Significance
India, harboring more than one-sixth of the world population, has been underrepresented in genome-wide studies of variation. Our analysis reveals that there are four dominant ancestries in mainland populations of India, contrary to two ancestries inferred earlier. We also show that (i) there is a distinctive ancestry of the Andaman and Nicobar Islands populations that is likely ancestral also to Oceanic populations, and (ii) the extant mainland populations admixed widely irrespective of ancestry, which was rapidly replaced by endogamy, particularly among Indo-European–speaking upper castes, about 70 generations ago. This coincides with the historical period of formulation and adoption of some relevant sociocultural norms.
Keywords: ancestry, admixture, haplotype blocks, endogamy, social stratification
Abstract
India, occupying the center stage of Paleolithic and Neolithic migrations, has been underrepresented in genome-wide studies of variation. Systematic analysis of genome-wide data, using multiple robust statistical methods, on (i) 367 unrelated individuals drawn from 18 mainland and 2 island (Andaman and Nicobar Islands) populations selected to represent geographic, linguistic, and ethnic diversities, and (ii) individuals from populations represented in the Human Genome Diversity Panel (HGDP), reveal four major ancestries in mainland India. This contrasts with an earlier inference of two ancestries based on limited population sampling. A distinct ancestry of the populations of Andaman archipelago was identified and found to be coancestral to Oceanic populations. Analysis of ancestral haplotype blocks revealed that extant mainland populations (i) admixed widely irrespective of ancestry, although admixtures between populations was not always symmetric, and (ii) this practice was rapidly replaced by endogamy about 70 generations ago, among upper castes and Indo-European speakers predominantly. This estimated time coincides with the historical period of formulation and adoption of sociocultural norms restricting intermarriage in large social strata. A similar replacement observed among tribal populations was temporally less uniform.
India has served as a major corridor for both Paleolithic and Neolithic migrations of anatomically modern humans (1). An early dispersal of modern humans from Africa into India through the southern coastal route (2–4) and migration from West and Central Asia through the northwest corridor (5–8) inferred by past genetic studies have been supported by archaeological evidence, admittedly scattered (2). This evidence fits with Reich et al.’s (9) proposed model that most extant populations of India are a result of admixture between two ancestral populations—Ancestral North Indian (ANI) and Ancestral South Indian (ASI) (9, 10). Anthropologists believe that some of Negrito hunter-gatherer tribes of the Andaman and Nicobar archipelago (A&N) in the Indian Ocean (such as the Jarawa and Onge included in this study) may hold the key to understand the peopling of eastern and southern Asia after anatomically modern humans came out to Africa. Reich et al. (9) also found a distinct component of ancestry among the tribals of A&N, and noted that these tribals are “unique in being ASI-related groups without ANI ancestry” (9). The process by which this archipelago was peopled is unknown but possibly holds the key to our understanding of peopling of South Asia, Pacific Islands, and Australia. Furthermore, multiple lines of evidence, including popularity of rice cultivation in East and Northeast India (11, 12), abundance of the Tibeto-Burman (TB) and Austro-Asiatic (AA) language speakers (13, 14), findings from past archeological and anthropometric (15) as well as genetic studies (6, 16), indicate major waves of migration through India’s northeast corridor.
Reich et al.’s (9) model that all populations of mainland India arose from admixture between two ancestral populations relied strongly on the finding of a north-to-south clinal arrangement of individuals drawn from various populations on a plot of the first two principal components (PCs). A decreasing proportion of “Middle Easterners, Central Asians, and Europeans-like” ancestry from north to south was noted (9). However, TB- and AA-speaking individuals, who were “off-cline” in the PC plot and excluded from further analysis (9, 10), represent additional ancestral components in the Indian population. By analyzing more representative population samples using robust statistical methods, here we provide a fine-grained reconstruction of India’s population history.
Contemporary populations of India are linguistically, geographically, and socially stratified (6, 16), and are largely endogamous with variable degrees of porosity. We analyzed high-quality genotype data, generated using a DNA microarray (Methods) at 803,570 autosomal SNPs on 367 individuals drawn from 20 ethnic populations of India (Table 1 and SI Appendix, Fig. S1), to provide evidence that the ancestry of the hunter-gatherers of A&N is distinct from mainland Indian populations, but is coancestral to contemporary Pacific Islanders (PI). Our analysis reveals that the genomic structure of mainland Indian populations is best explained by contributions from four ancestral components. In addition to the ANI and ASI, we identified two ancestral components in mainland India that are major for the AA-speaking tribals and the TB speakers, which we respectively denote as AAA (for “Ancestral Austro-Asiatic”) and ATB (for “Ancestral Tibeto-Burman”). Extant populations have experienced extensive multicomponent admixtures. Our results indicate that the census sizes of AA and TB speakers in contemporary India are gross underestimates of the extent of the AAA and the ATB components in extant populations. We have inferred that the practice of endogamy was established almost simultaneously, possibly by decree of the rulers, in upper-caste populations of all geographical regions, about 70 generations before present, probably during the reign (319–550 CE) of the ardent Hindu Gupta rulers. The time of establishment of endogamy among tribal populations was less uniform.
