Significance
The roseobacter constitute nearly 25% of marine bacteria in coastal areas. They have been shown to produce a potent antibiotic, tropodithietic acid (TDA), which confers far-reaching aquacultural and ecological benefits. We have examined the mode of action of TDA using a number of methods including bacterial cytological profiling. Surprisingly, TDA clusters with molecules that are structurally unrelated and highly cytotoxic. Subsequent screens demonstrate that, aside from its antibacterial activity, TDA is also a potent, broad-range, and potentially useful anticancer agent. Our studies highlight the power of profiling methods in repurposing bioactive compounds, reveal the mode of action of the unusual troponoid TDA, and uncover a previously unknown link between two metabolic cycles, which together provide TDA resistance to the producing roseobacter.
Keywords: tropodithietic acid, roseobacter, mode of action, antibiotic, anticancer agent
Abstract
While we have come to appreciate the architectural complexity of microbially synthesized secondary metabolites, far less attention has been paid to linking their structural features with possible modes of action. This is certainly the case with tropodithietic acid (TDA), a broad-spectrum antibiotic generated by marine bacteria that engage in dynamic symbioses with microscopic algae. TDA promotes algal health by killing unwanted marine pathogens; however, its mode of action (MoA) and significance for the survival of an algal–bacterial miniecosystem remains unknown. Using cytological profiling, we herein determine the MoA of TDA and surprisingly find that it acts by a mechanism similar to polyether antibiotics, which are structurally highly divergent. We show that like polyether drugs, TDA collapses the proton motive force by a proton antiport mechanism, in which extracellular protons are exchanged for cytoplasmic cations. The α-carboxy-tropone substructure is ideal for this purpose as the proton can be carried on the carboxyl group, whereas the basicity of the tropylium ion facilitates cation export. Based on similarities to polyether anticancer agents we have further examined TDA’s cytotoxicity and find it to exhibit potent, broad-spectrum anticancer activities. These results highlight the power of MoA-profiling technologies in repurposing old drugs for new targets. In addition, we identify an operon that confers TDA resistance to the producing marine bacteria. Bioinformatic and biochemical analyses of these genes lead to a previously unknown metabolic link between TDA/acid resistance and the γ-glutamyl cycle. The implications of this resistance mechanism in the context of the algal-bacterial symbiosis are discussed.
Naturally occurring small molecules represent a significant fraction of our current arsenal of antibiotics and anticancer drugs (1–3). Their structures have been selected over evolutionary time for the physiological functions they fulfill (4). That physiological function, however, is in many cases unknown. Research on natural products in general focuses on discovery, structural elucidation, and in some cases, in vitro biological activity. However, studies on mode of action (MoA), especially in a natural context, have received far less attention. Understanding the mechanisms of action of natural products critically informs how microbes interact with one another using small molecules and, in the case of antibiotics, provides insights into the emergence of resistance. One strategy for addressing this disconnect between structure and physiological function is to discover molecules in their natural context, rather than by a traditional grind-and-find approach. We recently provided one example of this search strategy by identifying molecules that modulate a naturally occurring algal–bacterial symbiosis (5). This symbiosis comprises members of a phylogenetically diverse set of marine bacteria, the roseobacter, and the microalgal haptophyte, Emiliania huxleyi.
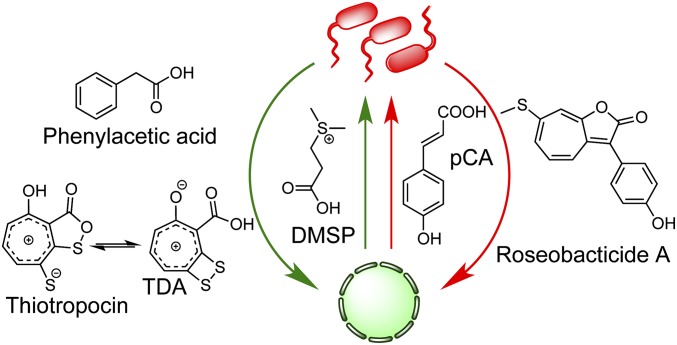
Members of the roseobacter clade are noted for occupying diverse marine niches spanning coastal and polar regions, making up nearly 25% of microbial life in the ocean (6–8). Geographically disparate marine isolates of one of the most well-studied roseobacter, Phaeobacter inhibens, have demonstrated a high degree of genomic synteny as well as efficacy in colonizing biotic and abiotic surfaces (9–12). The biotic surfaces include macro- and microalgal species. We have focused on the interaction of P. inhibens with E. huxleyi and discovered that it is dynamic in nature (5, 13). In our current model, the symbiosis is largely dictated by the health of the algal host (Fig. 1). When the alga is healthy, it provides an attachment surface and food in the form of dimethylsulfoniopropionate (DMSP), which roseobacter can use as a source of carbon and sulfur (14). The bacteria, in return, produce a growth promoter, phenylacetic acid, and a broad-spectrum antibiotic, tropodithietic acid (TDA) (15–18), which nurture and protect the host. However, a mutualist-to-parasite switch occurs when the algae senesce. Under these conditions, they release the phenylpropanoid, p-coumaric acid, which triggers production of potent algaecides, the roseobacticides from the bacteria.
Fig. 1.
Proposed model for the interaction between P. inhibens and E. huxleyi. In the mutualistic phase (green arrows), E. huxleyi provides a carbon and sulfur source (DMSP), while the bacteria generate algal growth-protecting (TDA) and promoting (phenylacetic acid) molecules. When the algae senesce, a proposed mutualist-to-parasite switch takes place. In the parasitic phase (red arrows), E. huxleyi releases p-coumaric acid, an inducer of bacterial roseobacticide production, which kills the algal host.
