Significance
Small RNAs (sRNAs) of 24 nt are associated with transcriptional gene silencing by targeting DNA methylation to complementary sequences. We demonstrated previously that sRNAs move from shoot to root, where they regulate DNA methylation of three endogenous transposable elements (TEs). However, the full extent of root DNA methylation dependent on mobile sRNAs was unknown. We demonstrate that DNA methylation at thousands of sites depends upon mobile sRNAs. These sites are associated with TE superfamilies found in gene-rich regions of the genome, which lose methylation selectively in an sRNA-deficient mutant. If the TEs were able to reactivate, they could cause genome instability and altered gene expression patterns, with negative effects on the plant. Consequently, mobile sRNAs may defend against these TEs.
Keywords: RNA-directed DNA methylation, plant grafting, transposable element, small RNA, transcriptional gene silencing
Abstract
RNA silencing at the transcriptional and posttranscriptional levels regulates endogenous gene expression, controls invading transposable elements (TEs), and protects the cell against viruses. Key components of the mechanism are small RNAs (sRNAs) of 21–24 nt that guide the silencing machinery to their nucleic acid targets in a nucleotide sequence-specific manner. Transcriptional gene silencing is associated with 24-nt sRNAs and RNA-directed DNA methylation (RdDM) at cytosine residues in three DNA sequence contexts (CG, CHG, and CHH). We previously demonstrated that 24-nt sRNAs are mobile from shoot to root in Arabidopsis thaliana and confirmed that they mediate DNA methylation at three sites in recipient cells. In this study, we extend this finding by demonstrating that RdDM of thousands of loci in root tissues is dependent upon mobile sRNAs from the shoot and that mobile sRNA-dependent DNA methylation occurs predominantly in non-CG contexts. Mobile sRNA-dependent non-CG methylation is largely dependent on the DOMAINS REARRANGED METHYLTRANSFERASES 1/2 (DRM1/DRM2) RdDM pathway but is independent of the CHROMOMETHYLASE (CMT)2/3 DNA methyltransferases. Specific superfamilies of TEs, including those typically found in gene-rich euchromatic regions, lose DNA methylation in a mutant lacking 22- to 24-nt sRNAs (dicer-like 2, 3, 4 triple mutant). Transcriptome analyses identified a small number of genes whose expression in roots is associated with mobile sRNAs and connected to DNA methylation directly or indirectly. Finally, we demonstrate that sRNAs from shoots of one accession move across a graft union and target DNA methylation de novo at normally unmethylated sites in the genomes of root cells from a different accession.
RNA silencing in plants and animals is a process that controls gene expression at both the transcriptional and posttranscriptional levels (1). In plants, small RNAs (sRNAs), 21–24 nt in length (2, 3), direct the RNA silencing machinery to target nucleic acids in a sequence-specific manner (4). The 21/22-nt sRNAs are primarily associated with mRNA cleavage and are involved in posttranscriptional gene silencing (PTGS) (3). The 24-nt sRNAs are primarily associated with RNA-directed DNA methylation (RdDM) and transcriptional gene silencing (2, 5). However, a recent study suggests that both 21- and 24-nt sRNAs are involved in deposition of DNA methylation (6). It is proposed that the 21-nt sRNAs may establish DNA methylation, whereas the 24-nt species are involved in its amplification and maintenance (7, 8).
RdDM involves methylation of cytosine residues in CG, CHG, and CHH sequence contexts (where H denotes any base except G) (9, 10). It is closely associated with repressive chromatin marks at target loci (11) and blocks gene transcription when present in promoter regions (12). RdDM maintains genome integrity by repression of transposable element (TE) activity, as well as contributing to environmental and developmental regulation of gene expression (4, 8, 13–17). The methylation status of DNA is heritable through both meiosis and mitosis, allowing it to persistently alter gene expression (18–20).
Initial establishment of RdDM involves cleavage of double-stranded RNA by DICER-LIKE (DCL) proteins to form 21- to 24-nt sRNAs, which load into ARGONAUTE (AGO) proteins (4, 21). These nucleoprotein complexes target chromatin-associated scaffold transcripts in a sequence-specific manner (22–24). The chromatin-bound complexes then recruit DOMAINS REARRANGED METHYLTRANSFERASES 1 and 2 (DRM1 and DRM2), which methylate DNA in CG, CHG, and CHH sequence contexts (8, 25). A complex set of maintenance mechanisms ensures persistence of established DNA methylation through cell division and even between generations. Most of these mechanisms are independent of RNA, and involve epigenetic histone marks. The VARIANT IN METHYLATION (VIM) family proteins 1, 2, and 3 and DNA METHYLTRANSFERASE 1 (MET1) efficiently maintain CG context methylation, resulting in near-complete methylation of target sequences (26–28). Non-CG context methylation (i.e., CHG, CHH) is maintained by a self-reinforcing loop involving KRYPTONITE family enzymes (29). These proteins recognize non-CG context methylated DNA and methylate lysine 9 of adjacent histone H3 (H3K9me2). CHROMOMETHYLASE (CMT) proteins CMT2 and CMT3 bind H3K9me2 and methylate adjacent non-CG sites (30) of the newly replicated DNA. Redundancy exists between target sites of CMT2 and CMT3, but their predominant functions are to maintain CHH and CHG context methylation, respectively (31). The activity of CMT2 is substantially less efficient than that of MET1, so that CMT2 target sites exhibit variable levels of DNA methylation. CMT3 efficiency is intermediate between MET1 and CMT2. Both CMT2 and CMT3 typically target long TEs and gene-distal TEs (31).
There are also RNA-dependent mechanisms to maintain DNA methylation that involve plant-specific DNA-dependent RNA polymerases IV and V (POL IV and POL V). These polymerases are recruited to chromatin by methyl-DNA–binding proteins Sawadee homeodomain homolog 1/DNA-binding transcription factor 1 (SHH1) SU(VAR)3–9 homolog 2 (SUVH2) and 9 (32). POL IV produces precursor RNAs that are processed into 24-nt sRNAs, whereas POL V produces chromatin-bound scaffold transcripts at sites of DNA methylation (33). Together, they ensure maintenance of CG, CHG, and CHH context DNA methylation through an AGO-dependent mechanism similar to the mechanism that establishes methylation, in which an AGO–sRNA recruits DRM1 and DRM2 to maintain non-CG DNA methylation (8, 25). The target sites of the DRM1/DRM2 DNA methylation maintenance pathway are largely nonoverlapping with those of CMT2 and CMT3, and tend to be short, gene-proximal TEs and the edges of long TEs (31, 34).
RdDM can operate cell-to-cell and systemically due to translocation of 23- to 24-nt sRNAs from shoots to roots (35). In our previous studies, we confirmed that these mobile 23- to 24-nt sRNAs target RdDM and TGS at one transgene and RdDM at three endogenous TEs (35–37). Depletion of shoot sRNAs corresponded to reduction of 23- to 24-nt sRNAs in wild-type roots, indicating that shoot-derived sRNAs contribute to the total root sRNA population (36). In this study, we investigate the extent to which mobile sRNAs mediate genome-wide RdDM. Our approach, as before, is to analyze sRNA and DNA methylation in roots of grafted plants that are defective for the production of the 24-nt sRNA species associated with RdDM. The sRNAs in these grafted plants move predominantly from shoot to root following source–sink gradients (36) and, by grafting different genotypes as shoots, we identify changes in DNA methylation and gene expression in the roots that are dependent on mobile sRNAs.
