Significance
The physiology and ecology of complex microbial communities are strongly dependent on the immediate surroundings of each microbe, including the identity of neighboring microbes; however, information on the micron-scale organization of microbiomes is largely lacking. Using sequencing data combined with spectral fluorescence imaging, we have discovered a multigenus, highly organized microbial consortium in human dental plaque. The spatial structure of the consortium reveals unanticipated interactions and provides a framework for understanding the organization, metabolism, and systems biology of the microbiome and ultimately, its effect on the health of the human host. Our synthesis of high-throughput sequencing data with spatial and structural information shows the informative value of microbial biogeography at the micron scale.
Keywords: biofilm, imaging, microscopy, microbial ecology
Abstract
The spatial organization of complex natural microbiomes is critical to understanding the interactions of the individual taxa that comprise a community. Although the revolution in DNA sequencing has provided an abundance of genomic-level information, the biogeography of microbiomes is almost entirely uncharted at the micron scale. Using spectral imaging fluorescence in situ hybridization as guided by metagenomic sequence analysis, we have discovered a distinctive, multigenus consortium in the microbiome of supragingival dental plaque. The consortium consists of a radially arranged, nine-taxon structure organized around cells of filamentous corynebacteria. The consortium ranges in size from a few tens to a few hundreds of microns in radius and is spatially differentiated. Within the structure, individual taxa are localized at the micron scale in ways suggestive of their functional niche in the consortium. For example, anaerobic taxa tend to be in the interior, whereas facultative or obligate aerobes tend to be at the periphery of the consortium. Consumers and producers of certain metabolites, such as lactate, tend to be near each other. Based on our observations and the literature, we propose a model for plaque microbiome development and maintenance consistent with known metabolic, adherence, and environmental considerations. The consortium illustrates how complex structural organization can emerge from the micron-scale interactions of its constituent organisms. The understanding that plaque community organization is an emergent phenomenon offers a perspective that is general in nature and applicable to other microbiomes.
Biogeography—the study of the distribution of organisms across the globe—seeks to recognize patterns in the spatial distribution of organisms and discover the forces that underlie those patterns. Bacteria are micron-sized, and many of the forces and factors that underlie their distributional patterns operate at micron scales and are qualitatively different from the large-scale factors, such as climate, that drive traditional biogeography. To frame the analysis of microbial distribution patterns at the scale that microbes themselves experience, we introduce the concept of micron-scale biogeography: the study of the distribution of microbes relative to micron-scale features of their environment. These features include the host or inanimate surfaces on which the microbes reside as well as local gradients of nutrients and oxygen. Key components of the micron-scale environment, particularly in biofilms and other densely populated habitats, are the microbes themselves, serving as substrates for attachment of other microbes, creating spatial structure, and acting as point sources for diffusible metabolites.
Micron-scale biogeography is critical to understanding the physiology and ecology of the community as well as its systems biology and its effects on human health and disease. Close proximity or physical contact between two microbes can substantially alter their physiology, for example conferring on an anaerobe the ability to survive in an aerobic environment (1) or dramatically altering the range of metabolites produced compared with those produced by the same organism in isolation (2–5). Thus, a mechanistic understanding of the physiology of key players depends on knowing the identity of the neighbors with which they commonly interact. When the microbiota is host-associated, its physiology and ecology become intimately connected with those of the host at both micron scales and host scale and are capable of critically influencing the promotion of health or the progression toward disease. Thus, it is necessary to know not only who is next to who but also, who is next to what.
Dental plaque is a human microbiome community with study that dates back to the initial observations of Leeuwenhoek over 300 years ago (6). Modern studies have analyzed taxon–taxon associations through pairwise binding interactions between members of different oral microbial species. These interactions, termed “coadhesion” or “coaggregation,” have been described in an extensive body of literature (7, 8) and form the basis for an influential model describing the structure and development of dental plaque as an ecological succession (9). This model begins with the salivary pellicle coating the teeth and the initial attachment of Streptococcus spp. and Actinomyces spp. to the pellicle. These attached microbes then serve as a substrate for the binding of a variety of other colonizers, including Fusobacterium nucleatum, which functions as a bridge between the early colonizers and the late-colonizing pathogens by virtue of its capacity to bind physically to both sets of microbes. This model synthesizes in vitro and in vivo observations to make testable predictions about the spatial structure of mature dental plaque, but a direct test of the model by high-resolution imaging has not previously been undertaken.
The study of microbial communities has been revolutionized by metagenomic and metatranscriptomic approaches, which have revealed enormous complexity (10). However, the groundbreaking methods that revealed the complexity have the drawback that the sample must be homogenized for nucleic acid extraction, thereby destroying any spatial structure at the micron scale that might have existed. The absence of detailed spatial information represents a fundamental gap in knowledge that precludes a full understanding of the assembly and interactions of complex microbial communities.
The integration of spatial information with high-throughput sequencing data by direct visualization of spatial structure opens an entirely different window into understanding community structure. Fluorescence in situ hybridization (FISH) targeting rRNA (11, 12) can be used to identify nearly any microbe, but because of technical limitations, it is generally used to differentiate only two or three microbial types simultaneously. The resulting images reveal distinctive distributions of individual organisms (13–15) but not the overall structure of the community. However, fluorescence spectral imaging allows the differentiation of many fluorophores and creates an opportunity to take a systems-level view of the spatial structure of the microbiota (16), simultaneously imaging and identifying all members of a complex microbial consortium. Here, we analyze sequencing data from the Human Microbiome Project (HMP) to identify the major bacterial taxa likely to be important in the structure and function of supragingival plaque, and by imaging the spatial organization of these most abundant taxa, we describe a complex, spatially organized, multigenus consortium. This synthesis of high-throughput sequencing data with spatial and structural information may serve as a case study in microbial biogeography at the micron scale.
Results
Identification of Bacterial Taxa Important in Supragingival Plaque.
The Human Oral Microbiome Database (HOMD) (17) contains 707 entries at the species level. This enormous diversity poses an enormous challenge for efforts to sort out the spatial and structural relationships of the taxa. In an attempt to reduce the complexity to manageable proportions, we sought guidance from the 16S rRNA gene sequencing data generated by the HMP (18). We previously applied an information theory approach to analysis of the oral microbiome at the single-nucleotide level, resulting in high-resolution sequence groups termed oligotypes (19). The oligotypes were assigned to HOMD species and analyzed for each of nine oral habitats defined by the HMP. This analysis showed that most species of oral bacteria are habitat specialists and that the complexity can be reduced simply by considering only the bacteria resident in the oral habitat of interest. In the following discussion, we consider plaque to mean specifically the biofilm that forms on teeth as opposed to other oral substrates, such as gums or tongue, and we focus on the microbiota resident in plaque above the gum line, supragingival plaque.
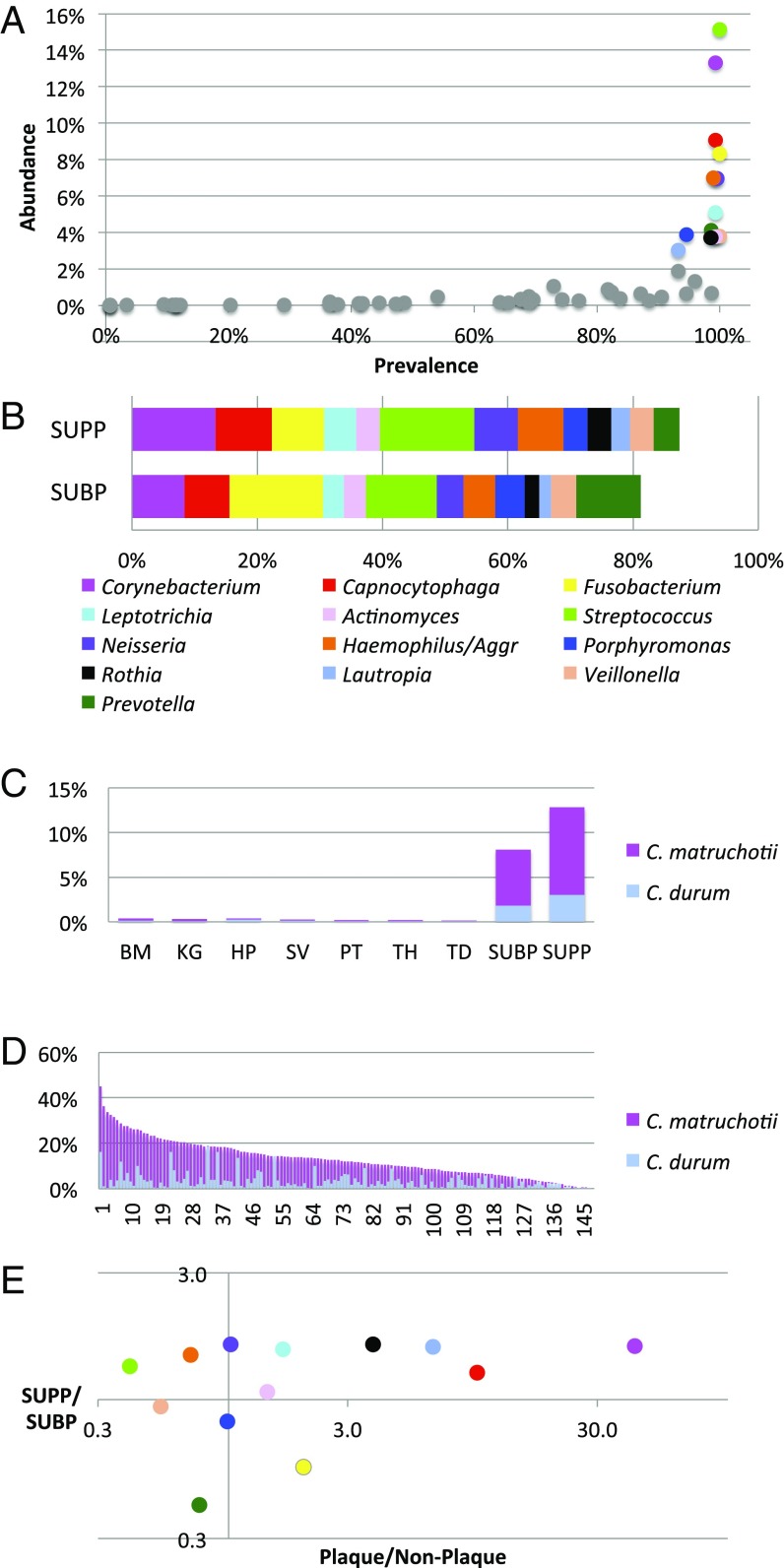
As an initial basis for identifying key taxa in supragingival plaque, we assessed the abundance and prevalence of the oligotypes grouped by genus. This analysis readily identified a group of bacterial genera that were both abundant and prevalent (Fig. 1A). Of 57 genera detected in supragingival plaque (SUPP), most had both low abundance and low prevalence. In contrast, 13 genera had at least 3% mean abundance and were also highly prevalent, each being detected in more than 90% of SUPP samples. Collectively, these 13 genera accounted for 85% of the sequencing data from SUPP. The same 13 genera also accounted for more than 80% of the subgingival plaque (SUBP) data (Fig. 1B), indicating a close relationship between the two plaque sites. Because of their abundance and prevalence, these taxa are likely to form both the spatial and the metabolic framework of the healthy plaque microbiome.
Fig. 1.
