Abstract
The isolation of single biological cells and their further cultivation in dedicated arrayed chambers are key to the collection of statistically reliable temporal data in cell-based biological experiments. In this work, we present a hydrodynamic single cell trapping and culturing platform that facilitates cell observation and experimentation using standard bio-lab equipment. The proposed design leverages the stochastic position of the cells as they flow into the structured microfluidic channels, where hundreds of single cells are then arrayed in nanoliter chambers for simultaneous cell specific data collection. Numerical simulation tools are used to devise and implement a hydrodynamic cell trapping mechanism that is minimally detrimental to the cell cycle and retains high overall trapping efficiency (∼70%) with the capability of reaching high fill factors (>90%) in short loading times (1–4 min) in a 400-trap device. A Monte Carlo model is developed using the design parameters to estimate the system trapping efficiencies, which show strong agreement with the experimentally acquired data. As proof of concept, arrayed mammalian tissue cells (MIA PaCa-2) are cultured in the microfluidic chambers for two days without viability problems.
I. INTRODUCTION
The recent alliance of microtechnologies with the biological and medical sciences has enabled the development of a wide range of applications in fundamental biology, such as the study of cell to cell/microenvironment interactions1 and intra/extracellular analysis;2,3 as well as in medicine, such as the development of rapid, low-cost, efficient diagnostic devices and drug discovery research platforms.4 The cell, as the smallest complex living unit that carries representative information of the tissue, organ, or organism it comes from, is commonly featured in ex-vivo experimentation. The challenge to interface the cell with sensing/actuating MEMS chips is solved by microfluidic cell manipulation5 and is accepted to be one of the major enabler technologies for the advancement of the fields of biology and medical research.4 Moreover, the microfluidic methods have empowered the scaling of biological experiments down to single cells,6–9 which enables researchers to see beyond the conventional statistically time- and population-averaged results and to better understand the cell behavior heterogeneity in both temporal and spatial dimensions.10 Moreover, the microfluidic platforms offer many advantages in cell culture experiments, such as flexibility, control, automation, and parallelization; the possibility to work with low cell numbers; reduced reagent consumption and contamination risk; and efficient high throughput experimentation over traditional macroscopic cell culturing.11 Thus, a microfluidic platform that allows for real-time, long-term, parallel microscopic observation of single-cells in confined, tagged, and arrayed chambers is essential to analyze the spatiotemporal data collected from sub-cell populations.
The hydrodynamic cell trapping is an adaptable and low-cost technique that can be implemented in standard bio-lab conditions.12 The two well-received hydrodynamic trapping systems are: (1) the serpentine method,13–16 where the cells are directed into the traps with flow paths offering much smaller flow resistance than the main flow channel, and (2) the DiCarlo method,17–20 where the flow-through cup-like traps are densely positioned in a wide channel in large arrays. While the serpentine method is widely exploited due to its high trapping efficiency, the cells are prone to mechanical deformations particularly after being trapped due to the large contrast of pressure across a trapped cell, exerting continuous force on the membrane under perfusion.21 The effects of mechanical stress on the living cell physiology are extensively studied and reported by the interdisciplinary field of mechanotransduction.22,23 Therefore, it is crucial to minimize the imposed experimental mechanical stress to reproduce bulk cell experiments at the single-cell level on the microfluidic chip. Moreover, the clogging of the trap positions with cell debris, aggregates, or cells that are above the average cell dimension is another inhibiting factor of the serpentine method.16 Recent reports on the serpentine systems16,21 rely only on gravity to load the cells into the serpentine channels to avoid the mechanical stress. In one of these studies,16 the cell culture up to 24-h was successfully observed, but the array size was limited to 20 cells/device to avoid clogging. On the other hand, the DiCarlo method does not exert detrimental shear stress on the cells.17 Recent applications using DiCarlo method propose dense trap arrays with minimum possible distance between the arrayed trap structures to keep trapping efficiency high while tolerating clogging.19 Thus, experiments with these densely arrayed trapped cells are in the minute/hour scale, unsuitable for studies requiring time-dependent cell growth observation. Therefore, it is crucial to adapt the gentle DiCarlo method into a more sparsely arrayed design to enable day-scale, on-chip cell culture that allows for cell growth and migration observation without sacrificing trapping efficiency or clogging tolerance.
