Significance
Epstein–Barr virus (EBV) was the first human tumor virus discovered. Although nearly all adults are infected with EBV, very few go on to develop disease, for reasons that we are only beginning to understand. Infection with EBV induces a period of very rapid cell division, which requires an increased supply of metabolites, such as nucleotides, amino acids, and lipids. We found that EBV-infected cells that are unable to meet this increased metabolic demand are forced to stop proliferating and undergo a permanent growth arrest called senescence.
Keywords: Epstein–Barr virus, oncogene-induced senescence, autophagy, B cell, metabolism
Abstract
Epstein–Barr virus (EBV) is an oncogenic herpesvirus that has been causally linked to the development of B-cell and epithelial malignancies. Early after infection, EBV induces a transient period of hyperproliferation that is suppressed by the activation of the DNA damage response and a G1/S-phase growth arrest. This growth arrest prevents long-term outgrowth of the majority of infected cells. We developed a method to isolate and characterize infected cells that arrest after this early burst of proliferation and integrated gene expression and metabolic profiling to gain a better understanding of the pathways that attenuate immortalization. We found that the arrested cells have a reduced level of mitochondrial respiration and a decrease in the expression of genes involved in the TCA cycle and oxidative phosphorylation. Indeed, the growth arrest in early infected cells could be rescued by supplementing the TCA cycle. Arrested cells were characterized by an increase in the expression of p53 pathway gene targets, including sestrins leading to activation of AMPK, a reduction in mTOR signaling, and, consequently, elevated autophagy that was important for cell survival. Autophagy was also critical to maintain early hyperproliferation during metabolic stress. Finally, in assessing the metabolic changes from early infection to long-term outgrowth, we found concomitant increases in glucose import and surface glucose transporter 1 (GLUT1) levels, leading to elevated glycolysis, oxidative phosphorylation, and suppression of basal autophagy. Our study demonstrates that oncogene-induced senescence triggered by a combination of metabolic and genotoxic stress acts as an intrinsic barrier to EBV-mediated transformation.
Epstein–Barr virus (EBV) is a gamma herpesvirus that establishes a lifelong, latent infection in >90% of adults worldwide. EBV is associated with a number of malignancies, including African endemic Burkitt’s lymphoma, posttransplant lymphoproliferative disease, nasopharyngeal carcinoma (NPC), and HIV-associated lymphomas (1). These malignancies primarily develop in immunocompromised patients, pointing to the critical role that the immune system plays in controlling infection. However, it has recently become appreciated that additional intrinsic responses limit the ability of EBV to transform cells.
In vitro stimulation of B cells either through EBV infection or mitogen treatment results in a transient period of hyperproliferation reminiscent of a germinal center reaction. EBV elicits entry into the cell cycle through the EBV latency proteins, EBNA2 and EBNA-LP, which up-regulate the expression of progrowth genes (2–4). This period of rapid proliferation leads to the activation of the DNA damage response (DDR), which can signal through p53 to induce either apoptosis or senescence (5). In contrast to mitogen-stimulated cells, EBV-infected cells are able to escape apoptosis and, instead, a subset undergo a G1/S-phase growth arrest (6). The specific cellular pathways that contribute to this growth arrest are poorly understood.
Oncogene-induced senescence (OIS) is a premature form of senescence in which cells undergo an irreversible growth arrest after chronic oncogene expression or the inactivation of tumor suppressors (7, 8). Current models suggest that the onset of OIS is a consequence of a persistent DDR resulting from replicative stress induced during oncogene-driven hyperproliferation (9–11). It is now appreciated that OIS plays an important role in suppressing tumorigenesis in a wide range of cell types (7). Additionally, studies suggest that OIS can suppress proliferation driven by the overexpression of viral proteins or after oncogenic virus infection (12, 13).
Increasing evidence suggests that there is a link between senescence and macroautophagy (hereafter referred to as autophagy) (7). Autophagy is a catabolic process in which organelles or proteins are targeted for lysosomal degradation and recycling (14, 15). Studies have demonstrated that autophagy promotes cell-cycle arrest and the production of senescence-associated interleukins (16). However, autophagy has also been linked to the progression of tumorigenesis by providing metabolic intermediates to fuel proliferation (14). Oncogene activation leads to a substantial increase in the need for ATP, biosynthetic intermediates, and reducing equivalents to maintain proliferation, thereby creating metabolic stress (17). Cancer cells have been shown to mitigate this stress by up-regulating the basal level of autophagy and by transitioning their metabolic profile from oxidative phosphorylation (OXPHOS) toward aerobic glycolysis, also known as the Warburg effect (18, 19).
The essential role of metabolism in driving virus replication has been hinted at since the 1950s and is now becoming fully appreciated with the advent of new technologies (20). It is now appreciated that many eukaryotic viruses alter host metabolism to provide the energetic and biosynthetic resources necessary to drive virus replication and virion production. Less intuitive is the observation that viruses also alter host cellular metabolism during latent infection despite the lack of need for biosynthetic intermediates to produce viral progeny. Kaposi’s sarcoma-associated herpesvirus (KSHV) induces a Warburg effect during latent infection of endothelial cells, which is necessary for the survival of infected cells (21). A detailed metabolomics study of cells latently infected with KSHV further confirmed the increased production of glycolytic metabolites and also found an up-regulation of long-chain fatty acids (21). Additionally, glycolysis and fatty acid synthesis were found to be up-regulated in KSHV-associated primary effusion lymphoma compared with uninfected B cells (22). EBV latency is also associated with an altered metabolic state. EBV-infected nasopharyngeal carcinoma (NPC) cells exhibit high levels of glycolysis, an effect that can be recapitulated by the expression of EBV latency protein, latent infection membrane protein 1 (LMP1), alone (23). This increased level of glycolysis could be attributed to the increased surface expression of GLUT1 that was shown to be associated with LMP1-mediated NF-κB signaling in B cells (24). However, LMP1 expression is low during EBV-induced B-cell hyperproliferation—a period in which the cell should have the greatest need for increased metabolic flux.
In the present study, we have developed a method to identify and isolate EBV-infected primary human B cells that initially undergo a period of hyperproliferation and then arrest. We have used this approach to define the metabolic demands of hyperproliferation that drives the majority of EBV-infected cells into permanent growth arrest.
Results
EBV Infection of Primary B Cells Induces a Senescence-Like Growth Arrest.
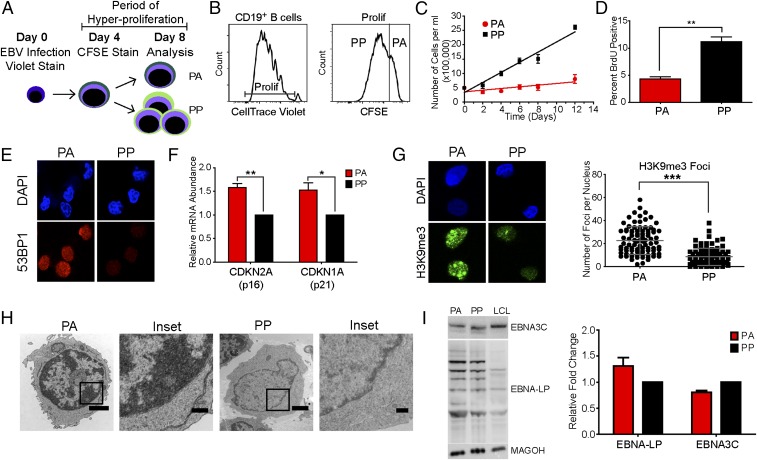
Early after EBV infection, B cells undergo a transient period of hyperproliferation that induces a G1/S-phase growth arrest in a subset of the population (5, 6). To functionally characterize this population, we have devised a protocol that allows us to identify and isolate cells that initially proliferate and then arrest. Peripheral blood mononuclear cells (PBMCs) were isolated from human blood and stained with CellTrace Violet followed by infection with the B95-8 strain of EBV. The cells were then stained with a second proliferation tracking dye, 6-carboxyfluorescein succinimidyl ester (CFSE), at day 4 after infection to coincide with the initial burst of hyperproliferation. The cells were then monitored over time, with those that were low for the Violet stain but high for the CFSE stain termed proliferated–arrested (PA) and those that were low for both stains designated proliferated–proliferated (PP) (Fig. 1 A and B).
Fig. 1.
