Significance
Juvenile hormone is unique to arthropods and controls numerous essential functions in these organisms. Its receptor, Methoprene-tolerant, turns on genes by directly binding to E-box–like motifs in their regulatory regions. However, how juvenile hormone represses genes has remained unclear. Here, we used the Aedes aegypti female mosquito, in which juvenile hormone is necessary for reproductive maturation, to show that the repressor Hairy is required for the gene-repressive action of juvenile hormone and Methoprene-tolerant. Moreover, we demonstrate that a corepressor, Groucho, supports this Hairy function. Hence, Methoprene-tolerant recruits the evolutionarily conserved Hairy/Groucho molecular system to mediate the juvenile hormone gene-repressive function.
Keywords: juvenile hormone, receptor, hairy, groucho, repressor
Abstract
The arthropod-specific juvenile hormone (JH) controls numerous essential functions. Its involvement in gene activation is known to be mediated by the transcription factor Methoprene-tolerant (Met), which turns on JH-controlled genes by directly binding to E-box–like motifs in their regulatory regions. However, it remains unclear how JH represses genes. We used the Aedes aegypti female mosquito, in which JH is necessary for reproductive maturation, to show that a repressor, Hairy, is required for the gene-repressive action of JH and Met. The RNA interference (RNAi) screen for Met and Hairy in the Aedes female fat body revealed a large cohort of Met- and Hairy-corepressed genes. Analysis of selected genes from this cohort demonstrated that they are repressed by JH, but RNAi of either Met or Hairy renders JH ineffective in repressing these genes in an in vitro fat-body culture assay. Moreover, this JH action was prevented by the addition of the translational inhibitor cycloheximide (CHX) to the culture, indicating the existence of an indirect regulatory hierarchy. The lack of Hairy protein in the CHX-treated tissue was verified using immunoblot analysis, and the upstream regions of Met/Hairy-corepressed genes were shown to contain common binding motifs that interact with Hairy. Groucho (gro) RNAi silencing phenocopied the effect of Hairy RNAi knockdown, indicating that it is involved in the JH/Met/Hairy hierarchy. Finally, the requirement of Hairy and Gro for gene repression was confirmed in a cell transfection assay. Thus, our study has established that Hairy and its cofactor Gro mediate the repressive function of JH and Met.
Arthropod-specific sesquiterpenoids, juvenile hormones (JH), control numerous essential physiological functions in immature and adult insects, including development, reproduction, pheromone production, and cast differentiation in social insects (1). Methoprene-tolerant (Met), a member of the family of basic helix–loop–helix (bHLH)–Per-Arnt-Sim (PAS) transcription factors, has been recognized as the JH receptor (1). In Drosophila melanogaster, JH action is mediated by two receptors, encoded by the paralogous genes Met and germ-cell-expressed (gce) (1). However, only a single Met gene, an ortholog of Drosophila gce, exists in the genome of the yellow fever mosquito Aedes aegypti, which is at present the most important vector of dengue and chikungunya viruses (2).
JH plays a critical role in reproduction of many insects by synchronizing sexual maturation as well as controlling vitellogenesis and egg production (3–5). JH III coordinates development of reproductive organs in newly eclosed female mosquitoes during the posteclosion (PE) phase which lasts 3–5 d (4, 6, 7). Ovaries become capable of receptor-mediated endocytosis of yolk protein precursors (8). Their primary follicles increase twofold in length, oocytes acquire endocytic organelles, and the follicular epithelium goes through differentiation (6, 9, 10). Similarly, the fat body, a tissue that is analogous to the vertebrate liver, undergoes JH-regulated cellular remodeling, building up secretory machinery for massive production of yolk protein precursors (11, 12). JH action is critical for a female mosquito in attaining responsiveness to 20-hydroxyecdysone (20E), an insect steroid hormone that is a major regulator of vitellogenic events in Dipteran insects. To accomplish this, JH induces the translation of the competence factor, nuclear orphan receptor betaFTZ-F1 (fushi tarazu binding factor 1) (13).
Microarray analysis has shown a high level of differential gene expression during the PE phase in the A. aegypti female fat body (14). The hierarchical clustering identified three major gene clusters: 1,843 early (EPE) genes maximally expressed at 6 h PE, 457 mid (MPE) genes at 24 h PE, and 1,815 late (LPE) genes at 66 h PE. The microarray screen with Met RNAi has demonstrated Met repression of EPE and MPE and activation of LPE genes. Bioinformatics analysis revealed the consensus for a 9-mer Met-binding motif—CACGC/TGA/GT/AG. Met-binding motif variants were overrepresented in promoters of Met up-regulated (LPE) genes, but not in Met down-regulated (EPE/MPE) genes. Electrophoretic mobility shift assays (EMSAs), using a combination of mutational and anti-Met antibody supershift analyses, confirmed binding properties of the Met consensus motif variants (14). Hence, it appears that the Met action on some activated genes is direct, whereas the repressive action of JH/Met requires additional factors, the identity of which has been unknown.
Considering that this question is of general importance for understanding the action of JH at the molecular level in insects, we used A. aegypti female mosquitoes to investigate this question. In these mosquitoes, JH and Met strongly activate transcriptional repressors Krüppel homolog-1 (Kr-h1) and Hairy (15, 16). The former has been implicated in mediating JH and Met signaling during development and metamorphosis (1), whereas the role of Hairy has remained unclear.
D. melanogaster hairy (DmHairy) is a pair-rule gene involved in early patterning of the embryo (17, 18). It plays a key role in the segmentation gene hierarchy by repressing a pair-rule gene, fushi tarazu (ftz) (19) As a long-range repressor, DmHairy has the ability to inhibit the action of activators located more than 1 kb away from the gene transcription start site (20). It can induce a widespread histone deacetylation and inhibit the recruitment of basal transcription machinery without triggering chromatin compaction in the repression of ftz gene (21).
In this work, we have shown that Hairy is an integral part of the JH/Met hierarchy, down-regulating genes in adult insects. Moreover, a corepressor Groucho (Gro) is required for this action.
Results
Identification of Genes Corepressed by Met and Hairy.
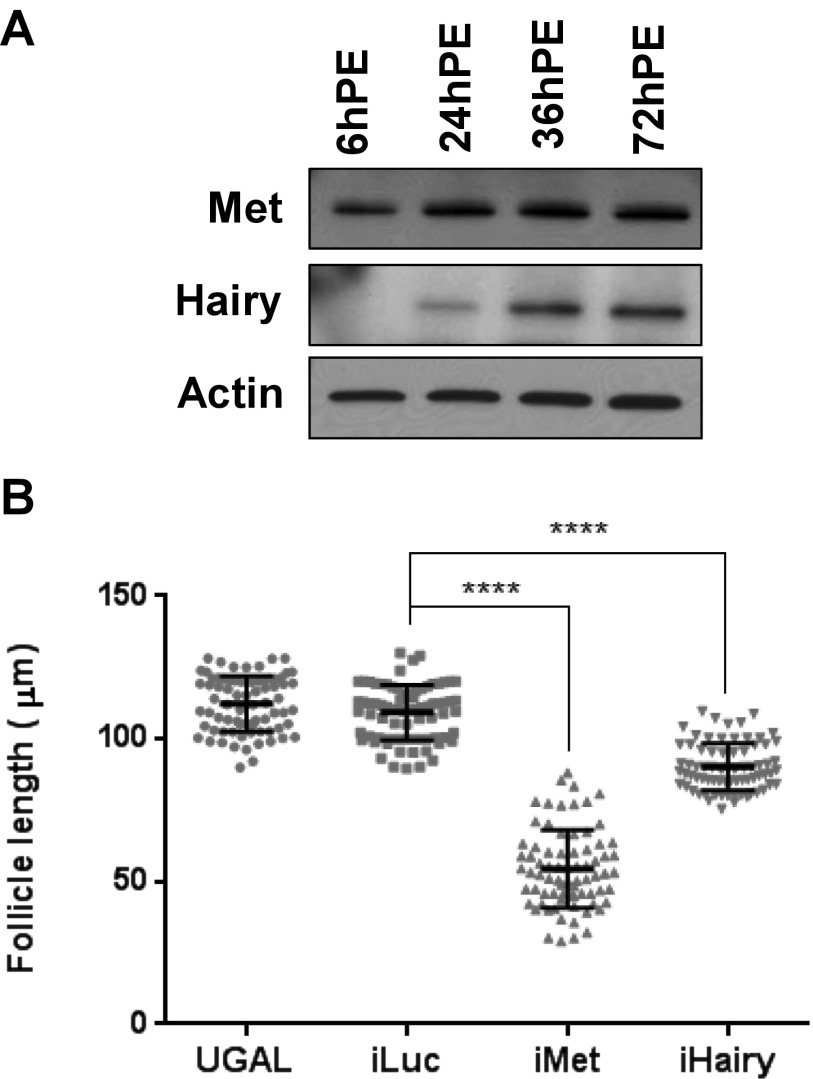
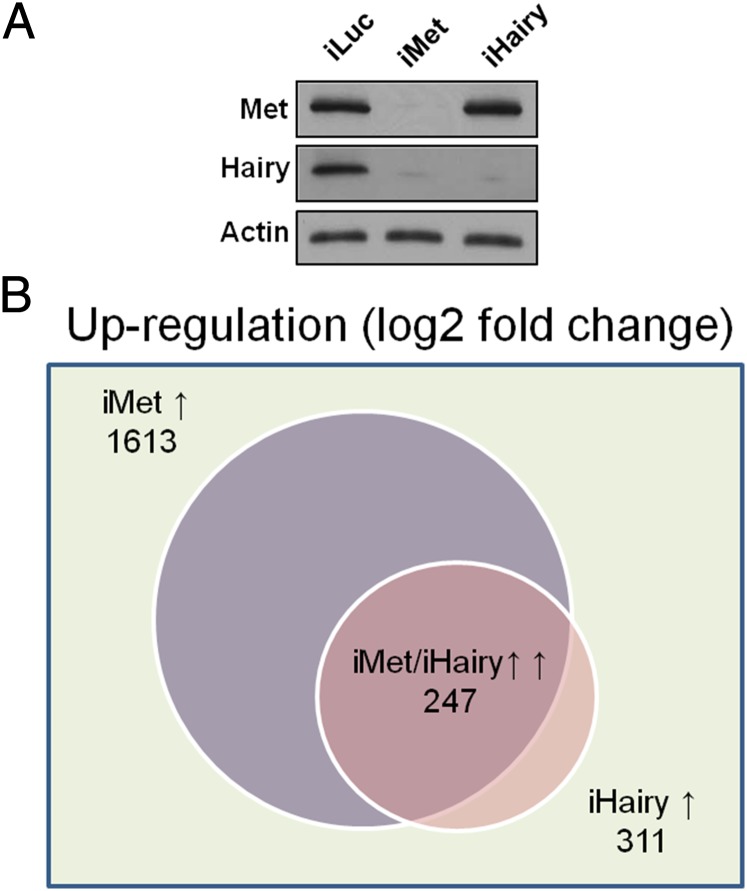
To investigate the role of Hairy as a possible intermediate factor downstream of Met, we performed RNAi-based transcriptomic screens for A. aegypti Met (AaMet) and Hairy (AaHairy). In preparation for this study, we investigated the endogenous levels of Met and Hairy in the A. aegypti female fat body at the protein levels by immunoblotting with anti-Met and anti-Hairy antibodies, respectively. This analysis showed that Met was constitutively present throughout the PE period (Fig. S1A). In contrast, the Hairy level raised significantly during the PE period; it was undetectable at 6 h PE, but was intensified between 24- and 72 h PE (Fig. S1A). The latter suggests that Hairy was regulated during the PE period. Indeed, the immunoblots revealed the absence of Hairy in fat-body samples from mosquitoes with RNAi-mediated depletions of Met (iMet), supporting the hypothesis that it is a factor downstream of Met (Fig. 1A). The level of Met was not altered in fat-body samples from mosquitoes with RNAi-mediated depletions of Hairy (iHairy), ruling out the presence of any feedback control loop (Fig. 1A). The Met and Hairy protein levels were negligible in fat-body samples with their respective RNAi depletions, indicating high efficiency of RNAi (Fig. 1A).
