Abstract
Background and Purpose
The 5‐HT transporter (SERT) is a target for antidepressant drugs. SERT possesses two binding sites: the orthosteric (S1) binding site, which is the presumed target for current SERT inhibitors, and an allosteric (S2) site for which potential therapeutic effects are unknown. The antidepressant drug citalopram displays high‐affinity S1 binding and low‐affinity S2 binding. To elucidate a possible therapeutic role of allosteric inhibition of SERT, a drug that specifically targets the allosteric site is required. The purpose of this study was to find a compound having higher selectivity towards the S2 site.
Experimental Approach
We performed a systematic structure–activity relationship study based on the scaffold of citalopram and the structurally closely related congener, talopram, which shows low‐affinity S1 binding in SERT. The role of the four chemical substituents, which distinguish citalopram from talopram in conferring selectivity towards the S1 and S2 site, respectively, was assessed by determining the binding of 14 citalopram/talopram analogous to the S1 and S2 binding sites in SERT using membranes of COS7 cells transiently expressing SERT.
Key Results
The structure–activity relationship study revealed that dimethyl citalopram possesses the highest affinity for the allosteric site relative to the S1 site in SERT and has approximately twofold selectivity for the allosteric site relative to the S1 site in SERT.
Conclusions and Implications
The compound could be a useful lead for future synthesis of drugs with high affinity and high selectivity towards the allosteric binding site.
Abbreviations
- SERT
serotonin transporter
- DAT
dopamine transporter
- NET
norepinephrine transporter
- 5‐HT
serotonin
- S‐CIT
Escitalopram
- SSRI
selective serotonin reuptake inhibitor
- SAR
structure‐activity relationship
- cpd
compound
- TM
transmembrane segment
- RT
room temperature
Tables of Links
These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Pawson et al., 2014) and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (Alexander et al., 2015).
Introduction
The serotonergic system controls a variety of functions in the human psychology and physiology such as mood, sleep, sexual drive and appetite (Mohammad‐Zadeh et al., 2008). One of the main regulators of synaptic 5‐hydroxytryptaminergic transmission is the 5‐HT transporter, SERT. SERT mediates reuptake of released 5‐HT into the presynaptic terminal and thereby terminates the synaptic signal. The process also ensures a replenishment of the intracellular 5‐HT stores. The important physiological role of SERT in 5‐HT signalling is substantiated by the fact that drugs targeting SERT are used in the treatment of a variety of diseases such as depression, anxiety, obsessive–compulsive disorder and neuropathic pain (Kristensen et al., 2011). Examples of the most widely prescribed selective 5‐HT reuptake inhibitors (SSRIs) include citalopram (Cipramil), the active (S)‐enantiomer of citalopram, escitalopram (Cipralex/Lexapro), fluoxetine (Prozac), sertraline (Zoloft) and paroxetine (Seroxat). Despite the pharmacological importance of SERT, there is still no direct structural information available for this protein.
SERT belongs to the family of neurotransmitter: sodium symporters (NSS) that also includes transporters for dopamine and noradrenaline as well as numerous bacterial homologues (Saier et al., 2006). The structure of three NSS proteins have been solved: the amino acid transporter, LeuT, from Aquifex aeolicus (Yamashita et al., 2005), the dopamine transporter from Drosophila melanogaster, dDAT (Penmatsa et al., 2013), and the hydrophobic l‐amino acid transporter, MhsT, from Bacillus halodurans (Malinauskaite et al., 2014). The X‐ray crystal structures of these proteins have provided much information about structural aspects of NSS proteins in general and have enabled the generation of reliable homology models of SERT (Celik et al., 2008; Andersen et al., 2010; Plenge et al., 2012; Andersen et al., 2014) and other NSS proteins (Beuming et al., 2008; Skovstrup et al., 2010; Hill et al., 2011; Stolzenberg et al., 2015). The structures of LeuT and dDAT are remarkably similar and revealed a protein with 12 transmembrane segments (TMs) organized in a Y‐shaped barrel‐like structure with a binding site for substrate, the S1 site, located in the centre of the protein (Yamashita et al., 2005; Penmatsa et al., 2013). The dDAT structure has been solved in complex with dopamine, the tricyclic antidepressants nortriptyline, reboxetine and nisoxetine as well as illicit drugs such as cocaine and amphetamine (Penmatsa et al., 2013; Penmatsa et al., 2015; Wang et al., 2015). Drug binding was in all cases located to the S1 site. This, together with data from crystal structures of LeuT mutants (Wang et al., 2013), molecular docking models of eukaryotic NSS (Beuming et al., 2008; Celik et al., 2008; Andersen et al., 2010; Skovstrup et al., 2010), and mutagenesis studies (Henry et al., 2006; Andersen et al., 2009; Sinning et al., 2010; Andersen et al., 2011), have provided compelling evidence that in NSS proteins, the primary high affinity binding site for inhibitors is the S1 site within the substrate binding pocket.