A subset of EBV-induced hyperproliferating cells exhibit characteristics of OIS. (A) Schematic representing the experimental protocol. (B) Histograms showing CD19+ B-cell division measured at 8 d after infection. (B, Left) Proliferation of CD19+ B cells was determined through the dilution of the CellTrace Violet stain. (B, Right) The cells labeled “Prolif” were further analyzed for dilution of the CFSE stain. Cells that dilute the CellTrace Violet stain, but not the CFSE stain, are considered arrested. (C) EBV-infected CD19+ B cells were sorted into PA and PP populations and recultured in fresh medium. Samples were counted by trypan blue exclusion every 48 h (n = 3). (D) The percent of BrdU incorporation in PA or PP cells at day 8 after infection. (E) IF of DAPI (blue) or 53BP1 (red) measured from sorted PA and PP cells (n = 3). (F) The expression level of CDKN2A and CDKN1A mRNA was measured from sorted PA and PP cells. Relative mRNA abundance was normalized to SETDB1. Data are represented as fold change relative to the PP cells (n = 5). (G, Left) IF of DAPI (blue) or H3K9me3 (green) measured from sorted PA and PP cells (n = 3). (G, Right) Quantification of IF. (H) Representative TEM images of sorted PA or PP cells (n = 2). (Magnification: PA, 3,300×; PP, 4,400×.) (Scale bars: 1 µm.) Insets show heterochromatic DNA. (Scale bar: 0.5 µm.) Error bars represent SEM. (I, Left) Representative immunoblot of sorted PA and PP cells or LCLs stained with the indicated antibody (n = 3). (I, Right) Quantitation of immunoblot normalized to MAGOH loading control (n = 3). Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 as determined by a paired t test. n, the number of independent donors tested.
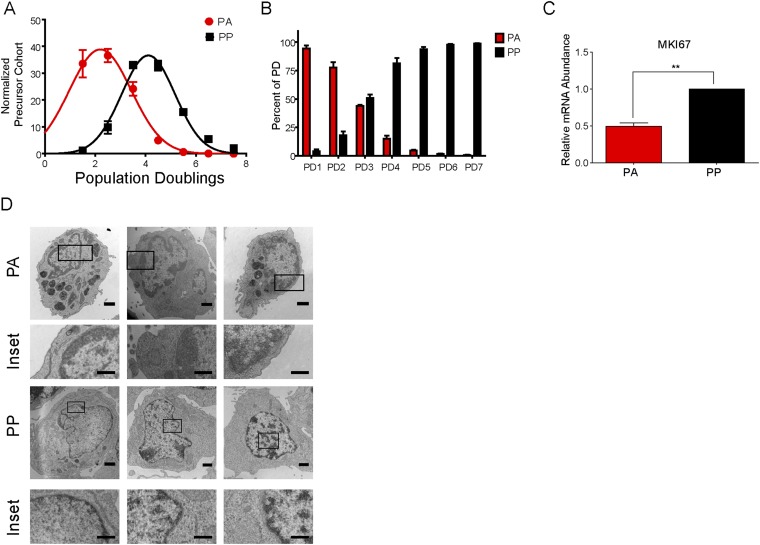
Precursor cohort analysis (25) of the double-stained cells on day 8 after infection demonstrated that the PA cells had a mean division number of 2, similar to the day 4 cell population and in contrast to the PP cells, which had a mean division number of 4 (Fig. S1A). Additionally, the double-stain experiment allows us to determine the percentage of cells that arrest after each initial division. The majority of the day 8 cells that divided three times or less, population doublings 1–3 (PD1–3), arrested (PA), whereas the cells that had divided more than four times were predominately in the PP population (Fig. S1B). Of note, the PD1–3 population was previously found to have elevated markers of the DDR, which is known to attenuate EBV-mediated B-cell transformation (5).
Fig. S1.
A subset of EBV-induced hyperproliferating cells exhibit characteristics of OIS. EBV-infected B cells were stained as described in Fig. 1A and analyzed at day 8 after infection. (A) The number of cells within each CellTrace Violet PD was determined for the PA and PP cells. The data were then precursor cohort normalized and plotted against PDs (n = 3). (B) The number of cells within each CellTrace Violet PD was further analyzed to determine the percentage that arrest (PA) vs. continue to proliferate (PP; n = 3). (C) The expression level of MKI67 mRNA was measured from sorted PA and PP cells. Relative mRNA abundance was normalized to SETDB1. (D) Representative TEM images of sorted PA or PP cells (n = 2). Inset shows heterochromatic DNA. (Magnification ranges from 5,600× to 7,100×.) (Scale bar: 1 μm; Inset, 0.5 μm.) Data are represented as fold change relative to the PP cells (n = 3). Error bars represent SEM. **P < 0.01 as determined by a paired t test. n, the number of independent donors tested.
The observed growth arrest could be a transient quiescence or senescence. To begin to address these possibilities, we sorted PA cells from three independent donors to purity and monitored their growth for 12 d. The number of PA cells remained constant with no new proliferation or cell death as determined by trypan blue exclusion. In contrast, the PP cells continued to divide and ultimately transformed into lymphoblastoid cell lines (LCLs) (Fig. 1C). Additionally, the PA cells had a significant decrease in the expression of the proliferation marker MKI67 (Fig. S1C), as well as decreased BrdU incorporation, further confirming the growth arrest (Fig. 1D). Senescent cells exhibit a number of distinctive characteristics in addition to growth arrest that allow for their identification in vitro (7). Unscheduled oncogene activation, resulting in DNA damage, leads to the activation of the DDR and the downstream p53-p21 and Rb-p16 signal transduction cascades, which can mediate the senescence growth arrest (7). The PA cells had a significant increase in the expression of the DDR marker 53BP1 (Fig. 1E) and the tumor suppressors, CDKN1A (p21) and CDKN2A (p16) (Fig. 1F). Senescent cells are also characterized by altered chromatin structure known as senescence-associated heterochromatic foci, which are enriched for trimethylated lysine 9 of histone H3 (H3K9me3) (7). Immunofluorescence (IF) analysis demonstrated that the PA cells have a significant increase in H3K9me3 staining relative to the PP cells (Fig. 1G), as well as an increase in overall heterochromatic DNA, as evidenced by transmission electron microscopy (TEM) (Fig. 1H and Fig. S1D).
EBV promotes and maintains B-cell proliferation through the concerted action of the EBV latency proteins. EBNA2 and EBNA-LP induce the expression of progrowth genes (2–4), whereas EBNA3C inhibits the expression of tumor suppressors such as p16 (26). We reasoned that the growth arrest could be due to the reduced expression of one of these proteins. However, we observed that there was a modest increase in the expression of EBNA-LP and only a slight decrease in EBNA3C in the PA cells relative to the PP population (Fig. 1I). This finding is consistent with our previous observations demonstrating that PD1–3 cells have a greater ratio of EBNA-LP to EBNA3C, reflective of an earlier state of viral latency-driven outgrowth (5) and is also consistent with the work of others indicating that proliferating B cells after EBV infection are uniformly EBNA2-positive (27).
Transcriptomic Analysis of PA vs. PP Indicates Heightened p53 Pathway and Decreased Cell Cycle and DNA Replication.
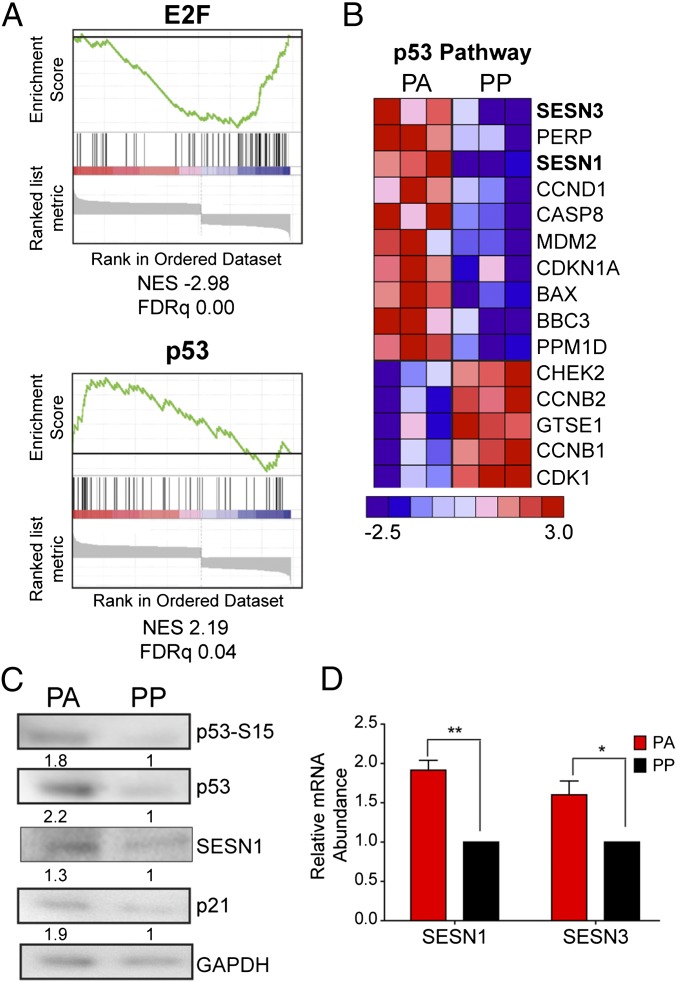
To delineate the effector pathways that mediate this EBV-induced growth arrest, we performed gene expression analysis using an Affymetrix Human U133 2.0 Plus microarray with three independent donors sorted into PA or PP populations. The resulting hybridization data were robust multi-array average (RMA)-normalized, and a total of 158 genes were expressed higher in the PA cells relative to PP cells (>1.5-fold, P < 0.05), whereas the expression of 160 genes was reduced (>1.5-fold, P < 0.05) (Dataset S1).