Here, we present a hybrid hydrodynamic cell trapping system of the two previously reported methods to address the demand for a facile and efficient microfluidic cell compartmentalization device that can be aligned on any substrate/chip of interest, enabling gentle manipulation and positioning of individual living cells. The proposed method allows for cell growth on the labelled microchamber arrays after cell trapping for day-scale culturing. In our design, traps, similar to DiCarlo's, are sparsely positioned in partially separated nanoliter chambers, which are serially located in serpentine-like channels. Relying on the stochastic nature of the cell arrival in the fluidic channel, and not on the high flow fraction into the trap, we carefully designed the trap positions to overcome the low trapping efficiency of each individual trap (13%), thus achieving a high system trapping efficiency (∼70%) and fast loading times (>90% fill factor reached in 1–4 min to fill 400-trap device). By doing so, we reduced the yield-degrading clogging and minimized mechanical stress on the cells during and after trapping, which allowed for cell viability in the day-scale incubation periods. Finally, the partially separated chambers allow for cell tracking during the static culturing period, as they stay well separated but free to migrate.
The high trapping efficiency was achieved with two crucial design steps. We first used the Comsol Multiphysics simulation tool to optimize the geometric parameters of the traps, taking into account cell dimension and hydrodynamic manipulation in the laminar flow regime. Subsequently, we performed Monte Carlo simulations to quantify the effects of the design parameters on the overall system trapping efficiency. We succeeded in experimentally reproducing the Monte Carlo efficiency estimations.
To verify the feasibility of single-cell culture in the fabricated devices, MIA PaCa-2 (pancreatic epithelial) cells were trapped and cultured in microfluidic platforms assembled on 6-well tissue culture plates. Based on the time-lapse data, the observed cell growth rates are comparable to the control bulk cell culture rates.
II. MATERIALS AND METHODS
A. Device fabrication
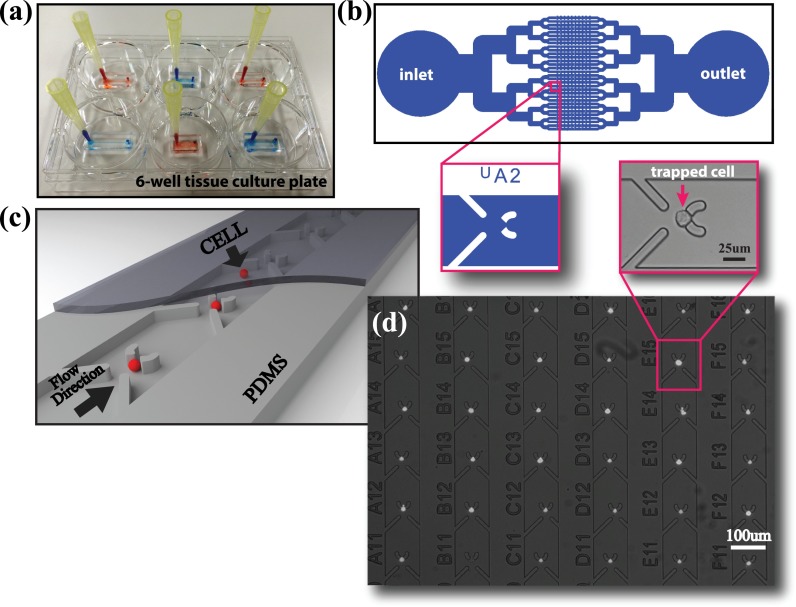
Single-level microfluidic cell isolation devices were fabricated using conventional Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning) micro-molding methods24 with total layer thickness of 2–3 mm. In order to adapt the use of PDMS for cell culture experiments, after curing the PDMS for 2 h in an 80 °C oven, we preserved the devices for 48 h at room temperature to complete the curing and eliminate the intrusion of uncrosslinked polymers into the cell culture medium.11 Next, we washed the cut and hole-punched PDMS devices in ethanol in an ultrasonic bath for 5 min to clean any small PDMS particles that may form during the punching and the cutting. We dried the devices with N2 gun and kept them in a vacuum chamber at 0.1 MPa for 15 min to allow the solvents to thoroughly evaporate from the pores of the PDMS. Finally, the PDMS microfluidic cell trapping devices were bonded either on cleaned and dried glass coverslips or 6-well tissue culture plates as seen in Fig. 1(a), which are commonly used for analytical research and are compatible with time-lapse experiments due to the abundance of multi-well holders for stage top incubators. While a single-step O2 plasma treatment on both the glass and the PDMS surfaces was sufficient to irreversibly bond PDMS on glass, the polystyrene culture dishes required preliminary treatment. They were surface modified using aminopropyltriethoxysilane (APTES) (10% in H2O)25 for 20 min, washed in MilliQ, N2 gun dried, and finally bonded to O2 plasma treated PDMS devices. Before cell loading, the devices were kept under UV for 1 h for sterilization, and the channels were pre-filled with sterile and filtered Phosphate-Buffered Saline (PBS) buffer.