In addition to the S1 site, the presence of an allosteric binding site (denoted S2 site) in SERT has been suggested (Wennogle and Meyerson, 1982; Plenge and Mellerup, 1985). We have obtained evidence that in SERT, this allosteric site is located at the extracellular vestibule of the transporter (Plenge et al., 2012) and thus in the same location as the low‐affinity binding site for antidepressants identified in LeuT (Singh et al., 2007; Zhou et al., 2007; Zhou et al., 2009). Notably, this site has also been suggested to be a possible second substrate binding site in LeuT (Shi et al., 2008). The binding of ligands to the allosteric site is characterized by impeded dissociation of a pre‐bound S1‐bound ligand, for example, the application of escitalopram (S‐CIT) can cause a dose‐dependent impairment of dissociation of pre‐bound [3H]‐escitalopram ([3H]‐S‐CIT), [3H]‐paroxetine and the cocaine analogue [125I]‐RTI‐55 (see Chen et al. (2005b for a detailed description of allosteric SERT ligands). Clomipramine can also inhibit the dissociation of [3H]S‐CIT (Plenge et al., 2012). A common finding for most published allosteric ligands to date is that (i) they bind to the allosteric site with low potency, typically in the micromolar range (Chen et al., 2005a, 2005b; Banala et al., 2013; Plenge et al., 2012), and (ii) they bind to the S1 site with considerably higher affinity, typically within the low nanomolar range. The most potent allosteric ligand reported so far is S‐CIT, which impairs [3H]‐S‐CIT dissociation with an IC50 value of ~5 μM (Plenge et al., 2012). Nonetheless, S‐CIT binds to the S1 site with an affinity around 1 nM (Owens et al., 2001), conferring >1000‐fold selectivity towards the S1 relative to the S2 site and complicates attempts to delineate any possible pharmacological effect of S2 binding using S‐CIT. Clearly, a compound with S2 selectivity is needed for further investigating the significance of inhibiting the S2 site.
The pharmacological implications of allosteric inhibition of SERT are largely unknown, mainly due to the current lack of ligands possessing selectivity and high affinity for the allosteric site. It has been suggested that the higher efficacy and faster onset of action by S‐CIT, relative to racemic citalopram, is due to its dual action at the two binding sites (Storustovu et al., 2004; Sanchez, 2006). However, in vivo microdialysis on transgenic mice expressing the human SERT failed to show any decrease in S‐CIT‐induced 5‐HT elevation by R‐citalopram (Jacobsen et al., 2014). To examine the effect of inhibitor binding to the allosteric S2 site, it is necessary to develop a ligand with both high affinity and selectivity towards the site relative to other binding sites at SERT. To the best of our knowledge, so far, no ligands have been demonstrated with preference for the S2 site over the S1 site.
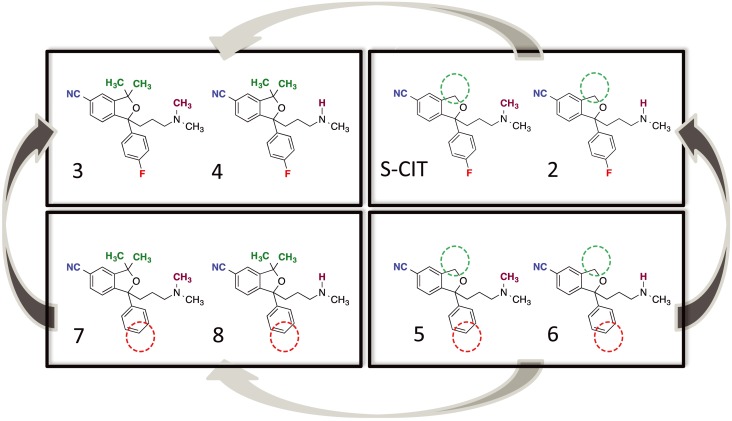
Here, we attempted to identify compounds having higher S2 selectivity by (i) decreasing S1 affinity and (ii) characterizing the molecular determinants, conferring affinity for the allosteric binding site in order to increase S2 affinity within the series. To this end, we took advantage of the fact that the citalopram analogue, talopram, only possesses SERT S1 affinity in the micromolar range. Citalopram and talopram can be distinguished by their substituent at only four positions, and we systematically assessed S1 and S2 binding ratios for compounds comprising all possible combinations of the differing substituents (Figure 1). Our results have identified a compound conferring selectivity (approximately twofold) for the S2 site relative to S1. This could be used as basis for the generation of additional compounds with high affinity and selectivity towards the S2 binding site.
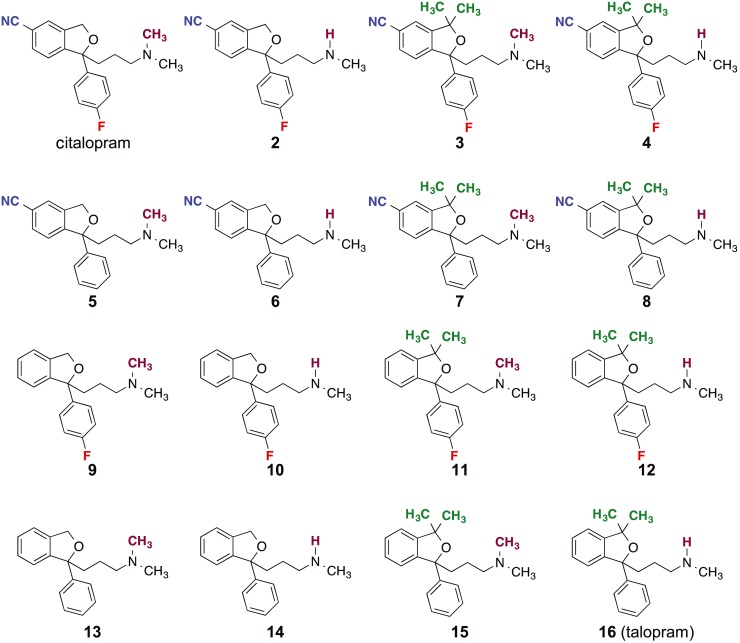
Figure 1.