The PA cells were depleted for mRNAs in pathways involved in cell-cycle progression and displayed lower levels of transcriptional targets of E2F indicative of G1/S cell-cycle arrest (Fig. 2A and Tables S1–S3). Additionally, the PA cells had elevated levels of p53 pathway transcriptional targets (Fig. 2 A and B and Tables S1–S3), which is activated in response to cellular stress and regulates the expression of genes involved in processes such as cell-cycle progression and metabolism (28). Indeed, we confirmed that PA cells displayed activated p53 pathway including p53 phosphorylation on serine 15, accumulation of total p53, and induction of the downstream target p21 (Fig. 2C). In addition, two notable p53 target genes, SESN1 and SESN3, were increased in the PA cells (29). The sestrins are an evolutionarily conserved group of genes induced in response to genotoxic stress and nutrient deprivation, leading to the inhibition of mammalian target of rapamycin complex 1 (mTORC1) signaling (29–31). We confirmed increased expression of sestrins in the PA population by quantitative RT-PCR (qRT-PCR) and immunoblot analysis (Fig. 2 C and D). These data suggest that EBV-induced hyperproliferation drives a subset of EBV-infected B cells into a state of metabolic or genotoxic stress that is then sensed by pathways that relay signals through the sestrins to induce growth arrest.
Fig. 2.
Transcriptomic changes between PA and PP cells indicate reduced cell-cycle progression and increased p53 activation in PA cells. EBV-infected B cells were stained as described in Fig. 1A and sorted at day 8 after infection. (A) GSEA of transcription factor use (MSigDB c3) in genes that exhibited significant change (two-way ANOVA, P < 0.05). Enrichment plots for E2F and p53 are shown. Normalized enrichment scores (NES) and false discovery rate q values (FDRq) are shown below each plot. (B) Heat map representing genes enriched in p53 Downstream Pathway (PID) obtained from GSEA. Genes exhibiting significant changes (two-way ANOVA, P < 0.05) were analyzed. The heat map shows genes that were changed at least 1.2-fold (n = 3). (C) Representative immunoblot of sorted PA and PP cells stained with the indicated antibody. Quantitation represents staining from three independent donors normalized to GAPDH. (D) The expression level of SESN1 and SESN3 mRNA was measured from sorted PA and PP cells (n = 3). Relative mRNA abundance was normalized to SETDB1. Data are represented as fold change relative to the PP cells. Error bars represent SEM; *P < 0.05; **P < 0.01 as determined by a paired t test. n, the number of independent donors tested.
Table S1.
Pathways enriched in PA cells/depleted in PP cells
| Name | NES | FDRq |
| GPCR Ligand Binding (REACTOME) | 2.82 | 0.00 |
| p53 Downstream Pathway (PID) | 2.26 | 0.02 |
| G alpha (i) signaling events (REACTOME) | 2.23 | 0.02 |
| Cytokine-cytokine receptor interaction (KEGG) | 2.13 | 0.03 |
| IL4 Pathway (PID) | 2.09 | 0.04 |
| NFAT pathway (PID) | 1.84 | 0.15 |
| IFNG pathway (PID) | 1.79 | 0.15 |
| cMyb pathway (PID) | 1.74 | 0.19 |
| Notch pathway (PID) | 1.72 | 0.19 |
| Lysosome (KEGG) | 1.72 | 0.18 |
| Cell adhesion molecules_CAMS (KEGG) | 1.70 | 0.16 |
| JAK-STAT signaling pathway (KEGG) | 1.64 | 0.19 |
| Calcium pathway (KEGG) | 1.61 | 0.20 |
| Wnt signaling pathway (KEGG) | 1.58 | 0.21 |
| HIF1a transcription factor pathway (PID) | 1.53 | 0.23 |
| Integration of energy metabolism (REACTOME) | 1.50 | 0.23 |
| TGF-beta signaling pathway (KEGG) | 1.49 | 0.23 |
Transcriptional analysis was performed on sorted PA and PP cells by using a Human Genome U133 Plus. 2.0 microarray. The resulting files were RMA-normalized, and a rank list of genes that exhibited significant changes (two-way ANOVA, P < 0.05) was generated for GSEA (Broad). The data are represented relative to the PA cells.
Table S3.
Transcription factor targets
| Name | NES | FDRq |
| Enriched in PA/depleted in PP | ||
| ETS1 | 2.33 | 0.09 |
| MAZ | 2.26 | 0.10 |
| ETS2 | 2.23 | 0.10 |
| FREAC2 | 2.15 | 0.09 |
| HSF1 | 2.14 | 0.08 |
| CEBPDELTA | 2.12 | 0.08 |
| AMEF2 | 2.10 | 0.09 |
| TGIF | 2.07 | 0.09 |
| FOXO4 | 2.05 | 0.09 |
| S8 | 2.04 | 0.09 |
| PITX2 | 2.02 | 0.10 |
| p53 | 2.01 | 0.10 |
| STAT6 | 1.98 | 0.11 |
| SRY | 1.96 | 0.12 |
| AP4 | 1.96 | 0.11 |
| AREB6 | 1.95 | 0.11 |
| STAT5A | 1.95 | 0.11 |
| MYOD | 1.94 | 0.11 |
| E47 | 1.94 | 0.11 |
| MMEF2 | 1.92 | 0.12 |
| Enriched in PP/depleted in PA | ||
| NRF1 | −3.37 | 0.00 |
| E2F | −3.31 | 0.00 |
| NFMUE1 | −2.70 | 0.00 |
| ELK1 | −2.60 | 0.00 |
| NFY | −2.52 | 0.00 |
| YY1 | −2.25 | 0.01 |
| ARNT | −2.17 | 0.01 |
| CREB | −1.80 | 0.05 |
| ATF | −1.80 | 0.06 |
| USF | −1.69 | 0.11 |
| ALPHACP1 | −1.69 | 0.14 |
| USF | −1.62 | 0.15 |
| SF1 | −1.57 | 0.18 |
| CEBPGAMMA | −1.55 | 0.19 |
| MAX | −1.54 | 0.19 |
| HNF4 | −1.53 | 0.20 |
Transcriptional analysis was done on sorted PA and PP cells using a Human Genome U133 Plus. 2.0 microarray. The resulting files were RMA normalized, and a rank list of genes that exhibited significant changes (two-way ANOVA, P < 0.05) was generated for GSEA (Broad). The data are represented relative to the PA cells.
Table S2.
Pathways depleted in PA cells/enriched in PP cells
| Name | NES | FDRq |
| Cell cycle (REACTOME) | −5.09 | 0.00 |
| Cell-cycle mitotic (REACTOME) | −4.99 | 0.00 |
| DNA replication (REACTOME) | −4.66 | 0.00 |
| PKL1 pathway (PID) | −3.63 | 0.00 |
| G1_S transition (REACTOME) | −3.36 | 0.00 |
| Aurora B pathway (PID) | −3.12 | 0.00 |
| Cell-cycle checkpoints (REACTOME) | −3.08 | 0.00 |
| mRNA processing (REACTOME) | −3.05 | 0.00 |
| mRNA splicing (REACTOME) | −2.94 | 0.00 |
| DNA repair (REACTOME) | −2.50 | 0.00 |
| Translation (REACTOME) | −2.43 | 0.00 |
| Metabolism of proteins (REACTOME) | −2.34 | 0.00 |
| Chromosome maintenance (REACTOME) | −2.26 | 0.00 |
| MHC class II antigen presentation (REACTOME) | −1.91 | 0.02 |
| TCA cycle and respiratory electron transport (REACTOME) | −1.85 | 0.03 |
| Transcription (REACTOME) | −1.77 | 0.05 |
| Myc-activated pathway (PID) | −1.66 | 0.08 |
| Downstream signaling of B-cell receptor (REACTOME) | −1.65 | 0.09 |
| Antiviral mechanism of IFN-stimulated genes (REACTOME) | −1.48 | 0.19 |
Transcriptional analysis was done on sorted PA and PP cells using a Human Genome U133 Plus. 2.0 microarray. The resulting files were RMA-normalized, and a rank list of genes that exhibited significant changes (two-way ANOVA, P < 0.05) was generated for GSEA (Broad). The data are represented relative to the PA cells.
