Abstract
Background
India is endemic to Japanese encephalitis virus (JEV) and recurrent outbreaks occur mainly in rice growing areas. Pigs are considered to be the amplifying host for JEV and infection in gestating pigs results in reproductive failure. Most studies conducted on JEV infection in Indian pigs have been serological surveys and very little is known about JEV genotypes circulating in pigs. So the potential risk posed by pigs in JEV transmission and the genetic relationship between viruses circulating in pigs, mosquitoes and humans is poorly understood.
Methodology/Principal Findings
This study was conducted in pigs with a history of reproductive failure characterized by stillborn piglets with neuropathological lesions. Japanese encephalitis (JE) suspected brain specimens inoculated intracerebrally into mice and Vero cells resulted in successful isolation of JEV/SW/IVRI/395A/2014. Clinicopathological observations in infected mice, demonstration of JEV antigen in brain, and analysis of the envelope protein identified the swine isolate as being neurovirulent. Phylogenetic analysis based on prM and E gene sequences showed that it belonged to genotype III. This swine isolate was closely related to JEV associated with the 2005 outbreak in India and JaoArS982 from Japan. Phylogenetic analysis of JEV strains collected between 1956 and 2014 in India categorized the GIII viruses into different clades blurring their spatial distribution, which has been discernible in the previous century.
Conclusions/Significance
Isolation of JEV from stillborn piglets and its close genetic relationship with viruses detected at least three decades ago in humans and mosquitoes in Japan suggests that the virus may have been circulating among Indian pigs for several decades. The close similarity between the present swine isolate and those detected in humans affected in the 2005 outbreak in Uttar Pradesh, India, suggests the need for more intensive surveillance of pigs and implementation of suitable strategies to control JE in India.
Introduction
Japanese encephalitis (JE) is an arthropod-borne zoonotic viral disease caused by Japanese encephalitis virus (Flaviviridae;Flavivirus; [JEV]). In the Australasian countries, where the disease is endemic, about 68,000 human JE cases and 10,000–15,000 deaths are estimated to occur every year [1–2]. In India, JE was first reported in 1955 from Vellore, in Tamil Nadu [3]. JE which was confined to south India spread to Burdwan and Bankura districts of West Bengal in 1973 and by 1978 it was recorded in 21 States of India [4–6]. Between 1978–2009, a dramatic increase in the number of JE cases was recorded in the Gorakhpur district of Uttar Pradesh [6].
The reliable diagnosis of JE is based on the isolation of the virus from2 to 4 day-old mice following intracerebral inoculation or on virus isolation from infected cell culture [7]. The neurovirulence of a JEV isolate is determined by intracerebral inoculation in 3–4 week old mice [8] and by molecular analysis of critical amino acid substitutions in the E protein [8–10]. Phylogenetic studies based on prM and E gene of JEV identified five different genotypes of JEV [10–11]. Genotype III is the predominant genotype found in the Indian subcontinent but the presence of genotype I in Gorakhpur region (Uttar Pradesh, India) in 2009 [12], and subsequently in 2010, in West Bengal [13] has also been reported.
Pigs act as amplifying hosts, and ardeid birds such as pond herons and egrets are the maintenance reservoirs for the virus. Mosquitoes, mainly Culex spp. act as vectors and human and equine are considered dead end hosts [7]. JEV infection in pregnant pigs can result in reproductive problems characterized by abortion, stillbirths and mummified fetuses [7, 14]. In India, extensive studies have been conducted on mosquitoes in JE endemic areas, and the virus has been recovered from 19 different mosquito species; but most studies on JEV infection in Indian pigs have been limited to seroprevalence in different parts of the country [15–16]. So far there is only one report of JEV isolation from a sentinel pig in India [17], but there is no information on the genotype involved and its genetic relationship with JEV strains detected in humans. This study was conducted in a pig herd in the State of Uttar Pradesh, which had a history of reproductive problems in pregnant sows. The pathological changes observed in stillborn piglets and the genetic characterization of the isolated virus are reported in this paper.
Methods
Ethics statement
The mice experiment in this study was conducted according to the guidelines of the Committee for the Purpose of Control and Supervision on Experiments on Animals (CPCSEA), India. Animals were handled in strict accordance with good animal practice, including humane endpoint treatment for mice, euthanasia, frequent monitoring of mice and reduced suffering and distress as defined by CPCSEA, Ministry of Environment and Forestry, Government of India. This study was approved by the Institute Animal Ethics Committee (IAEC), Indian Veterinary Research Institute (IVRI), Izatnagar, India [Permit number: F.1-53/2012-13/JD(R)]. The pig samples were drawn by the Swine Production Farm from naturally dead pigs submitted to the Post-Mortem Facility, Division of Pathology, IVRI, Izatnagar for disease investigation. The pig samples were drawn from naturally dead pigs (no experiment was carried out in live pigs), so ethical committee approval was not required.
Study population
This study was conducted in a pig herd in Uttar Pradesh, India, from April 2014 to October 2014. Systematic necropsy was conducted on stillborn piglets and the gross lesions were recorded. Representative tissue samples were collected in 10% neutral buffer formalin (NBF) for histopathological processing. Tissue specimens collected on ice were stored under -20°C until further use for molecular and virus isolation studies.