We show that mobile sRNAs influence genomic DNA methylation at thousands of loci, and that the affected loci are predominantly associated with transposons of specific classes. A very small number of protein-coding genes were influenced by this mobile RdDM. The mobile sRNA-dependent DNA methylation is associated with the DRM1/DRM2 RdDM pathway but not the CMT2/3 DNA methyltransferase pathway. Furthermore, we demonstrate that mobile sRNAs unique to one accession established DNA methylation de novo in unmethylated regions of the genome of a second accession.
Results
Identification of DNA Methylation Loci Targeted by Mobile sRNAs.
The primary aims of our study were to determine how many genomic loci in Arabidopsis may be targeted by RdDM due to the direct action of mobile sRNAs (Fig. 1) and to compare these loci with other sRNA-targeted and cytosine-methylated regions throughout the genome. We reasoned that mobile sRNAs that direct RdDM may be associated with specific features of the genome and depend upon specific genes in the RdDM pathway. In addition, we aimed to determine how many of these directly affected loci might influence gene expression. It could be, for example, that DNA methylation, leading to TGS, of a transposon target of mobile sRNA affects expression of an adjacent gene.
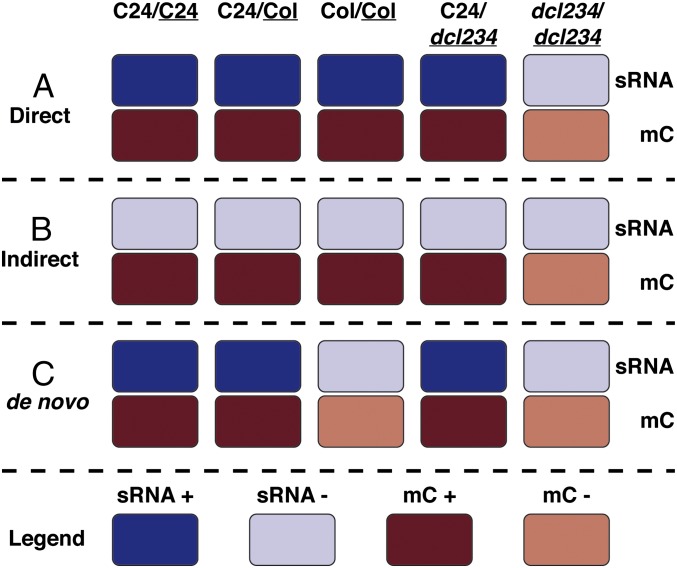
Fig. 1.
Genomic loci in Arabidopsis roots were classified according to their combination of sRNA and DNA methylation status. Grafts were made between shoots and roots of various Arabidopsis genotypes [combinations are indicated (Top), denoted shoot/root]. Target loci of sRNA and DNA methylation were identified by analyzing MethylC-seq and sRNA-seq data from roots of all graft combinations. Each locus classification was defined by a specific combination of sRNA and DNA methylation (mC) levels across the five graft combinations. “+” denotes a relatively high level of mC or sRNA, whereas “-” denotes a relatively low level. Classifications were designated A–C and are indicated (Left). See also SI Appendix, Fig. S1.
In previous work, we compared sRNA populations in roots of WT Arabidopsis thaliana (accessions Col-0, termed Col, and C24) and a dicer-like 2 dicer-like 3 dicer-like 4 triple mutant (Col background, termed dcl234) that had been grafted to C24, Col, and dcl234 shoots to identify loci producing mobile sRNAs in the Arabidopsis genome. The dcl234 triple mutant is unable to produce 22- to 24-nt sRNAs associated with RNA silencing (23). We reasoned that any 22- to 24-nt sRNAs present in dcl234 roots grafted to WT shoots must have moved from the grafted WT shoot (36). Loci similarly represented by sRNA reads in C24/C24, C24/Col, Col/Col, and C24/dcl234 but not dcl234/dcl234 datasets (notation shoot/root, with underlining indicating the analyzed plant part) could be confidently assigned as producing mobile sRNA, because the WT shoot complements the inability of dcl234 roots to produce sRNA. In contrast, the sRNAs from loci that were absent in C24/dcl234 and dcl234/dcl234 grafts and present in grafts with Col or C24 roots were interpreted as being dependent on DCL2,3,4 but not mobile. Loci where sRNAs were present in all datasets indicate sRNAs produced by DCL1 or in a DCL-independent manner.
We incorporated genome-wide MethylC-seq (sequencing) data in our analyses and applied the above reasoning to classify the genome-wide patterns of DNA methylation affected by graft-transmissible sRNAs. We used five genotype combinations in two-way grafts to define six locus classes (A–F) by their combinations of DNA methylation and sRNA representation across graft combinations (Fig. 1, where high and low sRNA abundance are indicated by dark and light blue, respectively, and high and low DNA methylation levels are indicated by dark and light red, respectively; also see SI Appendix, Figs. S1 and S2). Three of these classes were of primary interest:
-
A)
“Direct” loci correspond to overlapping regions of cytosine methylation and sRNAs present in C24/C24, C24/Col, Col/Col, and C24/dcl234 but not dcl234/dcl234. This model is consistent with direct targeting of DNA methylation by mobile sRNAs.
-
B)
“Indirect” loci show the same pattern of DNA methylation between grafts as direct loci but these were not associated with sRNAs. Here we infer that there are undetectable levels of mobile sRNAs or that the change in methylation is due to indirect effects of a mobile signal.
-
C)
“De novo methylated” loci correspond to overlapping regions of sRNAs and cytosine methylation present in C24/C24, C24/Col, and C24/dcl234 but not in dcl234/dcl234 or Col/Col. These loci likely correspond to sRNAs produced uniquely by C24 that target DNA methylation to previously unmethylated regions of the Col genome.
The class A (direct) and C (de novo) loci were of primary interest because these were most likely to be associated with the mobile sRNA. We compared their characteristics with those of class B (indirect) loci that are associated with a mobile signal but, by definition, do not correspond to detectable candidate sRNAs. Three classes of loci (D–F) not associated with regulation of DNA methylation by mobile sRNA were also identified, and are described in SI Appendix, Results.
Mobile sRNA and DNA Methylation.
We developed a statistical framework that could be applied to sRNA and MethylC-seq data to accurately assess the models described in Fig. 1 (see SI Appendix, Materials and Methods, sections 1, 3, and 4 for a detailed explanation). Our previous sRNA abundance data (36) were reanalyzed using this framework to ensure consistency with analyses of the MethylC-seq datasets generated in the current study (Table 1). DNA methylation associated with mobile sRNA was examined separately by sequence context (CG, CHG, CHH), because the genetic requirements for each are distinct.
Table 1.
