Abstract
Premise of the study:
Reconstructions of ancestral character states are among the most widely used analyses for evaluating the morphological, cytological, or ecological evolution of an organismic lineage. The software application Mesquite remains the most popular application for such reconstructions among plant scientists, even though its support for automating complex analyses is limited. A software tool is needed that automates the reconstruction and visualization of ancestral character states with Mesquite and similar applications.
Methods and Results:
A set of command line–based Python scripts was developed that (a) communicates standardized input to and output from the software applications Mesquite, BayesTraits, and TreeGraph2; (b) automates the process of ancestral character state reconstruction; and (c) facilitates the visualization of reconstruction results.
Conclusions:
WARACS provides a simple tool that streamlines the reconstruction and visualization of ancestral character states over a wide array of parameters, including tree distribution, character state, and optimality criterion.
Keywords: ancestral character state reconstruction, BayesTraits, Mesquite, Python, TreeGraph2, WARACS, wrapper
The reconstruction of ancestral character states is a central type of analysis in the evaluation of the cytological, ecological, metabolic, or morphological evolution of organismic lineages (Ronquist, 2004; Table 1). Numerous software applications have been developed that facilitate such reconstructions (Joy et al., 2014). The software application Mesquite (Maddison and Maddison, 2000) is a key tool in ancestral character state reconstruction (ACSR) and has been used in all branches of biology. Although it was introduced more than 15 years ago, it remains the most popular of such applications in the plant sciences. A search in the database PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) for all botanical investigations published between October 2012 and October 2015 that contain the term “ancestral character state reconstruction,” or any word combination thereof, in either the title, the key words, or the abstract, recovered a total of 27 publications (Table 1). An analogous search in the database ISI Web of Science (http://webofknowledge.com/) recovered a total of 93 publications. Of the 27 publications recovered via PubMed, 21 (78%) employed Mesquite, four (15%) employed functions in the R package APE (Paradis et al., 2004), two (7%) employed the software application BayesTraits (Pagel and Meade, 2004), and two (7%) employed the application SIMMAP (Bollback, 2006). The software applications RASP (Yu et al., 2010) and WinClada (Nixon, 2002) were employed in one publication (4%) each. While some publications used more than one of these applications, no less than two thirds conducted ACSR exclusively via Mesquite. Reconstructions that employed models specifically designed for biogeographic scenarios (e.g., Dillenberger and Kadereit, 2013; Salzman et al., 2015) or that evaluate character correlations (e.g., Soltis et al., 2013; Chartier et al., 2014) were not counted.
Table 1.
Details of recent botanical publications employing ACSR. This table displays the results of a literature search in PubMed for botanical investigations published between October 2012 and October 2015 that employ ACSR.
| Publicationa | Software | Character class | Character encoding | Optimality criterion/Reconstruction algorithm | Model of character evolutionb | Optimization conducted over |
| Brockington et al., 2015 | Mesquite | Metabolic | Discrete* | ML | Mk1 | Best tree of ML phyl. inf. |
| Pinheiro et al., 2015 | Mesquite | Ecological | Discrete* | MP | Best tree of ML phyl. inf. | |
| Wei et al., 2015 | Mesquite | Morphological | Discrete* | ML | Mk1 | Single tree (details n.s.) |
| Breitkopf et al., 2015 | Mesquite | Ecological | Discrete | ML | Mk1 | Post. tree dist. |
| Larson-Johnson, 2016 | APE (function n.s.) | Ecological | Discrete | ML | n.s. | Con. tree of ML phyl. inf. |
| Maguilla et al., 2015 | SIMMAP | Morphological | Discrete | SMM | Mk | Modified con. tree of post. tree dist. |
| Rodriguez-Riano et al., 2015 | Mesquite | Morphological | Discrete | MP, ML | Mk1 | Post. tree dist. |
| Salzman et al., 2015 | Mesquite, BayesTraits | Ecological | Discrete | ML, MCMC | ML: Mk1; MCMC: CTMM | Best tree of ML phyl. inf. |
| Cappa et al., 2015 | Mesquite | Metabolic | Discrete* | MP | Best tree of MP phyl. inf. | |
| Horn et al., 2014 | Mesquite | Metabolic | Discrete | ML | Mk | Post. tree dist. |
| Martins et al., 2014 | Mesquite | Morphological | Discrete | MP, ML | Mk1 | Con. tree of post. tree dist. |
| Panero et al., 2014 | Mesquite | Ecological, morphological | Discrete* | ML | Mk1 | Best tree of ML phyl. inf. |