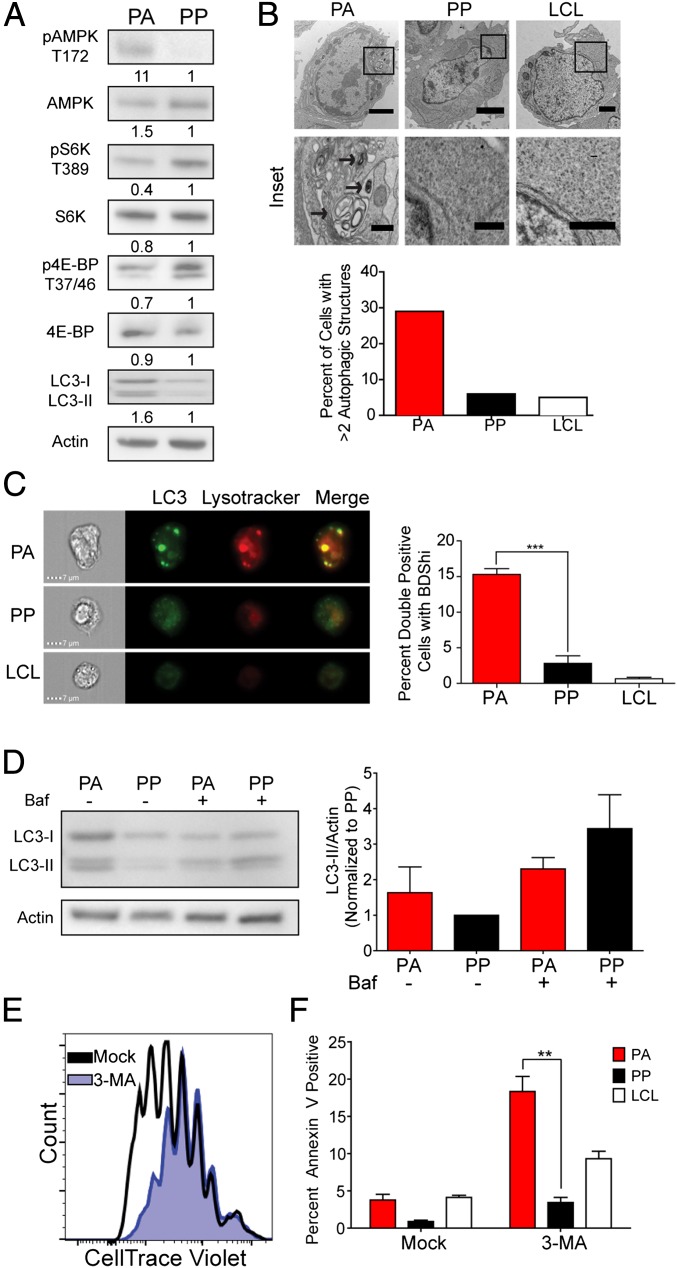
PA Cells Exhibit Reduced Activation of the mTORC1 Pathway and Inefficient Autophagic Flux.
The sestrins inhibit mTOR signaling through activation of the energy sensing protein AMP-activated protein kinase (AMPK) (29, 31). Suppression of mTOR signaling leads to a reduction in energy-consuming pathways, such as protein synthesis, and induces catabolic processes, such as autophagy (32). Consistently, we found that the PA cells had increased activation of AMPK and reduced activation of mTOR pathway components relative to the PP cells (Fig. 3A).
Fig. 3.
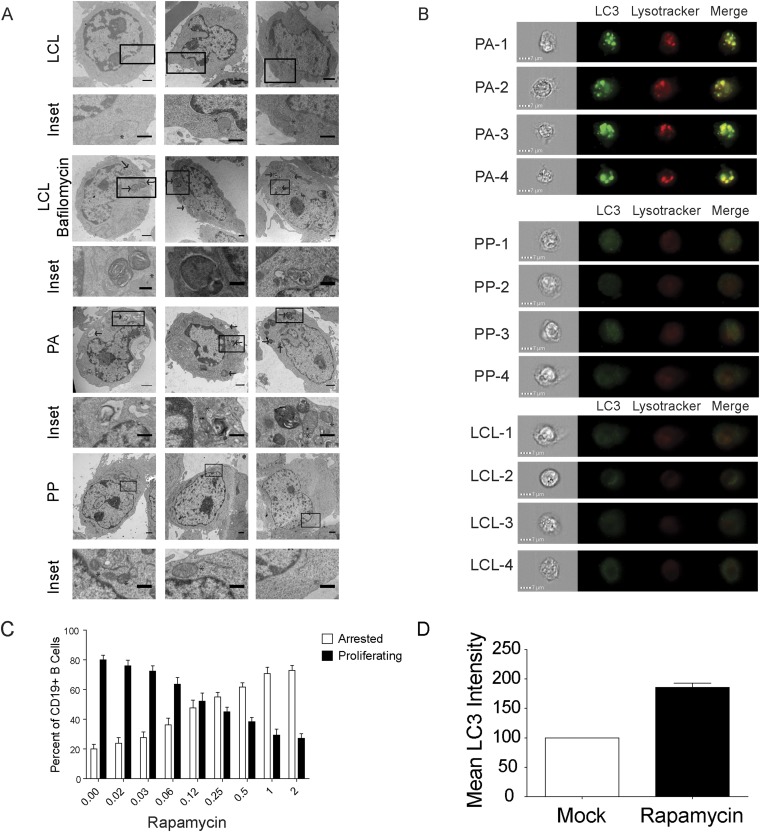
The PA cells exhibit suppression of the mTOR pathway and increased autophagy. EBV-infected B cells were stained as described in Fig. 1A and analyzed at day 8 after infection. (A) Representative immunoblot of sorted PA and PP cells stained with the indicated antibody (n = 3). Quantitation was done on three independent donors and normalized to actin. (B, Upper) Representative TEM image of sorted PA and PP cells as well as LCLs. Arrows indicate autolysosomes (n = 2). (Magnification: PA, 7,100×; PP, 8,800×; LCL, 4,400×.) (Scale bars: 2 µm; Inset, 0.5 µm.) (B, Lower) Quantitation of TEM data. The graph represents the percent of cells with greater than two autophagic structures. (C, Left) Imagestream analysis of PA, PP, and LCLs that were stained with Lysotracker and anti-LC3 (n = 3). (C, Right) Quantitation of cells showing high colocalization of Lysotracker with LC3 in double-positive cells. The data are represented as a measure of bright detail similarity as determined by IDEAS software (Version 3.0). (D, Left) Representative immunoblot of sorted PA and PP cells that were treated with Bafilomycin A or mock treated. (D, Right) Quantitation of LC3-II staining after normalization to the actin loading control (n = 3). (E) Histograms showing CD19+ B-cell division measured at 8 d after infection after treatment with 5 mM 3-MA for 48 h. (F) Percentage of Annexin positive PA or PP cells after treatment as in E. Error bars represent SEM. **P < 0.01; ***P < 0.001 as determined by a paired t test.
A consequence of decreased mTORC1 activation is the induction of autophagy, which has been linked to the onset of cellular senescence (16). We therefore assayed for markers of autophagy in our PA and PP cells. We observed an increase in the levels of the autophagy marker LC3-II in the PA cells relative to the PP population (Fig. 3A). We also observed an increase in the presence of autophagosomes and phagolysosomes in the PA cells relative to both the PP cells and LCLs as detected by TEM (Fig. 3B and Fig. S2A). Additionally, there was an increase in the colocalization of LC3 with lysosomes in the PA relative to the PP cells (Fig. 3C and Fig. S2B). LCLs exhibited a further reduction in colocalized LC3 with lysosomes relative to all populations (Fig. 3C). LC3-II is subject to autophagic degradation, and its accumulation in the PA cells could be the result of enhanced autophagy or reduced degradation due to a blockage in autophagic flux. To differentiate between these possibilities, we treated sorted PA and PP cells with bafilomycin A, which inhibits lysosomal acidification and autophagic degradation. We observed that there was only a modest increase in LC3-II staining in the PA cells after bafilomycin A treatment, in contrast to the PP cells, which exhibited a substantial increase in the levels of LC3-II (Fig. 3D). These data, combined with the increased colocalization of LC3 with lysosomes and increased number of autophagolysosomes in the PA cells, indicates that the PA cells have a blockage in autophagic flux that prevents them from efficiently degrading cellular material to drive proliferation.
Fig. S2.
The PA cells exhibit suppression of the mTOR pathway and increased autophagy. (A) Representative TEM images of sorted PA and PP cells as well as negative control LCLs and positive control serum starved and bafilomycin-treated LCLs. Arrows indicate autolysosomes and/or phagosomes (n = 2). (Magnification ranges from 3,400× to 11,500×.) (Scale bar: 1 μm; Inset, 0.5 μm.) (B) Imagestream analysis of PA, PP, and LCLs that were stained with Lysotracker and anti-LC3. (C) Percent of cells that arrest after treatment with the indicated dose of Rapamycin (n = 3). (D) Relative increase in LC3 mean fluorescence intensity in cells treated with 2 nM rapamycin or mock treated (n = 2). n, the number of independent donors tested.
