Summary
Finding an efficient and computationally feasible approach to deal with the curse of high-dimensionality is a daunting challenge faced by modern biological science. The problem becomes even more severe when the interactions are the research focus. To improve the performance of statistical analyses, we propose a sparse and low-rank (SLR) screening based on the combination of a low-rank interaction model and the Lasso screening. SLR models the interaction effects using a low-rank matrix to achieve parsimonious parametrization. The low-rank model increases the efficiency of statistical inference and, hence, SLR screening is able to more accurately detect gene-gene interactions than conventional methods. Incorporation of SLR screening into the Screen-and-Clean approach (Wasserman and Roeder, 2009; Wu et al., 2010) is also discussed, which suffers less penalty from Boferroni correction, and is able to assign p-values for the identified variables in high dimensional model. We apply the proposed screening procedure to the Warfarin dosage study and the CoLaus study. The results suggest that the new procedure can identify main and interaction effects that would have been omitted by conventional screening methods.
Keywords: Asymptotic normality, Gene-gene interactions, Low-rank approximation, Over-parametrization, Screen-and-Clean, Sparsity
1. Introduction
Usage of high-throughput data has revolutionized modern biological research and has yielded significant insights for cancer research, genetic diseases, and many genetic disorders. Yet the ever increasing data dimensions lead to the inevitable curse of high-dimensionality, where conventional methods in genetic association analysis lose power and often fail to detect meaningful signals. The problem is further exacerbated when the question of interest focuses on gene-gene interactions (G × G), where the dimensionality of the model explodes even faster. Let Y be the response of interest and g = (g1, · · · , gp)T be the genotypes at the p loci. The most widely used method to detecting G × G is marginal scan (MS) such as PLINK (Purcell et al., 2007) and BOOST (Wan et al., 2010), where the main effect terms were identified by testing for H0,j : cj = 0 in E(Y|gj) = c0 + cjgj, and the interaction terms were identified by testing for H0,jk : cjk = 0 in E(Y|gj, gk) = c0 + cjgj + ckgk + cjk(gjgk). MS has the advantages of using minimal parameters in model fitting and the ease of implementation. However, it has two major drawbacks: (i) MS is too conservative due to multiple testing correction when p is large; and (ii) MS has low detecting power due to the ignorance of joint effects.
To improve on (i), newly developed G × G detection methods adopt a multistage strategy where a screening step is used to reduce the number of variables (e.g., Wu et al., 2010). To improve on (ii), joint screening (based on a multi-locus model including all main and G × G effects) is used to identify candidate genes. Joint-screening can better identify loci that interact with each other but exhibit little marginal effects; it also improves the overall screening performance by reducing the unexplained variance in the model (Wu et al., 2010). A commonly used joint-screening method for G × G detection is to apply Lasso on the model
| (1) |
where ξj is the main effect of the jth locus, and ηjk is the G × G that corresponds to the jth and kth loci. The performance of Lasso depends on the number of parameters in model (1)
| (2) |
and the available sample size, n. Although it has been verified that Lasso performs well when mp is large, caution should be used when mp is ultra-large, e.g., on the order of exp{O(nδ)} for some δ > 0 (Fan and Lv, 2008). In addition, the mp encountered in modern biomedical studies are usually much larger than n even if p is of moderate size. In this situation, statistical inferences can become unstable and inefficient, which impacts the screening performance.
To improve the joint-screening of all main and G × G effects, we utilize the matrix nature of the interaction terms and consider a reduced model. In model (1), (gj gk) is the (j, k)th element of the symmetric matrix J = ggT , and it is natural to treat ηjk as the (j, k)th entry of the symmetric matrix parameter η. Thus, an equivalent expression of model (1) is
| (3) |
where ξ = (ξ1, . . . , ξp)T is the coefficient vector of main effects, and vecp(·) denotes the operator that stacks the lower half (excluding diagonals) of a symmetric matrix columnwisely into a long vector. With the equivalent model expression (3), we can utilize the structure of η to improve the inference procedure. Specifically, we posit the condition
| (4) |
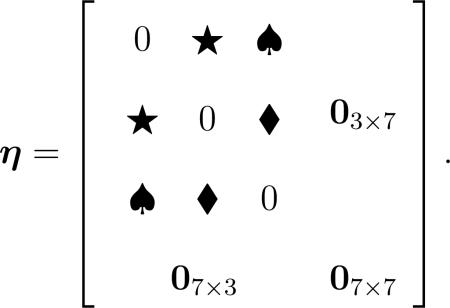
Condition (4) is typically satisfied in modern biomedical research. First, in a G × G scan, it is reasonable to assume that only a few pairs of G × G, say at most q pairs, are associated with the response, where q is much smaller than the number of genes p. This sparsity assumption is also the underlying rationale for applying Lasso for variable selection in conventional approaches. Second, the rank of such a symmetric G × G coefficient matrix η can be at most 2q, i.e., rank(η) ≤ 2q < < p, which motivates the low-rank assumption on η. Displayed below is an example of η with p = 10 and q = 3, and its rank is only three:
 |
(5) |
One key characteristic of our method is the consideration of the sparse and low-rank condition (4), which allows us to express η using far fewer parameters than (). In contrast, Lasso assumes that η is sparse, but without using the matrix low-rank structure, η still contains () parameters. From a statistical viewpoint, parsimonious parametrization can improve the efficiency of model inferences. The aim of this work is thus to use model (3) and condition (4) to propose an efficient joint-screening procedure, referred to as sparse and low-rank (SLR) screening. We will also demonstrate that the performances of existing multistage G × G detection methods can be enhanced by incorporating SLR screening.