| Wang et al., 2014 | RASP | Cytological, morphological | Discrete | BBM | Mk | Con. tree of post. tree dist. |
| Folk and Freudenstein, 2014 | Mesquite | Biogeographical, cytological, morphological | Discrete* | MP | Best tree of ML phyl. inf. | |
| Loriga et al., 2014 | APE (function “ace”) | Morphological | Discrete* | ML | ER | Con. tree of post. tree dist. |
| Schneider et al., 2014 | BayesTraits | Morphological | Discrete* | MCMC | CTMM | Post. tree dist. |
| Wagner et al., 2014 | Mesquite | Morphological | Discrete* | ML | Mk1 | Tree dist. (details n.s.) |
| Chartier et al., 2014 | Mesquite | Ecological, morphological | Discrete* | MP, ML | Mk1 | Modified best tree of ML phyl. inf. |
| Sundue and Rothfels, 2014 | Mesquite, Winclada | Morphological, cytological | Discrete* | MP, ML | Mk1 | Best tree of ML phyl. inf. |
| Fan et al., 2013 | Mesquite | Morphological | Discrete* | ML | Mk1 | Single tree (details n.s.) |
| Riser et al., 2013 | Mesquite, SIMMAP | Morphological | Discrete* | ML, SMM | Mk | Post. tree dist. |
| Bainard and Villarreal, 2013 | Mesquite | Cytological | Discrete* | MP | Single tree (details n.s.) | |
| Dillenberger and Kadereit, 2013 | Mesquite | Ecological | Discrete* | MP, ML | Mk1 | Tree dist. of MP phyl. inf., best tree of ML phyl. inf., post. tree dist. |
| He et al., 2013 | Mesquite | Genetic | Discrete* | MP | Best tree of ML phyl. inf. | |
| Soltis et al., 2013 | Mesquite, APE (function “ace”) | Ecological, morphological | Discrete* | MP, ML, SMM | ML: ER, ARD; SMM: Mk1 | Best tree of ML phyl. inf., tree dist. thereof generated by random polytomy resolution |
| Schaefer et al., 2012 | Mesquite | Ecological, morphological, cytological | Discrete* | ML | Mk1 | Best tree of ML phyl. inf. |
| Töpel et al., 2012 | APE (function “ace”) | Ecological | Continuous | ML | Brownian motion | con. tree of post. tree dist. |
Note: Model abbreviations: ARD = all-rates-different model (of function “ace” of the R package “ape”); CTMM = continuous-time Markov model; ER = equal-rates model (of function “ace” of the R package “ape”); Mk = class of Markov k-state models; Mk1 = Markov k-state one-parameter model (i.e., subset of “Mk”). Abbreviations for optimality criteria/reconstruction algorithms: BBM = Bayesian binary MCMC; MCMC = Markov chain Monte Carlo; ML = maximum likelihood; MP = maximum parsimony; SMM = stochastic mutational mapping. Other abbreviations: con. = consensus; dist. = distribution; n.s. = not specified; phyl. inf. = phylogeny inference; post. = posterior.
Publications are ordered by print date, beginning with the most recent.
Information on the model of character evolution for the likelihood and Bayesian reconstruction algorithms listed in column 5.
Asterisks indicate publications in which the authors explicitly state that a character state representing polymorphic or missing data was employed.
The plant sciences community would benefit from a tool that automates and streamlines the reconstruction of ancestral character states, particularly as conducted via Mesquite. Most software applications used in ACSR, except for those developed in the statistical scripting language R (R Core Team, 2015), feature designs that render the serialized execution of reconstructions challenging. Mesquite, for example, is primarily operated via a graphical user interface (GUI). Such a GUI, while comfortable for the transient user, impedes software operation in an automated workflow. Few users would choose to conduct import, export, analysis, and visualization operations by hand for dozens, or even hundreds of different data sets, even if their research questions warranted a replicated design. In addition, most software applications for the reconstruction of ancestral character states employ idiosyncratic input and output formats (e.g., SIMMAP) or lack the capability to visualize the results (e.g., BayesTraits). Consequently, it would be desirable for the plant sciences community to possess a tool that (a) standardizes the input and output feed to such software applications, (b) automates the reconstruction process, and (c) visualizes the reconstruction results in a publication-ready quality. The present publication introduces such a tool. A set of software scripts, collectively referred to as WARACS (Wrappers to Automate the Reconstruction of Ancestral Character States), was developed, which provides basic command line control to, and standardized input/output operations for, ACSR conducted in the applications Mesquite and BayesTraits. In addition, the script set provides a wrapper for the phylogenetic tree editor TreeGraph2 (Stöver and Müller, 2010), which facilitates the automatic visualization of ACSR results.
