Abstract
Background & Aims
Irritable bowel syndrome (IBS) is a stress-sensitive disorder. Environmental factors including stress can trigger epigenetic changes, which have not been well-studied in IBS. We performed a pilot study investigating genome-wide DNA methylation of IBS patients and healthy controls (HCs) to identify potential epigenetic markers and associated pathways. Additionally, we investigated relationships of epigenetic changes in selected genes with clinical traits.
Methods
Twenty-seven IBS patients (59% women; 10 IBS-Diarrhea, 8 IBS-Constipation, 9 IBS-Mixed) and 23 age and sex matched HCs were examined. DNA methylation from peripheral blood mononuclear cells (PBMCs) was measured using HM450 BeadChip and representative methylation differences were confirmed by bisulphite sequencing. Gene expression was measured using qPCR. Gastrointestinal (GI) and non-GI symptoms were measured using validated questionnaires. Associations were tested using non-parametric methods.
Results
Genome-wide DNA methylation profiling of IBS patients compared to HCs identified 133 differentially methylated positions (DMPs) (mean-difference ≥10%; p<0.05). These genes were associated with gene-ontology terms including glutathione metabolism related to oxidative stress and neuropeptide hormone activity. Validation by sequencing confirmed differential methylation of SCO-Spondin (SSPO), glutathione-S-transferases mu 5 (GSTM5), and tubulin polymerization promoting protein (TPPP) genes. Methylation of two promoter CpGs in GSTM5 was associated with epigenetic silencing. Epigenetic changes in SSPO gene were positively correlated with HAD depression scores in IBS patients (r>0.4 and FDR<0.05).
Conclusion
This study is the first to comprehensively explore the methylome of IBS patients. We identified DMPs in novel candidate genes which could provide new insights into disease mechanisms, however, these preliminary findings warrant confirmation in larger, independent studies.
Keywords: Irritable bowel syndrome, DNA methylation, Oxidative stress, Human Methylation 450 array, Bisulphite sequencing
Introduction
Irritable bowel syndrome (IBS) is a functional gastrointestinal (GI) disorder that is characterized by chronic or recurrent abdominal pain associated with diarrhea (IBS-D), constipation (IBS-C) or both (IBS-M).[1] IBS has a pooled worldwide prevalence of 11.2% [2] and is generally more prevalent in women.[3] IBS is thought to be a stress-sensitive disorder in which complex interactions between the host and environment are associated with altered brain-gut interactions. This in turn, can lead to alterations in visceral perception, gastrointestinal (GI) motility, intestinal permeability, hypothalamic-pituitary-adrenal (HPA) axis, neuroimmune and autonomic nervous system function.[4] [5]
Stress and other environmental factors (including adverse childhood experiences [ACEs], dietary factors and metabolites of gut microbiota[6] can trigger epigenetic changes, such as DNA methylation and histone modification, which have been implicated in the pathophysiology of several chronic diseases including cancer, chronic pain, and psychiatric diseases.[7][8][9] Epigenetic modifications can explain the variability observed in quantitative traits despite similarities in genetic background.[10] DNA methylation, in particular, has emerged as a leading candidate biological pathway linking gene-environment interactions to long-term behavioral development, particularly in complex disorders.[11] Epigenetic modifications are key regulators of important cellular events including development and differentiation.[12] Vertebrate genomes are methylated predominantly at the dinucleotide CpG. The globally methylated, CpG-poor genomic landscape is punctuated by CpG islands (CGIs), which are short interspersed DNA sequences (usually 1000 base-pairs) that deviate significantly from the average genomic pattern by having an elevated G+C base composition. CGIs are predominantly non-methylated and typically occur at or near the transcription start site of genes.[13] When a CGI in the promoter region of a gene is methylated, expression of the gene is repressed. Moreover, recent studies show that gene body methylation is positively correlated with gene expression and can be potential therapeutic targets.[14] The quantification of DNA methylation in diseased or environmentally impacted cells could provide useful information for detection and analysis of disease.
Chronic, sustained stressors experienced in childhood or adulthood have an increased prevalence in IBS and are associated with the onset and symptom flares.[15][16][17] In both animal and human studies, certain types of stress can increase pain sensitivity and alter the responsiveness of the HPA axis and autonomic nervous system, the latter having secondary effects on gut permeability, motility and secretion.[5] Based on a growing preclinical and clinical literature, the mechanisms underlying the long-term effects of ACEs are likely to result from epigenetic programming.[18] Also, epigenetic modifications are thought to link early life stress to later susceptibility to disorders, such as anxiety or depression, through interference with the development and functioning of the HPA axis early in life.[19] However, there are no studies on the global changes in DNA methylation patterns in IBS and traits such as bowel habits, ACEs, anxiety and depression in IBS patients compared to healthy controls, which can potentially serve as epigenetic markers.
At cellular and molecular levels, abnormalities in mucosal enteroendocrine system as well as mucosal and systemic immune responses have been postulated to participate in the brain-gut dysfunction.[20] An increased innate immune activity in the intestinal mucosa and in blood is found in subpopulations of patients with IBS.[21] A recent study demonstrated that peripheral blood mononuclear cell (PBMC) supernatants from IBS patients was associated with mechanical hypersensitivity on mouse colonic afferent endings, suggesting a role for immune dysfunction (specifically, increases in TNF-α, IL-1β and IL-6 in PBMC supernatants) on enhanced sensory pathways in IBS patients.[22]
Therefore, the aims of the current pilot study were to: 1) Determine genome-wide DNA methylation differences in peripheral blood mononuclear cells (PBMCs) in IBS patients and healthy controls using methylation arrays; 2) validate methylation levels of differentially methylated positions (DMPs) in selected genes using bisulfite sequencing; 3) correlate methylation levels of DMPs with the gene expression to determine DNA methylation mediated downregulation of gene expression (epigenetic silencing), and 4) identify DNA methylation patterns associated with clinical characteristics including IBS severity, bowel habit subtype, ACEs and HAD anxiety and depression.
Methods
Study Subjects and Recruitment
IBS patients 18–55 years of age who fulfilled Rome III diagnostic criteria[1] and 23 healthy controls recruited from community advertisements were included in this study. A gastroenterologist with expertise in IBS (LC) confirmed the diagnosis. At the screening visit, a history and physical examination and structured psychiatric interview (MINI) were performed.[23] A bowel symptom questionnaire was used to assess the presence and severity of IBS symptoms and duration of disease.[24] Healthy controls were recruited by newspaper or internet advertisement from the community. Controls had no history of IBS, other chronic pain conditions, or psychiatric illness, and were not taking medications that could affect GI function (anxiolytics, narcotics, antidepressants). No exclusion pertaining to medication was applied to IBS patients; however, three patients were taking antidepressant medications. Subjects were excluded if they smoked more than 0.5 packages of cigarettes daily, had 400 mg caffeine daily (equivalent to a 16oz cup of standard-brew coffee), or exercised 1 hour or more per day. Subjects were compensated for participating in the study. Informed consent was obtained from all subjects.
