Abstract
In this study, we analyzed the effects of the cloned embryo aggregation on in vitro embryo development and embryo quality by measuring blastocyst diameter and cell number, DNA fragmentation levels and the expression of genes associated with pluripotency, apoptosis, trophoblast and DNA methylation in the porcine. Zona-free reconstructed cloned embryos were cultured in the well of the well system, placing one (1x non aggregated group) or three (3x group) embryos per microwell. Our results showed that aggregation of three embryos increased blastocyst formation rate and blastocyst diameter of cloned pig embryos. DNA fragmentation levels in 3x aggregated cloned blastocysts were significantly decreased compared to 1x blastocysts. Levels of Oct4, Klf4, Igf2, Bax and Dnmt 1 transcripts were significantly higher in aggregated embryos, whereas Nanog levels were not affected. Transcripts of Cdx2 and Bcl-xl were essentially non-detectable. Our study suggests that embryo aggregation in the porcine may be beneficial for cloned embryo development and embryo quality, through a reduction in apoptotic levels and an improvement in cell reprogramming.
Introduction
Mammalian cloning by somatic cell nuclear transfer has an enormous potential in biotechnology and has opened new possibilities for specific genetic modifications in farm animals [1]. In addition to the potential application in agriculture and biomedicine, somatic cell nuclear transfer (SCNT) is one of the most powerful tools used to study events that occur during reprogramming and cellular differentiation [2,3]. Successful cloning using mammalian somatic cell indicates that epigenetic modifications in the differentiated nucleus can be remodeled to a totipotent state [2]. The first living cloned piglet obtained by nuclear transfer of adult granulosa cells was reported in 2000 [4], and subsequently, additional porcine clones have been obtained [5–7].
Recently, our laboratory has demonstrated the advantage of zona free cloned embryo aggregation in the equine and the feline [8–10]. Embryo aggregation consists of placing more than one zona free embryos in the same well; some of the reported benefits in mammals are: improvement in embryo development in pig [6], equine [8, 9], feline [10] and mouse [11], increasing the density of cells in the inner cell mass on cattle [12], blastocyst diameter in equine and in bovine [8, 13], reduction in apoptosis in bovine and pigs [13, 14, 15], normalization of gene expression in bovine [15]and in some cases, it has also improved in vivo embryo development and early pregnancy rates in mouse [11], equine [8] and in cattle [13, 16, 17]. As a single clone may have epigenetic defects [15], embryo aggregation could compensate the reprogramming deficiencies through the interactions between blastomeres [11] or by paracrine pathways [11]. Indeed, it has been proposed that such interaction could also improve post implantation development [18].
During normal embryogenesis a type of programmed cell death called apoptosis occurs as a physiological process and it has a role in the cellular response to suboptimal developmental conditions and stress [19]. Apoptosis helps in the removal of cells with chromosomal abnormalities or inappropriate developmental potentials [20]. However, embryo development could be compromised if apoptosis surpasses a certain threshold [19, 21, 22]. Moreover, its occurrence in preimplantation embryos has been considered one of the most important parameters for evaluation of embryo health and regulated embryo cell numbers [23–25]; the TUNEL assay (terminal deoxynucleotidyltransferase-mediated dUTP nick-end labelling) has been used for detection of apoptosis process in mammalian embryos [20].
Epigenetic mechanisms have shown to critically influence embryonic development through the control of gene expression and chromatin packaging [26] and several genes are reported to be associated with the pluripotency of the embryo: Oct4 [27], Sox2 [28] and Nanog [29] and with lineage segregation like Cdx2 [30]. However, porcine embryos present numerous differences in their expression profile of pluripotency markers compared to human or mouse. This may suggest that different mechanisms are implicated in the regulation of pluripotency in this species [31].
To improve the efficiency of porcine cloning, it is necessary to produce high-quality cloned blastocyst [12, 32, 33]. We hypothesized that embryo aggregation could improve the developmental competence and quality of cloned pig embryos. To this aim, we studied the embryo development rates, the total number of cells per blastocyst, the embryo diameter, the DNA fragmentation levels by TUNEL assay and the expression of genes associated with pluripotency, apoptosis, trophoblast and methylation in aggregated and non-aggregated cloned embryos.
Materials and Methods
Chemicals
Except otherwise indicated, all chemicals were obtained from Sigma Chemicals Company (St. Louis, MO, USA).
Recovery and in vitro maturation of oocytes
Ovaries were collected from gilts at a local slaughterhouse called "La Pompeya" located in the province of Buenos Aires and transported to the laboratory at around 25–30°C within 3h of collection. Cumulus-oocyte complexes (COCs) from follicles 3 to 6mm in diameter were aspirated using an 18-gauge needle attached to a 10mL disposable syringe. Compact COCs were selected and matured in 100μL drops of tissue culture medium (TCM-199- 31100–035; Gibco, Grand Island, NY, USA) under mineral oil (M8410), supplemented with 0.3mM sodium pyruvate (P2256), 100μM cysteamine (M9768), 5μg/ml myoinositol, 1μg/ml insulin transferrin selenium (ITS; product no. 51300–044; Gibco), 1% antibiotic-antimicotic (15240–096 Gibco), 10% (v/v) porcine follicular fluid, 5 ng/ml basic fibroblast growth factor (bFGF) and 10μg/ml of follicle-stimulating hormone (FSH) (NIH-FSH-P1Folltropin, Bioniche, Caufield Junction Caufield North, Victoria, Australia). Maturation was performed at 39°C in a humidified atmosphere of 6.5% CO2 in 90% air for 42-44h.
Preparation of oocytes for SCNT
The cumulus cells were removed by vortexing COCs for 3–4 min in hyaluronidase (H-4272) solution: 1 mg/ml in Tyrode’s Lactate-Pyruvate-HEPES medium (TALP-H). Nuclear maturation was confirmed by the presence of the first polar body. Matured oocytes were incubated in 1.5 mg/ml pronase (P-8811) in TALP-H for 1–2 min to remove the zona pellucida (ZP). Zona Free (ZF)-oocytes were kept in SOF until enucleation.