FIG. 1.
B. Device description
The hydrodynamic cell trapping takes place on the structured parallel microfluidic channels connecting the inlet to the outlet, as shown in Fig. 1(b). The inlet and the outlet were designed to provide enough media to the cells during day-scale static cell culturing. A 3D rendered model of the hydrodynamic cell-trapping channel, where the cells are sequentially trapped in the parallel microchannels, is illustrated in Fig. 1(c) (Multimedia view). Although the method we propose does not inherently impose a limit on the number of cells in a single device, we worked with 10 traps/channel (320 traps/device), 25 traps/channel (400 traps/channel), and 50 traps/channel (400 traps/device) to identify the optimum trapping and culturing conditions.
The design of the trapping system was undertaken based on the following three figures of merit, which are, in general, also relevant in the performance evaluation of hydrodynamic cell trapping methods:
-
(1)
Fill Factor (FF) = The ratio of the total number of traps filled with a cell to the total number of available traps at any given time.
-
(2)
Trap Flux Fraction (TFF) = The ratio of the flux that passes through the trap to the total flux in a channel.
-
(3)
Trapping Efficiency (TE) = Ratio of the total trapped cells to the minimum inserted cells necessary to achieve a FF of 90%.
Our goal is to achieve high TE that does not rely solely on increasing the TFF of each trap, which may cause mechanical stress on the cells.
C. Cell culture, loading, and observation
We used the cell line MIA PaCa-2 (ATCC CRL-1420) to test the operation of the proposed device. The cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% Fetal Bovine Serum (FBS), 1% penicillin-streptomycin, 1% MEM Non-Essential Amino Acids Solution, and 2.5% 1 M Hepes and incubated in a humidified atmosphere of 5% CO2 at 37 °C. For the TE characterization experiments, it is crucial to know the concentration of the cell suspension to control the inserted cell number by varying the inserted volume. After the cell suspension was prepared, its concentration was identified by counting the cells multiple times using a hemocytometer and averaging. We inserted the cell suspension using a pipet and left the pipet tip attached on the inlet while removing the excess fluid from the outlet by a glass capillary tube attached to an aspirator, commonly used on the bio-lab benches. The time necessary to insert 10 μl of fluid is roughly 1, 2, and 4 min for 32, 16, and 8 parallel channel devices, respectively, yielding 3 × 10−3 m/s average flow velocity for 100 μm width and 16 μm height channels. After cell loading, 20 μl of media was added into the reservoir at the inlet, and the device is kept in the incubator for 1 h to make sure the cells adhere at the trap sites under this gravity induced mild perfusion and wash away all the cells through the microchannels with traps towards the outlet. Then, the FF was calculated by counting the trapped cells using microscope images taken on a standard inverted microscope, as shown in Fig. 1(d). For the day-scale static cell culture and time-lapse experiments, we filled the well with cell medium after cell loading process, covering the 2–3 mm height PDMS device while making sure there were no air bubbles stuck at the inlet and the outlet. During the on-chip cell incubation/observation period, no perfusion was applied into the cell-hosting microfluidic channels. The time-lapse experiments were performed on a stage-top incubator assembled to an inverted microscope, coupled to a CMOS Camera Module (CCM) (ASTEC Real time cell monitoring system) at 10× magnification. In a field of view of 490 μm × 650 μm, we were able to observe 12 trap sites per frame and, by defining multiple positions, extend the total number of observed chambers.
