Summary
Older concepts of a hard-wired adult brain have been overturned in recent years by in vivo imaging studies revealing synaptic remodeling, now thought to mediate rearrangements in microcircuit connectivity. Using three-color labeling and spectrally resolved two-photon microscopy, we monitor in parallel the daily structural dynamics (assembly or removal) of excitatory and inhibitory postsynaptic sites on the same neurons in mouse visual cortex in vivo. We find that dynamic inhibitory synapses often disappear and reappear again in the same location. The starkest contrast between excitatory and inhibitory synapse dynamics is on dually innervated spines, where inhibitory synapses frequently recur while excitatory synapses are stable. Monocular deprivation, a model of sensory input-dependent plasticity, shortens inhibitory synapse lifetimes and lengthens intervals to recurrence, resulting in a new dynamic state with reduced inhibitory synaptic presence. Reversible structural dynamics indicates a fundamentally new role for inhibitory synaptic remodeling – flexible, input-specific modulation of stable excitatory connections.
Introduction
Historically, inhibitory synapses onto pyramidal cell dendrites were thought to reside mostly on the dendritic shaft, serving to locally modulate excitability within short dendritic segments. According to computational models of cortical circuit function, placement of inhibitory synapses along the dendritic shaft is strategic (Gidon and Segev, 2012), in that rearrangement of a small number of well-placed inhibitory synapses can significantly influence segment- or branch-specific computations (Chklovskii et al., 2004; Poirazi and Mel, 2001). Recently, the ability to directly visualize inhibitory synapses in vivo revealed that approximately 30% reside on dendritic spines, rather than on the dendritic shaft (Chen et al., 2012). Inhibitory synapses on dendritic spines are always adjacent to an excitatory synapse on the same spine, and can directly shunt excitation onto that synapse (Chiu et al., 2013). The prevalence of these dually innervated spines (DiS) with a compartmentalized mode of inhibitory modulation raises questions regarding how structural plasticity of this new class of inhibitory synapse could effectively act to alter local excitatory circuit properties. Yet, it is unknown how rapidly inhibitory synapses on dendritic spines or shafts can be assembled or removed in vivo, and how such changes in inhibitory synapses relate to changes in neighboring excitatory synapses.
Fluorescent proteins fused to postsynaptic scaffolding molecules have recently enabled direct visualization of synaptic remodeling in vivo (Cane et al., 2014; Chen et al., 2012; Kelsch et al., 2008; van Versendaal et al., 2012). However, the difficulty of simultaneously imaging both inhibitory and excitatory synapses in vivo has precluded a side-by-side comparison of their dynamic properties across subcellular compartments. At best, inhibitory synapses have been directly tracked in combination with a cell fill, where spine dynamics were assumed to represent excitatory synapse dynamics (Chen et al., 2012; van Versendaal et al., 2012). Here, we utilize in vivo, triple-color, two photon microscopy for side by side imaging of excitatory and inhibitory synapses on individual cortical pyramidal neurons tracked daily for eight consecutive days. This allowed the resolution of spines lacking PSD-95, spines containing a single excitatory PSD-95 positive synapse, termed singly innervated spines (SiS), and spines innervated by both an excitatory and an inhibitory synapse, termed dually innervated spines (DiS), as unique populations with different dynamic properties. Monitoring the short-term dynamics of excitatory synapses on SiS vs DiS showed that excitatory synapses on DiS were remarkably stable as compared to those on SiS. Further, side-by-side comparison of excitatory and inhibitory synapses residing on the same DiS, revealed a stark contrast between the stability of the excitatory synapse and the unusually dynamic nature of the inhibitory synapse on the same spine.
The percentage of dynamic structures seen with daily interval imaging were surprisingly high in comparison to previous studies imaging at 4-day intervals (Chen et al., 2012), indicating that many dynamic events are quickly reversed and therefore undetected with less frequent imaging. This was true for all inhibitory synapses, but was most pronounced for inhibitory synapses on DiS, which frequently disappeared and then reappeared in the same location. This new, recurrent type of synaptic structural dynamics provides a potential mechanism for reversible gating of specific excitatory connections.
Monocular deprivation (MD), a model of sensory input-dependent plasticity, significantly increased the number of dynamic inhibitory events, compared to normal experience (NE). After MD, dynamic inhibitory synapses are present for fewer consecutive days and inhibitory spine synapses are gone longer before reoccurring. These results show that the observed disinhibition previously seen by our lab and others (Chen et al., 2011; Chen et al., 2012; van Versendaal et al., 2012) in response to adult MD is not just a matter of a one-time inhibitory synaptic loss upon deprivation. Rather, MD results in a transition to a new dynamic state for inhibitory synapses where their net presence is reduced throughout the deprivation period.
Results
Triple-color labeling of pyramidal cells
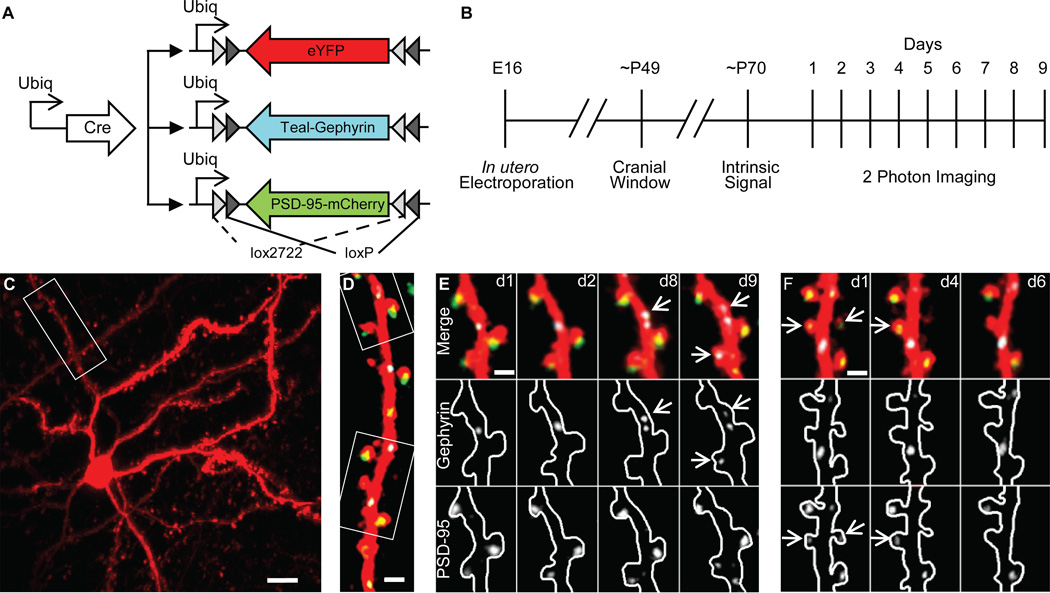
To visualize the full complement of inhibitory and excitatory postsynaptic sites onto individual L2/3 pyramidal cells in mouse V1, we extended our previous strategy for monitoring inhibitory synapse dynamics in vivo (Chen et al., 2012), to include excitatory synapses labeled with a third color. In our previous imaging studies, conducted at 4 or 7 day intervals, it was reasonable to assume that a dendritic spine that lasted for 2 consecutive imaging sessions contained an excitatory synapse. However, this is not necessarily the case when imaging at shorter intervals. Therefore, we co-electroporated three ‘double-floxed' inverted open reading frame (dio) based plasmids (Atasoy et al., 2008; Dhande et al., 2011), expressing either eYFP as a cell fill, Teal-gephyrin as a postsynaptic marker of inhibitory synapses (Essrich et al., 1998), or PSD-95-mCherry as a postsynaptic marker of excitatory synapses, at high molar ratios into E15.5 C57BL/6 mice, together with limiting amounts of a Cre construct (Fig. 1A). This favors high incidence of fluorophore co-expression, while providing the sparse labeling required for single neuron imaging and reconstruction.
Figure 1. Triple color labeling of L2/3 pyramidal cells in vivo.
(A) Plasmid combination for labeling cell fill (eYFP), inhibitory synapses (Teal-gephyrin), and excitatory synapses (PSD-95-mCherry). (B) Experimental time course. (C) Low-magnification maximum z-projection (MZP) of cell fill pseudo-colored red. (D) MZP of dendritic segment from (C) with labeled inhibitory synapses (cyan) and excitatory synapses (pseudo-colored green). (E and F) Examples of dynamic synapses from boxed regions in (D) on indicated days. Upper, middle, and lower panels show 3 channel merge, Teal-gephyrin alone, and PSD-95-mCherry alone, respectively. Arrows denote dynamic synapses. Inhibitory synapses in (E) appear on days 8 and 9. Excitatory synapse in (F) disappears on day 4 and its spine is removed on day 6. Scale Bars in µm: (C) 10; (D) 5; (E and F) 2.
In vivo triple color imaging of eYFP-labeled neuronal morphology, PSD-95-mCherry puncta, and Teal-gephyrin puncta in individual L2/3 neurons within adult V1 was performed through a cranial window with a custom built two-photon microscope. All three fluorophores were simultaneously excited using two excitation wavelengths and their emissions spectrally separated and collected with three PMTs. Bleed-through photons, due to overlapping emission spectra, were reassigned to their appropriate color channel by post-hoc spectral linear unmixing. To monitor synapse dynamics, neurons were imaged 9 times at 24-hour intervals (Fig. 1B), across a 200×200×200 µm volume capturing a large portion of the dendritic arbor at 250 nm/pixel XY and 0.9 µm/frame Z-resolution. Triple-labeled neurons showed clear arbor morphology with distinct resolution of dendritic spines, Teal-gephyrin puncta, and PSD-95-mCherry puncta (Fig. 1C–F).