Numbers of class A, B, and C loci in each DNA methylation context
| Class | mC context | mC loci (median length, nt) | sRNA loci (median length, nt) | mC overlaps to sRNA | sRNA overlaps to mC | Z | P |
| A | CG | 34 (33) | 925 (147) | 13* | 8 | 154.69 | 0 |
| CHG | 2,636 (94) | 398* | 257 | 77.173 | 0 | ||
| CHH | 2,980 (114) | 401* | 278 | 84.250 | 0 | ||
| B | CG | 34 (33) | 26,981 (1,759) | 13* | 12 | −11.686 | 7.55 × 10−32 |
| CHG | 2,636 (94) | 868* | 651 | −92.603 | 0 | ||
| CHH | 2,980 (114) | 1,029* | 787 | −105.48 | 0 | ||
| C | CG | 5 (1) | 63 (153) | 0* | 0 | NA | NA |
| CHG | 21 (48) | 2* | 2 | NA | NA | ||
| CHH | 125 (72) | 4* | 3 | 258.68 | 0 |
The columns “mC loci” and “sRNA loci” give the total numbers of mC and sRNA loci identified corresponding to each locus class A–C, as described in Fig. 1. The mC loci were counted individually by mC sequence context (CG, CHG, CHH). sRNA loci are not context-dependent, so only one set exists for each class A–C. The sRNA loci were intersected with the mC loci from the three sequence contexts within each class A–C. Comparisons were made in two directions: the number of mC loci overlapping sRNA loci (column “mC overlaps to sRNA”) and the number of sRNA loci overlapping mC loci (column “sRNA overlaps to mC”). Reversing the direction of comparison yielded different numbers of overlaps between sRNA and mC loci. This is possible because the sRNA and mC loci are of different sizes. Hence, one sRNA locus may overlap multiple mC loci.
Overlaps carried forward for downstream analyses of that class. A positive Z score indicates an enriched association between the sRNA and methylation loci, whereas a negative Z score indicates depleted association, relative to the overlap expected by chance. The final column gives the P values associated with the overlaps. NA (not applicable) indicates that no Z score, and hence no significance value, could be calculated from such small numbers of loci. Note that certain locus classes require low levels of mC or sRNAs in specific graft combinations (Fig. 1). For example, the number of overlaps columns (both mC overlaps to sRNA and sRNA overlaps to mC) for class B give the number of genomic loci where mC levels were high in roots of all graft combinations, except dcl234/dcl234, and where sRNA abundances were low in all graft combinations. Consequently, for this model, the sRNA loci column gives the number of loci where sRNA abundance was low in all graft combinations.
Our analyses revealed that non-CG DNA methylation is abundant at class A (direct) loci: We identified 13 CG, 398 CHG, and 401 CHH context class A loci with a false discovery rate (FDR) of 5% (Table 1 and Dataset S1). These included the three loci we identified previously as possessing mobile sRNA-associated DNA methylation (36). The association between regions of sRNA and DNA methylation that corresponded to the class A loci model was statistically significantly more frequent than expected by chance (Table 1, Z scores: CG, 154.69; CHG, 77.173; CHH, 84.250; P < 0.05). Furthermore, class A loci of all three DNA methylation contexts overlapped statistically significantly (SI Appendix, Fig. S3). Observation of colocalized DNA methylation in all three contexts is characteristic of RdDM. Fewer than 2.5% of the class A loci overlapped with regions identified as variable over 30 generations in Arabidopsis (18), and thus it is unlikely that the results are due to spontaneous epiallelic variation.
We investigated the genes potentially regulating DNA methylation at class A loci by analysis of MethylC-seq data from 86 Arabidopsis gene-silencing mutant lines (31). We examined the effects of mutation of a subset of genes with known roles in RdDM (dcl2/3/4, met1, drm1/2, ddm1, suvh4/5/6, vim1/2/3, cmt2, cmt3, and WT control) on DNA methylation at class A loci. We focused on non-CG methylation patterns because very few CG context class A loci were identified. Mutation of drm1/2 caused the strongest reduction in non-CG context methylation in class A loci (Fig. 2 and SI Appendix, Fig. S4). The other mutations examined showed weaker effects, of which cmt2 and cmt3 caused the least changes in DNA methylation: cmt3 reduced CHG context DNA methylation to levels intermediate between WT and drm1/drm2 but had no effect in the CHH context, whereas cmt2 had no effect in either context. DNA methylation was reduced to levels intermediate between WT and drm1/2 in both sequence contexts by all other mutations. It should be noted that the MethylC-seq data from gene-silencing mutant lines were generated using leaves of 3-wk-old plants, whereas our MethylC-seq data were generated using roots of 3-wk-old plants. We considered it appropriate to compare the two datasets because there is little evidence for tissue-specific variation in DNA methylation in Arabidopsis. We examined the similarity of DNA methylation in these two tissues by making comparisons between the likelihoods of methylation at loci identified as methylated in root tissue of the Col/Col graft from our dataset and in the 3-wk-old Col-0 (WT) leaf tissue of Stroud and colleagues (31). DNA methylation was broadly consistent between the two tissues (SI Appendix, Fig. S5 and Dataset S2). Additionally, a prior study demonstrated that DNA methylomes from two tissues in each of 11 Arabidopsis accessions cluster by accession, not tissue type (38).
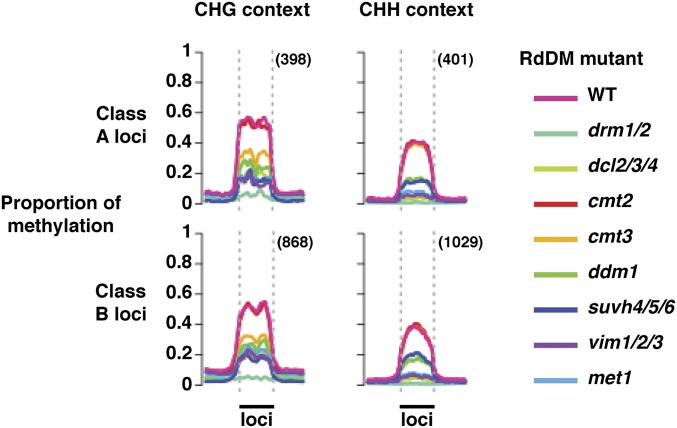
Fig. 2.
DNA methylation of mobile sRNA-associated loci (class A, direct; B, indirect) is reduced most strongly by drm1/2 mutation. The plots show the mean proportion of DNA methylation across class A and B loci in mutants of RNA-directed DNA methylation pathway components and wild-type (WT) plants. The legend (Right) indicates which color represents each mutant. Data are shown separately for non-CG (CHG and CHH) sequence contexts, which had the strongest association with mobile sRNAs. The proportion of methylation was calculated per base (between 0 and 1, unmethylated to fully methylated) for all loci within a class using published MethylC-seq data from RdDM mutants (31). Loci were then normalized to the same size, and the mean proportion of DNA methylation was calculated across them. The profiles of the mean proportion of methylation across the size-normalized loci are plotted between the dashed vertical lines (indicated by the solid black bars labeled “loci”). Mean proportions of methylation in flanking DNA, 4 kb upstream and downstream of the loci, are indicated to the left and right of the dashed vertical lines, respectively. Total numbers of loci assessed are given in parentheses.