To further define this symbiosis, we have been investigating the biosynthesis and detailed biological function of the attendant small molecules that characterize each phase of the interaction. While roseobacticides harbor somewhat specific algaecidal activities (5), TDA is a broad-spectrum antibiotic with an as-of-yet undetermined MoA (15, 18). Not only is the biological activity of TDA important for this algal–bacterial symbiosis, but more far-reaching ecological and aquacultural benefits have also been attributed to roseobacter and their ability to produce this potent antibiotic, which kills unwanted marine pathogens (19–21). TDA is a member of the rare troponoid family of natural products. Only about a handful of naturally occurring troponoids are known (22); P. inhibens produces two such molecules, TDA and roseobacticides. TDA contains a number of unusual structural features: It has been observed in two tautomeric forms, which have been given distinct names, TDA and thiotropocin (Fig. 1) (15–18). Bentley and colleagues have calculated that TDA is the more stable tautomer, by ∼4–5 kcal/mol, and that the activation energy for conversion of thiotropocin to TDA is fairly small, on the order of 2–8 kcal/mol (18). TDA contains a substituted tropone, which bears partial aromatic tropylium ion character in addition to a strained dithietene moiety. The importance of the carboxyl substituent, the dithietene, and the tautomer thiotropocin, to the biological activity of TDA remains unknown.
Here we examine the chemistry and biology underlying the function TDA in two complementary scenarios: how TDA vanquishes microbial life and how life triumphs over it. This approach is synergistic in that understanding the mechanism of action of TDA enabled our discovery of the mechanism of TDA resistance. Among other approaches, we have used the MoA-determining imaging technique, bacterial cytological profiling (BCP) (23), which not only provided a model for how TDA functions but also grouped it with potent anticancer molecules. Motivated by these results, we examined cytotoxic properties of TDA and report that it harbors potent anticancer activities. Our studies validate the use of BCP in finding the MoA of natural products and in repurposing known natural bioactive molecules for alternative targets.
Results and Discussion
TDA Rapidly Collapses the Proton Motive Force.
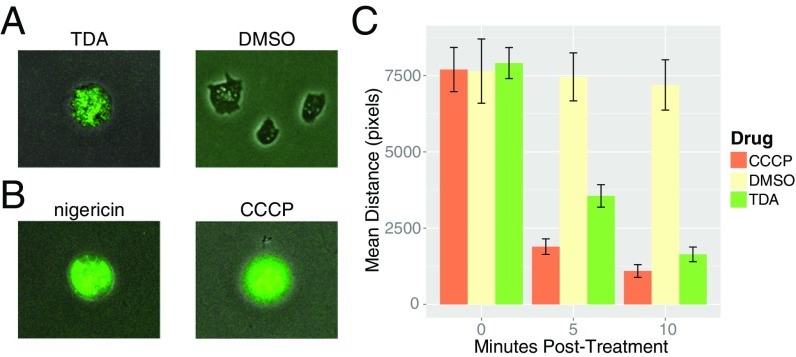
Previous studies showed that TDA is active against a broad spectrum of microorganisms, including both Gram-negative and Gram-positive bacteria as well as fungi (15). We sought to confirm TDA’s broad-spectrum activity by testing its effects on the eukaryotic amoeba Dictyostelium discoideum (SI Appendix, Fig. S1). Using an esterase-cleavable calcein green indicator as a marker for cell death, we observed that D. discoideum cells rapidly (within minutes) lost motility and displayed high green fluorescence signal, revealing their compromised outer membranes and thus cell death (Fig. 2A and Movies S1 and S2). Given TDA’s cross-domain activity and its small size, we deemed it unlikely that TDA would display different modes of action depending on the species tested. Instead, noting others’ failed attempts to select for TDA-resistant bacteria (24), we reasoned that its target must be a highly conserved, essential process maintained by some universal cellular component, which does not gain resistance by the basic mutations expected in a genetic screen. One such candidate target is the cell membrane, which maintains the universally conserved, essential process of ATP synthesis using the proton motive force (PMF). We tested the activity of two well-characterized PMF disruptors, carbonyl cyanide m-chlorophenyl hydrazine (CCCP) and nigericin, on D. discoideum and found that they too resulted in cell death (Fig. 2B and Movies S3 and S4), consistent with the idea that TDA could indeed be targeting the membrane.
Fig. 2.
Effect of PMF-collapsing drugs on the eukaryote D. discoideum and on flagellar rotation in E. coli. (A and B) D. discoideum was treated with TDA (200 μM) and DMSO control (A) as well as with known PMF disrupters nigericin and CCCP (200 μM each) (B). After treatment, cell wall integrity was visualized by calcein green. The fluorescence signal observed with antibiotics indicates compromised cellular membrane and cell death. (C) E. coli cells were treated with DMSO, CCCP, or TDA (each drug at 200 μM), and flagellar rotation subsequently quantified for a 45-s period at 0, 5, and 10 min after treatment. For the DMSO, CCCP, and TDA samples, 16, 20, and 22 replicates were averaged, respectively. Error bars represent SEM (Materials and Methods and SI Appendix).
In Escherichia coli the speed of the flagellar motor is directly coupled to the PMF (25). If TDA collapses the PMF, we would expect it to cause the motor to stop rotating the flagella. To address this prediction, we tethered E. coli KAF84 cells to BSA-treated glass bottom of a flow chamber as previously described and took time-course microscopy videos of the cells before and after treatment with CCCP, TDA, and DMSO control (25). We found that both in the presence of TDA and the known PMF disruptor CCCP, cells rapidly (>10 min) lose the ability to rotate their flagella (Movies S5–S7). We quantified this effect by measuring the average total distance that a tethered cell’s center of mass travels over a 5-min interval (Fig. 2C), and found that TDA and CCCP significantly reduced this distance in comparison with the untreated control (P values of 2.9 × 10−13 and 2.2 × 10−16, respectively, from Student’s t test).