The linear density per 10 µm of spines (4.62 ± 1.09), inhibitory shaft synapses (1.84 ± 0.36), and inhibitory synapses on DiS (0.79 ± 0.26) (Supplementary Fig. 1A) closely matched our two-color imaging data (Chen et al., 2012), and spine density was unaltered by co-expression of Teal-gephyrin and PSD-95-mCherry synaptic markers as compared to an eYFP cell fill (Supplementary Fig. 1C–E). These data indicate that in our system adding PSD-95-mCherry did not alter either inhibitory or excitatory synaptic numbers. Further, dendritic spine dynamics across imaging sessions were unaltered by addition of PSD-95-mCherry (Supplementary Fig. 1B). Since here PSD-95-mCherry served as an excitatory synaptic marker independent of the spine fill, we could discern that an average 21.6 ± 8.5% (335/1380) of spines lacked PSD-95-mCherry labeling (Fig. 2A). This is consistent with reports from dual color imaging of PSD-95-eGFP against DsRed Express fills of L2/3 pyramidal neurons imaged in vivo (Cane et al., 2014).
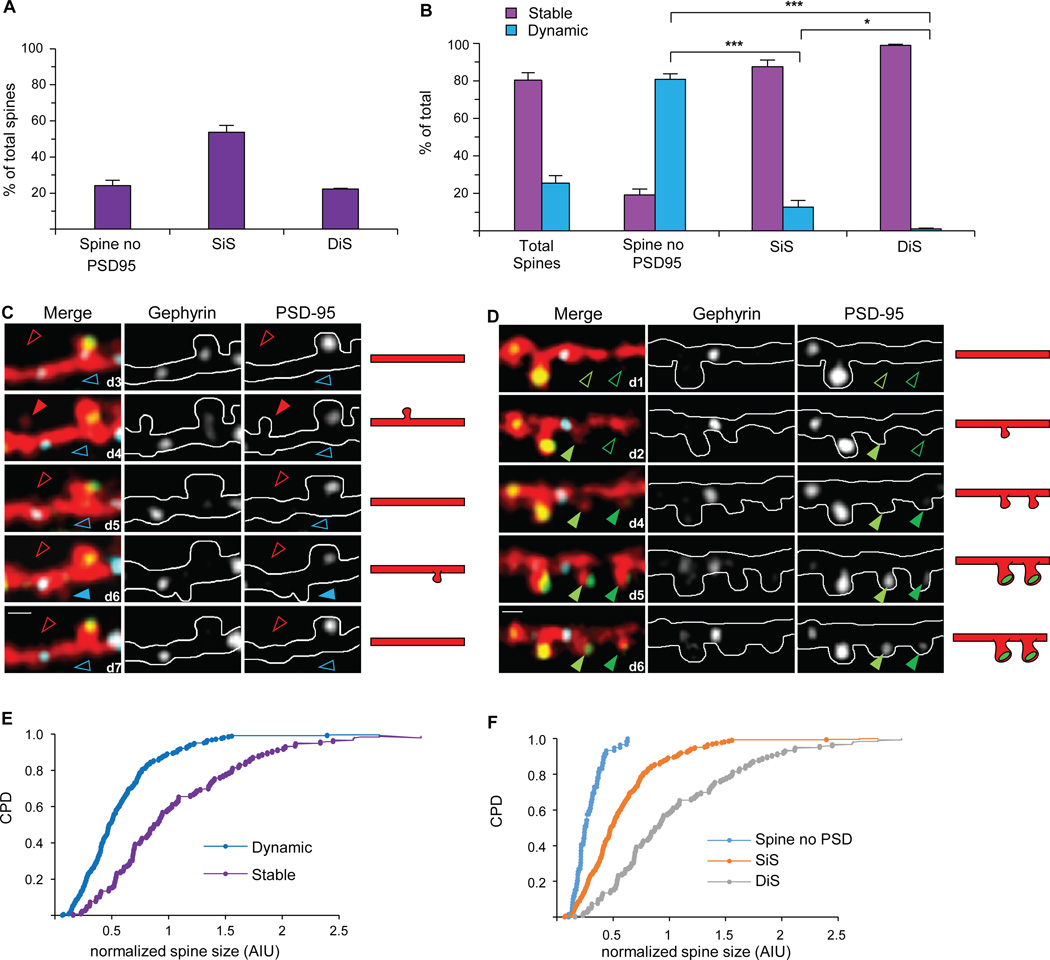
Figure 2. Triple color imaging resolves three spine types with distinct properties.
(A) Proportion of spines without PSD-95, singly innervated spines (SiS) containing only PSD-95, and dually innervated spines (DiS) containing both PSD-95 and gephyrin. (B) Fraction of each subclass which are dynamic or stable. The majority of spine dynamics are due to spines lacking PSD-95. (C and D) Examples of spine dynamics. Left, middle, and right panels show 3 channel merge, Teal-gephyrin alone, and PSD-95-mCherry alone, respectively. Arrows denote dynamic spines, filled when spine is present, empty when spine is absent. (C) Shows the brief appearance and removal of spines without PSD-95 at separate nearby locations. (D) Shows two spines that appear, gain PSD-95, and are stabilized. Scale bars = 2 µm. (E) Stable spines are larger than dynamic spines. Cumulative probability distribution (CPD) comparing between size of dynamic and stable spines, assessed as the ratio of YFP cell fill intensity in the spine head to the average intensity of the dendritic branch (n = 3 cells, 367 stable spines, 54 dynamic spines, p= 4.2×10−13 by KS test). (F) CPD comparing spine size between the 3 spine categories. DiS spines are larger than SiS and spines without PSD-95 (n= 3 cells with 119 DiS, 243 SiS, 59 no PSD-95. p= 6.4×10−13 by KS test).
To assess whether PSD-95 negative spines contain synaptic structures, we performed serial section electron microscopy (SSEM) on a dendritic segment from an in vivo imaged L2/3 pyramidal neuron fixed immediately after the last two-photon imaging session (Supplementary Fig. 2). Interestingly, we found 4 (of 13) spines on this segment that were negative for PSD-95, but contained small synapses visualized by EM. The imaging history of these small filapodia-like spines showed that three of them were recently formed. This suggests they are immature spines and is consistent with literature showing that immature spines don’t contain PSD-95, but rather MAGUKs such as SAP102 or SAP97 (Elias et al., 2008; Sans et al., 2000; Aoki et al., 2001). The fourth spine had PSD-95 for the first seven sessions but lost it 48 hours before the last session and fixation for EM, suggesting it was destined for removal.
Three color imaging resolves three spine types with different dynamic properties
Our triple-color labeling strategy now allows the classification of spines into 3 distinct categories, spines lacking PSD-95, singly innervated spines (SiS) which contain a PSD-95 puncta alone, and DiS that contain an excitatory synapse as well as an inhibitory synapse (Fig. 2A). Inhibitory synapses on spines were exclusively found on spines that also contained an excitatory synapse (DiS). In agreement with (Cane et al., 2014), we found that the stable spine population was mostly comprised of spines with PSD-95-mCherry puncta. Spines that never acquired a PSD-95-mCherry punctum were highly dynamic, with 80.7 ± 2.9% (288/359) dynamic over the entire imaging period (Fig. 2B). Typically, spines that failed to gain, or lost, a PSD-95-mCherry punctum disappeared and were unlikely to return to the same location (Figs. 2C and 1F, respectively). Only 12.6 ± 3.7% (104/848) of SiS were dynamic during this period (Fig. 2B). Many first lost their PSD-95-mCherry punctum and then disappeared (Fig. 1F), or appeared and then later gained PSD-95-mCherry (Fig. 2D). DiS were the most stable category of spines, with 99.0 ± 0.5% (345/348) stable over the entire imaging period (Fig. 2B).
Other studies have shown that larger spines are more likely to be stable (Holtmaat et al., 2005; Trachtenberg et al., 2002). Consistent with these results, we found that dynamic spines are significantly smaller than stable spines (Fig. 2E, p= 4.2 ×10−13 by KS test), and that spines lacking PSD-95 are smaller than spines containing PSD-95 (Fig. 2F, p=6.2×10−17 by KS test). With the addition of the Teal-gephyrin label, we were able to separate spines with PSD-95 into two populations, distinguishing SiS from DiS. This revealed that SiS are significantly smaller than DiS (Fig. 2F, p= 6.4×10−13 by KS test). Thus, the stability of DiS could be due to increased stabilization resulting from either the presence of two synapses or the larger size of DiS, although these two factors are likely related.
The contrast between excitatory and inhibitory synapse dynamics is greatest on DiS
We next examined the dynamics of PSD-95 puncta on spines and asked, ‘how do the dynamics of excitatory synapses differ from the dynamics of inhibitory synapses on spines?’ We found that, while PSD-95 puncta on stable SiS are occasionally dynamic (26.2 ± 4.7% dynamic, 177/744), PSD-95 puncta on DiS are extremely stable (97.8 ± 1.0% stable, 340/348), in stark contrast to inhibitory synapses on DiS, the majority of which (64.8 ± 5.3%, 215/348) are dynamic (Fig. 3A, p<0.0001 by one-way ANOVA with Tukey’s Multiple Comparison Test). Inhibitory synapses on DiS are by far the most dynamic population, significantly more dynamic than either spines or inhibitory synapses on the dendritic shaft (Fig. 3A, p<0.001 by ANOVA with Tukey’s Multiple Comparison Test). While this was qualitatively consistent with previous studies imaging at 4-day intervals (Chen et al., 2012), the percentage of dynamic synapses seen with daily interval imaging were surprisingly high in comparison, with 25.7 ± 3.9% (335/1380) of spines, 19.0 ± 2.3% (89/534) of inhibitory synapses on the dendritic shaft, and 64.2 ± 5.3% (175/304) of inhibitory synapses on DiS dynamic. This was not due to increased photobleaching, since dynamics were not significantly different in the first 2 days of imaging compared to the last 2 days of imaging (Supplementary Fig. 3A). To further rule out technical issues related to the shorter imaging interval, we restricted our analysis to data exclusively from day 0, 4, and 8, as if we had imaged at 4-day intervals, and found that dynamics were comparable to our previous study on a per day basis (Chen et al., 2012), and significantly lower than for the same cells analyzed daily (Fig. 3B, and Supplementary Fig. 3B for further clarification). This difference was particularly robust for inhibitory synapses on DiS, with 28.8 ± 7.7% dynamic per day when imaged and analyzed at daily intervals vs. 11.8 ± 2.9% dynamic per day when analyzed only for 4-day intervals. The increase in dynamic events when imaging at short intervals indicates that many dynamic events are quickly reversed and therefore undetected with less frequent imaging (examples in Fig. 3C and 3D).