Chemical structures of the compounds investigated in the SERT binding in this study. The compounds are citalopram, talopram and the 14 different possible combinations of the four substituents that distinguish citalopram from talopram. The differing substituents are coloured for clarity: the cyano‐group (blue), the dimethyl group (green), the fluorine atom (red) and the N‐methyl group (purple).
Methods
Site‐directed mutagenesis
hSERT was cloned into the pUbi1z vector using NotI/XbaI. Site‐directed mutagenesis was generated using either QuickChange PCR (adapted from Stratagene, San Diego, CA, USA) or ordered via GeneArt (ThermoFisher Scientific, Waltham, MA, USA). All SERT mutants were sequenced for confirmation.
Cell cultivation
COS‐7 cells were cultured in 175 cm2 flasks in 20 mL supplemented DMEM [1885, 1 g·L−1 glucose and 0.1 g·L−1 sodium‐pyruvate, 44 mM NaHCO3, 10% FBS, 2 mM l‐glutamine and penicillin (2000 IU penicillin)/streptomycin (5 mg·mL)] respectively. Cells were detached with 5 mM EDTA in PBS (pH 7.4), resuspended in DMEM 1885 and seeded into 175 cm2 flasks at a density of 6 × 106 cells per flask. The following day, the cells were transfected with a mixture if Opti‐MEM®, Lipofectamine™ 2000 regent (Invitrogen, Carlsbad, CA, USA) and pUbi1z‐hSERT plasmid following the manufacturer's instructions. The cells were harvested 3 days after transfection.
Membrane preparation
Cells were detached with 5 mM EDTA in PBS and pooled in 50 mL tubes. The suspensions were spun down at 4700 r.p.m. for 5 min at 4°C and subsequently resuspended in binding buffer (25 mM HEPES, 120 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, 1.2 mM MgSO4, 1 mM ascorbic acid and 5 mM glucose). Cells were lysed with a single shock ultrasound burst (Branson Sonifier 250, output 4) and pelleted at 4700 r.p.m. for 10 min at 4°C. The pellets were resuspended in binding buffer containing 0.3 M sucrose and homogenized using a syringe with a 21G 0.8 mm needle. The cell–membrane solutions were stored at −80°C until further use.
[3H]‐S‐CIT equilibrium binding experiments
[3H]‐S‐CIT was diluted in binding buffer to a final concentration of 5 nM. S‐CIT or one of the analogues was diluted using factor three dilution into final concentrations of 10 determinations spanning from 10−4 to 10−10 M. The ligands were added consecutively from low to high concentrations to the reaction tubes. All concentrations were added in triplicates. Binding buffer was used to measure maximal binding, while non‐specific binding was measured in the presence of paroxetine (10 μM). Membrane fragments were resuspended in binding buffer and transferred to a 96‐well plate (Almeco® Titerblock 96 deepwell Block PP square wells 1 mL) together with tracer and competing compound in a total volume of 400 μL. The assay was incubated for 2 h at room temperature. Binding was stopped by rapid loading of samples onto a 96‐well filter plate (Printed Filtermat B, Wallac; PerkinElmer, Waltham, MA, USA) soaked with 0.5% poly(ethyleneimine), using a 96‐well harvester (Tomtec Harvester 96 MACH III). Immediately after transfer to the filter, the samples were washed with ice‐cold 0.2 M NaCl buffer for 20 s and dried for 15 min at 90°C, followed by the addition of a scintillation plate (MeltiLex™ B/HS 1450–442; PerkinElmer). The filter was subsequently counted on a Microbeta2™ 2450 microplate counter (PerkinElmer). Competition binding were determined in at least three independent experiments.
[3H]‐S‐CIT dissociation rate assay
Membrane fragments expressing WT or mutant SERT were resuspended in binding buffer, with the addition of [3H]‐S‐CIT (9–12 nM), transferred to a 96‐well plate (sample size: 50 μL), placed in a water bath and kept at a stable temperature until binding equilibrium was obtained (approximately 30 min at room temperature). S‐CIT or one of the analogues was diluted in binding buffer with 1 μM paroxetine (to inhibit re‐association of [3H]‐S‐CIT). Note that paroxetine has no allosteric effect on [3H]‐S‐CIT binding (Plenge et al., 2012). Five hundred microlitres of the diluted compound was added to six consecutive reaction tubes, and the dissociation was assessed over a time period of 80 min. Free dissociation was determined using binding buffer containing 1 μM paroxetine. The assay was terminated by transfer of the samples onto a 96‐well filter plate followed by an immediate wash with ice‐cold NaCl buffer and counted as described for the binding experiments. Non‐specific binding was determined with 10 μM paroxetine at 37°C before adding [3H]‐S‐CIT. To determine a concentration‐dependent effect on the [3H]‐S‐CIT dissociation rate by the allosteric ligand, the dissociation experiment was performed with five concentrations of each ligand spanning from 100 to 1.2 μM following a consecutive factor three dilution row. The temperature for each mutant was adjusted for optimal measurement of [3H]‐S‐CIT dissociation rate. Dissociation rates were determined in at least three independent experiments.