III. RESULTS AND DISCUSSION
A. Simulation results
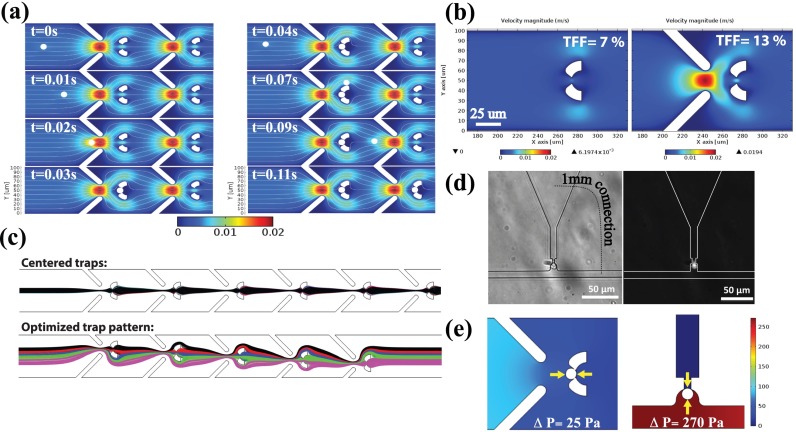
In the design of our cell trapping system, we used computational fluid dynamics simulations coupled with solid mechanics, implemented by the Finite Element Method (FEM)-based Comsol Multiphysics. Building the model, we adapted the previous work by Xu et al.26 The simulation results allowed us to predict the time dependent cell motion before trapping occurs, the flow velocity/flux distribution in the structured microfluidic channel, and the mechanical impact of the interaction of the cell with the fluid both before and after being trapped. Fig. 2(a) shows the time-lapse positions of a streaming cell being trapped when the trap is empty (left), and bypassing an occupied trap and being stopped at the adjacent trap site (right) (Multimedia view).
FIG. 2.
We optimized the geometry and the dimensions of the microfluidic system to achieve a high TE, while guaranteeing small mechanical deformation on the cells, short cell loading time to reach high FF, and high tolerance to clogging. The dimensions were determined considering average mammalian tissue cell dimensions with diameter in the range of 10–15 μm. The effect of the flow focusing “lead” structures was optimized to increase the TFF from 7% to 13%, increasing the hydrodynamic single cell trapping efficiency as shown in Fig. 2(b). Moreover, in order to further increase the overall trapping efficiency, we optimized the relative trap positions in the channel. In the laminar flow regime, the cells are expected to arrive in the channel stochastically with uniform probability in the transversal direction and follow the streamlines determined by the flow dynamics.27 Therefore, instead of all traps being centered, where the streamlines that pass through them overlap, the trap positions were varied so that a wider incoming flux range is covered. Fig. 2(c) shows the streamline patterns for the all-centered and optimized (±16, ±8, and 0 μm for a 100 μm width channel) trap positions, where the streamlines that pass through each trap are colored differently. With this configuration, the flux coverage in the channel increased from 13% to 44%. All the simulations were performed with an average channel inlet flow velocity of 3 × 10−3 m/s, which is approximately the average velocity used in the experiments.