We next wanted to elucidate the functional role of autophagy during hyperproliferation. We treated EBV-infected, CD19+ B cells for 48 h with the autophagy inhibitor 3-methyladenine (3-MA) and observed a substantial decrease in proliferation at 8 d after infection, with treated cells displaying a CellTrace Violet profile reminiscent of cells at the time of treatment (Fig. 3E). We also observed a substantial increase in apoptosis specifically in PA cells relative to both LCLs and the PP population (Fig. 3F). Conversely, treatment with the mTOR inhibitor rapamycin promoted autophagy as determined by an increase in LC3 staining and increased the percentage of arrested B cells (Fig. S2 C and D). Overall, these data demonstrate that EBV-infected, hyperproliferating cells need a balanced level of autophagy to produce the biosynthetic intermediates necessary for cell growth and proliferation.
Metabolic Analysis Reveals Decreased Mitochondrial Respiration in PA Cells.
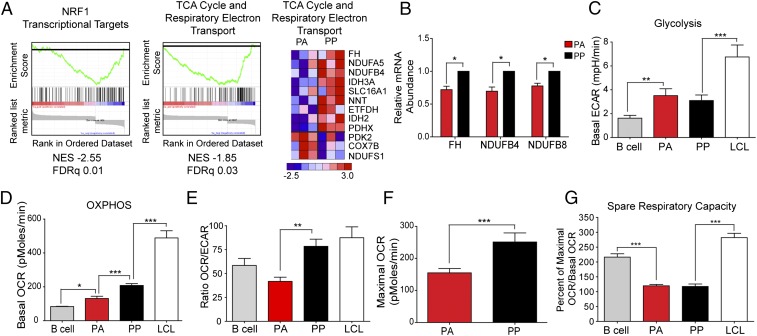
Autophagy is often triggered as a consequence of nutrient deprivation caused by a cellular metabolic imbalance (33). We therefore wanted to determine whether there was an altered metabolic state between the hyperproliferating cells (PA and PP) and other B-cell populations. Our gene expression analysis indicated that the PA cells had reduced levels of mRNAs in the canonical TCA cycle and respiratory electron transport pathways, as well as transcriptional targets of nuclear respiratory factor 1 (NRF1), which activates the expression of genes involved in mitochondrial biogenesis and OXPHOS (Fig. 4A). RT-PCR analysis confirmed that components of complex I of the electron transport chain and TCA cycle enzymes were decreased in the PA cells (Fig. 4B).
Fig. 4.
Integration of transcriptomic and metabolic profiling reveals key differences between uninfected B cells, PA, PP, and LCLs. (A, Left and Center) Enrichment plot of transcriptional targets of NRF1 (Left) or genes enriched in “TCA Cycle and Respiratory Electron Transport” pathway (Center) obtained from GSEA. (A, Right) Heat map representing genes enriched in TCA Cycle and Respiratory Electron Transport. Genes exhibiting significant changes (two-way ANOVA, P < 0.05) are represented (n = 3). (B) The expression levels of the indicated mRNAs were measured from sorted PA and PP cells. Relative mRNA abundance was normalized to SETDB1. Data are represented as fold change relative to the PP cells. (C–G) Metabolic analysis was performed on uninfected B cells; sorted PA and PP cells; or LCLs by using a Seahorse XF to measure ECAR (C), OCR (D), the ratio of OCR to ECAR (E), Maximal OCR (F), or the Spare Respiratory Capacity (G). Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 as determined by a paired t test. All analysis was performed on three independent donors.
EBV infection induces B cells to undergo a period of rapid proliferation combined with a concomitant increase in biomass, processes that require both energy and biosynthetic intermediates. The decreased expression of enzymes important for mitochondrial respiration and the TCA cycle could lead to a metabolic imbalance promoting autophagy and senescence. To look for metabolic changes that occur in B cells before and after primary B-cell infection with EBV, we used the Seahorse XF, which simultaneously measures the basal extracellular acidification rate (ECAR), a marker of glycolysis, and the oxygen consumption rate (OCR), an indicator of OXPHOS.
We found that the PA cells are metabolically distinct from the other B-cell populations. Although the PA cells are similar to PP cells in both the basal level and rate of glycolysis (ECAR) (Fig. 4C and Fig. S3A), they have a significantly lower OCR than PP cells, a level that is only slightly higher than that observed in resting B cells (Fig. 4D). Furthermore, the ratio of OCR to ECAR indicates that the PA cells are more heavily reliant on glycolysis to meet their energy needs (Fig. 4E). We also observed that there was a substantial increase in both glycolysis and OXPHOS as cells transition from the hyperproliferating state to transformed LCLs (Fig. 4 C–E).
Fig. S3.
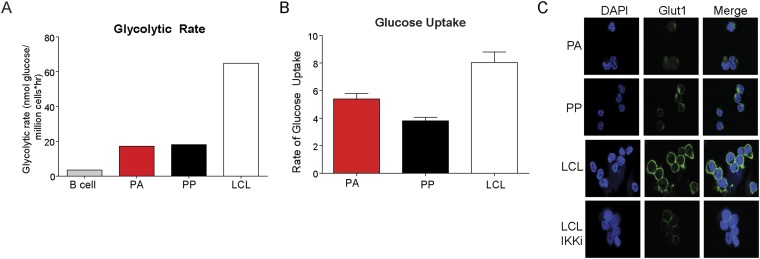
Metabolic profile reveals differences between uninfected B cells, PA, PP, or LCLs. (A) Glycolytic rate of uninfected B cells; CD19+, EBV-infected, sorted PA and PP cells, or LCLs. (B) Glucose uptake in PA, PP, or LCLs. Cells were starved in glucose-free medium for 1 h and then supplemented with 2-NBDG, a fluorescent glucose analog. (C) IF of DAPI (blue) or GLUT1 (green).
The lower basal level of mitochondrial respiration combined with the lower expression of genes involved in the TCA cycle and the electron transport chain suggests that the PA cells may have a reduced capacity to undergo mitochondrial respiration. To determine whether there was a difference in the potential maximal level of respiration, we uncoupled the electron transport chain from OXPHOS using carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), which dissipates the proton gradient across the mitochondrial inner membrane. FCCP causes the OCR to increase to the maximum level supported by the electron transport chain and substrate supply. We found that there is a significant decrease in the maximum level of OCR in PA cells relative to the PP cells (Fig. 4F). A comparison of the basal level of OCR relative to the maximal level can be used to calculate the spare respiratory capacity of the cells, which is defined as the amount of additional energy that the cell can make in times of stress. There is only a slight difference between the basal and maximal OCR in both the PA and PP cells (Fig. 4 D and F), leading to a significantly lower spare respiratory capacity relative to resting B cells and LCLs (Fig. 4G). These data suggest that the hyperproliferating cells have an overall reduced capacity to mitigate metabolic stress compared with LCLs and that the subset that arrest have a significant reduction in mitochondrial respiration.
Metabolic Stress and Genotoxic Stress Contribute to the Suppression of Early EBV-Induced B-Cell Proliferation.
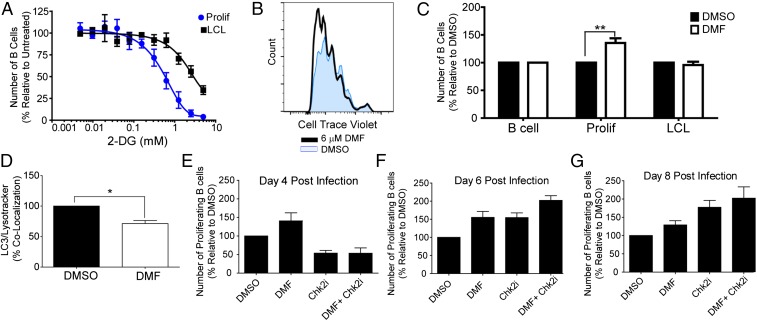
We next sought to functionally characterize this metabolic imbalance observed during early EBV infection. We found that early hyperproliferating cells were more sensitive to the nonhydrolyzable glucose analog, 2-DG, relative to LCLs (Fig. 5A). This result was not due to increased glucose import or glucose transporter expression, but rather a reduction in glycolysis (Fig. S3 B and C). As a corollary, we would also expect that increasing the ability of early proliferating cells to promote OXPHOS would stimulate early B-cell proliferation, while not affecting LCLs. Indeed, we found that supplementation of medium with a cell-permeable form of fumarate, dimethyl fumarate (DMF), significantly increased the number of cells that hyperproliferate, but had no effect on uninfected B cells or LCL proliferation (Fig. 5 B and C). Additionally, we found that DMF treatment reduced the accumulation of LC3 with lysosomes (Fig. 5D).
