Significance
Urinary tract infection is the most common nonepidemic bacterial infection in humans, with 150 million cases per year and a global health care cost above $6 billion. Because the urinary tract is not protected by mucus, mammals produce a molecular net that captures pathogenic bacteria in the urine and clears them from the body. By visualizing the 3D structure of its building block, glycoprotein uromodulin, we provide insights into how the net is built, and how it is compromised by mutations in patients with kidney diseases. Our work also explains nonsyndromic deafness due to mutations affecting the tectorial membrane, a similar filamentous structure in the human inner ear.
Keywords: uromodulin, ZP2, polymerization, zona pellucida domain, X-ray crystallography
Abstract
Uromodulin (UMOD)/Tamm–Horsfall protein, the most abundant human urinary protein, plays a key role in chronic kidney diseases and is a promising therapeutic target for hypertension. Via its bipartite zona pellucida module (ZP-N/ZP-C), UMOD forms extracellular filaments that regulate kidney electrolyte balance and innate immunity, as well as protect against renal stones. Moreover, salt-dependent aggregation of UMOD filaments in the urine generates a soluble molecular net that captures uropathogenic bacteria and facilitates their clearance. Despite the functional importance of its homopolymers, no structural information is available on UMOD and how it self-assembles into filaments. Here, we report the crystal structures of polymerization regions of human UMOD and mouse ZP2, an essential sperm receptor protein that is structurally related to UMOD but forms heteropolymers. The structure of UMOD reveals that an extensive hydrophobic interface mediates ZP-N domain homodimerization. This arrangement is required for filament formation and is directed by an ordered ZP-N/ZP-C linker that is not observed in ZP2 but is conserved in the sequence of deafness/Crohn’s disease-associated homopolymeric glycoproteins α-tectorin (TECTA) and glycoprotein 2 (GP2). Our data provide an example of how interdomain linker plasticity can modulate the function of structurally similar multidomain proteins. Moreover, the architecture of UMOD rationalizes numerous pathogenic mutations in both UMOD and TECTA genes.
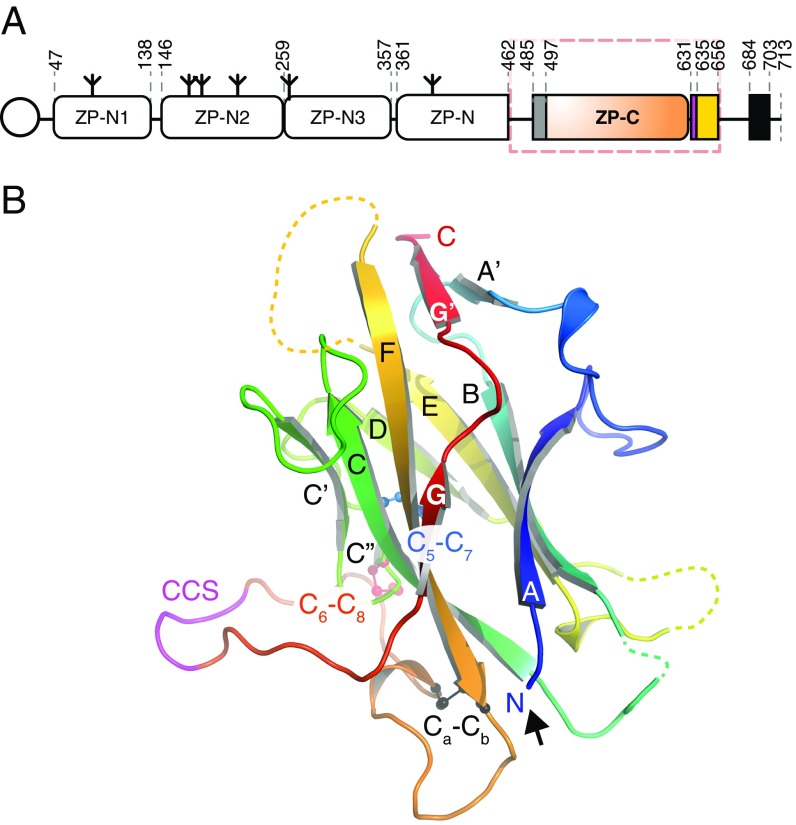
Uromodulin (UMOD) is expressed in the thick ascending limb of Henle’s loop as a GPI membrane-anchored precursor that consists of three EGF-like domains, a domain of unknown function (D8C), and a zona pellucida (ZP) module (1, 2) (Fig. 1A, Top). The latter, containing Ig-like domains ZP-N and ZP-C (3–5), is found in other medically important human glycoproteins linked to infertility (egg coat components ZP1–ZP4), nonsyndromic deafness [inner ear α- and β-tectorin (TECTA/B)], Crohn’s disease [glycoprotein 2 (GP2)], and cancer [TGF-β coreceptors betaglycan (BG) and endoglin (ENG)] (6, 7). Upon processing by Ser protease hepsin (8) at a consensus cleavage site (CCS) C-terminal to the ZP module (9), UMOD sheds a C-terminal propeptide (CTP) that contains a polymerization-blocking external hydrophobic patch (EHP), exposing an internal hydrophobic patch (IHP). This event triggers homopolymerization into filaments that are excreted into the urine (4, 10), where UMOD performs a plethora of biological functions, including protection against urinary tract infections, prevention of kidney stones, and activation of innate immunity (1, 2, 11, 12).
Fig. 1.
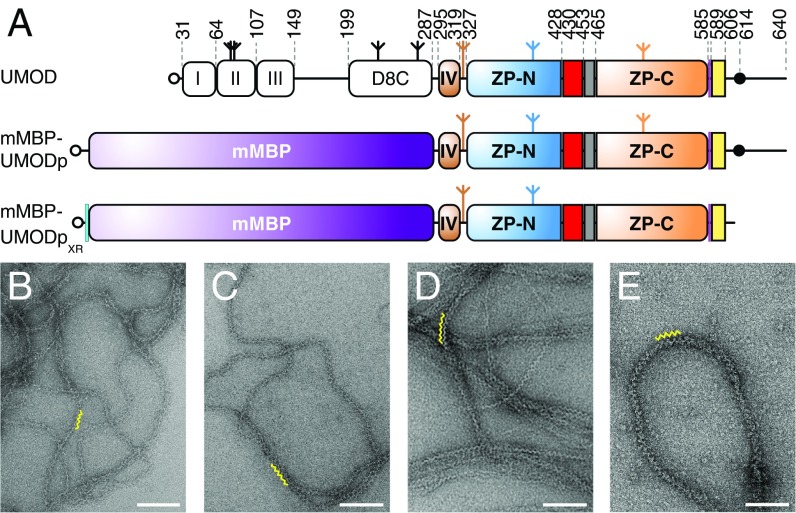
mMBP-fused UMODp forms filaments like native urinary UMOD. (A) Domain organization of urinary UMOD and recombinant constructs mMBP-UMODp and mMBP-UMODpXR. EGF domains are indicated by roman numerals. EGF IV identified by this study (brown), ZP-N/ZP-C linker (red), IHP (gray), CCS (magenta), CTP (yellow), and 6His-tag (cyan) are shown. Open circles, inverted tripods, and closed circles represent signal peptides, N-glycans, and GPI anchors, respectively. Electron micrographs of filaments of purified urinary UMOD (B), recombinant full-length UMOD from Madin–Darby canine kidney (MDCK) cells (C), purified elastase-digested urinary UMOD (D), and recombinant mMBP-UMODp from HEK293T cells (E). Yellow squiggles in B–E indicate the zigzag arrangement of UMOD repeats, which is most evident in samples lacking the N-terminal EGF I–III/D8C region. (Scale bars, 100 nm.)
Although UMOD activity is strictly linked to its supramolecular state (2), the mechanism of ZP module-dependent assembly remains unclear. Mass spectroscopy (MS) analysis of ZP-C disulfide linkages suggests that there are two types of ZP modules with different structures (13). Type II contains 10 conserved Cys (C1–7,a,b,8) and both homopolymerizes (UMOD, GP2, and TECTA) and heteropolymerizes (ZP1, ZP2, and ZP4), whereas type I (ZP3) includes eight conserved Cys (C1–8) and only heteropolymerizes with type II (7, 13, 14). However, MS studies of egg coat protein disulfides are contradictory (15), and type II disulfide linkages C5–C6, C7–Ca, and Cb–C8 are compatible neither with the fold of ZP3 (3) nor with structures of the ZP-C domain of BG, whose ZP module contains 10 Cys (16, 17). At the same time, interpretation of the latter data in relation to polymerization is complicated by the fact that, like ENG, BG remains membrane-associated and does not form filaments (7, 17).
To gain insights into the mechanism of ZP module protein assembly, we carried out X-ray crystallographic studies of the complete polymerization region of UMOD. The structure reveals that a rigid interdomain linker is responsible for maintaining UMOD in a polymerization-competent conformation. This rigid linker is conserved in homopolymeric ZP modules, but it is flexible in the structure of ZP2, also presented in this work, which, together with ZP3, forms heteropolymeric egg coat filaments. Furthermore, ZP module proteins that do not make filaments lack such a linker. Because UMOD and ZP2 show conservation of both disulfide pattern and fold, our data reveal that the interdomain linker, rather than a different ZP-C structure, underlies the ability of UMOD to self-assemble. Accordingly, polymerization-competent UMOD forms a dimer via β-sheet extension and hydrophobic interactions, and disruption of this dimer interface completely abolishes filament formation. Our study yields insights into the formation of an essential polymerization intermediate of UMOD and highlights how an interdomain linker can regulate the biological function of a multidomain protein.
Results and Discussion
Maltose-Binding Protein-Fused UMOD Secreted by Mammalian Cells Polymerizes Like Native UMOD.
To shed light on UMOD polymerization, we focused on a protease-resistant fragment (residues S292–F587) that contains the ZP module (Fig. 1A, Top), constitutes the core of UMOD filaments (18), and matches an alternatively spliced isoform of GP2 (19). UMODp (residues S292–Q640), a related construct that includes the C-terminal GPI-anchoring site, was expressed in mammalian cells as a fusion with a mammalianized version of bacterial maltose-binding protein (mMBP) (Fig. 1A, Middle). Electron microscopy (EM) revealed that secreted mMBP-UMODp forms native-like filaments with the characteristic zigzag structure of urinary UMOD (20), full-length recombinant UMOD, or elastase-treated UMOD (Fig. 1 B–E).