Metagenomic sequence analysis points to Corynebacterium as a key taxon in supragingival plaque. (A) Prevalence abundance plot for supragingival plaque. (B) Cumulative abundance of genera in both supra- and subgingival plaque. Genera with greater than 3% abundance in SUPP, mean across 148 subjects, are indicated by colored dots in A and bar segments in B; B also shows the abundance of these genera in SUBP. Data are from the HMP (18) V3–V5 region of 16S rRNA, analyzed by oligotyping (19), and grouped by genus. (C) Corynebacterium is far more abundant in plaque than in other oral sites. Mean abundances of C. matruchotii and C. durum are shown for each oral site analyzed by oligotyping (19). BM, buccal mucosa; HP, hard palate; KG, keratinized gingiva; PT, palatine tonsils; SV, saliva; TD, tongue dorsum; TH, throat. (D) Corynebacterium is a major component of most plaque samples. Relative abundance of Corynebacterium in the HMP SUPP samples from 148 individuals (19). C. matruchotii is usually more abundant, but C. durum dominates some samples. (E) Habitat analysis identifies genera that are strongly characteristic of SUPP. The plaque to nonplaque ratio measures the relative abundance of each genus in two plaque sites compared with seven nonplaque sites sampled by the HMP [calculated as (mean SUBP + mean SUPP)/(mean BM + mean KG + mean HP + mean SV + mean PT + mean TH + mean TD)]. This ratio identifies Corynebacterium and Capnocytophaga as the taxa most preferentially abundant in plaque. The SUPP to SUBP ratio identifies these genera as relatively more abundant in SUPP than in SUBP. Colors in E are the same as those in A and B.
Taxa that are present primarily or exclusively in one site may provide clues to the distinctive features of the habitat and the role that those taxa contribute to the site. Habitat analysis of the oral microbiome suggested that one genus, Corynebacterium, in particular was strikingly specific to supragingival and subgingival plaque. This genus was present in only trace amounts in saliva and on six of eight oral surfaces sampled (the tongue, buccal mucosa, keratinized gingivae, hard palate, tonsils, and throat) but made up 8% of the bacterial community in SUBP and more than 12% in SUPP (Fig. 1C) (19, 20). The HOMD recognizes six oral species within the genus Corynebacterium. However, of these six, only two, Corynebacterium matruchotii and Corynebacterium durum, were present at significant levels in plaque. Although C. matruchotii was the dominant species in most individuals, C. durum was dominant in some (Fig. 1D). Taking the two species together, the genus not only had a high mean abundance, but also, it was consistently abundant, with a relative abundance of 3% or more in 90% of the individuals. The abundance and prevalence of Corynebacterium suggest that it plays an important role in the plaque community, whereas its plaque specificity suggests that it occupies a niche that is dependent on properties of the tooth surface and/or the gingival crevice.
Habitat analysis of 12 other abundant plaque genera (Fig. 1E) showed large differences in their degree of specificity to plaque but only modest differences in their relative abundance in SUPP compared with SUBP. Genera with strong plaque specificity, in addition to Corynebacterium, included Capnocytophaga, which was 10-fold more abundant in plaque than at nonplaque sites, as well as Lautropia and Rothia. By contrast, genera such as Streptococcus occupied a broad range of habitats. Despite being the single most abundant genus in SUPP, Streptococcus was substantially more abundant at nonplaque sites than in plaque on average. This wide-ranging habitat preference likely reflects the capacity of Streptococcus to be an efficient colonizer of multiple oral surfaces. Additional genera with broad habitat range in the mouth include Haemophilus and Veillonella. Supragingival plaque is often characterized as being composed primarily of Gram-positive aerobes, whereas Gram-negative anaerobes come to dominate subgingival plaque, particularly in individuals affected by periodontitis (21). However, habitat analysis of genera shows that the similarities between the two plaques are more striking than their differences in the healthy individuals sampled by the HMP. Most of the abundant genera are enriched in SUPP compared with SUBP by a small and relatively constant factor of ∼1.3–1.6. Some genera (Actinomyces, Porphyromonas, and Veillonella) are equally abundant. A few predominantly anaerobic genera, notably Prevotella and Fusobacterium, are more abundant in SUBP but only by a factor of ∼2. Thus, the overall similarity of distribution suggests a close connection between these two spatially adjacent communities.
Plaque Microbiota Is Organized into Highly Structured, Multigenus Consortia.
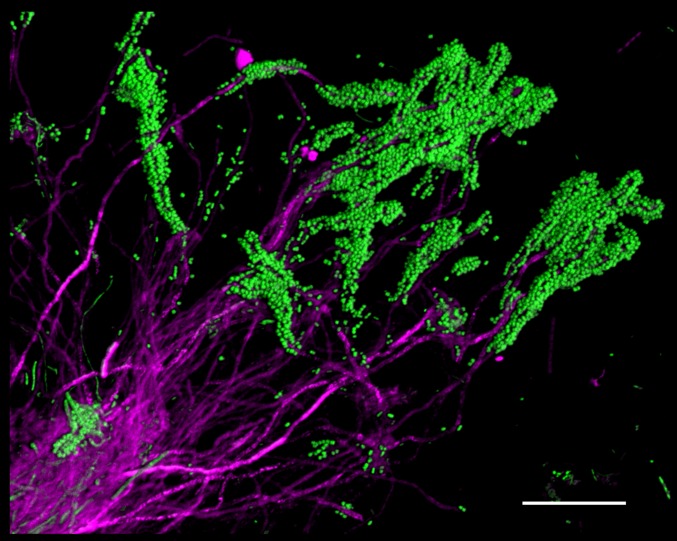
Bacteria are micron-sized and live in a chemical and structural environment with micron-scale heterogeneity. Therefore, an understanding of the micron-scale spatial organization of bacterial communities is necessary for understanding how these communities function. A striking degree of spatial organization can become visible, even with simple procedures, when plaque samples are prepared gently to retain their structure. For example, when plaque was spread out on a slide, conventional FISH with two probes revealed clumps of Corynebacterium, with long filaments that were coated at their tips by brightly staining cocci (Fig. 2). Associations between cocci and filaments in plaque were documented over 40 years ago and have been called “corncobs” (22, 23). However, the arrangement in dissected plaque, which we visualized with FISH, is striking in that the filaments are clearly continuous from the base of the clump to the tips, but the cocci are restricted to the tips or distal ends of the filaments (Fig. 2). This spatial arrangement suggests a role for Corynebacterium as a foundational taxon that structures the environment in a way that creates a microenvironment favorable to the growth of the cocci. Why the cocci are restricted to the distal ends is a key question, the answer to which requires more complete information about the surrounding structure.
Fig. 2.
Corncob structures formed by Corynebacterium and cocci in plaque. Corynebacterium cells (magenta) are visible as long filaments, with cocci (green) bound to the tips of the filaments. Partially disrupted plaque was hybridized with a probe for Corynebacterium and a universal bacterial probe. Image was acquired using a Zeiss AxioImager 63× Plan-Apochromat 1.4 N.A. objective and Apotome structured illumination. (Scale bar: 20 μm.)
We used two complementary methods designed to preserve and visualize the spatial structure of the plaque community: whole-mount preparations and methacrylate embedding. Whole mounts permitted the imaging of entire 3D structures, including long filaments, but at the expense of slight spatial distortion resulting from compression. Embedding and sectioning preserved micron-scale spatial relationships more accurately but at the expense of loss of 3D continuity. Regardless of the preparation method, we detected similar microbial consortia in all samples. For a systems-level analysis of the spatial organization of these samples, we used Combinatorial Labeling and Spectral Imaging FISH (CLASI-FISH) (16) to differentiate up to 15 taxa simultaneously. In our previous proof of concept of CLASI-FISH, we labeled plaque that was partially dispersed to single-cell thickness (16), so that spectral signatures created by binary combinations of fluorophores could be read unambiguously. For this study, we wished to analyze more intact 3D structures, in which multiple cells may lie on top of one another, even in a single optical plane of focus. In such samples, overlapping cells with different binary signals in the same pixel could generate ambiguity in taxon identification. To avoid this ambiguity, we used a simplified labeling strategy, in which a single fluorophore served as the spectral signature to identify each taxon, and as many as 10 distinct fluorophores were used simultaneously.
The combination of sequence analysis with imaging allowed an assessment of spatial organization that was taxonomically both wide-reaching and refined. We used FISH probes with broad coverage, using probes for four phyla, two classes, three families, and 15 genera (Table S1). More specificity was provided by HMP sequencing data, which showed that, for most plaque genera, a small number of species was dominant. Of the 13 most abundant genera, one genus was represented by only a single major plaque species (Lautropia mirabilis), and six were represented primarily by two or three major species or small clusters of species (Table S2). Collectively, the probes that we used targeted 96–98% of the cells in a healthy supragingival plaque microbiome as judged by rRNA tag sequencing data from the HMP (Table S2). Among these probes, 2 family- and 11 genus-level probes covered 88% of the sequencing data and are shown in Figs. 2–8 and Figs. S1–S4. When describing imaging results in the following section, we will use the taxon name as shorthand for cells in the image that are reactive with the taxon-specific probe, but it should be kept in mind that these organisms are likely to be members of the species shown in Table S2. The genera Haemophilus and Aggregatibacter are phylogenetically intertwined in the family Pasteurellaceae and targeted by probe Pas111, which we refer to as Haemophilus/Aggregatibacter. The genera Neisseria, Kingella, and Eikenella are likewise intertwined in the family Neisseriaceae and targeted by probe Nei1030, which we refer to as Neisseriaceae.
Table S1.
Probes used in this study
| Probe name | Target taxon | Probe sequence 5′–3′ | Source | |||||
| Domain | Phylum | Class | Family | Genus | Species | |||
| Eub338 | Bacteria | GCTGCCTCCCGTAGGAGT | 44 | |||||
| Actinobacteria | ||||||||
| Act381 | Actinobacteria | CGTCGCTGCATCAGGCTT | This paper | |||||
| Act692 | Actinobacteria | CTGATATCTGCGCATTCC | This paper | |||||
| Act476 | Actinomyces | ATCCAGCTACCGTCAACC | 45 | |||||
| Cor595 | Corynebacterium | CCGGAATTTCACAGACGACG | This paper | |||||
| Cor633 | Corynebacterium | AGTTATGCCCGTATCGCCTG | This paper | |||||
| Rot491 | Rothia | TAGCCGGCGCTTTCTCTG | 16 | |||||
| Cmat175 | Corynebacterium matruchotii | ACTAAACCATGGTCCTATCCG | This paper | |||||
| Bacteroidetes | ||||||||
| CFB286 | Bacteroidetes | TCCTCTCAGAACCCCTAC | 46 | |||||
| CFB563 | Bacteroidetes | GGACCCTTTAAACCCAAT | 46 | |||||
| Cap371 | Capnocytophaga | TCAGTCTTCCGACCATTG | 15 | |||||
| Prv392 | Prevotella | GCACGCTACTTGGCTGG | 32 | |||||
| Pg1160 | Porphyromonas | CCTCACGCCTTACGACGG | 16 | |||||
| Firmicutes | ||||||||
| LGC354A | Firmicutes | TGGAAGATTCCCTACTGC | 47 | |||||
| LGC354C | Firmicutes | CCGAAGATTCCCTACTGC | 47 | |||||
| Str405 | Streptococcus | TAGCCGTCCCTTTCTGGT | 48 | |||||
| Gem572 | Gemella | TAAACCACCTGCGCGCGCTT | 16 | |||||
| Vei488 | Veillonella | CCGTGGCTTTCTATTCCG | 49 | |||||
| Sel60 | Selenomonas | TCATTCGCTCCGTTCGAC | 16 | |||||
| Proteobacteria | ||||||||
| Bet42A | β-Proteobacteria | GCCTTCCCACTTCGTTT | 50 | |||||
| Lmir444 | Lautropia mirabilis | TGGCACAGTCCTTTTCGTTCC | This paper | |||||
| Nei1030 | Neisseriaceae | CCTGTGTTACGGCTCCCG | 16 | |||||
| Gam42A | γ-Proteobacteria | GCCTTCCCACATCGTTT | 50 | |||||
| Pas111 | Pasteurellaceae | TCCCAAGCATTACTCACC | 16 | |||||
| Cam1021 | Campylobacter | ATTTCTGCAAGCAGACACTC | 16 | |||||
| Fusobacteria | ||||||||
| Fus714 | Fusobacterium | GGCTTCCCCATCGGCATT | 16 | |||||
| Lep568 | Leptotrichia | GCCTAGATGCCCTTTATG | 16 | |||||
| Spirochaetes | ||||||||
| Trp684 | Treponema | TCTACAGATTCCACCCCTAC | 16 | |||||
| TM7 | ||||||||
| TM7550 | TM7 | CCCAGTCACTCCGGATAA | This paper | |||||
Probes are listed by phylum; the probe name, target taxon, and probe sequence are shown.
