktak AF. The use of artificial neural networks in decision support in cancer: a systematic review. Neural Netw. 2006;19(4):408–415. doi: 10.1016/j.neunet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Lisboa PJ. A review of evidence of health benefit from artificial neural networks in medical intervention. Neural Netw. 2002;15(1):11–39. doi: 10.1016/s0893-6080(01)00111-3. [DOI] [PubMed] [Google Scholar]
- 8.Karakitsos P, Megalopoulou TM, Pouliakis A, Tzivras M, Archimandritis A, Kyroudes A. Application of discriminant analysis and quantitative cytologic examination to gastric lesions. Anal Quant Cytol Histol. 2004;26(6):314–322. [PubMed] [Google Scholar]
- 9.McHugh ES, Shinn AP, Kay JW. Discrimination of the notifiable pathogen Gyrodactylus salaris from G. thymalli (Monogenea) using statistical classifiers applied to morphometric data. Parasitology. 2000;121(pt 3):315–323. doi: 10.1017/s0031182099006381. [DOI] [PubMed] [Google Scholar]
- 10.Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and Regression Trees. Belmont, CA: Wadsworth International Group; 1984. [Google Scholar]
- 11.Tzivras M, Megalopoulou TM, Pouliakis A, et al. The potential of tree classifiers on the cytological examination of gastric lesions. Arch Balk Med Union. 2008;43(4):239–244. [Google Scholar]
- 12.Saiti F, Naini AA, Shoorehdeli MA, Teshnehlab M. Thyroid Disease Diagnosis Based on Genetic Algorithms Using PNN and SVM; Paper presented at: The 3rd International Conference on Bioinformatics and Biomedical Engineering; June 11–13, 2009; Beijing. [Google Scholar]
- 13.Chen T, Chefd’hotel C. Deep learning based automatic immune cell detection for immunohistochemistry images. In: Wu G, Zhang D, Zhou L, editors. Machine Learning in Medical Imaging. Vol. 8679. Berlin: Springer International Publishing; 2014. pp. 17–24. [Google Scholar]
- 14.Lai M. Deep learning for medical image segmentation. arXiv. 2015;arXiv:1505–02000. [Google Scholar]
- 15.Yan X, Tao M, Qiwei F, Peilin Z, Maode L, Chang EIC. Deep learning of feature representation with multiple instance learning for medical image analysis; Paper presented at: 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 4–9 May, 2014; Shanghai, China. [Google Scholar]
- 16.Papanicolaou G. New cancer diagnosis; Paper presented at: 3rd Race Betterment Conference; Battle Creek: 1928. [Google Scholar]
- 17.Papanicolaou G, Traut H. The diagnostic value of vaginal smears in carcinoma of the uterus. Am J Obstet Gynecol. 1941;42:193–206. [PubMed] [Google Scholar]
- 18.Diamantis A, Magiorkinis E, Koutselini H. 50 years after the death of George Nicholas Papanicolaou 1883–1962 evaluation of his scientific work. Acta Med Hist Adriat. 2014;12(1):181–188. [PubMed] [Google Scholar]
- 19.Diamantis A, Beloukas AI, Kalogeraki AM, Magiorkinis E. A brief chronicle of cytology: from Janssen to Papanicolaou and beyond. Diagn Cytopathol. 2013;41(6):555–564. doi: 10.1002/dc.22887. [DOI] [PubMed] [Google Scholar]
- 20.Naylor B. The century for cytopathology. Acta Cytol. 2000;44(5):709–725. doi: 10.1159/000328553. [DOI] [PubMed] [Google Scholar]
- 21.Williams C, Rosenthal DL. Cytopathology in the 21st century. Am J Clin Pathol. 1993;99(4 suppl 1):S31–S33. [PubMed] [Google Scholar]
- 22.Dybowski R, Gant V. Artificial neural networks in pathology and medical laboratories. Lancet. 1995;346(8984):1203–1207. doi: 10.1016/s0140-6736(95)92904-5. [DOI] [PubMed] [Google Scholar]