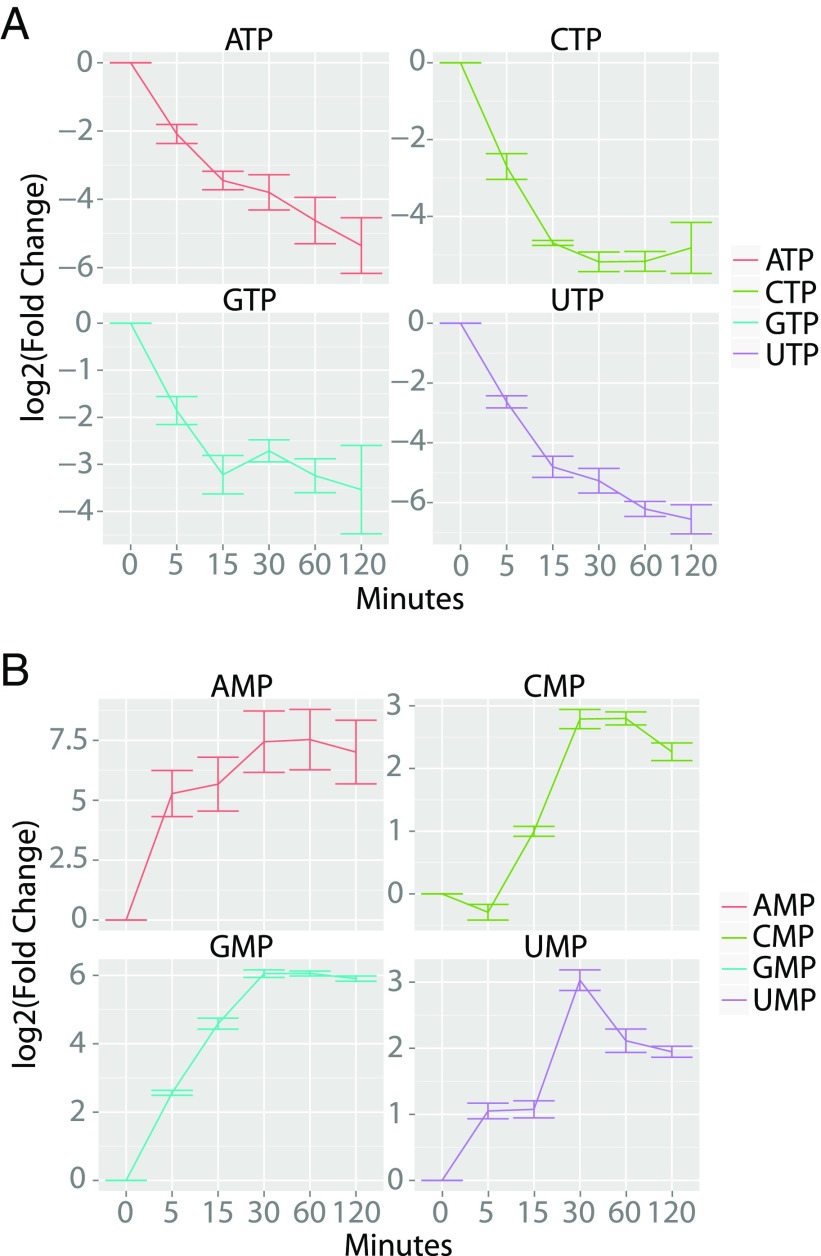
To further support our hypothesis that TDA targets the PMF, we performed a global metabolomics analysis of E. coli before and after drug treatment. The analysis of nucleotide levels in both samples as well as that of all metabolites monitored is shown (Fig. 3 and SI Appendix, Fig. S2). Relative to the untreated control, the nucleoside triphosphate levels fell 15.0 ± 6.7-fold [standard error of the mean (SEM)] upon TDA treatment for 2 h, while nucleoside monophosphate levels increased 122 ± 87-fold (Fig. 3 A and B). This result is characteristic of cells that lose the ability to regenerate high-energy nucleoside triphosphates as they accumulate in their monophosphorylated states. Collectively, the experiments above demonstrate that TDA’s broad-spectrum activity can be explained by its ability to collapse the PMF in prokaryotes and eukaryotes.
Fig. 3.
Metabolomics analysis of the effect of TDA on intracellular nucleotide pools. Quantification of nucleoside triphosphates (A) and nucleoside monophosphates (B) as a function of time after TDA treatment (100 μM). Fold change quantification is shown relative to the DMSO-treated control samples at t = 0. Four replicates were averaged for both the control and TDA-treated samples. Error bars represent SEM.
TDA Is an Electroneutral Proton Antiporter.
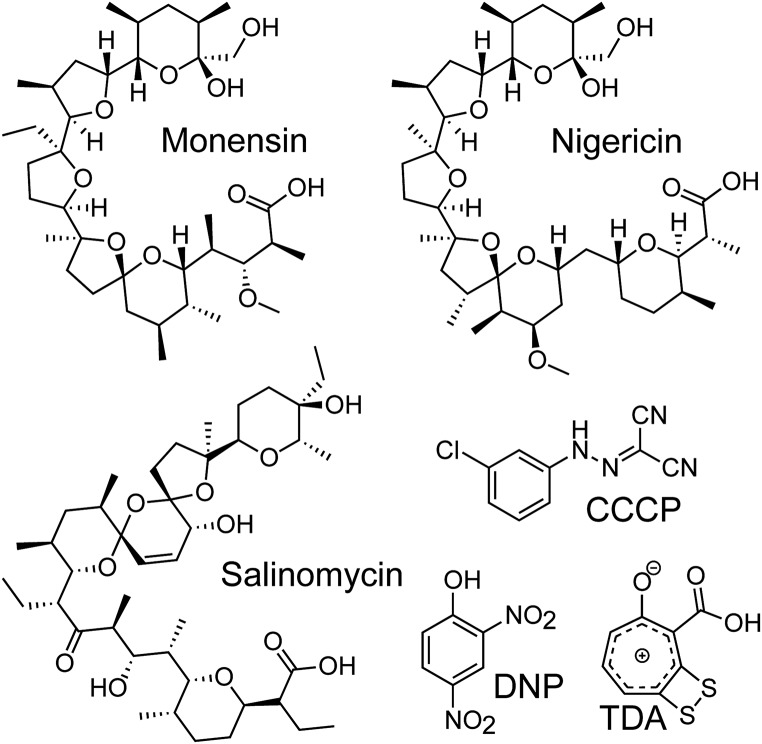
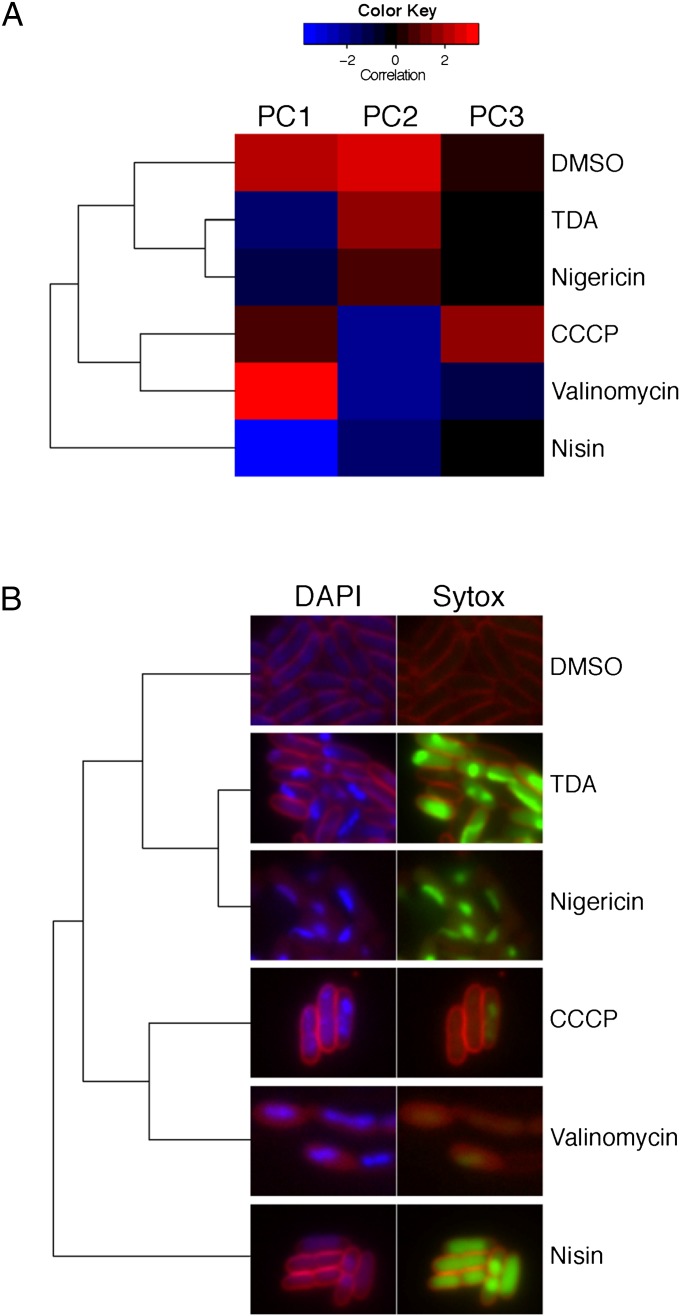
While the above experiments illustrate that TDA targets the PMF, they do not discern between the membrane potential (ΔΨ) and the transmembrane proton gradient (ΔpH) that together compose the PMF. Other PMF-collapsing drugs are known to work by destroying ΔΨ or ΔpH or both. Some examples are shown in Fig. 4. The polyether antibiotics nigericin, monensin, and salinomycin, for example, are electroneutral proton antiporters that largely affect the ΔpH across the cell membrane (26, 27). These molecules collapse the PMF by importing protons but do not significantly affect the ΔΨ, as they subsequently export a positively charged metal, such as K+ or Na+. On the other hand, small aromatic molecules such as CCCP or 2,4-dinitrophenol (DNP) only import a proton and thus modify both the ΔΨ and ΔpH (28). Pore formers, such as the lantipeptide