Table S2.
The supragingival plaque microbiota in health
| Genus | Species V1–V3 | % of total V1–V3 | % of genus V1–V3 | Species V3–V5 | % of total V3–V5 | % of genus V3–V5 |
| Actinomyces | A. naeslundii, A. sp. HOT 171 | 2.9 | 26 | A. naeslundii, A. oris, A. sp. HOT 171 | 3.3 | 87 |
| A. oris, A. sp. HOT 169 | 5.8 | 52 | Other Actinomyces | 0.5 | 13 | |
| Other Actinomyces | 2.4 | 22 | ||||
| Capnocytophaga | C. gingivalis* | 1.9 | 19 | C. gingivalis | 2.0 | 22 |
| C. granulosa* | 1.6 | 16 | C. granulosa | 1.6 | 17 | |
| C. leadbetteri | 2.4 | 24 | C. leadbetteri | 0.3 | 3 | |
| C. sputigena | 2.4 | 24 | C. sputigena | 2.6 | 29 | |
| C. sp. HOT 336, C. sp. HOT 864 | 0.5 | 5 | C. sp. HOT 335, C. sp. HOT 864 | 0.6 | 7 | |
| C. sp. HOT 323, C. sp. HOT 326 | 0.5 | 5 | C. sp. HOT 332 | 0.6 | 7 | |
| Other Capnocytophaga | 0.6 | 6 | C. sp. HOT 412 | 0.4 | 5 | |
| Other Capnocytophaga | 1.0 | 10 | ||||
| Corynebacterium | C. matruchotii | 5.7 | 67 | C. matruchotii | 10.3 | 78 |
| C. durum | 2.8 | 33 | C. durum | 3.0 | 22 | |
| Fusobacterium | F. nucleatum ss. polymorphum, F. nucleatum ss. vincentii† | 1.4 | 52 | F. nucleatum ss. polymorphum, F. nucleatum ss. nucleatum, F. sp. HOT 203 | 4.6 | 55 |
| F. nucleatum ss. vincentii† | 0.5 | 20 | F. nucleatum ss. vincentii | 1.7 | 20 | |
| F. nucleatum ss. animalis | 0.4 | 17 | F. nucleatum ss. animalis | 1.1 | 14 | |
| F. periodonticum, F. sp. HOT 370, F. nucleatum ss. nucleatum | 0.2 | 9 | F. periodonticum | 0.4 | 5 | |
| Other Fusobacterium | 0.1 | 2 | F. sp. HOT 370 | 0.6 | 7 | |
| Other Fusobacterium | 0.0 | 0 | ||||
| Haemophilus/Aggregatibacter | H. parainfluenzae‡ | 2.1 | 99 | H. parainfluenzae‡ | 4.9 | 68 |
| Other Haemophilus | 0.0 | 1 | A. paraphrophilus | 0.6 | 9 | |
| A. sp. HOT 458 | 0.8 | 11 | ||||
| A. segnis, A. sp. HOT 512, A. sp. HOT 513 | 0.4 | 6 | ||||
| Other Haemophilus/Aggregatibacter | 0.4 | 6 | ||||
| Lautropia | L. mirabilis | 4.9 | 100 | L. mirabilis | 3.0 | 100 |
| Leptotrichia | L. buccalis | 1.3 | 33 | L. buccalis | 0.6 | 11 |
| L. hofstadii | 0.2 | 5 | L. hofstadii | 0.8 | 16 | |
| L. hongkongensis | 0.6 | 15 | L. hongkongensis | 0.7 | 14 | |
| L. sp. HOT 212 | 0.8 | 21 | L. sp. HOT 212 | 0.4 | 8 | |
| L. sp. HOT 392 | 0.4 | 11 | L. sp. HOT 392 | 0.4 | 7 | |
| L. wadei | 0.3 | 7 | L. wadei | 0.5 | 9 | |
| Other Leptotrichia | 0.3 | 9 | Other Leptotrichia | 1.7 | 34 | |
| Neisseria/Kingella/Eikenella | N. sicca, N. mucosa, N. flava, N. oralis | 3.2 | 36 | N. sicca, N. mucosa, N. flava, N. sp. HOT 015 | 2.9 | 34 |
| N. elongata | 1.6 | 18 | N. elongata | 1.2 | 14 | |
| N. subflava | 0.9 | 10 | N. subflava, N. flavescens | 1.0 | 12 | |
| N. pharyngis | 0.4 | 5 | N. pharyngis | 1.4 | 16 | |
| K. oralis | 1.3 | 15 | K. oralis | 0.5 | 6 | |
| E. corrodens | 0.4 | 4 | E. corrodens | 0.3 | 3 | |
| Other Neisseria/Kingella | 1.2 | 14 | Kingella sp. HOT 459, Simonsiella muelleri | 0.5 | 6 | |
| Other Neisseria/Kingella | 0.6 | 7 | ||||
| Porphyromonas | P. catoniae, P. sp. HOT 284 | 0.7 | 33 | P. catoniae, P. spp. HOT 275, 277, 284 | 1.5 | 39 |
| P. pasteri (formerly P. sp. HOT 279) | 1.2 | 57 | P. pasteri (formerly P. sp. HOT 279) | 2.1 | 53 | |
| Other Porphyromonas | 0.2 | 10 | Other Porphyromonas | 0.3 | 8 | |
| Prevotella | P. nigrescens | 0.3 | 22 | P. nigrescens | 0.6 | 16 |
| P. oris | 0.2 | 15 | P. oris | 0.6 | 14 | |
| P. sp. HOT 317 | 0.1 | 9 | P. sp. HOT 317 | 0.6 | 16 | |
| P. sp. HOT 472 | 0.2 | 15 | P. sp. HOT 472 | 0.7 | 17 | |
| Other Prevotella | 0.5 | 38 | Other Prevotella | 1.6 | 38 | |
| Rothia | R. aeria | 9.8 | 98 | R. aeria | 1.9 | 53 |
| Other Rothia | 0.2 | 2 | R. dentocariosa | 1.8 | 47 | |
| Other Rothia | 0.0 | 0 | ||||
| Streptococcus | S. mitis, S. pneumoniae, S. cristatus, S. australis, S. spp. HOT 70, 71, 74 | 8.3 | 45 | S. mitis, S. oralis, S. peroris, S. spp. HOT 71, 423, 431 | 7.3 | 48 |
| S. oralis, S. infantis, S. spp. HOT 55, 58, 61 | 4.8 | 26 | S. infantis, S. spp. HOT 58, 61, 74, 486 | 1.4 | 9 | |
| S. sanguinis | 4.0 | 22 | S. sanguinis, S. agalactiae | 4.7 | 31 | |
| S. gordonii | 0.9 | 5 | S. gordonii, S. sp. HOT 056 | 0.8 | 5 | |
| S. parasanguinis | 0.0 | 0 | S. cristatus, S. sinensis, S. australis, S. parasanguinis | 0.5 | 3 | |
| Other Streptococcus | 0.5 | 3 | Other Streptococcus | 0.5 | 4 | |
| Veillonella | V. parvula, V. dispar | 3.3 | 99 | V. parvula, V. dispar, V. rogosae, V. atypica | 3.8 | 99 |
| Other Veillonella | 0.0 | 1 | Other Veillonella | 0.0 | 1 | |
| Subtotal | 87.7 | Subtotal | 89.0 | |||
| Other | Abiotrophia defectiva | 0.5 | Abiotrophia defectiva | 0.3 | ||
| Actinobaculum sp. HOT 183 | 0.5 | Actinobaculum sp. HOT 183 | 0.5 | |||
| Bacteroidales [G-2] sp. HOT 274 | 0.4 | Bacteroidales [G-2] sp. HOT 274 | 0.4 | |||
| Bergeyella sp. HOT 322 | 1.6 | Bergeyella sp. HOT 322 | 0.6 | |||
| Gemella haemolysans | 0.4 | Gemella sanguinis, G. haemolysans, G. morbillorum | 0.5 | |||
| Gemella morbillorum | 0.2 | |||||
| Granulicatella adiacens | 0.5 | Granulicatella elegans, G. adiacens | 0.7 | |||
| Propionibacterium propionicum | 0.5 | Propionibacterium propionicum, P. sp. HOT 194 | 0.6 | |||
| Propionibacterium sp. HOT 194 | 0.5 | |||||
| Treponema spp. | 0.6 | Treponema spp. | 1.1 | |||
| Campylobacter showae | 0.7 | Campylobacter showae, C. rectus | 0.3 | |||
| Campylobacter concisus | 0.5 | Campylobacter concisus | 0.2 | |||
| Campylobacter gracilis | 0.4 | Campylobacter gracilis | 0.3 | |||
| Selenomonas infelix, S. spp. HOT 479, 481 | 0.1 | Selenomonas infelix, S. dianae, S. spp. HOT 146, 479, 892, Centipeda periodontii | 1.0 | |||
| Selenomonas noxia | 0.7 | Selenomonas noxia, S. flueggei, S. spp. HOT 140, 388 | 0.8 | |||
| TM7 spp. | 2.4 | |||||
| Other taxa | 1.9 | Other taxa | 3.7 | |||
| Total | 100 | Total | 100 |
Species-level taxonomy of the HMP supragingival plaque samples from 16S rRNA gene sequencing data in the V1–V3 and V3–V5 regions as analyzed by oligotyping (19). Oligotypes from ref. 19 were assigned to the closest representative sequence in the HOMD to which they were at least 98% identical, except for C. matruchotii in V1–V3, for which all oligotypes were between 96.9% and 98.3% identical to the HOMD reference sequence. Where the species has not been formally named, it is listed as Human Oral Taxon (HOT) followed by its taxon number in the HOMD. Multiple taxon names separated by a comma (e.g., A. naeslundii, A. sp. HOT 171) indicate that the HOMD reference sequences for these taxa are identical or nearly identical in the region sequenced and cannot be differentiated by this data. Percentage abundance values given are the mean of 148 (V3–V5) or 77 individuals (V1–V3).
One oligotype was 99.1% identical to both C. gingivalis and C. granulosa in V1–V3 and assigned one-half to C. gingivalis and one-half to C. granulosa.
F. nucleatum ss. vincentii is listed twice under V1–V3, because it is represented by multiple reference sequences in the HOMD, one which is unique and one which is identical to F. nucleatum ss. polymorphum in the region sequenced.