Crystal Structure of the Polymerization Region of UMOD.
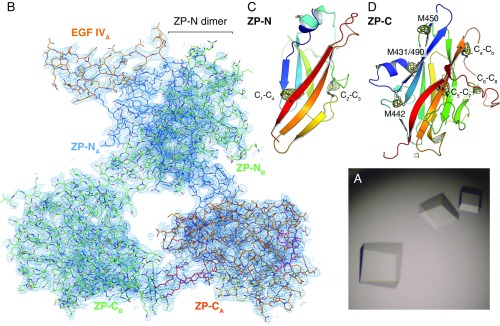
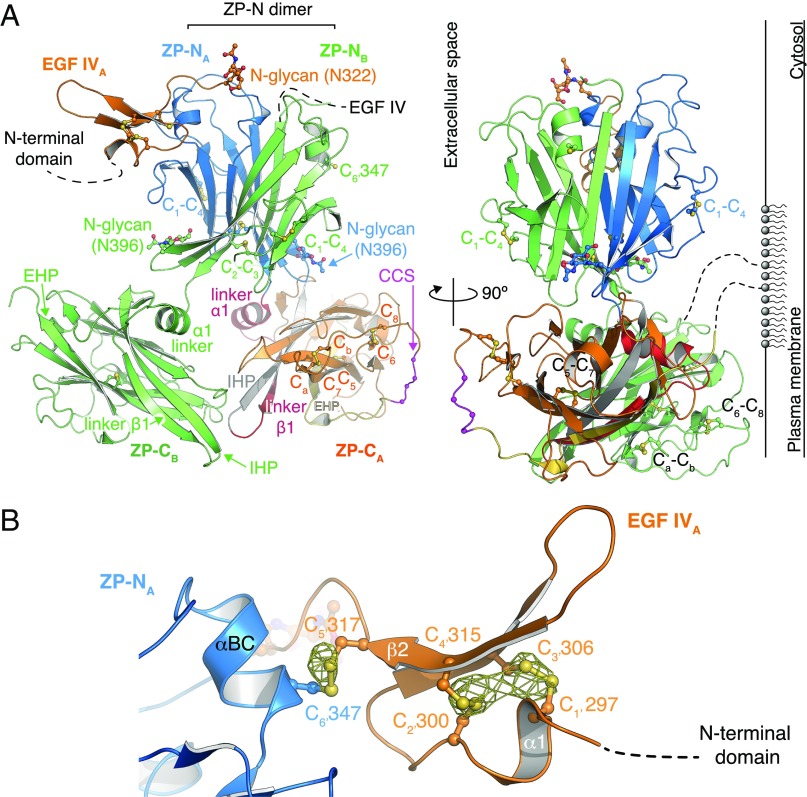
Despite extensive attempts, we could not obtain diffracting crystals of depolymerized native UMOD or unfused recombinant UMOD constructs. However, a soluble version of mMBP-UMODp (including UMOD residues G295–Q610) that cannot be cleaved at the CCS and carries a mutation of nonessential glycosylation site N513 (mMBP-UMODpXR; Fig. 1A, Bottom) formed crystals in high-salt conditions (Fig. S1A). The structure of mMBP-UMODpXR, with two molecules per asymmetrical unit, was solved by molecular replacement with MBP as a search model and refined to R = 22.1%, Rfree = 24.6% at a resolution of 3.2 Å (Fig. 2A and Table S1). The entire molecule A has well-defined electron density (Fig. S1B), which reveals that a fourth EGF-like domain precedes the ZP module of UMOD (Fig. 2A). This domain is structurally most similar to human TGF-α (21), with a root-mean-square deviation (rmsd) of 1.4 Å over 23 residues. Not visible in molecule B due to flexibility within the crystals rather than proteolytic degradation (Fig. S2), EGF IV consists of a short N-terminal α-helix and an antiparallel β-turn disulfide bonded with C1′–C3′ and C2′–C4′ connectivity (Fig. 2B). Mutations of the corresponding Cys are associated with autosomal dominant tubulointerstitial kidney disease (ADTKD) (Fig. S3 and Table S2). An additional C5′–C6′ disulfide tethers EGF IV C317 to ZP-N C347, which belongs to an α-helix that lies between strands B and C (Fig. 2B) and is absent in ZP3 (3, 5). Loss of either Cys is also associated with ADTKD, due to intracellular aggregation and impaired urinary secretion of UMOD (22, 23) (Fig. S3 and Table S2). Interestingly, human GP2 and TECTA, as well as chicken ZPD [a peripherally associated homopolymeric egg coat component (24)], also contain an EGF IV-like Cys-rich domain N-terminal to their ZP module (Fig. S3). Taken together, these data identify a subset of sequence-related but functionally diverse proteins that are characterized by EGF and ZP-N domains linked by a disulfide bond.
Fig. S1.
Crystal structure of mMBP-UMODpXR. (A) Prismatic crystals grown by hanging drop vapor diffusion using sodium/potassium tartrate as a precipitant. (B) B-factor sharpened 2mFo-DFc electron density map of the UMODpXR dimer, contoured at 1.0 σ. Structural elements are color-coded as in Fig. 2A. (C) ZP-N domain of UMOD, shown in a cartoon representation, rainbow-colored from blue (N terminus) to red (C terminus). Cys are depicted in a ball-and-stick representation, with C1, C2, C3, and C4 corresponding to human UMOD residues 335, 366, 389, and 425, respectively. A 4.0 σ anomalous difference map calculated from data collected at λ = 1.8 Å to confirm the identity of sulfur atoms is shown as a yellow mesh. (D) ZP-C domain, depicted as in C. Cys C5, C6, C7, Ca, Cb, and C8 correspond to residues 506, 527, 566, 571, 578, and 582, respectively. Met residues generating peaks in the anomalous difference map are also indicated.
Fig. 2.
Structure of the protease-resistant core of human UMOD. (A) Overall UMODpXR architecture, with molecule A colored as in Fig. 1A and molecule B in green. N-glycans and Cys are depicted in a ball-and-stick representation. (Right) Possible orientation relative to the plasma membrane due to GPI anchoring is depicted. (B) Close-up view of EGF IV and its connection to ZP-N. An anomalous difference map calculated with Bijvoet differences collected at λ = 1.8 Å and contoured at 3.5 σ is shown as a yellow mesh.
Table S1.
X-ray data collection and refinement statistics
| Crystal (PDB ID code) | mMBP-UMODpXR (4WRN) | ZP2 ZP-C (5BUP) |
| Experiment | ||
| Beamline | ESRF ID29 | DLS I02 |
| Wavelength, Å | 0.97625 | 0.97939 |
| No. of crystals | 1 | 1 |
| Data collection | ||
| Space group | H32/155 | P61/169 |
| Cell dimensions | ||
| a, b, c; Å | 242.32, 242.32, 258.86 | 105.21, 105.21, 40.55 |
| α, β, γ; ° | 90, 90, 120 | 90, 90, 120 |
| Molecules, A.U. | 2 | 1 |
| Solvent content, % | 73.7 | 54.5 |
| Mosaicity, ° | 0.104 | 0.183 |
| Wilson B factor, Å2 | 108.3 | 33.3 |
| Resolution, Å | 50.0–3.20 (3.28–3.20) | 45.56–2.25 (2.31–2.25) |
| Total reflections | 300,327 (20,429) | 41,295 (2,855) |
| Unique reflections | 47,986 (3,370) | 12,296 (866) |
| Completeness, % | 99.0 (94.5) | 99.4 (99.7) |
| Redundancy | 6.3 (6.1) | 3.4 (3.3) |
| I/σI | 16.20 (1.16) | 9.75 (1.13) |
| CC(1/2), % | 99.9 (43.2) | 99.6 (51.1) |
| Rpim, % | 3.8 (65.5) | 6.5 (67.7) |
| Refinement | ||
| Resolution, Å | 34.98–3.20 (3.31–3.20) | 45.56–2.25 (2.37–2.25) |
| Reflections | 47,792 (4,720) | 11,813 (1,536) |
| Free reflections | 2,431 (264) | 1,174 (149) |
| Rwork/Rfree, % | 22.10 (40.12)/24.59 (40.37) | 20.14 (34.30)/22.83 (36.25) |
| CCwork/CCfree | 0.938 (0.632)/0.915 (0.563) | 0.955 (0.617)*/0.934 (0.672)* |
| ML coordinate error, Å | 0.49 | 0.33 |
| ML phase error, ° | 28.21 | 26.02 |
| rmsd | ||
| Bond lengths, Å | 0.004 | 0.005 |
| Bond angles, ° | 0.840 | 0.859 |
| Ramachandran plot | ||
| Favored, % | 99.0 | 98.0 |
| Allowed, % | 1.0 | 2.0 |
| Outlier, % | 0.0 | 0.0 |
| No. of atoms | ||
| Total | 10,573 | 1,349 |
| Protein | 10,483 | 1,266 |
| Ligand/ion | 90 | 12 |
| Water | 0 | 71 |
| Protein residues | 1,356 | 159 |
| Average B factor, Å2 | ||
| Total | 141.3 | 46.9 |
| Protein | 141.2 | 47.0 |
| Ligand/ion | 152.1 | 81.8 |
| Water | — | 39.0 |
Parameters for the outermost shell are shown in parentheses. A.U., asymmetric unit; CC(1/2), percentage of correlation between intensities from random half-datasets; CCfree, correlation of the experimental intensities of free reflections excluded from the refinement with the intensities calculated from the refined molecular model; CCwork, correlation of the experimental intensities with the intensities calculated from the refined molecular model; DLS, Diamond Light Source; ESRF, European Synchrotron Radiation Facility; I/σI, signal-to-noise ratio; ML, maximum likelihood; Rpim, Σhkl √(1/n − 1) Σi|Ii(hkl) − I(hkl)|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity for an observation of a reflection and I(hkl) is the average intensity of all symmetry-related observations of a reflection.; Rwork, Σhkl‖Fobs| − k|Fcalc‖/Σhkl|Fobs|; Rfree, same as Rwork calculated from free reflections excluded from refinement.
Outer shell: 2.33–2.25 Å.
Fig. S2.