While the FEM simulation study reveals the details of cell motion and the hydrodynamic trapping capacity of a single trap, it is not trivial to analytically compute the total system TE due to the stochastic nature of the incoming cells to be trapped sequentially in the channel. Therefore, using the parameters derived from the FEM simulation, we built a Monte Carlo model to estimate the TE of the system. Uniformly distributed pseudorandom numbers were first generated to represent the transversal arrival positions of the cells in the channel. Each “cell” is individually evaluated subject to the dynamically updated trap occupancy array. A positive trapping event occurs when the arriving cell position is within 6.5 μm (which represents 13% TFF in 100 μm channel) of the center of an empty trap, upon which the trap occupancy array is updated and the trapped cell counter is incremented. For each number of total inserted cells, we estimated the average FF of the system using a large iteration number (104). Finally, the overall system TE is calculated using the number of inserted cells that achieve a FF of 90%.
B. Experimental results
To quantify the low-stress characteristic of our device, we calculated the mechanical pressure the cell experiences at the trap positions and compared it with the serpentine method. Numerical simulation results revealed that the pressure difference across the trapped cell is ten times higher with the serpentine method than with our design, as presented in Fig. 2(e). Moreover, we designed and fabricated a serpentine trap whose TFF is similar to our TE (∼70%). Under similar loading conditions, we observed that the cells in the serpentine traps were wedged into the trap openings as illustrated in Fig. 2(d), causing mechanical deformation.
In the hydrodynamic trapping systems, the available volume at the trap location and the cell dimension regulates the number of cells per trap. We optimized the channel height in addition to minimizing the available area at the cell-hosting opening of the trap, to ensure the trapping of only a single cell. We experimentally investigated three different channels with approximately 16, 21, and 30 μm height and found out that in the ∼16 μm channel more than 90% of the traps are occupied by only single cells, as shown in Fig. 3(a).
FIG. 3.
Statistical evaluation of the device performance based on both simulation and experiment derived data. (a) Effect of the channel height on the number of cells trapped at a single trap position showing 16 μm channel height is optimum for single cell trapping. (b) Monte Carlo simulation estimates of the trapping efficiency based on the trap positions (centered or optimized) and number of traps/channel (10, 25, and 50 traps/ch). (c) Monte Carlo simulation (continuous lines) and experiment (scattered points) trends for 6 combinations of the trap numbers and positions in microfluidic channels, indicating the highest efficiency ∼70% on the optimized pattern, 50 trap/ch device type.
The TEs estimated using Monte Carlo simulations to compare the optimized and centered trap positions for three different numbers of traps per channel (10, 25, and 50) are demonstrated in Fig. 3(b). According to the simulations, the TE of the optimized trap pattern should be significantly higher than that of the centered scheme, as expected. Moreover, the TE is estimated to increase for both patterns as the number of traps per channel increases. This can be explained by the higher interaction chance between the incoming cell and an empty trap. Furthermore, we characterized every combination of the centered and optimized trap patterns and the number of traps per channel and presented the data in Fig. 3(c). For each device type, we experimentally determined the FF for 15 different numbers of inserted cells to verify the estimated efficiency profiles. Despite the challenges in accurately calculating the inserted cell numbers from a cell suspension, the experiment and the simulation results showed strong agreement, which validates the accuracy of the design parameters and estimation precision. We consistently obtained ∼70% TE with the optimized and 50 trap/channel device.