H. parainfluenzae includes Terrahaemophilus aromaticivorans.
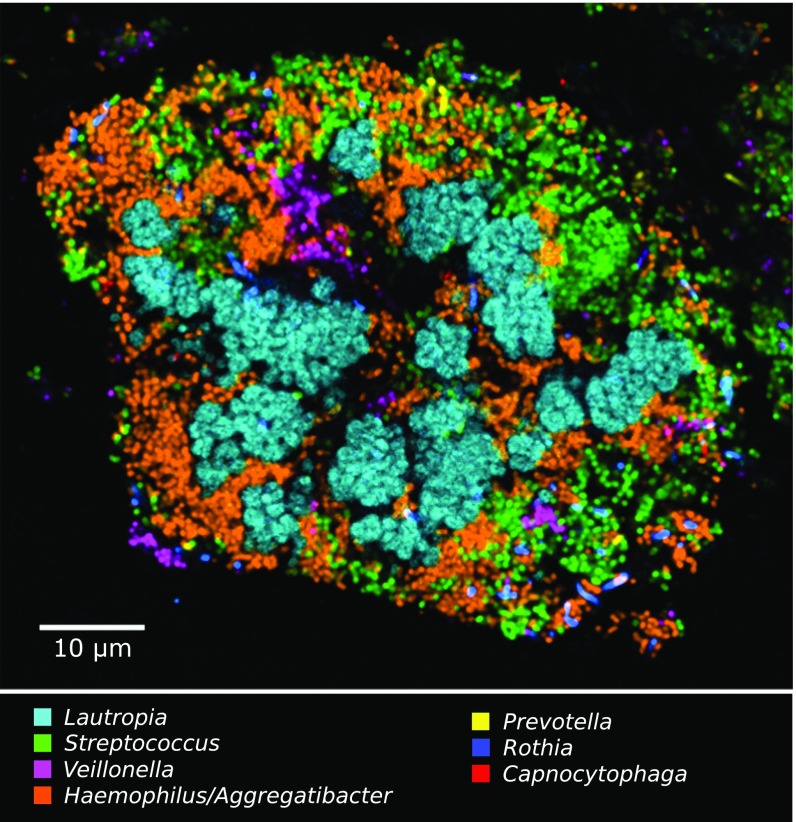
Fig. 8.
A cauliflower structure in plaque composed of Lautropia, Streptococcus, Haemophilus/Aggregatibacter, and Veillonella. Scattered cells of Prevotella, Rothia, and Capnocytophaga are also visible.
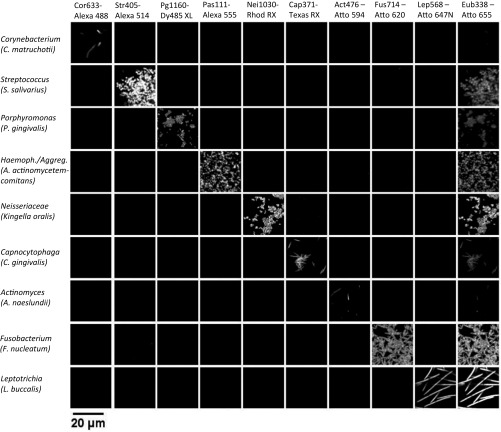
Fig. S1.
Probe set hybridizes as expected with pure cultures. The set of 10 probes, each labeled with a distinct fluorophore, was applied to pure cultures and subjected to imaging and linear unmixing under the same conditions used to image plaque samples. Each of nine taxon-specific probes hybridized with its target taxon and showed no significant hybridization to nontarget taxa. The near-universal probe Eub338 hybridized with all taxa, with variable intensity.
Fig. S4.
Tile scan of a hedgehog structure in plaque. Image is a composite of seven fields of view showing a plaque sample with three adjacent hedgehogs.
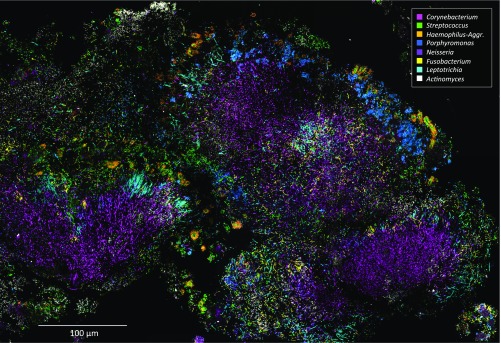
We detected in plaque a complex microbial consortium characterized by the presence of a mass of Corynebacterium filaments with Streptococcus at the periphery. We refer to this structure as a “hedgehog” because of its spiny, radially oriented filaments. We identified nine taxa as regular participants in hedgehog structures: Corynebacterium, Streptococcus, Porphyromonas, Haemophilus/Aggregatibacter, Neisseriaceae, Fusobacterium, Leptotrichia, Capnocytophaga, and Actinomyces. Other genera were detected rarely or inconsistently in the hedgehog structures. To visualize the regular constituents of the consortium simultaneously, we constructed a probe set consisting of 10 probes: the 9 probes targeting these taxa plus the universal probe Eub338 reactive with essentially all bacteria. Each of these 10 probes consisted of a unique oligonucleotide conjugated to a unique fluorophore (Table S3). To validate the probes for specificity, we applied the 10-probe set to pure cultures, which we hybridized and imaged under the same conditions as natural plaque samples. All probes reacted strongly with the target taxon and insignificantly with the nontarget taxa (Fig. S1).
Table S3.
Probe sets used in this study
| Oligonucleotide name | Probe set 1 | Probe set 2 | Probe set 3 | Probe set 4 | Probe set 5 | Probe set 6 | Probe set 7 |
| Eub338 | Eub338-Rhodamine Red X | Eub338-Atto 655 dual | Eub338-Alexa 647 | Eub338-Atto 655 dual | Eub338-Pacific Blue | ||
| Cor633 | Cor633-Alexa 488 | Cor633-Alexa 488 | Cor633-Alexa 610 | Cor633-Alexa 647 | Cor633-Alexa 488 | Cor633-Alexa 610 | |
| Str405 | Str405-Alexa 514 | Str405-Rhodamine Red X | Str405-Alexa 514 | Str405-Alexa 514 | Str405-Alexa 514 | ||
| Pg1160 | Pg1160-Dy485 XL | Pg1160-Alexa 555 | Pg1160-Rhodamine Red X | Pg1160-Texas Red X | |||
| Pas111 | Pas111-Alexa 555 | Pas111-Alexa 488 | Pas111-Alexa 488 | Pas111-Alexa 555 | Pas111-Dy615 | ||
| Nei1030 | Nei1030-Rhodamine Red X | Nei1030-Rhodamine Red X | Nei1030-Atto 633 dual | ||||
| Cap371 | Cap371-Texas Red X | Cap371-Texas Red X | |||||
| Act476 | Act476-Atto 594 dual | Act476-Atto 594 dual | Act476-Alexa 488 | ||||
| Fus714 | Fus714-Atto 620 dual | Fus714-Alexa 514 | Fus714-Atto 620 dual | ||||
| Lep568 | Lep568-Atto 647N dual | Lep568-Atto 647N dual | |||||
| Cmat175 | Cmat175-Alexa 514 | ||||||
| Act692 | Act690-Alexa 555 | ||||||
| Cor595 | Cor595-Rhodamine Red X | ||||||
| Act381 | Act382-Alexa 594 | ||||||
| Rot491 | Rot491-Alexa 647 | Rot491-Rhodamine Red X | |||||
| Prv392 | Prv392-Alexa 488 | ||||||
| Vei488 | Vei488-Alexa 555 | ||||||
| Lmir444 | Lmir444-Atto 655 dual |
For each set, the table lists the probes used (oligonucleotide and fluorophore). Dual indicates that the probe was labeled with the same fluorophore at both 3′ and 5′ ends; if dual is not specified, the probe was labeled only at the 5′ end. Probe set 1 is shown in Fig. 2. Probe set 2 is shown in Figs. 3, 4C, and 5, Upper and Figs. S1 and S2A. Probe set 3 is shown in Fig. 4A and Fig. S2C. Probe set 4 is shown in Fig. 4 B and C. Probe set 5 is shown in Figs. 5, Lower and 6 and Figs. S2B and S4. Probe set 6 is shown in Fig. 7 and Fig. S3. Probe set 7 is shown in Fig. 8.
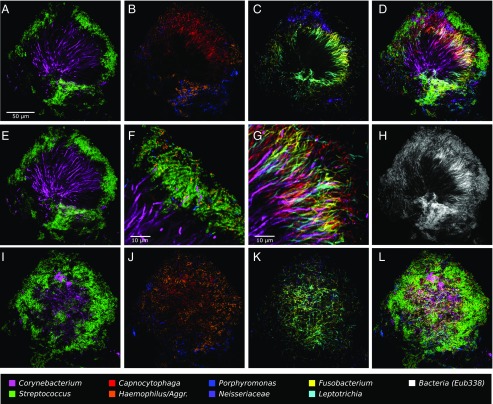
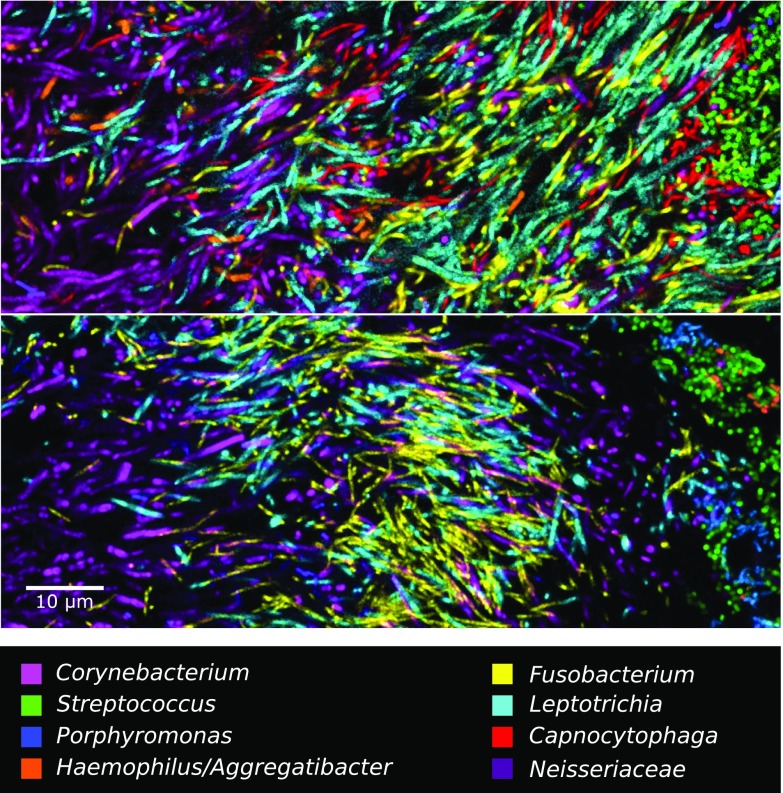
This 10-probe set revealed large, organized hedgehog structures with a generally consistent composition and spatial arrangement (Fig. 3). The fluorescence signal from each of the probes was acquired with a spectral, confocal microscope, was differentiated using a linear unmixing algorithm (Materials and Methods), and is presented in false color, with combinations of probes shown superimposed as detailed in Fig. 3. Fig. 3 A–D and F–H shows a single focal plane near the middle of the structure. Corynebacterium filaments radiate outward from near the center of the image. The coccoid Streptococcus cells are arranged around the distal tips of the Corynebacterium filaments (Fig. 3A). Also located at the periphery of the structure, in the same region as the Streptococcus, are cells of Haemophilus/Aggregatibacter and Porphyromonas (Fig. 3B). Capnocytophaga occupies a wide band just inside the periphery (Fig. 3B). Also occupying this band but forming a more complete ring or annulus between the periphery and the base are Fusobacterium and Leptotrichia (Fig. 3C). Neisseriaceae forms clusters in and near the periphery (Fig. 3C). Actinomyces, which was represented by only a small number of cells in this particular structure, tended to be located near the base. All taxa are shown superimposed in Fig. 3D.
