Significance
Recent estimates of mutation rates obtained by sequencing human pedigrees have challenged conceptions about split times between humans and our closest living relatives. In particular, estimates of human split times from chimpanzees and gorillas based on the new mutation rate estimates are more than twofold shorter than previously believed, seemingly at odds with the fossil record. Here we show that accounting for the effects of sex-specific life histories on mutation rates along the hominid phylogeny largely bridges this apparent gap and leads to more accurate split time estimates. Doing so can also explain other intriguing phylogenetic patterns in hominid and mammalian evolution.
Keywords: molecular clock, mutational slowdown, generation time effect, human–chimpanzee split, male mutation bias
Abstract
One of the foundational results in molecular evolution is that the rate at which neutral substitutions accumulate on a lineage equals the rate at which mutations arise. Traits that affect rates of mutation therefore also affect the phylogenetic “molecular clock.” We consider the effects of sex-specific generation times and mutation rates in species with two sexes. In particular, we focus on the effects that the age of onset of male puberty and rates of spermatogenesis have likely had in hominids (great apes), considering a model that approximates features of the mutational process in mammals, birds, and some other vertebrates. As we show, this model can account for a number of seemingly disparate observations: notably, the puzzlingly low X-to-autosome ratios of substitution rates in humans and chimpanzees and differences in rates of autosomal substitutions among hominine lineages (i.e., humans, chimpanzees, and gorillas). The model further suggests how to translate pedigree-based estimates of human mutation rates into split times among extant hominoids (apes), given sex-specific life histories. In so doing, it largely bridges the gap reported between estimates of split times based on fossil and molecular evidence, in particular suggesting that the human–chimpanzee split may have occurred as recently as 6.6 Mya. The model also implies that the “generation time effect” should be stronger in short-lived species, explaining why the generation time has a major influence on yearly substitution rates in mammals but only a subtle one in human pedigrees.
Most of our inferences about species split times on short phylogenetic timescales rely on the neutral molecular clock. According to the neutral theory, the number of substitutions K that accumulate in a lineage over T years (e.g., since the split from another species) is , where and are the average mutation rate per generation and average generation time, respectively (1). Inferring split times therefore requires estimates of the yearly mutation rate on the lineage in question. Such estimates generally derive from securely dated fossils on other lineages or from measurements of mutation rates in extant species (2–5). Using these estimates for dating thus necessitates an understanding of the way that yearly mutation rates may change over time.
Neutral substitution patterns in mammals offer some insights. Variation in yearly mutation rates on phylogenetic timescales can be assessed by comparing the number of neutral substitutions along two branches leading from a common ancestor to extant species. These comparisons show marked variation in yearly rates on autosomes. For example, there are 50% fewer substitutions on the human branch compared with rodents (6) and 25% fewer compared with baboons, with more moderate differences among hominine lineages (6–9). The average yearly rates are also negatively correlated with generation times (and their correlates) in extant mammals, leading to the notion of a “generation time effect” on the molecular clock (6, 10, 11).
Neutral substitutions rates vary not only among taxa but also between sex chromosomes and autosomes. For brevity, we consider the relative rates on X and autosomes, but these considerations extend naturally to Y (or ZW). Because autosomes spend the same number of generations in both sexes, whereas the X spends twice as many generations in females, rates of neutral substitutions on autosomes reflect a greater relative contribution of male mutations than on the X. In a wide range of taxa, neutral substitutions rates on autosomes are greater than on the X (or lower than on the Z), suggesting a male biased contribution to yearly mutation rates (12). Moreover, observed X-to-autosome ratios are extremely variable, ranging between 0.76 and 0.9 in hominids and up to 1.0 in surveyed mammals, indicating that the degree of male bias itself varies greatly on phylogenetic timescales (12, 13).
Our current understanding of mutation can help tie these observations together (5). Pedigree studies in humans and chimpanzees establish that most mutations are paternal in origin and that the paternal but not the maternal contribution increases strongly with age (4, 14). This has long been thought to be true because germ-cell division is arrested before birth in females but proceeds continuously postpuberty in males (5, 15–17). The same reasoning may extend to mammals, birds, and other vertebrate taxa in which oogenesis ceases before birth or hatching (18–20). These considerations suggest that maternal and paternal generation times should affect the molecular clock differently. They also imply that the age of puberty in males and the rate of spermatogenic germ cell divisions should affect yearly mutation rates (5). The variation observed among closely related extant species indicates that these parameters change over phylogenetic timescales. Here we ask how such changes would affect the molecular clock on X and autosomes.
Model
The Molecular Clock with Two Sexes.
We model the accumulation of neutral substitutions allowing for different generation times and mutation rates in males and females (cf. ref. 21). If, for example, the paternal generation time, (defined as the average time between births), is longer than the maternal, , then autosomes would spend a greater proportion of time (but not of generations) in males than in females [i.e., in males]. Therefore, the yearly mutation rate in males, (where is the rate per generation in males), would have a greater relative contribution to the autosomal mutation rate. Taking the corresponding weighted average for the expected number of substitutions on an autosomal lineage over T years yields
| [1] |
where and are the expected sex-averaged generation time and mutation rate per generation on an autosomal lineage. By the same token, on the X,
| [2] |
where in this case and are the expected sex-averaged generation time and mutation rate per generation on a X lineage (see SI Appendix, section 1, for rigorous derivations and the corresponding equations for Y and Z).