Somatic cell nuclear transfer
ZF- oocytes were incubated for 15 minutes in a SOF drop containing 1μg/mL Bisbenzimide H 33342 (B2261) and 0.5μg/ml of cytochalasin B. The metaphase phase was aspirated using a blunt pipette under UV light, and a closed holding pipette was used to support the oocyte during the enucleation. Successful enucleation was assessed by the observation, under UV light, of the entire metaphase plate inside the pipette [8]. ZF-enucleated oocytes were kept in SOF medium under culture conditions until nuclear transfer.
Landrace fetal fibroblast cells was established from a primary culture from fetal lungs of approximately 30 days and left to culture until 80% of confluence. Afterwards, cells were frozen in Dubelcco′s modified Eagle medium (DMEM) (11885, Gibco, Grand Island, NY, USA) containing 10% dimethyl sulfoxide. Three days before SCNT, cells were thawed, cultured until confluence and subsequently used between passages 1–4. Single cell suspension was prepared by trypsinization of the cultured cells and then resuspended in DMEM medium supplemented with 20% fetal bovine serum (FBS; 10499–044, Gibco) and 1% antibiotic–antimycotic (15240–096, Gibco) and stored in liquid nitrogen.
ZF-enucleated oocytes were individually transferred to a 40μl drop of 1mg/ml phytohemagglutinin (L8754) dissolved in TCM-199. After a few seconds, oocytes were quickly dropped over a single donor cell resting on the bottom of a 100 μl TALP-H drop. Afterward, the couplets were placed in fusion medium [0.3 M mannitol (M9546), 0.1 mM MgSO4 (M7506), 0.05 mM CaCl2 (C7902), and 1 mg/ml polyvinyl alcohol (P8136)] for 2–3 min and then transferred to a fusion chamber containing 2 ml of fusion medium. Electrofusion was performed using a BTX Electro-Cell Manipulator 830 (BTX, Inc., San Diego, CA) with a double-direct current pulse of 1.42 kV/cm, in which each pulse lasted for 30 μsec. Immediately after, couplets were individually placed in 5 μl microdrop of SOF medium and incubated under mineral oil at 39°C in 5% CO2 in air. Twenty to thirty minutes after the pulse, each couplet was considered fused when the donor cell was not observed in the droplet. Non-fused couplets were re-fused. Non-fused couplets after the second round of fusion were discarded.
Two hours after fusion, ZF-fused couplets were electrically activated by a single direct current pulse of 1.2 kV/cm for 80 μsec, followed by incubation for 3h of 2 mM 6-dimethylaminopurine (D2629) in a 100μl drop of SOF medium.
In vitro embryo culture and embryo aggregation
ZF-reconstructed embryos (ZFREs) were cultured in SOF medium in a humidified atmosphere of 5% CO2 and 95% air at 38.5°C for 7 days. A slightly modified well of the well system [34] was used to culture ZFREs [8]. Briefly, microwells were produced using a heated glass capillary slightly pressed to the bottom of a 35 mm x 10mm petri dish. Microwells were covered with a 50μl microdrop of SOF medium. Embryo aggregation was performed by placing randomly more than one ZFRE per microwell. Two different experimental groups were performed: group 1x, one ZFRE per microwell (non-aggregated embryos; control group); and group 3x, three ZFREs per microwell. The total number of ZFREs per microdrop was similar between groups. Embryos were cultured in a humidified gas mixture (5% CO2, 5%O2, 90% N2) at 38.5°C. Half of the medium was renewed at day 2 with fresh SOF, and at day 5 the medium was renewed again with SOF medium containing 10% FBS. Cleavage was assessed 48 h after activation, and blastocyst formation and blastocyst diameter were recorded on day 7 when the embryos were either fixed for TUNEL assay or stored in RNAlater (AM7020, Ambion Co., Austin, TX, USA) for gene expression study.
DNA fragmentation levels in blastocysts: TUNEL assay
Pig cloned blastocyst from both experimental groups were fixed in 4% paraformaldehyde (F1635) and then washed and stored in Dulbecco′s phosphate buffered saline (DPBS) (14287–072 Gibco, Gran Island, NY) solution with 1 mg/ml of bovine serum albumin (BSA). DNA fragmentation levels were detected in situ using the Dead End TM Fluorometric TUNEL System (terminal deoxynucleotidyltransferase (TdT)-mediated dUTP nick end labelling) (Promega G3250, Madison, WI, USA). Fixed embryos were permeabilized with 0.2% Triton X-100 in DPBS for 15 min at room temperature and rinsed in 0.4% DPBS-BSA. Then, samples were placed in incubation buffer consisting of equilibration buffer, a nucleotide mix containing fluorescein-dUTP and terminal deoxynucleotidyltransferase for 2 h at 39°C in dark. For negative controls, the terminal deoxynucleotidyltransferase was omitted from the reaction. For nuclei counterstain embryos were incubated with 0.5% propidium iodide for 10 min at room temperature, and then washed in BSA solution and finally mounted on a glass slide in 70% v/v glycerol under a coverslip. Blastocysts were analyzed on a Nikon Confocal C.1 scanning laser microscope. An excitation wavelength of 488 nm was selected for detection of fluorescein-12-dUTP and a 544 nm wavelength to excite propidium iodide. Images of serial optical sections were recorded every 1.5–2 μm vertical step along the Z-axis of each embryo. Three-dimensional images were constructed using software EZ-C1 3.9. Total cell numbers and DNA-fragmented nuclei were counted manually. The results were expressed as quantity from the relation between the number of cells with fragmentation of DNA and the number of total cells. The apoptotic index (IAp) was determined, using the following formula: IAp = TUNEL (+) cells * 100/ Total cell number. The apoptotic index in each blastocyst was calculated as the ratio of TUNEL-positive nuclei to total nuclei in each blastocyst and then an unpaired T-Student test was used to calculate the statistic differences between the index of different groups (p<0.05).