Histopathology and immunohistochemistry
The formalin-fixed tissues were processed for preparation of haematoxylin and eosin (H&E) stained slides for light microscopy following standard procedures.
For immunohistochemistry (IHC), 5 μm thick formalin-fixed paraffin-embedded tissue sections were cut and mounted on 3-Aminopropyltriethoxysilane (APES), (Sigma-Aldrich, USA)-coated slides and air dried for 30 min. Anti-JEV hyperimmune serum raised in rabbit (GeneTex, Irvine, CA, USA) was diluted to 1:150 in 1% BSA and Goat Anti-Rabbit IgG-HRPO conjugate (Genei, India) was used as the secondary antibody at 1:150 dilution in PBS. AEC (3-Amino-9-ethylcarbazole) (Sigma-Aldrich, USA) substrate was prepared as per manufacturer’s recommendation and counterstaining was done with Mayer’s hematoxylin. AEC stained sections were finally mounted with CC/Mount™ (Sigma-Aldrich, USA). For the negative control, primary antibody was substituted by 1% BSA and other procedures were followed as mentioned above.
RNA extraction and RT-PCR
Viral RNA was extracted from brain samples using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) as per manufacturer’s instruction. The cDNA was synthesized using SuperScript® III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The RT-PCR for the detection of E and prM genes of JEV was performed following previously published protocols (Table 1) [18–19].
Table 1. Gene targets and nucleotide sequences of primers used to amplify JEV gene segments.
| Gene | Primer | Sequence | Product size | Refs. |
|---|---|---|---|---|
| FLAVI NS5 | FLAVI-1 | 5’-AATGTACGCTGATGACACAGCTGGCTGGGACAC- 3’ | 863 bp | Ayers et al., 2006 |
| FLAVI-2 | 5’-TCCAGACCTTCAGCATGTCTTCTGTTGTCATCCA–3’ | |||
| JEV-prM | JEV-prMf | 5’-CGTTCTTCAAGTTTACAGCATTAGC-3’ | 675 bp | Wang et al., 2007 |
| JEV-prMr | 5’-CCYRTGTTYCTGCCAAGCATCCAMCC-3’ | |||
| JEV-E | JEV-Ef | 5’-TGYTGGTCGCTCCGGCTTA-3’ | 1582 bp | Wang et al., 2007 |
| JEV-Er | 5’-AAGATGCCACTTCCACAYCTC-3’ |
Cloning and sequencing
RT-PCR amplicons were purified using GeneJET Gel Extraction Kit (Thermo Scientific, Waltham, Massachusetts, USA) and cloning of the amplified PCR products was carried using InsTAclone PCR Cloning Kit (Thermo Scientific, Waltham, Massachusetts, USA). The plasmids were extracted with GeneJET Plasmid Miniprep Kit (Thermo Scientific, Waltham, Massachusetts, USA) and positive clones were sequenced from both ends (Eurofins Genomics India Pvt. Ltd., Bangalore, India).
Pathogenecity testing in mice
Mice for this study were obtained from the Laboratory Animal Resources (LAR) section, Indian Veterinary Research Institute (IVRI), Izatnagar, India. The inoculums were prepared as per OIE (7) recommendations (10% brain suspension) and 20 μl were inoculated intracerebrally into 3–4 week old mice (n = 6 mice). The mice were observed daily for neuro-pathological symptoms and sacrificed at 14 dpi (days post-infection). Systematic necropsy was conducted and samples were collected for histopathology, immunohistochemistry (IHC), virus isolation and molecular studies.
Propagation of JEV in Vero cell culture
Vero cells were obtained from Division of Virology, Indian Veterinary Research Institute (IVRI), Mukteswar Campus, Nainital (Uttaranchal), India. For the isolation of JEV in Vero cells, brain samples (10% suspensions in phosphate-buffered saline) from mice showing neuro-pathological symptoms and samples from stillborn piglets were processed according to the OIE Terrestrial Manual [7]. In Vero cells, four passages were carried out and the cytopathic effects were noticed and the presence of JEV was confirmed by RT-PCR and IHC.
Molecular analysis for neurovirulence of JEV
The critical amino acids in the E protein of JEV reported to be associated with neurovirulence [10] were analyzed by comparing the amino acid sequence of the present Indian isolate (JEV/SW/IVRI/395A/2014; from hereafter referred to as IVRI395A) with all available JEV (1956–2014) sequences (both GI and GIII viruses) from India and 150 viruses from other countries retrieved from the NCBI database.
Molecular characterization of the E protein in Indian JEV from 1956–2014 at sites under positive selection
The amino acids in the E protein of JEV reported to be associated with positive selection were analyzed by comparing the amino sequence of the present Indian isolate (IVRI395A) with all available JEV (1956–2014) sequences (both GI and GIII viruses) from India retrieved from the NCBI database. The haplotypes based on the four sites in the E protein (E-123, E-209, E-227 and E-408) were analyzed [20].