- 23.Dey P. Time for evidence-based cytology. Cytojournal. 2007;4:1. doi: 10.1186/1742-6413-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theodoridis S, Koutroumbas K. Pattern Recognition. 4th ed. London: Academic Press; 2009. [Google Scholar]
- 25.Haykin SS. Neural Networks: A Comprehensive Foundation. New York, NY: Macmillan; 1994. [Google Scholar]
- 26.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd ed. New York, NY: Wiley; 2001. [Google Scholar]
- 27.Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010;4:40–79. [Google Scholar]
- 28.Bengtsson E, Malm P. Screening for cervical cancer using automated analysis of PAP-smears. Comput Math Methods Med. 2014;2014:12. doi: 10.1155/2014/842037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouwer RK, MacAuley C. Classifying cervical cells using a recurrent neural network by building basins of attraction. Anal Quant Cytol Histol. 1995;17(3):197–203. [PubMed] [Google Scholar]
- 30.Hakama M, Pokhrel A, Malila N, Hakulinen T. Sensitivity, effect and overdiagnosis in screening for cancers with detectable pre-invasive phase. Int J Cancer. 2015;136(4):928–935. doi: 10.1002/ijc.29053. [DOI] [PubMed] [Google Scholar]
- 31.Nieminen P, Vuorma S, Viikki M, Hakama M, Anttila A. Comparison of HPV test versus conventional and automation-assisted Pap screening as potential screening tools for preventing cervical cancer. BJOG. 2004;111(8):842–848. doi: 10.1111/j.1471-0528.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 32.Irwig L, Macaskill P, Farnsworth A, et al. A randomized crossover trial of PAPNET for primary cervical screening. J Clin Epidemiol. 2004;57(1):75–81. doi: 10.1016/S0895-4356(03)00259-2. [DOI] [PubMed] [Google Scholar]
- 33.Polus E, Karttunen TJ. Maximal efficiency of PAPNET in the diagnosis of infections in cervicovaginal smears. Diagn Cytopathol. 2003;28(5):286–287. doi: 10.1002/dc.10283. [DOI] [PubMed] [Google Scholar]
- 34.Nieminen P, Hakama M, Viikki M, Tarkkanen J, Anttila A. Prospective and randomised public-health trial on neural network-assisted screening for cervical cancer in Finland: results of the first year. Int J Cancer. 2003;103(3):422–426. doi: 10.1002/ijc.10839. [DOI] [PubMed] [Google Scholar]
- 35.Jeronimo J, Castle PE, Herrero R, et al. Right-sided ectocervical lesions may be associated with false-negative cytology among women with histologic cervical intraepithelial neoplasia 2 or 3. J Low Genit Tract Dis. 2003;7(3):175–183. doi: 10.1097/00128360-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Mudu P, Migliore G, Alderisio M, et al. National Institute of Health Coordinating Group Papnet-assisted cytological diagnosis intensifies the already marked variability among cytological laboratories. Eur J Gynaecol Oncol. 2002;23(3):211–215. [PubMed] [Google Scholar]
- 37.Doornewaard H, van der Schouw YT, van der Graaf Y. Reproducibility in double scanning of cervical smears with the PAPNET system. Acta Cytol. 2000;44(4):604–610. doi: 10.1159/000328535. [DOI] [PubMed] [Google Scholar]
- 38.Losell K, Dejmek A. Comparison of Papnet-assisted and manual screening of cervical smears. Diagn Cytopathol. 1999;21(4):296–299. doi: 10.1002/(sici)1097-0339(199910)21:4<296::aid-dc14>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 39.Kok MR, Boon ME, Schreiner-Kok PG, Koss LG. Cytological recognition of invasive squamous cancer of the uterine cervix: comparison of conventional light-microscopical screening and neural network-based screening. Hum Pathol. 