Symptom Measures
At the screening visit, a bowel symptom questionnaire was used to assess the presence and severity of IBS symptoms and duration of disease.[24] It included the Rome III diagnostic questions for IBS, bowel habit subtypes, demographic characteristics, current abdominal pain severity (0–20), and usual IBS severity (“How bad are your symptoms usually?” None [0] to very severe [5]).
Validated questionnaires were administered to patients as well as controls to assess psychological and somatic symptoms. The Hospital Anxiety and Depression Scale (HAD) is a widely used 14-item questionnaire for assessing current symptoms of anxiety and depression.[25] The presence of ACEs before the age of 18 was measured using the Early Trauma Inventory- Self Report Form (ETI-SR).[26] It assesses ACEs in the following domains (number of items): general trauma (11), physical punishment (5), emotional abuse (5), and sexual abuse (6). Each of the 27 items was scored as “Yes”=1 or “No”=0 (total score range 0–27). The ETI-SR has been previously used in IBS and has shown higher total scores in IBS patients compared to controls.[17] ETI-SR emotional abuse score was the strongest predictor of IBS status.[17] The Perceived Stress Scale (PSS) [27] is a validated 10-item questionnaire used to evaluate the association of perceived stress over the past 1-month with disease severity in chronic conditions.
DNA methylation array
PBMCs were isolated from whole blood of study participants collected in anti-coagulant (EDTA) tubes, using Ficoll-Paque method. DNA was extracted using DNeasy Blood & Tissue Kit, Qiagen Inc., USA. For global methylation profiling, we used the Illumina Infinium HumanMethylation450 (HM450) BeadChip (Illumina, San Diego, CA), which interrogates DNA methylation status of > 450,000 CpGs and >99% of all genes. We performed bisulfite conversion on 1 μg of genomic DNA from each sample using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer's instructions. Bisulfite-converted DNA was whole genome amplified (WGA) and enzymatically fragmented prior to hybridization to BeadChip arrays. The oligomer probe designs of HM450 arrays follow the Infinium I and II chemistries, in which locus-specific base extension follows hybridization to a methylation-specific oligomer. The level of DNA methylation at each CpG locus was scored as beta (β) value calculated as (M/(M+U)), ranging from 0 to 1, with 0 indicating no DNA methylation and 1 indicating fully methylated DNA. The data were extracted and preprocessed using minfi package.[28] Data were normalized using subset-quantile within array normalization (SWAN). Of the 485,577 CpG probes on the array, we filtered out probes with high detection p values, cross reactive probes (probes with probes with at least 50 nucleotide homology [29]), probes with a SNP within 10 base pairs of the target CpG and repeat regions [30]) and probes on X and Y chromosomes, leaving 381,487 probes. Bata values were converted to M-values using beta2M function from minfi package. PCA plots were generated to assess batch differences. Since no batch effects were observed (Supplementary figure 1), no further correction was applied. Abundance measures for various cell types including, plasma blasts, CD8+CD28-CD45RA-T cells, naive CD8 T cells, and naive CD4 T cells CD8 T cells, CD4 T cells, natural killer cells, B cells, monocytes and granulocytes was estimated in PBMCs of IBS patients and healthy controls using `The epigenetic clock' software [31] which uses method and R code described by Houseman et al.[28][32] None of the estimated cell proportions were different between the two groups (Supplementary figure 2), therefore no adjustments were made. The top 10% autosomal probes with largest standard deviation were compared between IBS and HCs. The term `hyper-methylation' was used when there was an increased DNA methylation in IBS patients (or bowel habit subtypes) compared to controls and, the term `hypo-methylation' was used when we observed a decreased DNA methylation in IBS patients (or bowel habit subtypes) compared to controls. Differentially methylated regions (DMRs) were investigated using `DMRfind' function from `Charm' package.[28] DNA methylation β-values for the selected probes on IBS and control samples were represented graphically by plotting heatmaps, generated using the R package `heatmap.plus'.[33]
Targeted bisulphite sequencing
Bisulfite treatment was performed on the DNA using Zymo Research EZ Methylation kit (Cat.#D5002 or D5004) and the specific regions within the loci were then PCR amplified and sequenced using MiSeq Desktop Sequencer (Ilumina Inc.). Appropriate primers for targeted sequencing could only be designed for 4 of the 5 genes, SNCAIP, GSTM5, SSPO, and TPPP. Sequence reads from bisulfite-treated targeted bisulfite sequencing amplicon pools were identified using standard Illumina base-calling software and then analyzed using a Zymo Research Inc. proprietary analysis pipeline, which is written in Python. Low quality nucleotides and adapter sequences were trimmed off in a quality control (QC) step. Bismark [34] was used to perform the alignment. Paired-end alignment were used as default, requiring both read 1 and read 2 to be aligned within certain distance; otherwise, both read 1 and read 2 were discarded. Index files were constructed using the bismark_genome_preparation command and the entire reference genome. The `non_directional' parameter was applied while running Bismark. All other parameters were set to default. Nucleotides in primers were trimmed off from amplicons when doing methylation calling. The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T. CpG sites with at least 10 sequencing reads were included in the analysis.
However, very few reads were obtained for primers covering the DMP on SNCAIP gene on MySeq instrument. Therefore, new primers were designed and validation was repeated using pyrosequencing. Bisulfite treatment was performed on the DNA and the specific region within the locus was then PCR amplified and sequenced using PSQ™96HS system (Biotage). Data quality was assessed and % methylation was assessed using Pyrosequencing™Assay Design Software (Biotage). DNA methylation ratios (methylated signal/total; equivalent to methylation betas) for the differentially methylated CpGs between IBS and control samples were visualized graphically by heatmaps. The CpG coordinate positions are based on hg19 assembly and Genome Reference Consortium human (GRCh) 37.
Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and wikiPathways analyses on gene lists were performed using `Enrichr'.[35]
Quantitative PCR (qPCR)
qPCR for detection of gene expression was performed on RNA extracted from PBMCs. Total RNA was isolated from PBMCs using the column based, Qiagen RNeasy mini Kit (cat 74104), by the UCLA Clinical Microarray Core. Reverse Transcription was carried out on 200ng total RNA using Bio-Rad SuperScript First-Strand Synthesis System. Real time PCR was carried out using IQ SYBR Green supermix (170–8882, BioRad). Expression of genes was normalized to relative quantity of reference genes, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and B-ACTIN. All the experiments were performed in triplicate. ΔΔCT-based fold change calculations and Student's t-test were performed to assess differential expression. The sequences of the primers used are the following. GSTM5 F 5'- TGGATAACCACATGGAGCTG -3'; GSTM5 R 5' CTTTTCAGGGAGTTCCTCCA 3'; SNCAIP F 5'- GTCGGTCAGTCAGTCCCTTC -3'; SNCAIP R 5'- GGGGCTTCCATTATTCTTCC -3'; TPPP F 5'- GTGATCGACGGCAGGAAC -3'; TPPP R 5'- CTCCTGGAACTGCTCAAAGG -3'; SSPO F 5'- CTCTTCCTGCTGCTCTCGAC -3'; SSPO R 5'- AACTGAGGGGACAGTTGCAC -3'. B-ACTIN-F: 5'-CCCAGCACAATGAAGATCAA-3' B-ACTIN-R: 5'-ACATCTGCTGGAAGGTGGAC-3' GAPDH-F: 5'- ATGTTCGTCATGGGTGTGAA-3' GAPDH-R: 5'-GGTGCTAAGCAGTTGGTGGT-3'.