Real-time reverse transcription polymerase chain reaction
For gene expression analysis we used two groups (clone 1x and aggregated 3x clones). We pooled n = 12 blastocyst of each group and repeat this process three times because we did three biological replicates, each repetitions by triplicate. Embryos were washed twice in DPBS to eliminate the RNAlater in which were conserved. RNA was isolated using the RNeasy micro kit (Qiagen, Hilden, Germany, Cat. No.74004) according to the manufacturer’s instruction. All the samples were treated with DNase I (0.04 U/μl) for genomic DNA digestion. After that, RT PCR was performed in 20μl of final volume. Quantitative PCR was applied using SYBR according to the manufacturer’s instructions with MyiQ Single Color Real Time PCR detection System by BIO RAD cycler. Quantification of all gene transcripts was performed by real-time reverse transcription polymerase chain reaction (PCR) using ACTB as an internal standard. The reaction mixture (total 12.5 μL) contained 6.5 μl master mix, 0.25 μl of each primer (20 mmol/ul), 5 μL cDNA template (250 ng final concentration), and 0.5 μl Milliq water.
Primer design
Primers used for expression analysis were designed using Primer3 online version based on available sequences from the database of GenBank (NCBI). Primers and products sizes are shown in Table 1.
Table 1. Primers sequences and conditions for RT-qPCR.
| Genes | Primer sequences (5–3) | Size of PCR products (bp) | Tm (°C) | Reference or sequence accession numbers |
|---|---|---|---|---|
| Klf4 | CCATGGGCCAAACTACCCAC | 81 | 60 | NM_001031782.2 |
| Klf4 | GGCATGAGCTCTTGGTAATGG | 81 | 59 | NM_001031782.2 |
| Nanog | CCACTGGCCAAGGAATAGCA | 88 | 60 | NM_001129971.1 |
| Nanog | CAGGCATCCTTGGTGGTAGG | 88 | 60 | NM_001129971.1 |
| Oct4 | GCTCACTTTGGGGGTTCTCT | 80 | 59 | NM_001113060 |
| Oct4 | TGAAACTGAGCTGCAAAGCC | 80 | 59 | NM_001113060 |
| Bcl-xl | GTTGACTTTCTCTCCTACAAGC | 277 | 62 | SUN HWANG, 2008 |
| Bcl-xl | GGTACCTCAGTTCAAACTCATC | 277 | 62 | SUN HWANG, 2008 |
| Bax- α | ACTGGACAGTAACATGGAGC | 294 | 63 | SUN HWANG, 2008 |
| Bax- α | GTCCCAAAGTAGGAGAGGAG | 294 | 63 | SUN HWANG, 2008 |
| Dnmt1 | TTCTCACTGCCTGACGATGT | 79 | 59 | NM_001032355.1 |
| Dnmt1 | CCTTCACGCATTCCTTTTCTGT | 79 | 59 | NM_001032355.1 |
| Igf2 | GGCATCGTGGAAGAGTGCT | 128 | 60 | X56094.1 |
| Igf2 | CTGGGGAAGTTGTCCGGAAG | 128 | 60 | X56094.1 |
| Cdx2 | CAGCCAAGTGAAAACCAGGAC | 119 | 59 | NM_001278769.1 |
| Cdx2 | CGGCCTTTCTCCGAATGGT | 119 | 60 | NM_001278769.1 |
| ACTB | AGATCGTGCGGGACATCAAG | 93 | 59 | DQ452569.1 |
| ACTB | GCGGCAGTGGCCATCTC | 93 | 59 | DQ452569.1 |
Statistical analysis
In vitro embryo development, differences in blastocyst cell numbers and embryo diameter were compared by Fisher’s exact test analysis. TUNEL assay and dead cell index were compared using T-Student test. Differences were considered to be significant at p< 0.05 for both studies. We performed the real time PCR with ACTB as reference gene. The relative expression of each gene were calculated from the average Ct values of each triplicate using the 2 (-ΔCt) method. Differences in the level of gene expression on both groups were analyzed using a parametric T-Student test. Differences between groups were evaluated with a significance level of p< 0.05. The data obtained in this study were analyzed using the GraphPad Prism statistical program (GraphPad Software, San Diego, CA).
Results
In the present study, we aimed to elucidate if embryo aggregation improves in vitro cloning efficiency in the porcine, through a comprehensive study of the quality of the embryo. In order to evaluate this, embryo developmental rates, embryo diameter and cell number, DNA fragmentation levels and the pattern expression of different genes were analyzed.
In vitro embryo development records are shown in Table 2. We observed that cleavage rates per ZFREs were significantly higher in 3x aggregated cloned embryos than in the control group 1x, 92.17% vs 73.05% respectively, p<0.0001. However, blastocyst rate per ZFRE did not differ between groups. On the other hand, blastocyst rates per embryo (considering each well as an embryo) in the 3x group (37.39%) was more than three times that of the control group (11.38%) (p<0.0001). Zona free cloned embryo aggregation improved blastocyst rates, and did not involve the use of additional oocytes to obtain more embryos.
Table 2. Effects of clone porcine embryo aggregation on in vitro embryo development until day 7.
| Experimental groups | No. ZFREs* | No. Embryos (wells) | No. cleaved ZFREs (%) | No. blastocyst | % of blastocyst per ZFREs | % of blastocyst per embryo (well) |
|---|---|---|---|---|---|---|
| 1x | 167 | 167 | 122 (73.05) a | 19 | 11.38 a | 11.38 a |
| 3x | 345 | 115 | 318 (92.17) b | 43 | 12.46 a | 37.39 b |
| Total | 512 | 282 | 440 (85.94) | 62 | 12.11 | 21.99 |
*ZFREs: Zona Free Reconstructed Embryos (a,b) Values from the same method with different superscripts in a column are significantly different (p<0.05, Fisher’s exact test).