Fig. 5.
2-DG preferentially inhibits and DMF increases early EBV-infected B-cell proliferation in a manner distinct from Chk2i treatment. (A) EBV-infected cells were treated with the indicated concentration of 2-DG at day 4 after infection, and the number of proliferating CD19+ B cells was determined by FACS on day 8. LCLs were treated for an identical time period (n = 3). (B) Representative histogram showing CD19+ B cells proliferation at day 8 after infection. The cells were treated with either DMSO or 6 µM DMF at the time of infection. (C) The number of proliferating uninfected, CD19+ B cells (B cell); EBV-infected, CD19+ B cells that diluted CTV (Prolif) or LCLs was determined by FACS and graphed as a percentage of the DMSO-treated control. Prolif cells were treated with DMSO or 6 µM DMF at the time of infection and analyzed at day 8 after infection (n = 3). (D) EBV-infected B cells were treated with DMF as in B and analyzed for colocalization of LC3 with Lysotracker by Imagestream analysis. (E–G) The number of proliferating CD19+ B cells was determined for cells that were treated with DMSO, 6 µM DMF, 5 µM Chk2i, or a combination of Chk2i and DMF at the time of infection. The data were analyzed by FACS at day 4 (E), day 6 (F), or day 8 (G) after infection and are presented as a percentage of the DMSO-treated control. A minimum of three independent donors were analyzed for each time point. Error bars represent SEM. *P < 0.05; **P < 0.01 as determined by a paired t test. n, the number of independent donors tested.
We previously demonstrated that the DDR was a major suppressor of EBV-infected B-cell hyperproliferation and that inhibition of the Chk2 kinase would increase the number of proliferating B cells (5). We therefore assayed whether our observed metabolic imbalance was distinct from the DDR by infecting cells in the presence of a small molecule inhibitor of Chk2 (Chk2i) either with or without DMF. We observed a distinct difference in the proliferation kinetics between cells treated with DMF compared with Chk2i treatment. The proproliferative phenotype induced by DMF manifested by day 4 after infection/treatment, and the increased number of proliferating cells remained constant over time (Fig. 5 E–G). In contrast, treatment with Chk2i resulted in a suppression of proliferation on day 4 (Fig. 5E), followed by a strong increase in proliferation by day 6 after infection/treatment, which amplified over time (Fig. 5F). A combination of Chk2i and DMF further increased the number of proliferating cells at days 6 and 8 relative to single treatment alone (Fig. 5 F and G). These data indicate that both metabolic imbalance and DNA replicative stress contribute to restricting EBV-mediated B-cell proliferation.
Discussion
EBV is found ubiquitously within the adult population, but only leads to the development of malignancies in a small subset of the infected population. Although this discordance is largely due to the actions of an adaptive immune response, there is clearly an additional intrinsic response that suppresses the development of disease. It has also long been observed that the efficiency of EBV transformation is low, with only 1–10% of infected, primary human B cells becoming immortalized, further pointing to an innate barrier to transformation (34, 35). We have previously demonstrated that EBV infection drives human B cells into a transient period of hyperproliferation, which induces a DDR, leading to a G1/S-phase growth arrest (5, 6). We wanted to characterize this population further to gain an understanding of the host cell factors that mitigate transformation. We have developed a double-stain assay that allows us to identify and isolate the population of cells that undergo this early, rapid wave of proliferation and then arrest.
Senescence allows the host cell to inhibit the aberrant proliferation induced through the expression of viral latent oncoproteins. It is clear, however, that EBV has evolved mechanisms to bypass this growth arrest to allow for transformation (26). The viral latency protein EBNA3C is essential for transformation because cells infected with EBV lacking this gene undergo a cell-cycle arrest due to the up-regulation of p16 (36, 37). The need for EBNA3C is ablated in B cells that naturally lack functional p16 (38). However, cells infected with the EBNA3C-deficient virus do undergo the robust burst of proliferation and divide normally for the first week, albeit with a significant increase in markers of the DDR (5). Additionally, we only observed a modest decrease in the expression of EBNA3C in our PA cells relative to the PP population. Although this finding points to the need for additional latency genes to bypass the growth arrest, it is clear that EBNA3C plays a critical role in allowing EBV-infected cells to avoid OIS through its suppression of p16.
Autophagy has both beneficial and deleterious consequences during viral infection. Many viruses, including EBV, manipulate autophagy to complete lytic replication (39, 40). Additionally, autophagy has the potential to provide the metabolic intermediates needed to fuel the rapid proliferation necessary for the establishment of EBV latency. In contrast, autophagy has generally been viewed as a threat to many viruses because it can result in the degradation of viral proteins and leads to the onset of OIS (39). As such, many viruses have evolved elegant mechanisms of subverting autophagy (12, 39). Previously, very little was known about the role of autophagy during EBV latency. We found that the basal level of autophagy is increased in hyperproliferating cells relative to LCLs and that the inhibition of autophagy early during infection suppresses growth. We also found that the arrested cells had a reduced level of autophagic flux that could act as a barrier to the production of the biosynthetic intermediates necessary for proliferation.
The transient period of hyperproliferation requires a substantial increase in nucleotides, amino acids, and lipids for DNA replication and cell division. These needs can be met through a combination of metabolic reprogramming and an increase in the basal level of autophagy (17). We hypothesized that an inability to meet the metabolic demands of hyperproliferation could induce the EBV-infected B cells to undergo senescence. We demonstrate that the PA cells have a reduced ability to undergo OXPHOS and that complementing this defect with a soluble TCA intermediate increases the number of cells that hyperproliferate early after infection. We believe that the reduced ability to undergo mitochondrial metabolism points to the cause rather than a consequence of OIS, because senescent cells typically favor the more efficient OXPHOS over glycolysis (41).
In the transition from early hyperproliferation to transformation, EBV strongly up-regulates glycolysis and OXPHOS and also suppresses autophagy. LMP1 is an EBV latency protein that has been linked to the regulation of glycolysis and autophagy in LCLs. LMP1 can induce autophagy through the unfolded protein response as a way of regulating its own expression (42, 43). Conversely, LMP1 reduces autophagy in LCLs by up-regulating glucose import via NF-κB signaling (24). Additionally, LMP1 has been shown to up-regulate glycolysis in NPC cells (23). In this study, we found that LCLs have increased expression of GLUT1 as well as increased glucose uptake relative to the hyperproliferating cells, which we have previously shown to express much lower levels of LMP1 (44). We believe that the strong increase in glycolysis and suppression of autophagy that we observed in LCLs relative to the hyperproliferating cells is directly linked to the delayed expression of LMP1.
Our study characterizes the intrinsic pathways that contribute to the arrest of B cells early after EBV infection. Rapid cellular proliferation, as is seen after EBV infection, leads to replicative stress such as stalled or collapsed replication forks (45). Additionally, proliferating cells need to increase the production of biosynthetic intermediates such as nucleotides, amino acids, and lipids to promote cell growth and proliferation (17). An insufficient supply of nucleotides can contribute to the generation of stalled replication forks, linking replicative stress to metabolic stress (46–48). EBV-infected cells meet these metabolic demands through an increase in both glycolysis and mitochondrial respiration. The cells that arrest early after infection have a reduced capacity to undergo OXPHOS, which would lead to a reduction in the intermediates required for faithful DNA replication as well as cell division. Therefore, the combination of genotoxic and metabolic stress activates the p53 tumor suppressor, resulting in sestrin/AMPK-mediated permanent growth arrest of early infected cells, severely limiting EBV-driven B-cell immortalization.
Materials and Methods
Viruses and Cells.
B95-8 virus was produced from the B95-8 Z-HT cell line as described (49). Buffy coats were obtained from normal donors through the Gulf Coast Regional Blood Center, and PBMCs were isolated by Ficoll Histopaque gradient (Sigma, no. H8889). CD19+ B cells were purified from PBMCs by using the BD iMag Negative Isolation Kit (BD, no. 558007). Purity was routinely >90% as determined by flow cytometry. Primary cells were cultured in RPMI 1640 plus 15% (vol/vol) FCS, 2 mM l-glutamine, penicillin/streptomycin, and Cyclosporine A (0.5 µg/mL), whereas LCLs were cultured in similar medium containing only 10% (vol/vol) FCS (R10). All bulk infections were performed by incubating cells with B95-8 Z-HT supernatants 1 h at 37 °C in a CO2 incubator followed by washing in PBS and resuspending cells in R15 medium.
Chemicals.