2000;31(1):23–28. doi: 10.1016/s0046-8177(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 40.Doornewaard H, van der Schouw YT, van der Graaf Y, Bos AB, van den Tweel JG. Observer variation in cytologic grading for cervical dysplasia of Papanicolaou smears with the PAPNET testing system. Cancer. 1999;87(4):178–183. doi: 10.1002/(sici)1097-0142(19990825)87:4<178::aid-cncr3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Doornewaard H, van der Schouw YT, van der Graaf Y, Bos AB, Habbema JD, van den Tweel JG. The diagnostic value of computer-assisted primary cervical smear screening: a longitudinal cohort study. Mod Pathol. 1999;12(11):995–1000. [PubMed] [Google Scholar]
- 42.Cosentino A, Ghidoni D, Salemi M, et al. An interlaboratory study of the use of PapNet in the quality control of cervicovaginal cytology. Pathologica. 1999;91(2):101–106. [PubMed] [Google Scholar]
- 43.Assessment of automated primary screening on PAPNET of cervical smears in the PRISMATIC trial. PRISMATIC Project Management Team. Lancet. 1999;353(9162):1381–1385. [PubMed] [Google Scholar]
- 44.Mitchell H, Medley G. Detection of laboratory false negative smears by the PAPNET cytologic screening system. Acta Cytol. 1998;42(1):265–270. doi: 10.1159/000331556. [DOI] [PubMed] [Google Scholar]
- 45.Ghidoni D, Fabbris E, Folicaldi S, et al. The PAPNET system in the rescreening of negative cervical/vaginal smears. A study from the Imola cytology laboratory. Pathologica. 1998;90(4):357–363. [PubMed] [Google Scholar]
- 46.Abulafia O, Sherer DM. Automated cervical cytology: meta-analyses of the performance of the PAPNET system. Obstet Gynecol Surv. 1999;54(4):253–264. doi: 10.1097/00006254-199904000-00022. [DOI] [PubMed] [Google Scholar]
- 47.Sherman ME, Schiffman M, Herrero R, et al. Performance of a semiautomated Papanicolaou smear screening system: results of a population-based study conducted in Guanacaste, Costa Rica. Cancer. 1998;84(5):273–280. [PubMed] [Google Scholar]
- 48.Kok MR, Schreiner-Kok PG, Van Der Veen G, Boon ME. Potentially difficult smears of women with squamous cell carcinoma pose fewer problems when PAP-NET is used for primary screening. Cytopathology. 1999;10(5):324–334. doi: 10.1046/j.1365-2303.1999.00194.x. [DOI] [PubMed] [Google Scholar]
- 49.Cenci M, Giovagnoli MR, Olla SV, Drusco A, Vecchione A. Automation of cytological analysis of cervical smears] Minerva Ginecol. 1999;51(7–8):291–298. [PubMed] [Google Scholar]
- 50.Halford JA, Wright RG, Ditchmen EJ. Quality assurance in cervical cytology screening. Comparison of rapid rescreening and the PAPNET Testing System. Acta Cytol. 1997;41(1):79–81. doi: 10.1159/000332309. [DOI] [PubMed] [Google Scholar]
- 51.Keyhani-Rofagha S, Palma T, O’Toole RV. Automated screening for quality control using PAPNET: a study of 638 negative Pap smears. Diagn Cytopathol. 1996;14(4):316–320. doi: 10.1002/(SICI)1097-0339(199605)14:4<316::AID-DC7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Willis BH, Barton P, Pearmain P, Bryan S, Hyde C. Cervical screening programmes: can automation help? Evidence from systematic reviews, an economic analysis and a simulation modelling exercise applied to the UK. Health Technol Assess. 2005;9(13):1–207. iii. doi: 10.3310/hta9130. [DOI] [PubMed] [Google Scholar]
- 53.Brotzman G. Utility-and cost-effectiveness of PAPNET secondary screening technology for cervical cytology. J Low Genit Tract Dis. 1999;3(1):49. doi: 10.1097/00128360-199901000-00041. [DOI] [PubMed] [Google Scholar]