Fig. 3.
A hedgehog structure in plaque showing spatial organization of the plaque microbiome. Plaque was hybridized with a set of 10 probes each labeled with a different fluorophore. Each panel shows the superposition of several of these individual fluorophore channels. A–D and F–H show a single focal plane near the center of the structure, with two to three fluorophore channels shown in each of A–C and all nine specific probes superimposed in D. (E) Maximum intensity projection of three planes, representing a total of ∼2 μm of thickness, to visualize the continuity of Corynebacterium filaments from the center toward the edge of the structure. F is a detailed view of corncob structures. G is a detailed view of mixed filaments. H shows the fluorophore channel corresponding to the universal bacterial probe, showing that the specific probes (D) identify most of the cells that hybridize to the universal probe. I–L show a second focal plane near the periphery of the structure. Fluorophore channels shown correspond to the following genera in the figure: (A, E, and I) Corynebacterium and Streptococcus; (B and J) Capnocytophaga, Porphyromonas, and Haemophilus/Aggregatibacter; (C and K) Fusobacterium, Leptotrichia, and Neisseriaceae; (D and L) all nine specific probes; (F) Corynebacterium, Streptococcus, Porphyromonas, and Haemophilus/Aggregatibacter; (G) Corynebacterium, Fusobacterium, Leptotrichia, and Capnocytophaga; and (H) Bacteria. The plaque sample was fixed in 2% (wt/vol) paraformaldehyde, stored in 50% (vol/vol) ethanol, and spread onto the slide in 50% (vol/vol) ethanol in preparation for FISH.
The spatial arrangement of Corynebacterium relative to other taxa in the structure is detailed in Fig. 3 E–G. Long filaments that move in and out of the plane of focus can be only partially captured in a single optical section (∼1-µm thickness). To visualize the continuity of these filaments, we generated a maximum intensity projection of three adjacent optical sections (Fig. 3E), which shows single filaments that are continuous for more than 50 µm and reach from the core to the periphery of the structure. Some filaments remain visible after they enter the region that contains Streptococcus, whereas others apparently disappear when they enter this zone. A detail of the periphery (Fig. 3F) shows that the corncob structures are composed of a filamentous core (sometimes visualized as Corynebacterium but frequently not stained) surrounded primarily by Streptococcus but also by cells of two other taxa, Porphyromonas and Haemophilus/Aggregatibacter, both of which are in close contact with Streptococcus cells. On their way to this corncob region in the periphery, the Corynebacterium filaments traverse the annulus that is densely populated with elongated rods of Fusobacterium, Leptotrichia, and Capnocytophaga, with cells of all four taxa oriented in roughly the same direction (Fig. 3G).
Completing the overview of the structure, a comparison of the fluorescent signal from the universal probe (Fig. 3H) to the overlay of nine specific probes (Fig. 3D) shows that the taxon-specific probes identify nearly all of the cells in the structure. A second focal plane near the exterior of the structure (Fig. 3 I–L) shows a view of the outer shell composed primarily of corncobs. Toward the center of the image, the edge of the Fusobacterium–Leptotrichia annulus can be seen in end-on view (Fig. 3K). In summary, the plaque hedgehog is a radially organized, multigenus consortium with a framework composed primarily of Corynebacterium, a multitaxon filament-rich annulus, and a periphery of corncob structures.
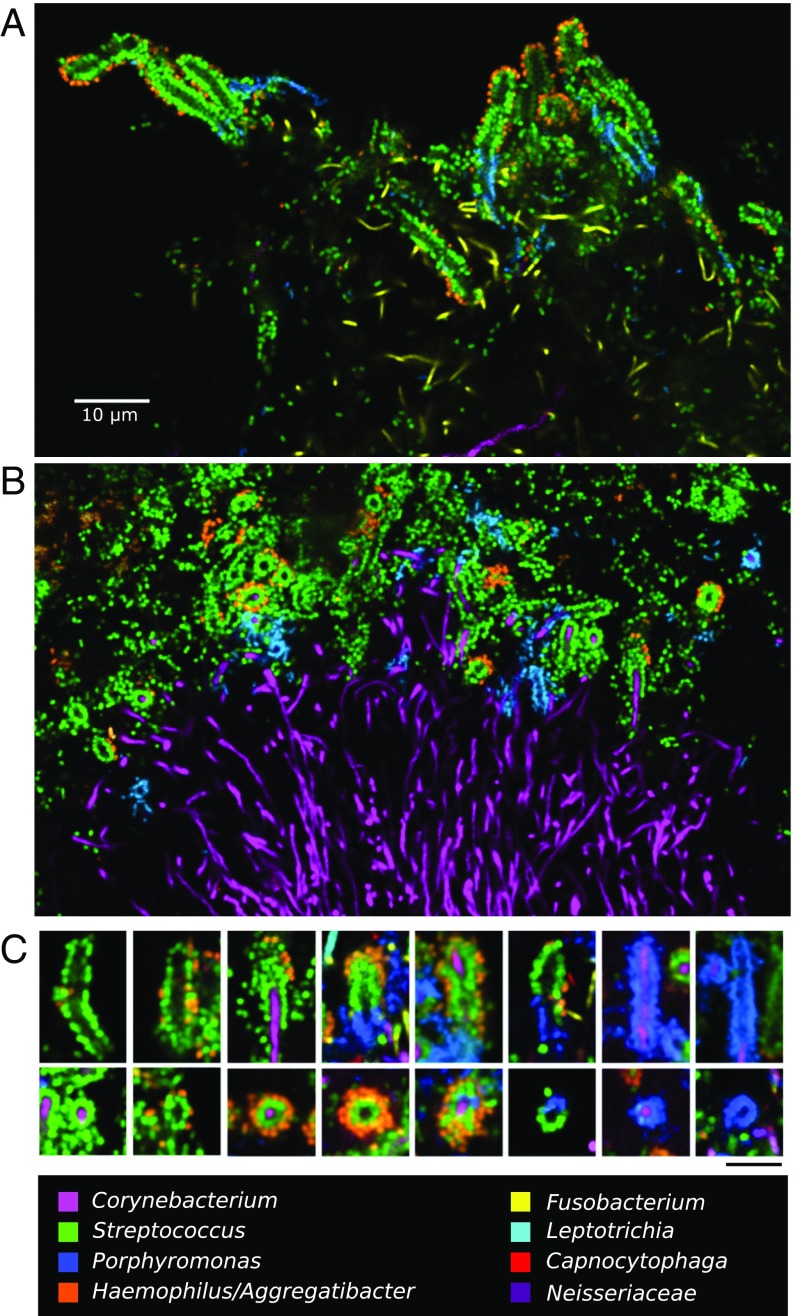
Corncobs are defined morphologically as structures in which coccoid cells, “kernels,” surround a central filament. Our CLASI-FISH results revealed that the kernels were of different taxonomic types and could be either single or double layer (Fig. 4). Single-layer corncobs had kernels of either Streptococcus or Porphyromonas; double-layer corncobs consisted of a combination of Streptococcus as the inner layer and Haemophilus/Aggregatibacter as the outer layer. The most common corncob had a single layer of Streptococcus kernels surrounded by a partial or complete layer of Haemophilus/Aggregatibacter. Porphyromonas kernels could be colinear with Streptococcus around the same filament, or could form entire corncobs of their own, but in either case were always organized in a single layer. In contrast, cells of Haemophilus/Aggregatibacter were never observed to form their own corncobs with Corynebacterium filaments. When present, they were always found adjacent to Streptococcus cells. The Haemophilus/Aggregatibacter–Streptococcus association was evidently specific, because Haemophilus/Aggregatibacter was not found adjacent to cells of Porphyromonas or other taxa in the absence of Streptococcus. Overall, the close spatial proximity of multiple taxa in corncobs suggests the possibility of significant competitive, exploitative, or mutualistic interactions among these taxa.
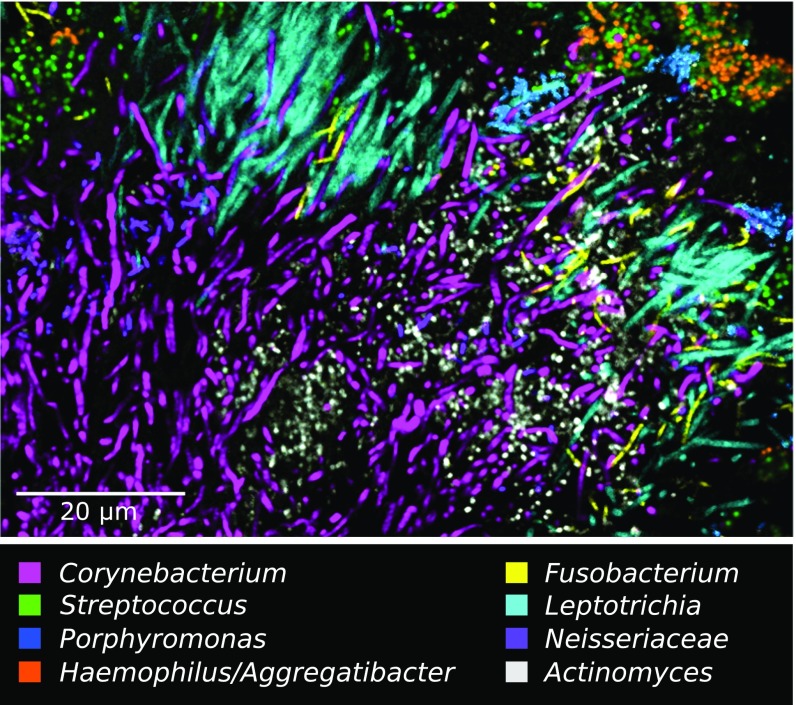
Fig. 4.
Complex corncob structures in SUPP. (A and B) Clusters of corncobs at the perimeter of hedgehog structures. (A) Whole mount of plaque hybridized with probes for Corynebacterium, Fusobacterium, Streptococcus, Porphyromonas, and Haemophilus/Aggregatibacter. (B) Methacrylate-embedded section hybridized with probes for Corynebacterium, Streptococcus, Porphyromonas, and Haemophilus/Aggregatibacter. (C) Gallery of representative images showing types of corncobs frequently observed. (Scale bar: C, 5 μm.)
In a substantial fraction of corncob structures, weak or no fluorescence signal was detected from the central filament in the region where the kernels were present. Lack of hybridization to the central filament was particularly frequent in whole-mount preparations (Fig. 4A). In embedded and sectioned preparations, the central filament was more consistently visualized (Fig. 4B). Higher magnification images of longitudinal and cross-section views (Fig. 4C) illustrate the visualization or lack thereof of the central filament. However, in all cases in which the central filament was clearly labeled, it hybridized with the Corynebacterium probe, even in cases in which cells of other taxa, including Fusobacterium, Leptotrichia, and Capnocytophaga, were present in abundance immediately adjacent to the corncobs (Fig. S2). This observation indicates that, rather than binding promiscuously to any available filamentous substrate, the cocci are engaged in a highly specific interaction with Corynebacterium.