Sex- and Age-Dependent Mutation Rates in Hominines.
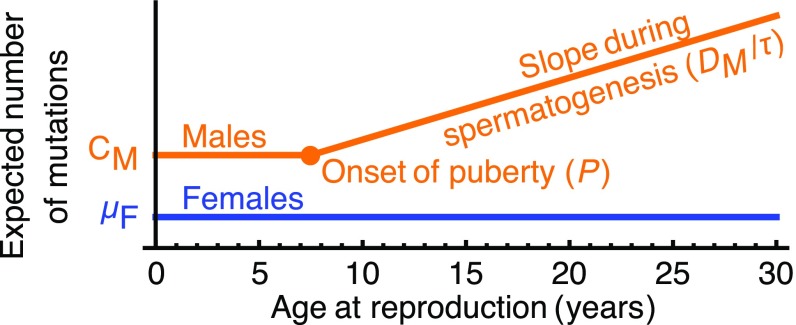
We model male and female mutation rates per generation, and , based on the current understanding of the process of accumulation of germ-line mutations (5, 16). Namely, we assume that mutations accumulate linearly with the number of germ-cell divisions, and the rate per division varies at different stages of development (cf. ref. 5). This is a natural assumption for replicative mutations and has recently been suggested to apply to nonreplicative mutations that are efficiently repaired (22) (see Discussion for other kinds of mutations).
In females, all oogonial mitotic divisions occur before birth, so the number of mutations should have no dependence on the age of reproduction (19). We therefore model the female per generation mutation rate as a constant.
In males, cell divisions in the germ line exhibit two main phases: prepuberty, starting from the zygote through the proliferation of germ cells in the growing testis, and postpuberty, with continuous divisions in the adult testis during spermatogenesis. Although age of puberty and testis mass vary considerably among hominids (13), the number of germ cell divisions before puberty should increase only logarithmically with mass and is similar in species exhibiting large differences in age of puberty [such as mice and humans (23)]. We therefore approximate the expected number of mutations prepuberty as constant (). Postpuberty, one germ cell division is thought to occur at each spermatogenic cycle [i.e., the seminiferous epithelial cycle (24)], and the length of the cycle () varies among hominines (Table 1). We therefore assume a mutation rate per year of in adult males, where is the expected number of mutations per spermatogenic division. We then model the average mutation rate per generation in males by
| [3] |
Table 1.
Estimates of spermatogenesis and life history parameters in hominines (see SI Appendix, section 4 for details)
| Populations | τ, days | P, years | GF, years | GM, years |
| Human | 16 | 13 | ||
| Hunter–gatherers | 26.9* | 33.8* | ||
| Chimpanzee | 14 | 7.5 | ||
| Western | 26.3 | 24.3 | ||
| Eastern | 24.8 | 24 | ||
| Average† | 25.2 | 24.1 | ||
| Gorilla | — | 7 | ||
| Mountain gorillas | 18.2 | 20.4 |
Revised from ref. 25 (SI Appendix, section 4).
Weighted by sample size (26).
where P is the expected age of puberty and GM-I is the expected generation time excluding gestation time (I). Both the constancy with age in females and the piecewise linearity in males (Fig. 1) are consistent with the observations of pedigree studies in humans and chimpanzees (4, 14). Moreover, a similar model [e.g., accounting for intermittent spermatogenesis (18)] may provide a reasonable approximation for vertebrate species in which oogenesis ceases before birth or hatching [e.g., mammals, birds, elasmobranchs, cyclostomes, and a few teleosts (19, 20)].
Fig. 1.
The mutational model.
In SI Appendix, sections 1 and 2, we consider how the molecular clock is affected by variation of mutational parameters (e.g., age of puberty and generation times) both within a population and over phylogenetic time. Specifically, we show that the expected number of substitutions is insensitive to the variance of the number of mutations or to the variance of the age at reproduction, thus justifying our consideration of only the means.
Parameter Values and Ranges.
To study how changes in life history traits and spermatogenesis should affect the rate of the molecular clock, we would like to assign realistic values to the parameters of the mutational model. Lacking evidence to the contrary, we assume that the parameters associated with the rates of mutation per germ-cell division at different developmental stages remained constant throughout the hominine phylogeny. We infer these parameters from the relationship between mutation rates and paternal ages in the largest human pedigree study published to date (4), which yields , , and per base pair (see SI Appendix, section 4, for details).
In contrast, life history and spermatogenesis parameters (, , , and ) are known to vary among species and even populations (Table 1 and SI Appendix, Tables S3–S7, and references therein). We use the variation among extant species to guide our choice of plausible ranges for these parameters on the phylogeny. These parameter ranges should be treated only as a rough guide because of the considerable uncertainty associated with estimates in extant species (see Discussion in SI Appendix, section 4) and the uncertainty about the extent to which they reflect the variation along the phylogeny.