- 54.Meerding WJ, Doornewaard H, van Ballegooijen M, et al. Cost analysis of PAPNET-assisted vs. conventional Pap smear evaluation in primary screening of cervical smears. Acta Cytol. 2001;45(1):28–35. doi: 10.1159/000327184. [DOI] [PubMed] [Google Scholar]
- 55.Mango LJ, Radensky P. Neural network-assisted (“NNA”) analysis of cervical smears: pooled effectiveness results and economic analysis. Prim Care Update Ob Gyns. 1998;5(4):162–163. doi: 10.1016/s1068-607x(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 56.Brown AD, Garber AM. Cost-effectiveness of 3 methods to enhance the sensitivity of Papanicolaou testing. JAMA. 1999;281(4):347–353. doi: 10.1001/jama.281.4.347. [DOI] [PubMed] [Google Scholar]
- 57.Brotzman GL, Kretzchmar S, Ferguson D, Gottlieb M, Stowe C. Costs and outcomes of PAPNET secondary screening technology for cervical cytologic evaluation. A community hospital’s experience. Arch Fam Med. 1999;8(1):52–55. doi: 10.1001/archfami.8.1.52. [DOI] [PubMed] [Google Scholar]
- 58.O’Leary TJ, Tellado M, Buckner SB, Ali IS, Stevens A, Ollayos CW. PAPNET-assisted rescreening of cervical smears: cost and accuracy compared with a 100% manual rescreening strategy. JAMA. 1998;279(3):235–237. doi: 10.1001/jama.279.3.235. [DOI] [PubMed] [Google Scholar]
- 59.Schechter CB. Cost-effectiveness of rescreening conventionally prepared cervical smears by PAPNET testing. Acta Cytol. 1996;40(6):1272–1282. doi: 10.1159/000334021. [DOI] [PubMed] [Google Scholar]
- 60.Duggan MA. Papnet-assisted, primary screening of cervicovaginal smears. Eur J Gynaecol Oncol. 2000;21(1):35–42. [PubMed] [Google Scholar]
- 61.Cenci M, Nagar C, Vecchione A. PAPNET-assisted primary screening of conventional cervical smears. Anticancer Res. 2000;20(5C):3887–3889. [PubMed] [Google Scholar]
- 62.Mango LJ. Clinical validation of interactive cytologic screening. Automating the search, not the interpretation. Acta Cytol. 1997;41(1):93–97. doi: 10.1159/000332312. [DOI] [PubMed] [Google Scholar]
- 63.Bergeron C, Masseroli M, Ghezi A, Lemarie A, Mango L, Koss LG. Quality control of cervical cytology in high-risk women. PAPNET system compared with manual rescreening. Acta Cytol. 2000;44(2):151–157. doi: 10.1159/000326353. [DOI] [PubMed] [Google Scholar]
- 64.Veneti S, Papaefthimiou M, Symiakaki H, Ioannidou-Mouzaka L. PAPNET for cervical cytology screening. Experience in Greece. Acta Cytol. 1999;43(1):30–33. doi: 10.1159/000330865. [DOI] [PubMed] [Google Scholar]
- 65.Wright RG. PAPNET superior to rapid rescreening. Med J Aust. 1998;168(5):253–254. doi: 10.5694/j.1326-5377.1998.tb140147.x. [DOI] [PubMed] [Google Scholar]
- 66.van Ballegooijen M, Beck S, Boon ME, Boer R, Habbema JD. Rescreen effect in conventional and PAPNET screening: observed in a study using material enriched with positive smears. Acta Cytol. 1998;42(5):1133–1138. doi: 10.1159/000332101. [DOI] [PubMed] [Google Scholar]
- 67.Sturgis CD, Isoe C, McNeal NE, Yu GH, DeFrias DV. PAPNET computer-aided rescreening for detection of benign and malignant glandular elements in cervicovaginal smears: a review of 61 cases. Diagn Cytopathol. 1998;18(4):307–311. doi: 10.1002/(sici)1097-0339(199804)18:4<307::aid-dc12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 68.Troni GM, Cipparrone I, Cariaggi MP, et al. Detection of false-negative Pap smears using the PAPNET system. Tumori. 2000;86(6):455–457. doi: 10.1177/030089160008600604. [DOI] [PubMed] [Google Scholar]
- 69.Kok MR, van Der Schouw YT, Boon ME, et al. Neural network-based screening (NNS) in cervical cytology: no need for the light microscope? Diagn Cytopathol. 2001;24(6):426–434. doi: 10.1002/dc.1093. [DOI] [PubMed] [Google Scholar]