Some notation is defined here for reference. Let be random copies of (Y, g), and let . Let Y = (Y1, · · · , Yn)T be an n-vector of observed responses, and let X be the n × mp matrix with the i-th row being . For any square matrix M, M+ is its Moore-Penrose generalized inverse. The operator that stacks a matrix columnwise into a vector is vec(·). The commutation matrix, Kp,k, is defined such that Kp,k vec(M) = vec(MT ) for any p × k matrix M (Magnus and Neudecker, 1979). The matrix P satisfies Pvec(M) = vecp(M) for any p × p symmetric matrix M, and can be chosen such that PKp,p = P. The symbol denotes the Kronecker product. For a vector, ∥ · ∥1 is its 1-norm, and ∥ · ∥ is its Euclidean norm (2-norm). The cardinality of a set is | · |.
This article is organized as follows. The inference procedures for the low-rank model are discussed in Section 2. The SLR screening and its extensions are discussed in Sections 3 and 4. Sections 5 and 6 provide numerical studies to demonstrate the utility of the proposed screening procedures. The article ends with a discussion in Section 7.
2. Inference Procedure for the Low-Rank Model
2.1 Model specification and estimation
To incorporate the low-rank property (4) for G × G into model building, we propose the following rank-r model, where r < < p is a pre-specified positive integer:
| (6) |
Although the above low-rank model expression is straightforward, it is not convenient for numerical implementation. Therefore, by spectral decomposition theorem, we use an equivalent parametrization, η(ϕ), that directly satisfies the constraint rank{η(ϕ)} ≤ r:
| (7) |
Note that the number of parameters required in (7) for interactions η(ϕ) can be much smaller than (). In the case of r = 1, for example, A = (a1, . . . , ap)T and it is equivalent to model the G × G of (gi, gj) as the product (uaiaj), i.e., using a total of p + 1 parameters to model G × G. See Remark 1 for further details. When model (6) is true, standard MLE arguments show that statistical inference based on model (6) must be the most efficient. Even if model (6) is incorrectly specified, we still favor the low-rank model when the sample size is small. We note that the concept of fitting a low-rank model is similar to applying singular value decomposition (SVD) and aiming to find the “best” rank-r approximation of the true η, “best” in the sense of maximizing the likelihood function. It thus provides a good “working” model that compromises between model approximation bias and efficiency of parameters estimation. With limited sample size, it is preferable to more efficiently estimate the approximated low-rank model than to obtain an unstable estimate of the full model.
Let the parameters of interest in rank-r model (6) be
| (8) |
which comprises an intercept, main effects, and interaction terms. Under model (6) and assuming independent and identically distributed errors from a normal distribution N(0, σ2), the log-likelihood function (apart from a constant term) is proportional to
| (9) |
To further stabilize the MLE, a common approach is to add a penalty on θ. We propose to estimate θ by maximizing the penalized log-likelihood function
| (10) |
where λl ≥ 0 is the penalty (the subscript l is for low-rank). Let be the penalized MLE from maximizing (10). The parameter of interest, β(θ), is estimated by
| (11) |
In practical implementation, we use 5-fold cross-validation to select λl. Detailed implementation algorithms for obtaining are described in Web Appendix A.
Remark 1
In (7), ϕ is over-parameterized. Indeed, only pr – r(r – 1)/2 identifiable parameters are required to specify a p × p rank-r symmetric matrix. One can impose the constraints ATA = Ir to make the parametrization identifiable. However, such identifiability constraints, which are merely to obtain a unique solution for A, unnecessarily complicate the numerical implementation. Since the parameters of interest are η(ϕ) but not ϕ itself, we keep the simple usage of over-parameterized ϕ without imposing any constraint on A.
2.2 Asymptotic properties
To derive the asymptotic distribution of in (11), we assume that the parameter space Θ of θ is bounded, open, and connected. Define V 0 = E[V n] with . Let β0 = β(θ0) for some θ0 ∈ Θ be the true parameter value of model (6), and define Δ0 = Δ(θ0), where
| (12) |
with Aj being the jth column of A. The result is summarized below, and the proof is given in the Web Appendix B.
Theorem 2
Assume model (6) and conditions (C1)-(C3) in the Web Appendix B. Assume also λl/n = o(n−1/2). Then, we have as n → ∞, where .
The asymptotic covariance matrix Σ0 can be estimated by
| (13) |
where and are estimators of σ2 and Δ0, and dr = 1 + p + {pr – r(r – 1)/2} is the number of parameters required to specify model (6). Here was added in (13) to stabilize the estimator, , and will not affect its consistency to Σ0. The over-parameterized nature of β(θ) can be observed from Theorem 2 that the asymptotic distribution of depends on θ0 only through the space span(Δ0). Theorem 2 further implies that, as mentioned in Remark 1, adding constraints on ϕ does not affect the asymptotic distribution of because the resulting space, span(Δ0), is unchanged.
Remark 3
We suggest to use dr to guide the determination of the maximum model rank r with given (n, p). That is, n – dr in (13) should be adequate for error variance estimation. One can further use cross-validation to select the rank within the proper range.