Table 1.
Sociocultural and linguistic characteristics of 20 population groups sampled from different geographical locations of India, with sample sizes
| Population name | Social hierarchy | Geography | Linguistic group | Primary occupation* | Sample size |
| Khatri (KSH) | Upper caste | North | Indo-European | Traditionally warrior* | 19 |
| Gujarati Brahmin (GBR) | Upper caste | Northwest | Indo-European | Traditionally priest* | 20 |
| West Bengal Brahmin (WBR) | Upper caste | East | Indo-European | Traditionally priest* | 18 |
| Maratha (MRT) | Upper caste | West | Indo-European | Traditionally warriors* | 7 |
| Iyer (IYR) | Upper caste | South | Dravidian | Traditionally priest* | 20 |
| Pallan (PLN) | Lower-middle caste | South | Dravidian | Agriculturist* | 20 |
| Kadar (KDR) | Tribe | South | Dravidian | Hunter-gatherer | 20 |
| Irula (IRL) | Tribe | South | Dravidian | Hunter-gatherer | 20 |
| Paniya (PNY) | Tribe | South | Dravidian | Hunter-gatherer | 18 |
| Gond (GND) | Tribe | Central | Dravidian/Austro-Asiatic | Agriculturist | 20 |
| Hunter-gatherer | |||||
| Ho (HO) | Tribe | Central and East | Austro-Asiatic | Agriculturist | 18 |
| Hunter-gatherer | |||||
| Santal (SAN) | Tribe | Central and East | Austro-Asiatic | Agriculturist | 20 |
| Hunter-gatherer | |||||
| Korwa (KOR) | Tribe | Central | Austro-Asiatic | Hunter-gatherer | 18 |
| Birhor (BIR) | Tribe | Central | Austro-Asiatic | Hunter-gatherer | 16 |
| Manipuri Brahmin (MPB) | Upper caste | Northeast | Tibeto-Burman | Traditionally warrior* | 20 |
| Tharu (THR) | Tribe | North | Indo-European | Agriculturist | 20 |
| Tripuri (TRI) | Tribe | Northeast | Tibeto-Burman | Agriculturist | 19 |
| Jamatia (JAM) | Tribe | Northeast | Tibeto-Burman | Agriculturist | 18 |
| Jarawa (JRW) | Tribe | Andaman and Nicobar | Ongan | Hunter-gatherer | 19 |
| Onge (ONG) | Tribe | Andaman and Nicobar | Ongan | Hunter-gatherer | 17 |
With the formation of the caste system, which is a system of social stratification, endogamous caste groups were traditionally attributed occupations that were to be hereditary. All of the caste groups in contemporary India are large populations and are engaged in a variety of occupations. The “Primary occupation” column describes the traditional occupation.
Islanders and Mainlanders: Exclusive Ancestries
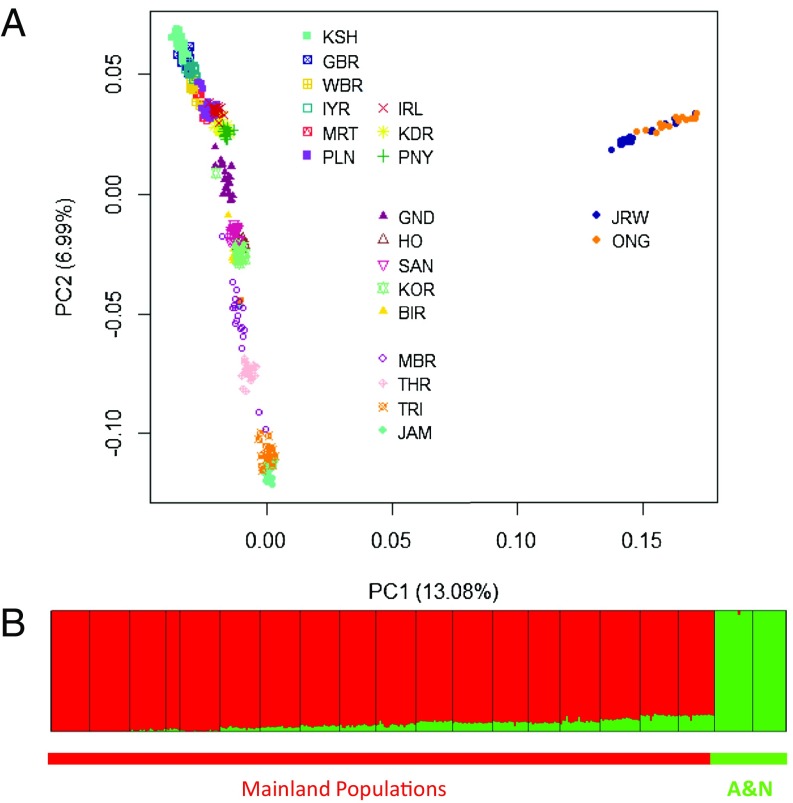
We determined the axes of human genomic variation using principal-components analysis (PCA), as implemented in EIGENSTRAT (17). Using a dynamic programming-driven unsupervised clustering algorithm, ADMIXTURE (18), we determined the genomic admixture at the individual level, by partitioning the genome of an individual into K components contributed by hypothetical ancestors and then estimating their relative contributions. The first principal component (PC-1) explained a high fraction (over 13%) of genomic variation and differentiated the populations of A&N Islands—JRW and ONG—from the mainland populations (Fig. 1), indicating long separation and negligible gene flow. This inference was strongly supported by ADMIXTURE analysis considering two ancestral populations (K = 2) that were found to have contributed disjointedly to the gene pools of the islanders and mainlanders (Fig. 1 and SI Appendix, section 1).