- 70.Hartmann KE, Nanda K, Hall S, Myers E. Technologic advances for evaluation of cervical cytology: is newer better? Obstet Gynecol Surv. 2001;56(12):765–774. doi: 10.1097/00006254-200112000-00003. [DOI] [PubMed] [Google Scholar]
- 71.Solomon HM, Frist S. PAPNET testing for HSILs. The few cell/small cell challenge. Acta Cytol. 1998;42(1):253–259. doi: 10.1159/000331554. [DOI] [PubMed] [Google Scholar]
- 72.Boon ME, Ouwerkerk-Noordam E, Meijer-Marres EM, Bontekoe TR. Switching from neural networks (PAPNET) to the Imager (Hologic) for computer-assisted screening. Acta Cytol. 2011;55(2):163–166. doi: 10.1159/000323310. [DOI] [PubMed] [Google Scholar]
- 73.Neofytou MS, Tanos V, Pattichis MS, Kyriacou EC, Pattichis CS, Schizas CN. Color multiscale texture classification of hysteroscopy images of the endometrium; Paper presented at: 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Vancouver, British Columbia, Canada. 2008. [DOI] [PubMed] [Google Scholar]
- 74.Karakitsos P, Kyroudes A, Pouliakis A, Stergiou EB, Voulgaris Z, Kittas C. Potential of the learning vector quantizer in the cell classification of endometrial lesions in postmenopausal women. Anal Quant Cytol Histol. 2002;24(1):30–38. [PubMed] [Google Scholar]
- 75.Clayton RD, Snowden S, Weston MJ, Mogensen O, Eastaugh J, Lane G. Neural networks in the diagnosis of malignant ovarian tumours. Br J Obstet Gynaecol. 1999;106(10):1078–1082. doi: 10.1111/j.1471-0528.1999.tb08117.x. [DOI] [PubMed] [Google Scholar]
- 76.Molnar B, Szentirmay Z, Bodo M, Sugar J, Feher J. Application of multivariate, fuzzy set and neural network analysis in quantitative cytological examinations. Anal Cell Pathol. 1993;5(3):161–175. [PubMed] [Google Scholar]
- 77.Karakitsos P, Stergiou EB, Pouliakis A, et al. Potential of the back propagation neural network in the discrimination of benign from malignant gastric cells. Anal Quant Cytol Histol. 1996;18(3):245–250. [PubMed] [Google Scholar]
- 78.Karakitsos P, Stergiou EB, Pouliakis A, et al. Comparative study of artificial neural networks in the discrimination between benign from malignant gastric cells. Anal Quant Cytol Histol. 1997;19(2):145–152. [PubMed] [Google Scholar]
- 79.Karakitsos P, Ioakim-Liossi A, Pouliakis A, et al. A comparative study of three variations of the learning vector quantizer in the discrimination of benign from malignant gastric cells. Cytopathology. 1998;9(2):114–125. [PubMed] [Google Scholar]
- 80.Karakitsos P, Pouliakis A, Koutroumbas K, et al. Neural network application in the discrimination of benign from malignant gastric cells. Anal Quant Cytol Histol. 2000;22(1):63–69. [PubMed] [Google Scholar]
- 81.Koss LG, Morgenstern N, Tahir-Kheli N, Suhrland M, Schreiber K, Greenebaum E. Evaluation of esophageal cytology using a neural net-based interactive scanning system (the PAPNET system): its possible role in screening for esophageal and gastric carcinoma. Am J Clin Pathol. 1998;109(5):549–557. doi: 10.1093/ajcp/109.5.549. [DOI] [PubMed] [Google Scholar]
- 82.Levine TS, Njemenze V, Cowpe JG, Coleman DV. The use of the PAPNET auto-mated cytological screening system for the diagnosis of oral squamous carcinoma. Cytopathology. 1998;9(6):398–405. doi: 10.1046/j.1365-2303.1998.00126.x. [DOI] [PubMed] [Google Scholar]
- 83.Hoda RS, Saccomanno G, Schreiber K, Decker D, Koss LG. Automated sputum screening with PAPNET system: a study of 122 cases. Hum Pathol. 1996;27(7):656–659. doi: 10.1016/s0046-8177(96)90394-9. [DOI] [PubMed] [Google Scholar]