Cloned blastocyst diameter was measured and embryos were divided in three different groups according to their diameter at day 7: 80 μm-199 μm (small), 200 μm-299 μm (medium) and > 300 μm (large). The numbers of embryos analyzed in each diameter category were: 1x (n = 16) and 3x (n = 28). Embryo diameters per group were: in the small 1x group n = 12: 99.6 μm (1), 199.2 μm (4), 132.8 μm (3), 166 μm (3) and 149.4 μm (1); in the medium 1x group n = 4: 1 of each value: 215.2 μm, 249 μm, 265.6 μm and 282.2 μm. In the small 3x group n = 7: 132.8 μm (3), 166 μm (3) and 199.2 μm (1); in the medium 3x group n = 12: 215.8 μm (2), 232.4 μm (2), 249 μm (4), 265.6 μm (2), 282.2 μm (1) and 298.8 μm (1). Finally, in the large 3x group n = 9: 365.2 μm (2), 381.8 μm (3), 415 μm (1), 448.2 μm (2) and 514.6 μm (1). Aggregated 3x cloned embryos were bigger than 1x non aggregated cloned embryos and these differences were statistically significant (p<0.011). Experimental group 1x had a higher proportion (75%) of small diameter embryos, and no blastocyst measured more than 300 μm. Conversely, 75% of cloned pig aggregated blastocysts (experimental group 3x) were of medium and large diameters. Results are shown in Table 3.
Table 3. Effects of embryo aggregation on in vitro porcine cloned embryo diameter.
| Experimental groups | No. Blastocyst | Blastocyst diameter | ||
|---|---|---|---|---|
| 80 μm- 199 μm (%) | 200 μm- 299 μm (%) | >/ = 300 μm (%) | ||
| 1x | 16 | 12(75,0)a | 4 (25,0) | 0 (0) a |
| 3x | 28 | 7 (25,0)b | 12 (42,86) | 9 (32,14)b |
| Total | 44 | 19 (43.18) | 16 (36.36) | 9 (20.45) |
Blastocyst diameter of cloned aggregated and non-aggregated embryos at day 7.Values with different superscripts in a column are significantly different (p<0.05, Fisher’s exact test).
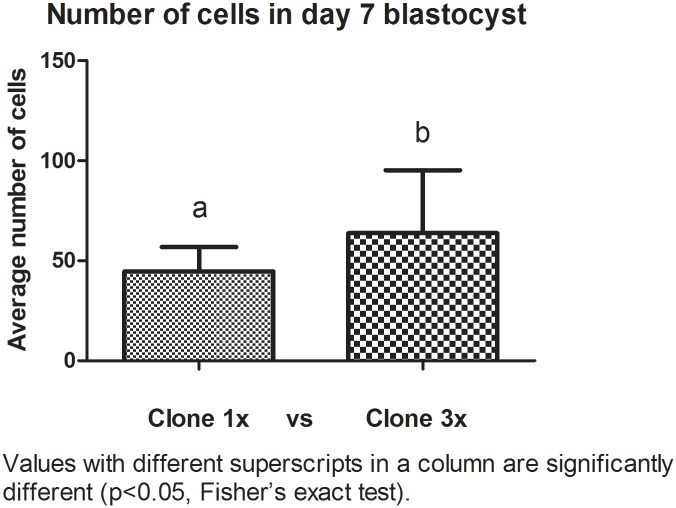
Cell numbers of day 7 cloned blastocysts were different among groups. The aggregated embryos had more cells than the non aggregated embryos. The average number of cells in 1x group was between 27–64 cells and in the 3x group between 26–108 cells (Fig 1). More of the 3x blastocysts were larger (> 300μm) compared with the 1x blastocysts (p< 0.0001) (Table 3).
Fig 1. Blastocyst number of cells of cloned aggregated and non-aggregated embryos at day 7.
Nuclei were counterstained with 30μg/mL propidium iodide (P4170) for 10 min in the dark.
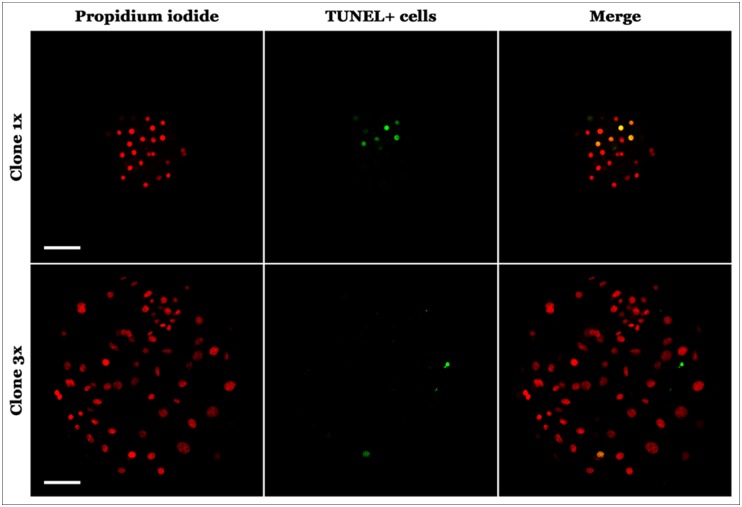
The levels of apoptotic cells were determined by detection of fragmented DNA by TUNEL assay. The 3x group embryos had lower apoptotic index cells than the control group1x (4.18% vs 13.18%) (p<0.05). Results are shown in Figs 2 and 3 and Table 4.