Compounds
The compounds used herein were synthesized as described previously (Eildal et al., 2008).
Compliance with design and statistical analysis requirements
The experimental design was randomized by performing the experiments in a non‐systematic order with respect to the compounds or mutants investigated. To minimize bias, data analysis was fully automated from retrieving data from scintillation counting to regression analysis. An uneven sample size is performed for the initial screen of compounds in Table 1. This is because, due to the high temperature dependence, a control [no compound (cpd)] dissociation was included in every experiment for comparison. Accordingly, the sample size became higher for this determination. For assessment of the compounds' allosteric potency (Figure 3 and Table 2), the effect on [3H]‐S‐CIT dissociation rate was determined for five compound concentrations. For every concentration, the dissociation rate constant was determined with six time points performed in duplicates, that is, 12 data points, and a mean k [cpd] was determined. This was then performed three independent times. For [3H]‐S‐CIT competitive binding, all experiments were performed at least three times, each with triplicate determination. The SEM is calculated based on the means from the triplicates within each experiment. For anova testing, a significant F ratio and homogeneity of sample variance were required to progress to post hoc tests. In Figure 3, an LSD post hoc test was chosen for assessment of significance between S‐CIT and compounds 3 and 4: due to the experimental setup, a higher uncertainty is associated with compounds having a high IC50 value (i.e. cpd #5, 6, 7 and 8). In a multiple comparisons test, the low‐affinity compounds will influence the statistical power of the high‐affinity compounds. When data normalization was performed, 100%, or 1, is defined as the top plateau of the regression analysis on the individual Datasets. zero is defined as the non‐specific binding.
Table 1.
Screening of allosteric effect of citalopram analogues on [3H]‐S‐CIT dissociation
| Cpd (50 μM) | t 1/2 for [ 3 H ]S‐CIT dissociation (min) | n |
|---|---|---|
| Control | 85 ± 6.9 | 10 |
| S‐CIT | 790 ± 160 | 4 |
| 2 | 380 ± 26 | 4 |
| 3 | 1090 ± 130 | 4 |
| 4 | 1170 ± 240 | 4 |
| 5 | 280 ± 70 | 4 |
| 6 | 260 ± 20 | 4 |
| 7 | 440 ± 30 | 4 |
| 8 | 570 ± 70 | 4 |
| 9 | 120 ± 10 | 4 |
| 10 | 110 ± 9.1 | 4 |
| 11 | 150 ± 18 | 4 |
| 12 | 150 ± 9.4 | 4 |
| 13 | 89 ± 6.5 | 4 |
| 14 | 85 ± 3.0 | 4 |
| 15 | 100 ± 15 | 4 |
| Talopram | 100 ± 3.8 | 4 |
Effect on t 1/2 (min) for [3H]‐S‐CIT dissociation by 50 μM of the indicated cpds. The t 1/2 values were calculated by non‐linear regression analysis of prebound [3H]‐S‐CIT dissociation on membrane preparations from COS7 cells transiently expressing SERT. Data are shown as mean ± SEM of n number of individual experiments. Due to the high temperature sensitivity of the assay, a control sample was always performed in parallel to the compounds, hence the high sample size for the control.
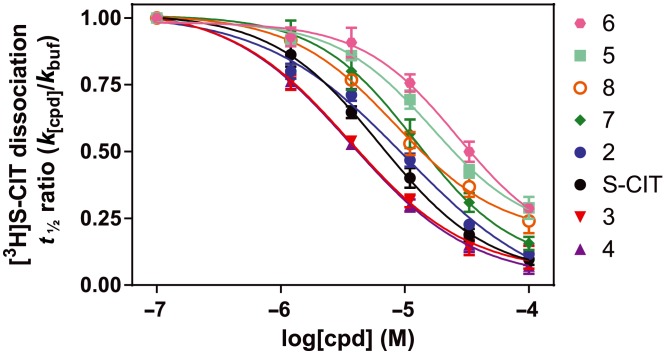
Figure 3.
Allosteric potency of the investigated cyano‐containing compounds. The allosteric potency is measured as the ability of a compound, in five dilutions ranging from 100 to 1.2 μM added at six different time points, to impede the dissociation of prebound [3H]‐S‐CIT from COS7 membranes transiently expressing the human SERT. The seven analogues and S‐CIT follow a pairing trend composed of compounds 3 and 4 (shown in red and purple triangles respectively), which both exhibited a higher allosteric potency than S‐CIT and 2 (black and blue circles respectively), followed by 7 and 8 (green diamonds and orange open circles) and lastly 5 and 6 (cyan squares and pink circles). This trend coincided with the different compound substituents starting with compounds 3 and 4, which both contain phenyl‐attached fluorine and the furan ring dimethyls. This is followed by S‐CIT and 2 (phenyl‐attached fluorine), 7 and 8 (furan di‐methyls) and ends with compound 5 and 6, which lack both the fluorine and the dimethyls. The data were calculated as [3H]‐S‐CIT dissociation rate in the presence of S‐CIT or an analogue (k [cpd]) relative to the [3H]‐S‐CIT dissociation rate in paroxetine buffer (k buf) plotted as a function of the added concentrations of allosteric bound compound. Each data point is an assessment of the t 1/2 of the compound at the indicated concentration, carried out as shown in Figure 1B with six time determinations performed in duplicate. Data are means with SEM (error bars), each based on three independent experiments.