Fig. S1.
(A) WBs showing developmental profiles of Met and Hairy protein throughout PE. Anti–β-actin antibody was used as a loading control. (B) Measurements of primary ovarian follicle length in iHairy, iMet, iLuc, and uninjected (UGAL) mosquito at 72 h PE. All measurements are in micrometers. Error bars represent ± SD. ****P < 0.0001.
Fig. 1.
Illumina RNA-seq analysis of Met and Hairy RNAi-depleted A. aegypti mosquito fat body. (A) WBs showing the presence of Met and Hairy proteins in iLuc, iMet, and iHairy samples. WBs also provided proof of the efficiency of dsRNA-mediated knockdowns. Anti–β-actin antibody was used as a loading control. (B) Venn diagram of Met and Hairy RNAi fat-body transcriptome analysis showing that 79% of iHairy up-regulated genes are also up-regulated by iMet. iMET↑, more than twofold up-regulation by dsRNA depletion of Met (AKA, Met-repressed); iHairy↑, more than twofold up-regulation by dsRNA depletion of Hairy (AKA, Hairy-repressed); iMet/iHairy↑↑, more than twofold up-regulation by dsRNA depletion of Met and Hairy (AKA, Met/Hairy-repressed). All comparisons were made with the control iLuc fat-body transcriptome.
The ovarian maturation of newly eclosed female mosquito is governed by JH and is manifested by the increase in length of the primary ovarian follicles to about 100–120 µm in length by 72 h PE (6), and this increase in follicular length is mediated by Met, providing a direct evidence for its involvement in reproductive maturation in adult female mosquito (14, 15). To investigate a possible effect of Hairy on this stage of reproduction, we evaluated the length of iHairy ovarian follicle compared with those of iMet and controls, iLuc and uninjected UGAL mosquitoes at 72 h PE. The average length of iMet primary follicles was 55 µm, whereas that of iLuc was 109 µm and that of wild-type UGAL was 112 µm (Fig. S1B). A more modest but significant decrease in follicular length was observed in ovaries of iHairy mosquitoes with an average length of 90 µm (Fig. S1B). This experiment suggests importance of Hairy for PE reproductive events.
Illumina RNA-seq technology was used to generate the transcriptional profiles of genes affected by RNAi-mediated depletions of Met (iMet) and Hairy (iHairy) in the fat body of A. aegypti female mosquitoes. RNAi for luciferase (iLuc) served as a control. Three biological replicates of RNA-seq libraries were constructed for each RNAi treatment. Each RNA-seq library contained between 62 and 241 million reads (read length, 101 nucleotides for first and second replicates; and 51 nucleotides for third) (Table S1). The reads were aligned with Bowtie2 against A. aegypti transcript sequence database (AaegL1.3 geneset; Vectorbase). The resulting count table was transformed into fragments per kilobase pair of transcript per million fragments mapped (FPKM) values. Relative transcript abundance of iLuc, iMet, and iHairy, defined as the sum of FPKM values of the three treatment samples, was sorted from highest to lowest.
Table S1.
Summary of RNA-seq experiments
| Replicates | Sample ID | Length | No. reads | % of ≥Q30 bases | Mean quality score |
| First | iLuc 1 | 101 | 241,496,528 | 89.76 | 34.97 |
| iMet 1 | 101 | 125,120,346 | 89.85 | 35.04 | |
| iHairy 1 | 101 | 214,169,272 | 88.58 | 34.66 | |
| Second | iLuc 2 | 101 | 71,899,072 | 95.90 | 37.18 |
| iMet 2 | 101 | 72,167,840 | 95.70 | 37.10 | |
| iHairy 2 | 101 | 66,299,014 | 95.82 | 37.16 | |
| Third | iLuc 3 | 51 | 120,748,264 | 90.13 | 35.21 |
| iMet 3 | 51 | 62,560,173 | 88.57 | 34.48 | |
| iHairy 3 | 51 | 107,084,636 | 91.89 | 35.77 |
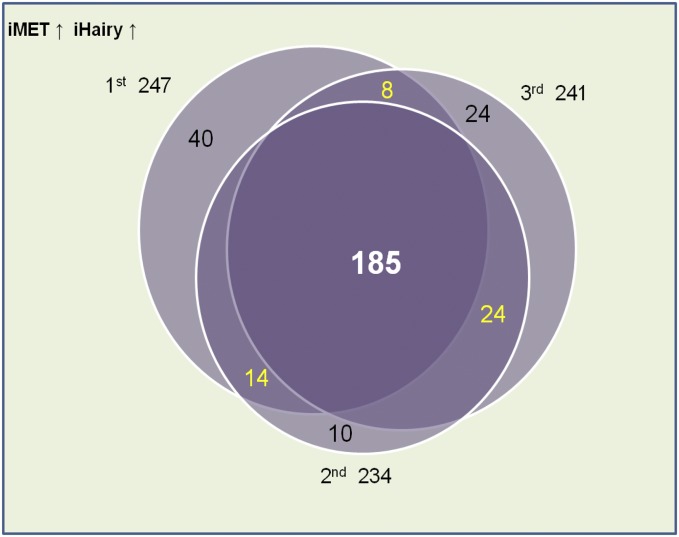
From the first set of RNA-seq experiments, 10,000 transcripts with the highest abundance were selected for further analysis (Dataset S1). The differentially expressed transcripts were defined by a twofold increase or decrease of FPKM values. Among these 10,000 abundantly present transcripts, a total of 2,151 transcripts was identified as having differential expression between iMet and control iLuc mosquitoes, with 1,613 transcripts being activated in iMet (Fig. 1B and Dataset S2). A total of 455 transcripts showed a more than twofold differential expression between iHairy and iLuc treatments, with 311 being up-regulated (Fig. 1B and Dataset S2). Significantly, 80% of iHairy-activated genes were also activated by iMet. In total, 247 common genes were activated by RNAi knockdowns of either AaMet or AaHairy. This large overlap between the two transcriptomes suggests that Met and Hairy are involved in the same hierarchy mediating the JH-repressive action (Fig. 1B). However, a relatively small number of Met-repressed genes corepressed by Hairy suggests the existence of an additional Hairy-independent mechanism for JH/Met gene repression. The reproducibility of results was verified by comparing genes coactivated by iMet and iHairy in three biological replicates. This comparison revealed that 185 genes were coactivated by iMet and iHairy in all three replicates (Fig. S2 and Dataset S3).
Fig. S2.
Venn diagram showing overlaps of up-regulated transcripts in dsRNA-mediated depletions of Met and Hairy from three different biological replicates. The differentially up-regulated transcripts were defined as more than twofold difference in 10,000 abundantly expressed transcripts. Of the up-regulated transcripts, 74% were common among the three RNA-seq libraries, clearly indicating the reproducibility of the parallel experiments.
Ontology Analysis of Genes Corepressed by Met and Hairy.
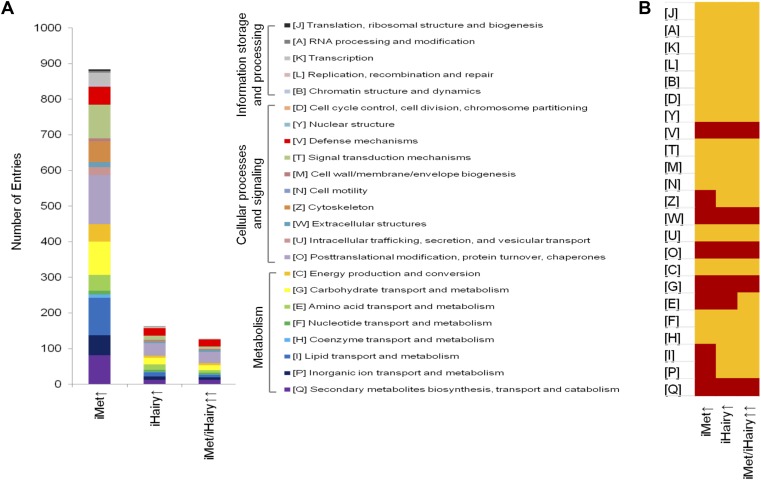
The analysis of orthologous groups (OGs) based on adjusted nonsupervised orthologous groups (NOG) (Table S2 and Dataset S4) revealed that Met- and/or Hairy-repressed transcripts were categorized mostly into two broad groups, one of Cellular Process and Signaling and another of Metabolism (Fig. S3A). The significantly overrepresented OGs (P < 0.01 in a hypergeometric distribution) in Hairy-repressed transcripts were as follows: three in the Cellular Process and Signaling category—[V] defense mechanisms, [W] extracellular structure, and [O] posttranslational modification, protein turnover, chaperones; and three in the Metabolism category—[G] carbohydrate transport and metabolism, [E] amino acid transport and metabolism, and [Q] secondary metabolite biosynthesis, transport, and metabolism (Fig. S3B). All of these OGs were also categorized as Met-repressed. Of these six overrepresented OGs, all but one—[E] Amino acid transport and metabolism—were also overrepresented among AaMet/AaHairy-coregulated transcripts, suggesting the existence of the Met–Hairy hierarchy in gene repression (Fig. S3B). Further, Met- and Hairy-coregulated genes were manually annotated, during which 54 isoform-specific and gene-duplicated transcripts were removed, resulting in 193-curated Met/Hairy-repressed genes (Dataset S5).