Statistical analysis
All the analyses were performed using R/Bioconductor packages unless specified otherwise. T-tests were performed to assess significance of differences in clinical characteristics such as age, overall IBS symptom severity, ETI-SR total and emotional abuse scores, HAD anxiety and depression symptom scores, and PSS scores between IBS and controls independently for each experiment as well as the entire cohort. Fisher's exact p value was computed to assess differences in distribution of sex between the groups. A p value <0.05 was considered significant in all the analyses.
Wilcoxon rank-sum tests were used to compare methylation array data between IBS and healthy controls. Magnitude of DNA methylation changes was assessed using methylation beta values. Correction for multiple comparisons was performed using FDR (Benjamini-Hochberg) approach. A corrected p value, denoted as, `q' ≤ 0.05 was considered significant in the initial analysis, however, genes with uncorrected p<0.05 were considered for further analyses. Fisher's exact test was used on counts obtained from bisulphite sequencing data, to identify DMPs. We used a Python module to perform the Fisher's exact test (https://pypi.python.org/pypi/fisher/). Correlations between CpG methylation and clinical variables were tested using Spearman rank correlation. Correlation analyses with HAD depression, HAD anxiety and ETI-SR were performed on the entire group including patients and controls, where as, the analysis with symptom severity measures was performed only in the IBS patient group.
Results
Clinical characteristics of study participants
Twenty-seven IBS patients and 23 healthy controls participated in the study. Genome-wide DNA methylation studies and subsequent gene expression studies were first performed in 12 IBS patients and 12 controls. The patients and controls were carefully selected for a narrow age range and matched for age and sex to avoid potential confounding effects on DNA methylation. Differential methylation was validated in a larger sample of subjects which was comprised of 25 IBS patients and 21 healthy controls. All but four samples (2 IBS and 2 controls, excluded due to insufficient amount of sample material) that were run on HM450 platform were included in the validation studies. The mean age, sex distribution (50–60% women), and race/ethnicity of the IBS and control groups were similar (Table 1). IBS patients reported significantly higher scores for HAD anxiety and depression symptoms, and PSS (all p's<0.05). Only one (<4%) IBS patient had a current psychiatric disorder (i.e., depression). A past history of a psychiatric condition was found in three IBS patients (11%; 1 depression, 1 substance abuse, 1 substance abuse + manic episode) and three controls (13%; 2 depression, 1 substance abuse). The clinical characteristics of the smaller group (12 IBS and 12 controls) were similar to the overall group of subjects.
Table 1.
Clinical Characteristics of IBS Patients and Healthy Controls
| Overall samples | HM450 methylation array | Targeted bisulphite sequencing | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IBS (N=27) | HC (N=23) | P | IBS (N=12) | HC (N=12) | P | IBS (N=25) | HC (N=21) | P | |
| Age [Mean (SD)] | 40.22 (9.12) | 36.48 (8.63) | 0.14 | 39.83 (3.43) | 39.83 (3.79) | 1 | 40.20 (9.49) | 36.52 (9.00) | 0.18 |
| Female % | 59.26 | 60.86 | 1 | 58.33 | 58.33 | 1 | 60.00 | 57.14 | 1 |
| Bowel habit [n (%)] IBS-C | 10 (37.03) | - | - | 3 (25) | - | - | 9 (36) | - | - |
| IBS-D | 8 (29.63) | - | - | 3 (25) | - | - | 8 (32) | - | - |
| IBS-M | 9 (33.33) | - | - | 6 (50) | - | - | 8 (32) | - | - |
| IBS usual severity (1–5) | 3.19 (0.49) | - | - | 3.18 (0.60) | - | - | 3.25 (0.44) | - | - |
| ETI-SR total score [Mean (SD)] | 6.78 (6.7) | 4.22 (3.98) | 0.1 | 8.33 (7.79) | 2.5 (1.57) | 0.03 | 6.72 (6.97) | 4.52 (4.11) | 0.19 |
| ETI-SR emotional abuse score [Mean (SD)] | 1.74 (1.93) | 1(1.76) | 0.16 | 2.08 (1.97) | 0.25 (0.45) | 0.01 | 1.6 (1.94) | 1.09 (1.81) | 0.37 |
| HAD-Anxiety [Mean (SD)] | 5.96 (5.63) | 2.52 (2.37) | 0.01 | 5.00(3.74) | 2.33 (1.87) | 0.04 | 6.04 (5.85) | 2.48 (2.46) | 0.01 |
| HAD-Depression [Mean (SD)] | 2.59 (3.39) | 0.74 (1.01) | 0.01 | 2.08 (1.97) | 0.75 (1.29) | 0.06 | 2.68 (3.48) | 0.71 (1.01) | 0.01 |
| PSS [Mean (SD)] | 12 (8.73) | 8.65 (7.29) | 0.15 | 10.33 (6.84) | 10.17 (7.84) | 0.96 | 12.40 (8.84) | 8.19 (7.19) | 0.08 |
| Psychiatric history -Current* [n (%)] | 1 (3.7%) | None | - | None | None | - | 1 (4%) | None | - |
| Psychiatric history - Past** [n (%)] | 3 (11.11%) | 3 (13.04%) | - | 2 (16.67%) | 1 (8.33%) | - | 3 (12.00%) | 3 (14.28%) | - |
| Race/ethnicity [n (%)] Caucasian | 7 (25.92) | 11 (47.83) | - | 3 (25.00) | 7 (58.33) | - | 7 (28.00) | 10 (47.62) | - |
| African American | 6 (22.22) | 4 (17.39) | - | 2 (16.67) | 3 (25.00) | - | 5 (20.00) | 4 (19.05) | - |
| Hispanic | 1 (03.70) | 1 (4.35) | - | 0 | 0 | - | 0 | 1 (4.76) | - |
| Asian | 4 (14.81) | 3 (13.04) | - | 2 (16.67) | 0 | - | 5 (20.00) | 3 (14.29) | - |
| Other (including multiracial) | 9 (33.33) | 4 (17.39) | - | 5 (41.67) | 2 (16.67) | - | 8 (32.00) | 3 (14.29) | - |
Abbreviations: IBS, irritable bowel syndrome; IBS-C, IBS with constipation; IBS-D, IBS with diarrhea; IBS-M, IBS with mixed bowel habit pattern; HC, healthy controls; SD, standard deviation; ETI-SR, emotional trauma inventory- self report; HAD, Hospital anxiety and depression scale; PSS, perceived stress score;
Current psychiatric history included 1 depression;
Past psychiatric history included 3 substance dependence or abuse (2 IBS, 1 control), 3 depression (1 IBS, 2 controls), 1 manic episode (1 IBS).