nisin, allow ions to pass through the membrane and thus have a similar effect on the PMF as CCCP or DNP (29). To examine the effect of TDA on the PMF, we performed a modified version of an imaging-based method that was recently pioneered by Pogliano and coworkers (23). This method, known as bacterial cytological profiling (BCP), allows identification of the cellular pathway(s) targeted by a small-molecule antibiotic using 13 features derived from quantitatively imaging E. coli cells that have been treated with three distinct fluorescent dyes. Specifically, upon treatment with an antibiotic, the cells are exposed to FM4-64, which discretely stains the cell membrane; Sytox, which reports on the permeability of the cell membrane; and DAPI, a DNA stain. These dyes collectively yield 13 parameters upon imaging, and assessment of these parameters by multivariate analysis has shown that antibiotics cluster into specific cytological profiles based on their mechanisms of action. Using this method, we determined the cytological profile of TDA-treated cells and found that it most closely resembles that of nigericin (Fig. 5A). Both TDA and nigericin-treated cells exhibit compact nuclei and high Sytox staining (Fig. 5B). It is well documented that nigericin acts by exchanging protons for K+ ions, thereby depleting the ΔpH while maintaining the ΔΨ (30). These data suggest that TDA operates like nigericin to disrupt the PMF by facilitating an electroneutral exchange of protons for positively charged metals across the cell membrane.
Fig. 4.
Divergent structures, similar modes of action. Shown are structures of polyether antibiotics, monensin, nigericin, and salinomycin, the protonophores CCCP and DNP, and the troponoid TDA. TDA is functionally similar to the polyether antibiotics despite structural dissimilarities compared to this compound family.
Fig. 5.
Clustered bacterial cytological profiles. (A) Averaged cell population characteristics after treatment with the indicated drug, clustered by the first three principal components. (B) Typical cell cytology of E. coli 2 h post-drug treatment. DAPI and FM4-64 are shown side-by-side with Sytox and FM4-64. Exposure times are the same across all treatments. Dendogram from A has been added for guided visual comparison.