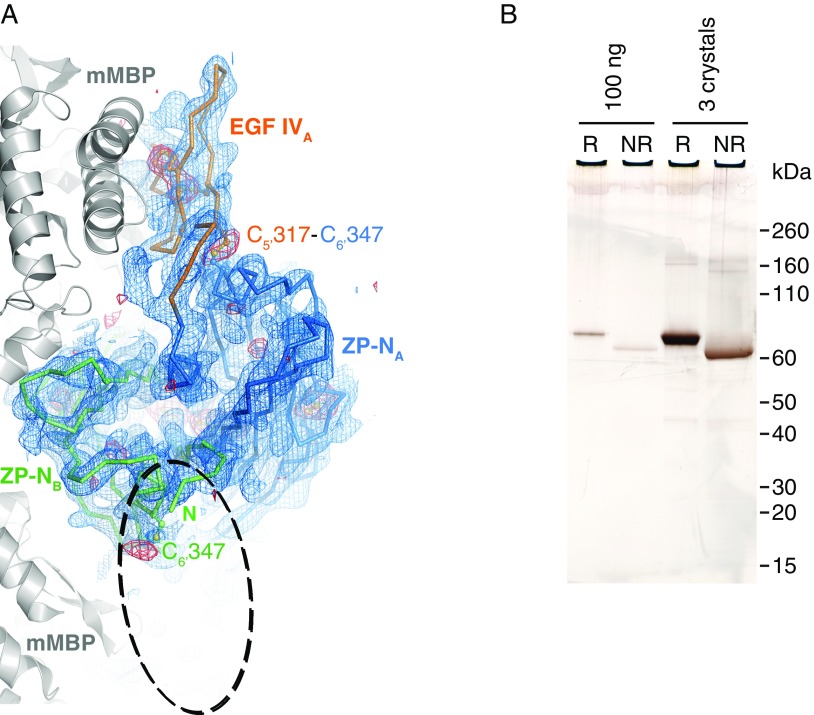
Analysis of crystallized mMBP-UMODpXR. (A) Top view of the ZP-N dimer, shown as a Cα-trace together with EGF IV, and mMBP molecules involved in crystal packing (gray cartoon). Blue mesh is a simulated-annealing composite omit map contoured at 1.0 σ. Red mesh is an anomalous difference map calculated with density-modified phases from the initial mMBP MR solution and contoured at 3.0 σ. The difference map, which is not biased by the structure of the UMOD moiety, shows peaks for all visible Cys residues. There is no density for the EGF IV domain in molecule B, which should be located in the area of the dashed ellipse. EGF IV in molecule A is stacked between ZP-N and mMBP, whereas in molecule B, it is protruding into the solvent, which allows it to be more flexible. Accordingly, the C-terminal part of mMBP in molecule B also shows a high degree of flexibility. (B) SDS/PAGE comparison of purified and crystallized material shows that mMBP-UMODpXR in the crystals is not degraded. Furthermore, the relative shift in migration between samples run in reducing (R) and nonreducing (NR) conditions, as well as the absence of additional bands, indicates that there is not even partial reduction of disulfide bonds. Crystallization at pH 7.0 and/or data collection in the presence of electron-scavenging ascorbate or oxidized glutathione did not result in clearly resolvable electron density for EGF IV in chain B. This observation excludes that flexibility is due to loss of the C5′–C6′ interdomain disulfide bond. Because molecule B represents 50% of the material in the crystal, protein degradation or disulfide bond reduction should be readily detectable by SDS/PAGE.
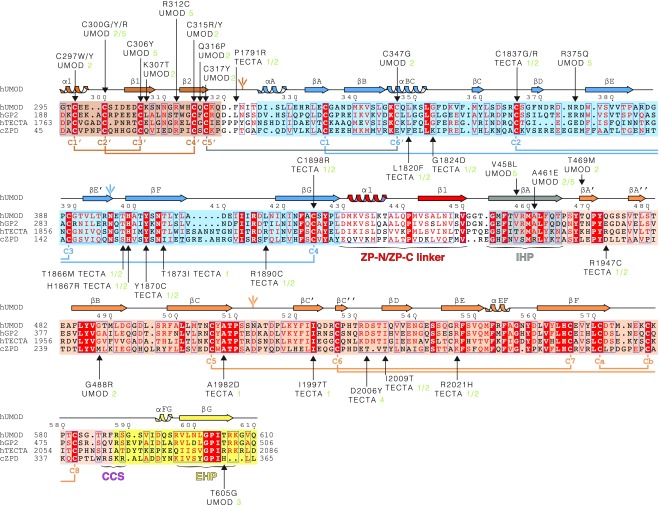
Fig. S3.
Sequence alignment of UMOD-like ZP module-containing proteins. The C-terminal extracellular regions of human GP2 and TECTA and the entire extracellular region of chicken ZPD are 55%, 37%, and 25% identical in sequence to human UMOD, respectively. Identical residues are highlighted in white and shaded in red, and conserved residues are indicated in red and marked by blue frames when clustered. Shaded boxes indicate EGF IV (brown), ZP-N (blue), and ZP-C (orange) domains, as well as IHP (gray) and external hydrophobic patch (EHP)-containing CTP (yellow). UMOD secondary structure elements, N-glycans, invariant Cys, and disulfide linkages are colored like the domains to which they belong, except for the C5′–C6′ disulfide that links EGF IV to ZP-N (gray). Arrows indicate residues affected by patient mutations, references for which are specified. Green numbers 1–4 and 5 next to pathogenic amino acid mutations indicate refs. 72–75 and the Uromodulin Kidney Disease Foundation UMOD mutation catalog (www.ukdcure.org/mutation_catalog), respectively.
Table S2.
Human UMOD and TECTA missense patient mutations annotated in Fig. S3
| Protein | Mutation | Predicted effect |
| UMOD | C297W/Y | Destroys conserved disulfide bond C1′–C3′ |
| C300G/Y/R | Destroys conserved disulfide bond C2′–C4′ | |
| C306Y | Destroys conserved disulfide bond C1′–C3′ | |
| K307T | Disrupts interaction with Q319 and Q316 | |
| C315R/Y | Destroys conserved disulfide bond C2′–C4′ | |
| R312C | Interferes with EGF IV disulfide bond formation | |
| Q316P | Disrupts interaction with K307 and Q316 | |
| C317Y | Destroys conserved disulfide bond C5′–C6′ between ZP-N and EGF IV, and thereby the coordination between the two domains | |
| C347G | Destroys conserved disulfide bond C5′–C6′ between ZP-N and EGF IV, and thereby the coordination between the two domains | |
| E375Q | Disrupts salt bridge with E304, and thereby the orientation between ZP-N and EGF IV | |
| V458L | Bulky residue in the hydrophobic core of ZP-C interrupts hydrophobic sheet stacking | |
| A461E | Bulky residue disrupts packing of linker α1 against the hydrophobic sheet of ZP-C, affecting ZP-N/ZP-C domain orientation | |
| T469M | Disrupts interaction with T465, and thereby orientation between IHP and βA′ | |
| G488R | Bulky residue disrupts packing of linker α1 against the hydrophobic sheet of ZP-C, affecting ZP-N/ZP-C domain orientation | |
| T605G | Disrupts EHP conformation due to loss of interaction with V477 backbone | |
| TECTA | P1791R | Disrupts the linker between EGF and ZP-N domains, thereby affecting their relative orientation |
| L1820F | Disrupts αBC that harbors the EGF/ZP-N disulfide | |
| G1824D | Disrupts the linker of αBC, harboring the EGF/ZP-N disulfide | |
| C1837G/R | Destroys conserved disulfide bond C2–C3 | |
| T1866M | Removes the highly conserved N-linked glycan site involved in dimerization | |
| H1867R | Disrupts interaction with conserved T1866 and N-glycan, thereby interfering with dimer formation | |
| Y1870C | Interferes with ZP-N C1–C4 disulfide bond | |
| T1873I | Destroys interaction with E1895 | |
| R1890C | Odd Cys interferes with disulfide bond formation and correct folding | |
| C1898R | Destroys conserved disulfide bond C1–C4 | |
| R1947C | Odd Cys interferes with disulfide bond formation and correct folding | |
| A1982D | Charged residue interferes with hydrophobic sheet-sheet packing in the ZP-C core | |
| I1997T | Polar residue interferes with hydrophobic sheet-sheet packing in the ZP-C core | |
| D2006Y | Bulky residue in the conserved βC′′–βD loop disrupts interactions with and conformation of the loop/disulfide N-terminal to the CTP | |
| I2009T | Polar residue interferes with hydrophobic sheet-sheet packing in the ZP-C core | |
| R2021H | Disrupts the interaction with conserved E2013 and interface between ZP-N, ZP-C, and ZP-N/ZP-C linker affecting ZP-N/ZP-C domain orientation |
The ZP-N domain of UMOD (Figs. S1C and S4A) is similar to the ZP-N domain of ZP3 (Fig. S4 B and C), including invariant disulfides (5) and a conserved Tyr (Fig. S4, arrow) whose mutation in TECTA is associated with hearing loss (25). Moreover, it contains an N-linked glycosylation site (N396; Fig. 2A) that is also found in GP2 and TECTA (Fig. S3) as well as additional ZP module proteins, including ZPD (26, 27) (Fig. S3), olfactorin (28), pirica (29), larval glycoprotein (30), and SPP120 (31).
Fig. S4.
Structural comparison of ZP-N domains. (A) Conformational rearrangements of the ZP-N domains of UMOD molecules A (blue) and B (green). A coiled segment (UMOD residues 323–332) in molecule A follows the EGF IV domain, whereas the same segment forms a β-strand in molecule B. In the presence of strand αA, helix αBC folds over toward strand βD. The different conformation adopted by residues 323–332, as well as the concomitant changes in the loops connecting βB/βC, βD/βE, and βF/βG, are most likely due to the flexibility of mMBP and EGF IV in molecule B (Fig. S2A). The invariant Tyr implicated in filament formation in TECTA is indicated by a black arrow. Cys are displayed in a ball-and-stick representation and labeled according to their connectivity. C6′, which staples the EGF IV domain to the ZP-N domain (omitted for clarity; Fig. 2B), is indicated by a gray sphere in αBC. (B) Superposition of the ZP-NA domain of UMOD (blue) and the ZP-N domain of mouse ZP3 (PDB ID code 3D4G; magenta), with rmsd = 2.6 Å. UMOD lacks the large FG loop of ZP3. (C) Superposition of the ZP-NA domain of UMOD (blue) and the ZP-N domain of chicken ZP3 (PDB ID code 3NK4; salmon), with rmsd = 2.0 Å.