Phylogenetic analysis of JEV
Phylogenetic analysis of JEV isolates was performed for the E and prM genes Nucleotide sequences available for partial prM gene of Indian isolates of JEV (1956–2014) were used to construct separate phylogenetic trees for close comparison with the present swine isolate. Partial E gene sequences (296 nts) available for JEV isolates associated with the major outbreak of 2005 in the State of Uttar Pradesh, India, [21] and viruses from Japan (M18370, AB028257, U70414) and Indonesia (U70397, JN375549) were analyzed separately to establish their genetic relationship. All trees were constructed with MEGA6 software using neighbor-joining method based on the Kimura-2 parameter model, substitutions to include transition and transversion and uniform rate among sites. The codon positions 1st, 2nd, 3rd, and non-coding were included. All positions containing gaps and missing data were eliminated. To determine internal node and better differentiation of different clades within genotype III, cutoff value of 50 was used with bootstrap support values expressed as percentage for 1,000 replicates.
Results
In total, 28 sows farrowed during the study period. Occurrence of stillbirth was recorded in 10 (7 crossbred and 3 Landrace) sows; with the number of stillborn piglets being 2 to 5 per sow (Table 2). Of 31 stillborn piglets (22 crossbred and 9 Landraces) delivered and tested by RT-PCR assay, seven (5 crossbred and 2 Landraces) piglets delivered by 2 sows were found positive for JEV.
Table 2. Sows which delivered JEV positive stillborn piglets in a herd.
Out of 28 sows that farrowed in a herd, 10 sows delivered stillborn piglets. Sow number, date of farrowing, breed, number of stillbirth and RT-PCR results for JEV are presented.
| S.No. | Sow No. | Date of Farrowing | CB/LR | No of Stillbirth | JEV RT-PCR |
|---|---|---|---|---|---|
| 1 | 64 | 01.04.14 | CB | 4 (3M;1F) | N |
| 2 | 768 | 23.04.14 | LR | 2 F | N |
| 3 | 160A/14 | 24.04.14 | CB | 2 F | N |
| 4 | 770 | 16.05.14 | LR | 5(4M; 1F) | N |
| 5 | 116 | 23.05.14 | CB | 2M | N |
| 6 | 754 | 29.05.14 | LR | 2(1M;1F) | P |
| 7 | 27 | 01.06.14 | CB | 2M | N |
| 8 | 146 | 05.06.14 | CB | 5 (3M;2F) | N |
| 9. | 115 | 08.09.14 | CB | 5 (3M; 2F) | P |
| 10 | 66 | 08.09.14 | CB | 2F | N |
CB- Crossbred; LR- Landrace; M-Male; F-Female; P- Positive; N-Negative
Gross and microscopic lesions
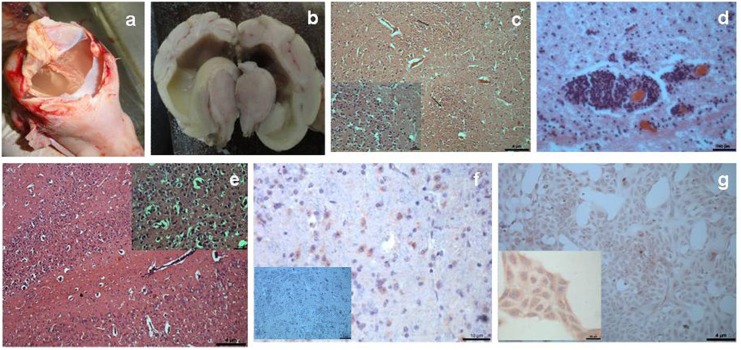
Five stillborn piglets positive for JEV showed following pathological lesions in the brain. The head appeared more rounded and subcutaneous hemorrhages were observed over the head. Upon opening the skull, one piglet revealed hydranencephaly with grayish-white flaky fluid filling the entire cranial cavity and only remnants of brain parenchyma seen as small strands (Fig 1a). Other piglets had swollen, rounded brains with complete loss of sulci and gyri. Cross sections of these brains revealed severe dilatation of ventricular spaces and thinning of the surrounding parenchyma (Fig 1b). The carcass lymph nodes were prominently congested. The most significant microscopic lesions found in the brains were widespread edema, congestion and microhemorrhages in the parenchyma. Neuronal degeneration, and focal to diffuse gliosis (Fig 1c), occasional finding of perivascular cuffing around small vessels particularly in the vicinity of the ventricular spaces (Fig 1d) and accumulation of glial cells in the subependymal region were the prominent microscopic lesions. IHC demonstrated the presence of JEV antigen mainly in the cytoplasm of cerebral neurons.
Fig 1.
1a-JEV infected stillborn piglet with hydranencephaly; 1b- Cross section of a brain with severe dilatation of ventricular spaces and thinning of the surrounding parenchyma; 1c- Diffuse gliosis in the cerebrum of piglet, (insert higher magnification), H& E; 1d-Perivascular cuffing in the subependymal region of piglet brain, H& E; 1e- Severe neuronal degeneration and necrosis in the mice brain, (insert higher magnification), H& E; 1f- JEV antigen in the cytoplasm of cerebral neurons (rose red color), IHC; 1g- JEV antigen in the cytoplasm of Vero cells (rose red color), insert higher magnification, IHC.