Fig. S2.
Corncob structures form around Corynebacterium and do not form around nearby Fusobacterium, Leptotrichia, or Capnocytophaga. A–C show methacrylate-embedded sections from three different donors. Rectangles indicate location of Insets; ovals highlight representative corncobs, each of which has a Corynebacterium core.
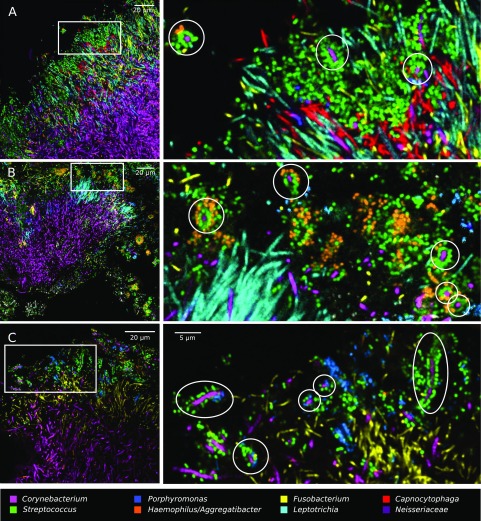
The filaments or elongated rods inhabiting plaque hedgehogs were striking in both their density and their spatial organization. Both Fusobacterium and Leptotrichia showed elongated morphology and were dispersed thinly at the periphery of the hedgehog but reached very high densities in the region immediately proximal to the periphery (Fig. 3C), a region that we call the filament-rich annulus. Capnocytophaga, likewise, reached high densities in the annulus but also extended into regions of the consortium that were rich in Neisseriaceae (Fig. 3 B and D). In whole-mount preparations, many cells in the filament-rich annulus overlapped in images in which all taxon channels were superimposed (Fig. 3D). This overlap was likely caused, in part, by compression of the 3D structure in whole-mount preparations, so that the cells were more densely packed than would occur in uncompressed material. In plaque embedded in methacrylate and sectioned, the compression was eliminated, and the images showed cells that were tightly packed but clearly resolved and distinct from one another (Fig. 5). Notably, these images showed that bacteria do not form large single-taxon clusters within hedgehogs. Instead, cells of at least four different taxa were intermingled at micron scales. These images show that the local environment of a cell in hedgehog consortia includes cells of several other taxa, even when we define local to mean within a radius of as little as 5–10 μm.
Fig. 5.
Filaments and rods of several genera intermingle at micron scales in an annulus of the hedgehog structure. The two images shown are from methacrylate-embedded, sectioned plaque from two different donors. Both samples were hybridized with probes for Corynebacterium, Fusobacterium, Leptotrichia, Streptococcus, Porphyromonas, Haemophilus/Aggregatibacter, and Neisseriaceae; the probe set in Upper also included a probe for Capnocytophaga.
By contrast, the localization of Actinomyces relative to Corynebacterium was characterized more by patchy clusters than by intermingling. Actinomyces cells were generally detected in clumps within the base of the hedgehog or adjacent to hedgehogs, as shown in Figs. 6 and 7. The presence of Actinomyces near the base of a hedgehog is suggestive of the possibility that Corynebacterium finds its attachment site in plaque not directly on the tooth but on a preexisting biofilm containing Actinomyces.
Fig. 6.
Localization of Actinomyces within hedgehogs, in patches within the base region of hedgehogs, and adjacent to them.
Fig. 7.
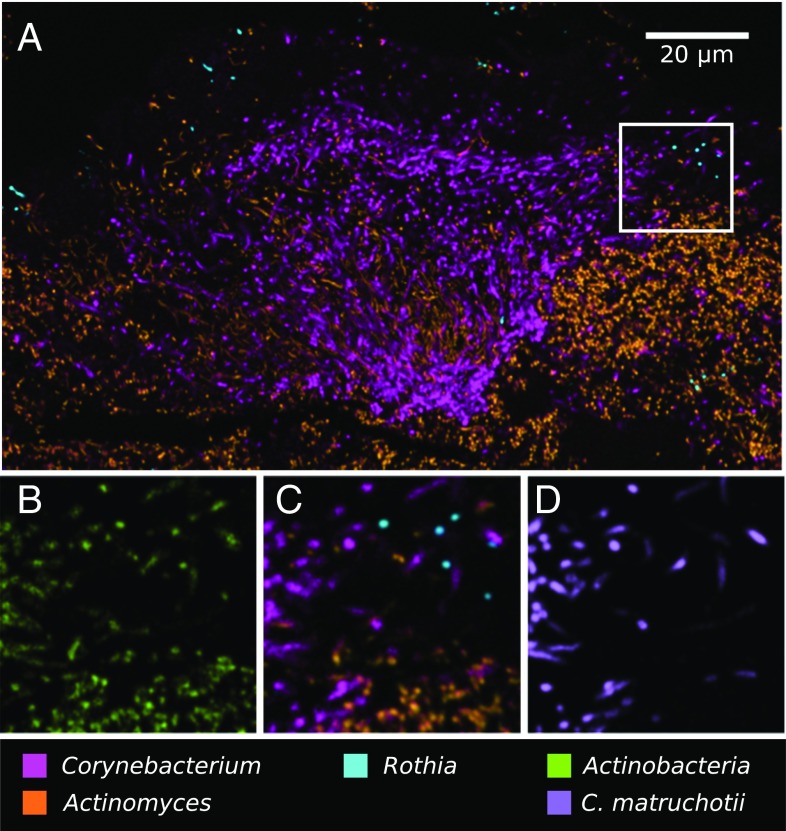
Nested probing for species-level identification of Corynebacterium. Methacrylate-embedded, sectioned plaque was hybridized with a nested probe set targeting cells at the taxonomic levels of phylum, genus, and species. (A) Low-magnification image shows the three major oral genera of phylum Actinobacteria: Corynebacterium, Actinomyces, and Rothia. High-magnification views show (B) all Actinobacteria, (C) the three genera, and (D) C. matruchotii.
Application of a nested probe set allowed identification of the framework Corynebacterium taxon to the species level. As shown in Fig. 7 and Fig. S3, only three genera, Corynebacterium, Actinomyces, and Rothia, comprised virtually all of the Actinobacteria present in plaque, and the species C. matruchotii comprised nearly all of the Corynebacterium in the hedgehog structure.
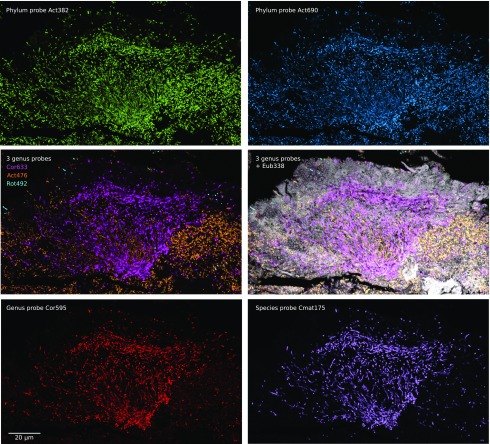
Fig. S3.
Nested probe set provides species-level identification of hedgehog Corynebacterium. Sample was hybridized with nine probes, each labeled with a different fluorophore, targeting cells at the level of kingdom, phylum, genus, and species. (Top) Two different probes targeting phylum Actinobacteria identify a consistent set of cells. Genus-level probes (Middle Left) identify these cells as the three genera Corynebacterium, Actinomyces, and Rothia and (Middle Right) are shown in the context of cells labeled with the universal probe Eub338 plus autofluorescence. (Bottom Left) A second genus-level probe validates the identity of Corynebacterium cells, and (Bottom Right) a species-level probe identifies them as Corynebacterium matruchotii.
Hedgehog structures showed near-universal prevalence among individuals, but the fraction of plaque consisting of hedgehogs was highly variable from sample to sample, even within a single individual. We detected hedgehogs in every individual who was sampled on multiple occasions and in ∼80% of individuals sampled only once. The most exposed surface sampled, the tooth surface on the buccal side, yielded hedgehogs; so did plaque from the gingival margin. Some samples contained multiple hedgehog structures adjacent to one another (Fig. S4). Other samples lacked hedgehogs but contained other consortia. For example, clusters of Lautropia formed the center of a structure that also contained Streptococcus, Haemophilus/Aggregatibacter, and Veillonella and was reminiscent of a cauliflower (Fig. 8). Most samples contained a mixture of hedgehogs and other consortia. Because of this extensive variability and the time-intensive nature of spectral imaging analysis, higher-throughput imaging methods will be required to conduct a comprehensive analysis of spatial, temporal, and individual variation in the abundance of hedgehogs and other consortia in plaque.
In summary, we have discovered distinctive, multigenus consortia in dental plaque, with each taxon localized in a precise and well-defined spatial zone. The precision and reproducibility of this spatial organization indicate that micron-scale organization reflects a finely tuned interaction among the cells comprising oral microbial communities.
Discussion
Organization of Hedgehog Structures.
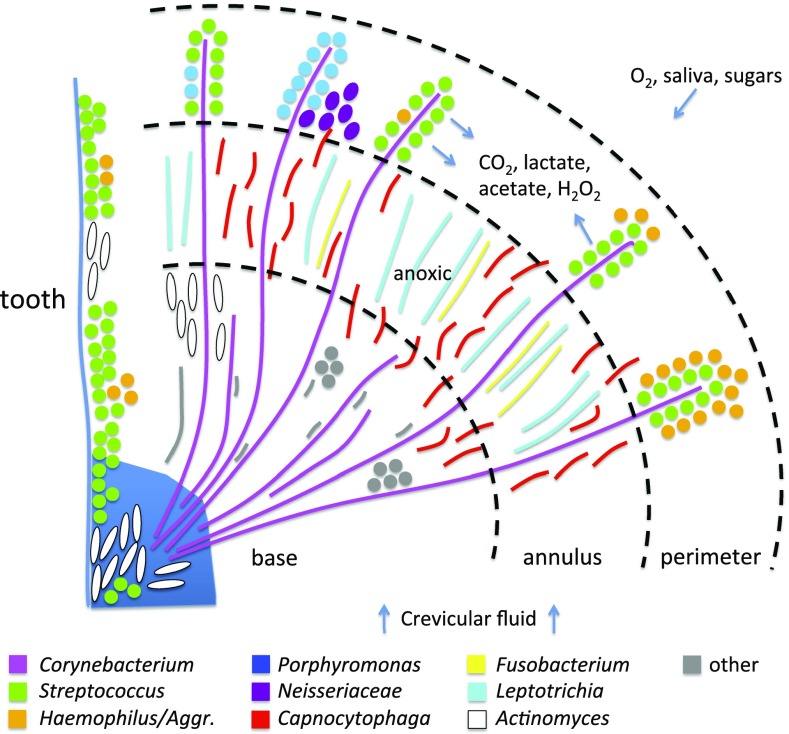
The spatial organization of hedgehog consortia provides a framework for understanding the community structure and metabolism of the plaque microbiome. A modest number of abundant taxa makes up the clear majority of the cells in the structure, and these taxa are arranged in an organized spatial framework, within which each microbe occupies a characteristic position. Based on the literature, we interpret the metabolic, adhesive, and environmental drivers of plaque spatial structure as diagrammed in Fig. 9.
Fig. 9.