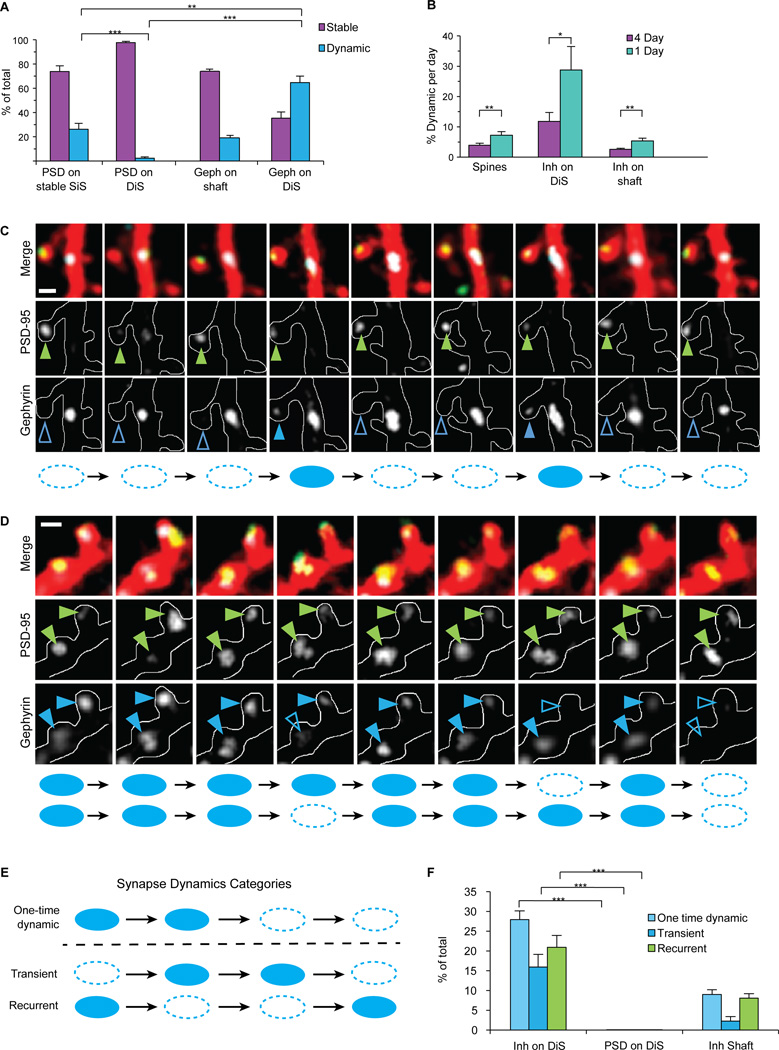
Figure 3. Inhibitory synapses disappear and appear again in the same location.
(A) Fraction of dynamic PSD-95 puncta on SiS and DiS, compared to dynamic gephyrin puncta on shaft or DiS (*p<0.05, **p<0.001 ***p<0.0001 by ANOVA). (B) Comparison of % dynamic structures per day with daily imaging vs 4-day imaging intervals (*p<0.02 **p<0.002 by two tailed student’s T-test) indicates that many events are short-term and go undetected with longer imaging intervals. (C and D) Daily imaging sessions showing examples of recurrent dynamic gephyrin puncta on DiS. Top, second, and third panels show 3 channel merge, PSD-95-mCherry alone, and Teal-gephyrin alone, respectively. Arrows on images denote dynamic synapses, filled when synapse is present, empty when synapse is absent. Scheme below summarizes when inhibitory synapses are present (filled circles) or absent (empty circles). Note that the PSD-95 puncta are stable over all sessions on these spines. Scale bars = 2 µm. (E) Schematic illustrating the classification of one-time dynamic, transient, and recurrent synapses. (F) Percentage of one time dynamic, transient and recurrent synapses (***p<0.0001 by ANOVA). All analyses in this figure were for 1555 spines and 955 inhibitory synapses from 63 dendrites, with statistics based on n= 8 cells All error bars represent SEM. Tukey’s Multiple Comparison Test was used for all ANOVA comparisons.
Inhibitory synapses disappear and appear again in the same location
Given the stunning prevalence of short-lived dynamics for the inhibitory synapse population on DiS, we classified synapses by their dynamic history across the entire 9-day imaging period. We defined synapses that change once (appear or disappear) and don’t change again within our imaging time frame as one-time dynamic, that appear once and disappear as transient, and those that disappear and reappear at least once as recurrent (Fig. 3E). From the inhibitory synapses on dendritic shafts, 9.2 ± 1.3% (44/517) were in the one-time dynamic category, 2.2 ± 0.9% (8/517) transient, and 7.7 ± 1.1% (36/517) recurrent (Fig. 3F). Inhibitory synapses on DiS exhibited significantly more of each dynamic category, with 28.8 ± 2.4% (98/348) one-time dynamic, 16.0 ± 3.5% (50/348) transient, and as many as 19.5 ± 2.5% (67/348) of total synapses exhibiting recurrent dynamics (Fig. 3F). The high dynamics, in particular recurrence, of inhibitory synapses on DiS was not due to fluorescence fluctuations around the scoring threshold, since it was unrelated to synapse size. Dynamic and recurrent inhibitory synapses showed a size distribution similar to stable inhibitory synapses on DiS (Supplementary Fig. 3C). The rapid insertion and removal of inhibitory synapses on DiS is in striking contrast to the stability of excitatory synapses on the same spines, which when broken down into these categories are 0.004 ± 0.004% (1/348) one-time dynamic, 0.008 ± 0.008% (2/348) transient and 0.008 ± 0.008% (4/248) recurrent (Fig. 3F, p<0.0005 by ANOVA with Tukey’s Multiple Comparison Test).
Teal-gephyrin puncta on dually innervated spines represent functional GABAergic synapses
Teal-gephyrin puncta have been previously validated as a faithful marker of inhibitory synaptic presence, with 100% correspondence between the presence and absence of synaptic puncta visualized by gephyrin imaging and by retrospective electron microscopy (EM) reconstruction ((Chen et al., 2012) and Supplementary Fig. S2). The absence of false positives or negatives indicate that when recurrent inhibitory synapses cannot be visualized by two-photon microscopy they are structurally gone. However, a limitation of validation by EM is that it cannot rule out the possibility of functional transmission in the absence of visible gephyrin clusters or a full synaptic structure.
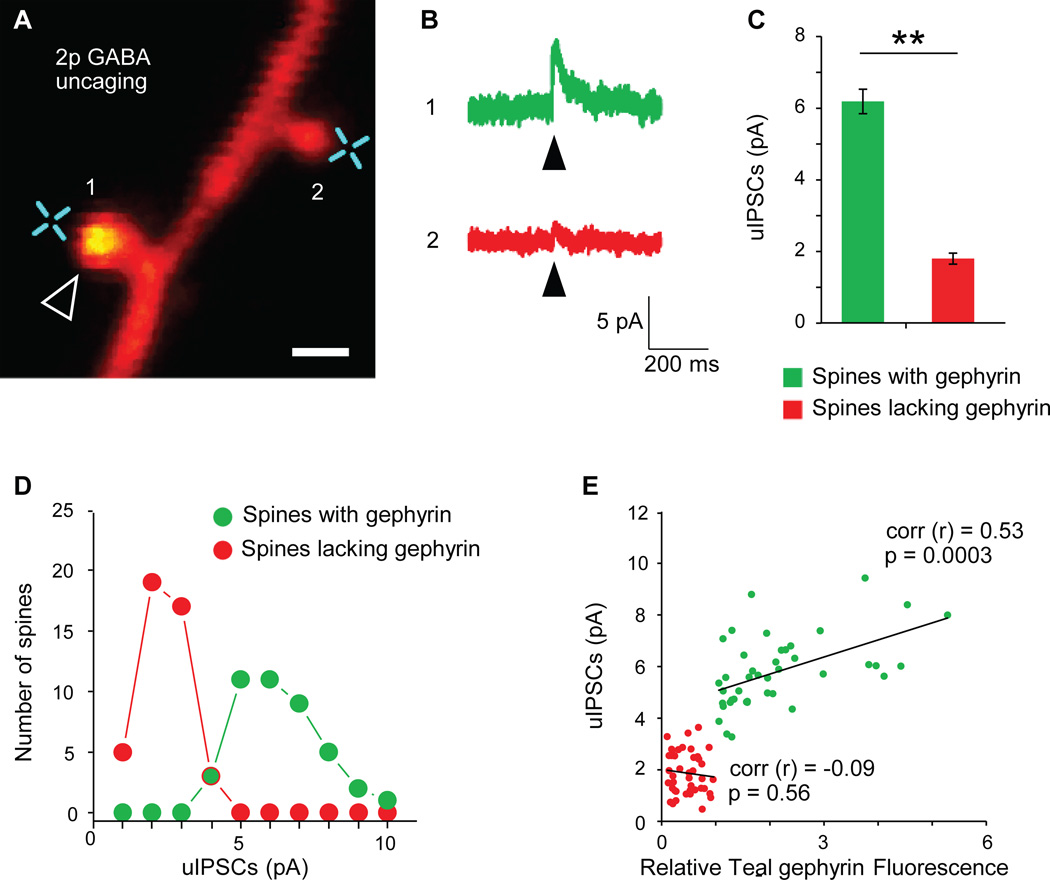
Given the importance of validating synaptic absence during recurrence, we addressed this limitation by performed two-photon GABA uncaging and electrophysiological recordings in organotypic slice cultures of L2/3 pyramidal neurons expressing tdTomato and Teal-gephyrin, to confirm that Teal-gephyrin puncta represent functional GABAergic synapses and that lack of a Teal-gephyrin punctum indicates the absence of a functional GABAergic synapse, 4-carboxymethoxy-5,7-dinitroindolinyl (CDNI)-GABA was uncaged adjacent to spines with or without Teal-gephyrin puncta (Fig. 4A). The average uncaging-evoked inhibitory postsynaptic current (uIPSC) from spines containing Teal-gephyrin puncta was 6.19 ± 0.34 pA, while the average uIPSC evoked from spines lacking Teal-gephyrin puncta was 1.80 ± 0.15 pA (Fig. 4B–C). Spines with and without gephyrin showed little overlap in terms of uIPSC amplitudes (Fig. 4D). The uIPSCs evoked by GABA uncaging adjacent to spines lacking Teal-gephyrin puncta were smaller than 4 pA, possibly due to nonsynaptic GABA receptors, known to be diffusely distributed across the cell membrane (Nusser et al., 1998; Thomas et al., 2005; Tretter et al., 2008; Yeung et al., 2003). The uIPSCs evoked from Teal-gephyrin containing spines were mostly 5 pA or higher, likely representing synaptic currents from clustered GABA receptors. Teal-gephyrin puncta size correlated with uIPSC amplitude, with larger synapses producing stronger uIPSC currents, while Teal background fluorescence on spines without teal-gephyrin puncta showed no correlation with uIPSC amplitude (Fig. 4E). Thus, gephyrin positive and gephyrin negative spines segregate as two distinct populations in terms of GABA evoked uIPSCs. Teal-gephyrin fluorescence imaged in vivo displayed a similarly clear threshold separating between spines with and without Teal-gephyrin puncta (Supplementary Fig. 3C), with the size distributions of teal-gephyrin puncta comparable between the in vivo imaging and organotypic slice data (Supplementary Fig. 4). These data are consistent with the requirement for gephyrin for synaptic GABA(A) receptor clustering (Kneussel et al., 1999) and indicate that Teal-gephyrin puncta on spines correspond to functional inhibitory synapses. Further, these experiments show that when gephyrin puncta are not visibly present, there is no inhibitory synaptic transmission.