Results
The Autosomal Molecular Clock.
We first consider the effects of the average and ratio of male and female generation times. In general, the impact of changing the ratio or average of generation times will depend on the way in which mutation rates vary with sex and age (see SI Appendix, section 3, for more details). If, for example, mutation rates increase more rapidly with paternal than maternal age, as is expected for mammals, then increasing the ratio of male-to-female generation times necessarily increases the mutation rate per year.
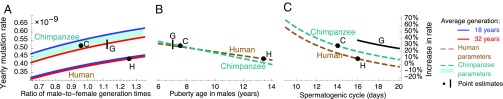
Predicting the effect of increasing the average generation time on yearly mutation rates requires additional assumptions. If we consider our mutational model (regardless of parameter values) and assume a constant proportion of the male generation time spent prepuberty and postpuberty (SI Appendix, Fig. S1), we find that an increase in the average generation time will decrease the rate of the molecular clock (SI Appendix, section 3), consistent with the generation time effect observed in mammals (6, 10, 11). If, instead, we assume that age of puberty remains constant, whereas the average generation time increases, which better describes the observations in extant hominids (Table 1 and SI Appendix, Table S3), then the effect on yearly mutation rates can go either way, depending on parameter values (i.e., on , , and ; see SI Appendix, section 3) (5, 22). Intriguingly, the parameter estimates from human pedigree studies are close to the boundary where the effect of the average generation time changes direction and is therefore negligible (Fig. 2A).
Fig. 2.
Predicted yearly mutation rates on autosomes as a function of the ratio of male-to-female generation times (A), male age of puberty (B), and spermatogenic cycle length (C). Rates are measured relative to the estimate of per bp per year reported by Kong et al. (4), whose data we used to fit our mutational model (Model). In each panel, we vary one parameter, while fixing others to their estimated values in extant humans (brown) or chimpanzees (green) (Table 1). Note that the point estimates for humans and chimpanzees (black points) do not coincide with those reported in pedigree studies, because ours account for the predicted effects of life history and spermatogenesis parameters. The estimated range for gorillas (black line), where the spermatogenic cycle length has not been measured, corresponds to cycles between 16 d and 20 d.
Within the ranges for the average and ratio of generation times estimated in hominines, changes to the ratio have a much greater impact (Fig. 2A). Replacing the standard (although often implicit) assumption of equal generation times in males and females with estimates of their ratio in current hunter–gatherer populations increases yearly mutation rates by ∼10%. Further considering that the ratios in extant hominines range between 0.92 for western chimpanzees and 1.26 for hunter–gatherers (SI Appendix, section 4) suggests that yearly rates could vary between 4% lower to 10% higher than the rate with a ratio of 1 (unless noted otherwise, when we vary a single parameter, other parameters are assigned their estimated values in humans). By comparison, varying the average generation time between 19 y, for gorillas, and 30.4 y, for hunter–gatherer populations, affects rates by less than 2% (see also ref. 22).
Both an earlier onset of male puberty and an increased rate of spermatogenesis increase the yearly mutation rates in males, resulting in an increased rate on autosomes (Fig. 2 B and C) (5, 22). Varying their values within the range known for hominines markedly affects yearly rates, by ∼18% for the male puberty age and ∼8% for the spermatogenic cycle length.
Considered jointly, these factors should generate differences in the branch lengths leading to extant hominines. Assuming parameter estimates for extant humans and chimpanzees, our model suggests that the human branch would be 15% shorter. Making the more plausible assumption that the yearly mutation rate in the ancestor was between extant estimates and that the rates on each lineage changed gradually, we would still expect the human branch to be somewhat shorter. The human branch has indeed been estimated to be 0.7–2.9% shorter (8, 9); these estimates include the contribution of ancestral polymorphism common to both branches, suggesting that the branch-specific difference is larger.
Our ability to make similar predictions about the gorilla branch is hindered by the lack of estimates for the spermatogenic cycle length. Nonetheless, spermatogenic cycle lengths are negatively correlated with relative testis mass in mammals (SI Appendix, Fig. S3), and the variation in both has been attributed to differences in the intensity of sperm competition in different mating systems (27, 28). In agreement with this hypothesis, the relative testis mass and rate of spermatogenesis in chimpanzees (0.27% of body weight and 14 d), which have a promiscuous mating system, are greater than in humans (0.07% of body weight and 16 d), which are largely monogamous, and the relative testis mass in polygynous gorillas (one male controls reproductive access to many females) is smaller than in both (0.02% of body weight) (13, 28). This suggests that the rate of spermatogenesis in gorillas is lower than in humans. Varying the length of the spermatogenic cycle between 16 d (its value for humans) and 20 d and assuming current estimates for life history yields predicted branch lengths 14–26% longer than the human branch, respectively. These predictions accord with current estimates (again, without accounting for the ancestral contribution), which suggest that the gorilla branch is between 8.7% and 11% longer than the human branch (8, 9).