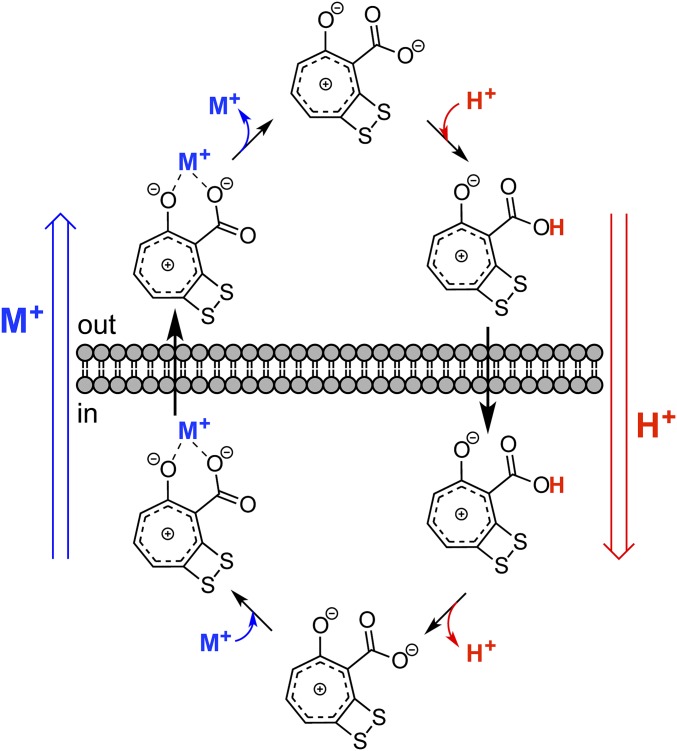
A model for how TDA can electroneutrally exchange H+s is shown (Fig. 6). The carboxyl group of TDA is protonated in the low-pH/high [H+] milieu of the outer membrane. The resulting neutral molecule diffuses across the cell membrane and releases its H+ in the pH-neutral environment of the cytosol. In this form, TDA can chelate a metal and the TDA–metal chelate diffuses out of the cell, thus facilitating a one-to-one exchange of H+ for M+ (a singly charged metal such as K+). The structural features of TDA are perfectly suited for this latter step. The basicity of tropone in general, and TDA specifically, derives from the aromatic tropylium ion character in these molecules. This property has recently been verified for TDA in computational studies (18). The juxtaposition of a carboxyl group alpha to a basic tropylium oxide allows efficient chelation of a metal resulting in a charge-neutral metal complex. The dithietene, which is presumably biosynthesized as a dithiol and oxidizes to a disulfide in the extracellular milieu, increases the electron density of the tropylium oxide and thus the propensity for metal chelation. This model provides a chemical underpinning for the biological activity of TDA.
Fig. 6.
Proposed mode of action of TDA. In E. coli, TDA acts as an electroneutral proton antiporter. At the elevated [H+] just outside the cell membrane, the TDA carboxyl group picks up a H+, and the neutral molecule diffuses into the cell. In the pH-neutral environment of the cytosol, TDA releases the H+. TDA’s basicity, resulting from the tropylium oxide and α-carboxyl group, allows chelation of a monovalent cation. This complex diffuses out of the cell, in aggregate resulting in an exchange of a H+ for a monovalent cation, like K+.
TDA Harbors Potent Anticancer Activities.
That TDA is structurally similar to CCCP and DNP, yet functions more akin to the structurally divergent polyether antibiotics, is surprising. The polyether compounds salinomycin, nigericin, and monensin were recently found to display anticancer activities (31). In a screen targeting epithelial cancer stem cells, salinomycin was found to be 100-fold more potent than taxol, a clinically used anticancer drug (32). Nigericin was also found to have antiproliferative properties in the same screen. Likewise, a half-maximal inhibitory concentration (IC50) of 0.5 μM has been determined for monensin against lymphoma, myeloma, and colon cancer cells (33, 34). We hypothesized that the functional similarities of TDA with these polyether drugs might extend to antiproliferative properties. Assays against a number of cancer cell lines showed this was true. Specifically, the NCI-60 screen revealed broad-spectrum lethal and growth-inhibitory activities of TDA (SI Appendix, Figs. S3 and S4). At a concentration of 10 μM, the strongest effect was observed against U251 CNS, SNB-19 CNS, and A498 renal cancer cell, with 91%, 76%, and 76% lethality, respectively. Broad-spectrum growth-inhibitory activities with >97% growth inhibition were also observed against LOX IMVI melanomas, HCT 116 colon cancer cells, and non-small cell lung cancers. Dose-dependent studies further validated these activities, exhibiting growth-inhibitory effects against many strains in the collection and lethality against some. Most notably, TDA exhibited LC50 values, the concentration at which 50% of the cells were killed, of 5.7 μM, 7.5 μM, and 6.3 μM against U251 CNS, SNB19 CNS, and A498 renal cancer, respectively. These studies also showed growth-inhibitory effects with IC50s in the range of 1–3 μM. In contrast, noncancerous MCF10A epithelial cells exhibited an IC50 of 19.5 μM (SI Appendix, Fig. S5). These data are summarized in SI Appendix, Figs. S3–S5 and demonstrate that, like the polyether H+/K+ antiporters, TDA also harbors potent anticancer activities. They further provide an example for the utility of BCP as a powerful drug-repurposing tool.
The tdaR Gene Region Is Necessary for TDA Resistance in P. inhibens.
Given its potent and broad-spectrum activity, how do cells that produce TDA avoid killing themselves? We initially noticed that P. inhibens was less sensitive to TDA when cells were grown to high cell densities than cultures prepared at low cell densities. Furthermore, having observed that peak TDA production occurs when cells are at high cell densities (SI Appendix, Fig. S6) and that the TDA biosynthetic genes are regulated by the LysR-type transcriptional regulator, tdaA (35, 36), we hypothesized that any putative TDA-resistance factors were likely to be coregulated by tdaA. To test this hypothesis we compared the sensitivity of two P. inhibens mutants, which carry transposon insertions in the tdaA gene locus. One of these transposon insertions is in the promoter region of tdaA (ΔtdaApr) and the other at the 3′ end of the tdaA ORF (ΔtdaAc). We found these insertions to dramatically increase the corresponding strain’s sensitivity to TDA (SI Appendix, Fig. S7), consistent with the idea that the resistance factor is up-regulated by TdaA.
After scrutinizing the tdaABCDE operon, we noticed three predicted ORFs of unassigned function adjoined to the end of the operon, 40 nucleotides after the stop codon of tdaE. We carried out bioinformatic analyses on these three genes, which we call tdaR1, tdaR2, and tdaR3. TdaR1 and R2 appear to encode transmembrane proteins. No significant homology by primary or predicted secondary sequence motifs can be found for TdaR1. TdaR2 shows high similarities to M23 Zn-dependent endopeptidases specific for Gly-Gly targets (37). TdaR3 shows similarities to ChaC, which harbors γ-glutamyl-cyclotransferase activity (see below) (38). In E. coli, the cha operon is involved in cation-proton exchange. The latter was especially striking, given TDA’s mode of action determined above. Because of their homology-based predicted functions and their likely coregulation with the rest of the TDA biosynthetic operon, we suspected that the tdaR1-R3 locus gives rise to TDA resistance in P. inhibens.