3. Sparse and Low-Rank (SLR) Screening for Genetic Main and G × G Effects
We propose a screening procedure called SLR screening based on the low-rank model (6). The main idea of SLR screening, as summarized below, is to filter out insignificant variables by first fitting a low-rank model and then fitting Lasso on the remaining variables.
The goal of Stage-(S1) in SLR screening is to utilize the low-rank property of η to efficiently identify important variables. By Theorem 2, the candidate set of variables, ILR, is formed as
| (14) |
for some αl > 0, where is the jth element of , and is the jth diagonal element of . Note that is the estimate of the intercept term, and is exempted from the screening procedure in (14). The threshold value, αl, controls the power of the low-rank screening. To ensure the selection-consistency of the multistage variable selection procedure, a critical condition is to require = 1, where denotes the true active set of variables. However, this condition holds for any fixed αl > 0, and thus cannot be used to determine αl. (That is, for any j ∈ I0, we have = ∞ under the validity of model (6), and thus j ∈ ILR with probability tending to one.) To simplify the procedure, we suggest to set αl such that
| (15) |
because the main purpose of low-rank screening is to reduce model size so that the subsequent Lasso screening can be more efficiently implemented, and because a linear model cannot identify more than n parameters. This rule shares the same idea as Fan and Lv (2008), which aims to reduce the model size from an ultrahigh scale to a relatively large scale. In practice, a smaller size, such as n/3, for |ILR| may also be considered; the choice can be made based on the size of n and the characteristics of the underlying study. The rule also implicitly assumes that |I0| ≤ n, which is commonly satisfied because |I0| is usually small comparing to n.
The goal of Stage-(S2) in SLR screening is to enforce sparsity. Based on the selected index set, ILR, we refit the model with a 1-norm penalty by minimizing
| (16) |
where XILR and βILR are the selected variables and parameters in ILR, respectively, b0 is the intercept, and λs ≥ 0 is the penalty for the sparsity constraint. The 5-fold cross-validation is applied to determine λs. Let the minimizer of (16) be with being its jth element. The final identified main effects and interaction terms from SLR screening are
| (17) |
Subsequent analysis is conducted on the variables in ISLR.
We close this section by providing two comments for SLR screening. First, the conventional single-staged Lasso screening has to deal with mp parameters. Typically, n < < mp and the fitting may be inefficient. However, SLR screening is a two-staged implementation that first fits a rank-r interaction model and then fits a sparse model. Fitting all main and G × G effects using a rank-r model requires only dr parameters. Because dr < n < < mp when r is small, the low-rank screening (14) is able to efficiently reduce the model size from mp to a relatively small number, so that the subsequent Lasso screening (17) can achieve higher detecting power. See Figure 1 for a detailed illustration. Second, although in theory the proposed SLR screening assumes the validity of the low-rank model (6), we observed that its practical performance is robust to model misspecification. As will be shown in our numerical studies, a small value of r suffices to filter out most irrelevant variables while keeping relevant ones. This is an appealing observation, which increases the applicability of SLR screening.
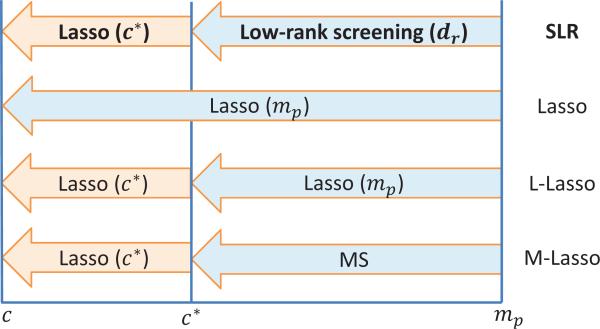
Figure 1.
Illustration of SLR for identifying c candidate variables from mp main and G × G effects. SLR first performs a low-rank screening using dr parameters to reduce the model size from mp to c* (e.g., c* = n), and then fits Lasso (with c* parameters) to select c candidates. Since dr < n < < mp, the numbers of required parameters in the two steps of SLR screening (i.e., dr and c*) can be smaller than n. In contrast, Lasso screening uses mp (> > n) parameters to select c candidates directly. L-Lasso and M-Lasso replace the low-rank screening by Lasso and marginal scan (MS), respectively.
4. Extensions of SLR screening
4.1 Extended Screen-and-Clean (ESC)
We discuss how SLR screening can be incorporated into the well-received Screen-and-Clean approaches (Wasserman and Roeder, 2009), with an aim to address two common issues encountered in real practice. First, in many practices it is of interest to obtain the p-values of the identified variables. Second, the number of parameters required for low-rank model (6), say dr, can still be very large when the original p is in the size of hundreds of thousand (i.e., n < < dr < < mp), which makes variable selection difficult. We tackle these issues in this section.
The Screen-and-Clean of Wasserman and Roeder (2009) is a novel variable selection procedure that is able to assign p-vales in high dimensional model. First, the data D are randomly split into two parts, where D1 is for screening and D2 is for cleaning. In the screening stage, Lasso is used to fit all covariates, from which zero estimates are dropped. In the cleaning stage, the least squares estimate (LSE) is applied on those variables survived through the screening step to identify significant covariates based on the p-values assigned to them (with Bonferroni correction). The reduction of the model size by Lasso guarantees the success of using LSE to identify important covariates. Recently, Screen-and-Clean is modified by Wu et al. (2010) to detect G × G (referred to as SC in the rest of discussion), where the authors propose to fit Lasso to model (1) in the screening stage, and use LSE to assign p-values in the cleaning stage. SC has the advantage of being a joint-screening method. SC also suffers less penalty from the Bonferroni correction, since the number of tests needed to be adjusted is the number of covariates entering the cleaning stage only. Similarly, the data splitting idea can be incorporated into SLR screening to obtain p-values. That is, one can use D1 for SLR screening and use D2 for obtaining LSEs and p-values.