- 84.Lai KC, Chiang HC, Chen WC, Tsai FJ, Jeng LB. Artificial neural network-based study can predict gastric cancer staging. Hepatogastroenterology. 2008;55(86–87):1859–1863. [PubMed] [Google Scholar]
- 85.Sharpe PK, Solberg HE, Rootwelt K, Yearworth M. Artificial neural networks in diagnosis of thyroid function from in vitro laboratory tests. Clin Chem. 1993;39(11 pt 1):2248–2253. [PubMed] [Google Scholar]
- 86.Karakitsos P, Cochand-Priollet B, Guillausseau PJ, Pouliakis A. Potential of the back propagation neural network in the morphologic examination of thyroid lesions. Anal Quant Cytol Histol. 1996;18(6):494–500. [PubMed] [Google Scholar]
- 87.Karakitsos P, Cochand-Priollet B, Pouliakis A, Guillausseau PJ, Ioakim-Liossi A. Learning vector quantizer in the investigation of thyroid lesions. Anal Quant Cytol Histol. 1999;21(3):201–208. [PubMed] [Google Scholar]
- 88.Cochand-Priollet B, Koutroumbas K, Megalopoulou TM, Pouliakis A, Sivolapenko G, Karakitsos P. Discriminating benign from malignant thyroid lesions using artificial intelligence and statistical selection of morphometric features. Oncol Rep. 2006;15 doi: 10.3892/or.15.4.1023. Spec no.:1023-1026. [DOI] [PubMed] [Google Scholar]
- 89.Shapiro NA, Poloz TL, Shkurupij VA, Tarkov MS, Poloz VV, Demin AV. Application of artificial neural network for classification of thyroid follicular tumors. Anal Quant Cytol Histol. 2007;29(2):87–94. [PubMed] [Google Scholar]
- 90.Ippolito AM, De Laurentiis M, La Rosa GL, et al. Neural network analysis for evaluating cancer risk in thyroid nodules with an indeterminate diagnosis at aspiration cytology: identification of a low-risk subgroup. Thyroid. 2004;14(12):1065–1071. doi: 10.1089/thy.2004.14.1065. [DOI] [PubMed] [Google Scholar]
- 91.Poloz TL, Shkurupii VA, Poloz VV, Demin AV. The results of quantitative cytological analysis of the structure of follicular thyroid tumors using computer and neural network technologies. Vestn Ross Akad Med Nauk. 2006;61(8):7–10. [PubMed] [Google Scholar]
- 92.Poloz TL, Tarkov MS. A neural network algorithm for automation of cytological diagnostics of a thyroid gland follicular tumors. Bull Nov Comp Center Comp Sci. 2006;25:59–62. [Google Scholar]
- 93.Daskalakis A, Kostopoulos S, Spyridonos P, et al. Design of a multi-classifier system for discriminating benign from malignant thyroid nodules using routinely H&E-stained cytological images. Comput Biol Med. 2008;38(2):196–203. doi: 10.1016/j.compbiomed.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 94.Varlatzidou A, Pouliakis A, Stamataki M, et al. Cascaded learning vector quantizer neural networks for the discrimination of thyroid lesions. Anal Quant Cytol Histol. 2011;33(6):323–334. [PubMed] [Google Scholar]
- 95.Ozolek JA, Tosun AB, Wang W, et al. Accurate diagnosis of thyroid follicular lesions from nuclear morphology using supervised learning. Med Image Anal. 2014;18(5):772–780. doi: 10.1016/j.media.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W, Slepcev D, Basu S, Ozolek JA, Rohde GK. A linear optimal transportation framework for quantifying and visualizing variations in sets of images. Int J Comput Vis. 2013;101(2):254–269. doi: 10.1007/s11263-012-0566-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen C, Wang W, Ozolek JA, Rohde GK. A flexible and robust approach for segmenting cell nuclei from 2D microscopy images using supervised learning and template matching. Cytometry A. 2013;83(5):495–507. doi: 10.1002/cyto.a.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wolberg WH, Mangasarian OL. Computer-aided diagnosis of breast aspirates via expert systems. Anal Quant Cytol Histol. 1990;12(5):314–320. [PubMed] [Google Scholar]