Figure 4. Presence or absence of Teal-gephyrin puncta on dually innervated spines reflects presence or absence of functional GABAergic synapses.
(A) Representative image of a dendritic segment from an L2/3 pyramidal neuron in organotypic slice culture expressing tdTomato (red) and Teal-gephyrin (green). Arrowhead marks a gephyrin positive spine. Crosses indicate two-photon GABA uncaging spots. Scale bar= 1µm. (B) Representative uIPSC traces evoked from a spine positive for gephyrin (1, green) and its neighboring gephyrin-negative spine (2, red). Arrowheads indicate onset of GABA uncaging. (C) Summary graph of uIPSC amplitudes from gephyrin positive spines (green bar; 42 spines, n = 18 cells) and spines lacking gephyrin puncta (red bar; 44 spines, n = 18 cells. **p < 0.01 by two-tailed student’s t-test). (D) Segregation of uIPSC amplitudes between spines with gephyrin puncta (green circles) and lacking puncta (red circles). (E) Gephyrin negative spines (red) clearly segregate from gephyrin positive spines (green). Positive correlation between uIPSCs and Teal-gephyrin signal intensity (r = 0.53 by Pearson’s correlation; p < 0.01) and no correlation between uIPSC and background gephyrin intensity of gephyrin negative spines (r=−0.09 by Pearson’s correlation; p=0.56; n = 42 spines, 18 cells). Error bars are S.E.M.
Kinetics of inhibitory synaptic dynamics are altered in experience dependent plasticity
We next investigated whether the rapid inhibitory synaptic dynamics revealed by short interval imaging reflect circuit modifications in response to visual experience. In the adult mouse, MD of at least 7 days produces a functional ocular dominance (OD) shift in binocular V1, a paradigm for sensory experience-dependent plasticity (Frenkel et al., 2006; Sato and Stryker, 2008; Spolidoro et al., 2009). We performed a MD immediately following the first imaging session and monitored synaptic dynamics at daily intervals for a total of 7 days (Fig. 5A). MD animals were compared to animals who had normal vision (normal experience, NE) for the same period. Previous studies have shown that dendritic spine dynamics on L2/3 pyramidal neurons are unaffected by MD (Chen et al., 2012; Hofer et al., 2009). We found this was true, regardless of whether or not the spine contained PSD-95 (SiS: NE 10.03% ±3.7% vs MD 13.87±2.34% P=.37 by two tailed student’s t-test; Spine without PSD-95: NE 79.0% ± 3.7% MD 78.9%±4.55% P=.99 by two tailed student’s t-test) (Fig. 5B). The PSD-95-mCherry puncta on DiS are also unaffected by MD (NE: .80% ±.50% vs MD: 1.8% ±.71% P=.30 by two tailed student’s t-test). MD caused a significant increase in the percent of dynamic inhibitory synapses on shafts (NE 16.9% ± 2.3% (89/534) vs. MD 25.8% ± 1.7% (186/717); p<.01 by two-tailed student’s t-test), but did not further increase dynamics of the inhibitory synapses on DiS (NE 61.3% ± 6.6% (175/304) vs. MD 58.7% ± 6.6% (283/556); p>.05 by two-tailed student’s t-test) (Fig. 5C).
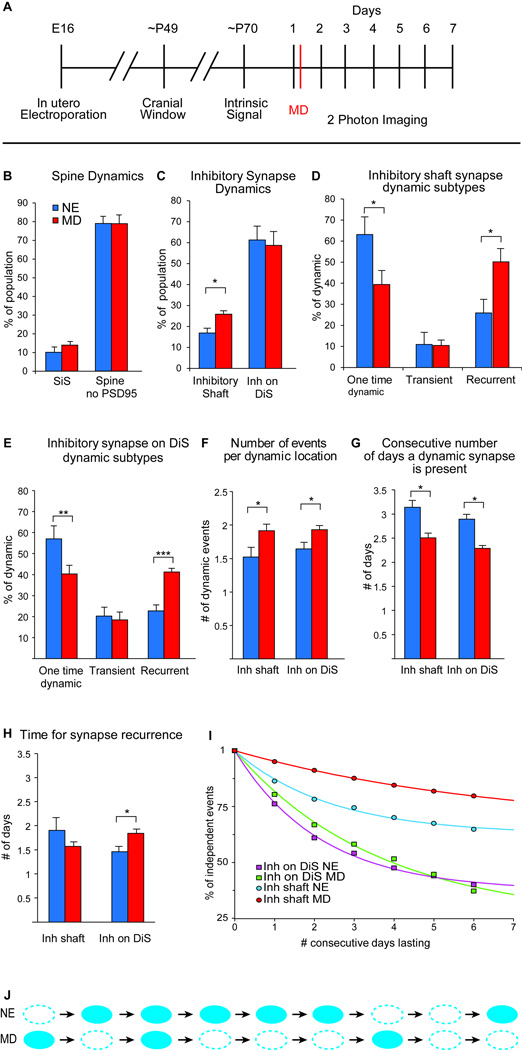
Figure 5. Kinetics of inhibitory synapse dynamics are altered during experience dependent plasticity.
(A) Experimental protocol, MD was performed immediately following the first imaging session. (B) Spine dynamics are unaffected by MD as compared to NE (p>.05). (C) MD increases the % of dynamic inhibitory shaft, but not inhibitory spine synapses. (D) Dynamic inhibitory shaft synapses are more recurrent after MD. (E) Dynamic inhibitory synapses on DiS are more recurrent after MD. (F) MD causes an increase in the number of dynamic events per location for both inhibitory shaft and inhibitory synapses on DiS. (G) MD decreases the number of consecutive days present for both dynamic inhibitory shaft and inhibitory spine synapses. (H) MD increases the number of days between synapse reappearance for inhibitory synapses on DiS. (B–H) Error bars = SEM. *p<0.05, **p<0.01, ***p<0.0005. All p-values from 2 tailed student’s T-test based on n=7 cells NE (304 inh on DiS, 534 inh shaft) and n=7 cells MD (556 inh on DiS, 717 inh on shaft). (I) Survival fraction of observed synaptic events (points), fit with the exponential and constant term, SF = fe–t/τ + s (lines), allowing visualization of how MD affects the survival of each population. (J) Schematic illustrating how inhibitory synaptic dynamics are altered by MD.
Breaking down the dynamic inhibitory population into dynamic categories showed a significant increase in the number of recurrent events after MD for both shaft and spine inhibitory synapses (% recurrent inhibitory synapses on DiS: NE 22.8 ± 2.8% (40/175) vs. MD 41.2 ± 1.8% (115/283); p<0.0002; on shaft: NE 25.9 ± 6.5% (24/89) vs. MD 50.2 ± 6.3% (91/186); p<0.02 by two-tailed student’s t-test) (Fig. 5D–E). This was at the expense of one-time changes, which were significantly lower for both populations (% one-time dynamic inhibitory synapses on DiS: NE 57.0 ± 6.2% (99/175) vs. MD 40.3 ± 4.1% (125/283); p<0.05; on shaft: NE 63.1 ± 8.4% (52/89) vs. MD 39.4 ± 6.7% (74/186); p<0.05 by two-tailed student’s t-test). Thus, inhibitory shaft synapses from a stable pool are recruited during MD to the dynamic population, while inhibitory synapses on DiS, which are already quite dynamic under naïve conditions, are not. For both inhibitory synapse types, MD shifts dynamic events from the onetime dynamic to the recurrent category. This increase in recurrence is reflected in more dynamic events per synaptic location (Fig. 5F) (inhibitory synapses on DiS: NE 1.6 ± 0.1 vs. MD 1.9 ± 0.06; p<0.03 by two-tailed student’s t-test; Inhibitory synapses on Shaft: NE 1.5 ± 0.2 vs. MD 1.9 ± 0.1; p<0.05 by two-tailed student’s t-test). Additionally, the average number of days dynamic synapses are present after MD is decreased in both synapse populations (Fig. 5G) (inhibitory synapses on DiS: NE 2.9 ± 0.2 vs. MD 2.3 ± 0.1; p<.05 by two-tailed student’s t-test; inhibitory synapses on shaft: NE 3.1 ± 0.3 vs. MD 2.5 ± 0.09; p<.05 by two-tailed student’s t-test). Recurrent inhibitory synapses on DiS were also absent for more days before reoccurring after MD (Fig. 5H) (NE 1.5 ± 0.1 vs. MD 1.8 ± 0.09; p<0.03 by two-tailed student’s t-test).