Table S2.
Adjusted nonsupervised orthologous groups (NOG) assignments used for functional analysis
| Major functional category | Simplified functional groups | Original functional groups by eggNOG (number) |
| Metabolism | [Q] Secondary metabolites biosynthesis, transport, and catabolism | Q |
| [P] Inorganic ion transport and metabolism | P, PT (6) | |
| [I] Lipid transport and metabolism | I, IJT (4), IKT (2), IM (1), IN (1), IO (1), IOTUV (1), IU (9) | |
| [H] Coenzyme transport and metabolism | H | |
| [F] Nucleotide transport and metabolism | F | |
| [E] Amino acid transport and metabolism | E, EG (1), EH (4), EI (2), EJ (1), ET (2), EU (1) | |
| [G] Carbohydrate transport and metabolism | G, CG (5) | |
| [C] Energy production and conversion | C, CD (3), CH (1), CO (5) | |
| Cellular processes and signaling | [O] Posttranslational modification, protein turnover, chaperones | O, KOT (2), OU (2) |
| [U] Intracellular trafficking, secretion, and vesicular transport | U, CU (3), KU (4), UZ (7) | |
| [W] Extracellular structures | W | |
| [Z] Cytoskeleton | Z | |
| [N] Cell motility | N, DN (1), NT (4), NTZ (1) | |
| [M] Cell wall/membrane/envelope biogenesis | M, MO (3), MOT (2), MU (1), MW (3) | |
| [T] Signal transduction mechanisms | T, IT (6), KT (5), TU (25), TUZ (2), TW (1), TZ (24) | |
| [V] Defense mechanisms | V, TV (25), VW (2) | |
| [Y] Nuclear structure | Y, UY (22) | |
| [D] Cell cycle control, cell division, chromosome partitioning | D, BD (2), BDL (1), BDLT (2), BDT (1), DO (12), DP (2), DT (5), DTUZ (1), DTZ (1), DU (3), DUZ (5), DZ (4) | |
| Information storage and processing | [B] Chromatin structure and dynamics | B, BL (2), BT (3) |
| [L] Replication, recombination, and repair | L | |
| [K] Transcription | K, AK (1), BK (12), DK (5), DKL (1), DKLT (2), JK (1), KL (2), KLO (1), KO (3) | |
| [A] RNA processing and modification | A, ABO (1), AT (2) | |
| [J] Translation, ribosomal structure, and biogenesis | J, JO (2), JT (2), JU (1) |
Fig. S3.
(A) Ontology analysis of gene cohorts differentially expressed in the female fat body after RNAi depletion of Met and Hairy. Three major categories and the functional groups with corresponding abbreviations and colors are indicated. (B) The OGs with significant overrepresentation (P < 0.01 in a hypergeometric distribution) are marked in red. The abbreviations used are same as that of A.
Hairy Acts as an Intermediate Component of the JH/Met Gene-Repressive Hierarchy.
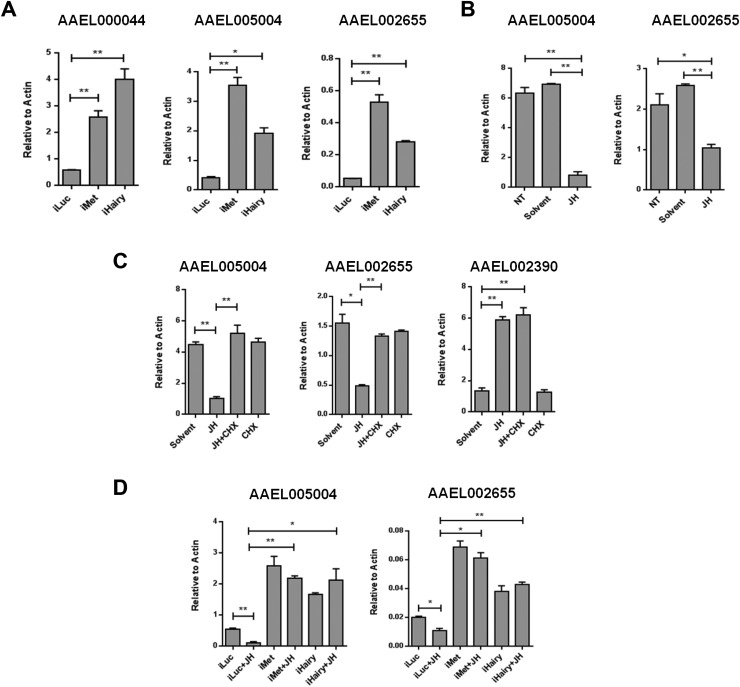
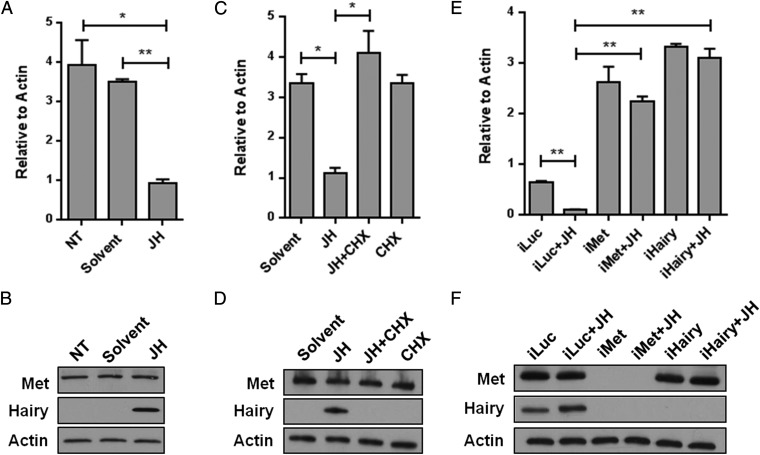
To test the hypothesis that Hairy is an intermediate component of the JH/Met gene-repressive hierarchy, we selected several highly up-regulated genes from 193 genes activated by both iMet and iHairy and cross-examined them using quantitative RT-PCR (qPCR). Compared with iLuc controls, the genes encoding AAEL000044 (Ornithine decarboxylase), AAEL005004 (Unknown), and AAEL002655 (Matrix metalloproteinase) were up-regulated in both iMet and iHairy samples (Fig. S4A). Next, we analyzed the effect of JH on these Met-Hairy–coregulated genes. JH was topically applied on female mosquitoes 3 h PE, when the JH titer is very low, and their fat bodies were collected 8 h later, as described previously (14). qPCR showed that treatment with JH dramatically repressed transcript levels of all of the tested genes (Fig. 2A and Fig. S4B). Immunoblots revealed the presence of a Hairy protein band only in fat-body samples from the JH-treated mosquitoes but not in controls treated with a solvent (acetone). Met protein, however, was detected in both JH- and solvent-treated samples (Fig. 2B).
Fig. S4.
(A) Validation of the expression of AAEL000044, AAEL005004, and AAEL002655 in Met- and Hairy-depleted A. aegypti female mosquito fat bodies. (B) Hormonal application experiments showing the effect of JH on the expression of AAEL005004 and AAEL002655. Both genes are repressed by JH, in addition to AAEL000044 shown in Fig. 2A. NT, no treatment. (C) In vitro fat-body culture experiments using JH and CHX. JH-mediated repression of AAEL005004 and AAEL002655 observed after the addition of JH was compromised in CHX-treated samples (JH+CHX), indicating the involvement of intermediate factors in this gene regulation process. Expression of the direct Met target Krüppel homolog 1 (AAEL002390) was not altered by the presence of CHX in the media. Solvent (acetone)- and CHX-treated samples were used as controls. (D) In vitro fat-body culture experiments with Met- and Hairy-depleted A. aegypti female mosquito fat bodies, showing the necessity of these two factors for the JH-mediated repression of AAEL005004 and AAEL002655. A similar expression of these two genes in iMet+JH and iHairy+JH samples established Hairy function as an intermediate factor downstream of Met in JH-mediated gene repression. All transcript measurements were performed using qPCR, and experiments were repeated thrice with similar results. Error bars represent ± SD. *P < 0.05; **P < 0.01.
Fig. 2.
Hairy acts as an intermediate component of JH/Met gene repression. (A) Effect of JH on the expression of the AAEL000044 gene. JH was applied topically onto the abdomen of newly eclosed female mosquitoes at 6 h PE, and the effect was determined by means of qPCR. NT, no treatment. (B) WB showing the levels of endogenous Met and Hairy proteins in the fat bodies under the same conditions as that of A. (C) In vitro fat-body culture experiments showing the effect of cycloheximide (CHX) on the JH-mediated induction of the expression of the AAEL000044 gene. The effect of JH on the AAEL000044 gene expression was compromised by the addition of CHX to the JH-containing tissue culture medium, as measured by qPCR. Minimal medium without JH and CHX (solvent) and CHX only (CHX) were used as controls. (D) WB showing the effect of JH and CHX on Met and Hairy protein levels under the same conditions as that of C. A clear induction of Hairy but not Met protein by JH is observed. CHX treatment results in the loss of Hairy but not Met protein. (E) Hairy is required for the JH-mediated repression of the AAEL000044 gene. Luc, Met, and Hairy dsRNA-treated fat bodies were cultured in medium with or without JH. The AAEL000044 gene was repressed by JH treatment in iLuc (control), but this repression was compromised in iMet+JH samples. The knockdown of Hairy phenocopied the effects of iMet samples, indicating that Hairy is a downstream component of the JH/Met-mediated gene repression hierarchy. (F) WB for Met and Hairy protein levels in the same sample sets described in E. All experiments were performed in triplicate, with similar results. Error bars represent ± SD. *P < 0.05; **P < 0.01. Additional genes are shown in Fig. S3.