Genome-wide methylation differences associated with IBS
DNA methylation of a total of 485,577 sites was analyzed in PBMCs from 12 IBS patients and 12 healthy controls using the Infinium HumanMethylation450 BeadChip. We examined if any individual sites exhibited differential DNA methylation in IBS patients vs healthy controls. One CpG site in the SNCAIP (Synphilin-1) gene was significantly different after multiple testing corrections (q<0.05). We also identified 133 DMPs between IBS patients and healthy controls (p<0.05 and mean difference ≥ 10%; Supplementary file 1). A 10% differential methylation cutoff was chosen based on previous studies in non-cancer diseases, such as types 1 and 2 diabetes,[36][37][38] that have reported modest differences in magnitude (ranging from 0.13–11%), in contrast to larger DNA methylation differences that are found comparing cancer to normal cells.[39] Of these, 50 (37%) CpG sites were hypo-methylated and 85 (63%) were hyper-methylated. Fifty-nine (43.7%) sites of these 133_belonged to CGIs. With regard to location within the gene, 42 were located in the 5' untranslated region (5' UTR)/ transcription start site (TSS)/promoter region while the rest were in the gene body.
Differentially methylated genes in IBS
Of the133 DMPs, we selected functionally important candidate genes for further study based on statistical significance (q <0.05) and presence of differentially methylated regions (DMRs). In contrast to a differentially methylated site, a DMR refers to a genomic region with multiple adjacent CpG sites that exhibit different methylation statuses among multiple samples. DMRs, one of the most important methylation variants in populations, have been described in various contexts, including imprinting-specific, tissue-specific, reprogramming-specific, cancer-specific and aging-specific functions.[40] Although no DMRs were identified, 14 genes had two or more differentially methylated CpG sites. Two genes, Glutathione-S-transferase mu 5 and 1 (GSTM5 and GSTM1) had the high number of differentially methylated CpG sites (Seven and three sites, respectively). The etiology of IBS has been attributed to dysregulation of the brain-gut axis/HPA axis. Communication between the central nervous system and enteric nervous system implies bidirectional interactions, i.e., the brain influences the function of the enteric nervous system and the gut influences the brain via neural pathways including vagal and sympathetic systems.[41] Therefore, we selected two other genes associated with neuronal/ brain function namely, subcommissural organ (SCO) Spondin (SSPO) and tubulin polymerization promoting protein (TPPP), for validation. SNCAIP was chosen since a CpG site in this gene was significantly differentially methylated between IBS patients and healthy controls after corrections for multiple comparisons.
Validation of differential methylation using targeted bisulphite sequencing
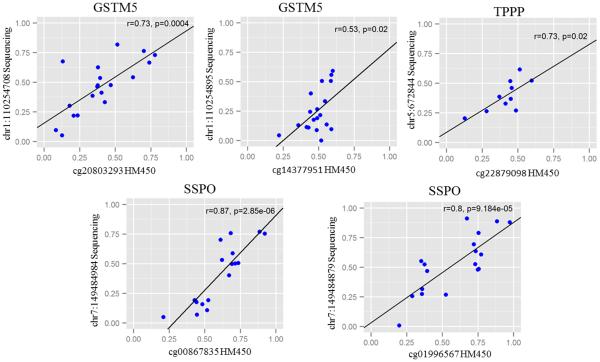
DMPs in SNCAIP, GSMT5, SSPO and TPPP genes were sequenced, along with the adjacent CpGs (200 bases per gene) in 25 IBS patients as well as 21 healthy controls. Figure 1 shows the correlations between the HM450 array and bisulphite sequencing data for selected CpG sites. Eight out of 13 CpG sites in the 4 genes that had data on both platforms showed significant correlations between the two platforms (r>0.5, p>0.05). Sequencing confirmed differential methylation of 3 out of 4 genes. We also identified other differentially methylated sites within these genes that were not included in HM450 array.
Figure 1.
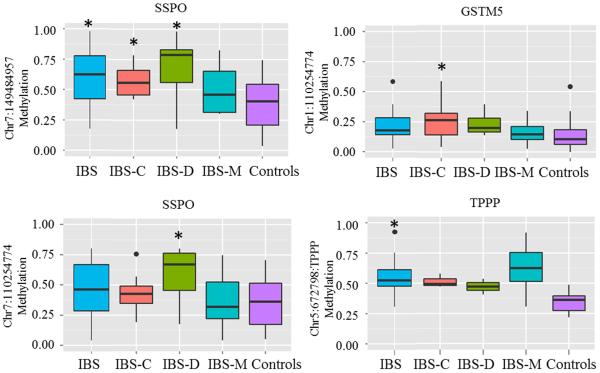
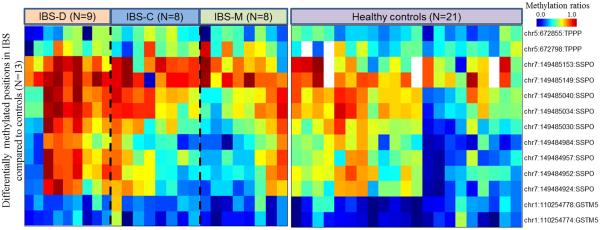
DNA methylation levels were significantly higher in some CpG sites of GSTM5, SSPO and TPPP genes in IBS patients (overall and bowel habit subtypes) compared to healthy controls (Table 2, Figure 2). A schematic of the location of these DMPs in genes is shown in Supplementary figure 3. Multiple CpGs in SSPO gene were hyper-methylated in IBS overall (chr7:149484957, chr7:149485153, chr7:149485034, chr7:149485030, chr7:149485149, chr7:149485040) as well in IBS-D (chr7:149484957, chr7:149484924, chr7:149485034, chr7:149485030, chr7:149484984, chr7:149484952, chr7:149485040) and IBS-C (chr7:149485040, chr7:149484957) patients compared to controls (p<0.05). A CpG site within the TPPP gene was significantly hyper-methylated in IBS overall and another CpG site was hyper-methylated in IBS-C vs controls (p=0.04 and 0.003, respectively). Two CpG sites in GSTM5 were significantly hyper-methylated in IBS-C patients compared to controls (p= 0.024 and 0.047). However, none of these sites were differentially methylated between IBS-M vs control groups. Figure 3 shows heatmap of hierarchically clustered, differentially methylated genes identified by bisulphite sequencing. Differential methylation of the CpG site in SNCAIP gene which was seen on the HM450 array was not validated by pyrosequencing.
Table 2.