To quantify how the differences in inhibitory synapse dynamics after MD impacted their overall lifetime, we calculated the survival fractions for each population, and fit an exponential curve to these values (Fig. 5I). The exponential fit is a convenient metric for the mean lifetime (τ) of each inhibitory synapse population. MD decreased the lifetime of inhibitory synapses on DiS (NE τ = 3.62 MD; τ = 2.15). The same is true for inhibitory synapses on the shaft (NE τ = 6.69 MD; τ = 2.40) (Fig. 5I). We hypothesized that the increase in the dynamic inhibitory shaft population after MD, combined with the shorter mean lifetimes of both dynamic inhibitory synapse populations, would result in a net disinhibition after MD. When we pool the number of imaging sessions each inhibitory synapse was present, there is a significant decrease in inhibitory synapse presence after MD (NE mean time present 5.95; N=838 inhibitory synapses; MD mean time present 5.75; N=1273 inhibitory synapses; p=0.0058 Wilcoxon Rank-Sum test, one tailed). Overall, these data show that rapid inhibitory synapse dynamics are responsive to sensory manipulations and likely reflect circuit adaptations to the visual environment. Further, disinhibition caused by MD, is not simply a result of one-time inhibitory synaptic loss, but rather a destabilization of inhibitory synapses, resulting in a new dynamic state where they are more dynamic and less likely to be present (Fig. 5J).
Discussion
The loss and gain of spines observed in vivo has long been assumed to represent the remodeling of excitatory connections: specifically, a permanent loss or gain of contact with the presynaptic cell. When a new spine is formed, if it lasts for at least 4 days, it usually gains an excitatory synapse and persists for months (Cane et al., 2014; Holtmaat et al., 2005). In agreement with recent observations (Cane et al., 2014), we find that the majority of spine dynamics are due to a short lived population of spines that never acquire PSD-95 puncta. These spine dynamics likely represent sampling or testing of potential partners rather than permanent changes in circuitry (Fig. 6A, example Fig. 2C). While EM reconstruction suggests that most PSD-95 negative spines contain small synaptic structures, their imaging history combined with the fact that these spines are smaller and more dynamic, suggest they constitute a distinct category of spines that are either immature and/or destined for removal.
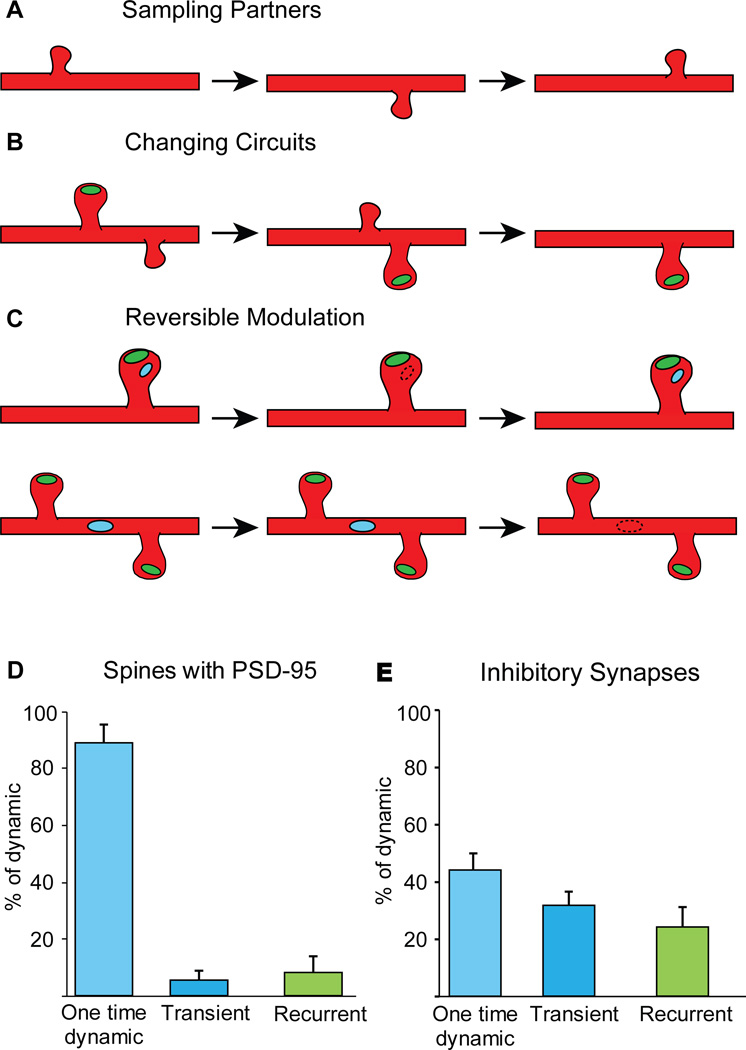
Figure 6. Different logic for excitatory vs. inhibitory synaptic changes.
(A–C) Schematics illustrating the most prevalent categories of dynamic events for spines without PSD-95, spines with PSD-95, and inhibitory synapses on DiS and on the shaft. (A) Dynamics of spines without PSD-95 are rapid and sample different locations, to test different potential partners. (B) Spines that lose PSD-95 are destabilized, while those that gain a PSD-95 are stabilized and persist. In both cases they represent a local rewiring of excitatory circuits. (C) Inhibitory synapses on the shaft or on DiS are removed and reassembled at stable locations, providing a mechanism for reversible inhibitory modulation of excitatory circuits. (D and E) Breakdown of spines carrying an excitatory synapse (D) and of all inhibitory synapses (E) broken down by dynamic category illustrate that recurrence is a feature of inhibitory synapses. (D) Of 71 dynamic spines with PSD-95 from the NE dataset, 64 (87%±6%) were dynamic once, 4 (5%±3%) were transiently present, and 3 (7.9%±5.6%) returned to the same location after the spine was eliminated. (E) Of 169 total inhibitory synapses from the NE dataset,110 (44%±6%) were one time dynamic, 76 (32%±5%) were transiently present, and 72 (24.2%±7.1%) were recurrent.
Changes to excitatory circuit structure, when spines with PSD-95 are removed or added (Fig. 6B, examples Fig. 1F and Fig. 2D), are almost exclusively one time events, with a spine rarely returning to the same location (Fig. 6E). The three-color imaging further enables distinction of spines with PSD-95 that are SiS vs DiS. We find that DiS are the largest and most stable of all spine classes. Since spine size is well known to correlate with stability (Prange and Murphy, 2001) (De Roo et al., 2008; Ehrlich et al., 2007) as is synaptic presence (Cane et al., 2014), the increased stability of DiS could be a function of their large size or the presence of two synapses, although these factors are likely related.
We have previously shown that inhibitory synapses on DiS are more dynamic than shaft inhibitory synapses or spines (Chen et al., 2012). Here we show that they are also more dynamic than excitatory synapses, in particular when compared against the extreme stability of excitatory synapses on the same DiS. Our short interval imaging protocol reveals that a large proportion of dynamic inhibitory synapses, in particular inhibitory synapses on DiS, are transient or recurrent. Even this large fraction may be an underestimate as events currently scored as one-time dynamic could potentially resolve as transient or recurrent if we were able to image longer than 9 daily sessions, or potentially when imaging at even shorter intervals. This repeated insertion and removal of inhibitory synapses at stable sites is unlikely to serve the purpose of rearranging circuitry in terms of testing or exchanging presynaptic partners, as has been the model for excitatory synapse remodeling (Chen and Nedivi, 2010; Nimchinsky et al., 2002) (Fig. 6B and 6E). Inhibitory synaptic remodeling on DiS may serve a fundamentally different purpose. It has been shown, in vivo, that GABA uncaging can reduce Ca2+ transients within spines (Chiu et al., 2013). Inhibitory synapse removal and insertion on DiS could enable flexible, input specific modulation of a stable excitatory synapse (Fig. 6C). Further, the size of individual inhibitory synapses can vary up to 3 fold across imaging sessions (Fig. S5A and B), suggesting that inhibitory spine synapses can potentially modulate their excitatory partner on a DiS beyond a yes/no switch. In any case, given that DiS are usually very large, and spine size is correlated with synaptic strength (Hendry and Jones, 1988; Huang et al., 1999; Kleindienst et al., 2011) an individual inhibitory synapse is unlikely to provide a complete shunt of excitation on a DiS. Considering that DiS often receive direct excitatory input from thalamus (Kubota et al., 2007; van Versendaal et al., 2012), feed forward connections are likely to be particularly influenced by this previously unsuspected mode of inhibitory modulation.
This interpretation would depend on the mechanism that enables inhibitory synapses to return to a stable location. If the presynaptic terminal remains in place and serves as a placeholder for their return, they are likely to maintain a stable partner, supporting a modulatory role for their dynamic assembly and removal. However, if there is a postsynaptic molecular placeholder that can trigger gephyrin assembly-disassembly at a constant site, this would open the possibility for perhaps ‘trading’ the presynaptic partner and enabling circuitry changes.