Table 2.
IC50 values for [3H]‐S‐CIT displacement and allosteric potency of S‐CIT and the cyano‐containing analogues
| Cpd | SERT S1 binding IC 50 (μM) | n | SERT allosteric potency IC 50 (μM) | n | S2: S1 ratio |
|---|---|---|---|---|---|
| S‐CIT | 0.010 [0.008; 0.013] | 3 | 5.8 [5.4; 6.3] | 3 | 580 |
| 2 | 0.041 [0.032; 0.051] | 4 | 10.1 [10.0; 10.2] | 3 | 250 |
| 3 | 6.4 [4.7; 8.8] | 4 | 3.6 [3.3; 3.8] | 3 | 0.56 |
| 4 | 0.50 [0.42; 0.58] | 4 | 3.6 [3.5; 3.7] | 3 | 7.2 |
| 5 | 0.16 [0.14; 0.18] | 4 | 17 [16; 18] | 3 | 110 |
| 6 | 0.11 [0.09; 0.13] | 4 | 27 [25; 29] | 3 | 250 |
| 7 | 10 [9.2; 12] | 4 | 12 [10; 13] | 3 | 1.1 |
| 8 | 1.4 [1.0; 2.0] | 4 | 8.8 [6.3; 12] | 3 | 6.3 |
IC50 values were calculated by non‐linear regression analysis either from [3H]‐S‐CIT binding inhibition assays (S1 binding) or from [3H]‐S‐CIT dissociation experiments (S2 binding) in the presence of increasing concentrations of the indicated compound. Data are shown as mean values calculated from means of pIC50 and the SEM interval from pIC50 ± SEM of n independent experiments performed in triplicates for the assessment of S1 binding affinity. See Methods section for the determination and sample size of allosteric potency. Experiments were performed on membrane preparations from COS7 cells transiently expressing SERT.
Data calculations
Dissociation rate constants (k [cpd]) at different S‐CIT or analogue concentrations were calculated and normalized in relation to the dissociation rate constant of unhindered radioligand dissociation in buffer (k buf). The dissociation rate ratio (k [cpd]/k buf) was then plotted as a function of the added concentration of allosteric bound compound. IC50 values, or the allosteric potency, were determined as compound concentration required to impair the dissociation rate by 50% relative to no compound present and were calculated from the normalized dissociation ratio (k [cpd]/k buf) versus the logarithmic concentration of the compound. Note that when calculating the allosteric potency, the ratio between k [cpd] and k buf was used. This eliminates the temperature‐dependency of the dissociation rate constants. Data are shown as mean values calculated from means of pIC50 and the (SEM interval) from pIC50 ± SEM. All data were processed with regression analysis using Prism 6.0 (GraphPad Software Inc., La Jolla, CA, USA). The data and statistical analysis comply with British Journal of Pharmacology guidelines (Curtis et al., 2015).
Results
Screening of citalopram/talopram analogues for allosteric binding properties
Higher selectivity towards the S2 site relative to the S1 site can be achieved by decreasing S1 affinity while maintaining or increasing S2 or vice versa. It is well established that minor modifications of S‐CIT causes a dramatic loss in S1 affinity, for example, by measuring the displacement of [125I]‐RTI‐55 (Andersen et al., 2011). The S‐CIT analogue talopram, for example, has approximately 1000‐fold lower affinity for SERT than S‐CIT (Andersen et al., 2011). In contrast, the search for ligands that bind to the S2 site has so far only provided low‐affinity ligands, and the S2 site seems promiscuous and favours weak binding as a diverse set of compounds show similar activity at the site (Chen et al., 2005b; Plenge et al., 2012; Banala et al., 2013). Thus, to potentially find a compound with similar or improved allosteric potency relative to S‐CIT, but without high affinity for the S1 site, we tested a series of citalopram analogues (Figure 1) that constituted all possible combinations of substituents between citalopram and talopram, displaying from high to low SERT S1 affinity (Eildal et al., 2008; Andersen et al., 2011). Furthermore, we envisioned that characterization of this compound selection might identify the molecular determinants responsible for the allosteric effect of S‐CIT on SERT. Accordingly, we assessed (i) allosteric effect at the S2 site by measuring the ability of the analogues to inhibit dissociation of prebound [3H]‐S‐CIT and (ii) S1 binding affinity for all analogous by measurement of their competitive displacement of [3H]‐S‐CIT. To assess the allosteric effect, we prepared membranes of COS‐7 cells transiently expressing the human SERT and pre‐incubated the membranes with [3H]‐S‐CIT. The samples were subsequently diluted 13 times to decrease the concentration of unbound [3H]‐S‐CIT, and a high concentration (50 μM) of S‐CIT, talopram or an analogue (cpds 2–15) was added. To block [3H]‐S‐CIT re‐association, paroxetine (1 μM) was added. Note that paroxetine has no allosteric effect on [3H]‐S‐CIT binding (Plenge et al., 2012). The dissociation rate was assessed by terminating the reaction at the indicated times and measuring remaining bound [3H]S‐CIT (see Methods section for details). The dissociation rates for the 16 compounds are summarized in Figure 2.