To establish the requirement of intermediate factor(s) and the necessity of de novo translation in the JH-dependent repression, we used an approach using the protein synthesis inhibitor cycloheximide (CHX). Fat bodies of newly eclosed female mosquitoes (6 h PE) were incubated in the JH-containing culture medium in the presence or absence of CHX. Transcript levels of tested genes were high after incubation with medium containing solvent (acetone), but were significantly less in the presence of JH. Addition of CHX to the culture medium rendered these genes unresponsive to the repressive action of JH (Fig. 2C and Fig. S4C). AaKr-h1 (AAEL002390), which is directly controlled by Met (1, 16), served as control in this experiment. CHX was ineffective in preventing the activating effect of JH on this gene (Fig. S4C). Western blot (WB) analysis showed that Hairy was reduced to an undetectable level as a result of CHX addition to the culture media (Fig. 2D). However, Met was not affected by this treatment. This result clearly established the role of intermediate factors downstream of Met in JH-mediated repression of the tested genes. The absence of Hairy in CHX-treated fat bodies strongly supports our hypothesis that JH/Met gene repression functions through Hairy de novo translation for a subset of Met/Hairy-corepressed genes.
To address specifically whether Hairy is involved as an intermediate factor in the JH/Met gene repression hierarchy, we carried out experiments combining RNAi silencing and in vitro fat-body culture, as previously described (14). Fat bodies from mosquitoes with RNAi-depleted Luc, Met, and Hairy were incubated in the presence or absence of JH. JH decreased transcript levels of the analyzed genes in Luc RNAi mosquito fat bodies, whereas RNAi depletion of Met rendered these genes insensitive to JH (Fig. 2E and Fig. S4D). Hairy RNAi silencing had a similar effect to that of Met RNAi on the expression of the tested genes (Fig. 2E and Fig. S4D). WB analysis using Met and Hairy antibodies showed that Hairy was absent after Met RNAi treatment, regardless of whether JH was present (Fig. 2F). This experiment establishes Hairy as the factor downstream of Met in the JH/Met repression hierarchy.
A Corepressor Gro1 Is Involved in the Met/Hairy-Mediated Gene Repression.
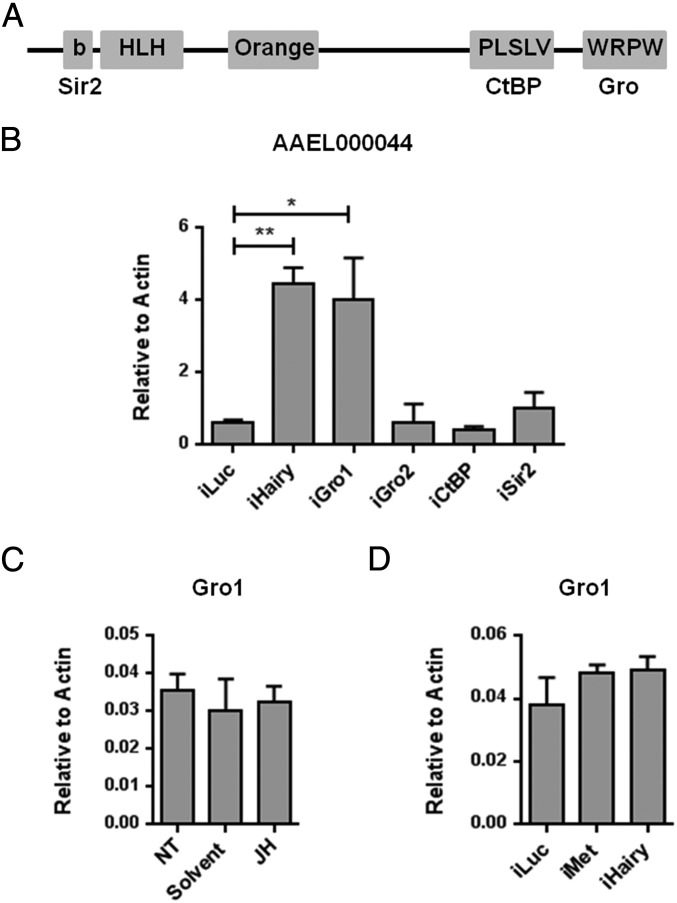
To act as a repressor, Hairy requires recruitment of corepressors (22–25). Hairy carries binding domains for three corepressors—silent information regulator 2 (Sir2), C terminus binding protein (CtBP), and Gro (Fig. 3A). There are two homologs of Drosophila gro in the A. aegypti genome—gro1 (AAEL010433) and gro2 (AAEL005009). To investigate the possible involvement of CtBP (AAEL005400), Sir2 (AAEL005816), and the two Gro in the Met/Hairy-mediated gene repression, we used their respective RNAi knockdowns and monitored the response of the Met/Hairy-regulated gene AAEL000044. RNAi silencing of gro1, but not gro2, CtBP or Sir2, phenocopied the effect of Hairy RNAi depletion on the expression of the AAEL000044 gene, indicating a possible corepressor role for Gro1 (Fig. 3B). Next, we tested the dependency of Gro1 expression on the JH/Met/Hairy cascade. The gro1 gene expression was insensitive to the JH topical application (Fig. 3C). Moreover, RNAi knockdowns of Met and Hairy had no effect on gro1 transcript levels (Fig. 3D). Taken together, these experiments suggest a corepressor role for Aedes Gro1 in the Hairy-mediated JH/Met gene repression hierarchy.
Fig. 3.
AaGro is a corepressor of AaHairy in the JH/Met gene repression hierarchy. (A) Schematic diagram showing the domain composition of Hairy protein and the potential binding sites for various cofactors (based on refs. 23–26). Hairy carries binding domains for three corepressors—silent information regulator 2 (Sir2), C-terminal binding protein (CtBP), and Groucho (Gro). (B) Involvement of putative Hairy corepressors was tested by their respective dsRNA-mediated knockdowns. Gro1 RNAi silencing phenocopied the effect of the Hairy RNAi depletion on the expression of the AAEL000044 gene. (C and D) The Gro1 expression is independent of the JH/MET/Hairy cascade. Gro1 is not regulated by JH as revealed by in vivo hormonal treatment assays (C). Depletions of either Met or Hairy do not affect the expression of Gro1 (D). Expression analyses were performed using qPCR. All experiments were done in triplicate with similar results. Error bars represent ± SD. *P < 0.05; **P < 0.01.
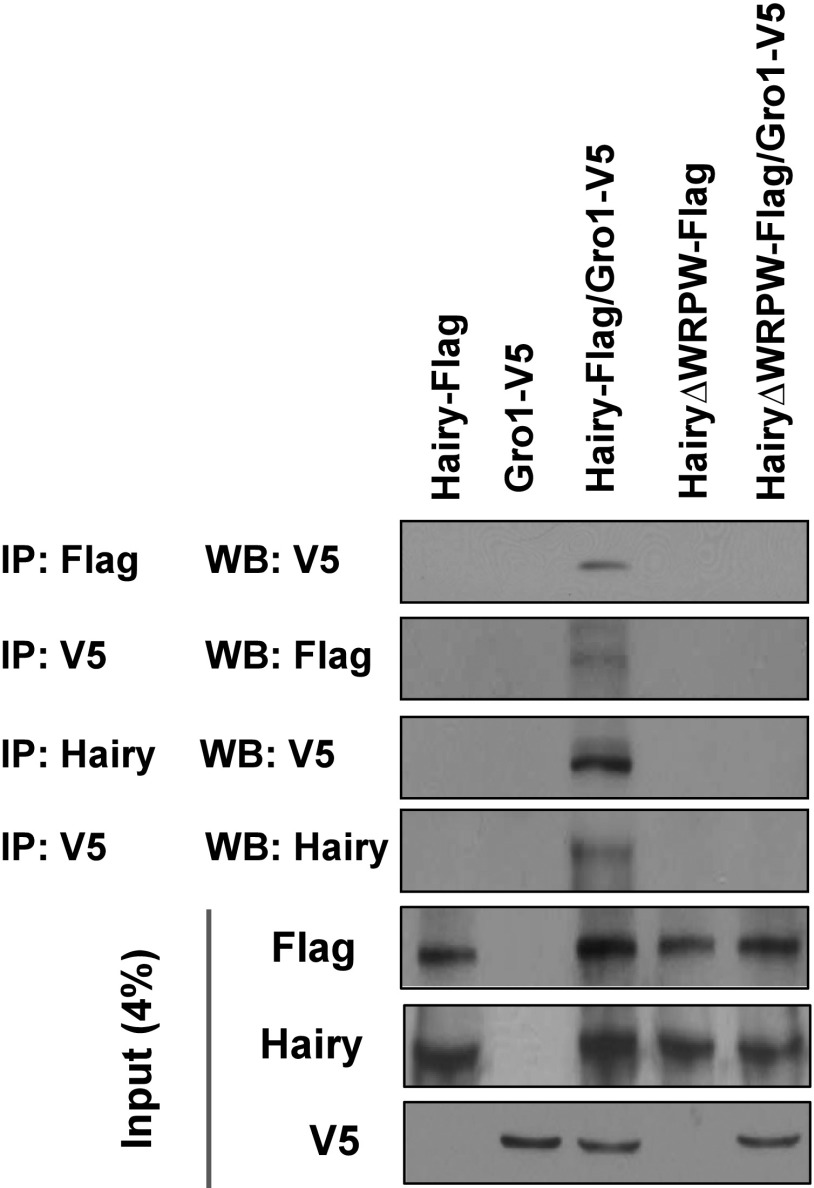
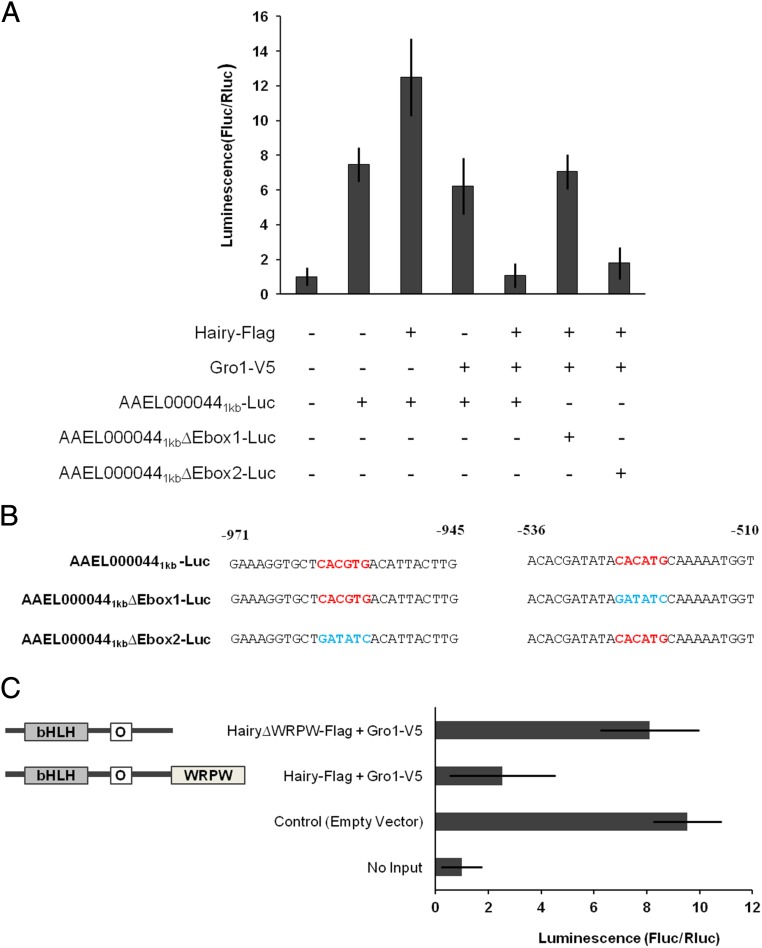
The tetra-amino acid WRPW (Trp-Arg-Pro-Trp) sequence that is located at the Hairy C terminus serves as a protein–protein interaction domain with Gro (26). To test physical interaction between Hairy and Gro1 from A. aegypti, we used coimmunoprecipitation (co-IP) experiments in a cell culture system. Flag-tagged, full-length Hairy (Hairy-Flag) and V5-tagged, full-length Gro1 (Gro1-V5) were cloned and expressed in D. melanogaster S2 cells. After immunoprecipitation (IP) with anti-Flag antibody, Gro1-V5 was detected as a co-IP product in WBs with anti-V5 antibody only when both fusion proteins were coexpressed in the cell culture system (Fig. S5). The protein–protein interaction between A. aegypti Hairy and Gro1 was further confirmed by additional co-IP experiments (IP:V5, WB:Flag; IP:Hairy, WB:V5; IP:V5, WB:Flag), where a clear band could only be detected when both proteins were present (Fig. S5). To test the domain specificity of this binding, a truncated Hairy fusion protein without the Gro1 interacting C terminus WRPW tail (Hairy∆WRPW-Flag) was cloned and transfected to D. melanogaster S2 cells in the presence or absence of Gro1-V5. The loss of Hairy WPRW domain completely compromised the Hairy–Gro1 interaction, as evidenced by the lack of a band in co-IP experiments (Fig. S5). Together, these experiments confirm the interaction between A. aegypti Hairy and Gro1 in a domain-specific manner.