METHODS AND RESULTS
General design
The script set WARACS is designed to wrap and connect three established and well-maintained software applications—Mesquite, BayesTraits, and TreeGraph2—to automate and standardize the task of ACSR. Each script wraps around an individual executable and performs tasks that would otherwise need to be conducted step-by-step through a human operator, such as the formatting of information blocks in NEXUS files, the selection of character models and optimality criteria, or the parsing of raw reconstruction results. Together, the scripts provide a pipeline that connects different application executables. The visualization of reconstruction results is hereby considered an integral part of the automated workflow. Specifically, WARACS connects the output handle of the reconstruction process to the input handle of the tree visualization software TreeGraph2 to enable a graphically consistent visualization of results across multiple analyses. The scripts are command line–based, enabling the user to conduct reconstructions iteratively over a series of different input files and parameters. Overall, the script set is consistent with the concept of glue code (Lapp et al., 2007) and, as such, is easily customizable (e.g., in case of changes to the input and output formats) and expandable (e.g., when novel applications are to be included into the pipeline). WARACS is controlled exclusively via command line parameters; to ensure their consistent use, the parameters have been standardized across the three applications. WARACS has been tested with several versions of the wrapped software applications, including Mesquite v.2.75, v.3.03, and v.3.04; BayesTraits v.2.0; and TreeGraph2 v.2.0, v.2.6, and v.2.7.
Input and output design
Several concepts govern the input and output specifications of the WARACS script set. (a) The wrappers follow an ad hoc compilation approach when formatting input data. Specifically, the scripts compile input files from individual components upon execution. Such a design avoids the need for idiosyncratic or noncompliant input formats, which tend to erode the interoperability of data standards (Vos et al., 2012). The application Mesquite, for example, appends its internal commands in an idiosyncratic information block to the main NEXUS file. Such customized NEXUS files are automatically saved to disk, even though they can rarely be imported by any other software application. The ad hoc compilation approach selected for WARACS can also circumvent shortcomings concerning input file names, such as the inability of certain versions of Mesquite to import files with underscores in their names via the command line (M. Gruenstaeudl, personal observation). (b) The wrappers were designed to generate output files whose names are informative about the underlying reconstruction process. By default, WARACS saves the name of the software application and the optimality criterion employed as well as the character selected as part of the output filename. (c) The wrappers were designed to print short error reports to the screen upon code execution to communicate to the user problems with input formats, the reconstruction process, or the employed applications. (d) The wrappers were designed to form a pipeline so that reconstructions can be saved automatically as publication-ready figures. Such a standardized visualization step is important because larger, better-annotated pie diagrams (e.g., Riser et al., 2013; Chartier et al., 2014; Salzman et al., 2015) and more information on the underlying phylogenetic trees (Table 1) would often be helpful in the presentation of reconstruction results. Effective visualizations portray reconstructed character states both graphically and numerically (Wong, 2011; Krzywinski, 2013); WARACS saves reconstruction results in precisely that form in both vector as well as raster graphic format.
Availability and compatibility
The script set WARACS was written in the interpreter language Python (Python Software Foundation, 2012) and is consequently platform independent. It is compatible with Python v.2.7 (https://www.python.org/download/releases/2.7/) as well as Python v.3.5 (https://www.python.org/downloads/release/python-350/). WARACS is available under a BSD open source license from a code-sharing repository at GitHub (https://github.com/michaelgruenstaeudl/WARACS). Installation and usage instructions (file “README.md”), as well as several example files (folder “examples”), are provided alongside the wrapper scripts. To use WARACS, a user must have a Python interpreter as well as the Python packages DendroPy (https://pypi.python.org/pypi/DendroPy), NumPy (http://www.numpy.org), and six (https://pypi.python.org/pypi/six) installed. Software applications that are wrapped by WARACS must also be installed; current links to their respective installation websites are provided in the usage instructions as well as via the command line parameter “-h”. WARACS has been tested on Ubuntu 14.04, ArchLinux 4.2.3, and Mac OSX 10.8.5.