Fig. 1.
(A) Scatterplot of the 367 individuals sampled from 20 Indian populations by the first two PCs extracted from genome-wide genotype data. The Andamanese populations (JRW and ONG) cluster together and are widely separated from mainland populations. (B) Ancestries of individuals estimated using ADMIXTURE with two ancestral components. The 367 individuals are clustered into two distinct groups: the mainlanders (red) and Andamanese islanders (green). (Ancestries of individuals estimated using ADMIXTURE for K = 2, 3, and 4 and related results are in SI Appendix.)
Mainlanders: Four Ancestral Components
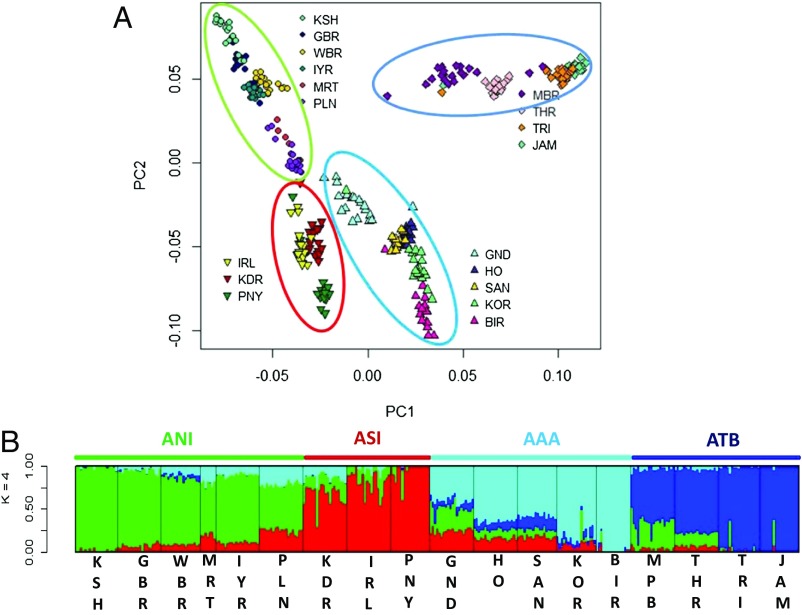
The analysis of genome-wide SNP data on 331 individuals from 18 mainland populations (excluding the 19 ONG and 17 JRW individuals), revealed four ancestral components that formed distinct clusters and clines, in contrast to two components inferred earlier (9) (Fig. 2A). The TB speakers formed a distinct clinal cluster along PC-1, representing descendants of ATB. Along PC-2, the dominant cline was the north-to-south ANI–ASI (9) cline. The AA speakers were also distributed along PC-2 but formed a separate cline indicative of a large contribution from a separate ancestral source (AAA). Central Indian tribal populations, such as Gond and Ho, occupying “central” positions in the PC-1 vs. PC-2 plot, have been noted to be extremely heterogeneous (19) and reported to be quite admixed. These features were also recapitulated by ADMIXTURE analysis (Fig. 2B and Table 2). Multiple runs of ADMIXTURE established the model with four ancestral components (K = 4) as the best-fitting model (SI Appendix, section 2). Model validation by optimum choice of the number of ancestral components (K) was achieved for each dataset by minimizing the cross-validation error (CVE) (18) considering different cutoff values for linkage disequilibrium (LD) and the proportion of data masked for CVE estimation (SI Appendix, section 2, Figs. S2 and S3 A–F). Detailed results for multiple runs of ADMIXTURE (provided in SI Appendix, section 2) show that the convergence is robust.
Fig. 2.