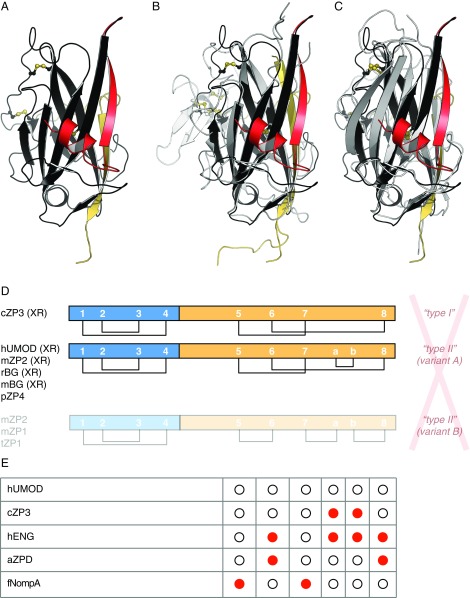
Surprisingly, our crystallographic data reveal that UMOD ZP-C (Figs. S1D and S5A) also shares the same fold and disulfide connectivity of ZP3 and BG ZP-Cs (3, 16, 17) (Fig. S5 B and C), except for the Ca–Cb disulfide not found in ZP3 proteins (15) (Fig. S5D). Accordingly, analysis of ZP-C Cys covariation based on multiple sequence alignments in Pfam (32) is consistent with C5–C7, C6–C8, and Ca–Cb connectivity (Fig. S5E).
Fig. S5.
Structural comparison and disulfide connectivity of ZP-C domains. (A) ZP-C domain of UMOD. ZP-N/ZP-C linker helix α1 and strand β1, which are coupled to ZP-C, are colored red. CTP is yellow, and Cys are shown in a ball-and-stick representation. (B) Superposition of the ZP-C domains of UMOD and avian ZP3 (PDB ID code 3NK4; gray/yellow), with rmsd = 2.6 Å. The ZP3 ZP-C subdomain is colored white. (C) Superposition of the ZP-C domains of UMOD and murine BG (PDB ID code 3QW9; gray/yellow), with rmsd = 2.1 Å. (D) X-ray crystallographic evidence (XR) indicates that, consistent with a common fold, all ZP modules share a single pattern of five conserved disulfide bonds. The only exception is disulfide Ca–Cb, which is not found in ZP3 homologs. Lowercase letters in front of protein names here and in E indicate common names of species (a, African clawed frog; c, chicken; f, fruit fly; h, human; m, mouse; p, pig; r, rat; t, trout), and previously used denominations (red) are italicized. (E) Cys covariation analysis of Pfam ZP domain family (PF00100) multiple sequence alignments shows that missing Cys (red) occur in pairs that match the disulfide connectivity observed in the X-ray structures of UMOD, ZP2, ZP3, and BG, as well as the disulfide connectivity proposed for pig ZP4, as shown in D (Top and Middle). On the other hand, they do not agree with the alternative connectivity proposed for mouse ZP1-2 and trout ZP1 (D, Bottom).
The Crystal Structure of ZP2 ZP-C Reveals That ZP Modules Have a Conserved Disulfide Connectivity.
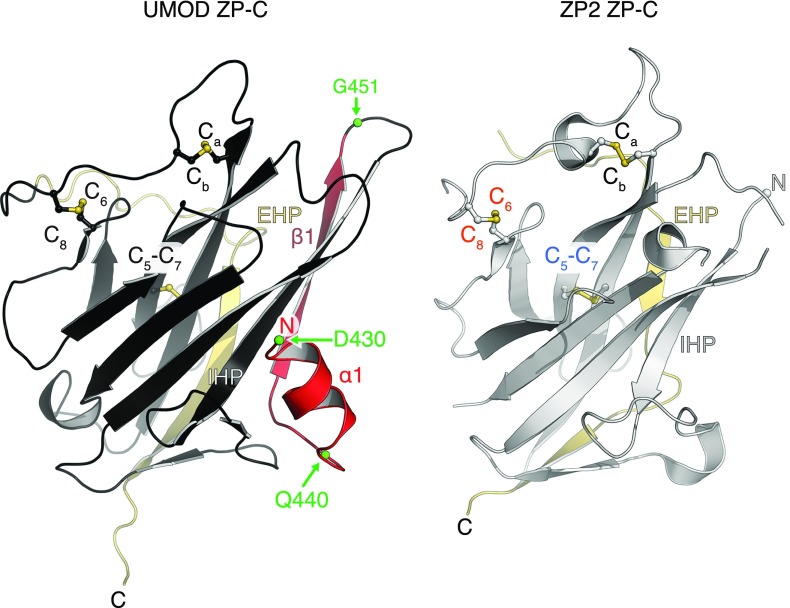
To confirm the existence of a single ZP module disulfide connectivity, we determined a 2.25-Å resolution structure of the ZP-C domain of mouse ZP2 (residues D463–D664; Fig. 3A and Fig. S6A). This molecule, which plays a key role in mammalian gamete recognition (33), has so far eluded structural determination but was reported to contain the alternative pattern based on C7–Ca, Cb–C8 MS assignments (13). The structure (R = 20.1%, Rfree = 22.8%; Fig. 3B, Fig. S6B, and Table S1) conclusively shows that ZP2 adopts the same disulfide linkages and overall fold as UMOD, ZP3, and BG (Fig. 4 A and B and Fig. S7). Collectively, these observations suggest that, contrary to what was previously thought, all ZP modules share a common architecture, so that other molecular features must regulate polymerization specificity.
Fig. 3.
The ZP-C domain of mouse ZP2 has a conserved fold. (A) Domain structure of mouse ZP2. Elements are depicted as in Fig. 1A, with the C-terminal transmembrane domain represented by a black rectangle. The region encompassed by the construct used for X-ray crystallography is indicated by a dashed red box. (B) Cartoon representation of ZP2 ZP-C, rainbow-colored from blue (N terminus) to red (C terminus). The CCS is magenta, disordered loops are depicted as dashed lines, and disulfide bonds are depicted in a ball-and-stick representation. The black arrow indicates the first ordered N-terminal residue, P485.
Fig. S6.
Crystal structure of mouse ZP2 ZP-C domain. (A) Needle-like crystals grown by hanging drop vapor diffusion using PEG 3350 as a precipitant. (B) 2mFo-DFc electron density map, contoured at 1.0 σ. Amino acids are colored according to Fig. 3A. As in Fig. 3B, the black arrow here and in D indicates the first ordered N-terminal residue, P485. (C) Anti-5His immunoblot of ZP2463–664 in conditioned medium (CM) and crystallized ZP2 ZP-C indicates that the latter is intact in the crystals. (D) Zoomed-in view of the N-terminal region of ZP2 ZP-C, showing the complete absence of electron density for ZP-N/ZP-C linker residues D463–G484.
Fig. 4.
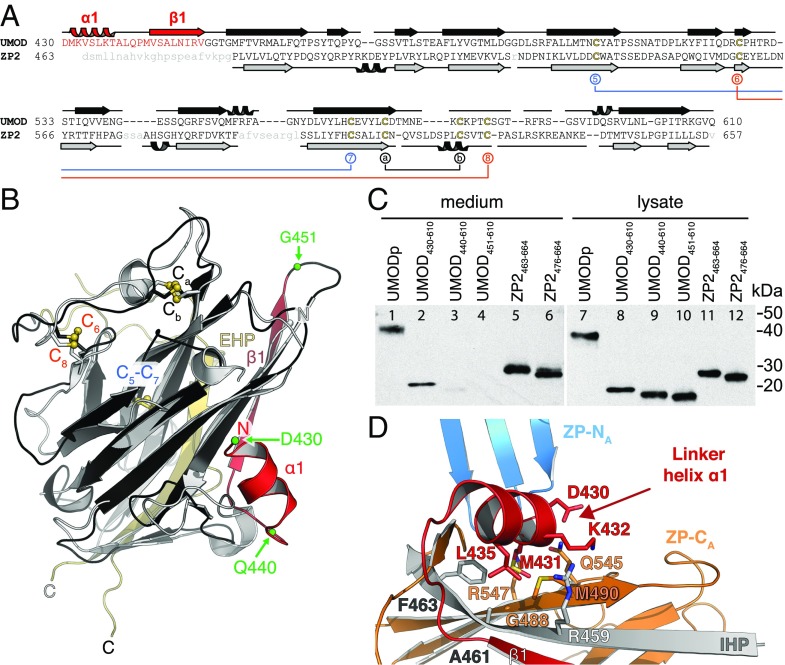
The structured ZP-N/ZP-C linker of UMOD is essential for ZP-C secretion and orients ZP-N relative to ZP-C. (A) Structure-based sequence alignment of UMOD and ZP2 ZP-Cs. The UMOD ZP-N/ZP-C linker is colored red, and ZP2 disordered residues are shown in lowercase gray. Disulfide bonds are colored as in Fig. 3B. (B) Structure comparison of the ZP-C domains of UMOD (black/red) and ZP2 (gray). The ZP-N/ZP-C linker of UMOD (red) is visible in the electron density, whereas the linker of ZP2 is flexible and not observed. Green spheres indicate truncation sites of the UMOD constructs analyzed in C. A side-by-side representation of this superposition can be found in Fig. S7. (C) Anti-5His immunoblot of conditioned medium and lysate of cells expressing different truncations of UMOD and ZP2. (D) UMOD ZP-C–associated α1 and β1 determine the relative position of ZP-N and ZP-C through hydrophobic interactions. Colors are as in Fig. 2A.
Fig. S7.
Side-by-side comparison of UMOD and ZP2 ZP-C. This figure is an alternative representation of the superposition shown in Fig. 4B, with the two molecules translated relative to each other.
A Structured Interdomain Linker Is Conserved in Self-Polymerizing ZP Modules.
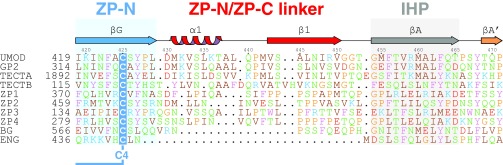
Structure comparison reveals a striking difference in the linker between ZP-N and ZP-C domains: Whereas this region is highly flexible in ZP3 (3), UMOD contains a rigid linker formed by α1 and β1 before the IHP (Fig. 4B and Fig. S5B). Analysis of UMOD ZP-C truncation constructs indicates that both of these secondary structure elements, which are also present in GP2, TECTA, and ZPD (Fig. S3), are essential for folding and secretion (Fig. 4C). Whereas UMOD430–610 starting with α1 is secreted comparably to UMODp (Fig. 4C, lanes 1–2), constructs beginning with β1 (UMOD440–610) or βA (UMOD451–610) are almost completely retained in the cell (Fig. 4C, lanes 3–4 and 9–10).