Usage
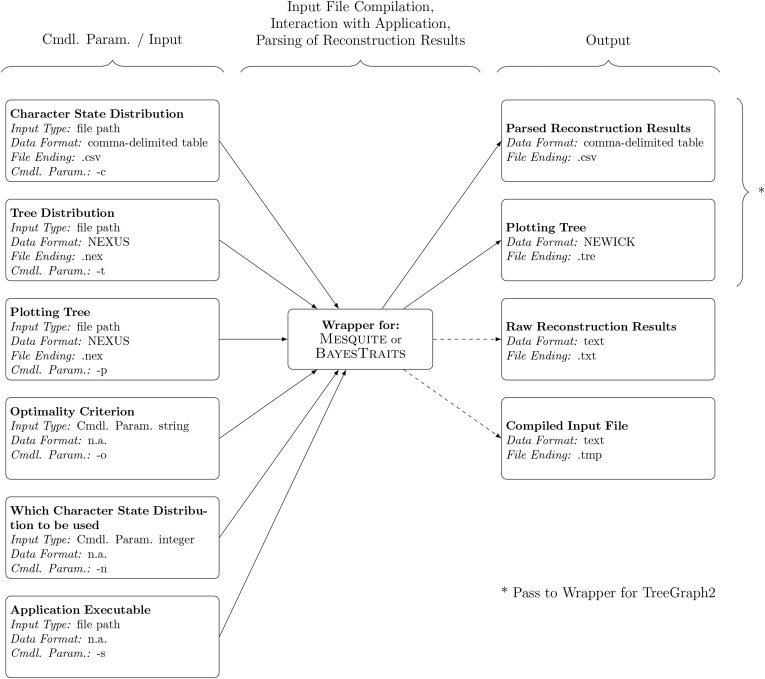
Use of the script set WARACS is driven exclusively via the specification of command line parameters. At a minimum, reconstructions of ancestral character states require five items of input: a distribution of character states for the organisms under study (hereafter “character state distribution”), one or more phylogenetic trees of the organisms under study on which the character state distribution is optimized (hereafter “tree distribution”), one phylogenetic tree on which the reconstruction results are plotted (hereafter “plotting tree”), an optimality criterion for the reconstruction, and a model of character evolution. Most applications for ACSR assign a default model of character evolution to the character state distribution under study; users of WARACS can select a different model by modifying the compiled input file. Multiple character state distributions can be supplied to WARACS, and users must select which of them to use in a particular reconstruction. In addition, users must specify the location of the application executable on the system. Hence, the WARACS wrappers for ACSR operate with six mandatory command line parameters (Fig. 1): (i) the character state distribution (command line parameter “-c”), specified as a file path to a comma-delimited table; (ii) the tree distribution (command line parameter “-t”), specified as a file path to a text file in NEXUS format (Maddison et al., 1997) containing one or more phylogenetic trees; (iii) the plotting tree (command line parameter “-p”), specified as a file path to a text file in NEXUS format containing a single phylogenetic tree; (iv) the optimality criterion (command line parameter “-o”), specified as a command line parameter string; (v) a specification of the character state distribution used in the reconstruction (command line parameter “-n”), specified as a command line parameter integer; and (vi) a file path to the application executable (command line parameter “-s”). For example, to conduct a reconstruction of character state distribution 2 on a distribution of phylogenetic trees under the maximum likelihood optimality criterion using a single-rate model (Mk1; Lewis, 2001) via Mesquite, a user on a Linux operating system would enter the following command into his command line shell of choice as a single, uninterrupted line:
python /path_to_WARACS/WARACS_Mesquite.py
-c /path_to_input/character_state_distribution.csv
-t /path_to_input/tree_distribution.tre
-p /path_to_input/plotting_tree.tre
-o likelihood
-n 2
-s /path_to_software/mesquite.sh
Fig. 1.
Schematic of the reconstruction of ancestral character states as facilitated by WARACS. Dashed lines indicate optional output. Cmdl. Param. = Command line parameter; n.a. = not applicable.
Upon execution, the wrapper script compiles a comprehensive input file in a modified NEXUS format, passes it to Mesquite, receives raw reconstruction results from Mesquite, parses these results, and saves up to four output files to the user’s working directory (Fig. 1). These output files are: (i) a comma-delimited table containing the parsed reconstruction results (saved with the file ending “.csv”), (ii) the plotting tree in NEWICK format (Maddison et al., 1997) (saved with the file ending “.tre”), (iii) the raw reconstruction results generated by Mesquite (saved with the file ending “.txt”), and (iv) the compiled input file (saved with the file ending “.tmp”). Files (iii) and (iv) are optional and only generated when command line parameter “-k” is invoked. The parsed results table consists of two columns: column 1 specifies the node numbers on the plotting tree, column 2 the corresponding reconstructed character states. A set of example files that illustrate the input and output of ACSR with WARACS is cosupplied with the scripts (folders “examples/example_Mesquite” and “examples/example_BayesTraits”). Currently, WARACS can facilitate reconstructions under discrete character states, which is the predominant type of character encoding in current botanical investigations (Table 1). Polymorphic or missing states can be included in analyses facilitated by WARACS, but their precise effect on the reconstructions is governed by the default settings of the wrapped software applications.