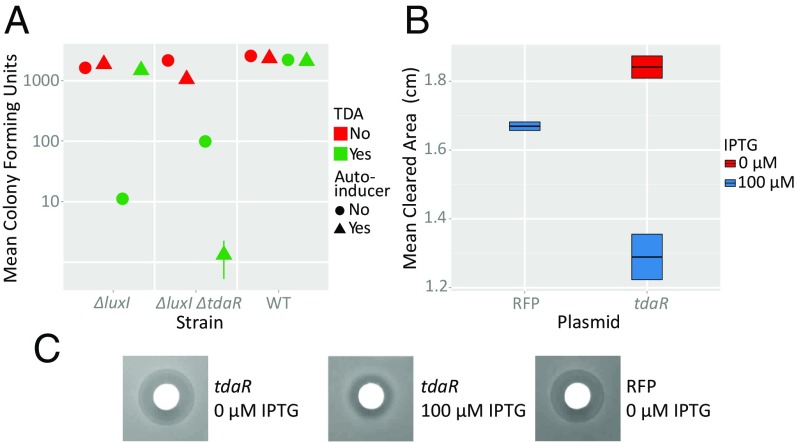
To test this hypothesis, we first examined a P. inhibens pgaI-null strain (ΔluxI), which is incapable of producing the autoinducer, N-3-hydroxydecanoylhomoserine lactone (3-OH-C10-HSL), required for induction of tdaA and thereby the TDA gene cluster. Then by deleting the tdaR1-R3 region in the ΔluxI background we were able to compare the effects of tdaR1-R3 gene expression on the strain’s sensitivity to TDA. We found that TDA sensitivity in the ΔluxI background depended on the presence of 3-OH-C10-HSL, whereas the ΔluxI ΔtdaR1-R3 strain was sensitive to TDA regardless of the presence of this autoinducer (Fig. 7A), indicating that the tdaR1-R3 gene locus is indeed necessary for TDA resistance. Furthermore, we noticed that the ΔluxI ΔtdaR1-R3 strain is more sensitive to TDA in the presence of autoinducer than in its absence. This likely reflects that strain’s ability to make additional TDA without simultaneously producing resistance.
Fig. 7.
Resistance conferred by the tdaR locus. (A) Comparison of mean colony-forming units of TDA-treated P. inhibens wild-type, ΔluxI, or ΔluxI ΔtdaR strains, with or without addition of saturating levels of N-acyl homoserine lactone autoinducer (3-OH-C10-HSL). Where indicated, TDA and 3-OH-C10-HSL were supplemented at final concentrations of 100 μM and 10 μM, respectively. Error bars, where not visible, are occluded by the symbols. (B) Quantification of the zones of clearing induced by TDA in E. coli with uninduced, IPTG-induced tdaR, or IPTG-induced RFP for control. The data in A and B represent the average of five and eight replicates, respectively. The error bars represent SEM. (C) Typical zones of clearing for each condition measured in B.
The tdaR Locus Is Sufficient for Conferring TDA Resistance to E. coli.
Having demonstrated that the tdaR gene region is necessary for conferring TDA resistance to P. inhibens, we sought to understand if this genetic locus alone contained the element(s) of resistance or if there were other essential components. We cloned tdaR1-tdaR3 into an IPTG-inducible plasmid and transformed it into wild-type E. coli NCM3722. First, we subjected this tdaR-gene-transplant strain to a classical antibiotic disk plate assay, where the zone of clearing was measured to reflect sensitivity to TDA. In comparison with our controls, E. coli bearing a red fluorescent protein (RFP) expressing vector or the E. coli tdaR1-R3 in the absence of IPTG, we observed significantly smaller zones of inhibition (Fig. 7 B and C; P values of 8.5 × 10−4 and 4.8 × 10−5, respectively). In addition, we performed a standard growth-curve assay with the abovementioned three conditions and found that only when the tdaR locus was induced did it confer TDA resistance (SI Appendix, Fig. S8). In particular, we found that cells expressing the tdaR genes were able to recover from TDA treatment earlier and reach greater population densities than either of the controls. Thus, the tdaR genes are sufficient for conferring TDA resistance.
A Working Model for TDA Resistance.
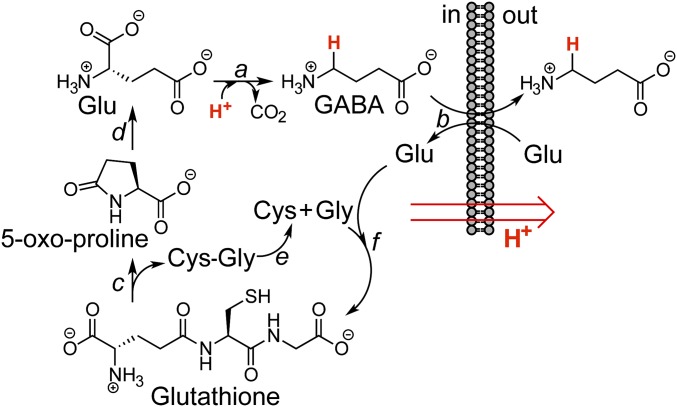
As mentioned above, TdaR3 likely encodes a γ-glutamyl-cyclotransferase, which catalyzes the aminolysis of glutathione to 5-oxo-proline, and a Gly-Cys dipeptide. 5-Oxo-proline is typically further hydrolyzed to glutamate via a 5-oxoprolinase. The predicted functions for these enzymes and their requirement in TDA resistance suggest a physiological link between the mode of action of TDA, glutathione, and glutamate. Bacterial cells deal with acid stress, imposed by acidic conditions or H+ importers like TDA, using three acid-response systems (39), the best-studied of which catalyzes decarboxylation of glutamate to γ-aminobutyric acid (GABA), which is then exchanged for extracellular glutamate, resulting in the export of 1 H+ per reaction cycle. We propose the working model in Fig. 8 for TDA resistance in P. inhibens. When TDA production is activated, via 3-OH-C10-HSL and tdaA, the tdaR locus is induced. TdaR3 facilitates the glutamate-dependent acid-response system by converting glutathione to glutamate. This glutamate is decarboxylated and the resulting GABA product exchanged for additional Glu. Consequently, P. inhibens can resist TDA by counteracting the TDA-induced proton influx with proton efflux mediated by the γ-glutamyl cycle. Our proposal thus links the γ-glutamyl cycle with acid response and the attendant mechanism of resistance to TDA (Fig. 8). Additional biochemical studies are necessary to assess this model.