(A) Scatterplot of 331 individuals from 18 mainland Indian populations by the first two PCs extracted from genome-wide genotype data. Four distinct clines and clusters were noted; these are encircled using four colors. (B) Estimates of ancestral components of 331 individuals from 18 mainland Indian populations. A model with four ancestral components (K = 4) was the most parsimonious to explain the variation and similarities of the genome-wide genotype data on the 331 individuals. Each individual is represented by a vertical line partitioned into colored segments whose lengths are proportional to the contributions of the ancestral components to the genome of the individual. Population labels were added only after each individual’s ancestry had been estimated. We have used green and red to represent ANI and ASI ancestries; and cyan and blue with the inferred AAA and ATB ancestries. These colors correspond to the colors used to encircle clusters of individuals in A. (Also see SI Appendix, Figs. S2 and S3.)
Table 2.
Estimates of ancestry proportions of 18 mainland Indian populations under the best-fitting ADMIXTURE model with (K = 4) four ancestral components
| Population | ANI | ASI | AAA | ATB |
| KSH | 0.9793 | 0.0149 | 0.0045 | 0.0013 |
| GBR | 0.8823 | 0.0759 | 0.0412 | 0.0006 |
| WBR | 0.7663 | 0.0994 | 0.101 | 0.0332 |
| MRT | 0.5751 | 0.2141 | 0.2105 | 0.0003 |
| IYR | 0.8046 | 0.111 | 0.0837 | 0.0007 |
| PLN | 0.4902 | 0.2761 | 0.2331 | 0.0006 |
| KAD | 0.0895 | 0.7681 | 0.1414 | 0.0011 |
| IRL | 0.0532 | 0.9255 | 0.0213 | 0 |
| PNY | 0.0252 | 0.9696 | 0.0052 | 0 |
| GND | 0.3697 | 0.193 | 0.3756 | 0.0617 |
| HO | 0.0475 | 0.1705 | 0.7116 | 0.0704 |
| SAN | 0.0347 | 0.1933 | 0.6398 | 0.1321 |
| KOR | 0.0181 | 0.0471 | 0.9091 | 0.0257 |
| BIR | 0.0082 | 0.0054 | 0.9864 | 0 |
| MPB | 0.2635 | 0.0512 | 0.0351 | 0.6502 |
| THR | 0.0935 | 0.0951 | 0.0447 | 0.7667 |
| TRI | 0.0156 | 0.0084 | 0.0117 | 0.9643 |
| JAM | 0.0149 | 0.0044 | 0.0031 | 0.9776 |
Names of the four populations in bold are identified with the four distinct ancestries.
The proportions of inferred ancestral components for each population estimated by ADMIXTURE (Table 2) were compared with maximum-likelihood estimates obtained using frappe (20); both sets of estimates were nearly identical (Table 2 compared with SI Appendix, section 2, Table S4). This concordant finding was further investigated using fineSTRUCTURE (21), which is robust to existing LD and is capable of identifying subtle population subdivisions. fineSTRUCTURE identified 69 subpopulations (SI Appendix, Fig. S4 A and B and section 2) from the data on 331 individuals drawn from 18 ethnic groups. These subpopulations were largely nonoverlapping and belonged to four major clades whose compositions were nearly identical to the four ancestral components identified by ADMIXTURE analysis (SI Appendix, Fig. S4A: depicting the close concordance between the coancestry matrix estimated by fineSTRUCTURE with proportions of ancestral components derived from ADMIXTURE analysis). Sixty (87%) of the 69 subpopulations identified by fineSTRUCTURE comprised individuals drawn from 1 of the 18 original ethnic groups. Viewed differently, only nine subpopulations contained individuals drawn from more than a single ethnic group; even in these rare instances, the individuals were always from closely related ethnic groups (for example, two AA-speaking tribal populations residing in the geographical region) (SI Appendix, section 2). Thus, the numerically larger ethnic groups are well differentiated, even though genomic subdivisions are discernible within them.
Compared with autosomes, the X chromosome has a smaller effective population size (hence more strongly affected by random genetic drift), lower mutation and recombination rates, and greater selective pressure in males. However, the X chromosome is the most informative source to evaluate sex bias in admixture. Sex bias in ancestry contribution was explored using the 107 females, identified unambiguously from genotype data (Methods), belonging to 15 mainland populations. ADMIXTURE analysis using data on these 107 females, with K = 4, separately for the X chromosome and autosomes (SI Appendix, Fig. S5A), reveals a sex bias in all ancestries, except AAA. Although the ATB shows clear excess of X-chromosomal component compared with autosomes, a reverse trend was observed for the ANI component (SI Appendix, Fig. S5B). We also combined the X-chromosome haplotypes of males with the inferred haplotypes (SI Appendix, section 6, Methods) of females and used them to construct a phylogenetic tree (SI Appendix, Fig. S6 A and B). The phylogenetic tree shows distinct clustering (SI Appendix, Fig. S6B) of the haplotypes in clades that belong to genetically closely related populations as inferred from the autosomal data.