Validation of differential methylation in selected genes using targeted bisulphite sequencing
| Gene Name | Chromosome: Start Coordinate | Location | Methylation status in IBS | Mean difference | P | |
|---|---|---|---|---|---|---|
| IBS (n=25) vs Controls (n=21) | ||||||
| SSPO | SCO-spondin | chr7:149485153 | Exon | Hyper-methylated | 0.15 | 0.016 |
| SSPO | SCO-spondin | chr7:149485149 | Intron | Hyper-methylated | 0.1 | 0.047 |
| SSPO | SCO-spondin | chr7:149485040 | Exon | Hyper-methylated | 0.11 | 0.044 |
| SSPO | SCO-spondin | chr7:149485034 | Exon | Hyper-methylated | 0.13 | 0.022 |
| SSPO | SCO-spondin | chr7:149485030 | Intron | Hyper-methylated | 0.13 | 0.041 |
| SSPO | SCO-spondin | chr7:149484957 | Exon | Hyper-methylated | 0.17 | 0.009 |
| TPPP | Tubulin polymerization promoting protein | chr5:672798 | Intron | Hyper-methylated | 0.11 | 0.040 |
| IBS-D (n=9) vs Controls (n=21) | ||||||
| SSPO | SCO-spondin | chr7:149485040 | Exon | Hyper-methylated | 0.17 | 0.030 |
| SSPO | SCO-spondin | chr7:149485034 | Exon | Hyper-methylated | 0.21 | 0.010 |
| SSPO | SCO-spondin | chr7:149485030 | Exon | Hyper-methylated | 0.21 | 0.017 |
| SSPO | SCO-spondin | chr7:149484984 | Exon | Hyper-methylated | 0.21 | 0.021 |
| SSPO | SCO-spondin | chr7:149484957 | Exon | Hyper-methylated | 0.27 | 0.006 |
| SSPO | SCO-spondin | chr7:149484952 | Exon | Hyper-methylated | 0.21 | 0.033 |
| SSPO | SCO-spondin | chr7:149484924 | Intron | Hyper-methylated | 0.22 | 0.025 |
| IBS-C (n=8) vs Controls (n=21) | ||||||
| SSPO | SCO-spondin | chr7:149485040 | Exon | Hyper-methylated | 0.16 | 0.025 |
| SSPO | SCO-spondin | chr7:149484957 | Exon | Hyper-methylated | 0.16 | 0.048 |
| TPPP | Tubulin polymerization promoting protein | chr5:672855 | Intron | Hyper-methylated | 0.15 | 0.003 |
Figure 2.
Figure 3.
Correlation of DNA methylation (measured by sequencing) and gene expression
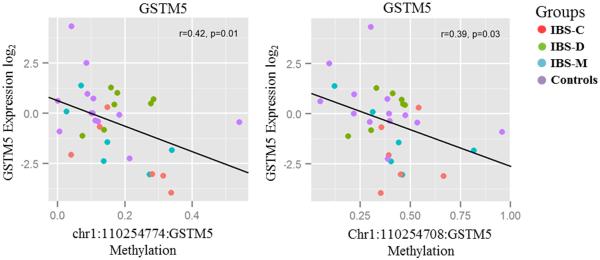
We measured gene expression in IBS patients and healthy controls that were sequenced (N=46) in order to assess epigenetic silencing of the 5 selected differentially methylated genes. There was a significantly negative correlation between DNA methylation in two promoter CpG sites of GSTM5 (chr1:110254708, corresponding to cg20803293 in the HM450 array and chr1:110254774) and gene expression (r=−0.39 & −0.42, p =0.03 & 0.01 respectively; Figure 4). There were no other statistically significant group differences in the expression of the remaining genes.
Figure 4.
Correlation of differentially methylated CpG sites (assessed by sequencing) and clinical traits
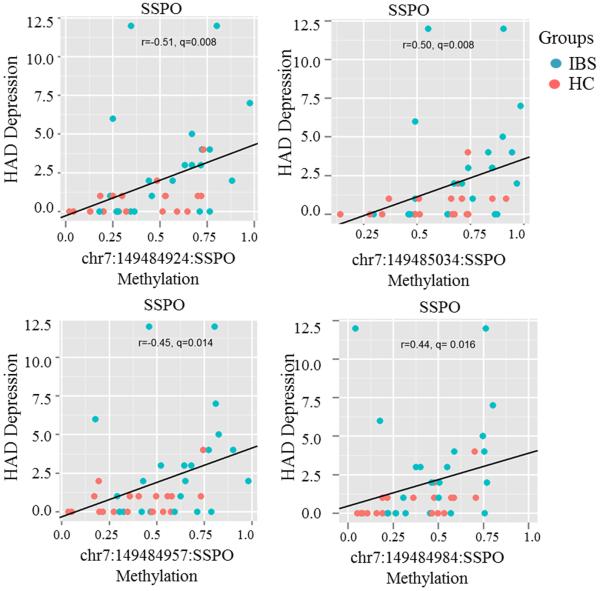
Methylation at 10 CpG sites in SSPO gene (chr7:149484924, chr7:149484952, chr7:149484957, chr7:149484984, chr7:149485016, chr7:149485030, chr7:149485034, chr7:149485040, chr7:149485071, chr7:149484873) positively correlated with HAD depression symptom scores after correcting for multiple comparisons (all r's >0.4; q's<0.05). Additionally, one CpG site in the SSPO gene (chr7:149485071) positively correlated with PSS scores (r=0.4; p= 0.009; q=0.19). Figure 5 shows correlation plots of methylation in 4 CpGs in SSPO gene with HAD depression scores. We did not find a correlation between methylation levels of CpG sites with IBS, ETI-SR scores, or HAD anxiety score.
Figure 5.
Identification of GO terms and functional pathways associated with differentially methylated genes in IBS
Table 3 shows the top five GO terms and functional pathways associated with the 133 differentially methylated genes in IBS. Glutathione transferase activity was the most significant GO term. Neuropeptide hormone activity was another functional annotation associated with the gene list. The gene associated with this GO term, pituitary adenylate cyclase-activating polypeptide (ADCYAP1), also known as PACAP, is implicated in stress-related disorders.[42] A CpG site in the body of this gene was significantly hypo-methylated in IBS patients compared to controls (mean difference=10.6%, p=0.04). Interestingly, when IBS patients were grouped according to their bowel habit, the IBS-D group had considerably lower methylation at that CpG site in PACAP gene compared to controls (mean difference =14.7%, p=0.0007). The KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways associated with the differentially methylated genes were Parkinson's disease, glutathione metabolism, notch signaling and xenobiotic metabolism.
Table 3.
Top 5 gene ontology (GO) terms and pathways associated with CpG sites differentially methylated between IBS patients and healthy controls.
| GO term | Symbol | P-value | Z-score | Combined Score |
|---|---|---|---|---|
| glutathione transferase activity (GO:0004364) | GSTM5, GSTM1 | 0.003 | −2.94 | 3.41 |
| protein kinase A binding (GO:0051018) | AKAP12; PRKAR1B | 0.006 | −2.70 | 3.13 |
| peptide binding (GO:0042277) | GPR37; GSTM1; HLA-DQB1 | 0.016036 | −2.41 | 2.79 |
| amide binding (GO:0033218) | GPR37; GSTM1; HLA-DQB1 | 0.017278 | −2.40 | 2.79 |
| steroid binding (GO:0005496) | ESRRA; PLA2G1B | 0.026984 | −2.37 | 2.75 |
| KEGG pathway | Symbol | P-value | Z-score | Combined Score |
|---|---|---|---|---|
| HSA05020 Parkinsons Disease | SNCAIP, GPR37 | 0.002301 | −1.83 | 4.95 |
| HSA00480 Glutathione Metabolism | GSTM5, GSTM1 | 0.0105 | −1.79 | 3.48 |
| HSA04330 Notch Signaling Pathway | MAML2, DLL1 | 0.01479 | −1.86 | 3.61 |
| HSA00980 Metabolism of Xenobiotics by Cytochrome P450 | GSTM5, GSTM1 | 0.03053 | −1.63 | 2.45 |
Discussion
To our knowledge, this is the first study to explore the entire methylome of IBS patients compared to healthy controls. Our preliminary findings suggest associations of DNA methylation in novel genes such as SSPO and GSTM5 with IBS. The main findings in the study are: 1) IBS is associated with changes in DNA methylation patterns in PBMCs, 2) These DNA methylation changes are associated with oxidative stress related pathways and neuronal function, and 3) There were positive correlations between methylation of several CpG sites in SSPO gene with HAD depression and PSS scores. If confirmed by larger studies, these previously unidentified epigenetic marks in IBS may serve as potential diagnostic markers and also provide insights into pathogenic mechanisms underlying IBS.