Like ZP3, ZP2 contains a ZP-N/ZP-C linker (Fig. S8); however, although this region was present in the crystals (Fig. S6C), ZP2 ZP-C is only defined from the IHP onward (Fig. 4B and Figs. S6D and S7). Moreover, unlike in the case of UMOD, the linker is not required for secretion of ZP2 ZP-C (Fig. 4C, lanes 5–6).
Fig. S8.
Sequence alignment of the ZP-N/ZP-C linker region of the ZP module. All sequences are human. Elements are depicted as in Fig. S3. Non-Cys residues are colored according to their physicochemical properties: positive, blue; negative, red; aromatic, purple; Pro/Gly, orange; hydrophobic, brown; polar, green. Note how the linker (red) between the ZP-N domain (blue) and the ZP-C domain (which starts with the IHP, gray) is either very short or completely absent in nonpolymerizing proteins BG and ENG.
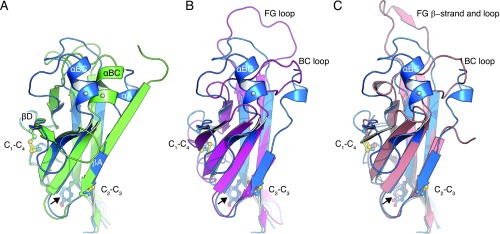
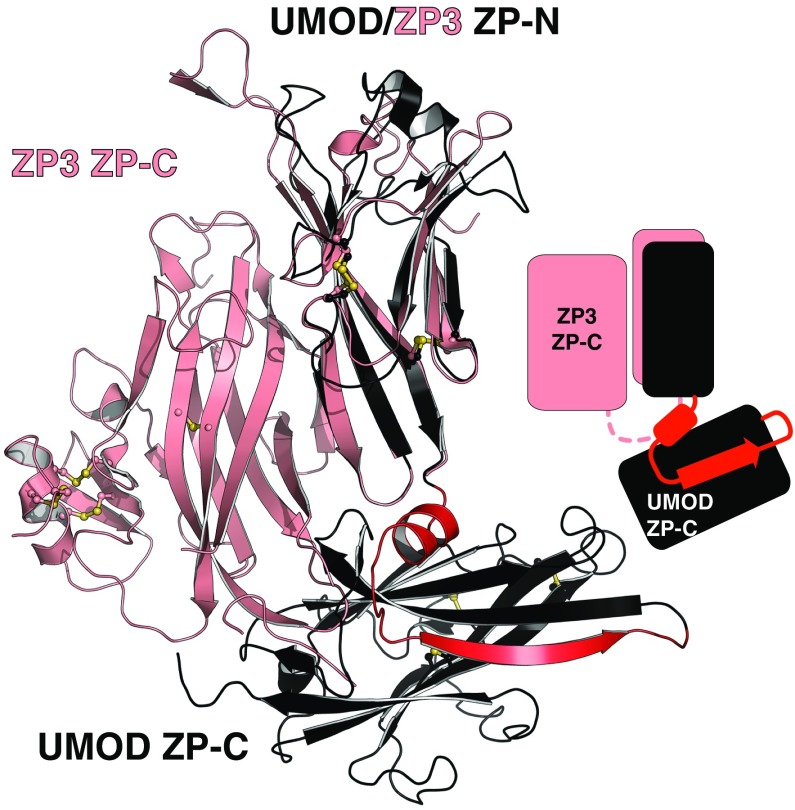
UMOD linker α1 packs tightly against the IHP-containing β-sheet (Fig. 4D and Fig. S7), shielding from the solvent hydrophobic residues also found in GP2, TECTA, and, to a lesser extent, ZPD (Fig. S3). Mutation of conserved α1 residues D430 and L435 causes trafficking and assembly defects of UMOD (10), whereas changes affecting amino acids located on the opposite side (A461E and G488R) are associated with kidney disease (Fig. S3). Thus, UMOD function is compromised upon disruption of contacts between α1/β1 and ZP-C. This interaction constrains the relative orientation between ZP-N and ZP-C, so that UMOD adopts an extended conformation that is significantly different from the conformation of ZP3 (Fig. 5). In the latter, as well as in ZP2, the linker lacks α1/β1 and the IHP-containing β-sheet surface is hydrophilic, resulting in a compact arrangement wherein ZP-N folds back onto ZP-C.
Fig. 5.
UMOD has a different ZP-N/ZP-C domain arrangement to ZP3. Comparison of the ZP modules of UMOD (black) and ZP3 (salmon). The structured linker between UMOD ZP-N and ZP-C is shown in red.
ZP-N Domain Dimerization Is Required for UMOD Polymerization.
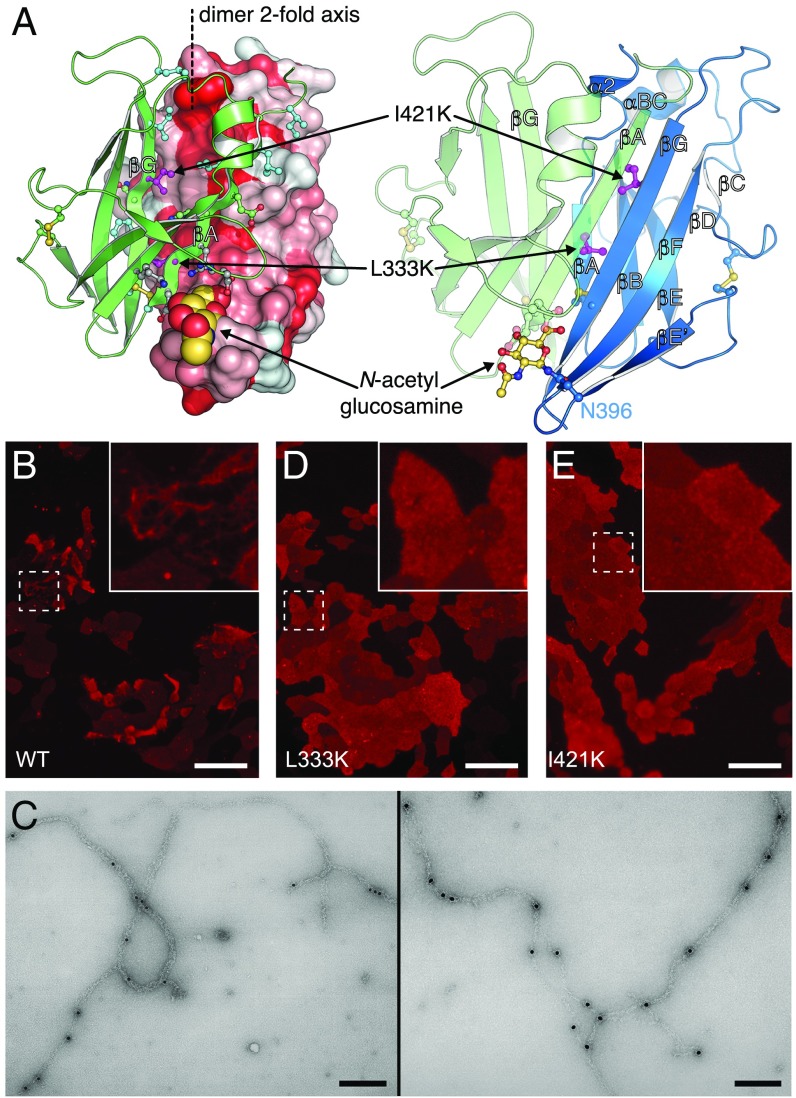
A major consequence of the extended configuration of the ZP module of UMOD is that the hydrophobic surface formed by ZP-N βA/βG is free to dimerize with the same region of a neighboring ZP-N through parallel β-sheet extension, burying a surface area of 2,148 Å2 (Figs. 2A and 6A). Computational analysis using PISA (34) scores this ZP-N/ZP-N interface as highly significant, and inward-facing hydrophobic residues in βA/βG are conserved across UMOD, GP2, TECTA, and ZPD (Fig. S3). Furthermore, the interface involves the N396 glycan, which forms intermolecular hydrogen bonds with the other UMOD molecule (Fig. 6A) and is also conserved among filament-forming ZP modules (Fig. S3). Notably, mutation of the corresponding N-glycosylation site of TECTA is associated with hearing loss (35), suggesting that this carbohydrate is important for tectorial membrane assembly.
Fig. 6.
UMOD ZP-N homodimerization is essential for filament formation. (A) UMOD ZP-N/ZP-N interface. Molecule A is in a solvent-accessible surface (Left: hydrophobic, red; hydrophilic, white) or depicted in a cartoon (Right, blue) representation; molecule B is depicted in a cartoon representation (green). Interface residues and disulfides are depicted in a ball-and-stick representation and are colored green (hydrophilic/charged), cyan (hydrophobic), gray (N396 glycan-interacting residues), and magenta (I421 and L333). (B) Immunofluorescence analysis of MDCK cells stably expressing full-length, HA-tagged, WT UMOD. (Scale bar, 50 μm.) (C) Immunogold labeling of full-length, HA-tagged, WT UMOD filaments produced in MDCK cells, with the same anti-HA primary Ab used in B. Two different areas are shown. (Scale bars, 0.2 μm.) (D and E) Immunofluorescence analysis of MDCK cells stably expressing full-length, HA-tagged UMOD dimerization interface mutants L333K and I421K. (Scale bars, 50 μm.)
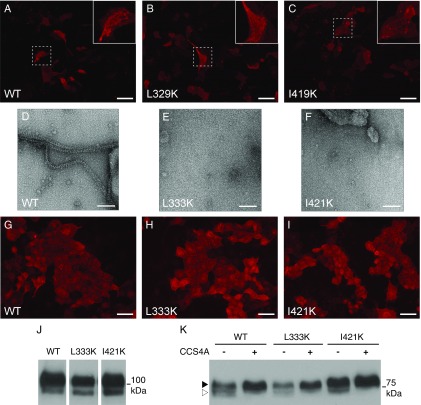
To evaluate the biological significance of the ZP-N homodimer, conserved interface residues (Fig. S3) were individually mutated to Lys to prevent edge-to-edge β-sheet interaction (36). Whereas mutation of peripheral residues L329 and I419 does not significantly affect UMOD assembly (Fig. S9 A–C), mutation of core residues L333 and I421 (Fig. 6A) completely abolishes filament formation (Fig. 6 D and E) compared with WT UMOD (Fig. 6 B and C). Accordingly, EM of corresponding mMBP-fused mutants of L333 and I421 detects no filaments (Fig. S9 D–F). Considering that neither mutation affects the trafficking (Fig. S9 G–I), secretion (Fig. S9J), or proteolysis (Fig. S9K) of UMOD, we conclude that the homodimer observed in our crystals represents a polymerization intermediate, whose formation is essential for the assembly of UMOD filaments.