RT-PCR
JEV nucleic acids were detected in the brains of seven stillborn piglets from two crossbred sows in RT-PCR using three sets of primers (Table 1). In total, out of 33 stillborn piglets tested by RT-PCR, seven stillborn piglets from two sows were found positive for JEV.
Pathogencity testing of JEV in mice
In mice (n = 6 mice) inoculated intracerebrally with brain suspensions from stillborn piglets neuro-pathological signs like reduced movement, circling and posterior paresis were observed between 11–14 dpi. At necropsy, no gross lesions were noticed. Microscopically, severe neuronal degeneration and necrosis, perivascular cuffing and gliosis were noticed (Fig 1e). IHC localized the JEV antigen in the cytoplasm of neurons in cerebrum (Fig 1f), cerebellum and midbrain.
Propagation of JEV in Vero cell culture
The cytopathatic effect (rounding and detachment of cells) in infected Vero cell line was noticed at 5th day of inoculation. Presence of JEV in infected Vero cells was confirmed by RT-PCR and IHC at 4th passage (Fig 1g).
Molecular analysis of neurovirulence
Neurovirulent specific critical amino acids in the E protein of IVRI395A and other Indian isolates from humans and mosquitoes are summarized in Table 3. The current swine isolate and all previous Indian isolates of JEV except KC526871 (human isolate from Malda, West Bengal, 2012) carried amino acids critical for neurovirulence.
Table 3. Comparative analysis of critical amino acids in the E protein associated with JEV neurovirulence.
Eight amino acids in the E protein critical for neurovirulence in the vaccine strain (SA14-14-2) and other JEV strains including current swine isolate.
| Amino acid position | Amino acids in | Swine isolate (JEV/SW/IVRI/395A/2014) | Other Indian isolates (81 G-III & 6 G-I) | |
|---|---|---|---|---|
| Vaccine Strain SA14-14-2 | Neurovirulent Strains | |||
| E-107 | F | L | L | L |
| E-138 | K | E | E | E |
| E-176 | V | I | I | I, V (Z34097) |
| T (AF080251; JX131374) | ||||
| E-177 | A | T | T | T |
| E-264 | H | Q | Q | Q, H (KC526871) |
| E-279 | M | K | K | K |
| E-315 | V | A | A | A, V (KC526871) |
| E-439 | R | K | K | K, R (KC526871) |
Molecular characterization of the E protein in Indian JEV from 1956–2014 at sites under positive selection
Five different haplotypes (E-123, E-209, E-227 and E-408) of JEV have been detected so far in India [20]. The predominant haplotype detected in humans and mosquitoes is SKSS. The present Indian swine isolate and all GI viruses reported from India belong to this haplotype. Four Indian JEV isolates, three from humans and one from mosquitoes, belong to the SKPS haplotype. The other three haplotypes, one each of RKSS, SRSS and SKTS, were detected in India. In comparison to other isolates, the E protein of IVRI395A had a unique amino acid substitution at E-169.