Fig. 8.
Model for self-resistance against TDA by P. inhibens. Acid stress is alleviated by the Glu-dependent acid response system (a and b). Proton export is facilitated by Glu decarboxylase (a), which generates GABA, and the subsequent antiport of GABA for Glu (b). In aggregate, this process results in removal of 1 equiv. of H+ per Glu. In addition, tdaR3 encodes a putative γ-glutamyl-cyclotransferase, which may cleave glutathione to give 5-oxo-proline and a Cys-Gly dipeptide (c). 5-Oxoprolinase hydrolyzes 5-oxo-proline to replenish Glu, which can be used to export additional protons (d). The Cys-Gly dipeptide may be hydrolyzed to monomeric amino acids, perhaps by TdaR2 (e). These three amino acids can be used to generate additional glutathione; this process requires ATP (f).
The studies above suggest that TdaR3 should confer resistance to any proton importer, not just TDA. We examined this possibility by performing experiments in which E. coli cells carrying an inducible, tagged tdaR3 were treated with sublethal concentrations of CCCP (4 μg/mL). Compared with the empty-vector control, we observed a 12.4 (±1.9 SEM)-min decrease in exponential doubling time of the strain with induced tdaR3 (SI Appendix, Fig. S9). The presence of induced tdaR3 also led to a faster recovery from CCCP treatment relative to control cultures. These results are fully consistent with our model and the role proposed for TdaR3 in TDA resistance.
Ecological Implications.
The tight association of marine bacteria and algae into microaggregates creates a miniature ecosystem sustained simply by sunlight and the relatively low nutrient concentrations of the pelagic region of the ocean. The circumstances of cooperativity between E. huxleyi and P. inhibens are mediated by a chemical correspondence between the two species. When P. inhibens senses deterioration in the health of its cohabitant algae, it preys on the senescing creatures by releasing potent algaecides, the roseobacticides. However, when both species are commensal E. huxleyi trades the climatically active nutrient DMSP to P. inhibens, which routes it into the marine food web and, in return, provides the algae with algal growth promoters as well as protection from other marine microbes via the production of TDA (5, 40). By confirming TDA’s broad-spectrum activity, we suggest that both in the wild and in aquaculture, TDA will confer protection to a tolerant symbiont from a wide range of pathogenic microbes.
Aside from its broad-spectrum activity, TDA’s mechanism of action is remarkably suited for thwarting opportunistic bacteria. It has been shown that a wide range of marine bacteria chemotax toward algal DMSP (41). By targeting the PMF using an electroneutral mechanism, TDA destroys both the proton gradient used by many bacteria to power flagellar-mediated locomotion (Fig. 2C), as well as the cation gradient used by some marine Vibrio pathogens (42). Thus, an established microaggregate producing TDA at steady state would be surrounded by a protective gradient, which would stop both potential invaders and pathogens. Furthermore, we speculate that the coproduction of a chemoattractant and an antibiotic could be a trap wherein invading microbes, or prey, are lured to the symbiotic microaggregate by DMSP but then blocked from chemotaxing and killed by TDA. This could be a mechanism to cull important nutrients, like Fe, in the nutrient-poor regions of the ocean, where this algal–bacterial symbiosis occurs.
Finally, as has been reported previously, the likelihood of other organisms gaining resistance to TDA by endogenous mutations like simple point mutations or deletions is low (24). However, here we have demonstrated that the tdaR genetic locus will indeed confer mild resistance to even as distant a bacterial relative as E. coli. This suggests a possible route for an invading pathogen to gain resistance to TDA and unlock the fruits of the algal–bacterial symbiosis through the exogenous route of horizontal gene transfer. Further, this resistance mechanism is likely general, as tdaR3 confers resistance to CCCP. Thus, as long as the tda cluster is expressed in P. inhibens, the bacteria will be resistant to PMF-disrupting biocides and exhibit acid tolerance.
Conclusions
We report that despite its drastic structural disparity from the polyether antibiotics, TDA has a mode of action very similar to salinomycin, nigericin, and monensin. In E. coli, TDA disrupts the PMF by a H+/M+ antiport mechanism. The structural features of TDA are well suited for this function. The H+ is carried on a carboxylic acid, as it is in the polyether antibiotics. Whereas the latter export metal ions via a polyoxo–metal complex, TDA’s basicity resulting from its tropylium ion character, reinforced by the dithiol substituent, allows efficient metal chelation and an electroneutral proton antiport mechanism. Thus, TDA is small and aromatic, like DNP and CCCP, but its MoA in E. coli mirrors those of the large polyether antibiotics. We show that this functional similarity extends beyond MoA and report that, like the polyether antibiotics, TDA also harbors lethal and growth-inhibitory activities against a number of cancer cell lines. Future studies should assess whether the anticancer activity of TDA can be useful. They may further delineate the role of glutathione and its link to the γ-glutamyl cycle as a “bacterial antacid,” which together help confer resistance to TDA.