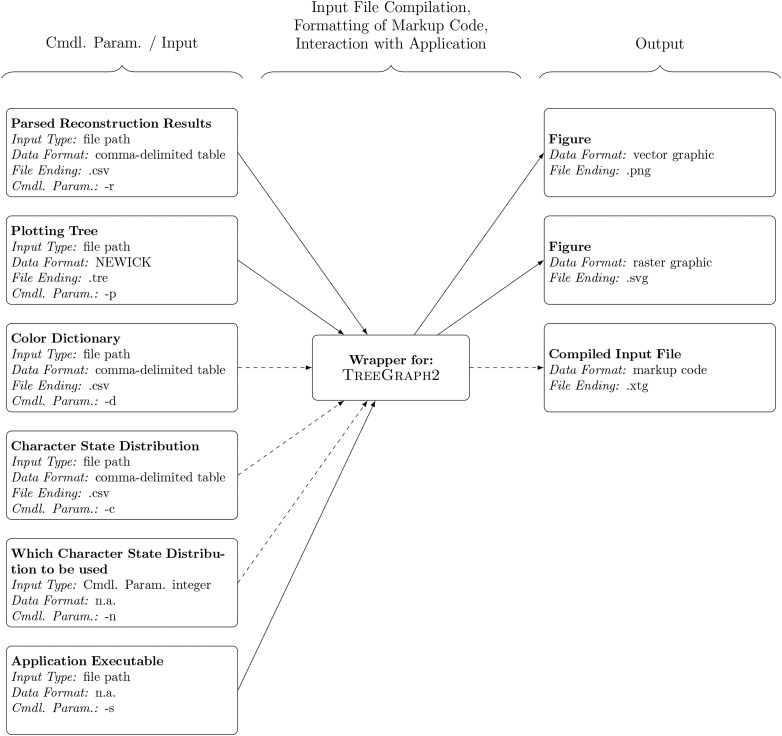
The visualization of an ACSR via TreeGraph2 requires a minimum of three items of information (Fig. 2): (i) a comma-delimited table containing the results of an ACSR (command line parameter “-r”), (ii) a plotting tree in NEWICK format (command line parameter “-p”), and (iii) a file path to the application executable (command line parameter “-s”). Moreover, a user can specify a comma-delimited table of color specifications (hereafter “color dictionary”; command line parameter “-d”) to link the states of the character state distribution to specific colors. Specifically, this table instructs the visualization engine about the colors that the pie diagram slices representing the reconstruction results must be filled with. A color dictionary is separated into two columns: column 1 specifies the character states, column 2 the corresponding colors in hexadecimal format. In the absence of a user-defined color dictionary, WARACS employs a default color palette as specified by http://colorbrewer2.org for qualitative characters. In addition to a color dictionary, a user can specify the character state distribution used during the reconstruction process to plot the character states of the terminal taxa (command line parameters “-c” and “-n”). Hence, to visualize the results of the ACSR listed above as well as the character states of the terminal taxa under a custom color dictionary, a user on a Linux operating system would enter the following command into his command line shell of choice as a single, uninterrupted line:
python /path_to_WARACS/WARACS_TreeGraph2.py
-r /path_to_input/treedistr__Mesquite_likelihood_char2.csv
-p /path_to_input/treedistr__Mesquite_likelihood_char2.tre
-d /path_to_input/color_dictionary.csv
-c /path_to_input/character_state_distribution.csv
-n 2
-s /path_to_software/TreeGraph.jar
Fig. 2.
Schematic of the visualization of ACSR results as facilitated by WARACS. Dashed lines indicate optional input and output. Cmdl. Param. = Command line parameter; n.a. = not applicable.
Upon execution, the wrapper script compiles a comprehensive input file in XML format and passes it to TreeGraph2, which generates a figure in two different image formats (in the vector graphic format “svg” and in the raster graphic format “png”) in the user’s working directory (Fig. 2). These figures represent the end product of the visualization process. If command line parameter “-k” is invoked, the compiled input file is saved to the working directory (with the file ending “.xtg”) to allow for a renewed visualization upon manual modification. A set of example files that illustrate the input and output of the visualization process with WARACS is cosupplied with the scripts (folder “examples/example_TreeGraph2”).
CONCLUSIONS
Despite the versatility of functions for ACSR in the statistical scripting language R, the plant sciences community holds a strong preference toward the software application Mesquite for reconstructing ancestral character states (Table 1). While Mesquite contains functionality to reproduce analyses via customized command scripts in an idiosyncratic scripting language, it lacks genuine command line support. Conducting serialized reconstructions of ancestral character states (e.g., iterating over a series of tree distributions or optimality criteria) is consequently challenging under Mesquite. Serialized reconstructions and subsequent result visualization could theoretically be achieved by combining several existing R packages, but this requires the knowledge to write customized R scripts and concatenate different functions into a pipeline. The wrapper scripts presented here constitute a fast and simple-to-use alternative to customized R scripts. Moreover, they support the community effort to make analyses more reproducible. If authors specify the command line parameters they selected alongside the original input data, others can reproduce their analyses and explore alternatives. In addition, the provision of interoperability between widely used software applications is an effective strategy to expand their functionality (e.g., Maddison and Maddison, 2014), because the wrapped applications have likely undergone extensive testing and follow good coding practices (Leprevost et al., 2014).