Fig. S5.
Coimmunoprecipitation (co-IP) experiments showing the protein–protein interaction between AaHairy and AaGro1. Hairy-Flag, HairyΔWRPW-Flag, and Gro1-V5 were overexpressed in Drosophila S2 cells in various combinations. The following combinations of antibodies were used for different co-IPs—IP:Flag, WB:V5; IP:V5, WB:Flag; IP:Hairy, WB:V5; IP:V5, WB:Hairy. IP, immunoprecipitation; WB, WB.
Identification of Hairy-Binding Motifs in Met/Hairy-Repressed Genes.
Hairy belongs to the Hairy/Enhancer of split/Deadpan (HES) subclass of repressor bHLH proteins (27). bHLH proteins bind to a canonical E-box motif (CANNTG) that occurs in two classes—class A with consensus sequence CAG/CCTG and class B with consensus sequence CACG/ATG (28). D. melanogaster Hairy binds specifically to a canonical class B site CACGTG, but also interacts with noncanonical site CGCGTG, resulting in a Hairy-binding consensus sequence CA/GCGTG (29, 30). Additionally, previous studies have shown that bHLH proteins interacting with CACGTG site can recognize and bind to an E-box variant with an adenine in place of guanine at the fourth position (28). Based on this information, we used EMSA to test the interaction of in vitro-translated AaHairy with these motifs.
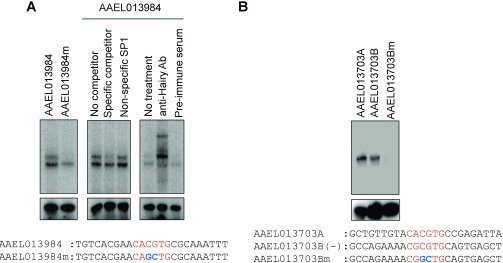
The CA/GCGTG motifs along with their flanking sequences from Met/Hairy-regulated genes AAEL013984 and AAEL013703 interacted specifically with AaHairy (Fig. S6). Hairy formed a band with the CACGTG core and its flanks from the 5′ upstream region of the AAEL013984 gene (Fig. S6A). The binding was abolished after a 2-nt mutation in the E-box sequence to CAGCTG. Excess of the unlabeled specific probe, but not the nonspecific one, competed out the labeled probe resulting in the abolition of the band. A supershift was observed using anti-Hairy polyclonal antibodies, indicating the presence of Hairy in the complex (Fig. S6A). Hairy also formed a band interacting with the CACGTG (AAEL013703A) and CGCGTG (AAEL013703B) cores and their respective flanking sequences from the AAEL013703 gene promoter (Fig. S6B). However, a 2-base mutation in the core CGCGTG to CGGCTG (AAEL013703Bm) abolished the binding, indicating specificity of the E-box–like motif (Fig. S6B).
Fig. S6.
(A) The binding of in vitro-translated Hairy with E-box motif “CACGTG” and its flanks from the promoter region of the Met/Hairy-repressed gene AAEL013984; mutation of CACGTG sequence to “CAGCTG” (AAEL013984m) abolishes DNA–protein interaction. The specificity of the binding was confirmed by competition with unlabeled specific probe, whereas the presence of Hairy in the DNA–protein complex was verified by a supershift with anti-AaHairy polyclonal antibody. (B) The binding of in vitro-translated Hairy with the E-box motifs CACGTG (AAEL013703A) and “CGCGTG” (AAEL013703B) and their corresponding flanking regions from the promoter of Met/Hairy-regulated gene AAEL013703. Mutation of CGCGTG site to “CGGCTG” (AAEL013703Bm) results in disruption of DNA–protein interaction, indicating specificity.
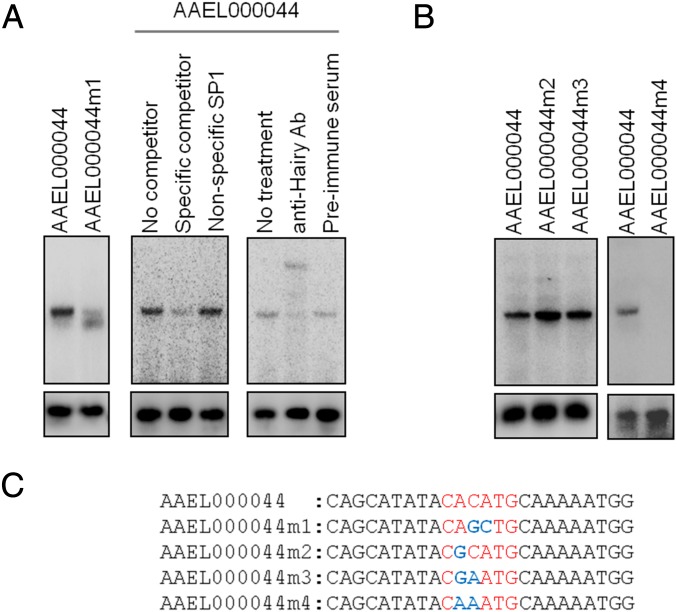
To test AaHairy binding to CA/GCATG motifs, we used oligonucleotide sequences from the promoter region of the Met/Hairy-repressed gene AAEL000044 in EMSA. A strong binding was observed between the CACATG along with its flanking sequence and in vitro-translated Aedes Hairy protein (Fig. 4 A and C). The mutation of the CACATG sequence of AAEL000044 to CAGCTG (AAEL000044m1) almost completely abolished this binding, indicating the binding specificity and verifying the fact that AaHairy does not recognize class A E-box motif (CAG/CCTG). Competition assay with unlabeled specific probe, but not with a nonspecific competitor SP1, resulted in elimination of the band, providing further evidence for specificity of this interaction (Fig. 4A). Addition of anti-Hairy antibody resulted in the supershift of the band, showing that Hairy is a component of the DNA–protein complex (Fig. 4A). Then, we tested the binding of the E-box–like motif CGCATG to AaHairy by mutating the AAEL000044 (CACATG) to AAEL000044m2 (CGCATG), which also resulted in a clear binding between this motif-variant and AaHairy (Fig. 4 B and C). Interestingly, a mutated core-binding motif CGAATG could also be recognized by Hairy. However, the CAAATG sequence was incapable of interaction with Hairy, as evidenced by the lack of DNA–protein complex in EMSA (Fig. 4 B and C). Thus, we have identified previously unknown Hairy-interacting motifs, CA/GCATG and CGAATG, in addition to the canonical CA/GCGTG motif in A. aegypti. The detailed number of Hairy-interacting motifs found in upstream regions of Met/Hairy-repressed genes is provided in Dataset S6.