Materials and Methods
Detailed descriptions of materials and methods, including isolation of TDA from P. inhibens, D. discoideum experiments, E. coli flagellar rotation assays, metabolomics measurements, bacterial cytological profiling, and other procedures used are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Joshua D. Rabinowitz for use of his HPLC-MS instrument for primary metabolite quantitation, Prof. Bonnie L. Bassler for the kind gift of 3-OH-C10-HSL, and Leah B. Bushin for helpful discussions. We also gratefully acknowledge grants from the National Institutes of Health (GM098299 to M.R.S. and 1DP2OD004389 to Z.G.) for support of this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518034113/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Microbial antitumor drugs: Natural products of microbial origin as anticancer agents. Curr Opin Investig Drugs. 2009;10(12):1280–1296. [PubMed] [Google Scholar]
- 3.Cragg GM, Newman DJ. Natural products: A continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432(7019):829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 5.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3(4):331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 7.Buchan A, González JM, Moran MA. Overview of the marine roseobacter lineage. Appl Environ Microbiol. 2005;71(10):5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkhoff T, Giebel HA, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: A short overview. Arch Microbiol. 2008;189(6):531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 9.Rao D, Webb JS, Kjelleberg S. Microbial colonization and competition on the marine alga Ulva australis. Appl Environ Microbiol. 2006;72(8):5547–5555. doi: 10.1128/AEM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porsby CH, Nielsen KF, Gram L. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl Environ Microbiol. 2008;74(23):7356–7364. doi: 10.1128/AEM.01738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng H, Belas R. Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr Opin Biotechnol. 2010;21(3):332–338. doi: 10.1016/j.copbio.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Thole S, et al. Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 2012;6(12):2229–2244. doi: 10.1038/ismej.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seyedsayamdost MR, Carr G, Kolter R, Clardy J. Roseobacticides: Small molecule modulators of an algal-bacterial symbiosis. J Am Chem Soc. 2011;133(45):18343–18349. doi: 10.1021/ja207172s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65(9):3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kintaka K, Ono H, Tsubotani S, Harada S, Okazaki H. Thiotropocin, a new sulfur-containing 7-membered-ring antibiotic produced by a Pseudomonas sp. J Antibiot (Tokyo) 1984;37(11):1294–1300. doi: 10.7164/antibiotics.37.1294. [DOI] [PubMed] [Google Scholar]
- 16.Bruhn JB, et al. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl Environ Microbiol. 2005;71(11):7263–7270. doi: 10.1128/AEM.71.11.7263-7270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng H, Bruhn JB, Nielsen KF, Gram L, Belas R. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol. 2008;74(5):1535–1545. doi: 10.1128/AEM.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greer EM, Aebisher D, Greer A, Bentley R. Computational studies of the tropone natural products, thiotropocin, tropodithietic acid, and troposulfenin. Significance of thiocarbonyl-enol tautomerism. J Org Chem. 2008;73(1):280–283. doi: 10.1021/jo7018416. [DOI] [PubMed] [Google Scholar]
- 19.D’Alvise PW, Melchiorsen J, Porsby CH, Nielsen KF, Gram L. Inactivation of Vibrio anguillarum by attached and planktonic Roseobacter cells. Appl Environ Microbiol. 2010;76(7):2366–2370. doi: 10.1128/AEM.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas M, et al. Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Lostonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture. 2006;255(1-4):323–333. [Google Scholar]
- 21.Prado S, Montes J, Romalde JL, Barja JL. Inhibitory activity of Phaeobacter strains against aquaculture pathogenic bacteria. Int Microbiol. 2009;12(2):107–114. [PubMed] [Google Scholar]
- 22.Bentley R. A fresh look at natural tropolonoids. Nat Prod Rep. 2008;25(1):118–138. doi: 10.1039/b711474e. [DOI] [PubMed] [Google Scholar]
- 23.Nonejuie P, Burkart M, Pogliano K, Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci USA. 2013;110(40):16169–16174. doi: 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porsby CH, Webber MA, Nielsen KF, Piddock LJ, Gram L. Resistance and tolerance to tropodithietic acid, an antimicrobial in aquaculture, is hard to select. Antimicrob Agents Chemother. 2011;55(4):1332–1337. doi: 10.1128/AAC.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabel CV, Berg HC. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc Natl Acad Sci USA. 2003;100(15):8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutton CJ, Banks BJ, Cooper CB. Polyether ionophores. Nat Prod Rep. 1995;12(2):165–181. doi: 10.1039/np9951200165. [DOI] [PubMed] [Google Scholar]
- 27.Mollenhauer HH, Morré DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim Biophys Acta. 1990;1031(2):225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terada H. The interaction of highly active uncouplers with mitochondria. Biochim Biophys Acta. 1981;639(3-4):225–242. doi: 10.1016/0304-4173(81)90011-2. [DOI] [PubMed] [Google Scholar]
- 29.Moll GN, Konings WN, Driessen AJ. Bacteriocins: Mechanism of membrane insertion and pore formation. Antonie van Leeuwenhoek. 1999;76(1-4):185–198. [PubMed] [Google Scholar]
- 30.Harold FM. Antimicrobial agents and membrane function. Adv Microb Physiol. 1969;4(1):45–104. [Google Scholar]
- 31.Huczyński A. Polyether ionophores-promising bioactive molecules for cancer therapy. Bioorg Med Chem Lett. 2012;22(23):7002–7010. doi: 10.1016/j.bmcl.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 32.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park WH, Kim ES, Kim BK, Lee YY. Monensin-mediated growth inhibition in NCI-H929 myeloma cells via cell cycle arrest and apoptosis. Int J Oncol. 2003;23(1):197–204. [PubMed] [Google Scholar]
- 34.Park WH, Kim ES, Jung CW, Kim BK, Lee YY. Monensin-mediated growth inhibition of SNU-C1 colon cancer cells via cell cycle arrest and apoptosis. Int J Oncol. 2003;22(2):377–382. [PubMed] [Google Scholar]
- 35.Geng H, Belas R. TdaA regulates Tropodithietic acid synthesis by binding to the tdaC promoter region. J Bacteriol. 2011;193(15):4002–4005. doi: 10.1128/JB.00323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger M, Neumann A, Schulz S, Simon M, Brinkhoff T. Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J Bacteriol. 2011;193(23):6576–6585. doi: 10.1128/JB.05818-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar A, et al. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13(12):1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 40.Moran MA, Reisch CR, Kiene RP, Whitman WB. Genomic insights into bacterial DMSP transformations. Annu Rev Mar Sci. 2012;4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 41.Seymour JR, Simó R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science. 2010;329(5989):342–345. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 42.Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J Bacteriol. 1999;181(6):1927–1930. doi: 10.1128/jb.181.6.1927-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.