More Robust Identification of the Ancestral Components
To more robustly identify and characterize the ancestral components, we combined our data on mainland populations of India with Europe (Eur), Middle Easterners (ME), Central-South Asians (CS-Asian), East Asians (E-Asian) included in Human Genome Diversity Panel (HGDP) (22, 23). The resultant dataset comprised a common set of 630,918 markers. Reich et al. (9) have characterized the ANI ancestry as “genetically close to Middle Easterners, Central Asians, and Europeans.” Similar to Li et al. (22), our PCA plot shows the Eur and ME cluster distinctly, despite being genetically close to the CS-Asians and populations that have high proportion of ANI ancestry (SI Appendix, Fig. S7).
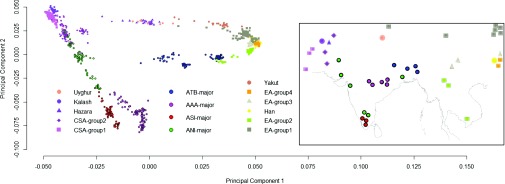
In Fig. 3, PC-1 represents the systematic variation broadly separating the CS-Asian ancestry from E-Asian ancestry, whereas PC-2 represents the systematic variation broadly between the combined AAA plus ASI ancestry and others. The separation of the CS-Asians and E-Asians broadly recapitulated the findings of Li et al. (22). The populations of India with a large proportion of ANI component; particularly the KSH with ∼97% ANI ancestry is inseparable from the CS-Asian, particularly Burusho, Pathan, and Sindhi. The hypothesis that the root of ANI is in Central Asia is further bolstered by the recent evidence derived from analysis of ancient DNA samples (24) and linguistic studies (25). Similarly, the JAM and TRI who have more than 95% ATB ancestry are inseparable from E-Asian populations, e.g., Dai, Lahu, and Cambodian, who live in or near southwestern China and have the lowest “northern” Chinese ancestry (22). Fig. 3 reveals concordance of geographical residence and genetic axes of variation between populations (SI Appendix, section 3).
Fig. 3.
Approximate “mirroring” of genes and geography. Genomic variation of individuals, represented by the first two PCs, sampled from 18 mainland Indians combined with the CS-Asians) and E-Asians from HGDP, compared with the map of the Indian subcontinent showing the approximate locations from which the individuals and populations were sampled.
The Indian dataset, including the JRW and ONG data (A&N), when combined with the HGDP populations of CS-Asia, E-Asia, and Oceania, reveal discernable components of genetic variation that distinguish the CS-Asians from E-Asians, and the Oceanic from other populations (SI Appendix, Fig. S8A). The A&N populations also appear to share a common ancestry with the Oceanic PIs, particularly the Papuans (SI Appendix, Fig. S8A). Owing probably to geographical separation and random genetic drift due to isolation of the island populations, they also separate along the third PC (SI Appendix, Fig. S8 B and C).
Admixture to Endogamy
The extent of borrowed Dravidian and AA linguistic elements (26, 27) in the Rigveda, the earliest of the Vedic texts (dated between 1500 and 1000 BCE), has prompted historians and linguists to argue in favor of a “fair degree” of mixing of the populations (15, 25, 27). Earlier genetic studies have also argued that India was a “relatively” pan-mixing society that embraced endogamy between 1,900 and 4,200 y (9, 10). We reinvestigated the extent of ancient admixture, using a model where individuals could derive their ancestries, at varying degrees, from four genetically distinct components (ANI, ASI, AAA, ATB), instead of three (ANI, ASI, AAA) as the linguists have proposed (26, 27) or two (ANI, ASI) as inferred from previous genetic studies (9, 10).
At homologous genomic regions, distinct ancestral populations are expected to possess distinctive DNA sequences. In other words, different ancestral populations possess a large number of distinguishable haplotype blocks. Meiotic recombination results in exchange of homologous segments between the chromosomes of individuals. Therefore, for an individual with multiple ancestral contributions, distinctive haplotype blocks corresponding to the ancestral populations get fragmented with each event of recombination. When a recipient population (P2) receives, in each generation, a small proportion of haplotypes from a donor ancestral population (P1), the haplotypes of P2 will contain a mixture of fragmented haplotypes and intact haplotypes from P1. If the influx of genetic material from P1 to P2 suddenly ceases, in each subsequent generation, intact haplotypes of P1 in P2 will get fragmented due to recombination. Recombination events, on an average, occur at a rate of one per morgan per generation, and can be appropriately modeled as a Poisson process. Therefore, in the recipient population P2, the distribution of the lengths of haplotype (chromosomal) segments of the donor population P1 will follow an exponential distribution with mean (28, 29), where α (small) is the proportion of admixture per generation of genes from P1 to P2 and T is the number of generations before present (GBP) when this admixture stopped. It is to be noted here that α, if large, that is, if the major portions of the haplotypes are from a particular ancestry, will imply that even if haplotypes break down by recombination into smaller blocks these will not be identifiable because of their similarities with background haplotypes (NA in Table 3). Thus, the time and extent of admixture can be estimated from the distribution of the length of haplotype tracts identified with distinct ancestries in admixed genomes.