When the number of main effect terms is large (i.e., n < < dr) which makes SLR screening less efficient, we follow the idea of SC to start with a pre-screening step using Lasso on main effects only before applying SLR screening (denote gLasso as the identified set). The underlying rationale is that interactive factors tend to exhibit marginal effects even when the interaction terms are not modeled (Cordell, 2002; Hirschhorn and Daly, 2005) and, hence, a Lasso pre-screening is used to rule out loci with no effects. This pre-screening, however, may miss interactions with weak marginal main effects. A remedy to this situation is to further include genes identified by MS. Specifically, we suggest to apply PLINK (based on D1) to produce a list gMS, where gi ∈ gMS if either gi or (gigj) for some j are identified by PLINK. Then, take g = gLasso to enter SLR screening.
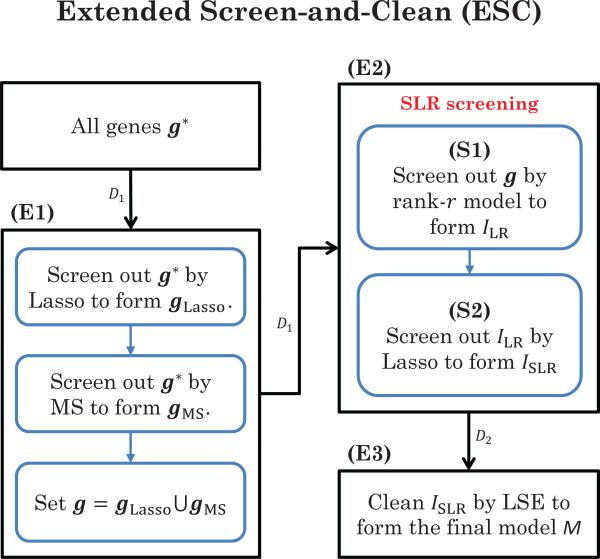
The above two ideas are summarized in the Extended Screen-and-Clean (ESC), which is able to assign p-values for SLR screening. Let g* be the set of whole genes under consideration, and let α be the family-wise error rate.
To determine the 1-norm penalty λm of Lasso in Step-(E1), the StARS (Stability Approach to Regularization Selection) of Liu, Roeder, and Wasserman (2010) is applied, and this criterion is adopted in the R code of Screen & Clean (http://wpicr.wpic.pitt.edu/WPICCompGen/). We remind the readers that Step-(E1) of ESC is only required when dr is large. For moderate size of p, we can start from Step-(E2) directly. Moreover, when p-values are not of interest, Steps-(E1)-(E2) (if dr is large) or Step-(E2) (if dr is moderate) can be directly applied on the entire data, D, without Step-(E3). In practical implementation, we suggest to include in G* only the top, say 5000, covariates identified by the sure independence screening (SIS) of Fan and Lv (2008). It is our experience that this pre-selection procedure will save computational cost without sacrificing the detecting power. Note that the main difference between ESC and SC is that in Step-(E2), ESC fits SLR screening on model (6), while SC fits Lasso screening on model (1). See Figure 2 for the flowchart of ESC.
Figure 2.
Flowchart of ESC ((E1)-(E3)) to detecting G × G, where SLR screening ((S1)-(S2)) is implemented in (E2). The arrows indicate which part of the data is used, where D1 is for screening and D2 is for cleaning. The SC procedure ignores the low-rank screening (S1). The LSC and MSC procedures replace the low-rank screening (S1) by Lasso and marginal scan (MS), respectively.
We close this section by emphasizing the differences between the multistage joint-screening based G × G detection methods (e.g., ESC and SC) and MS. The MS, as mentioned in Section 1, has the drawbacks of ignoring joint effects and being too conservative due to Bonferroni correction. Multistage joint-screening method is thus more preferable, since the number of variables entering the cleaning stage can be rather small and, hence, suffers less penalty from Bonferroni correction. SC, however, will suffer the problem of inefficiency when fitting model (1). The proposed ESC is an intermediate method between MS and SC. On one hand, ESC is a multistage joint-screening method which avoids the drawbacks of MS. On the other hand, ESC overcomes the ineffciency problem of SC by fitting the low-rank model (6). The superiority of ESC will be demonstrated by simulations in Section 5.
4.2 ESC with aggregated p-values via multi-split technique
The data partition in ESC allows the calculations of p-values and the control of the error rates, but the resulting performance is sensitive to the random partition of D. To reduce the impact of random partition, one possible solution is to apply the multi-split technique of Meinshausen et al. (2009) to obtain an aggregated p-value as summarized below:
-
(i)
For b = 1, . . . , B, obtain a random partition, and perform ESC. For the variables selected in the final model for the bth replicate, output the p-values, , using the LSEs in the cleaning stage. Otherwise, set = 1.
-
(ii)
Calculate , where m(b) is the number of variables entering LSE cleaning in the bth replicate.
-
(iii)
Fix smin ∈ (0, 1). Calculate pj = min {1,(1 – log smin) infs∈(smin,1) Qj(s)} , where and qs(·) is the s-quantile function.