Hyper-methylation of a promoter CpG in GSTM5 was observed in IBS patients compared to healthy controls and was associated with decreased expression, although higher methylation was more prominent in IBS-C and IBS-M patients compared to IBS-D group. GSTM5 was associated with the GO term `glutathione transferase activity'. Glutathione plays an important role in antioxidant defense, nutrient metabolism, regulation of gene expression, DNA and protein synthesis, cell proliferation and apoptosis, and cytokine production and immune response.[43] Glutathione deficiency contributes to oxidative stress, which plays a key role in aging and the pathogenesis of many diseases, including seizure disorder, Alzheimer's disease, Parkinson's disease, and liver disease.[43] It has been shown that L-glutathione levels in cells can be dramatically depleted by excessive oxidative stress, which, in turn contributes to further oxidative stress and disease, resulting in a vicious cycle. Increased oxidative stress has been reported in chronic fatigue syndrome (CFS),[44][45] which often coexists with IBS.[45] Thus, oxidative stress may be a common molecular mechanism underlying stress-sensitive disorders such as CFS and IBS. The associated epigenetic silencing of GSTM5 warrants further investigation into the role of glutathione and oxidative stress pathways in IBS.
Epigenetic changes were found in another neuronal gene, TPPP/p25, a brain-specific protein, which induces tubulin polymerization and microtubule bundling and is enriched in Lewy bodies characteristic of Parkinson's disease.[46] This is particularly interesting in the light of a recent study that showed that patients with IBS had an increased incidence of Parkinson's disease (Hazards Ratio [HR]: 1.67).[47] Also, synphilin-1 protein encoded by SNCAIP gene whose hyper-methylation in PBMCs in IBS patients could not be confirmed by bisulfite sequencing, has been implicated in Parkinson's disease.[48][49]
We found that methylation of SSPO was significantly higher in IBS than controls and correlated with HAD depression symptom and PSS scores. SCO-spondin (SSPO) corresponds to glycoproteins secreted by the subcommissural organ (SCO), an ependymal differentiation of the vertebrate brain located at the entrance to the Sylvian aqueduct. SSPO, a member of the “thrombospondin” super-family, is strongly expressed during mammalian central nervous system development.[50] The most prominent neural system involved the regulation of the SCO secretory activity is the serotonin (5-HT) system.[51] Richter et al reported that 5-HT downregulates SCO-spondin biosynthesis, and suggested that 5-HT may exert its effect on the SCO via the cerebrospinal fluid CSF.[51] Serotonin-related mechanisms have been shown to be important pathophysiologic processes in IBS and associated conditions such as depression.[52] Only a few of our subjects had a history of depression and/or an elevated HAD depression score. Subjects with a past history of depression (1 patient and 2 controls) did not show high mean methylation of SSPO gene (<30%). One IBS patient had current depression as well as an elevated HAD depression score and had a high mean methylation (70%). Thus, although there was a significant correlation of methylation between SSPO and HAD depression symptom scores in the overall group, the association of SSPO in IBS does not appear to be related to a coexistence of depression in this subject sample. However, it is conceivable that its association to IBS is related to serotonergic mediated pathways. The lack of IBS associated changes in SSPO mRNA levels in PBMCs may be due to its functional role in neuronal tissues rather than PBMCs. Given the limitations in obtaining neuronal tissues, PBMCs may be a good surrogate for these studies.
One of the GO terms associated with the list of differentially methylated genes in IBS was “neuropeptide hormone”, defined as “any peptide hormone that acts in the central nervous system”. This finding is consistent with existing literature that symptoms of IBS result from dysregulation of the “brain-gut axis”, [41] which includes the bidirectional communication via autonomic (including the enteric nervous system) and central nervous systems, and the stress response systems. The gene associated with this term, ADCYAP1, encodes neuropeptide hormone PACAP. A CpG site in the gene body of ADCYAP1 gene was significantly hypomethylated in IBS patients compared to controls. PACAP is a peptide homologous to vasoactive intestinal peptide (VIP) as they are both members of a family of regulatory peptides and are the main endogenous ligands of a class of G-protein coupled receptors, known as PACAP/VIP receptors. [53] VIP and PACAP are widely distributed in the body including the GI tract where they regulate secretion and motility.[54] VIP has been reported to be upregulated in the colonic mucosa of IBS-D patients compared to healthy controls.[55] The upregulation of VIP in the colon in IBS-D is not surprising given that VIP stimulates enterochromaffin-like cells, relaxes smooth muscle, and stimulates intestinal secretion.[56] The products of this gene are thought to be key mediators of the neuroendocrine stress response.[57][42] Hypo-methylation of the PACAP gene in IBS-D patients could be associated with increased expression, which is interesting in light of the finding that VIP levels in the serum [58] and colonic mucosa of patients with IBS-D are higher than that in healthy controls.[55] However, further studies are needed.
This study has limitations. We attempted to overcome the limitation of sample size by careful patient selection and by validating differential gene methylation using targeted next generation bisulphite sequencing. HM450 array data did not yield many differences when corrected for multiple tests. We tried to address this by validation of carefully selected genes based on multiple occurrences of DMPs within one gene. We observed that some of the DMPs identified by HM450 array did not overlap with the DMPs identified by sequencing in the same gene. This may be due to the smaller numbers of samples, including IBS bowel habit subtypes, used in the HM450 array study. In order to identify genome-wide DMPs associated with bowel habit subtypes, larger sample sizes are needed. As the peripheral blood transcriptome shares >80% homology with genes expressed in the brain, heart, liver, spleen, colon, kidney, prostate and stomach, [59] it could be possible that the effect size is diluted by cell types. A recent study reported that there is an unexpectedly broad movement of leukocyte subsets to and from the gut at steady state, encompassing all lymphoid and myeloid populations, suggesting that PBMCs can reflect the molecular events in the gut.[60] Nonetheless, further studies on colon biopsies may reveal novel insights into gut related disease mechanism of IBS. Replication in a larger population is necessary to establish the role of the proposed candidates as biomarkers or therapeutic targets for IBS. A small subset of subjects had a history of a psychiatric disorder; however this is unlikely to have significantly affected the results.