- 99.Dawson AE, Austin RE, Jr, Weinberg DS. Nuclear grading of breast carcinoma by image analysis. Classification by multivariate and neural network analysis. Am J Clin Pathol. 1991;95(4 suppl 1):S29–S37. [PubMed] [Google Scholar]
- 100.Ravdin PM, Clark GM, Hilsenbeck SG, et al. A demonstration that breast cancer recurrence can be predicted by neural network analysis. Breast Cancer Res Treat. 1992;21(1):47–53. doi: 10.1007/BF01811963. [DOI] [PubMed] [Google Scholar]
- 101.Markopoulos C, Karakitsos P, Botsoli-Stergiou E, et al. Application of back propagation to the diagnosis of breast lesions by fine needle aspiration. Breast. 1997;6(5):293–298. [Google Scholar]
- 102.Markopoulos C, Karakitsos P, Botsoli-Stergiou E, et al. Application of the learning vector quantizer to the classification of breast lesions. Anal Quant Cytol Histol. 1997;19(5):453–460. [PubMed] [Google Scholar]
- 103.Ohno-Machado L, Bialek D. Diagnosing breast cancer from FNAs: variable relevance in neural network and logistic regression models. Stud Health Technol Inform. 1998;52(pt 1):537–540. [PubMed] [Google Scholar]
- 104.Reigosa A, Hernandez L, Torrealba V, et al. Automatic intraductal breast carcinoma classification using a neural network-based recognition system. Breast J. 1998;4(4):238–244. doi: 10.1046/j.1524-4741.1998.440238.x. [DOI] [PubMed] [Google Scholar]
- 105.Einstein AJ, Wu HS, Sanchez M, Gil J. Fractal characterization of chromatin appearance for diagnosis in breast cytology. J Pathol. 1998;185(4):366–381. doi: 10.1002/(SICI)1096-9896(199808)185:4<366::AID-PATH122>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 106.Dey P, Logasundaram R, Joshi K. Artificial neural network in diagnosis of lobular carcinoma of breast in fine-needle aspiration cytology. Diagn Cytopathol. 2013;41(2):102–106. doi: 10.1002/dc.21773. [DOI] [PubMed] [Google Scholar]
- 107.Subbaiah RM, Dey P, Nijhawan R. Artificial neural network in breast lesions from fine-needle aspiration cytology smear. Diagn Cytopathol. 2014;42(3):218–224. doi: 10.1002/dc.23026. [DOI] [PubMed] [Google Scholar]
- 108.Abbod MF, Catto JW, Linkens DA, Hamdy FC. Application of artificial intelligence to the management of urological cancer. J Urol. 2007;178(4 pt 1):1150–1156. doi: 10.1016/j.juro.2007.05.122. [DOI] [PubMed] [Google Scholar]
- 109.Schwarzer G, Schumacher M. Artificial neural networks for diagnosis and prognosis in prostate cancer. Semin Urol Oncol. 2002;20(2):89–95. doi: 10.1053/suro.2002.32492. [DOI] [PubMed] [Google Scholar]
- 110.Koss LG, Bartels PH, Bibbo M, Freed SZ, Taylor J, Wied GL. Computer discrimination between benign and malignant urothelial cells. Acta Cytol. 1975;19(4):378–391. [PubMed] [Google Scholar]
- 111.Moallemi C. Classifying cells for cancer diagnosis using neural networks. IEEE Expert. 1991;6(6):8–12. [Google Scholar]
- 112.Pantazopoulos D, Karakitsos P, Iokim-Liossi A, Pouliakis A, Botsoli-Stergiou E, Dimopoulos C. Back propagation neural network in the discrimination of benign from malignant lower urinary tract lesions. J Urol. 1998;159(5):1619–1623. doi: 10.1097/00005392-199805000-00057. [DOI] [PubMed] [Google Scholar]