Fig. S9.
Immunofluorescence, EM, and Western blot analysis of UMOD ZP-N homodimerization mutants. (A–C) Immunofluorescence analysis of MDCK cells transiently expressing WT UMOD or mutants L329K and I419K. (Scale bars, 50 μm.) (D–F) Electron micrographs of mMBP-UMODp WT as well as L333K and I421K mutant material, processed following the same protocol used for Fig. 1E. (Scale bars, 100 nm.) (G–I) Immunofluorescence analysis of permeabilized MDCK cells stably expressing WT UMOD or mutants L333K and I421K. No intracellular aggregation or retention of mutant isoforms is observed. (Scale bars, 50 μm.) (J) Western blot analysis of MDCK cell lysates. UMOD is present as two isoforms of 100 kDa and 80 kDa, corresponding to the mature protein and its endoplasmic reticulum (ER) precursor. No accumulation of the ER precursor form is observed for either mutant. (K) Western blot analysis of PNGase F-treated UMOD secreted by MDCK cells. Two isoforms of UMOD are found in the conditioned medium of transfected cells: a short isoform correctly processed at the CCS (white arrowhead) and a longer isoform spuriously processed at a distal site (black arrowhead). Neither L333K nor I421K affects protein processing. Moreover, as in the case of WT UMOD, substitution of the CCS with four Ala residues (CCS4A) abolishes the secretion of the short isoform of both mutants.
Sequence alignments and structural data indicate that the two moieties of the ZP module can be joined by very few residues (BG and ENG) or connected by a linker that is either unstructured (ZP1–ZP4, and TECTB) or structured (UMOD, GP2, and TECTA) (Figs. S3 and S8). Remarkably, these combinations coincide with the different polymerization abilities of the corresponding proteins: BG and ENG do not polymerize; ZP1–ZP4 and TECTB heteropolymerize; and UMOD, GP2, and TECTA homopolymerize (7, 17, 37, 38). This observation is consistent with the idea that in the last set of proteins, coupling of an α1/β1-containing linker to ZP-C induces an extended conformation of the ZP module. This conformation, in turn, exposes the βA/βG surface of ZP-N to form a dimer that initiates homopolymerization. On the other hand, the presence of a flexible linker may allow ZP1–ZP4 to adopt a secretion-competent conformation, such as the conformation observed in the structure of full-length ZP3 (3), which could require additional factors to trigger heteropolymerization and incorporation into the egg coat (39).
Conclusion
First isolated more than 60 years ago (40) and redescribed 35 years later as UMOD (41), UMOD has been recognized as a guardian against urinary tract infection and a crucial player in innate immunity; kidney disease; and, more recently, hypertension (1, 2, 42, 43). Our work gives mechanistic insights into how UMOD and other ZP module proteins assemble into their biologically active form, and how their structure and polymerization can be perturbed by pathogenic human mutations.
Materials and Methods
For structural studies, mMBP-UMODpXR and ZP2 ZP-C proteins were transiently expressed in HEK293S and HEK293T cells, respectively, based on published protocols (44–46); immunofluorescence studies were performed using stably transfected MDCK cell lines, essentially as described (10). Construct information and detailed methods for protein purification, deglycosylation, crystallization, and structure determination; UMOD filament preparation; and EM and immunofluorescence analyses are provided in SI Materials and Methods. X-ray data collection and refinement statistics are summarized in Table S1. Atomic coordinates and structure factors for human UMODpXR and mouse ZP2 ZP-C have been deposited in the Protein Data Bank (ID codes 4WRN and 5BUP, respectively). Urine for EM analysis of native UMOD was kindly donated by M. Bokhove.
SI Materials and Methods
DNA Constructs.
Expression constructs were generated by PCR using PfuTurbo DNA polymerase (Agilent Technologies); mutations were introduced by overlap extension PCR or with a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). Oligonucleotides were from Sigma–Aldrich, and all constructs were verified by DNA sequencing (Eurofins Genomics).
Except for immunofluorescence studies, which relied on previously described plasmids encoding HA-tagged WT UMOD and CCS4A mutants (586-RFRS-589 to AAAA) (10), all constructs were based on mammalian expression vector pHLsec (44). For expression of secreted fusions, a codon-optimized gene encoding an N-terminally 6His-tagged mMBP was synthesized (GenScript), which includes a combination of mutations increasing maltose affinity and crystallizability (47–50). The mMBP gene was cloned in-frame to the chicken CRYPα1 R2G signal peptide-encoding sequence of pHLsec, and includes a 3′ NotI restriction site that results in a three-Ala linker between mMBP and passenger proteins. Nonpolymerizing constructs for X-ray crystallographic studies were generated using cDNA fragments carrying mutations of the CCS-encoding sequences of human UMOD (R586A/R588A) and mouse ZP2 (K634N/R635A). UMOD N-glycosylation site N513, which is not essential for secretion, was also mutated in UMODpXR, as well as the UMOD truncation mutants analyzed in Fig. 4C.
Protein Expression.
DNA for small- and large-scale transfections was prepared using HiSpeed Plasmid Midi Kits and EndoFree Plasmid Maxi/Giga Kits, respectively (both from QIAGEN). HEK293 cells were cultivated using DMEM supplemented with 4 mM l-glutamine (Life Technologies) and 10% (vol/vol) FBS (Saveen & Werner) at 37 °C in 5% CO2. Subsequently, they were transiently transfected in DMEM with 4 mM l-glutamine using 25-kDa branched polyethylenimine (Sigma–Aldrich), essentially as described (44). Small-scale transfections were performed in six-well plates (10 cm2 per well; Corning), large-scale transfections were carried out in T-flasks (150 cm2; BD Biosciences) or ribbed roller bottles (2,125 cm2; Greiner).
Production of glycosylated mMBP-UMODpXR was performed using HEK293S cells (CRL-3022; American Type Culture Collection), which secrete homogeneous Man5GlcNAc2-glycosylated proteins that can be treated with endoglycosidase H (Endo H) to remove N-glycans (45, 46). ZP2 ZP-C, which does not contain N-linked glycosylation sites, was expressed in HEK293T cells. In both cases, conditioned medium was harvested 3 d after transfection. Notably, fusion to mMBP was absolutely essential to obtain suitable amounts of well-behaved UMODp, as well as for its crystallization.
MDCK cell lines stably expressing HA-tagged constructs of UMOD were generated as described previously, and conditioned media and cell lysates were prepared as published (10).
mMBP-UMODp filaments were produced by transiently cotransfecting HEK293T cells with constructs expressing WT or mutant mMBP-fused UMODS292–Q640 in a 10:1 molar ratio with a pcDNA3.1(+)-derived vector expressing human hepsin (8).
Protein Analysis.
Cell lysates were obtained by resuspending cells from a 10-cm2 culture well in lysis buffer [50 mM Tris⋅HCl/50 mM 3-(N-morpholino)propanesulfonic acid (pH 7.7), 0.1% SDS, 1 mM EDTA] supplied with protease inhibitors (Roche), followed by centrifugation for 10 min at 18,000 × g at 4 °C and filtration using a 0.22-μm syringe filter (Millipore).
Samples were separated on SDS/PAGE gels and transferred to nitrocellulose membranes (GE Healthcare). Immunoblotting was performed with Penta-His mouse mAb (1:1,000; QIAGEN) or anti-HA mouse Ab (1:1,000; Covance). Chemiluminescence detection was performed with Western Lightning ECL Plus (PerkinElmer) or using an Immobilon Western Chemiluminescent Horseradish Peroxidase Substrate Kit (Millipore).
Protein Purification and Deglycosylation.
Conditioned medium was adjusted to 5 mM imidazole, 150 mM NaCl, 20 mM Na-Hepes (pH 8.0) [immobilized metal affinity chromatography (IMAC) binding buffer]. Ten mL of preequilibrated nickel-nitrilotriacetic acid (Ni-NTA) agarose slurry (QIAGEN) was then added per L of medium and allowed to incubate overnight at 4 °C on a shaker. Ni-NTA beads were collected, washed with IMAC binding buffer, and batch-eluted with 500 mM imidazole, 150 mM NaCl, and 20 mM Na-Hepes (pH 8.0). The IMAC elution fraction was concentrated using centrifugal filtration devices (Amicon) with an appropriate molecular weight cutoff (MWCO). In the case of mMBP-UMODpXR expressed in HEK293S cells, concentrated fusion protein was deglycosylated with Endo H (1:10 mass ratio) for 1 h at 37 °C in 120 mM Na/K phosphate (pH 6.0). Concentrated material was applied to a Superdex 200 26/600 size exclusion chromatography (SEC) column attached to an ÄKTAFPLC system (GE Healthcare) and preequilibrated with 100 mM NaCl, 20 mM Na-Hepes (pH 8.0), and 10 mM d-maltose. For purification of ZP2 ZP-C, all buffers contained 500 mM NaCl and a Superdex 75 26/600 column was used. SEC fractions containing purified proteins were pooled, concentrated, and used for crystallization trials.
Protein Crystallization.
Purified mMBP-UMODpXR was concentrated to 6.5–15.0 mg/mL in 100 mM NaCl, 20 mM Na-Hepes (pH 8.0), and 1.5 mM maltose and was crystallized at room temperature (RT) by hanging drop vapor diffusion against mother liquor containing 900 mM Na/K tartrate and Tris⋅HCl (pH 7.0–8.6) (Fig. S1A). Crystals could only be obtained in the presence of Zn(OAc)2, which was added in a 1:1.5 molar ratio to the mMBP-UMODpXR solution before crystallization and was eventually found to mediate crystal packing by interacting with the 6His-tag of mMBP-UMODpXR. Crystals grew to 200–400 μm and were subsequently slowly accommodated to 4 °C. Finally, they were transferred in six steps to mother liquor containing 20–30% (vol/vol) glycerol and flash-cooled in liquid nitrogen before data collection at 100 K.