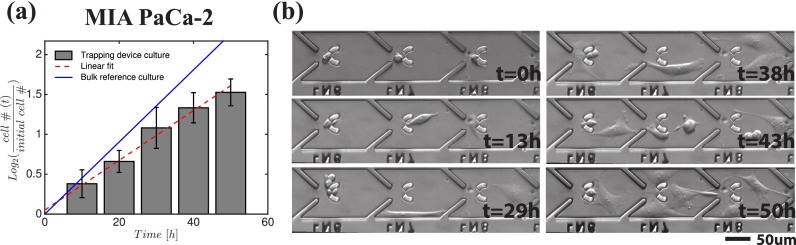
As a proof of principle, we evaluated the cell culture conditions in our device based on the proliferation rate of the cells. The MIA PaCa-2 cell division times were extracted from the time-lapse images of 73 single-cell hosting traps collected every 15 min for 50 h from the microfluidic devices bonded on a 6-well tissue culture plate. In Fig. 4(a), each bar shows the 10-h binned average proliferation rates along with its standard deviation, and the linearly fitted line approximates the average doubling frequency as 0.8 day−1. As a control experiment, we seeded 6.3 × 104 MIA PaCa-2 cells directly in 6-well plates so that each cell has 1.5 × 10−8 m2 area (similar to the area of one trap chamber in the microfluidic device), and calculated the average doubling frequency in 48 h as 1.1 day−1, also plotted in Fig. 4(a) for comparison. The small difference in the doubling rates can be accounted by unequal conditions between the control and on-chip cell culture experiments, such as medium access and cell-to-cell interactions. Fig. 4(b) (Multimedia view) shows six representative pictures from the time-lapse observation of MIA PaCa-2 cells, where the temporal behavior of the cells in the trap chambers can be seen in detail, particularly typical cell migration, cell-cell interaction, and the cell division events can be observed.
FIG. 4.
For the purpose of this study, we built a device that can trap and host at most 400 cells, but it is important to note that our method was designed to be highly scalable (∼thousands). Moreover, longer incubation periods can be achieved by standard medium exchange protocols.
IV. CONCLUSIONS
In response to the growing need for manipulating single cells in microfluidic platforms, we designed, fabricated, and verified hydrodynamic single cell trapping and culturing devices. Our main goals were simplicity, high trapping efficiency, low mechanical intervention, short cell-loading time, long-term microscopic observation compatible with stage-top incubators, and biological experimentation compatibility to facilitate the use of the method with standard bio-lab tools. We achieved ∼70% trapping efficiency leveraging the stochastic nature of the cell arrivals in a microfluidic channel. Our average cell loading times were in minute scale (1–4 min). We cultured ∼400 isolated single cells per device and monitored them using a stage-top incubator microscope. We did not observe any viability issue during the day-scale (∼2 days) static cell culture periods. Moreover, we measured the time dependent proliferation rate of 73 individual MIA PaCa-2 cells (doubling frequency = 0.8 day−1), which was comparable with the control bulk cell culture proliferation rate (doubling frequency = 1.1 day−1). We are convinced that our single cell microarraying method will stimulate and open the way to the studies of many biological questions in life sciences, particularly in the cell-cell and cell-microenvironment interaction, and cell heterogeneity research.
ACKNOWLEDGMENTS
This research was partially supported by the European Commission funded NanoVista Project (FP7-ICT-2011-7, under Grant Agreement No. 288263) and EUJO-LIMMS Project (No. 295089). The authors would like to acknowledge the support of Professor Renaud (EPFL STI IMT LMIS4) and Dr. Buchanan (EPFL SV IBI-SV LLCB) for their invaluable contribution to this paper.