Conclusion
This is the first study to comprehensively explore the methylome in IBS patients. This approach in PBMCs could result in a potential diagnostic marker for IBS as well as provide a novel molecular framework associated with the pathogenesis of IBS. Our study suggests an association of novel epigenetic targets involved in oxidative stress and neuronal function with IBS. Differentially methylated gene profiles support that IBS is a complex neurobiologic disorder with overlapping molecular signatures with diseases such as CFS, Parkinson's disease, and depression. Overall, we propose a role for epigenetic changes in IBS pathogenesis, while further studies are needed to establish their mechanistic implications and the potential of epigenetic therapies for IBS patients.
Supplementary Material
Key Messages.
Our study suggests that IBS is associated with DNA methylation changes in genes including those related to neuronal and oxidative stress pathways in peripheral blood mononuclear cells (PBMCs). This is the first study to comprehensively explore the methylome in IBS patients.
IBS is a stress related disorder and stress has been associated with epigenetic changes. Hence, the aim of the study was to explore DNA methylation changes in PBMCs associated with IBS.
Genome-wide methylation was assessed using Illumina HM450 arrays and differential methylation was confirmed using targeted bisulphite sequencing.
Epigenetic modifications in novel targets such as SCO-Spondin (SSPO) and glutathione-S-transferase M5 (GSTM5) were associated with IBS.
Glutathione metabolism was among the key pathways associated with the list of differentially methylated genes.
Acknowledgements
We acknowledge Zymo Research, Irvine, CA, for targeted bisulfite sequencing assays and GenoSeq Core facility at University of California, Los Angeles, for the pyrosequencing services.
Grant support: NIH/NIDDK and ORWH P50 DK64539 (EAM, LC), RO1 DK048351 (EAM), R21 DK104078-01A1 (LC), P30 DK 41301 (PI: Rozengurt) and 1UL1 RR033176 (PI: Dubinett).
Abbreviations
- IBS
(irritable bowel syndrome)
- DNA
(deoxyribo nucleic acid)
- RNA
(ribo nucleic acid)
- PCR
(polymerase chain reaction)
- RT
(reverse transcription)
- QC
(quality control)
- FDR
(false discovery rate)
- GO
(gene ontology)
- GI
(gastrointestinal)
- HPA
(hippocampus-pitutary-adrenal)
- KEGG
(Kyoto Encyclopedia of Genes and Genomes)
- ETI
(emotional trauma inventory)
- HAD
(hospital anxiety depression)
- VSI
(visceral sensitivity index)
- PSS
(perceived stress score)
- UTR
(untranslated region)
- TSS
(transcription start site)
- CFS
(chronic fatigue syndrome)
Footnotes
Disclosures: No conflicts of interest exist.
Author contributions: LC designed the study and concept, obtained funding, acquired the data, interpreted the data, and critically revised the manuscript. SM designed the study and concept, performed the research, statistical analyses and interpretation of data, drafted and critically revised the manuscript. EM obtained funding and critically revised the manuscript. CPol interpreted the data and critically revised the manuscript. CPot and DI critically revised the manuscript.
References
- [1].Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–91. doi: 10.1053/j.gastro.2005.11.061. doi:10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- [2].Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2012;10:712–21. e4. doi: 10.1016/j.cgh.2012.02.029. doi:10.1016/j.cgh.2012.02.029. [DOI] [PubMed] [Google Scholar]
- [3].Adeyemo MA, Spiegel BMR, Chang L. Meta-analysis: do irritable bowel syndrome symptoms vary between men and women? Aliment Pharmacol Ther. 2010;32:738–55. doi: 10.1111/j.1365-2036.2010.04409.x. doi:10.1111/j.1365-2036.2010.04409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mayer EA, Bradesi S, Chang L, Spiegel BMR, Bueller JA, Naliboff BD. Functional GI disorders: from animal models to drug development. Gut. 2008;57:384–404. doi: 10.1136/gut.2006.101675. doi:10.1136/gut.2006.101675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chang L. The role of stress on physiologic responses and clinical symptoms in irritable bowel syndrome. Gastroenterology. 2011;140:761–5. doi: 10.1053/j.gastro.2011.01.032. doi:10.1053/j.gastro.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang T, Owen JL, Lightfoot YL, Kladde MP, Mohamadzadeh M. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med. 2013;19:714–25. doi: 10.1016/j.molmed.2013.08.005. doi:10.1016/j.molmed.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. doi:10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- [8].Denk F, McMahon SB. Chronic pain: emerging evidence for the involvement of epigenetics. Neuron. 2012;73:435–44. doi: 10.1016/j.neuron.2012.01.012. doi:10.1016/j.neuron.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Radley JJ, Kabbaj M, Jacobson L, Heydendael W, Yehuda R, Herman JP. Stress risk factors and stress-related pathology: neuroplasticity, epigenetics and endophenotypes. Stress Amst Neth. 2011;14:481–97. doi: 10.3109/10253890.2011.604751. doi:10.3109/10253890.2011.604751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet TIG. 2004;20:350–8. doi: 10.1016/j.tig.2004.06.009. doi:10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- [11].Roth TL. Epigenetic mechanisms in the development of behavior: Advances, challenges, and future promises of a new field. Dev Psychopathol. 2013;25:1279–91. doi: 10.1017/S0954579413000618. doi:10.1017/S0954579413000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kiefer JC. Epigenetics in development. Dev Dyn Off Publ Am Assoc Anat. 2007;236:1144–56. doi: 10.1002/dvdy.21094. doi:10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- [13].Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. doi:10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–90. doi: 10.1016/j.ccr.2014.07.028. doi:10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mayer EA, Naliboff BD, Chang L, Coutinho SV. V. Stress and irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2001;280:G519–24. doi: 10.1152/ajpgi.2001.280.4.G519. [DOI] [PubMed] [Google Scholar]
- [16].Chitkara DK, van Tilburg MAL, Blois-Martin N, Whitehead WE. Early life risk factors that contribute to irritable bowel syndrome in adults: a systematic review. Am J Gastroenterol. 2008;103:765–74. doi: 10.1111/j.1572-0241.2007.01722.x. quiz 775. doi:10.1111/j.1572-0241.2007.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bradford K, Shih W, Videlock EJ, Presson AP, Naliboff BD, Mayer EA, et al. Association between early adverse life events and irritable bowel syndrome. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2012;10:385–90. e1–3. doi: 10.1016/j.cgh.2011.12.018. doi:10.1016/j.cgh.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. doi:10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- [19].Xiong F, Zhang L. Role of the hypothalamic-pituitary-adrenal axis in developmental programming of health and disease. Front Neuroendocrinol. 2013;34:27–46. doi: 10.1016/j.yfrne.2012.11.002. doi:10.1016/j.yfrne.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barbara G, Cremon C, Carini G, Bellacosa L, Zecchi L, De Giorgio R, et al. The Immune System in Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2011;17:349–59. doi: 10.5056/jnm.2011.17.4.349. doi:10.5056/jnm.2011.17.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ohman L, Simrén M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol. 2010;7:163–73. doi: 10.1038/nrgastro.2010.4. doi:10.1038/nrgastro.2010.4. [DOI] [PubMed] [Google Scholar]