Last, we consider how accounting for life history affects estimates of the split time between humans and chimpanzees. Current estimates of this parameter are obtained by dividing the neutral divergence on the human lineage [∼0.4% per bp after subtracting the estimated contribution of ancestral polymorphism (29)] by estimates of the yearly mutation rate. This approach yields a split time of ∼10 Mya (30) (SI Appendix, section 5), given pedigree-based estimates of the mutation rate of ∼0.4 × 10−9 per bp per year (4).
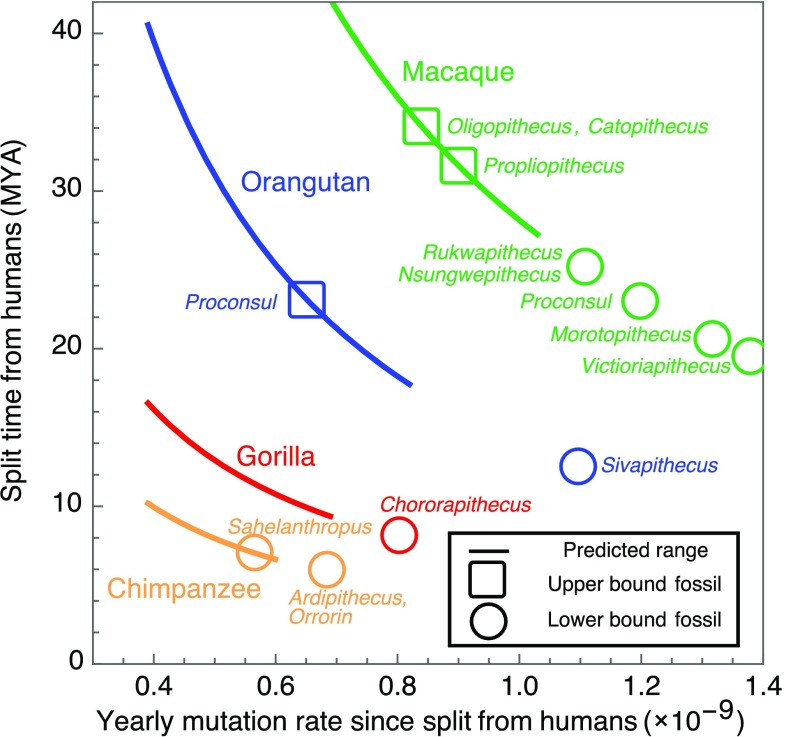
These estimates, however, do not account for most of the effects that we have considered (also see Discussion). Notably, rather than assuming an equal generation times in males and females, one may want to consider the ratio estimated in extant hunter–gatherers. Moreover, the earlier age of puberty in chimpanzees and possible earlier puberty in Homo erectus [e.g., from Nariokotome boy dated ∼1.5 Mya (31)] suggest that the onset of puberty may have occurred at younger ages during most of the human lineage. Similarly, the longer spermatogenesis cycle in humans than in chimpanzees (and all other surveyed primates; SI Appendix, Fig. S2) indicates that it may have been shorter along the human lineage. Although it is difficult to translate these considerations into point estimates, they all suggest a higher yearly mutation rate (Fig. 2) and a split time that is more recent than suggested by the standard pedigree-based estimates. As an illustration, if the average mutation rate on the human lineage were between the point estimates in extant humans and chimpanzees, then our model would suggest a split time between 7.7 Mya and 9.1 Mya (Fig. 2 and SI Appendix, section 5). If we further allow individual life history parameters to vary between their values in extant humans and chimpanzees, then the lower bound on the split time is further reduced to ∼6.6 Mya (see Fig. 5).
Fig. 5.
Estimated ranges of split times from humans (y axis) and the yearly mutation rates since the split (x axis). The inference is detailed in SI Appendix, section 5. Upper (squares) and lower (circles) bounds on split times are based on the hypothesized phylogenetic positioning of fossils (SI Appendix, section 5) (38).
The Relative Rates of the Molecular Clock on X and Autosomes.