References
- 1. Adutler-Lieber S., Zaretsky I., Platzman I., Deeg J., Friedman N., Spatz J. P., and Geiger B., J. Autoimmun. 54, 100 (2014). 10.1016/j.jaut.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 2. Mu X., Zheng W., Sun J., Zhang W., and Jiang X., Small 9, 9 (2013). 10.1002/smll.201200996 [DOI] [PubMed] [Google Scholar]
- 3. Lautenschläger F. and Piel M., Curr. Opin. Cell Biol. 25, 116 (2013). 10.1016/j.ceb.2012.10.017 [DOI] [PubMed] [Google Scholar]
- 4. Sackmann E. K., Fulton A. L., and Beebe D. J., Nature 507, 181 (2014). 10.1038/nature13118 [DOI] [PubMed] [Google Scholar]
- 5. Nguyen T. A., Yin T.-I., Reyes D., and Urban G. A., Anal. Chem. 85, 11068 (2013). 10.1021/ac402761s [DOI] [PubMed] [Google Scholar]
- 6. Grünberger A., Wiechert W., and Kohlheyer D., Curr. Opin. Biotechnol. 29, 15 (2014). 10.1016/j.copbio.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 7. Love K. R., Bagh S., Choi J., and Love J. C., Trends Biotechnol. 31, 280 (2013). 10.1016/j.tibtech.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 8. Yin H. and Marshall D., Curr. Opin. Biotechnol. 23, 110 (2012). 10.1016/j.copbio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 9. White A. K., VanInsberghe M., Petriv I., Hamidi M., Sikorski D., Marra M. A., Piret J., Aparicio S., and Hansen C. L., Proc. Natl. Acad. Sci. 108, 13999 (2011). 10.1073/pnas.1019446108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weaver W. M., Tseng P., Kunze A., Masaeli M., Chung A. J., Dudani J. S., Kittur H., Kulkarni R. P., and Di Carlo D., Curr. Opin. Biotechnol. 25, 114 (2014). 10.1016/j.copbio.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halldorsson S., Lucumi E., Gómez-Sjöberg R., and Fleming R. M. T., Biosens. Bioelectron. 63, 218 (2015). 10.1016/j.bios.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 12. Karimi A., Yazdi S., and Ardekani A. M., Biomicrofluidics 7, 021501 (2013). 10.1063/1.4799787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan W.-H. and Takeuchi S., Proc. Natl. Acad. Sci. 104, 1146 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobel S., Valero A., Latt J., Renaud P., and Lutolf M., Lab Chip 10, 857 (2010). 10.1039/b918055a [DOI] [PubMed] [Google Scholar]
- 15. Chung J., Kim Y.-J., and Yoon E., Appl. Phys. Lett. 98, 123701 (2011). 10.1063/1.3565236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y.-C., Allen S. G., Ingram P. N., Buckanovich R., Merajver S. D., and Yoon E., Sci. Rep. 5, 9980 (2015). 10.1038/srep09980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlo D. D., Wu L. Y., and Lee L. P., Lab Chip 6, 1445 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- 18. Kim M.-C., Wang Z., Lam R. H. W., and Thorsen T., J. Appl. Phys. 103, 044701 (2008). 10.1063/1.2840059 [DOI] [Google Scholar]
- 19. Dura B., Dougan S. K., Barisa M., Hoehl M. M., Lo C. T., Ploegh H. L., and Voldman J., Nat. Commun. 6, 5940 (2015). 10.1038/ncomms6940 [DOI] [PubMed] [Google Scholar]
- 20. Huebner A., Bratton D., Whyte G., Yang M., deMello A. J., Abell C., and Hollfelder F., Lab Chip 9, 692 (2009). 10.1039/B813709A [DOI] [PubMed] [Google Scholar]
- 21. Jin D., Deng B., Li J. X., Cai W., Tu L., Chen J., Wu Q., and Wang W. H., Biomicrofluidics 9, 014101 (2015). 10.1063/1.4905428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iskratsch T., Wolfenson H., and Sheetz M. P., Nat. Rev. Mol. Cell Biol. 15, 825 (2014). 10.1038/nrm3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shemesh J., Jalilian I., Shi A., Yeoh G. H., Tate M. L. K., and Warkiani M. E., Lab Chip 15, 4114 (2015). 10.1039/C5LC00633C [DOI] [PubMed] [Google Scholar]
- 24. Xia Y. and Whitesides G. M., Annu. Rev. Mater. Sci. 28, 153 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- 25. Sunkara V., Park D.-K., Hwang H., Chantiwas R., Soper S. A., and Cho Y.-K., Lab Chip 11, 962 (2011). 10.1039/C0LC00272K [DOI] [PubMed] [Google Scholar]
- 26. Xu X., Li Z., and Nehorai A., Biomicrofluidics 7, 054108 (2013). 10.1063/1.4822030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan A., Shenoy A., Smith R., and Li Z., Biomicrofluidics 9, 024103 (2015). 10.1063/1.4914469 [DOI] [PMC free article] [PubMed] [Google Scholar]