Purified ZP2 ZP-C was concentrated to 5.0 mg/mL in 20 mM Na-Hepes (pH 8.0) and 200 mM NaCl and was crystallized at RT by hanging drop vapor diffusion against 25% (wt/vol) PEG 3350, 100 mM NaOAc (pH 5.5), 200 mM (NH4)2SO4, and 20% (vol/vol) glycerol. Microseeding was necessary to grow single crystals of adequate size for analysis, and produced well-ordered crystals up to 0.5 mm long (Fig. S6A). Crystals were flash-cooled in liquid nitrogen before data collection at 100 K.
X-Ray Diffraction Data Collection.
Native datasets were collected using PILATUS 6M-F detectors at European Synchrotron Radiation Facility beamline ID29 (51) (mMBP-UMODpXR) or Diamond Light Source beamline I02 (ZP2 ZP-C). Data collection statistics can be found in Table S1. Anomalous difference data were obtained from multiple passes collected at a wavelength of 1.8 Å.
Data Processing and Structure Determination.
All datasets were integrated and scaled with XDS (52).
The structure of mMBP-UMODpXR was solved by molecular replacement (MR) with PHASER (53), using MBP coordinates extracted from PDB ID code 3D4G (5) as a search model. Successful MR runs showed a clear positive different density peak in the MBP binding site for maltose, which was not included in the search model. After obtaining initial MR phases, density modification was performed with RESOLVE (54) in the PHENIX package (55). The model of mMBP-UMODpXR was generated by combining multiple rounds of PHENIX AutoBuild (56) with Buccaneer (57) and manual building in Coot (58).
The crystal structure of ZP2 ZP-C was solved by MR with PHASER, using the ZP-C domain of ZP3 (PDB ID code 3NK4) (3), BG (PDB ID code 3QW9) (17) or UMOD as a search model. After autotracing with PHENIX AutoBuild (56), manual rebuilding was performed with Coot (58).
Refinement of both structures was performed with the phenix.refine module of the PHENIX package (59). Protein geometry was validated with MolProbity (60), and carbohydrate structure validation was carried out using pdb-care (61) and Privateer (62).
Structure Analysis.
Structure comparisons were performed using PDBeFold (63). Oligomeric state was analyzed using PISA (34). PISA analysis shows that ZP-N/ZP-N dimer formation buries 16.3% and 9.0% of the total ZP-N domain and ZP module surface, respectively, resulting in a ΔG of −11.6 kcal/mol. Together with a complexation significance score of 1.0 and the lack of any additional interaction scored as significant, this analysis suggests that the interface observed in the crystals is biologically relevant. Furthermore, the relative orientation of mMBP and UMODpXR is different in the two molecules, indicating that the mMBP fusion is flexible and mMBP does not affect the overall structure of UMODpXR itself.
UMOD Filament Preparation.
Native UMOD filaments were purified from healthy male urine as described (64), and elastase-resistant filaments were prepared as previously reported (18).
To purify recombinant filaments, 1 mL of conditioned medium of UMOD-expressing HEK293T cells was harvested and spun for 5 min at 500 × g to remove cell debris. The supernatant was dialyzed at 4 °C against 10 mM Na-Hepes (pH 8.0) and 10 mM NaCl using a 30-kDa MWCO Slide-A-Lyzer dialysis cassette (Thermo Scientific). After dialysis, the material was centrifuged for 30 min at 18,000 × g to remove aggregates, followed by the addition of 150 mM NaCl to the supernatant and incubation on ice for 1 h to gelify UMOD filaments. The supernatant was then spun for 2 h at 18,000 × g, resulting in a small pellet of filaments that was finally vigorously resuspended in 20 μL of ultrapure water.
EM.
The sample (5 μL) was applied to a glow-discharged 400 mesh copper grid with a carbon support film. After 2 min, the solution was removed with filter paper, followed by washing with 20 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl. A final wash step was performed with ultrapure water, followed by negative staining with 2% (wt/vol) uranyl acetate. Samples were analyzed using a CM120 electron microscope (Philips) equipped with a LaB6 electron source. Images were recorded on a 1K Tietz camera or with SO-163 electron film (Kodak). Films were digitized using an Epson Perfection 4990 PHOTO flatbed scanner.
For on-grid immunogold labeling, 5 μL of diluted filament solution was applied to glow-discharged 400 mesh gold grids with a carbon support film. After 5 min, the grids were washed by inverting them onto a drop of ultrapure water for 2 min and then moved onto a 30-μL drop of blocking buffer [20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.1% BSA, or 0.25% blotting grade nonfat dry milk] for 15 min. Grids were then moved onto a 10-μL drop of blocking buffer containing a 1:20 or 1:40 dilution of anti-HA Ab [HA.11 clone 16B2 purified mouse monoclonal primary Ab (Covance)] for 1 h. Grids were washed by inverting them three times for 5 min onto 30-μL drops of washing buffer [20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl] and moved onto a 20-μL drop of blocking buffer containing a 1:40 dilution of goat anti-mouse Ab [18 nm of gold-conjugated purified goat polyclonal Ab against mouse IgG-Fc (Abcam)] for 1 h. After washing as previously detailed, followed by three 5-min washes in ultrapure water, grids were negatively stained and imaged as described above.
Immunofluorescence Analysis.
Immunofluorescence studies were performed essentially as described (10). MDCK cells grown on coverslips were fixed in 4% (wt/vol) paraformaldehyde for 20 min at RT. When needed, cells were permeabilized for 20 min at RT with PBS solution-0.5% (vol/vol) Triton X-100 [note that this procedure essentially removes UMOD filaments from the cell surface (65)]. Permeabilized or unpermeabilized cells were incubated with 10% (vol/vol) preimmune donkey serum (Abcam) for 30 min at RT and then labeled for 1 h at RT with anti-UMOD primary Ab diluted in PBS with 1% (vol/vol) donkey serum (1:500; MP Biomedicals). Cells were washed in PBS and incubated with the appropriate Alexa Fluor 594-conjugated secondary Ab [1:500 in PBS with 1% (vol/vol) donkey serum; Life Technologies]. Nuclei were stained with DAPI (Life Technologies), and slides were mounted using fluorescent mounting medium (DAKO). Slides were visualized with a DM 5000B fluorescence upright microscope (Leica DFC480 camera, Leica DFC Twain Software, 40×/0.75 lens; Leica Microsystems). All images were imported in Photoshop CS (Adobe Systems) and adjusted for brightness and contrast.
Sequence Analysis.
Except for D8C (66), EGF IV, and ZP-N/ZP-C (this study), domain boundaries reported in Fig. 1A were based on SMART (67). The sequence alignment reported in Fig. S3 was generated using T-Coffee (68) and assembled using ESPript (69). The alignment in Fig. S8 was generated with PROMALS3D (70), manually edited, and colored using TEXshade (71). Cys covariation analysis was performed by manual inspection of ZP-like domain (PF00100) seed sequence alignments from Pfam (32).
Acknowledgments
This work is dedicated to the memory of Franca Serafini-Cessi, whose studies increased our understanding of uromodulin biology. Correspondence with Dr. Serafini-Cessi directly inspired this work. We thank M. Monné (Università degli Studi della Basilicata) for initial work on the project, H. Hebert and the Department of Biosciences and Nutrition (Karolinska Institutet) for access to the Center for High Resolution Electron Microscopy, the European Synchrotron Radiation Facility (ESRF; Grenoble) and Diamond Light Source (DLS; Oxford) for beam time (ESRF: mx1416/mx1551/mx1639, DLS: mx8492-18/mx8492-34), and G. Wallis (University of Otago) for comments and discussion. We are grateful to R. Aricescu and Y. Zhao (University of Oxford) for mammalian expression vector pHLsec and HEK293T cells, to D. Leahy (Johns Hopkins University School of Medicine) for Escherichia coli expression vector pProEX HT-endoglycosidase H, and to D. Waugh (National Cancer Institute) for E. coli strain BL21(DE3)-RIL/pRK793. This research was supported by the Karolinska Institutet, the Center for Innovative Medicine, Swedish Research Council Grant 2012-5093, the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine, the Sven and Ebba-Christina Hagberg Foundation, a European Molecular Biology Organization Young Investigator award, the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 260759 (to L.J.); and the Fondazione Telethon (GGP14263), the Italian Ministry of Health (RF-2010-2319394), and Fondazione Cariplo (2014-0827) (to L.R.). Crystallographic data collection was also supported by FP7/2007-2013 under BioStruct-X (Grant Agreement 283570).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.W. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4WRN and 5BUP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519803113/-/DCSupplemental.