Differences in neutral rates of substitutions on sex chromosomes and autosomes have generally been attributed to differences in mutation rates per generation between sexes. In fact, X-to-autosome ratios have been widely used to infer the male mutation bias, , in lineages of mammals, birds, flies, fish, and plants (10, 12). These inferences rely on Miyata’s formula (32),
| [4] |
where . Critically, Miyata’s formula assumes equal male and female generation times. When this assumption is relaxed (using Eqs. 1 and 2), the X-to-autosome ratio also depends on the ratio of male-to-female generation times:
| [5] |
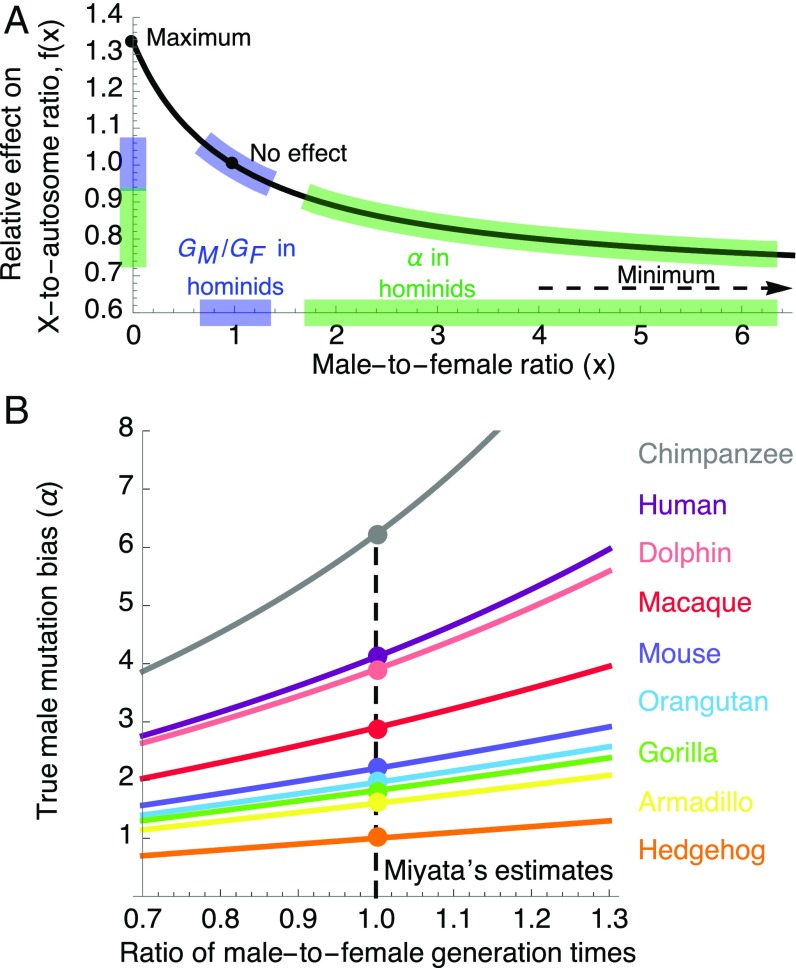
(the corresponding relationships for other sex chromosomes are provided in SI Appendix, section 1). This suggests a theoretical range between 1/2 and 2 compared with a range between 2/3 and 4/3 based on Miyata’s formula. We note that this predicted ratio applies to the accumulation of substitutions after species split but not to the contribution of ancestral polymorphism, because the average TMRCA also differs between X and autosomes.
What is made clear by this derivation is that ignoring differences in generation times could introduce substantial biases in estimates of the male mutation bias, (Fig. 3). Notably, when is large, a greater increase in is required to explain an incremental increase in the X-to-autosome ratio (because of the form of f; Fig. 3A). In this case, even moderate differences in the ratio of generation times translate into large biases in estimates of (Fig. 3B and SI Appendix, Fig. S3). As differences in generation times between sexes are common (26), this consideration suggests that current estimates of the male mutation bias might suffer from substantial bias and uncertainty (Fig. 3B and SI Appendix, Fig. S3), and that information about the ratio of generation times (e.g., from extant species) is key to assessing this uncertainty.
Fig. 3.
The effects of the male-to-female ratio of generation times on estimates of the male mutation bias, . (A) The function f mediating the effects of the ratios of male-to-female generation times and mutation rates on the X-to-autosome ratio of substitution rates. (B) The potential biases in estimates of in nine mammalian species, as a function of the ratio of male-to-female generation times. The curve for each species is based on the X-to-autosome divergence ratio reported along its lineage (12, 13) (these ratios likely underestimate the X-to-autosome ratios of substitutions due to the contribution of ancestral polymorphism).
X-to-Autosome Ratios in Hominines.
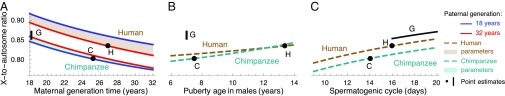
We use our model and parameter estimates to investigate how life history and spermatogenesis affect the X-to-autosome ratio in hominines. Increasing the paternal generation time has opposing effects on the ratio (Eq. 5) because it increases both the male mutation bias (; SI Appendix, Fig. S4) and the ratio of generation times (). In contrast, changing the maternal generation time affects only the ratio of generation times, resulting in a greater effect on the X-to-autosome ratio (Fig. 4A). In turn, both earlier male puberty and an increased rate of spermatogenesis increase mutation rates in males, resulting in a lower ratio (Fig. 4 B and C). Based on the known parameter ranges in hominines, the X-to-autosome ratio varies by ∼6.9% due to maternal age, whereas the variation due to each of the other parameters is smaller (∼1.2–2.7%).
Fig. 4.
Predicted X-to-autosome ratios of substitution rates as a function of male and female generation times (A), male age of puberty (B), and rate of spermatogenesis (C). Other details are the same as in Fig. 2.