Phylogenetic analysis of JEV
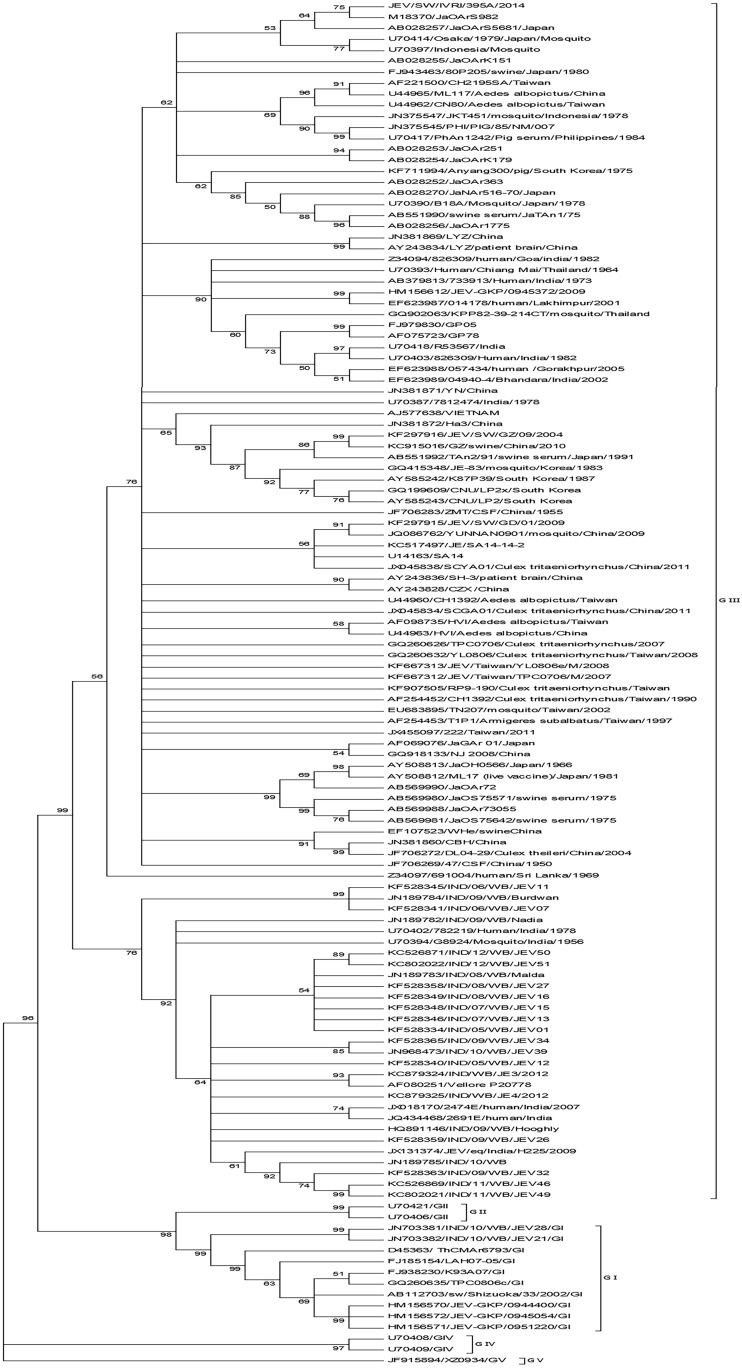
Phylogenetic analysis based on E gene of IVRI395A showed that it belonged to genotype III. Maximum identity of 99.9% was seen with Japanese isolate JaOArS982 and 98% with GP78 from India. Viruses closely related to the present swine isolate from India have been detected in humans, mosquitoes and pigs in JEV endemic areas of Asia (Fig 2). The Indian GIII isolates grouped into two distinct clades, Clade-I with GP78 as the prototype virus and Clade-II with Vellore-P20778 as the prototype virus (Fig 2). IVRI395A grouped with Clade-I viruses.
Fig 2. Phylogenetic analysis for the E gene of JEV.
The tree was constructed with MEGA6 software using neighbor-joining method based on the kimura-2 parameter model. Data are based on a bootstrap value of 1000 and cutoff value of 50.
When the partial E gene sequences of the virus strain pertaining to the 2005 outbreak in Uttar Pradesh, India, was compared to the corresponding fragment of the Indian swine isolate, only one nucleotide substitution was noticed. Similar identity was shared by JaOArS982, JaOArS5681 and Osaka/1979 from Japan and JKT220507 from Indonesia.
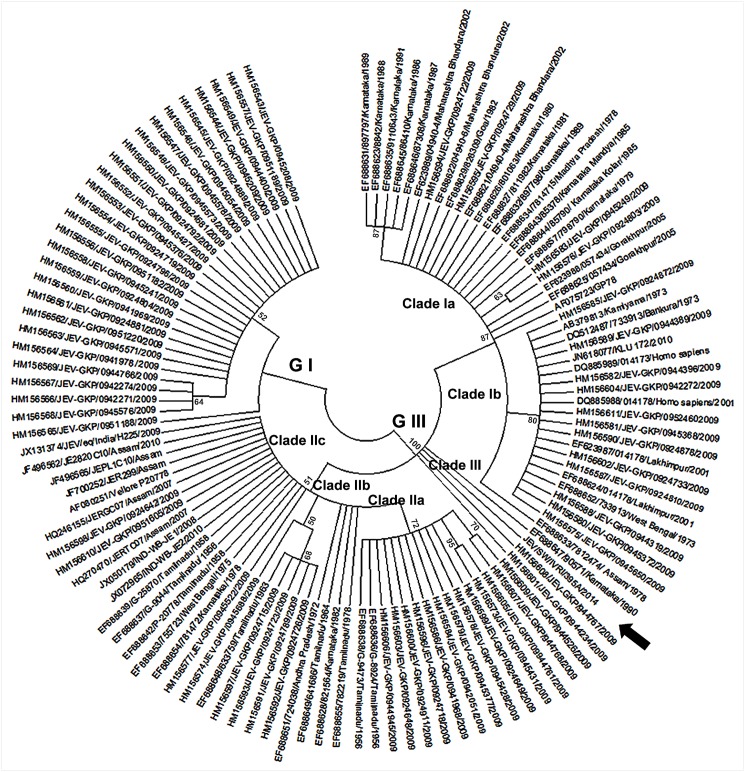
Phylogenetic analysis of the prM gene also showed that IVRI395A belonged to genotype III (Fig 3). Maximum identity of 99.6% was seen with Japanese strain JaOArS982and 96% with IND-WB-JE2 (JX072965) and IND-WB-JE1 (JX050179) from West Bengal, India. The phylogenetic tree constructed with prM gene sequences from India separated into three distinct clades, clade-I, clade-II and clade-III. At the nucleotide level, 5% divergence was noticed between the clade I and clade-II, and their spatial distribution was not restricted to particular regions in the country. The IVRI395A belonged to Clade-III, as were two other isolates (HM156608/944767 and HM156601/944234) collected in 2009 from Gorakhpur, Uttar Pradesh, India. Further it was noted that each of the clade I and clade II could be further subdivided into different subclades: Ia (AF075723/GP78 from Uttar Pradesh as the prototype virus), Ib (EF688652/733913 from West Bengal as the prototype virus), IIa (EF688636/G-8924 from Tamil Nadu as the prototype virus), IIb (EF688648/633759 from Tamil Nadu as the prototype virus), IIc (AF080251/Vellore P20778 from Tamil Nadu as the prototype virus), and clade III (EF688647/90571 from Karnataka as the prototype virus) (Fig 4). Among the various clades and subclades of GIII viruses circulating in India, clade Ia (6 States) and clade IIc (7 States) are more widespread in geographical distribution (Table 4).