The 2-deoxy-d-glucose (Sigma, no. D8375) and Metformin (hydrochloride) (Cayman Chemicals, no. 13118) were resuspended directly in R15 medium. Rapamycin (MP, no. 159346), Bafilomycin A1 (Sigma, no. B1793), DMF (Sigma, no. 242926), and Chk2i II (EMD, no. 220486) were resuspended in DMSO.
Antibodies.
Mouse anti-human CD19 antibody (clone 33-6-6; gift from Tom Tedder, Department of Immunology, Duke University Medical School, Durham, NC) conjugated with either APC or PE was used as a surface B-cell marker in flow cytometry.
Additional antibodies used within this study are shown in Table S4.
Table S4.
Antibodies used
| Name | Company | Cat no. | Conc. |
| Phospho-AMPKα (Thr-172) (40H9) | Cell Signaling | 2535 | 1:1,000 |
| AMPK (D5A2) | Cell Signaling | 5831 | 1:1,000 |
| Phospho-p70 S6 kinase (Thr-389) (108D2) | Cell Signaling | 9234 | 1:1,000 |
| p70 S6 kinase (49D7) | Cell Signaling | 2708 | 1:1,000 |
| Phospho-4E-BP1 (Thr-37/46) (236B4) | Cell Signaling | 2855 | 1:1,000 |
| 4E-BP1 | Cell Signaling | 9452 | 1:1,000 |
| β-Actin | Rockland | 600-401-886 | 1:1,000 |
| H3K9me3 | Millipore | 07-442 | 1:100 |
| GLUT1 | Abcam | Ab115730 | 1:250 |
| 53BP1 | Cell Signaling | 4937 | 1:50 |
| LC3 | MBL | PM036 | 1:500 |
| Sestrin 1 | Novus Biologics | NP68677 | 1:500 |
| Phospho-p53 (Ser-15) | Cell Signaling | 9286 | 1:1,000 |
| p53 | Cell Signaling | 2525 | 1:1,000 |
Cat, catalog; Conc., concentration.
Double Staining Protocol to Capture Early Proliferating and Arresting B Cells.
PBMCs were isolated from a buffy coat and stained with CellTrace Violet (Invitrogen, no. C34557) followed by infection with EBV. The cells were grown in R15 medium for 4 d before staining with CFSE (Sigma, no. 21888). The samples were resuspended in fresh R15 medium, and proliferation was monitored as described below.
Cell Proliferation Assays.
PBMCs were EBV-infected and stained with CellTrace Violet and CFSE as described above. Proliferation was monitored in CD19+ B cells by the dilution of the CellTrace Violet stain at day 8 after infection on a BD FACS Canto II. The percent arrested population was determined by calculating the percentage of cells that diluted the CellTrace Violet stain but did not dilute the CFSE stain. Data were analyzed by using FlowJo software (Version 10.0).
Cell Sorting.
CD19+ B cells were sorted for the PA and PP populations based on the CellTrace Violet and CFSE profile by using either a Beckman Coulter Astrios or Beckman Coulter MoFlo XDP sorter.
Lysotracker.
One million PBMCs or LCLs were stained with 50 nM Lysotracker Deep Red (Molecular Probes, no. L12492) for 2 h at 37 °C in R15 medium. The stain was washed out with PBS, and the data were collected on a BD FACS Canto II or Imagestream.
Imagestream.
CFSE and CellTrace Violet labeled and EBV-infected PBMCs or LCLs were stained with Lysotracker as described above. The cells were then fixed and permeabilized with BD Cytofix/Cytoperm buffer (BD, no. 554714) according to the manufacturer’s instructions. The cells were blocked in Perm/Wash buffer containing 5% (vol/vol) goat serum for a minimum of 60 min followed by incubation with the LC3 antibody for 1 h at 4 °C. The samples were then washed and incubated with Alexa Fluor 568 goat anti-rabbit IgG (Life Technologies, no. A11036) for 30 min at 4 °C. Images were acquired by using an ImageStream multispectral imaging flow cytometer (Amnis Corporation). Data were analyzed by using IDEAS software (Version 3.0; Amnis Corporation) as described (51). Single cells were first gated by size and CellTrace Violet staining to identify the proliferating population. The proliferating cells were then further subdivided based on CFSE staining as described in Fig. 1B. For colocalization studies, the PA and PP subsets were then plotted for log intensities of LC3 and Lysotracker. LCLs were not stained with proliferation dyes but were analyzed in an identical fashion in all other respects. Cells that stained high for both markers were further analyzed for colocalization by looking for cells with a high Bright Detail Similarity (BDS) score. BDS calculates the degree of overlapping pixels taken from different channels of fluorescence imagery (52). The BDS is the log-transformed Pearson’s correlation coefficient that is nonmean normalized and is applied to the image. At least 10,000 cells were used for each analysis.
Immunofluorescence.
Samples were pelleted, resuspended in 25 μL of PBS, spread on a microscope slide, and dried at 37 °C for 15 min. Cells were fixed in 4% (vol/vol) paraformaldehyde for 15 min at 4 °C, washed in PBS, permeablized in PBS containing 0.5% Triton X-100 for 10 min and then blocked in PBS with 0.2% Triton X-100 containing 5% (vol/vol) normal goat serum for 1 h. Primary antibodies were incubated overnight at 4 °C followed by secondary antibody incubation with Alexa Fluor 488 goat anti-rabbit IgG (Life Technologies, no. A11034) for 2 h. Slides were mounted in Vectashield (Vector Laboratories, H-1200) containing DAPI. All IF slides were visualized by using Zeiss 780 upright confocal microscope. The above IF method was modified for visualization of GLUT1. In the case of GLUT1 IF, the cells were not permeabilized.
Western Blot.
Cells were washed one time with PBS before being pelleted and frozen at −80 °C. Pellets were resuspended in LDS Sample Buffer (NuPAGE, no. NP0008) containing 1 mM DTT, NaF, sodium pyrophosphate, sodium orthovanadate, beta-glycerophophate, sodium molybdate, and complete protease inhibitors without EDTA and incubated on ice for 30 min before sonication. Protein concentration was determined by BCA assay (Thermo, no. 23225) according to the manufacturer’s protocol. Samples were separated on a 4–12% Bis-Tris gel (NuPAGE, NP no. 0322) run in Mops-SDS running buffer (NuPAGE, NP no. 0001) followed by transfer to PVDF membrane. The PVDF membrane was blocked for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C before washing and staining with a secondary anti-rabbit horse radish peroxidase (HRP)-conjugated antibody (Sigma-Aldrich, no. A0545). Quantification was performed by using Gene Tools software with normalization to β-actin after imaging using the G-box gel imaging system.
RNA Isolation and qRT-PCR.
Total RNA was isolated by using the RNeasy kit (Qiagen, no. 74106) and reverse-transcribed by using the High Capacity cDNA Reverse Transcription kit (Life Technologies, no. 4368814) according to the manufacturer’s instructions. Relative mRNA abundance was measured by using a SYBR green-based real time PCR assay with 5 ng of cDNA per reaction. All primers were used at a concentration of 1 µM per reaction. qRT-PCR was carried out by using the Step One Plus Real Time PCR light-cycler (Applied Biosytems), and data were analyzed by using the supplied Step One software. All expression levels were first normalized to SETDB1 as a control and then to PPs. All primers were purchased from Sigma-Aldrich, with the exception of SETDB1, which was purchased from IDT. Primers used in this study are shown in Table S5.
Table S5.
Primers used
| Target | Orientation | Sequence |
| SESN1 | Forward | CAGATGCATGCTTTATTTGC |
| Reverse | AATATTGTGGGTGGAAAACC | |
| SESN3 | Forward | AAAAGTTTCGGATGGTCTAC |
| Reverse | ACCTGATTCCAAACATACAG | |
| FH | Forward | CCTCATATAGGGTATGACAAGG |
| Reverse | GTAAATCACTTTGGACCCAG | |
| NDUFB4 | Forward | TGTCTATCCTAATTTCAGACCC |
| Reverse | TTCTTTCCTATCCCTCTCAG | |
| NDUFB8 | Forward | CTTGGCATGTCATGTGTATG |
| Reverse | TAAGGATACTGCTTTGGTCC | |
| SETDB1 | Forward | TCCATGGCATGCTGGAGCGG |
| Reverse | GAGAGGGTTCTTGCCCCGGT | |
| CDKN2A | Forward | CCCCTTGCCTGGAAAGATAC |
| Reverse | AGCCCCTCCTCTTTCTTCCT | |
| CDKN1A | Forward | TGTCCGTCAGAACCCATGC |
| Reverse | AAAGTCGAAGTTCCATCGCTC | |
| MKI67 | Forward | CAGACTCCATGTGCCTGAGA |
| Reverse | CTGCACAGATTTGCTCTCCA |
Microarray Analysis.