Table 3.
Estimates of time (in GBP) of contribution of each of the ancestral components to the populations considered
| Population | ANI | AAA | ASI | ATB |
| GBR | NA* | 69.3833 | 69.3265 | † |
| WBR | NA | 69.5409 | 68.3778 | 63.3518 |
| MRT | NA | 48.7989 | 48.92 | † |
| IYR | NA | 69.1751 | 71.699 | † |
| PLN | NA | 74.3893 | 76.1979 | † |
| KAD | 47.5509 | 60.7911 | NA | † |
| IRL | 39.4951 | 49.8475 | NA | † |
| GND | 77.6637 | 91.9575 | 70.509 | 58.1287 |
| HO | 54.0405 | NA | 67.8753 | 52.9333 |
| SAN | 54.8661 | NA | 71.5929 | 61.5647 |
| KOW | 46.5407 | NA | 55.7532 | 46.6478 |
| MPB | 69.7002 | 67.6769 | 70.4008 | NA |
| THR | 62.7826 | 65.2317 | 72.9749 | NA |
| TRI | 65.1124 | 69.6447 | 70.5565 | NA |
This table pertains to the 14 populations that are considered as admixed and excludes the four populations (KSH, PNY, BIR, JAM) that are considered as representatives of the ancestral components (ANI, ASI, AAA, ATB, respectively).
See text for an explanation of “NA” (not applicable).
The contribution of the ancestral component is too low for reliable estimation of time depth.
We inferred local ancestries and reconstructed each individual’s genome as a potential mosaic of the four components. Individual haplotypes were inferred using Shapeit2 (30, 31) and ancestry of each block was identified using PCAdmix (32) (Methods). Owing to their near nonadmixed status, KSH (98% ANI), PNY (97% ASI), BIR (99% AAA), and JAM (98% ATB) were chosen as best representatives of the ANI, ASI, AAA, and ATB populations.
In each population, the distribution of the ancestral block lengths (ABLs) thus identified, fitted well with the exponential distribution expected under the assumption of sudden cessation of admixture (SI Appendix, section 5). For each population, the times, in generations before present, at cessation of admixture with distinct ancestries were estimated by the method of moments (Table 3).
We estimated that all upper-caste populations, except MPB from Northeast India, started to practice endogamy about 70 generations ago (Table 3). The length distributions of the AAA blocks and the ASI blocks within any one of these populations (GBR, WBR, IYR) were very similar (SI Appendix, section 5). The most parsimonious explanation of this is that the practice of gene flow between ancestries in India came to an abrupt end about 1,575 y ago (assuming 22.5 y to a generation). This time estimate belongs to the latter half of the period when the Gupta emperors ruled large tracts of India (Gupta Empire, 319–550 CE).
Except WBR, with whom the northeast populations are geographically proximal, we found that there is significant ATB ancestry only among AA speakers. Even though the AA speakers presently occupy fragmented geographical regions in India, their presence in Northeast India (Khasis inhabiting Assam and Riang inhabiting Tripura) may indicate a more shared habitat with TB speakers in earlier times. Consistent with an earlier estimate (33), we estimated that the extant TB speakers freely admixed until more recently, 1,500–1,000 y ago (Table 3). Our results indicate that tribal populations may have practiced admixture until more recent times compared to upper-caste populations.
An asymmetry of admixture was also revealed; ABLs attributable to ANI among AA speakers, Dravidian tribes, and TB speakers are longer than those attributable to other ancestries (Table 3), indicating that the ancestral North Indian population continued to provide genomic inputs into these populations (Table 3) well after inputs from other ancestries had ceased.
Discussion
By sampling populations, especially the autochthonous tribal populations, which represent the geographical, ethnic, and linguistic diversity of India, we have inferred that at least four distinct ancestral components—not two, as estimated earlier (9, 10)—have contributed to the gene pools of extant populations of mainland India. The Andaman archipelago was peopled by members of a distinct, fifth ancestry.
The absence of significant resemblance with any of the neighboring populations is indicative of the ASI and the AAA being early settlers in India, possibly arriving on the “southern exit” wave out of Africa. Differentiation between the ASI and the AAA possibly took place after their arrival in India (ADMIXTURE analysis with K = 3 shows ASI plus AAA to be a single population in SI Appendix, Fig. S2). The ANI and the ATB can clearly be rooted to the CS-Asians and E-Asians (Fig. 3 and SI Appendix, Fig. S7B), respectively; they likely entered India through the northwest and northeast corridors, respectively. Ancestral populations seem to have occupied geographically separated habitats. However, there was some degree of early admixture among the ancestral populations (ref. 9 and this study) as evidenced by extant populations possessing multiancestral components and some geographical displacements as well (6).