The script set WARACS enables several types of analyses that are not or only partially available through the application of Mesquite alone. WARACS was designed to provide basic command line control to the software applications Mesquite, BayesTraits, and TreeGraph2 for the purpose of automating and streamlining the reconstruction and visualization of ancestral character states. By using these wrappers, several types of analyses become available without the need to write intricate analysis scripts. First, researchers can automatically iterate over multiple trees or tree distributions to compare the reconstruction results. This option could be desirable for investigations anticipating a different placement of taxa across different gene tree distributions (e.g., Folk and Freudenstein, 2014) or such attempting to accommodate phylogenetic uncertainty among the target organisms (e.g., Dillenberger and Kadereit, 2013). Second, researchers can automatically iterate over multiple optimality criteria and reconstruction algorithms to avoid bias caused by individual algorithm implementations. Ricklefs (2007), for example, pointed out that it is currently unclear if random-walk models such as those implemented in stochastic character mapping truly reflect natural processes. Instead of relying on any one reconstruction method, scientists should compare the results of an ACSR under different optimality criteria (e.g., Ekman et al., 2008; Soltis et al., 2013). Third, the application of WARACS simplifies the process to visualize reconstruction results on phylogenetic trees that are different from those used in the reconstruction process. For example, researchers who wish to infer ancestral character states over a posterior tree distribution, but visualize their results on the best phylogenetic tree inferred under maximum likelihood, can feed such independent input files to WARACS as long as they share the same taxon set. Likewise, WARACS simplifies the process to combine the results of different reconstructions on a single tree (e.g., Larridon et al., 2015). For example, researchers who wish to plot ancestral character states inferred under different data partitions onto a particular consensus tree (e.g., de Villiers et al., 2013) can visualize their results jointly through concatenation of either the compiled TreeGraph2 input files or the vector graphics. Without this functionality, researchers are forced to present near-identical figures that differ only in the reconstructed character states (e.g., Schaefer et al., 2012). In summary, the script set WARACS provides an easily accessible interface to popular software applications for ACSR and makes several intricate types of character state reconstruction available to the average user.
LITERATURE CITED
- Bainard J. D., Villarreal J. C. 2013. Genome size increases in recently diverged hornwort clades. Genome 56: 431–435. [DOI] [PubMed] [Google Scholar]
- Bollback J. P. 2006. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkopf H., Onstein R. E., Cafasso D., Schlüter P. M., Cozzolino S. 2015. Multiple shifts to different pollinators fuelled rapid diversification in sexually deceptive Ophrys orchids. New Phytologist 207: 377–389. [DOI] [PubMed] [Google Scholar]
- Brockington S. F., Yang Y., Gandia-Herrero F., Covshoff S., Hibberd J. M., Sage R. F., Wong G. K. S., et al. 2015. Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytologist 207: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappa J. J., Yetter C., Fakra S., Cappa P. J., DeTar R., Landes C., Pilon-Smits E. A. H., Simmons M. P. 2015. Evolution of selenium hyperaccumulation in Stanleya (Brassicaceae) as inferred from phylogeny, physiology and X-ray microprobe analysis. New Phytologist 205: 583–595. [DOI] [PubMed] [Google Scholar]
- Chartier M., Gibernau M., Renner S. S. 2014. The evolution of pollinator-plant interaction types in the Araceae. Evolution 68: 1533–1543. [DOI] [PubMed] [Google Scholar]
- de Villiers M. J., Pirie M. D., Hughes M., Moller M., Edwards T. J., Bellstedt D. U. 2013. An approach to identify putative hybrids in the ‘coalescent stochasticity zone’, as exemplified in the African plant genus Streptocarpus (Gesneriaceae). New Phytologist 198: 284–300. [DOI] [PubMed] [Google Scholar]
- Dillenberger M. S., Kadereit J. W. 2013. The phylogeny of the European high mountain genus Adenostyles (Asteraceae-Senecioneae) reveals that edaphic shifts coincide with dispersal events. American Journal of Botany 100: 1171–1183. [DOI] [PubMed] [Google Scholar]