Fig. 4.
Characterization of Hairy-interacting motifs in A. aegypti JH/Met-repressed genes. (A) EMSAs showing the binding of in vitro-translated Hairy with E-box–like motif “CACATG” and its flanks from the promoter region of the Met/Hairy-repressed gene AAEL000044. The specificity of the binding was confirmed by competition with unlabeled specific probe; the presence of Hairy in the DNA–protein complex was verified by a supershift with anti-AaHairy polyclonal antibody. (B) The binding capacity of in vitro-translated Hairy to variants of E-box–like motif was tested by mutating the CACATG core. Hairy binds to “CGCATG” and CGAATG, but not to “CAAATG” sites. (C) The motifs along with corresponding flanking sequences used for the EMSA assays.
Analysis of Gene Promoters Containing AaHairy-Interacting Motifs in the Cell Transfection Assay.
To test whether Hairy can transrepress gene expression, a 1-kb 5′ upstream regulatory region of the Met/Hairy-repressed AAEL000044 gene harboring the Hairy binding site was cloned into the luciferase reporter vector pGL3basic (AAEL0000441kb-Luc). A sixfold to eightfold basal induction in the reporter activity was observed when AAEL0000441kb-Luc was transfected into D. melanogaster S2 cells (Fig. 5A), rendering this experimental system feasible to address gene repression. Surprisingly, cotransfection of AAEL0000441kb-Luc and the Hairy-Flag expression vector into the cell culture system resulted in an induction of luciferase activity, which was elevated with increasing concentrations of the Hairy vector (Fig. S7A). This result suggested that Hairy by itself was not sufficient to repress the promoter activity and raised the possibility of involvement of additional cofactors. Therefore, the AaGro1-V5 expression vector was cotransfected along with AaHairy-Flag into S2 cells. The dual-luciferase assay revealed a clear repression of the promoter activity in the presence of 200 ng of AaHairy and of AaGro1 expression vectors (Fig. 5A). However, no decrease in luciferase activity by Hairy/Gro1 was observed after the Hairy binding E-box–like motif CACATG in AAEL000044 promoter (AAEL0000441kb∆Ebox1-luc) was mutated, demonstrating the necessity of the DNA–protein interaction for this repression (Fig. 5 A and B). Interestingly, mutation of E-box CACGTG keeping the E-box–like motif CACATG intact (AAEL0000441kb∆Ebox2-luc) did not alter the reporter activity when supplied with Hairy-Flag and Gro1-V5 (Fig. 5 A and B). There could be several possible reasons for this observation including the distance of the “CACGTG” site from the core promoter and the lack of sufficient number of nucleotides on the 5′ end of the motif. No change in the reporter activity was observed when only AaGro1-V5 was transfected in the cell culture system, indicating that AaGro1 by itself is incapable of gene repression (Fig. S7A). Furthermore, the gene repression was compromised when the expression vector with the truncated Hairy lacking a C-terminal Gro-binding WPRW domain was used (Hairy∆WPRW-Flag) along with Gro1-V5 instead of the full-length Hairy (Fig. 5C). Taken together, these experiments prove that AaHairy and AaGro1 are necessary for repression of luciferase activity by the 1-kb promoter of the AAEL000044 gene harboring the Hairy-binding E-box–like motif CACATG.
Fig. 5.
Gro1 is required for the Hairy-mediated gene repression from the AAEL000044 promoter. (A) Luciferase reporter assays after cotransfection of expression vectors Hairy-Flag and/or Gro1-V5 along with the desired reporter constructs. Treatments with no input DNA and empty expression vector served as controls. (B) The putative Hairy binding sites along with flanking regions harbored within the 1-kb region of the AAEL000044 promoter and the mutated sequences for experiments in A are indicated. (C) Luciferase reporter assay with truncated Hairy lacking the C-terminal Gro binding domain. The experimental conditions are similar to above. A clear loss of repression from the 1-kb AAEL000044 promoter was observed when HairyΔWPRW-Flag and Gro1-V5 were cotransfected into the cell culture system. Empty expression vector pAc5.1 was added to normalize the final concentration of DNA. In the graph, readings from no input DNA are represented as 1, with corresponding adjustments for other sample readings. Luminescence represents Firefly to Renilla luciferase readings. Data represent means ± SD (n = 3).
Fig. S7.
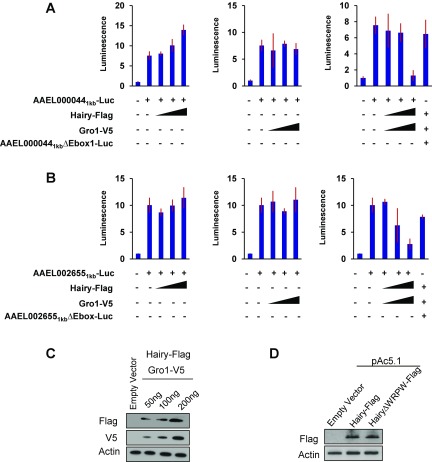
(A) Luciferase assays with reporter construct with 1-kb promoter harboring the E-box–like binding site “CACATG” (AAEL0000441kb-Luc) and increasing concentrations of expression vectors Hairy-Flag or Gro1 or both Hairy-Flag and Gro1-V5. The concentrations of each plasmid used are 50, 100, and 200 ng. A reporter construct with mutated CACATG site (AAEL00441kbΔEbox1-Luc) was also transfected with 200 ng of both Hairy-Flag and Gro1-V5 to validate the necessity of the motifs for Hairy recruitment. (B) Similar experiments as above, performed with reporter constructs with 1-kb promoter region of AAEL002655 harboring the E-box–like binding site “CGAATG” (AAEL0026551kb-Luc) and mutated CGAATG site (AAEL0026551kbΔEbox1-Luc), verify the Hairy-mediated transrepression capacity of a CGAATG site and its flanking promoter. Empty expression vector pAc5.1 was added to normalize the final concentration of DNA. In the graphs, a reading from no-input DNA (left-most column) is represented as 1, with corresponding adjustments for other sample readings. Luminescence represents Firefly to Renilla luciferase readings. Data represent means ± SD (n = 3). (C) WBs showing the expression of tagged proteins Hairy and Gro1. (D) WBs showing the expression of tagged truncated Hairy protein lacking Gro-interacting domain (HairyΔWPRW-Flag). Actin was used as loading control.
Similarly, the 1-kb promoter region of the AAEL002655 gene harboring the Hairy-binding motif variant “CGAATG” was also cloned into a reporter vector and transfected to S2 cells along with increasing concentrations of Hairy-Flag, Gro1-V5, or both. A clear repression could only be observed when both factors were present (Fig. S7B). The repression of luciferase activity was lost when CGAATG was mutated to CGGCTG, indicating this motif’s capacity to act as a functional Hairy-binding element leading to gene repression (Fig. S7B). The expression of the tagged proteins AaHairy-Flag, AaGro1-V5, and AaHairy∆WPRW-Flag in the cell culture system was confirmed by WBs using anti-Flag and anti-V5 antibodies (Fig. S7 C and D).
The Met/Hairy/Gro Repression Hierarchy Is Conserved in Anopheles gambiae Mosquitoes.
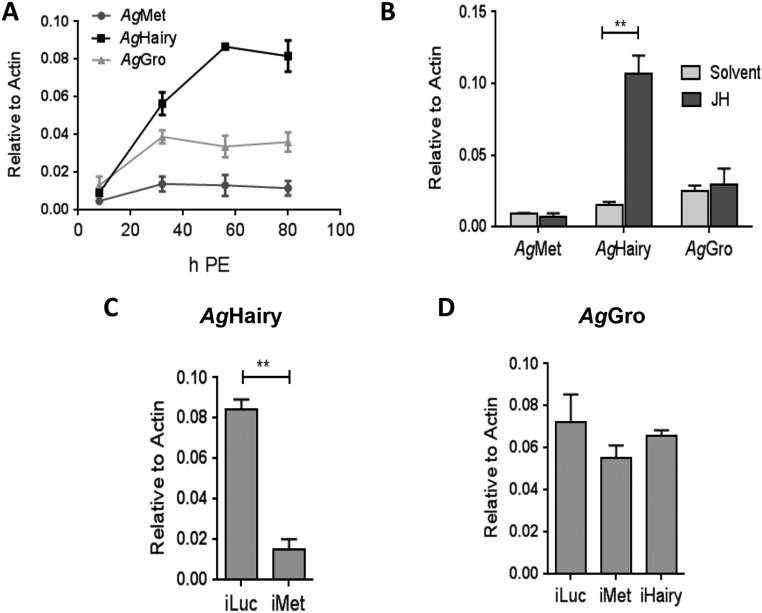
We investigated whether the Met/Hairy/Gro signaling cascade was conserved. To test this, we used another major vector mosquito, A. gambiae, which transmits malaria pathogens. First, we characterized whether Hairy is a downstream factor of the JH/Met pathway in A. gambiae. We examined temporal expression patterns of AgHairy, AgMet, and AgGro during the PE development of female A. gambiae mosquitoes. We found that expression patterns of these factors were similar to those of A. aegypti; the AgHairy transcript showed a gradual increase in expression throughout the PE phase, reaching a peak at 60 h PE, whereas transcript levels of AgMet and AgGro remained constant during this phase (Fig. S8A). JH induced expression of AgHairy but had no effect on AgMet or AgGro (Fig. S8B). RNAi-mediated AgMet knockdown resulted in a dramatic decrease in the abundance of AgHairy mRNA (Fig. S8C). AgGro showed no change in expression in either iMet or iHairy mosquitoes (Fig. S8D). These results reveal that AgHairy is a downstream factor of the JH/Met pathway.
Fig. S8.