Fig 3. Phylogenetic analysis for the prM gene of JEV.
The tree was constructed with MEGA6 software using neighbor-joining method based on the kimura-2 parameter model. Data are based on a bootstrap value of 1000 and cutoff value of 50.
Fig 4. Phylogenetic analysis for prM gene of JEV including sequences data from Indian JEV isolates only.
The tree was constructed with MEGA6 software using neighbor-joining method based on the kimura-2 parameter model. Data are based on a bootstrap value of 1000 and cutoff value of 50.
Table 4. Distribution of various clades of JEV Genotype III in different States of India (1956–2014).
Genotype III was further classified into three clades and distribution of different subclades in different states of India, year of detection and number of sequences (in parenthesis) are presented.
| Clade | State | Year of Isolation |
|---|---|---|
| Ia | Assam | 1978(1) |
| Madhya Pradesh | 1978(1) | |
| Uttar Pradesh | 1978(1), (2005(2), 2009(2) | |
| Karnataka | 1979(1), 1980(1), 1981(1), 1985(2), 1986(1), 1987(1), 1988(1), 1989(2), 1991(1) | |
| Goa | 1982(1) | |
| Maharashtra | 2002(3) | |
| Ib | West Bengal | 1973(2) |
| Uttar Pradesh | 2001(2), 2009(12) | |
| IIa | Tamil Nadu | 1956(2) |
| Uttar Pradesh | 2009(12) | |
| IIb | Tamil Nadu | 1963(1) |
| Uttar Pradesh | 2009(5) | |
| IIc | Tamil Nadu | 1956(1), 1958(2), 1964(1), 1978(1) |
| Andhra Pradesh | 1972(1) | |
| West Bengal | 1975(1), 2008(1), 2010(1) | |
| Karnataka | 1978(1) | |
| Assam | 2007(2) | |
| Haryana | 2009(1) | |
| Uttar Pradesh | 2009(4) | |
| III | Karnataka | 1990(1) |
| Uttar Pradesh | 2009(2), 2014(swine) |
Discussion
In India, JE is an endemic disease particularly in States where rice cultivation is done extensively and pig rearing is practiced by poor, landless farmers with limited resources [22]. Pigs which are known to be amplifiers of JEV should be actively monitored in the country. Moreover pigs infected with JEV may suffer from reproductive problems and this has an adverse effect on profitability of pig farming. But most studies on JEV infection in pigs have been limited to serological surveys in JE endemic areas [15–16] and to the best of our knowledge none have focused on reproductive failure in pigs and genetic characterization of JEV circulating in the Indian pig population.
Investigation of reproductive failure manifested in an increased number of stillborn piglets with neuropathological lesions led to the detection and isolation of JEV as the causative agent. JEV related reproductive failure and stillbirth was first recorded in pigs in Japan (1947–1948), and subsequently JEV was isolated from stillborn piglets [23–24]. JEV can cause stillbirth, mummification, embryonic death and abortion in sows and aspermia in boars [25–28]. In the present study, naturally infected stillborn piglets showed brain lesions as reported in experimentally infected pigs [29].
Presence of serine (S) in E at position 227 in the present swine isolate corroborates earlier observations of amino acids critical for adaptation to Vero cells. It has been reported that amino acid substitution S227P in E can slightly impair the growth rate in Vero cells [20]. The current swine isolate which grows easily in Vero cells may facilitate the development of an inactivated JE vaccine for use in pigs.
Mice inoculated with brain suspensions prepared from stillborn piglets resulted in characteristic neuropathological symptoms and molecular analysis of E identified critical amino acid residues attributable for neurovirulence of JEV [10]. Comparison of E of the current swine isolate with SA14-14-2 (vaccine strain) revealed 10 amino acid substitutions out of which 8 have been reported to be critical for neurovirulence. E-244 has been linked to neurovirulence [9], but the significance of amino acid substitution at E-169 is not known. Interestingly, IVRI395A had a E169I substitution in E as in JaOArS982 isolated in Japan, while all other Indian isolates analyzed in this study had E169V. Further studies are required to ascertain whether this substitution has any significance for virus replication in pigs. In the present study, analysis of the envelope protein of JEV revealed that 5 different haplotypes are circulating in India. While 4 of them have been reported earlier [20] the SKTS haplotype identified in this study pertains to JEV detected in a human case from Chandigarh, India, in 2007. The RKSS haplotype has been associated with positive selection in mosquitoes [20] but our analysis showed that in one instance, this haplotype was detected from a human case of JE in West Bengal, India in 2011 This study revealed that the SKSS haplotype is the predominant haplotype of GIII and GI viruses circulating in humans and mosquitoes in India, and the current Indian swine isolate also belongs to this haplotype.