- Ekman S., Andersen H. L., Wedin M. 2008. The limitations of ancestral state reconstruction and the evolution of the ascus in the Lecanorales (lichenized Ascomycota). Systematic Biology 57: 141–156. [DOI] [PubMed] [Google Scholar]
- Fan D.-M., Chen J.-H., Meng Y., Wen J., Huang J.-L., Yang Y.-P. 2013. Molecular phylogeny of Koenigia L. (Polygonaceae: Persicarieae): Implications for classification, character evolution and biogeography. Molecular Phylogenetics and Evolution 69: 1093–1100. [DOI] [PubMed] [Google Scholar]
- Folk R. A., Freudenstein J. V. 2014. Phylogenetic relationships and character evolution in Heuchera (Saxifragaceae) on the basis of multiple nuclear loci. American Journal of Botany 101: 1532–1550. [DOI] [PubMed] [Google Scholar]
- He L., Zhao M., Wang Y., Gai J., He C. 2013. Phylogeny, structural evolution and functional diversification of the plant PHOSPHATE1 gene family: A focus on Glycine max. BMC Evolutionary Biology 13: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn J. W., Xi Z., Riina R., Peirson J. A., Yang Y., Dorsey B. L., Berry P. E., et al. 2014. Evolutionary bursts in Euphorbia (Euphorbiaceae) are linked with photosynthetic pathway. Evolution 68: 3485–3504. [DOI] [PubMed] [Google Scholar]
- Joy J. B., Mccloskey R. M., Liang R. H., Nguyen T., Poon A. F. Y. 2014. PLoS Computational Biology Ancestral reconstruction topic page. Website http://compbiolwiki.plos.org/wiki/Ancestral_reconstruction [accessed 23 October 2015]. [DOI] [PMC free article] [PubMed]
- Krzywinski M. 2013. Points of view: Labels and callouts. Nature Methods 10: 275. [DOI] [PubMed] [Google Scholar]
- Lapp H., Bala S., Balhoff J. P., Bouck A., Goto N., Holder M., Holland R., et al. 2007. The 2006 NESCent Phyloinformatics Hackathon: A field report. Evolutionary Bioinformatics Online 3: 287–296. [Google Scholar]
- Larridon I., Walter H. E., Guerrero P. C., Duarte M., Cisternas M. A., Hernandez C. P., Bauters K., et al. 2015. An integrative approach to understanding the evolution and diversity of Copiapoa (Cactaceae), a threatened endemic Chilean genus from the Atacama Desert. American Journal of Botany 102: 1506–1520. [DOI] [PubMed] [Google Scholar]
- Larson-Johnson K. 2016. Phylogenetic investigation of the complex evolutionary history of dispersal mode and diversification rates across living and fossil Fagales. New Phytologist 209: 418–435. [DOI] [PubMed] [Google Scholar]
- Leprevost F., Da V., Barbosa V. C., Francisco E. L., Perez-Riverol Y., Carvalho P. C. 2014. On best practices in the development of bioinformatics software. Frontiers in Genetics 5: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Loriga J., Schmidt A. R., Moran R. C., Feldberg K., Schneider H., Heinrichs J. 2014. The first fossil of a bolbitidoid fern belongs to the early-divergent lineages of Elaphoglossum (Dryopteridaceae). American Journal of Botany 101: 1466–1475. [DOI] [PubMed] [Google Scholar]
- Maddison D. R., Swofford D. L., Maddison W. P. 1997. NEXUS: An extensible file format for systematic information. Systematic Biology 46: 590–621. [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R. 2000. onward. Mesquite: A modular system for evolutionary analysis, version 3.04, August 2015, Website http://mesquiteproject.org [accessed 23 October 2015].
- Maddison W. P., Maddison D. R. 2014. onward. Zephyr: A Mesquite package for interacting with external phylogeny inference programs, version 1.1, January 2015, Website https://mesquitezephyr.wikispaces.com [accessed 23 October 2015].
- Maguilla E., Escudero M., Waterway M. J., Hipp A. L., Luceno M. 2015. Phylogeny, systematics, and trait evolution of Carex section Glareosae. American Journal of Botany 102: 1128–1144. [DOI] [PubMed] [Google Scholar]
- Martins A. C., Scherz M. D., Renner S. S. 2014. Several origins of floral oil in the Angelonieae, a southern hemisphere disjunct clade of Plantaginaceae. American Journal of Botany 101: 2113–2120. [DOI] [PubMed] [Google Scholar]
- Nixon K. C. 2002. WinClada, version 1.00.08. Published by the author, Ithaca, New York, USA. Website http://www.cladistics.com/aboutWinc.htm [accessed 23 October 2015].
- Pagel M., Meade A. 2004. onward. BayesTraits, version 2.0, September 2014, Website http://www.evolution.reading.ac.uk/BayesTraits.html [accessed 23 October 2015].