In hominines, X-to-autosome ratios have garnered considerable attention, in part because of what seems like puzzlingly low ratios (8, 13, 33). Estimates of the ratios of neutral divergence are ∼0.76 for the chimpanzee lineage, ∼0.8 for the human lineage, and ∼0.9 for gorillas (13). These estimates, however, do not correspond directly to the ratios that we are considering. Notably, they include the contributions of ancestral polymorphism, which are greater on autosomes (8, 33), suggesting that the ratio after the species splits is closer to 1. Also, they do not control for differences in the average mutability of base pairs between X and autosomes [e.g., due to local effects of base composition (34)]. In that regard, the standard practice of dividing by divergence to an outgroup only complicates the interpretation by compounding estimates of the X-to-autosome ratios in hominines with the ratio on the outgroup lineage. For all these reasons, we consider existing estimates to be rough.
These caveats notwithstanding, we ask whether the observed values can be explained by considering our mutational model. Using parameter estimates from extant species and assuming a spermatogenic cycle between 16 d and 20 d in gorillas, the model predicts an X-to-autosome ratio of 0.80 on the chimpanzee lineage, 0.83 on the human lineage, and between 0.85 and 0.87 on the gorilla lineage. These estimates recover the ordering of ratios among lineages as well as the rough magnitude of the reduction below 1. Further allowing individual mutational parameters to vary within their ranges in extant species can explain both a smaller ratio in chimpanzees and a greater difference between chimpanzees and gorillas (SI Appendix, Table S9). Thus, it is plausible that observed X-to-autosome ratios could be explained by differences in rates of spermatogenesis (as first suggested by ref. 13) and life history, without recourse to elaborate demographic scenarios. We note, however, that these factors cannot explain the greater variation in divergence levels on the X compared with autosomes, which has been suggested to reflect the footprints of linked selection in the ancestral population or during speciation in the presence of gene flow (8, 33).
Discussion
Pedigree-based estimates of the mutation rate in humans should allow for improved inferences about the timing of species’ splits and other evolutionary events (e.g., out-of-Africa migrations). To translate estimates from pedigrees into yearly mutation rates, the standard approach is to divide the sex-averaged mutation rate per generation in a study by estimates of the sex-averaged generation time along the lineage under consideration. This calculation assumes that the mutation rate per generation remains constant, implicitly reflecting an extreme interpretation of the generation time effect (5). It also ignores other life history traits and rates of spermatogenesis that differ between pedigree studies and the lineage of interest, and whose effects are likely to be substantially greater than those of the sex-averaged generation time. Thus, translating the results of pedigree studies into yearly rates is not as straightforward as it seems and requires consideration of these life history factors.
A more appropriate approach is to fit the results of pedigree studies to a mutational model that reflects the dependency of yearly rates on age and sex. The fitted model can then be used to estimate yearly mutation rates and the uncertainty associated with them, based on estimates of life history parameters for the lineage under consideration (e.g., based on estimates for extant species). We illustrate this approach with a simple mutational model for hominines and show that it leads to substantial changes in estimates of yearly rates and corresponding split times. Importantly, it revises split times for the human–chimpanzee split downward, from ∼10 Mya (30) (SI Appendix, section 5) to as low as ∼6.6 Mya (Fig. 5 and SI Appendix, Table S9).
Our mutational model will surely be refined as we learn more about germ-line mutations. For example, molecular evolutionary patterns suggest that rates of certain types of mutations [notably, transitions at CpG sites (7, 34)] may track some combination of absolute time and number of cell divisions (5). If so, maternal age could affect the accrual of these mutations (5). In addition, there is still uncertainty about the number of cell divisions in the male germ line during spermatogenesis. In particular, it has been suggested that cells—the stem cells from which sperm are generated—are replenished by, or experience turnover with, spermatogonial stem cells throughout adulthood, resulting in fewer cell divisions in the germ line between puberty and reproduction (24, 35, 36). If this hypothesis is true, it would suggest that the mutation rate per spermatogenic cell division ( is higher than our estimates and closer to the (considerably higher) rates per division in females, in males prepuberty (5), and in other taxa (SI Appendix, Table S2) (37). In terms of the hominine mutational model, such a revision would introduce an additional parameter for turnover, which could lead to greater variation among species in male mutation rates postpuberty (36). These refinements notwithstanding, our mutational model already suggests several predictions that align with observations. For instance, our predicted mutation rates in chimpanzees fall within confidence intervals of a recent pedigree study (14). We also predict the reduction in branch length leading to humans compared with gorillas and provide a plausible explanation for the X-to-autosome ratios of substitutions rates observed in hominines.
Our results also bear on the recently invigorated discussion about split times in the catarrhine (i.e., old world monkeys and apes) phylogeny (30). For instance, assuming that current pedigree-based estimates reflect yearly rates along the hominid phylogeny places the human–orangutan split at ∼40 Mya, much earlier than all hominid or even hominoid fossils (SI Appendix, section 5) (29, 30), putting results from human genetics at odds with those from paleontology. Attempts to reconcile pedigree and fossil-based evidence appear to be moving from both ends. From one end, it has been suggested that mutation rates may have experienced a slowdown in the hominoid phylogeny toward the present (cf. refs. 7, 29, 30). From the other, the new mutation rate estimates triggered a reevaluation of the (already controversial) phylogenetic positioning of key catarrhine fossils and, specifically, those used to place bounds on the hominine–orangutan and hominoid–cercopithecoid (i.e., old world monkeys) split times (38). More realistic models of the molecular clock can inform this discussion by suggesting the plausible extent of the mutational slowdown on different phylogenetic timescales and, as a result, by better delineating which fossil positions would need to be revised for their ages to be consistent with pedigree studies.