We have provided evidence that gene flow ended abruptly with the defining imposition of some social values and norms. The reign of the ardent Hindu Gupta rulers, known as the age of Vedic Brahminism, was marked by strictures laid down in Dharmaśāstra—the ancient compendium of moral laws and principles for religious duty and righteous conduct to be followed by a Hindu—and enforced through the powerful state machinery of a developing political economy (15). These strictures and enforcements resulted in a shift to endogamy. The evidence of more recent admixture among the Maratha (MRT) is in agreement with the known history of the post-Gupta Chalukya (543–753 CE) and the Rashtrakuta empires (753–982 CE) of western India, which established a clan of warriors (Kshatriyas) drawn from the local peasantry (15). In eastern and northeastern India, populations such as the West Bengal Brahmins (WBR) and the TB populations continued to admix until the emergence of the Buddhist Pala dynasty during the 8th to 12th centuries CE. The asymmetry of admixture, with ANI populations providing genomic inputs to tribal populations (AA, Dravidian tribe, and TB) but not vice versa, is consistent with elite dominance and patriarchy. Males from dominant populations, possibly upper castes, with high ANI component, mated outside of their caste, but their offspring were not allowed to be inducted into the caste. This phenomenon has been previously observed as asymmetry in homogeneity of mtDNA and heterogeneity of Y-chromosomal haplotypes in tribal populations of India (6) as well as the African Americans in United States (34). In this study, we noted that, although there are subtle sex-specific differences in admixture proportions, there are no major differences in inferences about population relationships and peopling whether X-chromosomal or autosomal data are used. We have also found our inferences to become more robust when our data are jointly analyzed with HGDP data.
We surmise that the number of ancestral components in the populations of India may have been underestimated by Reich et al. (9) because of (i) lack of inclusion of tribal populations, who are considered by anthropologists to be the autochthones of India, and (ii) inadequate representation of the geocultural diversity of India in the set of sampled populations, and (iii) selective removal of some populations based on deviance of their genomic profiles. Our study has corrected this deficiency and has provided a more robust explanation of the genomic diversities and affinities among extant populations of the Indian subcontinent, elucidating in finer detail the peopling of the region.
Methods
Ethical Approval and Informed Consent.
DNA samples were collected with informed consent and after obtaining approvals of institutional ethics committees of the Indian Statistical Institute and the National Institute of BioMedical Genomics.
DNA Isolation, Assessment of Quality and Quantity.
DNA was isolated by the salting-out method (35). Quantity and quality of isolated DNA were assessed using NanoDrop 8000 spectrophotometer.
DNA Microarray Analysis and Data Curation.
Genotyping of each DNA sample was done using Illumina Omni 1-Quad, version 1.0, DNA analysis bead chip on IlluminaiScan, using the manufacturer’s protocol as described in Infinium HD Assay Super Protocol Guide, catalog WG-901–4002. Genotype calling was done using Illumina Genome Studio following Genotyping Module, version 1.0, part 11319113. Quality metric, Gen Call score threshold was set to 0.25 to determine higher stringency in genotype calling. Markers with genotype calls for >90% individuals were included only (details in SI Appendix, section 6).
Because there was no information available about the sex of the individuals sampled, we inferred sex from the X-chromosome genotype. If the inbreeding (homozygosity) estimate (F) was more than 0.8, the individual was inferred to be a male; she was inferred to be a female if F was less than 0.2 (36) (SI Appendix, section 6).
Population Structure.
An unsupervised clustering algorithm, ADMIXTURE (18), was run on our high-density dataset to explore global patterns of population structure varying the number of ancestral clusters (K = 2 through 6) and were successively tested. As LD can adversely affect the inferences of ADMIXTURE (18), the program was run on multiple datasets after pruning SNPs at LD (SI Appendix, sections 1 and 2). Cross-validation errors for each K are available in SI Appendix, sections 1 and 2. PCA was applied to both datasets using EIGENSOFT 4.2 (17) and plots were generated using R 2.12.2 (https://www.r-project.org/). fineSTRUCTURE (21) and frappe (20) were run using the default parameters.
Phasing.
Haplotype estimation both for the autosomes and X chromosome from genome-wide data of unrelated individuals was separately done using segmented haplotype estimation and imputation tool (Shapeit2) (30, 31). Shapeit2 uses a modified hidden Markov model. The algorithm was run only on genotypes with no missing data. Both the model parameters and the number of iterations were set as the default options in Shapeit2.
ABL Estimation.