Meinshausen et al. (2009) showed that selecting variables by {j : pj ≤ α} controls the family-wise error rate at level α, and can achieve higher detecting power than the single-split method. They also showed that a direct method (e.g., apply SLR screening on the whole data D to identify variables directly) usually achieves a higher true positive rate than its multi-split version, but at the cost of having a higher false positive rate. Thus, either the direct method or the multi-split method has its own merits, and the choice depends on the purpose of the underlying study.
5. Simulation Studies
5.1 Simulation settings
Our simulation models are based on the design considered in Wu et al. (2010) with some extensions, where the genotypes g is generated using the CoLaus dataset (see Section 6.2 for detailed description). Given g, Y is generated from two different models, M1 and M2, where β ∈ {0.25, 0.5, . . . ,1.5} is the effect size and ε ~ N(0, 1):
M1: Y = β(g5g6 + 0.8g5g10 + 0.6g6g10 + g11g16 + g11g21 + 2g10 + 2g11) + ε.
M2: Y = βvecp(η)T vecp(J) + ε, where we randomly generate ηjk = sign(u1) · u2 with u1 ~ U(−1, 9) and u2 ~ U(0.5, 1) for 1 ≤ j ≠ k ≤ 7, and ηjk = 0 for j, k > 7.
M1 contains both main and interaction effects, and M2 only contains interaction effects among (g1, g2, . . . , g7). Additional simulation results under other model can be found in Web Appendix C. In each simulation, we generated two independent data sets D1 and D2, each with sample size n. Set D1 is used to evaluate SLR, and set {D1, D2} is used to evaluate ESC. We choose αl such that |ILR| = n for SLR screening, and use the significance level α = 0.05 for all procedures. Let M0 be the index set of nonzero G × G coeffcients of the true model and let M be the index set of nonzero G × G coeffcients of the estimated model. For SLR screening, we report the numbers of true positive (TP), , and false positive (FP), . For ESC, we report the true positive rate (TPR), , and the false positive rate (FPR), . Simulation results are reported based on 100 replications.
Because SLR contains two steps for variable selection (i.e., low-rank screening and Lasso), we consider three other strategies as benchmarks as illustrated in Figure 1: (1) Lasso (i.e., skipping the low-rank screening), (2) L-Lasso (i.e., replacing low-rank screening by Lasso), which is also known as relaxed Lasso (Meinshausen, 2007), and (3) M-Lasso (i.e., replacing low-rank screening by MS, where MS is based on the PLINK procedure with modifications to accommodate the continuous traits). Following this naming system, SLR is in essence ”LowRank-Lasso” although we stay with the term ”SLR”. Finally, to compare with ESC, Lasso, L-Lasso and M-Lasso are also combined with an independent cleaning stage (denoted by SC, LSC and MSC, respectively; see also Figure 2 for details).
5.2 Simulations with mp in the size of 106
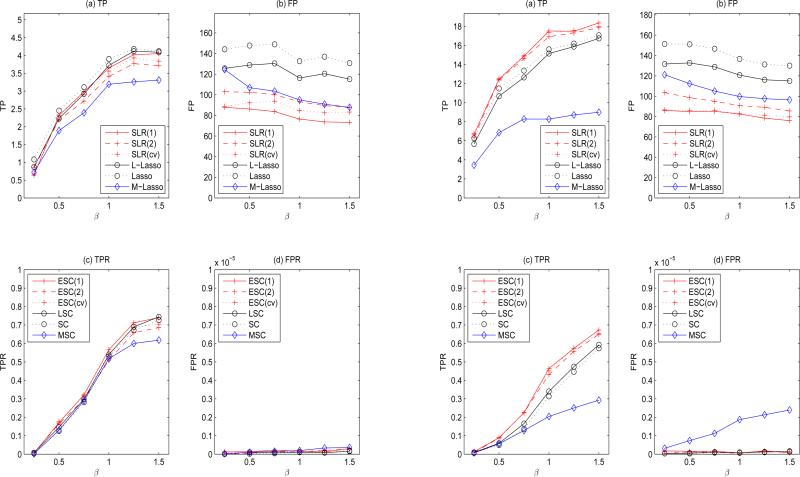
Chromosome 11 (with 1465 SNPs) is used to generate g, which gives about 106 pairs of candidate G × G for each of n = 300 subjects. The number of genes entering the screening stage ranges from 55 to 70, suggesting (by Remark 3) that a low-rank model can be properly fitted (using n = 300) with r ≤ 2. We thus implement SLR with r = 1, 2, which is denoted by SLR(r). We also implement SLR with r being selected by cross-validation, which is denoted by SLR(cv). The corresponding ESC procedures are denoted by ESC(r) and ESC(cv). Simulation results are placed in Figure 3.
Figure 3.
Simulation results with (n, mp) = (300, 106) under model M1 (left panel; the true model size is |M0| = 5) and model M2 (right panel; the true model size is |M0| = 21). (a)-(b): results of SLR screening using D1; (c)-(d): results of ESC using {D1,D2}.