Fig 2. Index of apoptosis of aggregated and non-aggregated pig cloned blastocyst, showing statistically differences (p<0.05, T-Student test).
Fig 3. Photomicrographs of day 7porcine cloned embryo expression of TUNEL.
Above non-aggregated cloned porcine embryo, 40x zoom. Below (day 7) 3x aggregated cloned embryo, 40x zoom.
Table 4. Index of apoptosis of aggregated and non-aggregated pig cloned blastocyst at day 7.
| Cloned 1x Blastocyst | Cloned 3x Blastocyst | ||||
|---|---|---|---|---|---|
| Total cell number | Apoptotic cell number | IAP (Index of Apoptosis) | Total cell number | Apoptotic cell number | IAP (Index of Apoptosis) |
| 27 | 4 | 14,81 | 26 | 2 | 7,69 |
| 42 | 4 | 9,52 | 24 | 4 | 16,67 |
| 54 | 4 | 7,41 | 66 | 3 | 4,54 |
| 54 | 5 | 9,26 | 38 | 2 | 5,26 |
| 64 | 9 | 14,06 | 68 | 8 | 11,76 |
| 27 | 4 | 14,81 | 89 | 1 | 1,12 |
| 49 | 12 | 24,49 | 108 | 3 | 2,78 |
| 43 | 10 | 23,25 | 103 | 1 | 0,97 |
| 42 | 1 | 2,38 | 52 | 0 | 0 |
| 402 | 53 | 13,18a | 574 | 24 | 4,18b |
Values with different superscripts in a row are significantly different (T-Student test p<0.05).
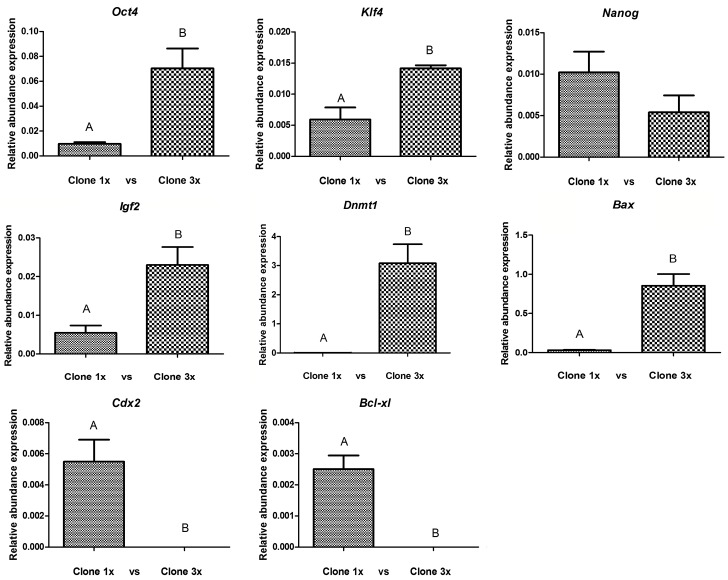
In order to evaluate the effect of cloned embryo aggregation on cellular reprogramming, we measured mRNA of pluripotency genes: Oct4, Klf4 and Nanog; two differentiation related markers Cdx2 and Igf2; two apoptosis markers Bcl-xl and Bax and finally Dnmt1, a key modulator of DNA methylation. Primers designed for this study are listed in materials and methods. We observed a significant statistical increase in the relative expression of Oct4, Klf4, Igf2, Bax and Dnmt1 genes. On the other hand, Nanog expression was not affected by embryo aggregation, whereas the relative amount of mRNA of Cdx2 and Bcl-xl genes were not detected in the 3x aggregate group in 40 cycles of the qPCR. Results of gene expression are shown in Fig 4.
Fig 4. Relative transcript abundance of Oct4, Klf4, Nanog, Igf2, Cdx2, Dnmt1, Bcl-xl, and Bax genes in pig day 7 blastocysts generated by SCNT.
All genes were normalized with the ACTB gene. (A, B) different letters are significantly different within each gene expression (p<0.05).
Discussion
In this study, we combined the assessment of developmental competence in terms of cleavage, blastocyst rate, number of cells, embryo diameter and DNA fragmentation levels to assess the effects of cloned pig embryo aggregation. Additionally, we performed a quantitative analysis of relative mRNA abundance of different genes involved in cell reprogramming (Klf4, Nanog, Oct4, Bcl-xl, Bax, Igf2, Dnmt 1 and Cdx2 compared with an endogenous gene: ACTB).
There are some limitations for cloning technique, such as the need of several good quality embryos to produce and maintain a pregnancy [1]. Furthermore, porcine oocytes and embryos are even more sensitive to temperature fluctuations than those from other domestic species [1]. It is believed that for a successful cloning, the pattern of epigenetic modifications in the donor cells must be remodeled to be similar to the pattern present in the fertilized embryos [35].As a result, we performed embryo aggregation and obtained a significant increase in in vitro embryo development and cell number, 20% cleavage and a three-fold increase in blastocyst rate. Similar observations were also reported following embryo aggregation in domestic animals such as the pig following aggregation of 2 cells [33], or of 4 cells [11, 14, 36, 37] and in the bovine [17], the horse [8, 9] and the feline [10]. Increased cleavage, development rates and cell number proved that the medium used was adequate for normal development. This is the first report where SOF medium was used for porcine embryos; it demonstrated a comparable rate of embryo development to other commonly used media.