Summary hypothesis for interpretation of hedgehog structures. Corynebacterium filaments bind to an existing biofilm containing Streptococcus and Actinomyces. At the distal tips of the Corynebacterium filaments, corncob structures form in which the filaments are surrounded by cocci, including Streptococcus and Porphyromonas, in direct contact with the Corynebacterium filament as well as Haemophilus/Aggregatibacter in contact with Streptococcus. Clusters of Neisseriaceae also occupy the periphery of the hedgehog. The Streptococcus cells create a microenvironment rich in CO2, lactate, and acetate, containing peroxide, and low in oxygen. Elongated filaments of Fusobacterium and Leptotrichia proliferate in this low-oxygen, high-CO2 environment in an annulus just proximal to the corncob-containing peripheral shell of the hedgehog. The CO2-requiring Capnocytophaga also proliferates abundantly in and around this annulus. The base of the hedgehog is dominated by Corynebacterium filaments and thinly populated by additional rods, filaments, and/or cocci.
The radial organization of hedgehogs, built on a framework of Corynebacterium, suggests that Corynebacterium is the foundation taxon of the consortium: it structures the environment, thereby creating habitat for other organisms and nucleating a plaque-characteristic consortium. Consistent with this view, our habitat analysis showed that Corynebacterium was the genus most characteristic of plaque. The physical environment of plaque is distinctive from all other oral habitats because of the tooth surface itself: the tooth represents a solid surface permanently exposed in the mouth, whereas all other oral surfaces are covered in epithelial cell layers that frequently shed. Our model suggests that Corynebacterium proliferates in plaque and structures the plaque environment because it has adopted a strategy of filamentous growth outward from the tooth, anchored in a base cemented to that permanent, exposed surface. By embedding itself in a biofilm matrix attached to the tooth, Corynebacterium could anchor the entire structure and create a protected reservoir from which it can regrow after its removal by abrasion or oral hygiene procedures, thus accounting for how this organism maintains a high relative abundance and a prominent position specifically in the microbiota associated with the tooth surface, i.e., dental plaque.
Other taxa involved in the hedgehog are stratified into distinct zones within the structure, so that different sets of taxa are characteristic of the perimeter, the filament-rich annulus, and the base. A consequence of this stratification is to reduce the number of taxa likely to be the major drivers of community metabolism in each of the zones. Of the nine taxa that we show to be abundant participants in hedgehogs, the three that are most likely to be key participants in the aerobic metabolism at the perimeter are Streptococcus, Haemophilus/Aggregatibacter, and Porphyromonas. In addition, Neisseriaceae may contribute, although it is less regular in its presence at the perimeter. Key participants in metabolism of the filament-rich annulus are likely to be the three taxa that proliferate most specifically and abundantly there: Fusobacterium, Leptotrichia, and Capnocytophaga. The base is likely dominated by the metabolism of Corynebacterium itself, modulated by other taxa, such as Actinomyces, when they are present. Imaging the spatial organization of the structure focuses attention on small sets of taxa that are highly likely to interact with one another in the healthy human mouth, and can serve to guide studies of metabolic interaction in the most promising direction.
The selection and spatial stratification of taxa in the hedgehog are likely driven by environmental and biochemical gradients. Plaque is bathed in a flow of saliva that presents an oxidizing environment, but microbes at the surface of a biofilm can rapidly consume the O2, resulting in a hypoxic environment within 10–20 μm of the surface (24). Streptococcus, a facultative aerobe, can thrive in an oxygen-rich environment, consistent with its location at the periphery of the consortium. In agreement with our localization of the Streptococcus-containing corncobs to the periphery of the hedgehog, earlier electron micrographs of the plaque colonizing teeth and enamel crowns clearly show the localization of corncob structures at the periphery of plaque, on the side away from the enamel surface (23, 25), although these earlier studies were unable to assign a taxonomic identity to the participants in the corncob structures. The location of Streptococcus at the periphery, in turn, drives biochemical gradients. Streptococci consume sugars and oxygen and generate lactate, acetate, CO2, and hydrogen peroxide (26, 27). Both lactate and hydrogen peroxide can be inhibitory to susceptible microbes but not to other participants in corncobs: hydrogen peroxide is detoxified by catalase-producing members of the commensal microbiota, including Corynebacterium and Aggregatibacter, and lactate is used preferentially as a growth substrate by Aggregatibacter (28). Capnocytophaga requires CO2 for growth, and its abundance just inside the corncob shell suggests that it is making use of a CO2-rich environment generated by Streptococcus. Fusobacterium and Leptotrichia are generally considered anaerobes, but strains of both have recently been found to grow efficiently as microaerophiles as well (1, 29–31), and their proliferation in the annulus proximal to the corncob shell suggests a low-oxygen environment there. In summary, the precise localization of taxa within this complex structure is consistent with the modulation of the chemical environment that we would expect to be carried out by Streptococcus but likely includes other, as yet unknown metabolic interactions among neighboring cells. In contrast, the localization of Porphyromonas in the outer shell of the hedgehog structure, in a presumably aerobic environment, is unexpected. Both of the widely studied species of Porphyromonas, Porphyromonas gingivalis and Porphyromonas endodontalis, are strict anaerobes. However, those two species are rare in the plaque of healthy individuals, whereas two additional species, Porphyromonas catoniae and Porphyromonas pasteri (previously Porphyromonas sp. HOT 279), are abundant (19). It is likely that the Porphyromonas spp. that we detect in hedgehogs are representatives of these less well-known taxa and are at least moderately aero-tolerant.
Within each of the strata in the hedgehog, cells of each taxon are arranged not as single-taxon microcolonies but in close proximity to cells of other genera. Corncob structures, for example, are characterized by direct physical contact between the central Corynebacterium filament and the surrounding Streptococcus (23). We detected Porphyromonas cells sometimes forming separate corncobs, but also present in mixed corncobs immediately adjacent to Streptococcus. This arrangement raises the question of whether the Porphyromonas cells are in direct competition with Streptococcus, for example competing for attachment sites on the Corynebacterium filament that allow the attached cell to be bathed in the surrounding nutrient-rich saliva. Alternatively, it is conceivable that Streptococcus and Porphyromonas may facilitate each other’s attachment in the corncob and thereby gain some cross-feeding metabolic advantage. The positioning of Haemophilus/Aggregatibacter in corncobs, by contrast, was not directly adjacent to the central filament in the absence of Streptococcus. This arrangement could arise simply from the binding properties of the cells, if they find attachment sites only on Streptococcus, or alternatively could reflect a metabolic need for close proximity to Streptococcus. Whether each taxon–taxon interaction in corncobs is primarily mutualistic, commensal, or parasitic is a subject for future research, as is the degree to which the relationships might change depending on local conditions, and the degree to which active competition shapes the composition and spatial organization of the assemblage. Similar questions arise for the cells in the filament-dense annulus and the hedgehog base.
From the point of view of microenvironments inhabited by the component cells of the hedgehog, one taxon stands out as being part of multiple communities: a single filament of Corynebacterium may experience several distinctive microenvironments along its length. The proximal part of the filament inhabits the hedgehog base, which is largely dominated by Corynebacterium but may contain Actinomyces and is also populated by a scattering of other cells. Farther along its length, the Corynebacterium filament traverses the filament-rich annulus, where its neighbors consist of Fusobacterium, Leptotrichia, and Capnocytophaga. At its distal end, the Corynebacterium filament is encased in a corncob shell of Streptococcus and Porphyromonas, with frequently abundant Haemophilus/Aggregatibacter and Neisseriaceae. These different environments may alter the local physiology of Corynebacterium, even within a single filament.
Relation to Previous Models of Plaque Structure and Development.
The prevalence of hedgehog structures alters our understanding of the dynamics of colonization of oral surfaces and the successional development of plaque. Clean enamel, glass, or hydroxyapatite surfaces in the mouth are initially colonized by a mixed community in which Streptococcus and Actinomyces are prominent (32–34). Previous models of development and succession in plaque, after initial colonization, assign a central role to Fusobacterium spp. in physically linking early and late colonizers (9, 35) or creating the conditions necessary for colonization of plaque by pathogens (1, 36, 37). Whether these models were meant to describe interactions in supragingival as opposed to subgingival plaque is not entirely clear; the work on initial colonization generally used substrates mounted supragingivally in the mouth, whereas the pathogens in the climax community were subgingival anaerobes. The genus Corynebacterium is conspicuously absent from the early microbiota colonizing enamel and from these models but is one of the more abundant plaque taxa detected in cultivation-independent analyses based on sequencing of rRNA genes (18, 20, 38). These cultivation-independent analyses represent neither the very earliest stages in colonization nor the highly mature and complex subgingival biofilm associated with periodontitis, but instead represent ordinary daily plaque accumulation sampled from healthy subjects. The results that we present here, using HMP sequencing data and samples of ordinary plaque from healthy volunteers, show Corynebacterium as the taxon that provides a physical link to each of the other taxa in the hedgehog structure. Our results do not suggest a central role for Fusobacterium in physically connecting members of the consortium. Although it may contribute to consortium organization, Fusobacterium is only one of four filamentous taxa in hedgehogs, and it is neither the most abundant nor the most spatially extensive. Bioinformatic analysis suggests that the genus Fusobacterium as a whole is substantially more abundant in subgingival than in supragingival plaque (20). Our FISH probe detects the entire Fusobacterium genus, but within the genus, only F. nucleatum has high abundance in plaque. Furthermore, several distinct subspecies of F. nucleatum are recognized in the HOMD, and in HMP sequencing data, these subspecies showed distinctive habitat preferences: some for subgingival and some for supragingival plaque (19). We conclude that the Fusobacterium in hedgehogs is likely one of the supragingival-abundant subspecies of F. nucleatum.
As a slowly growing taxon that is not prominent or even frequently represented among initial colonizers of enamel, how does Corynebacterium and the hedgehog structures that it apparently organizes come to be so abundant in plaque? It seems likely that Corynebacterium finds attachment sites on the preexisting biofilm consisting of Streptococcus and Actinomyces, which are among the early colonizers (33) and can be found near the base of hedgehog structures. EM examination of the microbial community colonizing removable enamel chips worn inside the mouth showed scattered filamentous cells oriented perpendicularly to the primarily coccus-covered surface at 24 hours and a mixed community of abundant filamentous organisms by 48 hours (39), suggesting that colonization with Corynebacterium may take place around the 24-hour stage in plaque development. In the subgingival crevice or within the dental calculus that precipitates around these filamentous aggregates (40), the attachment sites of Corynebacterium would be protected, and when distal portions of the structure are lost, by abrasion or oral hygiene procedures similar to our sampling methods, the filaments could rapidly regrow from these reservoirs.
Understanding Microbiomes: Genes Vs. Organisms.
The imaging approach cuts through the overwhelming complexity of detail in microbial communities and allows common patterns to shine through. With deep sequencing, it has become clear that many taxa in the oral microbiota are shared across individuals but are abundant in some samples and almost vanishingly rare in others (19, 38). These differences in abundance may result from real differences between individuals, fluctuations within a single individual over time, or a combination of the two. Whatever its cause, the lack of a consistently abundant microbial “core” has led to the idea that perhaps it is not organisms but genes and functions that are conserved within the microbiome, distributed across a variety of organisms whose identities are irrelevant. The discovery of hedgehog consortia argues against this idea, at least for some microbiomes. The consistency of the composition and structure of the hedgehog across many individuals suggests that organisms are highly relevant to understanding the roles, organization, and dynamics of the members of the consortium. We suggest that it is neither necessary nor desirable to disregard taxonomy and reduce the microbiome to a collection of genes or metabolites unmoored from their source organism. Instead, we suggest the converse, that an understanding of the ecology and physiology of the organisms in the consortium will provide an organizing principle for understanding and interpreting metagenomic and metatranscriptomic data.