Under M1 (the left panel of Figure 3), we see while SLR screening yields similar TPs with Lasso, the FPs of SLR are largely reduced. Recall the only difference between ESC and SC is the method used in the screening stage. Comparing to SC, the variable set of ESC entered into the cleaning stage had fewer variables and a higher signal-to-noise ratio; consequently, ESC had higher TPRs than SC while the FPRs were adequately controlled (subplots (c)-(d)). The same phenomenon is observed when comparing SLR with the two-staged screening method L-Lasso. Although M-Lasso also has two-staged nature in screening, it has the lowest TPs (and larger FPs than SLR); consequently MSC has a poor performance. This result also reveals the drawback of MS which ignores the joint effects in screening. The gain of SLR is more obvious under M2 (the right panel of Figure 3), which contains a substantial amount of interaction effects. Under M2, SLR screening, which uses a low-rank model, can better identify significant interactions in η (subplot (a)). In contrast, conventional methods (Lasso, L-Lasso, and M-Lasso), which do not utilize the low-rank structure of η, tend to incorrectly filter out significant interactions (e.g., fewer TPs than SLR in subplot (a)) and retain many insignificant terms (e.g., more FPs than SLR in subplot (b)). Therefore, the subsequent LSEs were obtained from a model that was further deviated away from the correct model, and had smaller TPRs than ESC (subplot (c)). Generally, ESC has the best performance, followed by LSC, SC, and MSC.
Note that the rank of η in models M1-M2 ranges from 5 to 7, and using SLR with r ≤ 2 is suffcient to achieve good performance. This result indicates the robustness and applicability of low-rank model (6), even with an under-specified rank, r. Moreover, we observed that ESC(cv) and ESC(1) have comparable performance, and ESC(1) outperforms ESC(2) in most of the settings. The observations suggest that the key component for the performance gain of SLR and ESC is the effciency of the low-rank model fitting (as opposed to the precision in model approximation). Because the goal of the low-rank screening is to reduce the model size so as to stabilize the subsequent Lasso, an approximation of η, e.g., the rank-1 model, was able to remove non-important terms. In contrast, although the rank-2 model approximates η more precisely, it also requires more parameters in the model fitting. With limited sample size, the improvement in approximation accuracy cannot compensate for the loss in estimation effciency, and thus ESC(2) did not have a better performance than ESC(1) when the sample size is not adequate. See Remark 3 for a discussion of selecting r.
5.3 Simulations with mp in the size of 107
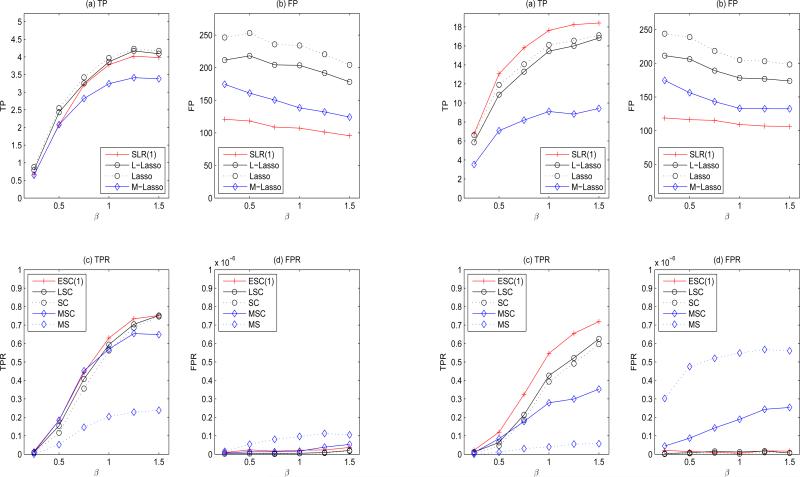
In this simulation, Chromosomes 10-12 (with 4666 SNPs) are used to generate g, which gives about 107 pairs of candidate G × G. We consider the sample size to be n = 400. Since the available sample size is small in comparison with the number of SNPs, we only implement SLR with r = 1. For comparison, we also conduct MS (using the whole data {D1, D2} directly). Simulation results are placed in Figure 4.
Figure 4.
Simulation results with (n, mp) = (400, 107) under model M1 (left panel; the true model size is |M0| = 5) and model M2 (right panel; the true model size is |M0| = 21). (a)-(b): results of SLR screening using D1; (c)-(d): results of ESC using {D1,D2}.
Comparing SLR with other screening methods (subplots (a)-(b)), we observed a similar pattern of TPs and FPs as in Figure 3 under both M1 and M2. That is, comparing to SLR, screening methods without using the low-rank nature of η identified a similar number of TPs but at the price of incorporating many additional FPs. The number of FPs of Lasso and L-Lasso were often twice of that of SLR in the high dimensional setting. Consequently, it is not surprising to see that ESC outperformed SC and LSC (subplots (c)-(d)). Comparing to the case of (n, mp) = (300, 106), ESC exhibited comparable or better performance, which suggested the potential scalability of the proposed methods.
MS yielded lowest TPRs and highest FPRs among all methods. Unlike the joint-screening based ESC, MS examined variables one at a time and ignored the joint effects of variables. In addition, the multiple testing correction conducted in MS also led to severe loss in detecting power. Specifically, the total number of tests to be adjusted in ESC is the number of positive findings obtained from the D1 analysis, while the total number of tests to be adjusted in MS is the number of all main effects and pairwise interactions from 4644 SNPs, i.e., about 107. As a result, MS cannot have better performance (i.e., low TPRs and high FPRs), even when the effect size β is moderate or large. In summary, the proposed ESC procedure improved upon SC, avoided the drawbacks of MS, and was able to perform satisfactorily in the case when mp was large in size.