Rapid inhibitory synapse dynamics are responsive to sensory manipulations. However, rather than abrupt inhibitory synaptic loss with later recovery observed in previous studies with long imaging intervals (Chen et al., 2011; Chen et al., 2012; van Versendaal et al., 2012), we find that MD causes a destabilization of inhibitory synapses, and transition to a new dynamic state where they are more dynamic and less likely to be present. Inhibitory shaft synapses are recruited from the stable population into the dynamic pool, both dynamic inhibitory synapse populations show shorter mean lifetimes, and inhibitory synapses on DiS show longer time to recurrence. Disinhibition through inhibitory synaptic loss after sensory deprivation is consistent with studies showing that whisker stimulation leads to an increase in the number of inhibitory synapses on spines (Knott et al., 2002). Inhibitory synapse dynamics are clearly sensitive to activity levels in the excitatory circuits they modulate.
It is interesting to note that for inhibitory synapses imaged in vivo, as well as in organotypic slice culture there is a clear threshold for scoring Teal-gephyrin positive puncta that corresponds to the emergence of GABA evoked uIPSCs. This suggests that GABA receptor clustering by gephyrin may require a critical aggregation step that specifies emergence of an inhibitory synapse. Since the reversible inhibitory dynamics we see in vivo are sensitive to visual experience, this aggregation step is likely to be regulated and influenced by activity. Cell culture studies show that two days of TTX treatment results in the loss of postsynaptic inhibitory synaptic sites (Kilman et al., 2002), and this loss of inhibition is completely and rapidly reversible (Rutherford et al., 1997). This ability of inhibitory synapses to rapidly remodel in response to changes in activity levels in cell culture (Kilman et al., 2002; Marty et al., 1997; Rutherford et al., 1997), and in vivo (Chen et al., 2012; van Versendaal et al., 2012) is an important homeostatic mechanism for maintaining a stable excitatory/inhibitory balance in cortical circuits (Nelson and Turrigiano, 2008; Turrigiano, 2011). Thus, the rapid addition and removal of inhibitory synapses at stable sites that we show for the first time in vivo may serve to not only to reversibly regulate individual synaptic contacts on DiS and dendritic excitability locally, but to more broadly regulate homeostatic drive onto individual pyramidal neurons (Kilman et al., 2002; Turrigiano and Nelson, 2004).
Experimental Procedures
Generation of Expression Plasmids
The Cre-dependent eYFP and Teal-gephyrin plasmids (pFUdioeYFPW, pFUdioTealGephyrinW) have been described previously in (Chen et al., 2012), and the Cre plasmid in (Subramanian et al., 2013). To generate a Cre-dependent PSD-95-mCherry within a dio cassette, PSD-95 lacking a stop codon was first amplified with an added 5’Nhe1 site from pFuPSD-95-TealW (gift from Jerry Chen), then cloned into pcDNA3 (Invitrogen) between the KpnI and EcoRI sites, to create pcDNA3-PSD-95. mCherry was amplified from pcDNA3.3-mCherry (Addgene) with an added 3’AgeI site, then cloned into pcDNA3-PSD-95 between the EcoRI and XhoI sites to make pcDNA3-PSD-95-mCherry. PSD-95-mCherry was then removed by NheI and AgeI digestion and subcloned into the Cre dependent plasmid pFudio-AscI-AgeI-NheIW, generated by replacing eYFP in pFudio-eYFPW with a linker sequence containing the AgeI restriction site. The vector backbone used for all our expression constructs is the lentivirus transfer vector pFUGW. This is not a strong expression vector and therefore likely expresses at lower levels than more conventional expression constructs. This is an advantage when expressing synaptic markers, and explains the lack of synaptic artifacts frequently seen in other systems, in particular with PSD-95 expression, such as increases in synapse number or stability.
In Utero Electroporation
All animal experiments were approved by the Massachusetts Institute of Technology Committee on Animal Care and meet the National Institute of Health guidelines for the use and care of vertebrate animals. In utero electroporation on E15.5 timed pregnant C57BL/6J mice was performed to label L2/3 cortical pyramidal neurons, as previously described (Tabata and Nakajima, 2001). Animals were co-electroporated with Cre-dependent constructs expressing eYFP, PSD-95-mCherry, or Teal-gephyrin along with a plasmid expressing Cre recombinase at a ratio of 10:10:5:1, respectively (total DNA concentration 2 µg/µL), with 0.1% Fast Green for visualization. A total of 0.75 µl of the plasmid solution was injected into the right lateral ventricle with a 32 gauge Hamilton syringe (Hamilton Company, Reno, NV, USA). Five pulses of 36V (duration 50 ms, frequency 1 Hz) targeting the visual cortex were delivered from a square wave electroporator (ECM830; Harvard Apparatus, Holliston, MA, USA) using 5 mm diameter platinum electrodes (Protech International, Boerne, TX, USA).
Cranial Window Implantation
After in utero electroporation, pups were reared to adulthood (P42-57) and implanted with a 5 mm cranial window over the right hemisphere, as described (Lee et al., 2008).
Sulfamethoxazole (1 mg/ml) and trimethoprim (0.2 mg/ml) were chronically administered in the drinking water to maintain optical clarity of implanted windows.
Optical Intrinsic Signal Imaging
To identify binocular visual cortex, optical imaging of intrinsic signal and data analysis were performed as described previously (Kalatsky and Stryker, 2003). Mice were anesthetized and maintained on 0.25%−0.75% isofluorane and secured in a stereotaxic frame. A horizontal bar (5° in height and 73° in width) drifting upward with a periodicity of 12 s was presented for 60 cycles on a high refresh rate monitor 25 cm in front of the animal. Optical images of visual cortex were acquired continuously under 610 nm illumination with an intrinsic imaging system (LongDaq Imager 3001/C; Optical Imaging Inc., New York, NY, USA) through a 2.5x/0.075 NA objective (Zeiss, Jena, Germany). Images were spatially binned by 4×4 pixels for analysis and cortical intrinsic signal was computed by extracting the Fourier component of light reflectance changes matched to stimulus frequency. Response magnitude was the fractional change in reflectance, and the magnitude maps were thresholded at 30% of peak response amplitude. Binocular visual cortex was delineated upon stimulation of the ipsilateral eye only.
Two-photon imaging
Animals were allowed 2 weeks for recovery after cranial window surgery. When windows cleared, labeled cells in binocular visual cortex were screened for the presence of all 3 fluorescent labels. Cells that exhibited high background labeling with synaptic labels or any ectopic clumps of synaptic proteins were not used for experiments. This selection process ensured that all imaged cells had similar levels of fluorescent labeling. Only one cell was imaged per animal. For eight mice with normal visual experience in vivo two-photon imaging was performed daily for 9 consecutive sessions using a custom-built microscope with custom acquisition software to enable triple color imaging. For each imaging session, mice were anesthesized with isofluorane (0.75%-1.25%) and secured in a stereotaxic frame. Fluorophores were simultaneously excited with a commercial Mai Tai HP Ti: Sapphire laser (Spectra-Physics, Santa Clara, CA, USA) at 915 nm to excite eYFP and Teal, and a Chameleon Compact OPO (Coherent, Santa Clara, CA, USA) at 1085 nm to excite mCherry. The outputs of the two excitation lasers were combined at a polarized beam splitter with orthogonal polarization directions, and delivered to the two-photon microscope through the same beam path. After scanning with a galvonometric XY-scanning mirrors (6215H, Cambridge Technology) and a piezo actuator Z-positioning system (Piezosystem Jena, Jena, Germany), the two laser beams were focused by a 20x/1.0 NA water immersion objective lens (W Plan-Apochromat; Zeiss, Jena, Germany) to the same focal volume location in the specimen, within one pixel size accuracy. These lasers produce ~100 fs unsynchronized pulses at a rate of 80 MHz. The power delivered by each laser to the specimen ranged from approximately 35–50 mW depending on imaging depth. The emission signals from the three fluorophores were collected by the same objective lens, passed through an IR blocking filter (E700SP; Chroma Technology, Bellows Falls, VT, USA), and were separated according to their emission spectra by dichroic mirrors at 520 nm and 560 nm. After passing through three independent bandpass filters (485/70m, 550/100m, and 605/75m) the three emission signals were collected simultaneously onto 3 separate PMTs. Imaging for synapse monitoring was performed at high resolution (250 nm/pixel XY-resolution, 0.9 µm/frame Z-resolution). Two-photon raw scanner data was processed for spectral linear unmixing and converted into a RGB image z-stack using Matlab and ImageJ (National Institutes of Health, Bethesda, MD, USA).
Monocular Deprivation
Monocular deprivation was performed by eyelid suture immediately after the first imaging session. Mice were anesthetized with 2% isoflurane, lid margins were trimmed and triple antibiotic ophthalmic ointment (Bausch & Lomb, Rochester, NY, USA) was applied to the eye. Four to five individual stitches were placed using 6-0 vicryl along the extent of the trimmed lids. Daily imaging sessions were performed for six additional sessions after MD. Suture integrity was inspected directly prior to each imaging session. Animals whose eyelids did not seal fully or had reopened were excluded from further experiments. A total of seven mice with successful MD and no photobleaching were used for analysis.
Spectral Linear Unmixing and Image Processing
Spectral linear unmixing for three colors was performed using a similar approach as previously described for two color unmixing (Chen et al., 2012). Briefly, spectral linear unmixing is based on the fact that the total photon count at each pixel in a given channel is the linear sum of the spectral contribution of each fluorophore weighted by its abundance. For a triple channel detection system, the contribution of three fluorophores can be represented by the following equations.