DNA Methylation Under Indirect Regulation by the Mobile Signal.
Class B (indirect) loci show the same pattern of DNA methylation between grafts as do class A (direct) loci. However, they are distinguished by the lack of associated sRNA. We found that class B and class A loci had extremely similar characteristics. Hundreds of class B loci were detected almost exclusively in non-CG contexts (13 CG, 868 CHG, 1,029 CHH; Table 1 and Dataset S1). These loci were unambiguously identified and located in the genome. We found that fewer than 0.5% of the class B loci overlapped with regions previously identified as spontaneously variable between parent and offspring plants, suggesting our results are not a consequence of this phenomenon (18).
The distribution of the class B loci across DNA methylation contexts mirrors that of the class A loci (13 CG, 398 CHG, and 401 CHH), although substantially more class B loci were identified. We focused on non-CG class B loci because so few CG loci were identified. The association between regions lacking sRNAs and possessing DNA methylation, corresponding to the class B loci model, was lower than would be expected by chance (Table 1; CHG, P = 0, Z = −92.603; CHH, P = 0, Z = −105.48). This pattern suggests that methylation loci regulated by a mobile signal are negatively associated with regions lacking in small RNAs.
The class A and B loci were also influenced similarly by mutations in RdDM pathway genes (31) (Fig. 2 and SI Appendix, Fig. S4). Mutation of drm1/2 caused the strongest reduction in non-CG context DNA methylation (Fig. 2 and SI Appendix, Fig. S4). All other mutations examined reduced non-CG context methylation at class B loci except cmt2. Notably, cmt3 had the next-weakest effect, reducing CHG context DNA methylation but not affecting CHH context methylation.
In summary, there was very little to distinguish class A and class B loci, suggesting they are ultimately under the regulation of the same mobile signal. In principle, this signal could be an as-yet unidentified secondary regulatory factor dependent on the DCL234-dependent sRNA at the class A loci. Alternatively, it could be caused by the very low levels of DCL234-dependent sRNA that fail to pass the significance thresholds in our model (SI Appendix, Figs. S6–S8), or that are not well-sequenced.
We used an independent statistical approach to estimate the total number of class A and B loci in the dataset, as a complement to the results above (see SI Appendix, Materials and Methods, section 5 for a detailed explanation). This approach evaluates all loci that may exist without specifying genomic coordinates. We estimate that there exist 72 CG (59–85 with 95% confidence), 1,557 CHG (1,506–1,608 with 95% confidence), and 2,238 CHH (2,170–2,303 with 95% confidence) loci of class A, and 526 CG (485–568 with 95% confidence), 5,699 CHG (5,597–5,802 with 95% confidence), and 10,943 CHH (10,785–11,099 with 95% confidence) loci of class B by this approach. From these data, we conclude that thousands of DNA methylation loci found predominantly in the non-CG context may be regulated by the mobile signal.
DNA Methylation Regulated by the Mobile Signal Associates with Specific Features of the Genome.
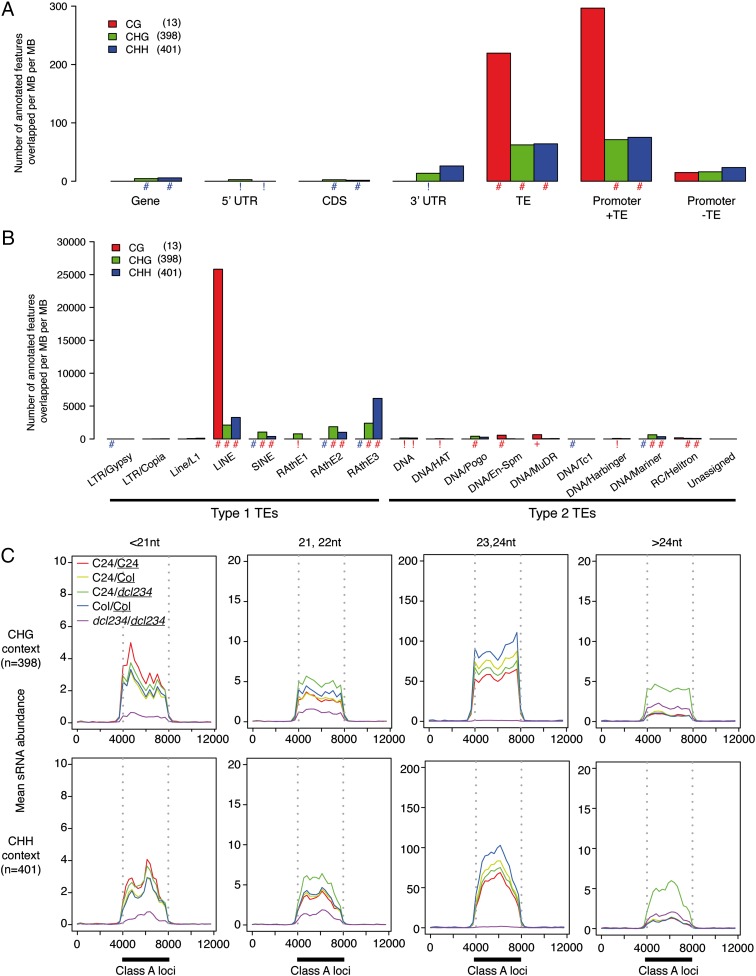
Non-CG context class A loci were significantly associated with TEs and promoters containing TEs but not with promoters that do not contain TEs (Fig. 3A). In contrast, they were significantly depleted in genes, coding regions, and 5′ UTRs (untranslated regions). CG context class A loci were significantly associated with TEs only. Further analysis showed that class A loci associated significantly with many superfamilies of TEs (Fig. 3B). RAth elements and LINE (long interspersed nuclear element) and SINE (short interspersed nuclear element) superfamily TEs were targeted more strongly than other TEs, most clearly so by the non-CG context class A loci. TEs are broadly grouped according to their replication strategies as type 1 (retroelements) or type 2 (DNA elements), and the targeted superfamilies are all short, gene-proximal type 1 TEs. Class A loci were targeted most highly by 23- to 24-nt mobile sRNAs, eliminated in dcl234/dcl234 grafts (Fig. 3C). These results indicate that DNA methylation directly targeted by the shoot–root mobile sRNA is associated with specific TE superfamilies.
Fig. 3.
Mobile sRNA directly targets DNA methylation at promoters containing transposable elements (TEs). (A) CHG and CHH (non-CG) context class A loci are enriched in TEs and in promoters containing TEs but are depleted in genes and coding sequences (CDSs). Very few CG context loci were identified. (B) Non-CG context class A loci preferentially target classes of type 1 retroelement, indicated by significant enrichment. They also, less strongly, target some type 2 DNA element classes. y-axis units in A and B normalize the feature/locus overlap by both sum of genome feature size and sum of methylation locus size, which permits comparison between columns. These units are the number of annotated features per total megabase (MB) of named feature per total MB of methylation in the locus class. Numbers of loci in each DNA methylation context are shown in parentheses. Significance levels: #, 0 < P < 10−5; +, 10−4 < P < 10−5; *, 10−3 < P < 10−4; !, 10−2 < P < 10−3; blue, underrepresented;red, overrepresented (relative to background). (C) Class A loci are targeted by graft-transmissible 23- to 24-nt sRNAs (the known mobile class). Loci were normalized to the same size on the graph, indicated by the dark bar on the position axis, flanked by sRNA coverage 4 kb upstream and downstream of the locus. Numbers of loci assessed are given in parentheses.