Local ancestry assignment was performed using PCAdmix (https://sites.google.com/site/pcadmix/) (32) with K = 4 ancestral groups. This approach relies on phased data from reference panels and the admixed individuals. The populations Khatri (KSH), Paniya (PNY), Birhor (BIR), and Jamatia (JAM) with more than 97% ancestry from the ANI, ASI, AAA, and ATB, respectively, were used as the reference panel. Each chromosome is analyzed independently, and local ancestry assignment is based on loadings from PCA of the four putative ancestral population panels. PCAdmix partitions the genomic data into nonoverlapping windows, and for each of these windows the distribution of individual scores within a population is modeled by fitting a multivariate normal distribution (32). Given an admixed chromosome, these distributions are used to compute likelihoods of belonging to each panel. We only considered local ancestry assignments using a greater than 0.85 posterior probability threshold for each window (SI Appendix, section 6).
Data curation, statistical analysis, and graphical representations were done using PLINK (36), version 1.07 (pngu.mgh.harvard.edu/∼purcell/plink/download.shtml), and R, version 2.12.2 (https://www.r-project.org/).
Supplementary Material
Acknowledgments
We thank all of the individuals who volunteered to donate their DNA for the analysis. In addition to some of the authors of this study, sample collection with informed consent was done by C. S. Chakraborty, R. Lalthantluanga, M. Mitra, A. Ramesh, N. K. Sengupta, S. K. Sil, J. R. Singh, C. M. Thakur, and M. V. Usha Rani. We thank B. Dey and B. Bairagya for the sample curation; I. Bagchi and R. Dhar for assistance in generating DNA microarray data; and S. Bhattacharjee, A. Mukherjee, N. K. Biswas, D. Tagore, and S. Chakraborty for assistance in preparing figures. The sample collection and some DNA analyses were partially supported by agencies of the Government of India, including Department of Biotechnology, Department of Science and Technology, and the Indian Council of Medical Research (primarily to P.P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513197113/-/DCSupplemental.
References
- 1.Cann RL. Genetic clues to dispersal in human populations: Retracing the past from the present. Science. 2001;291(5509):1742–1748. doi: 10.1126/science.1058948. [DOI] [PubMed] [Google Scholar]
- 2.Mellars P. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313(5788):796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- 3.Macaulay V, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308(5724):1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 4.Quintana-Murci L, et al. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23(4):437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 5.Sengupta S, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78(2):202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basu A, et al. Ethnic India: A genomic view, with special reference to peopling and structure. Genome Res. 2003;13(10):2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordaux R, et al. Independent origins of Indian caste and tribal paternal lineages. Curr Biol. 2004;14(3):231–235. doi: 10.1016/j.cub.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Bamshad M, et al. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11(6):994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorjani P, et al. Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93(3):422–438. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond J, Bellwood P. Farmers and their languages: The first expansions. Science. 2003;300(5619):597–603. doi: 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 12.Kumar V, et al. Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations. BMC Evol Biol. 2007;7:47. doi: 10.1186/1471-2148-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaubey G, et al. Population genetic structure in Indian Austroasiatic speakers: The role of landscape barriers and sex-specific admixture. Mol Biol Evol. 2011;28(2):1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaubey G, Metspalu M, Kivisild T, Villems R. Peopling of South Asia: Investigating the caste-tribe continuum in India. BioEssays. 2007;29(1):91–100. doi: 10.1002/bies.20525. [DOI] [PubMed] [Google Scholar]
- 15.Thapar R. Early India: From the Origins to AD 1300. Univ of California Press; Berkeley, CA: 2004. [Google Scholar]
- 16.Abdulla MA, et al. HUGO Pan-Asian SNP Consortium Indian Genome Variation Consortium Mapping human genetic diversity in Asia. Science. 2009;326(5959):1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 17.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell RV. The Tribes and Castes of the Entral Provinces of India. Vol 1 Macmillan; London: 1916. [Google Scholar]
- 20.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28(4):289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 21.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8(1):e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 24.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522(7555):207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang W, Chundra C, Hall D, Garrett A. Ancestry-constrained phylogenetic analysis supports the Indo-European steppe hypothesis. Language. 2015;91(1):194–244. [Google Scholar]
- 26.Kuiper PBJ, editor. 1991. Aryans in the Rigveda. Leiden Studies in Indo-European I (RODOPI, Amsterdam)
- 27.Witzel M. Substrate languages in Old Indo-Aryan (Rigvedic, Middle and Later Vedic) Electron J Vedic Stud. 1999;5:1–97. [Google Scholar]
- 28.Pool JE, Nielsen R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics. 2009;181(2):711–719. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin W, Li R, Zhou Y, Xu S. Distribution of ancestral chromosomal segments in admixed genomes and its implications for inferring population history and admixture mapping. Eur J Hum Genet. 2014;22(7):930–937. doi: 10.1038/ejhg.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 31.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 32.Brisbin A, et al. PCAdmix: Principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 2012;84(4):343–364. doi: 10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson EK, et al. Natural selection in a Bangladeshi population from the cholera-endemic Ganges River delta. Sci Transl Med. 2013;5(192):192ra86. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lind JM, et al. Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet. 2007;120(5):713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 35.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.