J1(x,y)=s1.1 x I1 (x,y) + s1.2 x I2 (x,y) + s1.3 x I3 (x,y)
J2(x,y)=s2.1 x I1 (x,y) + s2.2 x I2 (x,y) + s2.3 x I3 (x,y)
J3(x,y)=s3.1 x I1 (x,y) + s3.2 x I2 (x,y) + s3.3 x I3 (x,y)
Where J is the total signal per channel, I is the fluorophore abundance, and S is the contribution of that fluorophore. These equations can be expressed as a matrix:
[J]=[S][I],
whereby the unmixed image [I] can be calculated using the inverse matrix of S:
[I]=[S]−1 [J].
Assuming the detected signal in both channels represents the total spectral contribution for all three fluorophores:
s1.1 + s2.1 + s3.1 = 1
s1.2 + s2.2 + s3.2 = 1
s1.3 + s2.3 + s3.3 = 1
[S] was determined experimentally from two-photon images of HEK cell cultures expressing single fluorophores and excited by both lasers at the same wavelengths used in vivo. Average laser power was adjusted to achieve photon count levels approximating in vivo signal intensity. We have previously shown that S derived from cell culture and in vivo data is interchangeable (Chen et al., 2012). The mean contribution for each fluorophore into each channel (s1.1 – 3.3) was calculated using Matlab (Mathworks, Natick, MA, USA). These values were subsequently used for spectral linear unmixing of triple channel two-photon raw scanner data into a RBG image z-stack using Matlab and ImageJ (National Institutes of Health). For all figures, images are filtered and interpolated for optimal visualization. 3D image stacks are converted into maximum intensity z-projections where noted in figure legends.
Data Analysis
Dendritic spines, PSD-95 containing excitatory synapses, and inhibitory synapses were scored manually with a custom-written 4D point tracking system implemented in Fiji (Schindelin et al., 2012) using a modified version of the ObjectJ plugin (https://sils.fnwi.uva.nl/bcb/objectj/index.html). To avoid individual scoring bias, each cell was independently scored by two investigators. Dendritic spine analysis criteria was defined as previously described (Chen et al., 2012; Holtmaat and Svoboda, 2009). Because of the bright labeling on the soma and axon initial segment, individual contacts in these regions could not be resolved and analysis was restricted to dendrites starting approximately 40 microns from the soma to the most distal tips. Because z-projecting spines lacking PSD-95 could not be visualized, for consistency, all z-projecting spines were excluded from analysis, even those which contained PSD-95. Gephyrin puncta were scored as synapses if they were at least 3×3 pixels, or 8 – 9 clustered pixels (0.56 µm2) in size, with a minimal average signal intensity of at least four times above shot noise background levels, and were present in 2 consecutive z planes. PSD-95 puncta were scored as synapses if they were least 2×2 pixels, or 4–5 clustered pixels (0.27 µm2) in size with a minimal average signal intensity of at least four times above shot noise background levels and were present in 2 consecutive z planes. Previous EM validation confirmed that these criteria represent inhibitory and excitatory synapses, respectively (Cane et al., 2014; Chen et al., 2012). In general, the PSD-95-mCherry fluorophore was more prone to bleaching than YFP or Teal-gephyrin. Cells that lost PSD-95-mCherry were not analyzed further. One-time dynamic changes were defined as any structure that changed one time and never changed again. Changes were scored as transient if a structure was added and later removed in a subsequent imaging session. Changes were scored as recurrent if structures appeared, then disappeared, and then appeared again at the same site; or conversely, ever disappeared and then reappeared more than once in the same location. At least 25 spines and synapses were counted per branch and at least 200 structures were counted per animal. Dendritic arbors were manually traced in Neurolucida (MicroBrightField, Inc., Williston, VT, USA) to quantify the length of scored branches.
In the normal experience dataset, we tracked a total of 1555 spines and 955 inhibitory synapses on 62 dendritic segments from 8 animals. These included 30 basal, and 32 apical dendrites, with a combined branch length of 3.01 mm, 1.63mm basal and 1.42mm apical. Analysis for 1 animal was restricted to sessions 1–7 and another to sessions 1–6 due to photo-bleaching of PSD-95-mCherry in later sessions. The animal with 6 sessions was not included in the comparisons with the MD cells.
In the MD dataset, 1500 spines and 1273 inhibitory synapses were tracked over 7 consecutive imaging sessions. In all, 43 dendritic segments from 7 animals, 22 basal and 21 apical dendrites, with a combined branch length of 2.83mm, 1.54mm basal and 1.29mm apical. All comparisons between MD and Normal Experience datasets were done using only the first 7 imaging sessions of all cells.
For statistical analyses of all the imaging data related to dynamics (Fig. 2A–B; 3A–B, F; 6D–E,) each “n” represents a cell (each from an individual animal). Spine size calculations were performed by placing a 5×5 pixel box in the most prominent z frame around the spine head, subtracting background fluorescent levels and normalizing to the average dendritic shaft intensity, as performed in (Holtmaat et al., 2005). Cumulative probability distributions were calculated in Matlab and significance was determined by a two-sample Kolmogorov-Smirnov test.
Serial Section Electron Microscopy
To demonstrate that Teal-gephyrin and PSD-95-mCherry puncta visualized in vivo correspond to synapses, we performed serial section immuno-EM on in vivo imaged L2/3 pyramidal neuron dendrites labeled with eYFP, Teal-gephyrin, and PSD-95-mCherry. Immediately after two-photon imaging, the brain was fixed with 4% paraformaldehyde, 1% glutaraldehyde and 0.2 % picric acid in 0.1 M PB, cut to 50 µm sections, and stained with an antiserum to GFP (1:2000 dilution, rabbit antiserum, gift from Dr. Tamamaki, Kyoto University, RRID: AB 2314554) followed by a biotin-conjugated secondary (Vector Laboratories Cat# BA-1000, RRID: AB_2313606) and then detected with nickel diaminobenzidine (Ni-DAB). Tissue was then flat-embedded in Epon. Four dendritic segments with identifiable DAB staining were relocated and then further investigated using a combined FIB/SEM (Focused Ion Beam / Scanning Electron Microscope)(Kubota et al., 2011). For EM observation, the Epon block containing the dendritic segments was glued to a stainless-steel sample holder using silver paste to avoid charging the epoxy. The top surface was coated by several 10 nm thick layers of iridium using a sputter coater. The mounted block was transferred in a FIB/SEM, (Hitachi MI4000L, Tokyo, Japan) which contains two beams that intersect at a right angle.The dendritic segments were identified by SEM imaging on the top surface of the block using guidance lines intersecting at right angles that were etched shallowly using the FIB, and by comparing this to light microscopy images obtained earlier. The top region was protected by ion beam induced deposition of platinum (52um*20um area, 1nA ion beam current, 900 sec deposition time). After the coarse milling process by the FIB, the freshly exposed surface of the block was imaged at 1 kV acceleration potential and 1nA beam current using the in-lens detector with 10 µsec dwell time/pixel. High resolution imaging was achieved by low kV imaging and detection of the secondary electrons of the stained tissue surface. Using contrast inversion, TEM-like contrast and comparable imaging information was obtained. Using the ‘Multi-Cut & See’ function, seven adjacent images of 2000 × 2000 store resolution were acquired serially after milling the block surface at 12 nm z-steps. Between 1183 and 1641 serial section images were acquired for each image stack. The milling time was 26 seconds/slice and, the imaging time was about 40 sec/image. In total, the image acquisition took 7 days. The serial image alignment was done using a homemade script for Matlab (kindly provided by Dr. Shawn Mikula, Max Planck Institute, Heidelberg, Germany). The dendritic and synaptic structures were rendered using the 3D reconstruction software, Reconstruct(Fiala, 2005) (available at http://synapses.clm.utexas.edu/tools/index.stm). Synapses were scored according to 3 criteria, the presence of a postsynaptic density, the aggregation of small synaptic vesicles within the presynaptic terminal, and a clear synaptic cleft structure between the pre and postsynaptic membranes at a distance of approximately 20 nm between two parallel membranes. Contacts with DAB staining obscuring the postsynaptic compartment were only categorized as synapses if at least two of these criteria were present, in three consecutive serial ultrathin sections.
Preparation of organotypic slice cultures and DNA transfection
Organotypic slice cultures from mouse visual cortex were prepared from P3-P4 C57BL/6 mice (Stoppini et al., 1991), and transfected 3–4 days before imaging/uncaging experiments using biolistic gene transfer (180 psi)(Woods and Zito, 2008). The same Cre-dependent Teal-gephyrin, and Cre plasmids used for the in vivo studies were coated onto 6–7 mg of gold particles together with a tdTomato plasmid (Kwon et al., 2012) for cell fill (12 µg of tdTomato, 18 µg of Teal-gephyrin, and 16 µg of Cre).
Two-photon slice imaging and GABA uncaging
Imaging and uncaging were performed at 19–22 days in vitro (DIV) on transfected layer 2/3 pyramidal neurons within 40 µm of the slice surface at room temperature in recirculating artificial cerebrospinal fluid (ACSF; in mM: 127 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 25 D-glucose, aerated with 95% O2 /5 % CO2) in the presence of 2 mM CaCl2, 1 mM MgCl2, 1 mM CDNI-GABA, and 0.001 mM TTX. For each neuron, image stacks (512 × 512 pixels; 0.035 µm / pixel) with 1 µm z-steps were collected from one segment of secondary or tertiary apical dendrites 30–50 µm from the soma using a two-photon microscope (Prairie Technologies, Inc) with a pulsed Ti::sapphire laser (Mai Tai DeepSee, Spectra Physics) tuned to 930 nm (2–2.5 mW at the sample). To record uncaging-evoked inhibitory postsynaptic currents (uIPSCs), layer 2/3 pyramidal neurons were patched in voltage-clamp configuration (electrode resistances 5–8 MΩ, Vhold = +10 mV) using cesium-based internal solution (in mM: 135 Cs-methanesulfonate, 10 HEPES, 10 Na2 phosphocreatine, 4 MgCl2, 4 Na2-ATP, 0.4 Na-GTP, 3 Na L-ascorbate, 0.02 Alexa 594, ~300 mOsm, ~pH 7.25) in ACSF. For GABA uncaging, 720 nm light was delivered 0.5 µm away from the target spine with a power of 18~20 mW for 3 ms. uIPSC amplitudes from individual spines were quantified as the average of 8–10 pulses at 0.15 Hz.