Total mRNA was isolated from sorted PA and PP cells by using an RNeasy kit (Qiagen, no. 74106). The RNA was processed by using an Ambion MessageAmp Premier Package (Life Technologies, no. AM1792) and hybridized to a Human Genome U133 Plus 2.0 Chip (Affymetrix, no. 900466) by the Duke Center for Genomic and Computational Biology Microarray Core. The resultant CEL files were RMA-normalized (Partek), and the data were analyzed with GenePattern (53) and GSEA (Version 2; ref. 54).
Electron Microscopy.
B cells were pelleted and washed in serum-free RPMI medium, and 2% (vol/vol) glutaraldehyde was overlaid onto the undisturbed cells. The pellets were scraped into 1- to 1.5-mm piles on parafilm and encased in 1% molten agar. The agar-embedded pellets were washed three times with 0.1 M phosphate buffer and further fixed and stained in 1% osmium tetroxide in phosphate buffer for 30 min by microwave processing or 1–2 h at room temperature. They were then dehydrated in a graded series of acetone and infiltrated with EmBed 812 epoxy resin using a 50:50 mixture of 100% acetone for 1 h and two changes of 100% resin for 1 h each. After baking at 60 °C for 48 h, ultrathin sections were cut on a diamond knife. Sections were poststained with 2% (vol/vol) aqueous uranyl acetate, washed in water, stained with 1% aqueous lead citrate, and washed again in water. Sections were viewed in an FEI CM 12 electron microscope, and micrographs were recorded on an AMT 2.6K digital camera.
Metabolic Assays.
Glycolysis and glucose uptake assays by using 3H-glucose or 3H-2-deoxyglucose have been described (55, 56). All values were normalized to cell number. OCR and ECAR were measured with an XF24 extracellular flux analyzer (Seahorse Bioscience) as described (50). Suspension cells were attached to culture plates by using Cell-Tak (BD Bioscience). OCR and ECAR were measured in unbuffered DMEM (Sigma-Aldrich) supplemented with 10 mM d-glucose (Sigma-Aldrich) and 10 mM l-glutamine, as indicated. OCR and ECAR values were normalized to cell number. For certain experiments, OCR was measured over time after injection of oligomycin, FCCP, rotenone, and antimycin A. Maximal OCR is defined as the OCR value after FCCP injection. Spare respiratory capacity is defined as the difference between basal OCR and OCR after FCCP injection.
Glucose Uptake.
Cells were starved in glucose-free medium for 1 h before the addition of 25 µM 2-[N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxyglucose (2-NBDG) (Thermo, N13195) for 5–120 min. The mean fluorescence intensity was determined by flow cytometry and fit to linear regression model. The slope of the line was defined as the rate of glucose uptake.
Supplementary Material
Acknowledgments
We thank Lynn Martinek, Nancy Martin, and Mike Cook for extensive help in flow-based cytometry experiments; Sara Miller for TEM experiments; Matthias Gromeier and Mike Brown for reagents; Ashley Chi for helpful discussions; and Jorn Coers and Arun Haldar for reagents and helpful discussions. We give special thanks to Alex Price for his wit and ingenuity with Adobe Illustrator. This work was supported by National Institutes of Health (NIH) Grants R01-CA140337 (to M.A.L.) and R01-DK105550 (to J.C.R.) and Duke Center for AIDS Research Grant 5P30 AI064518 (to M.A.L.). K.M. was supported by NIH Grants T32-CA009111 and T32-AI007392. A.Y.H. and J.E.M. were supported by NIH Grant T32-CA009111.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE76137).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517141113/-/DCSupplemental.
References
- 1.Rickinson A, Kieff E. In: Epstein-Barr Virus Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Lippincott, Williams, and Wilkins; Philadelphia: 2007. pp. 2603–2654. [Google Scholar]
- 2.Sinclair AJ, Palmero I, Peters G, Farrell PJ. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13(14):3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Kikutani H, Tsang SF, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65(8):4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181(2):595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 5.Nikitin PA, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8(6):510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikitin PA, Price AM, McFadden K, Yan CM, Luftig MA. Mitogen-induced B-cell proliferation activates Chk2-dependent G1/S cell cycle arrest. PLoS One. 2014;9(1):e87299. doi: 10.1371/journal.pone.0087299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192(4):547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 10.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 11.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21(1):43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leidal AM, Pringle ES, McCormick C. Evasion of oncogene-induced senescence by gammaherpesviruses. Curr Opin Virol. 2012;2(6):748–754. doi: 10.1016/j.coviro.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Koopal S, et al. Viral oncogene-induced DNA damage response is activated in Kaposi sarcoma tumorigenesis. PLoS Pathog. 2007;3(9):1348–1360. doi: 10.1371/journal.ppat.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yee J, White RE, Anderton E, Allday MJ. Latent Epstein-Barr virus can inhibit apoptosis in B cells by blocking the induction of NOXA expression. PLoS One. 2011;6(12):e28506. doi: 10.1371/journal.pone.0028506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Invest. 2015;125(1):47–54. doi: 10.1172/JCI73942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23(7):798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palsson-McDermott EM, O'Neill LA. The Warburg effect then and now: From cancer to inflammatory diseases. BioEssays. 2013;35(11):965–973. doi: 10.1002/bies.201300084. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez EL, Lagunoff M. Viral activation of cellular metabolism. Virology. 2015;479-480:609–618. doi: 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado T, Sanchez EL, Camarda R, Lagunoff M. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 2012;8(8):e1002866. doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatt AP, et al. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc Natl Acad Sci USA. 2012;109(29):11818–11823. doi: 10.1073/pnas.1205995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, et al. Targeting Epstein-Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene. 2014;33(37):4568–4578. doi: 10.1038/onc.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommermann TG, O’Neill K, Plas DR, Cahir-McFarland E. IKKβ and NF-κB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res. 2011;71(23):7291–7300. doi: 10.1158/0008-5472.CAN-11-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawkins ED, et al. Measuring lymphocyte proliferation, survival and differentiation using CFSE time-series data. Nat Protoc. 2007;2(9):2057–2067. doi: 10.1038/nprot.2007.297. [DOI] [PubMed] [Google Scholar]
- 26.Allday MJ. EBV finds a polycomb-mediated, epigenetic solution to the problem of oncogenic stress responses triggered by infection. Front Genet. 2013;4:212. doi: 10.3389/fgene.2013.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon-Lowe C, et al. Epstein-Barr virus-induced B-cell transformation: Quantitating events from virus binding to cell outgrowth. J Gen Virol. 2005;86(Pt 11):3009–3019. doi: 10.1099/vir.0.81153-0. [DOI] [PubMed] [Google Scholar]
- 28.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469(3):325–346. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 29.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327(5970):1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Budanov AV, Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013;18(6):792–801. doi: 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunlop EA, Tee AR. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 2012;4(9):a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson E, Miller G, Robinson J, Heston L. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 1977;76(1):152–163. doi: 10.1016/0042-6822(77)90292-6. [DOI] [PubMed] [Google Scholar]
- 35.Sugden B, Mark W. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J Virol. 1977;23(3):503–508. doi: 10.1128/jvi.23.3.503-508.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruo S, et al. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103(51):19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010;6(6):e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skalska L, et al. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog. 2013;9(2):e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams LR, Taylor GS. Autophagy and immunity - insights from human herpesviruses. Front Immunol. 2012;3:170. doi: 10.3389/fimmu.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granato M, et al. Epstein-barr virus blocks the autophagic flux and appropriates the autophagic machinery to enhance viral replication. J Virol. 2014;88(21):12715–12726. doi: 10.1128/JVI.02199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, et al. Oncogene-induced cellular senescence elicits an anti-Warburg effect. Proteomics. 2013;13(17):2585–2596. doi: 10.1002/pmic.201200298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DY, Sugden B. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood. 2008;111(4):2280–2289. doi: 10.1182/blood-2007-07-100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27(20):2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 44.Price AM, et al. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J Virol. 2012;86(20):11096–11106. doi: 10.1128/JVI.01069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hills SA, Diffley JF. DNA replication and oncogene-induced replicative stress. Curr Biol. 2014;24(10):R435–R444. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aird KM, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Reports. 2013;3(4):1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aird KM, et al. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Reports. 2015;11(6):893–901. doi: 10.1016/j.celrep.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johannsen E, et al. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci USA. 2004;101(46):16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caro-Maldonado A, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192(8):3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phadwal K, et al. A novel method for autophagy detection in primary cells: Impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8(4):677–689. doi: 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beum PV, et al. Quantitative analysis of protein co-localization on B cells opsonized with rituximab and complement using the ImageStream multispectral imaging flow cytometer. J Immunol Methods. 2006;317(1-2):90–99. doi: 10.1016/j.jim.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 53.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 54.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plas DR, Talapatra S, Edinger AL, Rathmell JC, Thompson CB. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J Biol Chem. 2001;276(15):12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- 56.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18(4):1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.