- Panero J. L., Freire S. E., Ariza Espinar L., Crozier B. S., Barboza G. E., Cantero J. J. 2014. Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Molecular Phylogenetics and Evolution 80: 43–53. [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. 2004. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pinheiro F., Cafasso D., Cozzolino S., Scopece G. 2015. Transitions between self-compatibility and self-incompatibility and the evolution of reproductive isolation in the large and diverse tropical genus Dendrobium (Orchidaceae). Annals of Botany 116: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Python Software Foundation. 2012. Python Language Reference, versions 2.7 and 3.5. Website http://www.python.org [accessed 23 October 2015].
- R Core Team. 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Website http://www.R-project.org/ [accessed 23 October 2015].
- Ricklefs R. E. 2007. Estimating diversification rates from phylogenetic information. Trends in Ecology & Evolution 22: 601–610. [DOI] [PubMed] [Google Scholar]
- Riser J. P., Cardinal-McTeague W. M., Hall J. C., Hahn W. J., Sytsma K. J., Roalson E. H. 2013. Phylogenetic relationships among the North American cleomoids (Cleomaceae): A test of Iltis’s reduction series. American Journal of Botany 100: 2102–2111. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Riaño T., Valtueña F. J., López J., Navarro-Pérez M. L., Pérez-Bote J. L., Ortega-Olivencia A. 2015. Evolution of the staminode in a representative sample of Scrophularia and its role as nectar safeguard in three widespread species. Naturwissenschaften 102: 37. [DOI] [PubMed] [Google Scholar]
- Ronquist F. 2004. Bayesian inference of character evolution. Trends in Ecology & Evolution 19: 475–481. [DOI] [PubMed] [Google Scholar]
- Salzman S., Driscoll H. E., Renner T., Andre T., Shen S., Specht C. D. 2015. Spiraling into history: A molecular phylogeny and investigation of biogeographic origins and floral evolution for the genus Costus. Systematic Botany 40: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H., Hechenleitner P., Santos-Guerra A., Menezes De Sequeira M., Pennington R. T., Kenicer G., Carine M. A. 2012. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evolutionary Biology 12: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. V., Bissiengou P., Do M., Amaral C. E., Tahir A., Fay M. F., Thines M., Sosef M. S. M., et al. 2014. Phylogenetics, ancestral state reconstruction, and a new infrafamilial classification of the pantropical Ochnaceae (Medusagynaceae, Ochnaceae s.str., Quiinaceae) based on five DNA regions. Molecular Phylogenetics and Evolution 78: 199–214. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Mort M. E., Latvis M., Mavrodiev E. V., O’Meara B. C., Soltis P. S., Burleigh J. G., Rubio de Casas R. 2013. Phylogenetic relationships and character evolution analysis of Saxifragales using a supermatrix approach. American Journal of Botany 100: 916–929. [DOI] [PubMed] [Google Scholar]
- Stöver B. C., Müller K. F. 2010. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundue M. A., Rothfels C. J. 2014. Stasis and convergence characterize morphological evolution in eupolypod II ferns. Annals of Botany 113: 35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpel M., Antonelli A., Yesson C., Eriksen B. 2012. Past climate change and plant evolution in Western North America: A case study in Rosaceae. PLoS ONE 7: e50358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos R. A., Balhoff J. P., Caravas J. A., Holder M. T., Lapp H., Maddison W. P., Midford P. E., et al. 2012. NeXML: Rich, extensible, and verifiable representation of comparative data and metadata. Systematic Biology 61: 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S. T., Hesse L., Isnard S., Samain M.-S., Bolin J., Maass E., Neinhuis C., et al. 2014. Major trends in stem anatomy and growth forms in the perianth-bearing Piperales, with special focus on Aristolochia. Annals of Botany 113: 1139–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang X.-Q., Sun H., Yu Y., He X.-J., Hong D.-Y. 2014. Evolution of the platycodonoid group with particular references to biogeography and character evolution. Journal of Integrative Plant Biology 56: 995–1008. [DOI] [PubMed] [Google Scholar]
- Wei L., Xiang X.-G., Wang Y.-Z., Li Z.-Y. 2015. Phylogenetic relationships and evolution of the Androecia in Ruteae (Rutaceae). PLoS ONE 10: e0137190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong B. 2011. Points of view: Points of review (part 2). Nature Methods 8: 189. [DOI] [PubMed] [Google Scholar]
- Yu Y., Harris A. J., He X. J. 2010. onward. RASP (Reconstruct ancestral state in phylogenies), version 3.2, July 2015. Website http://mnh.scu.edu.cn/soft/blog/RASP [accessed 23 October 2015].