- 113.Pantazopoulos D, Karakitsos P, Pouliakis A, Iokim-Liossi A, Dimopoulos MA. Static cytometry and neural networks in the discrimination of lower urinary system lesions. Urology. 1998;51(6):946–950. doi: 10.1016/s0090-4295(98)00024-7. [DOI] [PubMed] [Google Scholar]
- 114.Pantazopoulos D, Karakitsos P, Iokim-Liossi A, Pouliakis A, Dimopoulos K. Comparing neural networks in the discrimination of benign from malignant lower urinary tract lesions. Br J Urol. 1998;81(4):574–579. doi: 10.1046/j.1464-410x.1998.00587.x. [DOI] [PubMed] [Google Scholar]
- 115.Van Biesen W, Sieben G, Lameire N, Vanholder R. Application of Kohonen neural networks for the non-morphological distinction between glomerular and tubular renal disease. Nephrol Dial Transplant. 1998;13(1):59–66. doi: 10.1093/ndt/13.1.59. [DOI] [PubMed] [Google Scholar]
- 116.Vriesema JL, van der Poel HG, Debruyne FM, Schalken JA, Kok LP, Boon ME. Neural network-based digitized cell image diagnosis of bladder wash cytology. Diagn Cytopathol. 2000;23(3):171–179. doi: 10.1002/1097-0339(200009)23:3<171::aid-dc6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 117.Truong H, Morimoto R, Walts AE, Erler B, Marchevsky A. Neural networks as an aid in the diagnosis of lymphocyte-rich effusions. Anal Quant Cytol Histol. 1995;17(1):48–54. [PubMed] [Google Scholar]
- 118.Barwad A, Dey P, Susheilia S. Artificial neural network in diagnosis of metastatic carcinoma in effusion cytology. Cytometry B Clin Cytom. 2012;82(2):107–111. doi: 10.1002/cyto.b.20632. [DOI] [PubMed] [Google Scholar]
- 119.Deng L, Yu D. Deep learning: methods and applications. Found Trends Sig Process. 2014;7(3–4):197–387. [Google Scholar]
- 120.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013;16(pt 2):411–418. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 121.Schmidhuber J. Deep learning in neural networks: an overview. Neural Networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 122.Picot J, Guerin CL, Le Van Kim C, Boulanger CM. Flow cytometry: retrospective, fundamentals and recent instrumentation. Cytotechnology. 2012;64(2):109–130. doi: 10.1007/s10616-011-9415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O’Leary TJ. Flow cytometry in diagnostic cytology. Diagn Cytopathol. 1998;18(1):41–46. doi: 10.1002/(sici)1097-0339(199801)18:1<41::aid-dc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 124.Frankel DS, Frankel SL, Binder BJ, Vogt RF. Application of neural networks to flow cytometry data analysis and real-time cell classification. Cytometry. 1996;23(4):290–302. doi: 10.1002/(SICI)1097-0320(19960401)23:4<290::AID-CYTO5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 125.Boddy L, Wilkins MF, Morris CW. Pattern recognition in flow cytometry. Cytometry. 2001;44(3):195–209. doi: 10.1002/1097-0320(20010701)44:3<195::aid-cyto1112>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 126.Zong N, Adjouadi M, Ayala M. Artificial neural networks approaches for multidimensional classification of acute lymphoblastic leukemia gene expression samples; Proceedings of the 9th WSEAS International Conference on Computers; Athens, Greece: 2005. [Google Scholar]
- 127.Zong N, Adjouadi M, Ayala M. Optimizing the classification of acute lymphoblastic leukemia and acute myeloid leukemia samples using artificial neural networks. Biomed Sci Instrum. 2006;42:261–266. [PubMed] [Google Scholar]
- 128.Rossman M. FIU Electronic Theses and Dissertations. Miami, FL: Florida International University; 2011. Automated Detection of Hematological Abnormalities through Classification of Flow Cytometric Data Patterns. [Google Scholar]