6. Data Analysis
6.1 Application to the Warfarin study (moderate mp
Warfarin is one of the most widely used oral anticoagulant agents, and the appropriate dose of Warfarin varies substantially among individuals. There is great interest in identifying the genetic and non-genetic factors that contribute to the variation in dosages, and in using these factors to determine appropriate dosages. The Warfarin data were compiled by the International Warfarin Pharmacogenetics Consortium (2009), and can be applied from the Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB, www.pharmgkb.org). We focused on a homogeneous subset of the data comprising 265 Caucasian individuals who had complete information on two candidate genes (VKORC1 and CYP2C9 ) and basic demographic variables. There were 13 variables in the analysis: variables 1-7 are from VKORC1, variables 8-9 are from CYP2C9, and variables 10-13 are Age, Sex, Height, and Weight.
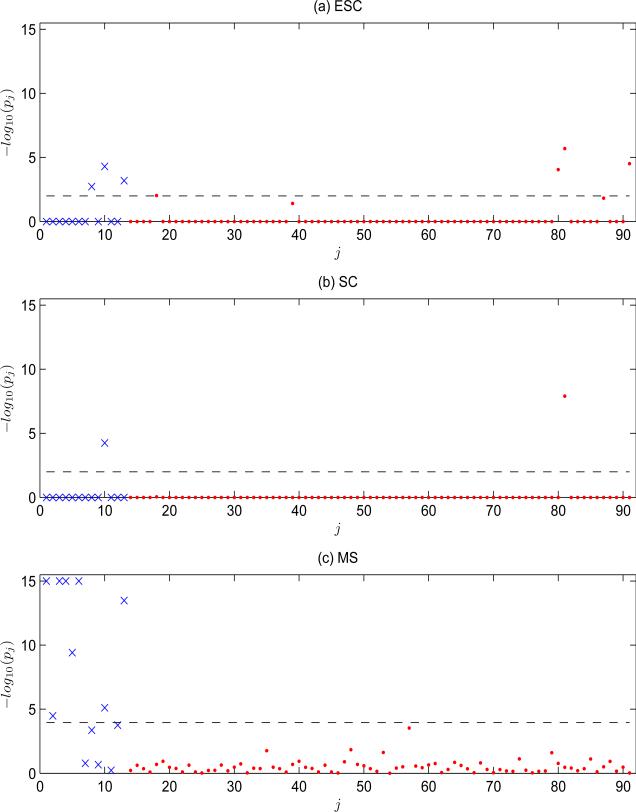
Figure 5 shows the p-values for each variable obtained using ESC (with r = 1 being determined by 5-fold cross-validation and |ILR| = n/3), SC, and MS. The p-values of ESC and SC are obtained as discussed in Section 4.2; the p-values of MS are obtained as described in Section 1. A family-wise error rate of 0.01 was used to identify important variable (see Web Appendix D for detailed results). Comparing ESC to SC, both methods identified the main effect of Age and an interaction effect between CYP2C9*1 and Weight. In addition, ESC also identified interaction effects among CYP2C9*1, Height, and Weight, an interaction between VKORC1*1 and VKORC1*6 (i.e., 2 SNPs within VKORC1), and 2 main-effect terms (CYP2C9*1 and Weight). In contrast to the findings of ESC and SC, MS identified no interaction terms but 8 main effect terms, among which Age and Weight overlap with the findings of ESC. While the influence of VKORC1 and CYP2C9 variations on warfarin dosages has been well established in the literature (e.g., the International Warfarin Pharmacogenetics Consortium (2009)), SC missed VKORC1 effects and MS missed CYP2C9 effects. In contrast, ESC identified effects from both genes. Though typical Warfarin dosage research focuses on main effects, our analysis suggests that interaction effects may play a role in Warfarin dosage.
Figure 5.
The p-values (in the scale of – log10) of the variables in the Warfarin data from ESC, SC, and MS. The x-axis represents the variable number, j, where j = 1, . . . , 13 are for main effects (the ×), and j = 14, . . . , 91 are for interactions (the ●). The horizontal dash line indicates the critical value under 0.01 family-wise error rate. The identified variables by each approach are listed in Web Appendix D.
6.2 Application to the CoLaus study (large mp
Triglyceride concentration is an important risk factor for cardiovascular diseases. Understanding how the genetic variants modulate triglyceride would facilitate the development of therapeutic interventions to cardiovascular disease risk. In this data application, we used data from the Cohorte Lausannoise (CoLaus) study (Firmann et al., 2008). In our analysis, we focused on the sub-samples of 883 male subjects with available information on ages, triglycerides and GWAS data (genotyped with the Affymetrix 500K SNP chip). As did in Wu et al. (2008), we restricted the interaction analysis on those SNPs with “promising” univariate effects on log-triglycerides. Specifically, we performed a single-SNP screening on the GWAS SNPs with minor allele frequencies (MAF) > 0.01 and selected 1780 SNPs with p-values < 0.005 after adjusting for age and population substructures. We performed an interaction analysis using ESC, SC, and MS to identify important predictors among the main age effect, the main SNPs effects, and the pairwise interactions among age and SNPs.
Under 0.05 family-wise error rate, ESC identified a significant interaction between rs6589567 and age; SC did not identify any significant main or interaction effects; MS identified a significant main effect from rs6589567. Figure 6 shows the relationships between age and logtriglyceride by different genotype groups of rs6589567. While the triglyceride levels typically increase with age (Miller et al., 2011), we observed a decreasing trend for the CC genotype group, suggesting a potential rs6589567×age effect as identified by ESC.