Phylogenetic analysis based on complete E gene sequences indicated that IVRI395A belonged to genotype III and is closely related to certain isolates from Japan. However, a close analysis of the partial E gene sequences available for the 2005 outbreak in humans in Uttar Pradesh, India, suggested high identity with the present swine isolate. Further, viruses sharing similar identity were detected in 1979 from mosquitoes and humans in Japan and from mosquitoes in Indonesia. This suggested that the present swine isolate is closely related to JEV strains detected in the Asian continent at least three decades ago.
In India, GIII continues to be the predominant genotype [12] but incursion of GI viruses has also been reported [12–13]. Detailed analysis of prM gene sequences of Indian JEV collected from 1956 to 2014 categorized the GIII viruses into three clades. While the clade II viruses were confined to Southern India until 1975, the clade I viruses were mostly detected in North India except for one south Indian State, Karnataka, where it was first detected in 1979 and repeatedly thereafter until 1991. The clade III contained the present Indian swine isolate and three other Indian viruses detected in humans; two from Gorakhpur in North India (2009) and one from Karnataka in the South (1990). This indicates that similar clade III viruses are circulating in human populations in geographically distant locations.
The detection of clade IIc in south India preceded clade Ia in North India. Clade IIc consists of some of the oldest strains (Vellore, P20778 and similar viruses) detected in the South Indian States such as Tamil Nadu, Andhra Pradesh and Karnataka between 1956 and 1978. The clade IIc viruses were first detected in North India in the State of West Bengal (1975) and subsequently in Assam, Haryana and Uttar Pradesh between 2007 and 2009. Clade Ia viruses (GP78 and similar viruses) were first detected simultaneously during 1978 in the North India in the States of Uttar Pradesh, Madhya Pradesh and Assam. Soon after, in 1979, clade Ia viruses were detected in the South India, in Karnataka, where they continued to be detected intermittently until 1991. The 2009 outbreak of JE in the Northern State of Uttar Pradesh showed co-circulation of all three clades of GIII viruses and GI viruses for the first time in India. This suggests that some poorly understood epidemiological factors were responsible for the change in spatial distribution of different genotypes and their clades in India in recent years.
In conclusion, JEV associated with reproductive failure in Indian pigs caused severe neuropathological lesions in stillborn piglets, and the isolated virus belonged to the predominant haplotype of GIII circulating in humans and mosquitoes in India. Analysis of the E protein of JEV and neuropathology in experimentally infected mice characterized IVRI395A as neurovirulent strain. It was phylogenetically related to JEV associated with the 2005 outbreak in India. The spatial distribution of different genotypes and their clades in India has changed in recent years.
Acknowledgments
We appreciate the technical assistance from Mr. Jas Ram and Mr Vinod K Khokar.
Data Availability
JEV sequences generated and used for analysis in this paper are available in GenBank, Accession NO. KP164498.
Funding Statement
The authors are thankful to ICAR-IVRI, Izatnagar, India, for financial support to conduct this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Solomon T. Control of Japanese encephalitis—within our grasp? N Engl J Med. 2006; 355:869–871. . [DOI] [PubMed] [Google Scholar]
- 2.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, et al. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ. 2011; 89:766–774. 10.2471/BLT.10.085233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namachivayam V, Umayal K. Proceedings of National Conference on Japanese Encephalitis,New Delhi: Indian Council of Medical Research. 1982; 30–33.
- 4.Dhillon GP, Raina VK. Epidemiology of Japanese encephalitis in context with Indian scenario. J Indian Med Assoc. 2008; 106:660–3. . [PubMed] [Google Scholar]
- 5.Kumari R, Joshi PL. A review of Japanese encephalitis in Uttar Pradesh, India. WHO South-East Asia J Public Health. 2012; 1:374–395. [DOI] [PubMed] [Google Scholar]
- 6.Tiwari S, Singh RK, Tiwari R, Dhole TN. Japanese encephalitis: A review of the Indian perspective. Braz J Infect Dis. 2012; 16:564–573. 10.1016/j.bjid.2012.10.004 . [DOI] [PubMed] [Google Scholar]
- 7.OIE. OIE Terrestrial Manual 2010. 2010; Chapter 2.1.7.—Japanese encephalitis.