We observed positive effects of embryo aggregation. This situation could be due to a) an epigenetic compensation since some epigenetic defects during cell reprogramming could be overcome by the combination of three embryos derived from the same cell line but with epigenetic differences; b) an increase in embryo cell number at day 0 of their development and a larger embryonic volume in the microwell system that may help embryos compact more easily, or c) a microenvironment generated by the interaction of beneficial and paracrine factors liberated by the aggregate embryo could increase rates of embryonic development; or d) a combination of all those situations. Additionally, embryo aggregation has been proposed as an alternative to avoid the increased level of heteroplasmy caused by the fusion of enucleated oocytes during the handmade cloning procedure [12, 16, 18]. In general, the term "heteroplasmy" refers to different mtDNA genomes in a single cell. Besides the minimal heteroplasmy produced by fusing a somatic cell into an enucleated oocyte during SCNT, embryo aggregation does not imply higher levels of heteroplasmy [38]. However, the potential effect of different mitochondrial DNA among cells but not within a cell generated by aggregated cloned embryos needs further investigation.
Based on results from the TUNEL assays, apoptotic cell indices were statistically lower in the 3x aggregated group compared with non-aggregated group. Lower levels of apoptotic cells in aggregated embryos were also reported for the pig [39] and the cow [40]. However, in other species, apoptotic levels do not change with embryo aggregation [9]. This situation may highlight that embryo aggregation effects depend on the embryo physiology characteristics of each species. Additional recent observations agree with our study, indicating that pig embryo aggregation has an anti-apoptotic effect due to fewer numbers of apoptotic cells as seen by TUNEL assay [14]. Embryo aggregation at day 0 increased embryo quality not only by increasing blastocyst diameters and initial number of cells during early embryo development but also by reducing the amounts of cells with fragmented DNA.
Unexpectedly, the relative expression of apoptotic related genes in the 3x experimental group is not necessary correlated with the observations obtained with the TUNEL assay. Bcl-xl, an anti apoptotic associated gene in cloned pig aggregated embryos was extremely low and could not be detected in 40 cycles of RT-qPCR and Bax (pro apoptotic associated gene) expression was significantly increased.
mRNA degradation is controlled by rates of synthesis and of decay [41]. However, recent results indicate that numerous untranslated mRNAs are assembled into “P bodies”, this consist in numerous messenger ribonucleoproteins that could accumulate in cytoplasmic foci and can be also degraded or return to translation [42– 45]. In eukaryotes, two general pathways of mRNA decay have been described and both pathways share the de-adenylation (removal of the polyA tail) [43, 46]. In aggregate embryos we saw increased numbers of cells and decreased protein translation showing less apoptosis in larger embryos. Mechanisms of this nature might be occurring in aggregated embryos. It remains unclear how this process is regulated in embryos, and how embryo aggregation could be involved. Further studies are required to understand the complete process.
As we mentioned previously, pluripotent related gene expression plays a critical role during cellular reprogramming and affects subsequent embryonic development [46]. Cloned embryos are reported to have lower expression of pluripotent genes when compared to in vivo embryos [47]. We observed higher expression of Oct4, Klf4, Igf2 in aggregated blastocyst, suggesting that embryo aggregation at one cell stage could improve the pluripotent status at the blastocyst stage.
It has been reported in mice that aggregated clones have an improved expression of Oct-4 and greater developmental potential compared with single clones [11]. Similarly, our results are in agreement with other reports in porcine aggregated blastocysts derived from SCNT embryos [33, 48]. Interestingly, embryo aggregation did not change Nanog expression while in previous studies expression levels of Nanog showed to be downregulated in cloned bovine blastocysts compared with their IVF counterparts [49, 50]. Surprisingly, we could not detect expression of Cdx2 in aggregated embryos, possibly due to extremely low expression levels in this experimental group. Contradictory, it has been reported an increased expression of this gene in aggregated pig embryos [14]. The reported benefits of embryo aggregation in the establishment of pregnancies and subsequent in vivo embryo development [51], suggest that if Cdx2 expression is affected in aggregated blastocyst, this may not affect future placentation. However, experiments focusing on placentation and in vivo embryo development of aggregated embryos are needed to determine the effects of embryo aggregation on blastocyst gene expression and its subsequent post implantation development.
Finally, Dnmt1 (DNA cytosine-5 methyl-transferase) catalyzes the production and regulation of the dynamics of mammalian patterns of global genomic DNA methylation [52, 53, 54].Our results could involve redirection of methylase Dnmt 1 in de novo methylation, involving reorganization of maternal methylation. The only way to verify if the rescheduling was successful, is to have pigs born alive, because the process of reprogramming the genome to turn into embryo is complicated and involves several steps, a process still not completely understood [54].
In conclusion, our data suggest that an incomplete reprogramming of porcine cloned embryos could be partially compensated by embryo aggregation, possibly by the combination of three different epigenetic embryos at the one cell stage supported by the higher expression of pluripotency genes observed in 3x aggregated embryos. Moreover, embryo aggregation improved the pre implantation in vitro developmental potential and increased blastocyst quality since diameter and cell number was higher while the level of apoptosis was lower.