Figure 6.
The scatter plots of log-triglyceride and age within different categories of rs6589567 in the CoLaus study. The line represents the fitted regression line that regresses log-triglyceride on age.
7. Discussion
The proposed method can be extended. First, in this article, we only consider the case of a normal error model with continuous response. It is important to extend our ESC procedure to the case of GLM. Although the idea is straightforward, the extension is not trivial due to the over-parameterization when modeling G × G as η = AU AT , which further complicates the derivation of the asymptotic properties. Second, it is of interest to extend our method to a more complicated situation to approximate biological interaction. For example, consider gj ∈ {aa, Aa, AA} that represents a categorical random variable (instead of the numeric 0/1/2), which gives 3 × 3 possible interactions. To fit our model, we can encode gj as , where gjk's are binary random variables: if gj = {aa}, if gj = {Aa}, and if gj = {AA}. Let be a (2p)-vector. The similar inference procedure for the low-rank model (6) developed in this paper can be applied by using the newly defined g, where the definition of vecp(·) and the selection criterion of ESC need some modifications accordingly. Finally, the effciency gain of our method comes from treating G × G as a matrix, η, and imposing a low-rank constraint on it. In view of low-rank, we can alternatively use the trace norm penalty ∥η∥* on η to identify G × G. Trace norm is a convex surrogate of the low-rank constraint and has deserved many advantages. It is of interest to investigate these extensions in future studies.
Supplementary Material
Sparse and Low-Rank Screening (SLR Screening).
(S1) Low-Rank Screening: Fit the low-rank model (6). Examine the significance of to form the index set ILR.
(S2) Sparse (Lasso) Screening: Fit Lasso on ILR. Variables with non-zero estimates are included in the index set ISLR.
Extended Screen-and-Clean (ESC) (see Figure 2).
(E1) Based on D1, fit Lasso on (Y, g*) to obtain gLasso (i.e., genes with non-zero estimates), and fit MS on (Y, g*) to obtain gMS (i.e., significant gene pairs). Set g = .
(E2) Based on D1, implement SLR screening on (Y, g) to obtain ISLR. Let S consist of the main effects and interaction terms in ISLR.
(E3) Based on D2, fit LSE on (Y, S) to obtain the p-values of main effect ξj and interaction ηkl as and . The chosen model is .
Acknowledgments
The authors thank the International Warfarin Pharmacogenetics Consortium and the PharmGKB resources for supplying the Warfarin data, thank Drs. Peter Vollenweider and Gerard Waeber, PIs of the CoLaus study, and Drs. Meg Ehm and Matthew Nelson, collaborators at GlaxoSmithKline, for providing the CoLaus phenotype and genetic data, and thank Dr. Shannon Holloway for input to improve the manuscript. This work was partially supported by National Science Council of Taiwan (to H.H. and S.Y.H.), by National Institutes of Health grant 5U01-HL-114494 (to P.C.), and by National Institutes of Health grants R01 MH084022 and P01 CA142538 (to J.Y.T.).
Footnotes
8. Supplementary Materials
Web Appendices, Tables, and Figures referenced in Sections 2, 5, and 6, and a Matlab code to implementing the low-rank model fitting are available with this paper at the Biometrics website on Wiley Online Library.
Contributor Information
Hung Hung, Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taiwan.
Yu-Ting Lin, Institute of Statistical Science, Academia Sinica, Taiwan.
Penweng Chen, Department of Applied Mathematics, National Chung Hsing University, Taiwan.
Chen-Chien Wang, Yahoo, Sunnyvale, CA, USA.
Su-Yun Huang, Institute of Statistical Science, Academia Sinica, Taiwan.
Jung-Ying Tzeng, Department of Statistics and Bioinformatics Research, Center, North Carolina State University, USA; Department of Statistics, National Cheng-Kung University, Taiwan.
References
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Human Molecular Genetics. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nature Reviews Genetics. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. Journal of the Royal Statistical Society: Series B. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, Hayoz D, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovascular Disorders. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. The New England Journal of Medicine. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Roeder K, Wasserman L. Stability approach to regularization selection (StARS) for high dimensional graphical models. arXiv:1006.3316v1. 2010 [PMC free article] [PubMed] [Google Scholar]
- Magnus JR, Neudecker H. The commutation matrix: some properties and applications. Annals of Statistics. 1979;7:381–394. [Google Scholar]
- Meinshausen N. Relaxed Lasso. Computational Statistics & Data Analysis. 2009;52:374–393. [Google Scholar]
- Meinshausen N, Meier L, Bühlmann P. p-values for high-dimensional regression. Journal of the American Statistical Association. 2009;104:1671–1681. [Google Scholar]
- Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, et al. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123:2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Yang C, Yang Q, Xue H, Fan X, Tang NL, Yu W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. The American Journal of Human Genetics. 2010;10:325–40. doi: 10.1016/j.ajhg.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L, Roeder K. High-dimensional variable selection. Annals of Statistics. 2009;37(5A):2178–2201. doi: 10.1214/08-aos646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Devlin B, Ringquist S, Trucco M, Roeder K. Screen and clean: a tool for identifying interactions in genome-wide association studies. Genetic Epidemiology. 2010;34:275–285. doi: 10.1002/gepi.20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.