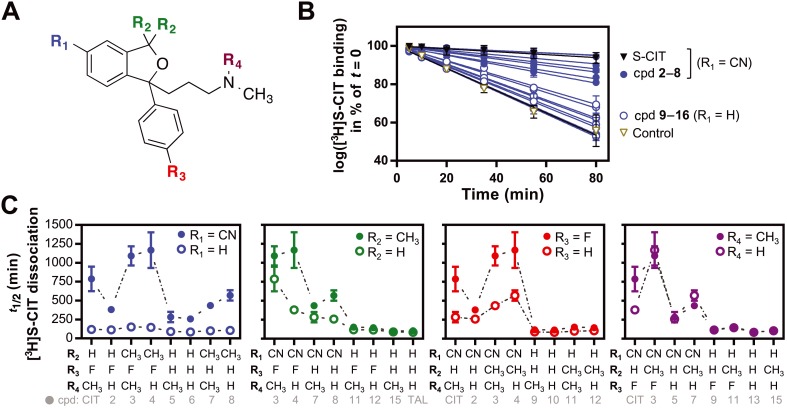
Figure 2.
Allosteric effect of the 16 compounds consisting of all possible substituents between citalopram and talopram. (A) Depiction of the four substituents, constituting the 16 tested compounds. The variables, R1 to R4, are colour coded for clarity. The colour coding is consistent in (B) and (C). (B) Allosteric effect of the 15 compounds divided into the ones containing the cyano‐group (cpd 2–8) and not containing a cyano‐group (cpd 19–16). Depicted is the inhibitory effect of 50 μM of the compound on the dissociation of [3H]‐S‐CIT bound to membranes of COS7 cells transiently expressing the human SERT. S‐CIT and control data are shown for comparison. (C) Effect of the different substituents on the t 1/2 of [3H]‐S‐CIT dissociation. The difference in t 1/2 (min) of each substituent (filled circles) relative to hydrogen (open circles) at the same position and then related to the combinations of substituents on the other positions. Panels from far left: R1, cyano‐group (blue). Middle left: R2, dimethyl group (green). Middle right: R3, fluor atom (red). Far right: R4, methyl group (purple). The bottom line in grey refers to the compound depicted with filled circle. Data are shown as means ± SEM of 4 to 10 individual experiments (Table 1).
Only the compounds containing the cyano‐group, including S‐CIT itself and cpds 2–8, showed a significant (P < 0.05, one‐way anova with Dunnett's multiple comparisons test) increase in t 1/2 relative to the control dissociation and thereby any allosteric activity (Figure 2B). On average, the compounds showed a fivefold increase in the inhibition of the [3H]‐S‐CIT off‐rate relative to their congeners not containing the cyano‐group (i.e. S‐CIT vs. cpd 9; 2 vs. 10; 3 vs. 11; 4 vs. 12; 5 vs. 13; 6 vs. 14; 7 vs. 15 and 8 vs. talopram, Figure 2C, far left panel). A similar comparison of the compounds with or without the phthalane dimethyl substituent (cpd 3 vs. S‐CIT; 4 vs. 2; 7 vs. 5; 8 vs. 6; 11 vs. 9; 12 vs. 10; 15 vs. 13 and talopram vs. 14, Figure 2C, middle left panel) showed that the dimethyl substituents cause a decreased [3H]‐S‐CIT dissociation rate, if they also contain the cyano‐group, relative to the analogues with hydrogens in the same position. The effect was modest with ~1.5‐fold (Table 1) decrease in dissociation rate. A comparison of the compounds with and without the fluorine atom (S‐CIT vs. cpd 5; 4 vs. 8; 2 vs. 16; 12 vs. talopram and 10 vs. 14, Figure 2C, middle right panel) showed that a fluorine attached to the phenyl ring causes an approximately twofold decrease in the [3H]‐S‐CIT dissociation rate (Table 1) compared with compounds not containing the fluorine. Here, the difference is also dependent on the presence of the cyano‐group. In contrast, a comparison of compounds, which differ in the number of N‐methyl substituents (mono or dimethyl), showed an almost complete overlap in dissociation rates for all pairs (S‐CIT vs. cpd 2; 3 vs. 4; 5 vs. 6; 11 vs. 12 and 13 vs. 14, Figure 2C, far right panel). Only compound 2 had a faster dissociation rate than its congener.
Taken together, the initial structure–activity relationship (SAR) screen on compounds with systematic combinations of the four substituents that distinguish S‐CIT from talopram suggested that the cyano‐group has the highest influence on the allosteric effect as measured by their inhibition of [3H]‐S‐CIT dissociation. The fluorine and the phthalane dimethyl group also influenced the allosteric effect, whereas a mono or a dimethyl amino group did not seem to play any significant role.
Allosteric potency of the cyano‐containing citalopram analogues
The screen of allosteric binding properties yielded dissociation times from 85 to 1170 min. A measured t 1/2 of, for example, 1000 min is imprecise because the fraction of dissociated [3H]‐S‐CIT was very small within the experimental time period of 80 min. To obtain a more precise measurement of the allosteric interaction of a compound, it was necessary to determine its allosteric potency by assessing the change in t 1/2 as a function of the concentration of the compound. The screening showed that the cyano‐group was an important determinant for the allosteric effect. Thus, to further investigate the allosteric inhibition of [3H]‐S‐CIT dissociation, we measured the IC50 value of the cyano‐containing compounds (cpds 2–8 compared with S‐CIT) for inhibition of [3H]‐S‐CIT dissociation as described previously (Plenge et al., 2012). The IC50 value is a means of obtaining a binding potency for the compounds to the allosteric site. In short, it is obtained by determining the [3H]‐S‐CIT dissociation rates in the presence of increasing concentrations of allosteric inhibitor (Figure 3).