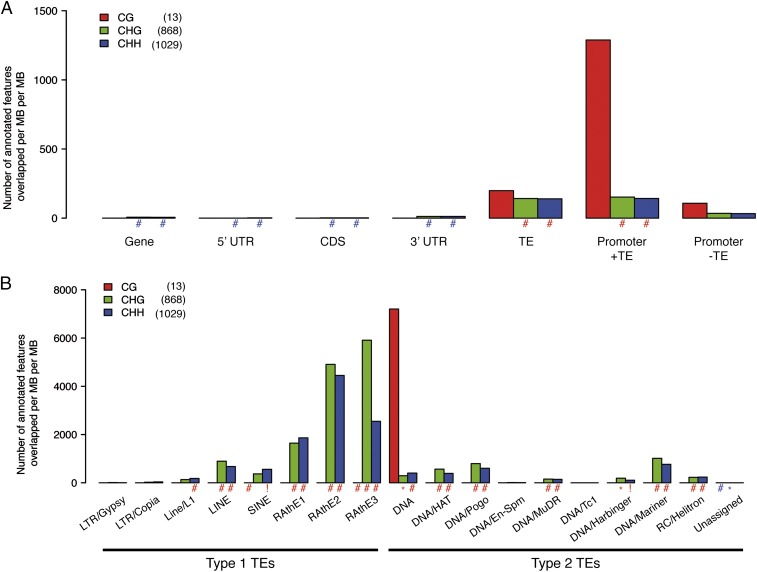
Non-CG class B loci were also associated significantly with TEs and promoters containing TEs (Fig. 4A and SI Appendix, Fig. S2), as were non-CG class A loci. They were associated with many different TE superfamilies, most strongly with certain type 1 retroelement superfamilies (RAths, LINE, SINE). They also showed clear associations with specific type 2 retroelement superfamilies (DNA elements) including DNA Mariner, Pogo, and HAT.
Fig. 4.
Loci where DNA methylation is indirectly targeted by mobile sRNAs (class B) exhibit similar characteristics to loci that are directly targeted (class A). (A) Non-CG (CHG and CHH) context B loci are significantly enriched in TEs and in promoters containing TEs but are depleted in promoters that do not contain TEs, genes, and CDSs. Very few CG context loci were identified. (B) Non-CG context B loci target most classes of TEs but show most significant association with classes of type 1 retroelement. They also, less strongly, target some type 2 DNA element classes. y-axis units normalize the feature/locus overlap by both sum of feature size and sum of methylation locus size, which permits comparison between columns. These units are the number of annotated features per total MB of named feature per total MB of methylation in the locus class. Significance levels: #, 0 < P < 10−5; +, 10−4 < P < 10−5; *, 10−3 < P < 10−4; !, 10−2 < P < 10−3; blue, underrepresented; red, overrepresented (relative to background). Numbers of loci assessed are given in parentheses.
The genome features associated with class A and B loci might either be specific to these locus classes and to the mobile signal or they could be driven by dependence of the loci upon DCL234-derived sRNAs. To address this point, we compared the features associated with class A and B loci with all features that lose DNA methylation in dcl234/dcl234 grafts. The features that lost DNA methylation in dcl234/dcl234 grafts were highly similar to those associated with class A and B loci (SI Appendix, Fig. S9). The data indicate that a specific subset of TEs lose DNA methylation in the dcl234 mutant. Furthermore, they suggest that dependency upon DCL234 drives the features associated with class A and B loci rather than these features being specific to the mobile signal.
The Influence of the Mobile Signal on Gene Expression.
We examined whether DNA methylation regulated by the mobile signal may influence gene expression. Root transcriptomes of the graft combinations described by Fig. 1 and SI Appendix, Fig. S1 (excluding C24/C24) were profiled by RNA-seq. A total of 23 transcripts were identified as significantly differentially regulated in a manner that indicated association with the mobile signal (Dataset S3). These included transcripts coding for proteins involved in cell-wall formation, membrane transport, and regulation of osmotic potential. Three of the genes encoding these were associated with DNA methylation regulated either directly or indirectly by the mobile signal: AT2G01880, PURPLE ACID PHOSPHATASE 7; AT2G36490, REPRESSOR OF SILENCING 1 (ROS1); and AT3G43270, a pectin methylesterase. We measured the number of differentially expressed genes between C24/Col and dcl234/dcl234 roots as a control. Twenty-seven transcripts were identified as significantly differentially regulated in roots of dcl234 compared with WT roots (Dataset S4). Differential expression of ROS1 (AT2G36490.1) was confirmed in roots of dcl234/dcl234 versus C24/dcl234 and C24/Col by quantitative real-time PCR (Q-RT-PCR) (SI Appendix, Fig. S10). These findings demonstrate that loss of DCL2, DCL3, and DCL4 causes differential expression of a small number of genes, consistent with data in previous reports (39).
We investigated whether transcripts dependent upon DCL234 (Dataset S4) or associated with the mobile signal (Dataset S3) might also depend upon DRM1/2. This was done by identifying transcripts differentially regulated between WT and the drm1/2 mutant using published RNA-seq data. These data were generated from leaves of 3-wk-old plants in parallel with the MethylC-seq data discussed in Fig. 3 and SI Appendix, Fig. S4 (31). Eighty-six transcripts were significantly differentially regulated between WT and drm1/2 plants (Dataset S5). The transcript of ROS1 (AT2G36490) was shared between these and transcripts associated with the mobile signal (Dataset S3B). The transcript AT1G53480.1 was dependent upon DRM1/2 (Dataset S5) and DCL234 (Dataset S4A). We attribute the low degree of overlap between DRM1/2-dependent and mobile signal-associated transcripts to tissue specificity of gene expression. Our sRNA-seq and MethylC-seq experiments were conducted on roots of 3-wk-old plants. As discussed above, comparison between DNA methylomes from different tissues is appropriate because there is little evidence for tissue-specific variation in DNA methylation. However, transcript abundance differs more between tissues. This was demonstrated by comparison of RNA-seq data from two tissues of multiple Arabidopsis accessions, where transcript abundance correlated more highly between like tissues of different accessions than different tissues of one accession (38).
Epialleles Are Transmitted Across Accessions by Mobile sRNAs.