Teal-gephyrin expression level in individual spines was measured from background-subtracted and bleed-through-corrected green fluorescence intensities using the integrated pixel intensity of a boxed region of interest (ROI) surrounding the spine head, as described (Woods et al., 2011). In brief, relative Teal-gephyrin enrichment in spines was calculated by normalizing the green fluorescence intensities (as described above) for each individual spine to the mean green fluorescence intensities measured from four ROIs on the dendritic shaft: “spine with gephyrin” (expression level > mean ) vs. “spine lacking gephyrin” (expression level < mean). Statistical analysis was done with ‘n’ representing number of cells
Supplementary Material
Acknowledgments
We thank Dr. Charles Jennings, Dr. Jeff Hoch, and members of the Nedivi lab, especially Dr. Sven Loebrich, for comments on the manuscript. We thank Dr. Nelson Spruston for helpful discussions. We also thank Ms. Sayuri Hatada (NIPS) for assistance with EM histology, Dr. Xin Man (Hitachi) for help with the FIB/SEM imaging, and Ms. Chihiro Shiozu, Ms. Hiroko Kita (NIPS), and Mr. Alsayed Abdelhamid Mohamed for work on the 3D reconstruction of the electron microscopy. This work was sponsored by National Eye Institute grant RO1 EY017656, and partly RO1 EY011894 to EN; NIH P41EB015871-26A1, 4R44EB012415-02, NSF CBET-0939511, the Singapore-MIT Alliance 2, and the Singapore-MIT Alliance for Science and Technology Center to J.W.C. and P.T.C.S; and F31AG044061 to K.V. Partial support for K.V. and K.B. was provided by NIH Pre-Doctoral Training Grant T32GM007287. Y.K. was supported by grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (B) 25290012, on Innovative Areas-Adaptive circuit shift (No. 3603), 26112006 and 15H01456; and the Imaging Science Project of CNSI, National Institutes of Natural Sciences (NINS) IS261004. H.B.K. was supported by R01 MH107460.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: K.V and E.N. conceived the project. K.V. and K.B. developed and performed the majority of the in vivo 2-photon imaging experiments and analysis. J.S created and validated the dio-PSD-95-mCherry plasmid and contributed to the 2-photon imaging experiments and analysis. W.C.O. and H.B.K. performed GABA uncaging experiments. J.W.C and P.S. developed and built the 2-photon microscope with multispectral capabilities and custom acquisition software. Y.K. performed EM reconstruction and analysis. K.V. and E.N. wrote the paper.
References
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane M, Maco B, Knott G, Holtmaat A. The relationship between PSD-95 clustering and spine stability in vivo. J Neurosci. 2014;34:2075–2086. doi: 10.1523/JNEUROSCI.3353-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Lin WC, Cha JW, So PT, Kubota Y, Nedivi E. Structural basis for the role of inhibition in facilitating adult brain plasticity. Nat Neurosci. 2011;14:587–594. doi: 10.1038/nn.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Nedivi E. Neuronal structural remodeling: is it all about access? Curr Opin Neurobiol. 2010;20:557–562. doi: 10.1016/j.conb.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Villa KL, Cha JW, So PT, Kubota Y, Nedivi E. Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron. 2012;74:361–373. doi: 10.1016/j.neuron.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Lur G, Morse TM, Carnevale NT, Ellis-Davies GC, Higley MJ. Compartmentalization of GABAergic inhibition by dendritic spines. Science. 2013;340:759–762. doi: 10.1126/science.1234274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chklovskii DB, Mel BW, Svoboda K. Cortical rewiring and information storage. Nature. 2004;431:782–788. doi: 10.1038/nature03012. [DOI] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Mendez P, Poglia L, Muller D. Activity-dependent PSD formation and stabilization of newly formed spines in hippocampal slice cultures. Cereb Cortex. 2008;18:151–161. doi: 10.1093/cercor/bhm041. [DOI] [PubMed] [Google Scholar]
- Dhande OS, Hua EW, Guh E, Yeh J, Bhatt S, Zhang Y, Ruthazer ES, Feller MB, Crair MC. Development of single retinofugal axon arbors in normal and beta2 knock-out mice. J Neurosci. 2011;31:3384–3399. doi: 10.1523/JNEUROSCI.4899-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Klein M, Rumpel S, Malinow R. PSD-95 is required for activity-driven synapse stabilization. Proc Natl Acad Sci U S A. 2007;104:4176–4181. doi: 10.1073/pnas.0609307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat Neurosci. 1998;1:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- Fiala JC. Reconstruct: a free editor for serial section microscopy. J Microsc. 2006;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Frenkel MY, Sawtell NB, Diogo AC, Yoon B, Neve RL, Bear MF. Instructive effect of visual experience in mouse visual cortex. Neuron. 2006;51:339–349. doi: 10.1016/j.neuron.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Gidon A, Segev I. Principles governing the operation of synaptic inhibition in dendrites. Neuron. 2012;75:330–341. doi: 10.1016/j.neuron.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Jones EG. Activity-dependent regulation of GABA expression in the visual cortex of adult monkeys. Neuron. 1988;1:701–712. doi: 10.1016/0896-6273(88)90169-9. [DOI] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Kalatsky VA, Stryker MP. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–545. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- Kelsch W, Lin CW, Lois C. Sequential development of synapses in dendritic domains during adult neurogenesis. Proc Natl Acad Sci U S A. 2008;105:16803–16808. doi: 10.1073/pnas.0807970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman V, van Rossum MC, Turrigiano GG. Activity deprivation reduces miniature IPSC amplitude by decreasing the number of postsynaptic GABA(A) receptors clustered at neocortical synapses. J Neurosci. 2002;22:1328–1337. doi: 10.1523/JNEUROSCI.22-04-01328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleindienst T, Winnubst J, Roth-Alpermann C, Bonhoeffer T, Lohmann C. Activity-dependent clustering of functional synaptic inputs on developing hippocampal dendrites. Neuron. 2011;72:1012–1024. doi: 10.1016/j.neuron.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Kneussel M, Brandstatter JH, Laube B, Stahl S, Muller U, Betz H. Loss of postsynaptic GABA(A) receptor clustering in gephyrin-deficient mice. J Neurosci. 1999;19:9289–9297. doi: 10.1523/JNEUROSCI.19-21-09289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GW, Quairiaux C, Genoud C, Welker E. Formation of dendritic spines with GABAergic synapses induced by whisker stimulation in adult mice. Neuron. 2002;34:265–273. doi: 10.1016/s0896-6273(02)00663-3. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Hatada S, Kondo S, Karube F, Kawaguchi Y. Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J Neurosci. 2007;27:1139–1150. doi: 10.1523/JNEUROSCI.3846-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Karube F, Nomura M, Gulledge AT, Mochizuki A, Schertel A, Kawaguchi Y. Conserved properties of dendritic trees in four cortical interneuron subtypes. Sci Rep. 2011;1:89. doi: 10.1038/srep00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Kozorovitskiy Y, Oh WJ, Peixoto RT, Akhtar N, Saulnier JL, Gu C, Sabatini BL. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15:1667–1674. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Chen JL, Huang H, Leslie JH, Amitai Y, So PT, Nedivi E. A dynamic zone defines interneuron remodeling in the adult neocortex. Proc Natl Acad Sci U S A. 2008;105:19968–19973. doi: 10.1073/pnas.0810149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty S, Berzaghi Mda P, Berninger B. Neurotrophins and activity-dependent plasticity of cortical interneurons. Trends Neurosci. 1997;20:198–202. doi: 10.1016/s0166-2236(96)01026-0. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron. 2001;29:779–796. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- Prange O, Murphy TH. Modular transport of postsynaptic density-95 clusters and association with stable spine precursors during early development of cortical neurons. J Neurosci. 2001;21:9325–9333. doi: 10.1523/JNEUROSCI.21-23-09325.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford LC, DeWan A, Lauer HM, Turrigiano GG. Brain-derived neurotrophic factor mediates the activity-dependent regulation of inhibition in neocortical cultures. J Neurosci. 1997;17:4527–4535. doi: 10.1523/JNEUROSCI.17-12-04527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Distinctive features of adult ocular dominance plasticity. J Neurosci. 2008;28:10278–10286. doi: 10.1523/JNEUROSCI.2451-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolidoro M, Sale A, Berardi N, Maffei L. Plasticity in the adult brain: lessons from the visual system. Exp Brain Res. 2009;192:335–341. doi: 10.1007/s00221-008-1509-3. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Subramanian J, Dye L, Morozov A. Rap1 signaling prevents L-type calcium channel-dependent neurotransmitter release. J Neurosci. 2013;33:7245–7252. doi: 10.1523/JNEUROSCI.5963-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu Rev Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- van Versendaal D, Rajendran R, Saiepour MH, Klooster J, Smit-Rigter L, Sommeijer JP, De Zeeuw CI, Hofer SB, Heimel JA, Levelt CN. Elimination of inhibitory synapses is a major component of adult ocular dominance plasticity. Neuron. 2012;74:374–383. doi: 10.1016/j.neuron.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Woods G, Zito K. Preparation of gene gun bullets and biolistic transfection of neurons in slice culture. J Vis Exp. 2008 doi: 10.3791/675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods GF, Oh WC, Boudewyn LC, Mikula SK, Zito K. Loss of PSD-95 enrichment is not a prerequisite for spine retraction. J Neurosci. 2011;31:12129–12138. doi: 10.1523/JNEUROSCI.6662-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.