By plotting the [3H]‐S‐CIT dissociation rate in the presence of inhibitor relative to the absence of inhibitor (k [cpd]/k buf) as a function of the concentration of added compound, we obtained a dose–response curve reflecting the allosteric potency of a compound. Consistent with previous observations (Plenge et al., 2012), S‐CIT displayed an allosteric potency of 5.8 [5.4; 6.3] μM (Figure 3, black‐filled circles). Most of the analogues did not confer a higher allosteric potency than S‐CIT itself, but gave IC50 values in the range of 9–27 μM (Table 2).
Interestingly, compound 3, had a significantly higher allosteric potency than S‐CIT (P = 0.03. one‐way anova with Fisher's LSD test, F ratio: 47.8, P = 0.0035). We observed a similar tendency, although not significant, for compound 4 (P = 0.07). The allosteric potency of compounds 3 and 4 were 3.6 [3.3; 3.8] and 3.6 [3.5; 3.7] μM, respectively [mean (SEM interval), Figure 3 and Table 2]. In agreement with the observations in the screen, the difference in N‐substituted methyl groups had an insignificant effect on the allosteric potency, with an average of 1.3‐fold loss in allosteric potency [difference in allosteric potency (fold change) cpd 2 vs. S‐CIT = 1.9; 4 vs. 3 = 1.0; 6 vs. 5 = 1.6; 8 vs. 7 = 0.75, Table 2]. In contrast, the phthalane dimethyl had a significant (P < 0.05, Student's paired t‐test) positive effect on the allosteric potency causing, on average, a 2.3‐fold increase [difference in allosteric potency (fold change): S‐CIT vs. cpd 3 = 1.6; 2 vs. 4 = 2.8; 5 vs. 7 = 1.5; 6 vs. 8 = 3.1, Table 2]. Following the cyano‐group, the fluorine atom had the highest contribution to the allosteric potency causing an average of 2.8‐fold increase relative to the analogue compound without the fluorine [difference in allosteric potency (fold change): cpd 5 vs. S‐CIT = 2.9; 6 vs. 2 = 2.8; 7 vs. 3 = 3.2 and 8 vs. 4 = 2.4, Table 2].
To summarize, the data suggest that both the fluorine and the phthalane dimethyl substituent contribute to the increased allosteric potency of the compounds. In contrast, the number of N‐methyl substituents does not seem to play a marked role on the allosteric effect.
Binding of the cyano‐containing compounds to the orthosteric (S1) binding site
In order to assess S2 selectivity relative to S1, we investigated the ability of the cyano‐containing compounds to competitively inhibit [3H]‐S‐CIT binding (Figure 4).
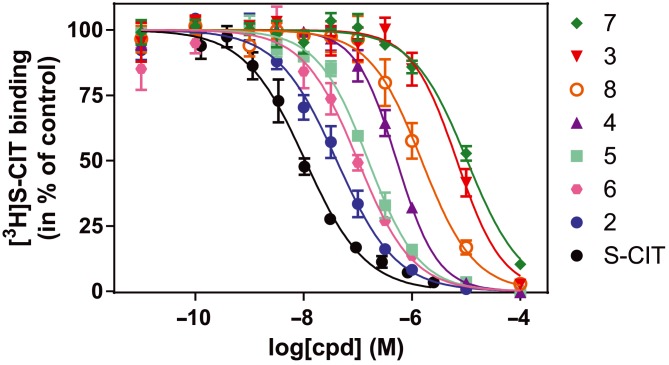
Figure 4.
[3H]‐S‐CIT competition binding to the S1 binding site for S‐CIT and its cyano‐containing analogues. All the tested analogous possess a lower IC50 than S‐CIT itself ranging from 41 [32; 51] nM for compound 2 to 6400 [4700; 8800] nM and 10 400 [9200; 11 800] nM for compounds 3 and 7 respectively. Experiments were performed as inhibition of [3H]‐S‐CIT binding by addition of increasing concentration of S‐CIT or analogues in 11 consecutive concentrations on intact COS7 cells transiently expressing hSERT WT. Data are means ± SEM (error bars) based on three independent experiments each carried out in triplicate.