The consortium that we have described is composed of organisms of not only different species but nine different genera classified into eight families in five phyla. Thus, it is a consortium not of close relatives but of highly disparate organisms. The intimate association of widely diverged taxa may be a general property of spatially organized microbial consortia. Darwin (41) was the first to recognize that the “struggle for existence” was most severe between closely allied forms and that “divergence of character” would support the greatest amount of life. In his words, “the advantages of diversification of structure, with the accompanying differences of habit and constitution, determine that the inhabitants, which thus jostle each other most closely, shall, as a general rule, belong to what we call different genera and orders” (41). Although our imaging studies to date have mostly been restricted to genus-level analysis, we suggest that the members of each genus are not interchangeable with one another. Future, more taxonomically refined analyses will reveal which representatives of each genus participate in hedgehog structures. The likely identity of these representatives can be inferred from sequencing data and is generally small in number, as noted above for genus Corynebacterium with only two major oral species.
Potential Significance of Plaque Spatial Structure for Health and Disease and for Modeling Using Synthetic Communities.
The sources of both spatial and temporal heterogeneity in the plaque community and their significance are a rich field for additional investigation. The existence of heterogeneity is clearly visible in the sequencing data, and its functional significance is suggested by morphological studies showing, for example, that primarily filamentous and primarily coccoid communities coexist side by side in supragingival plaque, and that the filamentous communities are more likely to form calculus (40). Would plaque rich in hedgehog structures, compared with other alternate consortia, make the preservation of health or transition to disease more or less likely? Critical topics for future study include whether there are features of host biology, behavior, or immune system that encourage the development of the hedgehog consortium over other microbial assemblages in plaque and what, if any, role hedgehog structures play in the maintenance of oral health or progression toward disease. The ability to create realistic in vitro models of the plaque biofilm is critical for advancing the testing of antimicrobial or microbe-modulating compounds as well as exploring biochemical and metabolic interactions among taxa. Plaque as currently reconstituted in flow cells or other in vitro systems bears little resemblance to the structures that we see in plaque harvested from the mouth. Improving these reconstituted models, with success judged by their ability to recapitulate the spatial structure of natural plaque, will open up a vast area of mechanistic inquiry.
In summary, the biogeography of a microbial community on the micron scale reveals a spatial organization critical to understanding the physiology, ecology, and functional significance of the community. Whether the community in question is host-associated or environmental, these insights are generalizable to the study of any microbiome.
Materials and Methods
Sample Collection, Fixation, and Storage.
We collected supragingival plaque from 22 healthy volunteers, each of whom had given informed consent. For 10 volunteers, oral health was confirmed by clinical examination; the rest were self-reported as both orally and systemically healthy. Volunteers refrained from oral hygiene for 12–48 h before sample collection. Plaque was collected using toothpicks to scrape visible plaque from the gingival margin or tooth surface, or using floss to collect plaque from throughout the mouth. Samples were prepared in three ways. (i) Plaque was applied directly to slides and air-dried, and the samples were fixed directly on the slide, washed, dehydrated through an ethanol series, and subjected to FISH. (ii) Plaque was fixed in paraformaldehyde, stored in 50% (vol/vol) ethanol, spread onto slides in 50% (vol/vol) ethanol, and air-dried immediately before FISH. (iii) Plaque was fixed in paraformaldehyde followed by immediate embedding in methacrylate resin, which was later sectioned, and the sections were applied to slides and then subjected to FISH directly on the slide. Samples were handled gently with a minimum of agitation so as to preserve as much spatial structure as possible. Fixation was carried out in 2% (wt/vol) paraformaldehyde in PBS or 10 mM Tris (pH 7.5) for at least 1.5 h on ice. For samples not fixed directly on the slides, sample was allowed to sediment by gravity, and the supernatant was removed by pipetting. Samples were washed in 1× PBS or 10 mM Tris (pH 7.5) for 15 min and again sedimented by gravity, and the supernatant was removed. Samples were resuspended in PBS or 10 mM Tris, mixed with an equal volume of 100% ethanol, and stored at −20 °C, or they were immediately dehydrated through an ethanol series, washed with acetone, and embedded in Technovit 8100 methacrylate resin (EMSdiasum.com). Embedded blocks were stored at room temperature and sectioned dry using a triangular tungsten carbide knife (Delaware Diamond Knives, Inc.).
Sequencing and Sequence Analysis.
Sequence analysis was carried out on a previously published oligotyping analysis (19) using publicly available data from the HMP (18); details of sample collection, sequencing, and human subjects research approvals are contained in the original publications.
Spectral Imaging FISH, Image Acquisition, and Analysis.
Fluorophore-labeled oligonucleotides were purchased from ThermoFisher.com or www.Biomers.net. FISH was carried out using standard protocols (42) on sections or whole-mount samples mounted onto UltraStick Slides (Thermo Scientific). Hybridization solution [900 mM NaCl, 20 mM Tris, pH 7.5, 0.01% SDS, 20% (vol/vol) formamide, each probe at a final concentration of 2 nM] was applied to samples and incubated at 46 °C for 2–4 h in a chamber humidified with 20% (vol/vol) formamide. Slides were then washed in wash buffer (215 mM NaCl, 20 mM Tris, pH 7.5, 5 mM EDTA) at 48 °C for 15 min, dipped in cold water, air-dried, mounted in ProLong Gold Antifade Solution (ThermoFisher), covered with a #1.5 coverslip, and allowed to cure in the dark overnight. Slides were imaged using a Zeiss LSM 780 Confocal Microscope with a 40× 1.4 N.A. Plan-Apochromat objective. Each field of view was imaged using sequential excitation with the 633-, 594-, 561-, 514-, 488-, and 405-nm laser lines. Linear unmixing was performed using the Zeiss ZEN software using reference spectra acquired from cultured cells hybridized as above with the Eub338 probe labeled with the appropriate fluorophore. Unmixed images were assembled and false-colored using Fiji (43).
Acknowledgments
We thank Alex Valm for probe design; Katherine Lemon and Matthew Ramsey for helpful discussions; and Jake Casper, Louie Kerr, Carissa McKinney, Janina Schuhmann, Braden Tierney, Steven Wilbert, Liping Xun, and the members of the MBL 2014 Physiology Summer Course. This research was supported by NIH National Institute of Dental and Craniofacial Research Grant DE022586 (to G.G.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522149113/-/DCSupplemental.
References
- 1.Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–472. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- 2.Mahowald MA, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106(14):5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4(4):e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakubovics NS. Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J Mol Biol. 2015;427(23):3662–3675. doi: 10.1016/j.jmb.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 5.Wright CJ, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28(2):83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobell C, editor. Antony Van Leeuwenhoek and His “Little Animals,” Being Some Account of the Father of Protozoology and Bacteriology and His Multifarious Discoveries in These Disciplines. Harcourt, Brace, and Co.; New York: 1960. [Google Scholar]
- 7.Kolenbrander PE, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 8.Palmer RJ., Jr Composition and development of oral bacterial communities. Periodontol 2000. 2014;64(1):20–39. doi: 10.1111/j.1600-0757.2012.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolenbrander PE, London J. Adhere today, here tomorrow: Oral bacterial adherence. J Bacteriol. 1993;175(11):3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segata N, et al. Computational meta’omics for microbial community studies. Mol Syst Biol. 2013;9:666. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: Ribosomal RNA-based probes for the identification of single cells. Science. 1989;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 12.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vartoukian SR, Palmer RM, Wade WG. Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl Environ Microbiol. 2009;75(11):3777–3786. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drescher J, et al. Molecular epidemiology and spatial distribution of Selenomonas spp. in subgingival biofilms. Eur J Oral Sci. 2010;118(5):466–474. doi: 10.1111/j.1600-0722.2010.00765.x. [DOI] [PubMed] [Google Scholar]
- 15.Zijnge V, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 2011;108(10):4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111(28):E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H, et al. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182(24):7067–7069. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones SJ. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch Oral Biol. 1972;17(3):613–616. doi: 10.1016/0003-9969(72)90081-7. [DOI] [PubMed] [Google Scholar]
- 23.Listgarten MA, Mayo HE, Tremblay R. Development of dental plaque on epoxy resin crowns in man. A light and electron microscopic study. J Periodontol. 1975;46(1):10–26. doi: 10.1902/jop.1975.46.1.10. [DOI] [PubMed] [Google Scholar]
- 24.Wessel AK, et al. Oxygen limitation within a bacterial aggregate. MBio. 2014;5(2):e00992. doi: 10.1128/mBio.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47(1):1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7(3):e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189(17):6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard K, Cooper C, Tessier S, Ewan EP. Use of chemotaxonomy as an aid to differentiate among Capnocytophaga species, CDC group DF-3, and aerotolerant strains of Leptotrichia buccalis. J Clin Microbiol. 1991;29(10):2263–2265. doi: 10.1128/jcm.29.10.2263-2265.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz PI, Zilm PS, Rogers AH. The response to oxidative stress of Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol Lett. 2000;187(1):31–34. doi: 10.1111/j.1574-6968.2000.tb09132.x. [DOI] [PubMed] [Google Scholar]
- 31.Woo PCY, et al. Leptotrichia hongkongensis sp. nov., a novel Leptotrichia species with the oral cavity as its natural reservoir. J Zhejiang Univ Sci B. 2010;11(6):391–401. doi: 10.1631/jzus.B1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz PI, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72(4):2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dige I, Raarup MK, Nyengaard JR, Kilian M, Nyvad B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009;155(Pt 7):2116–2126. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol. 2004;97(6):1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 35.Lancy P, Jr, Dirienzo JM, Appelbaum B, Rosan B, Holt SC. Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect Immun. 1983;40(1):303–309. doi: 10.1128/iai.40.1.303-309.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 37.Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23(3):196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- 38.Zaura E, Keijser BJF, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyvad B, Fejerskov O. Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95(4):287–296. doi: 10.1111/j.1600-0722.1987.tb01844.x. [DOI] [PubMed] [Google Scholar]
- 40.Friskopp J, Hammarström L. A comparative, scanning electron microscopic study of supragingival and subgingival calculus. J Periodontol. 1980;51(10):553–562. doi: 10.1902/jop.1980.51.10.553. [DOI] [PubMed] [Google Scholar]
- 41.Darwin C. 2013. The Origin of Species by Means of Natural Selection (Forgotten Books, London), Vol 1.
- 42.Pernthaler J, Gloeckner FO, Schoenhuber W, Amann R. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001;30(3):207–226. [Google Scholar]
- 43.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Env Microbiol. 1990;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gmür R, Lüthi-Schaller H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J Microbiol Methods. 2007;69(2):402–405. doi: 10.1016/j.mimet.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Weller R, Glöckner FO, Amann R. 16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides. Syst Appl Microbiol. 2000;23(1):107–114. doi: 10.1016/S0723-2020(00)80051-X. [DOI] [PubMed] [Google Scholar]
- 47.Meier H, Amann R, Ludwig W, Schleifer KH. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G + C content. Syst Appl Microbiol. 1999;22(2):186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 48.Paster BJ, Bartoszyk IM, Dewhirst FE. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 1998;20(1):223–231. [Google Scholar]
- 49.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190(24):8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: Problems and solutions. Syst Appl Microbiol. 1992;15(4):593–600. [Google Scholar]