- [22].Hughes PA, Harrington AM, Castro J, Liebregts T, Adam B, Grasby DJ, et al. Sensory neuroimmune interactions differ between irritable bowel syndrome subtypes. Gut. 2013;62:1456–65. doi: 10.1136/gutjnl-2011-301856. doi:10.1136/gutjnl-2011-301856. [DOI] [PubMed] [Google Scholar]
- [23].Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33. quiz 34–57. [PubMed] [Google Scholar]
- [24].Munakata J, Naliboff B, Harraf F, Kodner A, Lembo T, Chang L, et al. Repetitive sigmoid stimulation induces rectal hyperalgesia in patients with irritable bowel syndrome. Gastroenterology. 1997;112:55–63. doi: 10.1016/s0016-5085(97)70219-1. [DOI] [PubMed] [Google Scholar]
- [25].Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–70. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- [26].Bremner JD, Bolus R, Mayer EA. Psychometric properties of the Early Trauma Inventory-Self Report. J Nerv Ment Dis. 2007;195:211–8. doi: 10.1097/01.nmd.0000243824.84651.6c. doi:10.1097/01.nmd.0000243824.84651.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav. 1983;24:385–96. [PubMed] [Google Scholar]
- [28].Aryee MJ, Wu Z, Ladd-Acosta C, Herb B, Feinberg AP, Yegnasubramanian S, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostat Oxf Engl. 2011;12:197–210. doi: 10.1093/biostatistics/kxq055. doi:10.1093/biostatistics/kxq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. doi:10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Byun H-M, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–17. doi: 10.1093/hmg/ddp445. doi:10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. doi:10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. doi:10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].R Development Core Team . R: A language and environment for statistical computing. Vienna, Austria: 2010. [Google Scholar]
- [34].Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma Oxf Engl. 2011;27:1571–2. doi: 10.1093/bioinformatics/btr167. doi:10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. doi:10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–83. doi: 10.1093/hmg/ddr472. doi:10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. doi:10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. doi:10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. doi:10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mayer EA, Tillisch K. The brain-gut axis in abdominal pain syndromes. Annu Rev Med. 2011;62:381–96. doi: 10.1146/annurev-med-012309-103958. doi:10.1146/annurev-med-012309-103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dias BG, Ressler KJ. PACAP and the PAC1 receptor in post-traumatic stress disorder. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2013;38:245–6. doi: 10.1038/npp.2012.147. doi:10.1038/npp.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu G, Fang Y-Z, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–92. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- [44].Sido B, Hack V, Hochlehnert A, Lipps H, Herfarth C, Dröge W. Impairment of intestinal glutathione synthesis in patients with inflammatory bowel disease. Gut. 1998;42:485–92. doi: 10.1136/gut.42.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lakhan SE, Kirchgessner A. Gut inflammation in chronic fatigue syndrome. Nutr Metab. 2010;7:79. doi: 10.1186/1743-7075-7-79. doi:10.1186/1743-7075-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tirián L, Hlavanda E, Oláh J, Horváth I, Orosz F, Szabó B, et al. TPPP/p25 promotes tubulin assemblies and blocks mitotic spindle formation. Proc Natl Acad Sci U S A. 2003;100:13976–81. doi: 10.1073/pnas.2436331100. doi:10.1073/pnas.2436331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lai S-W, Liao K-F, Lin C-L, Sung F-C. Irritable bowel syndrome correlates with increased risk of Parkinson's disease in Taiwan. Eur J Epidemiol. 2014;29:57–62. doi: 10.1007/s10654-014-9878-3. doi:10.1007/s10654-014-9878-3. [DOI] [PubMed] [Google Scholar]
- [48].Engelender S, Kaminsky Z, Guo X, Sharp AH, Amaravi RK, Kleiderlein JJ, et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–4. doi: 10.1038/8820. doi:10.1038/8820. [DOI] [PubMed] [Google Scholar]
- [49].Wakabayashi K, Engelender S, Yoshimoto M, Tsuji S, Ross CA, Takahashi H. Synphilin-1 is present in Lewy bodies in Parkinson's disease. Ann Neurol. 2000;47:521–3. [PubMed] [Google Scholar]
- [50].Gobron S, Creveaux I, Meiniel R, Didier R, Herbet A, Bamdad M, et al. Subcommissural organ/Reissner's fiber complex: characterization of SCO-spondin, a glycoprotein with potent activity on neurite outgrowth. Glia. 2000;32:177–91. doi: 10.1002/1098-1136(200011)32:2<177::aid-glia70>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [51].Richter HG, Tomé MM, Yulis CR, Vío KJ, Jiménez AJ, Pérez-Fígares JM, et al. Transcription of SCO-spondin in the subcommissural organ: evidence for down-regulation mediated by serotonin. Brain Res Mol Brain Res. 2004;129:151–62. doi: 10.1016/j.molbrainres.2004.07.003. doi:10.1016/j.molbrainres.2004.07.003. [DOI] [PubMed] [Google Scholar]
- [52].Crowell MD. Role of serotonin in the pathophysiology of the irritable bowel syndrome. Br J Pharmacol. 2004;141:1285–93. doi: 10.1038/sj.bjp.0705762. doi:10.1038/sj.bjp.0705762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Felley CP, Qian JM, Mantey S, Pradhan T, Jensen RT. Chief cells possess a receptor with high affinity for PACAP and VIP that stimulates pepsinogen release. Am J Physiol. 1992;263:G901–7. doi: 10.1152/ajpgi.1992.263.6.G901. [DOI] [PubMed] [Google Scholar]
- [54].Mahavadi S, Bhattacharya S, Kim J, Fayed S, Al-Shboul O, Grider JR, et al. Caveolae-dependent internalization and homologous desensitization of VIP/PACAP receptor, VPAC2, in gastrointestinal smooth muscle. Peptides. 2013;43:137–45. doi: 10.1016/j.peptides.2013.03.008. doi:10.1016/j.peptides.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Camilleri M, Carlson P, Acosta A, Busciglio I, Nair AA, Gibbons SJ, et al. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol. 2014;306:G1089–98. doi: 10.1152/ajpgi.00068.2014. doi:10.1152/ajpgi.00068.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zeng N, Athmann C, Kang T, Lyu RM, Walsh JH, Ohning GV, et al. PACAP type I receptor activation regulates ECL cells and gastric acid secretion. J Clin Invest. 1999;104:1383–91. doi: 10.1172/JCI7537. doi:10.1172/JCI7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Smith CB, Eiden LE. Is PACAP the major neurotransmitter for stress transduction at the adrenomedullary synapse? J Mol Neurosci MN. 2012;48:403–12. doi: 10.1007/s12031-012-9749-x. doi:10.1007/s12031-012-9749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Han B. Correlation between gastrointestinal hormones and anxiety-depressive states in irritable bowel syndrome. Exp Ther Med. 2013;6:715–20. doi: 10.3892/etm.2013.1211. doi:10.3892/etm.2013.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liew C-C, Ma J, Tang H-C, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–32. doi: 10.1016/j.lab.2005.10.005. doi:10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- [60].Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, Mathis D. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci U S A. 2014;111:6696–701. doi: 10.1073/pnas.1405634111. doi:10.1073/pnas.1405634111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.