As an illustration, we use our mutational model, with estimates of life history and spermatogenesis parameters from extant species, to suggest plausible ranges for yearly mutation rates on the hominoid phylogeny (Fig. 5 and SI Appendix, Table S9 and section 5). We allow each of the parameters on branches following a split to vary independently within the range observed in descendant species; in the few cases in which we lack estimates in extant species, we also rely on information from an outgroup (SI Appendix, section 5). Under these assumptions, ranges for individual parameters and the resulting yearly mutation rates can only become larger when we go farther back in time. Interestingly, only the upper bound on the range of mutation rates increases as we go farther back in time, supporting the notion of a slowdown. Moreover, the inferred ranges could, in theory, reconcile the apparent discrepancy between yearly rates of per bp observed in pedigree studies and yearly rates of per bp inferred from the fossil record (Fig. 5 and SI Appendix, Table S11). The assumption that life history and spermatogenesis parameters vary independently might be overly permissive, however, because combinations of these traits could be under stabilizing selection.
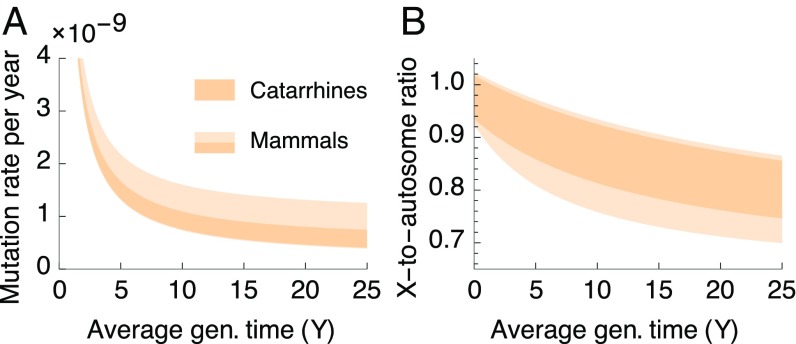
Moving to broader phylogenetic contexts, how can we reconcile our results suggesting that the (sex-averaged) generation time has a moderate effect in hominines (Fig. 2) (also cf. refs. 5, 22) with phylogenetic studies showing that it is a major predictor of the rates of neutral substitutions in mammals (6, 10, 11)? Assuming that the number of mutations tracks the number of cell divisions in the germ line, a possible answer may arise from the way the number of divisions relates to the generation time. Comparing mice, with a generation time of ∼9 mo, and humans, with a generation time of ∼30 y, the estimated numbers of cell divisions in the germ line are 25 and 31 in females, 27 and 34 prepuberty in males, and 35 and 390 during male spermatogenesis, respectively (23) (SI Appendix, Table S1). If we assume more generally that only the number of cell divisions during spermatogenesis increases rapidly with the generation time, then many of the mutations in hominines occur during spermatogenesis because of their exceptionally long generation times. In species with short generation times, the relative contribution of spermatogenesis would be much smaller and therefore the rate per generation would be roughly constant; equivalently, in such species, the yearly mutation rate would be roughly inversely proportional to the generation time (39).
To draw out these implications, we extrapolate our hominine mutational model to a wider range of life history and spermatogenesis parameter values roughly corresponding to mammals (Fig. 6), acknowledging that such an extrapolation provides only a qualitative depiction because underlying mutational parameters may vary among mammals (37) (SI Appendix, Table S2). We find that the sex-averaged generation time dominates the variation in yearly mutation rates when species with shorter generation times are included in the comparison (Fig. 6A). The average generation time also affects the expected X-to-autosome ratios [with shorter generation times corresponding to lower α and a ratio closer to 1 (39)], although its effect is expected to be much more moderate and comparable with those of other life history traits (Fig. 6B and SI Appendix, Fig. S5). Both of these predictions accord with observations in mammals (6, 10, 11).
Fig. 6.
The generation time effect on the molecular clock in a broader phylogenetic context. The yearly mutation rates (A) and X-to-autosome ratios (B) as a function of the sex averaged generation time, based on our mutational model. Other parameter ranges roughly correspond to catarrhines and mammals (SI Appendix, Table S4).
Supplementary Material
Acknowledgments
We thank M. Przeworski for helpful discussions throughout this work and D. Pilbeam for helping us navigate the catarrhine fossil record. We also thank P. Moorjani, M. Wyman, D. Conrad, A. Scally, and D. Reich for helpful discussions; P. Moorjani for sharing her unpublished results; and M. Crist, D. Murphy, L. Hayward, G. Coop, B. Charlesworth, D. Pilbeam, M. Przeworski, and two anonymous reviewers for comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515798113/-/DCSupplemental.