(A–D) AgHairy is a downstream factor of the JH/Met pathway. Expression profiles of AgMet, AgHairy, and AgGro during the PE development as measured by qPCR (A). Hormonal application experiments showing the effect of JH on the mRNA levels of AgMet, AgHairy, and AgGro (B). The effect of dsRNA-mediated depletion of AgMet on AgHairy transcript levels (C). The effect of dsRNA-mediated depletion of AgMet or AgHairy on the transcript level of AgGro (D). All expression analyses were done using qPCR. All experiments were done in triplicate with similar results. Error bars represent ± SD. *P < 0.05; **P < 0.01.
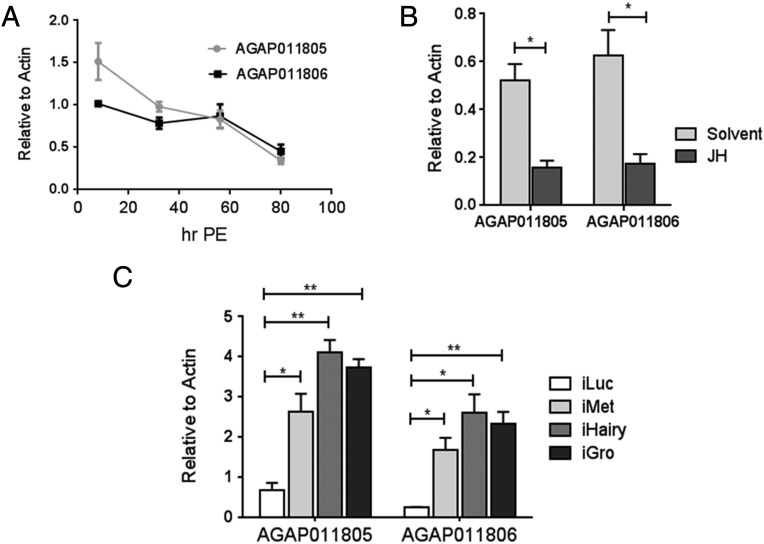
Next, we probed whether this AgMet/AgHairy cascade was involved in the repression of JH-regulated genes. A. gambiae orthologs of the gene encoding A. aegypti Ornithine decarboxylase (AAEL000044) was used to address this question. There are two paralogs of this gene in the A. gambiae genome (AGAP0011805 and AGAP0011806), both of which were assayed in this study. Transcripts of both these genes showed a similar pattern of expression, being highly up-regulated after adult eclosion and gradually decreasing to basal levels by late PE (Fig. 6A). Expression of both genes was down-regulated by the ectopic JH application in a manner similar to that of their A. aegypti ortholog (Fig. 6B). Thus, these duplicated genes were good candidates to test the existence of the Met/Hairy-Gro gene repression hierarchy in A. gambiae. Both AGAP0011805 and AGAP0011806 were transcriptionally up-regulated in A. gambiae iMet mosquitoes (Fig. 6C). RNAi-mediated knockdown of either AgHairy or AgGro phenocopied this induction, demonstrating conservation of the role of Hairy and Gro in mediating the JH/Met signaling in gene repression during reproductive development of adult female mosquitoes (Fig. 6C).
Fig. 6.
JH/Met/Hairy-Gro signaling pathway regulates Anopheles ortholog of Aedes AAEL000044 gene. (A) Expression profiles of AGAP011805 and AGAP011806 during PE development, as measured using qPCR. (B) Hormonal application experiments showing that both AGAP011805 and AGAP011806 genes are repressed by JH. (C) The effect of dsRNA-mediated depletion of AgMet or AgHairy or AgGro on transcript levels of AGAP011805 and AGAP011806 compared with iLuc controls. All expression analyses were done using qPCR. All experiments were done in triplicate with similar results. Error bars represent ± SD. *P < 0.05; **P < 0.01.
Discussion
In this study, we found that Hairy and its corepressor Gro mediate the action of JH and Met in gene repression. The initial evidence of Hairy involvement originated from the RNAi screen for Met and Hairy showing that 80% of Hairy-repressed genes were also found in the Met-repressed transcriptome. Additional support came from the immunoblot analysis using anti-AaMet and anti-AaHairy antibodies that revealed the absence of Hairy in iMet-treated fat bodies, suggesting that Hairy is a factor downstream of Met. Using topical application of JH onto female mosquitoes, we demonstrated that this hormone repressed selected genes from the gene cohort, which were corepressed by Met and Hairy. Similar to AaMet RNAi, AaHairy RNAi silencing rendered this JH action ineffective in the in vitro fat-body culture assay, once again showing that Hairy mediates JH action along with Met. A classic approach using CHX, described by Ashburner et al. (31) for the ecdysone gene regulatory hierarchy, established the requirement of an intermediate factor in the JH-dependent repression in female mosquitoes. JH repression of tested genes was prevented by CHX in the fat-body culture, and Hairy protein was reduced to an undetectable level as a result of the CHX treatment. This further substantiates our hypothesis that Hairy serves as a downstream factor in the JH/Met gene repression hierarchy.
As mentioned above, D. melanogaster Hairy binds to a canonical E-box class B site CACGTG but also interacts with a noncanonical site CGCGTG, resulting in a Hairy-binding consensus sequence CA/GCGTG (29, 30). Using EMSA, we have identified different Hairy-interacting motifs, CA/GCATG and CGAATG, in addition to the canonical CA/GCGTG motif, in A. aegypti. Functionality of the newly identified Hairy-interacting motifs was confirmed in the cell transfection assay.
Hairy recruits corepressors essential for the down-regulation of target genes during D. melanogaster embryo and larval development. The best-studied Hairy corepressor is Gro, which directly interacts with Hairy through its conserved C-terminal tetrapeptide, WPRW domain (22, 26). Other than Gro, alternative Hairy-related proteins can also interact with another cofactor, CtBP, through its PLSLV domain situated immediately upstream of the WPRW Gro binding domain (23). Studies in D. melanogaster indicate that CtBP and Gro regulate separate sets of Hairy-mediated genes affecting different pathways of transcriptional repression (32, 33). Here, we provide evidence that implicates AaGro1 as a corepressor in the Met/Hairy-mediated gene hierarchy, required for Hairy to function. We established this Gro1 role using RNAi knockdowns for Sir2, CtBP, and Gro1 and Gro2, and monitoring a response of the Met/Hairy-regulated gene AAEL000044. Additionally, the cell transfection assay demonstrated that Gro1 is required for the Hairy repressive action. Moreover, we demonstrated by means of co-IP and cell transfection assays that AaHairy interacts with Gro1 via the conserved C terminus WPRW domain. During D. melanogaster development, Gro is broadly present and it is the availability of Hairy that dictates the target gene repression (34). When Hairy is expressed, it associates with Gro resulting in the repression of a target gene. We found that the AaHairy level increased by the end of the female mosquito PE phase under the control of JH and Met, whereas AaGro is constitutively present, suggesting existence of the similar Hairy/Gro regulatory system in A. aegypti.
In addition to its role as a pair-rule gene that is involved in early patterning of the D. melanogaster embryo (17–19), Hairy also regulates other developmental processes: sex determination (22), peripheral nervous system development during larval development (22, 29, 30), and morphogenetic furrow progression in the developing eye (35). It has likewise been implicated in metabolic suppression of hypoxia tolerance in D. melanogaster (36). This Hairy–Gro repression molecular axis is conserved in vertebrates, in which Gro homologs, known as Transducin-like enhancers (TLEs), physically interact with the Hairy homolog HES (37).
In this work, we have identified another role of Hairy as a downstream factor of Met in the JH gene repression hierarchy in A. aegypti mosquitoes. We also uncovered that this role is conserved in A. gambiae mosquitoes. The A. gambiae orthologs of the AAEL000044 gene are repressed by JH/AgMet indirectly through AgHairy. Furthermore, Gro is involved as a Hairy corepressor in this hierarchy in A. gambiae. Thus, our findings indicate that Met employs an evolutionarily conserved Hairy–Gro molecular system to mediate the JH action in gene repression. Future work should establish how widely this system occurs in insects of other orders.
Experimental Procedures
Experimental Animals.
A. aegypti and A. gambiae mosquito larvae were cultured at 27 °C in water supplemented with a mixture of yeast and rat chow (1:1 ratio). Adult mosquitoes were maintained in specifically designed cages at 27 °C, 86% humidity, and supplied with unlimited access to 10% (wt/vol) sucrose solution and water. All dissections were performed in Aedes physiological solution (38). Four-day-old female A. aegypti and A. gambiae mosquitoes were blood fed on white rats and white Leghorn chickens, respectively. All procedures for the use of vertebrate animals were approved by University of California, Riverside Institutional Animal Care and Use Committee.
dsRNA-Mediated Gene Silencing.
dsRNA synthesis was performed using the MEGAscript kit (Ambion), as previously described (14). The bacterial luciferase gene was used to generate control iLuc dsRNA. Microinjection of dsRNA into the thorax of CO2-anesthetized, 1-d-old female mosquitoes PE was performed using Picospritzer II (General Valve). Mosquito fat bodies were collected 4 d postinjection. Primers used for dsRNA synthesis are listed in Dataset S7.
Illumina RNA-seq Library Preparation.
At 4 d postinjection, 10 fat bodies were collected from female mosquitoes injected with Luc (control), Met, or Hairy dsRNA. Total RNA was extracted using TRIzol (Gibco/BRL) and RNA-seq libraries prepared using TruSeq RNA Library Preparation Kit (Illumina). Three independent biological replicates of the experiment were performed, resulting in three RNA-seq libraries per treatment.
Ontology Analyses of Aedes Transcriptome.
To examine the ontology of the genes, Evolutionary Genealogy of Genes: Nonsupervised Orthologous Groups (eggNOG) database, version 3.0, was used, which is constructed through identification of reciprocal best BLAST matches and triangular linkage clustering (39). The OGs are annotated with functional descriptions along with functional categories, which were derived from the original Clusters of Orthologous Groups/Eukaryotic Orthologous Groups categories.
In Vivo Hormonal Treatment.
A 0.3-µL aliquot of 1 µg/mL JH III (Sigma) or solvent (acetone) was topically applied to the abdomen of newly emerged female mosquitoes (3 h PE). Samples were collected 8 h posttreatment and subjected to qPCR and WB analysis. The efficiency of the hormonal treatment was tested by induction of established JH-activated genes.