Acknowledgments
The authors wish to thank to the technician Sørensen A L of the Stem Cell Epigenetics Laboratory, Institute of Basic Medical Sciences, Faculty of Medicine, University of Oslo, Norway. Also to Dr. MV San Martín for providing the material from the slaughterhouse using in this study. Finally, thanks to Professor Ornella Buemo for the English revision.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1.Nagashima H, Fujimura T, Takahagi Y, Kurome M, Wako N, Oichiai T, et al. Development of efficient strategies for the production de genetically modified pigs. Theriogenology 2003. 59 (1): 95–106. [DOI] [PubMed] [Google Scholar]
- 2.Tamada H, Kikyo N. Nuclear reprogramming in mammalian somatic cell nuclear cloning. Cytogenet Genome Res 2004. 105: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prather RS. Nuclear remodeling and nuclear reprogramming for making transgenic pigs by nuclear transfer.Adv Exp Med Biol 2007. 591: 1–13. [DOI] [PubMed] [Google Scholar]
- 4.Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 2000. 407 (6800): 86–90. [DOI] [PubMed] [Google Scholar]
- 5.Hyun S, Lee G, Kim D, Kim H, Lee S, Nam D, et al. Production of nuclear transfer-derived piglets using porcine fetal fibroblasts transfected with the enhanced green fluorescent protein. Biol Reprod 2003. 69 (3): 1060–8. [DOI] [PubMed] [Google Scholar]
- 6.Huan YJ, Zhu J, Xie BT, Wang JY, Liu SC, Zhou Y, et al. Treating cloned embryos, but not donor cells, with 5-aza-2'-deoxycytidine enhances the developmental competence of porcine cloned embryos. J Reprod Dev 2013. 59 (5):442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordignon V, El-Beirouthi N, Gasperin BG, Albornoz MS, Martinez-Diaz MA, Schneider C, et al. Production of cloned pigs with targeted attenuation of gene expression. PLoS One 2013. 8 (5): e 64613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambini A, Jarazo J, Olivera R, Salamone DF. Equine cloning: in vitro and in vivo development of aggregated embryos. Biol Reprod 2012. 87:15 1–9. [DOI] [PubMed] [Google Scholar]
- 9.Gambini A, De Stefano A, Bevacqua RJ, Karlanian F, Salamone DF. The aggregation of four reconstructed zygotes is the limit to improve the developmental competence of cloned equine embryos. PLoS One 2014. 9 (11): e 110998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moro LN, Hiriart MI, Buemo C, Jarazo J, Sestelo AJ, Veraguas D et al. Cheetah Embryo aggregation in interspecific SCNT improves development but not gene expression. Reproduction 2015. 150 (1):1–10. [DOI] [PubMed] [Google Scholar]
- 11.Boiani M, Eckardt S, Leu NA, Scholer HR, McLaughlin KJ Pluripotency deficit in clones overcome by clone-clone aggregation: epigenetic complementation? EMBO J 2003. 22: 5304–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misica-Turner PM, Oback FC, Eichenlaub M, Wells DN, Oback B. Aggregating embryonic but not somatic nuclear transfer embryos increases cloning efficiency in cattle. Biol Reprod 2007. 76: 268–278. [DOI] [PubMed] [Google Scholar]
- 13.Bang JI, Jin JI, Ghanem N, Choi BH, Fakruzzaman M, Ha AN, et al. Quality improvement of transgenic cloned bovine embryos using an aggregation method: Effects on cell number, cell ratio, embryo perimeter, mitochondrial distribution, and gene expression profile. Theriogenology 2015. 84 (4): 509–23. 10.1016/j.theriogenology.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 14.Siriboon C, Tu CF, Kere M, Liu MS, Chang HJ, et al. Production of viable cloned miniature pigs by aggregation of handmade cloned embryos at the 4-cell stage. Reprod Fertil Dev 2014. 26: 395–406. 10.1071/RD12243 [DOI] [PubMed] [Google Scholar]
- 15.Zhou W, Xiang T, Walker S, Abruzzese RV, Hwang E, Farrar V, et al. Aggregation of bovine cloned embryos at the four-cell stage stimulated gene expression and in vitro embryo development. Mol Reprod Dev 2008. 75 (8): 1281–9. 10.1002/mrd.20875 [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro ES, Gerger RP, Ohlweiler LU, Ortigari I Jr, Mezzalira JC, Forell F, et al. Developmental potential of bovine hand-made clone embryos reconstructed by aggregation or fusion with distinct cytoplasmic volumes. Cloning and Stem Cells 2009. 11: 377–386. 10.1089/clo.2009.0022 [DOI] [PubMed] [Google Scholar]
- 17.Akagi S, Yamaguchi D, Matsukawa K, Mizutani E, Hosoe M, Adachi N, et al. Developmental ability of somatic cell nuclear transferred embryos aggregated at the 8-cell stage or 16- to 32-cell stage in cattle. J Reprod Dev 2011. 57(4):500–6. [DOI] [PubMed] [Google Scholar]
- 18.Vajta G, Krugh PM, Mtango NR, Callesen H. Hand-made cloning approach: potentials and limitations. Reprod Fertil Dev 2005. 17, 97–112. [DOI] [PubMed] [Google Scholar]
- 19.Betts DH, King WA. Genetic regulation of embryo death and senescence. Theriogenology 2001. 55(1):171–91. [DOI] [PubMed] [Google Scholar]
- 20.Hardy K. Cell death in the mammalian blastocyst. Mol Hum Reprod 1997. 3: 919–925. [DOI] [PubMed] [Google Scholar]
- 21.Fabian D, Koppel J, Maddox-Hyttel. Apoptotic processes during mammalian preimplantation development. Theriogenology 2005. 64 (2): 221–31. [DOI] [PubMed] [Google Scholar]
- 22.Ju S, Rui R, Lu Q, Lin P, Guo H. Analysis of apoptosis and methyltransferase mRNA expression in porcine cloned embryos cultured in vitro. J Assist Reprod Genet 2010. 27 (1): 49–59. 10.1007/s10815-009-9378-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brison DR, Schultz RM. Apoptosis during mouse blastocyst formation: evidence for a role for survival factors including transforming growth factor alpha. Biol Reprod 1997. 56 (5): 1088–96. [DOI] [PubMed] [Google Scholar]
- 24.Hardy K, Spanos S. Growth factor expression and function in the human and mouse preimplantation embryo. J Endocrinol 2002. 172(2): 221–36. [DOI] [PubMed] [Google Scholar]