There is natural variation in RdDM and its associated sRNAs in Arabidopsis, and the term “epiallele” describes genomic locations where methylation differs between accessions (38, 40, 41). We investigated whether sRNAs produced uniquely by C24, and not by Col, could direct DNA methylation de novo at previously unmethylated sites in the Col genome (Fig. 1, class C loci). Two class C loci were unambiguously identified in the CHG DNA methylation context, and four in the CHH context (Dataset S1). Three distinct sites of de novo RdDM existed after taking into account overlap of these loci between DNA methylation contexts (Fig. 5). None of these loci had been previously identified as spontaneously variable between parent and offspring plants (18). These results indicate that exogenous sRNAs supplied by shoots can target de novo DNA methylation at unmethylated sites in the genome of root cells, thereby transmitting epiallelic states from one Arabidopsis accession to another.
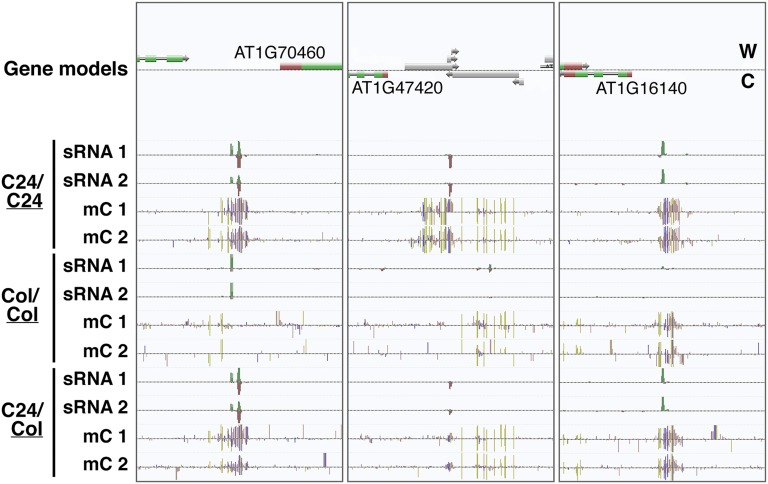
Fig. 5.
C24-derived sRNAs can target DNA methylation de novo at unmethylated regions of the Col-0 genome. Three such de novo loci are shown in AnnoJ genome browser (www.annoj.org) screenshots. The screenshots display sRNA reads and methylated cytosine residues (mCs) for graft combinations C24/C24, Col/Col, and C24/Col. Results from two independent biological replicates, suffixed 1 and 2, are shown for each graft combination. Note the groups of sRNA reads present only in C24/C24 and C24/Col, which correlate with the presence of mC. Red and green sRNAs indicate that they map to the Watson (W) and Crick (C) strands, respectively. Gold, blue, and pink mC positions indicate methylation in the CG, CHG, and CHH contexts, respectively.
Discussion
In this study, we have characterized the genome-wide distribution of 23- to 24-nt shoot/root mobile sRNAs and identified regions of DNA methylation that they target directly (class A loci). We also identified regions of DNA methylation dependent indirectly upon mobile sRNAs (class B loci). We found that loss of mobile sRNAs generated in the shoots disrupts DNA methylation at thousands of sites in the roots. The class B (indirect) loci were most numerous, but the two classes were otherwise indistinguishable. Both were almost entirely in the non-CG (CHG/CHH) DNA methylation context. The class A and B loci were also significantly associated with the same TE superfamilies. We found that these TE superfamilies also lost DNA methylation in the dcl234 mutant, which is deficient in 22- to 24-nt sRNAs, indicating that the feature association of class A and B loci was driven by the dependency of mobile sRNAs on DCL234. Our results suggest that the function of mobile sRNAs is to reinforce silencing of these TEs. Furthermore, we have provided mechanistic insights into the specific RdDM pathways regulating mobile sRNA-dependent methylation. Data can be visualized on our interactive genome browser (neomorph.salk.edu/mobile_methylome.php).
Our data confirm that mobile sRNA-dependent DNA methylation requires DRM1 and DRM2, key components of the 24-nt RdDM pathway. There was only limited dependence upon CMT3 and CMT2, both of which belong to a distinct DNA methylation maintenance pathway. Furthermore, class A and B loci were associated with the superfamilies containing the shortest (on average) TEs (Figs. 2 and 4 and SI Appendix, Table S1), including type 1 retroelements, such as RAth elements (E1, E2, E3), SINEs, and LINEs, as well as certain type 2 DNA elements (Mariner, Pogo, RC Helitron). The DNA methylation pathways containing DRM1/2 and CMT2/3 are distinct but overlapping, and both deposit non-CG methylation (8, 34). The DRM1/2 pathway methylates small TEs and TE edges, whereas the CMT2/3 pathway is responsible for methylation of long TEs (31, 34). Our data are in concordance with this pattern, and establish a relationship of mobile sRNA-dependent methylation with a specific RNA silencing mechanism.
The direct and indirect (class A and B) loci are essentially identical according to our data, except that no mobile sRNAs were identified as being associated with indirect class B loci. These findings lead to three hypotheses for further investigation. The first is that the mobile sRNAs from the shoot regulate an unidentified secondary signal in the roots. This secondary signal would then directly regulate DNA methylation at the B loci. The second is that mobile sRNAs can direct the RNA silencing machinery to sites that they match imperfectly (i.e., with which they have mismatches in sequence homology). We required perfect matching between sRNAs and genomic sequence when identifying sRNA target loci, but it is possible that a certain degree of mismatch between mobile sRNAs and their targets is either tolerated or required in certain circumstances. However, allowing mismatches during sRNA mapping (and therefore in targeting) permits individual sRNAs to target multiple sites due to their inherently short sequence, many of which are presumably spurious (42). Therefore, detailed analysis of individual methylation regions and thorough experimental validation are required to test this hypothesis. Moreover, it appears unlikely that the conclusions of our study would be altered by the outcome of such experiments, because we find the characteristics of class A (direct) and class B (indirect) loci to be essentially identical. The third hypothesis is that these loci are targeted by mobile sRNAs of very low abundance, which could not be detected in our sRNA-seq libraries.
Class A and B loci were substantially more numerous than the other classes (Fig. 1, Table 1, and SI Appendix, Fig. S1 and Table S2). The class D–F loci described loci not methylated by mobile sRNAs, not remethylated by mobile sRNAs, and whose methylation was independent of DCL234-derived sRNAs, respectively. Class F loci were the least numerous. This observation suggests there may be few mobile sRNA-targeted loci at which that sRNA does not regulate DNA methylation.
sRNAs of 24-nt length were proportionally the most common sRNA species detected in our sRNA-seq experiments and demonstrated the clearest shoot–root transmission (Fig. 2) (36). This study and another indicate that 21- to 22-nt sRNAs are also mobile and associated with DNA methylation (37). These data are consistent with previous proposals of a role for the 21- to 22-nt size class in RdDM (6, 7, 37). Furthermore, the type 1 retroelements that we find to be the predominant targets of mobile sRNA-dependent DNA methylation produce RNA intermediate replication stages, making it possible that if transcriptionally active they are also targets of PTGS via mobile 21-nt sRNAs (43). However, fewer 21- to 22-nt sRNAs were mobile than 24-nt sRNAs, suggesting 24-nt sRNAs are the primary shoot–root signal. The design of our experiment did not permit us to completely eliminate the possibility that “mobile” 21-nt sRNAs are produced in the roots. This is because the dcl234 mutant contains functional DCL1, which generates 21-nt sRNAs, and the dcl234 mutant eliminates their production only partially (23, 44–46). Detailed investigation of their contribution would involve generation of an additional series of Arabidopsis mutant grafts and is complicated by the lethality of a dcl1 null plant (47).