In Vitro Fat-Body Culture.
Fat-body tissue culture experiments were performed as previously described (16). The complete medium was supplemented with 10 nM JH III or solvent (acetone). For CHX treatments, 20 µM CHX (dissolved in DMSO) was added to the culture medium, with or without JH. The controls were treated with DMSO only. All incubations lasted 6 h, after which samples were collected for qPCR and WB analysis.
RNA Extraction and qPCR Analysis.
RNA extraction, cDNA synthesis, and qPCR analysis were performed as previously described (14). All of the primers used for qPCR are listed in Dataset S7.
WB Analysis.
cDNAs encoding the full-length AaHairy (AAEL005480) and C-terminal region of AaMet (AAEL001746) were subcloned into the prokaryotic expression vector pGEX-4T-1. The plasmids were expressed in an Escherichia coli expression system and, following protein purification, antibodies were produced in rabbits. Antisera against both AaHairy and AaMet were purified by affinity chromatography using the ImmunoPure IgG purification kit (Pierce). Fat-body protein sample collections and WB analysis were performed as previously described (40, 41). β-Actin was used as a loading control; it was detected by the specific mouse monoclonal antibody (Sigma).
Coimmunoprecipitation.
Aedes Hairy and Gro1 cDNA were PCR amplified and cloned into pAc5.1 vectors (Invitrogen) with a C-terminal Flag (Hairy-Flag) and V5 (Gro1-V5) tag, respectively. These two plasmids were transfected separately or together in Drosophila S2 cells using FuGENE reagent (Promega). Cells were harvested after 48 h incubation, and co-IP experiments were performed using Protein G Immunoprecipitation Kit (Roche), as previously described (16). IPs and WBs were performed with various combinations of the following antibodies: mouse monoclonal anti-Flag (GeneScript), mouse monoclonal anti-V5 (Invitrogen), and rabbit polyclonal anti-AaHairy. To address the domain specificity of the Hairy–Gro interaction, Flag-tagged truncated Hairy lacking the Gro binding WRPW domain (Hairy∆WRPW-Flag) was also cloned, transfected with or without Gro1-V5, in S2 cells. For input control, Hairy-Flag, Gro1-V5, and Hary∆WRPW-Flag were transfected separately or together in D. melanogaster S2 cells and detected using WB with the antibodies mentioned above.
EMSA.
Full-length A. aegypti Hairy cDNA was cloned into pcDNA3.1/Zeo(+) (Invitrogen). Hairy protein was synthesized using a coupled in vitro transcription–translation system (Promega). The specific synthesis of Hairy protein was confirmed by labeling it with 35S-methionine followed by detection of the appropriate-sized band in autoradiography. EMSAs were performed with [γ-32P]ATP-labeled annealed oligonucleotide and the in vitro-translated Hairy using a gel shift assay system (Promega). The DNA–protein complex was separated on 5% (wt/vol) TBE Criterion Precast Gel (Bio-Rad) and visualized by means of autoradiography. For competition assays, 50-fold unlabeled specific motif or unlabeled SP1 motif was incubated with Hairy for 10 min and then further incubated with labeled motif for 20 min. For supershift assay, polyclonal antibodies against AaHairy were used.
Luciferase Reporter Assay.
The 1-kb promoter regions of AAEL000044 and AAEL002655 were PCR amplified and subcloned into reporter vector PGL3basic (Promega), and their sequences were confirmed. The putative Hairy-binding site was mutated by incorporating a restriction site in place of E-box or E-box–like sequence in reporter plasmids. Expression plasmids Hairy-Flag, Gro1-V5, and Hairy∆WPRW-Flag were used. Transient transfections were carried out in Drosophila S2 cells, as mentioned above. Samples of 200 ng of desired reporter plasmid along with 50 ng of the control Renilla luciferase reporter vector pRLCMV (Promega). Renilla luciferase reporter was cotransfected with 200 ng of Hairy-Flag and Gro1-V5, either separately or together. For concentration gradients of Hairy and Gro1, increasing amounts of the expression vectors (50, 100, and 200 ng) were transfected. For validation of the necessity of Hairy–Gro1 interaction for gene repression, 200 ng of expression plasmid with truncated Hairy that lacks Gro-interacting motif (Hairy∆WPRW-Flag) was used, keeping all experimental conditions the same as above. Accordingly, the empty expression vector pAc5.1 was added to normalize the final concentration of DNA in each well. Luciferase assays were performed using the Dual Luciferase Assay Kit (Promega), as described previously (42).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 AI036959 (to A.S.R.), Strategic Priority Research Program of the Chinese Academy of Sciences XDB11030600, National Basic Research Program of China 2014CB138405 (to Z.Z.), and Fellowship 201208500073 from China Scholarship Council, Chinese Ministry of Education (to W.D.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1474.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523838113/-/DCSupplemental.
References
- 1.Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- 2.Baumann A, Fujiwara Y, Wilson TG. Evolutionary divergence of the paralogs Methoprene tolerant (Met) and germ cell expressed (gce) within the genus Drosophila. J Insect Physiol. 2010;56(10):1445–1455. doi: 10.1016/j.jinsphys.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt GR, Davey KG. Cellular and molecular actions of juvenile hormone. II. Roles of juvenile hormone in adult insects. Adv Insect Physiol. 1996;26:1–155. [Google Scholar]
- 4.Raikhel AS, Brown MR, Belles X. Hormonal control of reproductive processes. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol 3. Elsevier; Boston: 2005. pp. 433–491. [Google Scholar]
- 5.Bilen J, Atallah J, Azanchi R, Levine JD, Riddiford LM. Regulation of onset of female mating and sex pheromone production by juvenile hormone in Drosophila melanogaster. Proc Natl Acad Sci USA. 2013;110(45):18321–18326. doi: 10.1073/pnas.1318119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gwadz RW, Spielman A. Corpus allatum control of ovarian development in Aedes aegypti. J Insect Physiol. 1973;19(7):1441–1448. doi: 10.1016/0022-1910(73)90174-1. [DOI] [PubMed] [Google Scholar]
- 7.Clements AN. 1992. The Biology of Mosquitoes: Development, Nutrition, and Reproduction (Chapman and Hall, London), Vol 1.
- 8.Raikhel AS, Dhadialla TS. Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol. 1992;37:217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- 9.Raikhel AS, Lea AO. Previtellogenic development and vitellogenin synthesis in the fat body of a mosquito: An ultrastructural and immunocytochemical study. Tissue Cell. 1983;15(2):281–299. doi: 10.1016/0040-8166(83)90023-x. [DOI] [PubMed] [Google Scholar]
- 10.Raikhel AS, Lea AO. Control of follicular epithelium development and vitelline envelope formation in the mosquito; role of juvenile hormone and 20-hydroxyecdysone. Tissue Cell. 1991;23(4):577–591. doi: 10.1016/0040-8166(91)90015-l. [DOI] [PubMed] [Google Scholar]
- 11.Raikhel AS, Lea AO. Hormone-mediated formation of the endocytic complex in mosquito oocytes. Gen Comp Endocrinol. 1985;57(3):422–433. doi: 10.1016/0016-6480(85)90224-2. [DOI] [PubMed] [Google Scholar]
- 12.Raikhel AS, Lea AO. Juvenile hormone controls previtellogenic proliferation of ribosomal RNA in the mosquito fat body. Gen Comp Endocrinol. 1990;77(3):423–434. doi: 10.1016/0016-6480(90)90233-c. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, Chen L, Raikhel AS. Posttranscriptional control of the competence factor betaFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100(23):13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou Z, et al. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci USA. 2013;110(24):E2173–E2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, Busche JM, Zhang X. Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem Mol Biol. 2010;40(1):23–29. doi: 10.1016/j.ibmb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Shin SW, Zou Z, Saha TT, Raikhel AS. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc Natl Acad Sci USA. 2012;109(41):16576–16581. doi: 10.1073/pnas.1214209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll SB, Laughon A, Thalley BS. Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes Dev. 1988;2(7):883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- 18.Hooper KL, Parkhurst SM, Ish-Horowicz D. Spatial control of hairy protein expression during embryogenesis. Development. 1989;107(3):489–504. doi: 10.1242/dev.107.3.489. [DOI] [PubMed] [Google Scholar]
- 19.Ingham PW, Pinchin SM, Howard KR, Ish-Horowicz D. Genetic analysis of the hairy locus in Drosophila melanogaster. Genetics. 1985;111(3):463–486. doi: 10.1093/genetics/111.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16(10):2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li LM, Arnosti DN. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr Biol. 2011;21(5):406–412. doi: 10.1016/j.cub.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paroush Z, et al. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79(5):805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 23.Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17(7):2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phippen TM, et al. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J Biol Chem. 2000;275(48):37628–37637. doi: 10.1074/jbc.M004234200. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109(4):447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- 26.Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16(6):2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushlow CA, et al. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989;8(10):3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV, Dolde C, Gillison ML, Kato GJ. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8(22):2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 30.Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: Hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8(22):2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, Chihara C, Meltzer P, Richards G. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:655–662. doi: 10.1101/sqb.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- 32.Bianchi-Frias D, et al. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2(7):E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96(2):535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18(5):435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Brown NL, et al. daughterless is required for Drosophila photoreceptor cell determination, eye morphogenesis, and cell cycle progression. Dev Biol. 1996;179(1):65–78. doi: 10.1006/dbio.1996.0241. [DOI] [PubMed] [Google Scholar]
- 36.Zhou D, et al. Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: Hairy as a metabolic switch. PLoS Genet. 2008;4(10):e1000221. doi: 10.1371/journal.pgen.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12(13):1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 38.Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37(12):1317–1326. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell S, et al. eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40(Database issue):D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen IA, Attardo GM, Roy SG, Raikhel AS. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J Biol Chem. 2005;280(21):20565–20572. doi: 10.1074/jbc.M500712200. [DOI] [PubMed] [Google Scholar]
- 41.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mane-Padros D, Cruz J, Cheng A, Raikhel AS. A critical role of the nuclear receptor HR3 in regulation of gonadotrophic cycles of the mosquito Aedes aegypti. PLoS One. 2012;7(9):e45019. doi: 10.1371/journal.pone.0045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.