- 25.Kamjoo M, Brison DR, Kimber SJ. Apoptosis in the preimplantation mouse embryo: effect of strain difference and in vitro culture. Mol Reprod Dev 2002. 61 (1):67–77. [DOI] [PubMed] [Google Scholar]
- 26.Jones PL, Wolffe AP. Relationships between chromatin organization and DNA methylation in determining gene expression. Semin Cancer Biol 1999. 9 (5): 339–47. [DOI] [PubMed] [Google Scholar]
- 27.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe- Nebenius D, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998. 95:379–91. [DOI] [PubMed] [Google Scholar]
- 28.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003. 17:126–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003. 113:643–55. [DOI] [PubMed] [Google Scholar]
- 30.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005. 132:2093–102. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Jammes H, Rasmussen MA, Oestrup O, Beaujean N, Hall V, et al. Epigenetic regulation of gene expression in porcine epiblast, hypoblast, trophectoderm and epiblast-derived neural progenitor cells. Epigenetics 2011. 6 (9): 1149–61. 10.4161/epi.6.9.16954 [DOI] [PubMed] [Google Scholar]
- 32.Tang PC, West JD. The effects of embryo stage and cell number on the composition of mouse aggregation chimaeras. Zygote 2000. 8: 235–243. [DOI] [PubMed] [Google Scholar]
- 33.Lee SG, Park CH, Choi DH, Kim HS, Ka HH, Lee CK.In vitro development and cell allocation of porcine blastocysts derived by aggregation of in vitro fertilized embryos. Mol. Reprod Dev 2007. 74, 1436–1445. [DOI] [PubMed] [Google Scholar]
- 34.Vajta G, Peura TT, Holm P, Paldi A, Greve T, Trounson AO, et al. Method for culture of zona-included or zona-free embryos: the Well of the Well (WOW) system. Mol Reprod Dev 2000. 55: 256–264. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Whyte J, Prather RS. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res 2010. 341 (1): 13–21. 10.1007/s00441-010-1000-x [DOI] [PubMed] [Google Scholar]
- 36.Balbach ST, Esteves TC, Brink T, Gentile L, McLaughlin KJ, Adjaye JA, et al. Governing cell lineage formation in cloned mouse embryos. Dev Biol 2010. 343: 71–83. 10.1016/j.ydbio.2010.04.012 [DOI] [PubMed] [Google Scholar]
- 37.Kurosaka S, Eckardt S, Ealy AD, McLaughlin KJ. Regulation of blastocyst stage gene expression and outgrowth interferon tau activity of somatic cell clone aggregates. Cloning Stem Cells 2007. 9: 630–641. [DOI] [PubMed] [Google Scholar]
- 38.Hao Y, Lai L, Mao J, Im GS, Bonk A, Prather RS. Apoptosis and In Vitro Development of Preimplantation Porcine Embryos Derived In Vitro or by Nuclear Transfer. Biology of reproduction 2003. 69, 501–507. [DOI] [PubMed] [Google Scholar]
- 39.Burgstaller JP, Schinogl P, Dinnyes A, Müller M, Steinborn R. Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Dev Biol. 2007. 7:141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon SW, Park CH, Lee SG, Kim HM, Park JK, Uh KJ, et al. Anti-apoptotic effect of aggregation on preimplantation development of bovine in vitro fertilized embryos. Reprod Fertil Dev 2009. 21, 210–211. [Google Scholar]
- 41.Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 2013. 153 (5): 1000–11. 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 42.Meyer S, Temme C, and Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 2004. 39, 197–216. [DOI] [PubMed] [Google Scholar]
- 43.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of micro RNA genesis. EMBO Rep 2006. 7, 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker R and Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Struct Mol Biol 2004. 11:121–127. [DOI] [PubMed] [Google Scholar]
- 45.Parker R, Sheth U. P Bodies and the Control of mRNA Translation and Degradation. Molecular Cell 2007. 25 (5): 635–46. [DOI] [PubMed] [Google Scholar]
- 46.Eckardt S, McLaughlin KJ. Interpretation of reprogramming to predict the success of somatic cell cloning. Anim Reprod Sci 2004. 82–83:97–108. [DOI] [PubMed] [Google Scholar]
- 47.Jeong YI, Park CH, Kim HS, Jeong YW, Lee JY, Park SW,et al. Effects of Trichostatin A on In vitro Development of Porcine Embryos Derived from Somatic Cell Nuclear Transfer. Asian-Australasian Journal of Animal Sciences 2013, 1680–1688. 10.5713/ajas.2013.13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terashita Y, Sugimura S, Kudo Y, Amano R, Hiradate Y, Sato E. Improving the quality of miniature pig somatic cell nuclear transfer blastocysts: aggregation of SCNT embryos at the four-cell stage. Reprod Domest Anim 2011. 46: 189–196. 10.1111/j.1439-0531.2010.01614.x [DOI] [PubMed] [Google Scholar]
- 49.Beyhan Z, Forsberg EJ, Eilertsen KJ, Kent-First M, First NL. Gene expression in bovine nuclear transfer embryos in relation to donor cell efficiency in producing live offspring. Mol Reprod Dev 2007. 74: 18–27. [DOI] [PubMed] [Google Scholar]
- 50.Aston KI, Li GP, Hicks BA, Sessions BR, Davis AP, Rickords LF. Abnormal levels of transcript abundance of developmentally important genes in various stages of preimplantation bovine somatic cell nuclear transfer embryos. Cell Reprogram 2010. 12: 23–32. 10.1089/cell.2009.0042 [DOI] [PubMed] [Google Scholar]
- 51.Park SK, Roh S, Park JI. A simplified one-step nuclear transfer procedure alters the gene expression patterns and developmental potential of cloned porcine embryos. J Vet Sci 2014. 15 (1): 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet 2000. (16): 2395–402. Review. [DOI] [PubMed] [Google Scholar]
- 53.Leonhardt H, Page AW, Weier HU, Bestor TH.A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 1992. 27; 71 (5): 865–73. [DOI] [PubMed] [Google Scholar]
- 54.Golding MC, Williamson GL, Stroud TK., Westhusin ME, Long CR. Examination of DNA Methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol Reprod Dev 2011. 78 (5): 306–317. 10.1002/mrd.21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.