Although mobile sRNAs have minimal effects on gene expression in Arabidopsis, we predict that their influence would be much greater in TE-rich genomes. In such genomes, including those of many common crops, mobile sRNAs may be an important mechanism of genome defense. The TE superfamilies targeted by mobile sRNAs are enriched in euchromatic regions, typically within c. 500 nt of the nearest gene (48, 49). TEs inserted near genes can have dramatic effects on gene expression (15, 49). Expression of a small number of transcripts was differentially regulated in our study, consistent with previous observations (39). However, the A. thaliana genome contains substantially fewer TEs than related outcrossing species, such as Arabidopsis lyrata (50, 51). Furthermore, TEs in the A. thaliana genome show a lower rate of active transposition (50). This suggests that TEs in the A. thaliana genome may have relatively less influence on gene expression than TEs in outcrossing species, which would result in fewer mobile sRNA-dependent regulated genes. Nonetheless, those genes that did exhibit mobile sRNA-dependent regulation had diverse functions, indicating they may have significant influence in the correct conditions. Moreover, mobile sRNAs are able to move into both meristematic and meiotically active tissues, where they can alter DNA methylation and gene expression (37, 52). In these tissues it is essential to protect genome stability by repression of TEs, so that gametes and developing organs are not harmed (53, 54).
Grafting is routinely used in agriculture to combine rootstocks and shoots (scions) with desirable characteristics, such as for grapevine, apples, and tomatoes (55, 56). We have demonstrated that mobile sRNAs regulate patterns of DNA methylation genome-wide, and that expression of specific genes exhibits mobile regulation. Base-resolution methylomes are being actively generated in multiple species, including crops and nonmodel plants (38, 57–59). Clear diversity in the epigenomes of closely related subspecies and accessions has been observed from these data (38, 57). With our demonstration that site-specific transmission of epiallelic states from one accession to another can be achieved by grafting and by de novo methylation of unmethylated DNA, it is likely that at least some effects of grafting are due to the movement of RNA. Our findings also indicate that DNA methylomes may provide a potential new resource to growers who use grafting. They could consider potential modification of gene expression patterns in sink tissues via sRNA transmission from source tissues. The mobile sRNAs can alter DNA methylation in germ-line tissue (37), so that DNA methylation patterns altered by grafting may be heritable by progeny plants (37, 60).
In summary, we have shown that transmission of sRNAs from shoots to roots of Arabidopsis regulates DNA methylation at thousands of sites genome-wide. Mobile sRNA-dependent methylation is predominantly in the non-CG context (CHG and CHH), and is associated with short type 1 retroelements found in gene-rich regions of the genome. We confirm that deposition of mobile sRNA-dependent methylation is dependent upon the DRM1 and DRM2 RdDM pathway and largely independent of the CMT2 and CMT3 methylation pathway. Our conclusions underpin future research into why plants possess a system for communicating methylation status from shoot to root tissues.
Materials and Methods
See also SI Appendix, Materials and Methods.
Plant Materials and Grafting.
Plants of A. thaliana accessions Col-0 and C24 were used in our experiments, as well as a previously described dcl2-1/dcl3-1/dcl4-2 triple mutant (Col-0 background) (23). The C24 plants had a GFP transgene silenced by a partial GFP inverted repeat, termed GxGF-IR, as previously described (36). Arabidopsis plants were grown under 10-h supplemental fluorescent lighting at 20 °C on vertical plates of 0.8% agar, 1∕2 Murashige and Skoog media (pH 5.7). Micrografting was conducted 7 d after germination as previously described (56, 61). Plant tissue was harvested 5 wk after grafting, taking care to separate root from shoot and exclude tissue 0.3–0.5 cm surrounding the graft junction. Two independent bioreplicates were conducted for all RNA-seq samples, and two to four for each MethylC-seq sample (SI Appendix, Table S3).
MethylC-Seq and RNA-Seq Library Construction.
DNA for the MethylC-seq libraries was prepared from three or four pooled plants per sample using the Gentra Puregene Tissue Kit (Qiagen) according to the manufacturer’s instructions. MethylC-seq libraries were constructed as described previously (62). RNA-seq libraries were prepared from total RNA extracted as previously described (36). RNA-seq libraries were generated with the ScriptSeq RNA-Seq Library Preparation Kit (Epicentre) according to the manufacturer’s instructions.
Sequencing Operations.
MethylC-seq and RNA-seq libraries were sequenced using Illumina HiSeq 2000 and 2500 platforms, respectively, according to the manufacturer’s instructions. MethylC-seq libraries were sequenced for 101 cycles, and RNA-seq libraries for 100 cycles. Image analysis and base calling were carried out using manufacturer-supplied software and standard parameters.
Sequencing Analyses.
See SI Appendix, Materials and Methods for a detailed description of analytical methods and software. In brief, sRNA data were aligned to the TAIR9 reference genome (63) requiring perfect matching. When analyzing MethylC-seq data, we used YAMA (Yet Another Methylome Aligner), based upon Bowtie 2 (64), to map reads to the genome (source code is available at https://github.com/tjh48/YAMA). Mapping statistics of libraries are given in SI Appendix, Table S3. Methylation loci were then identified independently in each context (CG, CHG, CHG) using the segmentSeq R package (65), accounting for nonconversion rates in each sample (66). Subsequently, loci exhibiting various models of differential methylation were identified using the empirical Bayesian methods for analysis of methylation data (65) with the baySeq R package (67, 68). Average methylation profiles were calculated by dividing defined regions into an equal number of windows and calculating methylation across these windows. Gene annotations were retrieved from the Arabidopsis Information Portal (https://www.araport.org) (69) and transposon annotations from the Arabidopsis Information Resource (70). Estimations of the numbers of loci expected to fit a given model were calculated using the sum of the posterior likelihoods for that model, and confidence intervals were calculated by simulating 10,000 sets of true and false positives based on the posterior likelihoods. Unambiguous lists of specific differentially represented loci were identified by applying a false discovery rate of lower than 0.05 to the model, which in general represents a more conservative analysis. Associations between DMRs, genome features, and sRNAs were assessed using a block-bootstrap method (71). Intersections between loci and genome features were normalized for abundance of loci and features, the sizes of loci and features, and how loci and features might cluster within the genome when plotting. We did so by calculating the number of features overlapped by the DNA methylation/sRNA loci per megabase total feature length per megabase total locus length. This approach gave a visually accurate representation of significance of association. Independent measurements of statistical significance based upon the block-bootstrap analyses are presented beneath the bars also (Fig. 3 A and B, Fig. 4, and SI Appendix, Figs. S3, S11, and S12). Promoters were defined as 2,000 nt preceding a transcriptional start site. Data were deposited in the European Nucleotide Archive (accession no. E-MTAB-3473).
Q-RT-PCR.