References
- 1.Rampoldi L, Scolari F, Amoroso A, Ghiggeri G, Devuyst O. The rediscovery of uromodulin (Tamm-Horsfall protein): From tubulointerstitial nephropathy to chronic kidney disease. Kidney Int. 2011;80(4):338–347. doi: 10.1038/ki.2011.134. [DOI] [PubMed] [Google Scholar]
- 2.Serafini-Cessi F, Malagolini N, Cavallone D. Tamm-Horsfall glycoprotein: Biology and clinical relevance. Am J Kidney Dis. 2003;42(4):658–676. doi: 10.1016/s0272-6386(03)00829-1. [DOI] [PubMed] [Google Scholar]
- 3.Han L, et al. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143(3):404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 4.Jovine L, Qi H, Williams Z, Litscher ES, Wassarman PM. A duplicated motif controls assembly of zona pellucida domain proteins. Proc Natl Acad Sci USA. 2004;101(16):5922–5927. doi: 10.1073/pnas.0401600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monné M, Han L, Schwend T, Burendahl S, Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456(7222):653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Lett. 1992;300(3):237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 7.Jovine L, Darie CC, Litscher ES, Wassarman PM. Zona pellucida domain proteins. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- 8.Brunati M, et al. The serine protease hepsin mediates urinary secretion and polymerisation of Zona Pellucida domain protein uromodulin. eLife. 2015;4:e08887. doi: 10.7554/eLife.08887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santambrogio S, et al. Urinary uromodulin carries an intact ZP domain generated by a conserved C-terminal proteolytic cleavage. Biochem Biophys Res Commun. 2008;370(3):410–413. doi: 10.1016/j.bbrc.2008.03.099. [DOI] [PubMed] [Google Scholar]
- 10.Schaeffer C, Santambrogio S, Perucca S, Casari G, Rampoldi L. Analysis of uromodulin polymerization provides new insights into the mechanisms regulating ZP domain-mediated protein assembly. Mol Biol Cell. 2009;20(2):589–599. doi: 10.1091/mbc.E08-08-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo L, et al. Ablation of the Tamm-Horsfall protein gene increases susceptibility of mice to bladder colonization by type 1-fimbriated Escherichia coli. Am J Physiol Renal Physiol. 2004;286(4):F795–F802. doi: 10.1152/ajprenal.00357.2003. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. Progressive renal papillary calcification and ureteral stone formation in mice deficient for Tamm-Horsfall protein. Am J Physiol Renal Physiol. 2010;299(3):F469–F478. doi: 10.1152/ajprenal.00243.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boja ES, Hoodbhoy T, Fales HM, Dean J. Structural characterization of native mouse zona pellucida proteins using mass spectrometry. J Biol Chem. 2003;278(36):34189–34202. doi: 10.1074/jbc.M304026200. [DOI] [PubMed] [Google Scholar]
- 14.Darie CC, Biniossek ML, Jovine L, Litscher ES, Wassarman PM. Structural characterization of fish egg vitelline envelope proteins by mass spectrometry. Biochemistry. 2004;43(23):7459–7478. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- 15.Kanai S, et al. Disulfide linkage patterns of pig zona pellucida glycoproteins ZP3 and ZP4. Mol Reprod Dev. 2008;75(5):847–856. doi: 10.1002/mrd.20836. [DOI] [PubMed] [Google Scholar]
- 16.Diestel U, et al. Identification of a Novel TGF-β-Binding Site in the Zona Pellucida C-terminal (ZP-C) Domain of TGF-β-Receptor-3 (TGFR-3) PLoS One. 2013;8(6):e67214. doi: 10.1371/journal.pone.0067214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SJ, Hu Y, Zhu J, Woodruff TK, Jardetzky TS. Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-β binding. Proc Natl Acad Sci USA. 2011;108(13):5232–5236. doi: 10.1073/pnas.1010689108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovine L, Qi H, Williams Z, Litscher E, Wassarman PM. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat Cell Biol. 2002;4(6):457–461. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- 19.Fukuoka S. Molecular cloning and sequences of cDNAs encoding α (large) and β (small) isoforms of human pancreatic zymogen granule membrane-associated protein GP2. Biochim Biophys Acta. 2000;1491(1-3):376–380. doi: 10.1016/s0167-4781(00)00057-9. [DOI] [PubMed] [Google Scholar]
- 20.Delain E, Thiery JP, Coulard D, Joliviene A, Hartman L. Étude chimique et ultrastructurale de la glycoprotéine de Tamm et Horsfall ou uromucoïde. Biol Cell. 1980;39:31–42. French. [Google Scholar]
- 21.Moy FJ, et al. Solution structure of human type-α transforming growth factor determined by heteronuclear NMR spectroscopy and refined by energy minimization with restraints. Biochemistry. 1993;32(29):7334–7353. doi: 10.1021/bi00080a003. [DOI] [PubMed] [Google Scholar]
- 22.Rampoldi L, et al. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet. 2003;12(24):3369–3384. doi: 10.1093/hmg/ddg353. [DOI] [PubMed] [Google Scholar]
- 23.Tinschert S, et al. Functional consequences of a novel uromodulin mutation in a family with familial juvenile hyperuricaemic nephropathy. Nephrol Dial Transplant. 2004;19(12):3150–3154. doi: 10.1093/ndt/gfh524. [DOI] [PubMed] [Google Scholar]
- 24.Okumura H, et al. Identification of distinctive interdomain interactions among ZP-N, ZP-C and other domains of zona pellucida glycoproteins underlying association of chicken egg-coat matrix. FEBS Open Bio. 2015;5:454–465. doi: 10.1016/j.fob.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legan PK, et al. A deafness mutation isolates a second role for the tectorial membrane in hearing. Nat Neurosci. 2005;8(8):1035–1042. doi: 10.1038/nn1496. [DOI] [PubMed] [Google Scholar]
- 26.Lindsay LL, Yang JC, Hedrick JL. Identification and characterization of a unique Xenopus laevis egg envelope component, ZPD. Dev Growth Differ. 2002;44(3):205–212. doi: 10.1046/j.1440-169x.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- 27.Okumura H, et al. A newly identified zona pellucida glycoprotein, ZPD, and dimeric ZP1 of chicken egg envelope are involved in sperm activation on sperm-egg interaction. Biochem J. 2004;384(Pt 1):191–199. doi: 10.1042/BJ20040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Schiavi E, Riano E, Heye B, Bazzicalupo P, Rugarli EI. UMODL1/Olfactorin is an extracellular membrane-bound molecule with a restricted spatial expression in olfactory and vomeronasal neurons. Eur J Neurosci. 2005;21(12):3291–3300. doi: 10.1111/j.1460-9568.2005.04164.x. [DOI] [PubMed] [Google Scholar]
- 29.Mori T, et al. Identification of a novel uromodulin-like gene related to predator-induced bulgy morph in anuran tadpoles by functional microarray analysis. PLoS One. 2009;4(6):e5936. doi: 10.1371/journal.pone.0005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallis LJ, Wallis GP. Extreme positive selection on a new highly-expressed larval glycoprotein (LGP) gene in Galaxias fishes (Osmeriformes: Galaxiidae) Mol Biol Evol. 2011;28(1):399–406. doi: 10.1093/molbev/msq208. [DOI] [PubMed] [Google Scholar]
- 31.Gerrard DT, Meyer A. Positive selection and gene conversion in SPP120, a fertilization-related gene, during the East African cichlid fish radiation. Mol Biol Evol. 2007;24(10):2286–2297. doi: 10.1093/molbev/msm159. [DOI] [PubMed] [Google Scholar]
- 32.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg’s zona pellucida protein. Science. 2010;329(5988):216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Sagong B, et al. Two novel missense mutations in the TECTA gene in Korean families with autosomal dominant nonsyndromic hearing loss. Ann Clin Lab Sci. 2010;40(4):380–385. [PubMed] [Google Scholar]
- 36.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99(5):2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legan PK, et al. A targeted deletion in α-tectorin reveals that the tectorial membrane is required for the gain and timing of cochlear feedback. Neuron. 2000;28(1):273–285. doi: 10.1016/s0896-6273(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 38.Russell IJ, et al. Sharpened cochlear tuning in a mouse with a genetically modified tectorial membrane. Nat Neurosci. 2007;10(2):215–223. doi: 10.1038/nn1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiménez-Movilla M, Dean J. ZP2 and ZP3 cytoplasmic tails prevent premature interactions and ensure incorporation into the zona pellucida. J Cell Sci. 2011;124(Pt 6):940–950. doi: 10.1242/jcs.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamm I, Horsfall FLJ., Jr Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med. 1950;74(1):106–108. [PubMed] [Google Scholar]
- 41.Muchmore AV, Decker JM. Uromodulin: A unique 85-kilodalton immunosuppressive glycoprotein isolated from urine of pregnant women. Science. 1985;229(4712):479–481. doi: 10.1126/science.2409603. [DOI] [PubMed] [Google Scholar]
- 42.Graham LA, et al. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension. 2014;63(3):551–558. doi: 10.1161/HYPERTENSIONAHA.113.01423. [DOI] [PubMed] [Google Scholar]
- 43.Trudu M, et al. Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19(12):1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 45.Chang VT, et al. Glycoprotein structural genomics: Solving the glycosylation problem. Structure. 2007;15(3):267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Center RJ, et al. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 1998;7(7):1612–1619. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laganowsky A, et al. An approach to crystallizing proteins by metal-mediated synthetic symmetrization. Protein Sci. 2011;20(11):1876–1890. doi: 10.1002/pro.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: Combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19(5):901–913. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker IH, Hsieh PC, Riggs PD. Mutations in maltose-binding protein that alter affinity and solubility properties. Appl Microbiol Biotechnol. 2010;88(1):187–197. doi: 10.1007/s00253-010-2696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Sanctis D, et al. ID29: A high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J Synchrotron Radiat. 2012;19(Pt 3):455–461. doi: 10.1107/S0909049512009715. [DOI] [PubMed] [Google Scholar]
- 52.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11(Pt 1):49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lütteke T, von der Lieth CW. pdb-care (PDB carbohydrate residue check): A program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics. 2004;5:69. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agirre J, et al. Privateer: Software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol. 2015;22(11):833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- 63.Krissinel E, Henrick K. Lecture Notes in Computer Science. Springer-Verlag; Berlin Heidelberg: 2005. Multiple alignment of protein structures in three dimensions; pp. 67–78. [Google Scholar]
- 64.Serafini-Cessi F, Bellabarba G, Malagolini N, Dall’Olio F. Rapid isolation of Tamm-Horsfall glycoprotein (uromodulin) from human urine. J Immunol Methods. 1989;120(2):185–189. doi: 10.1016/0022-1759(89)90241-x. [DOI] [PubMed] [Google Scholar]
- 65.Schaeffer C, et al. Urinary secretion and extracellular aggregation of mutant uromodulin isoforms. Kidney Int. 2012;81(8):769–778. doi: 10.1038/ki.2011.456. [DOI] [PubMed] [Google Scholar]
- 66.Yang H, Wu C, Zhao S, Guo J. Identification and characterization of D8C, a novel domain present in liver-specific LZP, uromodulin and glycoprotein 2, mutated in familial juvenile hyperuricaemic nephropathy. FEBS Lett. 2004;578(3):236–238. doi: 10.1016/j.febslet.2004.10.092. [DOI] [PubMed] [Google Scholar]
- 67.Letunic I, Doerks T, Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 69.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pei J, Grishin NV. PROMALS3D: Multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beitz E. TEXshade: Shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics. 2000;16(2):135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- 72.Fokkema IF, et al. LOVD v.2.0: The next generation in gene variant databases. Hum Mutat. 2011;32(5):557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 73.Stenson PD, et al. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei X, et al. Novel uromodulin mutation in familial juvenile hyperuricemic nephropathy. Am J Nephrol. 2012;36(2):114–120. doi: 10.1159/000339752. [DOI] [PubMed] [Google Scholar]
- 75.Bai H, et al. A rare novel mutation in TECTA causes autosomal dominant nonsyndromic hearing loss in a Mongolian family. BMC Med Genet. 2014;15:34. doi: 10.1186/1471-2350-15-34. [DOI] [PMC free article] [PubMed] [Google Scholar]