References
- 1.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; London: 1983. [Google Scholar]
- 2.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21(1):12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 3.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156(1):297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 6.Wu C-I, Li W-H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA. 1985;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Elango N, Warden C, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2(10):e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441(7097):1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 9.Elango N, Thomas JW. NISC Comparative Sequencing Program, Yi SV Variable molecular clocks in hominoids. Proc Natl Acad Sci USA. 2006;103(5):1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson Sayres MA, Venditti C, Pagel M, Makova KD. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution. 2011;65(10):2800–2815. doi: 10.1111/j.1558-5646.2011.01337.x. [DOI] [PubMed] [Google Scholar]
- 11.Ohta T. An examination of the generation-time effect on molecular evolution. Proc Natl Acad Sci USA. 1993;90(22):10676–10680. doi: 10.1073/pnas.90.22.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson Sayres MA, Makova KD. Genome analyses substantiate male mutation bias in many species. BioEssays. 2011;33(12):938–945. doi: 10.1002/bies.201100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Presgraves DC, Yi SV. Doubts about complex speciation between humans and chimpanzees. Trends Ecol Evol. 2009;24(10):533–540. doi: 10.1016/j.tree.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venn O, et al. Strong male bias drives germline mutation in chimpanzees. Science. 2014;344(6189):1272–1275. doi: 10.1126/science.344.6189.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penrose LS. Parental age and mutation. Lancet. 1955;269(6885):312–313. doi: 10.1016/s0140-6736(55)92305-9. [DOI] [PubMed] [Google Scholar]
- 16.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1(1):40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 17.Haldane JB. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen. 1947;13(4):262–271. doi: 10.1111/j.1469-1809.1946.tb02367.x. [DOI] [PubMed] [Google Scholar]
- 18.Pudney J. Spermatogenesis in nonmammalian vertebrates. Microsc Res Tech. 1995;32(6):459–497. doi: 10.1002/jemt.1070320602. [DOI] [PubMed] [Google Scholar]
- 19.Franchi L, Mandl AM, Zuckerman S. The development of the ovary and the process of oogenesis. In: Zuckerman S, editor. The Ovary. Vol 1. London: Academic Press; 1962. pp. 1–88. [Google Scholar]
- 20.Nieuwkoop PD, Sutasurya LA. Primordial Germ Cells in the Chordates: Embryogenesis and Phylogenesis. Cambridge Univ Press; London: 1979. [Google Scholar]
- 21.Charlesworth B. Evolution in Age-Structured Populations. Cambridge Univ Press; Cambridge, UK: 1980. [Google Scholar]
- 22.Gao Z, Wyman MJ, Sella G, Przeworski M. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 2015. 14(1):e1002355. [DOI] [PMC free article] [PubMed]
- 23.Drost JB, Lee WR. Biological basis of germline mutation: Comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 1995;25(S2):48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- 24.Ehmcke J, Wistuba J, Schlatt S. Spermatogonial stem cells: Questions, models and perspectives. Hum Reprod Update. 2006;12(3):275–282. doi: 10.1093/humupd/dmk001. [DOI] [PubMed] [Google Scholar]
- 25.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128(2):415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 26.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci USA. 2012;109(39):15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramm SA, Stockley P. Sperm competition and sperm length influence the rate of mammalian spermatogenesis. Biol Lett. 2010;6(2):219–221. doi: 10.1098/rsbl.2009.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin RD. The evolution of human reproduction: A primatological perspective. Am J Phys Anthropol. 2007;134(Suppl 45):59–84. doi: 10.1002/ajpa.20734. [DOI] [PubMed] [Google Scholar]
- 29.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scally A, Durbin R. Revising the human mutation rate: Implications for understanding human evolution. Nat Rev Genet. 2012;13(10):745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 31.Graves RR, Lupo AC, McCarthy RC, Wescott DJ, Cunningham DL. Just how strapping was KNM-WT 15000? J Hum Evol. 2010;59(5):542–554. doi: 10.1016/j.jhevol.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: A model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 33.Dutheil JY, Munch K, Nam K, Mailund T, Schierup MH. Strong selective sweeps on the X chromosome in the human-chimpanzee ancestor explain its low divergence. PLoS Genet. 2015;11(8):e1005451. doi: 10.1371/journal.pgen.1005451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 2004;101(39):13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forster P, et al. Elevated germline mutation rate in teenage fathers. Proc R Soc B. 2015;282(1803):20142898. doi: 10.1098/rspb.2014.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scally A. 2015. Mutation rates and the evolution of germline structure. bioRxiv, dx.doi.org/10.1101/034298.
- 37.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107(3):961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen Seaman MI, Hooper Boyd KA. Molecular clocks: Determining the age of the human–chimpanzee divergence. eLS. 2013 doi: 10.1002/9780470015902.a0020813.pub2. [DOI] [Google Scholar]
- 39.Li W-H, Ellsworth DL, Krushkal J, Chang BH-J, Hewett-Emmett D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996;5(1):182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.