Transcript abundance was assessed in roots of independent graft plants as described previously (37). Fold changes in abundance of test transcripts relative to the ACTIN 2 transcript (AT3G18780), a stable control (72), were calculated using delta delta CT methodology (73, 74). Primer sequences were 5′-GGATATTGTAGCAAGCCACAG-3′ (ROS1 forward), 5′-CAAAGTTCATCTGGAGAAAGAC-3′ (ROS1 reverse), 5′-GCCATCCAAGCTGTTCTCTC-3′ (ACTIN 2 forward), and 5′-CCCTCGTAGATTGGCACAGT-3′ (ACTIN 2 reverse).
Supplementary Material
Acknowledgments
We thank Matthew D. Schultz and Yupeng He for assistance with analyses; Huaming Chen for assistance with web browser development; and Roberto Solano, Robert J. Schmitz, Taiji Kawakatsu, Chongyuan Luo, Ryan Lister, Ronan C. O’Malley, and Lindsay Robinson for helpful discussions. M.G.L. was funded by an EU Marie Curie FP7 International Outgoing Fellowship (252475). Work in J.R.E.’s laboratory is funded by the Gordon and Betty Moore Foundation (3034) and the National Science Foundation (MCB-1024999). J.R.E. is an investigator of the Howard Hughes Medical Institute. C.W.M. was funded by a Clare College (Cambridge, United Kingdom) Junior Research Fellowship. Work in the D.C.B. laboratory was supported by the Gatsby Charitable Foundation, European Union FP7 Collaborative Project Grant AENEAS, and European Research Council Advanced Investigator Grant ERC-2013-AdG 340642. D.C.B. is the Royal Society Edward Penley Abraham Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (accession no. E-MTAB-3473).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515072113/-/DCSupplemental.
References
- 1.Martínez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: A diversity of pathways. Biochim Biophys Acta. 2013;1829(12):1300–1308. doi: 10.1016/j.bbagrm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21(17):4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102(33):11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi Y, et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443(7114):1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 6.Nuthikattu S, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013;162(1):116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112(3):917–922. doi: 10.1073/pnas.1413053112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19(5):320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 10.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447(7143):418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 11.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SW-L, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303(5662):1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 13.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109(32):E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31(2):154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 15.Gent JI, et al. CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2013;23(4):628–637. doi: 10.1101/gr.146985.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zilberman D, et al. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14(13):1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 17.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334(6054):369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker C, Weigel D. Epigenetic variation: Origin and transgenerational inheritance. Curr Opin Plant Biol. 2012;15(5):562–567. doi: 10.1016/j.pbi.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Richards EJ. Inherited epigenetic variation—Revisiting soft inheritance. Nat Rev Genet. 2006;7(5):395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 21.Papp I, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132(3):1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41(5):630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38(6):721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 24.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105(8):3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong X, et al. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157(5):1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4(8):e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan SW-L, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6(5):351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Kim JH, Richards EJ, Chung KM, Woo HR. Arabidopsis VIM proteins regulate epigenetic silencing by modulating DNA methylation and histone modification in cooperation with MET1. Mol Plant. 2014;7(9):1470–1485. doi: 10.1093/mp/ssu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507(7490):124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12(8):483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 34.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21(1):64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30(17):3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328(5980):872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 37.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21(19):1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495(7440):193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laubinger S, et al. Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc Natl Acad Sci USA. 2010;107(41):17466–17473. doi: 10.1073/pnas.1012891107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bond DM, Baulcombe DC. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 2014;24(2):100–107. doi: 10.1016/j.tcb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Groszmann M, et al. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA. 2011;108(6):2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338(6108):758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 44.Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19(3):926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15(16):1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37(12):1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 47.Golden TA, et al. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130(2):808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenoir A, et al. The evolutionary origin and genomic organization of SINEs in Arabidopsis thaliana. Mol Biol Evol. 2001;18(12):2315–2322. doi: 10.1093/oxfordjournals.molbev.a003778. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Weigel D, Smith LM. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 2013;9(2):e1003255. doi: 10.1371/journal.pgen.1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Chaux N, Tsuchimatsu T, Shimizu KK, Wagner A. The predominantly selfing plant Arabidopsis thaliana experienced a recent reduction in transposable element abundance compared to its outcrossing relative Arabidopsis lyrata. Mob DNA. 2012;3:2. doi: 10.1186/1759-8753-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Wessler SR. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA. 2004;101(15):5589–5594. doi: 10.1073/pnas.0401243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, et al. Graft-transmissible movement of inverted-repeat-induced siRNA signals into flowers. Plant J. 2014;80(1):106–121. doi: 10.1111/tpj.12622. [DOI] [PubMed] [Google Scholar]
- 53.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melnyk CW, Meyerowitz EM. Plant grafting. Curr Biol. 2015;25(5):R183–R188. doi: 10.1016/j.cub.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 56.Melnyk CW, Schuster C, Leyser O, Meyerowitz EM. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol. 2015;25(10):1306–1318. doi: 10.1016/j.cub.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics. 2012;13:300. doi: 10.1186/1471-2164-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitz RJ, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23(10):1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takuno S, Gaut BS. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA. 2013;110(5):1797–1802. doi: 10.1073/pnas.1215380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu R, et al. Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants. PLoS One. 2013;8(4):e61995. doi: 10.1371/journal.pone.0061995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnbull CGN, Booker JP, Leyser HMO. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002;32(2):255–262. doi: 10.1046/j.1365-313x.2002.01419.x. [DOI] [PubMed] [Google Scholar]
- 62.Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat Protoc. 2015;10(3):475–483. doi: 10.1038/nprot.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hardcastle TJ. 2015. Discovery of methylation loci and analyses of differential methylation from replicated high-throughput sequencing data. bioRxiv:021436.
- 66.R Development Core Team . R: A Language and Environment for Statistical Computing. R Found Stat Comput; Vienna: 2008. [Google Scholar]
- 67.Hardcastle TJ, Kelly KA. Empirical Bayesian analysis of paired high-throughput sequencing data with a beta-binomial distribution. BMC Bioinformatics. 2013;14:135. doi: 10.1186/1471-2105-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hardcastle TJ. Generalized empirical Bayesian methods for discovery of differential data in high-throughput biology. Bioinformatics. October 1, 2015 doi: 10.1093/bioinformatics/btv569. [DOI] [PubMed] [Google Scholar]
- 69.International Arabidopsis Informatics Consortium Taking the next step: Building an Arabidopsis information portal. Plant Cell. 2012;24(6):2248–2256. doi: 10.1105/tpc.112.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huala E, et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29(1):102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bickel PJ, Freedman DA. Some asymptotic theory for the bootstrap. Ann Stat. 1981;9(6):1196–1217. [Google Scholar]
- 72.Havecker ER, Wallbridge LM, Fedito P, Hardcastle TJ, Baulcombe DC. Metastable differentially methylated regions within Arabidopsis inbred populations are associated with modified expression of non-coding transcripts. PLoS One. 2012;7(9):e45242. doi: 10.1371/journal.pone.0045242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuan JS, Wang D, Stewart CN., Jr Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol J. 2008;3(1):112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]
- 74.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.