- 8.Liu WJ, Zhu M, Pei JJ, Dong XY, Liu W, Zhao MQ, et al. Molecular phylogenetic and positive selection analysis of Japanese encephalitis virus strains isolated from pigs in China. Virus Res. 2013; 178:547–552. 10.1016/j.virusres.2013.09.002 . [DOI] [PubMed] [Google Scholar]
- 9.Arroyo J, Guirakhoo F, Fenner S, Zhang ZX, Monath TP, Chambers TJ. Molecular basis for attenuation of neurovirulence of a yellow fever virus/Japanese encephalitis virus chimera vaccine (ChimeriVax-JE). J Virol. 2001; 75:934–942. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao QS, Li XM, Zhu QY, Wang DD, Chen HC, Qian P. Isolation and molecular characterization of genotype 1 Japanese encephalitis virus, SX09S-01, from pigs in China. Virol J. 2011; 8:472 10.1186/1743-422X-8-472 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004; 10:S98–109. . [DOI] [PubMed] [Google Scholar]
- 12.Fulmali PV, Sapkal GN, Athawale S, Gore MM, Mishra AC, Bondre VP. Introduction of Japanese encephalitis virus genotype I, India. Emerg Infect Dis. 2011; 17 10.3201/eid1702.100815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar A, Taraphdar D, Mukhopadhyay SK, Chakrabarti S, Chatterjee S. Molecular evidence for the occurrence of Japanese encephalitis virus genotype I and III infection associated with acute encephalitis in patients of West Bengal, India, 2010; Virol J. 2012; 9:271 10.1186/1743-422X-9-271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morimoto T, Miura Y, Sugimori T, Kurogi H, Fujisaki Y. Isolation of Japanese Encephalitis-Virus and a hemagglutinating DNA virus from brain of stillborn piglets. Natl Inst Anim Health Q. 1972; 12:127–136. . [PubMed] [Google Scholar]
- 15.Ratho RK, Sethi S, Prasad SR. Prevalence of Japanese encephalitis and West Nile viral infections in pig population in and around Chandigarh. J Commun Dis. 1999; 31:113–6. . [PubMed] [Google Scholar]
- 16.Kalimuddin MD, Narayan KG, Choudhary SP. Serological evidence of Japanese encephalitis virus activity in Bihar. Int J Zoonoses. 1982; 9:39–44. . [PubMed] [Google Scholar]
- 17.Geevarghese G, George S, Bhat HR, Prasanna Y, Pavri KM. Isolation of Japanese encephalitis virus from a sentinel domestic pig from Kolar district in Karnataka. Indian J Med Res. 1987; 86:273–5. . [PubMed] [Google Scholar]
- 18.Ayers M, Adachi D, Johnson G, Andonova M, Drebot M Tellier R. A single tube RT-PCR assay for the detection of mosquito-borne flaviviruses. J Virol Methods. 2006; 135:235–239. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX, et al. Molecular epidemiological analysis of Japanese encephalitis virus in China. J Gen Virol. 2007; 88:885–894. . [DOI] [PubMed] [Google Scholar]
- 20.Han N, Adams J, Chen P, Guo ZY, Zhong XF, Fang W, et al. Comparison of genotypes I and III in Japanese encephalitis virus reveals distinct differences in their genetic and host diversity. J Virol. 2014; 88:11469–11479. 10.1128/JVI.02050-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parida M, Dash PK, Tripathi NK, Ambuj, Sannarangaiah S, Saxena P, et al. Japanese encephalitis outbreak, India, 2005. Emerg Infect Dis. 2006; 12:1427–1430. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh D, Basu A. Japanese encephalitis—A pathological and clinical perspective. PLoS Negl Trop Dis. 2009; 3(9): e437 10.1371/journal.pntd.0000437 PMCID: PMC2745699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoya H, Matumoto M, Iwasa S. Epizootiological studies on stillbirth of swine occurred in Japan during summer months of 1948. Japanese J Exp Med. 1950; 20:587–595. [Google Scholar]
- 24.Burns K. Congenital Japanese B encephalitis infection of swine. Proceedings of the Society for Experimental Biology and Medicine. 1950; 75, 621–625. [DOI] [PubMed] [Google Scholar]
- 25.Salmon H. Immunity in the fetus and the newborn infant: A swine model. Reprod Nutr Dev. 1984; 24:197–206. . [PubMed] [Google Scholar]
- 26.Joo HS, Chu RM. Japanese B encephalitis In: Disease of Swine, ed. Straw BE, D’Allaire S, Mengeling WL, and Taylor DJ, 8th ed., pp. 173–178. Iowa State University Press, Ames, IA, 1999. [Google Scholar]
- 27.Jubb KVF, Huxtable CR. The nervous system In: Pathology of Domestic Animals, ed. Jubb KVF, Kennedy PC, and Palmer N, 4th ed., vol. 1, p. 412 Academic Press, San Diego, CA, 1992. [Google Scholar]
- 28.Gresham A. Infectious reproductive disease in pigs, In Practice, 2003; 25, 466–473. [Google Scholar]
- 29.Yamada M, Nakamura K, Yoshii M, Kaku Y. Nonsuppurative encephalitis in piglets after experimental inoculation of Japanese encephalitis flavivirus isolated from pigs. Vet Pathol. 2004; 41:62–67. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
JEV sequences generated and used for analysis in this paper are available in GenBank, Accession NO. KP164498.