Accordingly, membranes were prepared from COS‐7 cells transiently expressing SERT, and the IC50 values of the compounds were determined (Figure 4). The IC50 value for S‐CIT was in this setup found to be 10 [8; 13] nM [Mean (SEM interval)]. This is in accord with previous observations (Plenge et al., 2012). All the cyano‐containing analogues exhibited lower affinity for the S1 site of SERT relative to S‐CIT itself. In general, the dimethylamino moiety seems to increase affinity for S1 binding relative to the monomethyl congeners. The effects are in line with previous observations using displacement of [125I]‐RTI‐55 binding (Andersen et al., 2011). Interestingly, the combination of an N‐substituted methyl group and the phthalane dimethyl group resulted in a marked decrease in the S1 site affinity (difference in IC50 value for S1 binding (fold change): 3:4 = 12.8 and 7:8 = 7.4, Table 2). In contrast, the combination without the phthalane dimethyl resulted either in no change (difference in IC50 for S1 binding, compound 5 vs. 6) or in a decrease (difference in IC50 for S1 binding, compound 2 vs. S‐CIT: 4.1‐fold, Table 2). The phthalane substituted dimethyl group (compounds 3, 4, 7 and 8) caused a pronounced contribution to the loss of S1 affinity with the largest effect in the compounds with a dimethylamino group (difference in IC50 for S1 binding: 3 vs. S‐CIT = 640‐fold and 7 vs. 5 = 65‐fold, Table 2). The effect of the monomethylamino group was less pronounced, but still a more than 10‐fold decrease in affinity was seen (four with 12‐fold and eight with 13‐fold decrease relative to two and six respectively, Table 2). Lastly, the fluorine had very little effect on the S1 affinity (less than threefold for most compounds). The only exception was compound 5 relative to S‐CIT where the analogue with the fluorine atom resulted in a 16‐fold increase in affinity when compared with the analogue without fluorine (Table 2).
Altogether, the phthalane dimethyl had the most profound effect on S1 affinity by causing a 12‐fold to 640‐fold decrease in affinity. The N‐substituted methyl also caused a decrease in affinity, in particular in combination with the phthalane dimethyl group. The fluorine had only a minor if any effect on S1 affinity.
Mutation of residues in the S2 binding site decreases the allosteric potency of compound 3
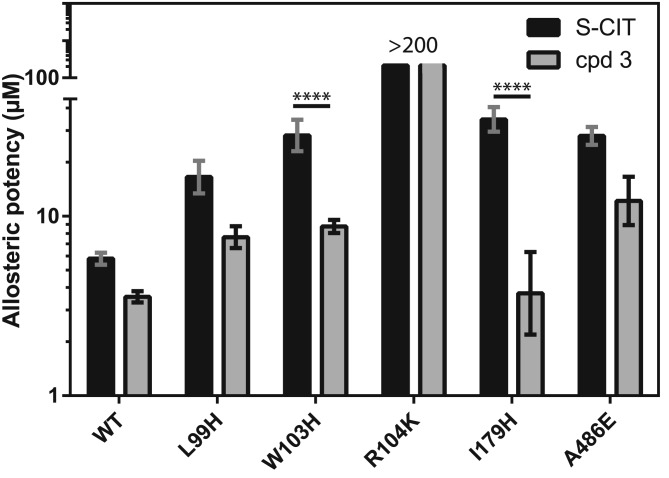
The data above suggest that compounds 3 and 4 have the highest allosteric potency of the tested compounds. Of the two, compound 3 with the extra N‐substituted methyl had the highest effect on S1 affinity with a 640‐fold decrease relative to S‐CIT. We have previously located the allosteric binding site for S‐CIT to the extracellular vestibule (S2 site) (Plenge et al., 2012). Accordingly, we wanted to assess whether compound 3 could bind in a similar manner within the allosteric S2 site. To this end, we performed mutagenesis of selected residues predicted to be located in the allosteric vestibule and which previously have been shown to decrease the allosteric potency of S‐CIT (Plenge et al., 2012). The investigated mutants were: L99H, W103H and R104K in TM1, I179H in TM3 and A486E in TM10 (Figure 5 and Table 3).
Figure 5.
Effect of SERT mutations on allosteric potency of S‐CIT and compound 3. The bars indicate the allosteric potency of S‐CIT and compound 3 on SERT WT and selected mutations in TM 1 (L99H, W103H and R104K), TM3 (I179H) and TM10 (A486E), all suggested to line the S2 site. When comparing the change in allosteric potency for S‐CIT in the SERT WT to the mutants, it is significantly changed by W103H, I179H and A486E. For cpd 3, W103H and A486E significantly changed the allosteric potency relative to WT. When comparing the allosteric potency of S‐CIT and cpd 3 for each mutant, W103H and I179H have significantly different effects, whereas the changes observed in L99H and A486E were not significant. Experiments were performed as in Figure 3 and show means ± SEM (error bars) based on at least three independent experiments each performed in triplicate. Statistical analyses were performed as one‐way anova with Bonferroni's multiple comparisons test (F value: 22.73; P < 0.0001). Due to the profound effect of the R104K mutation, we were not able to assess a precise allosteric potency, which made statistical analysis impossible. **** P < 0.0001.
Table 3.
Effect of SERT mutations on the allosteric potency of cpd 3 relative to S‐CIT
| Construct | S‐CIT allosteric potency IC 50 (μM) | n | Cpd 3 allosteric potency IC 50 (μM) | n |
|---|---|---|---|---|
| WT | 5.8 [5.4; 6.3] | 10 | 3.6 [3.3; 3.8] | 6 |
| L99H | 17 [13; 20] | 4 | 7.6 [6.7; 8.8] | 3 |
| W103H | 28 [23; 34] | 6 | 8.8 [8.1; 9.5] | 5 |
| R104K | >200 | 6 | >200 | 5 |
| I179H | 35 [30; 41] | 4 | 3.7 [2.2; 6.3] | 3 |
| A486E | 28 [25; 31] | 3 | 12 [8.9; 17] | 3 |
The allosteric potencies are the IC50 values obtained from non‐linear regression analysis of data from [3H]‐S‐CIT dissociation experiments in the presence of increasing concentrations of S‐CIT or cpd 3. Data were calculated and experiments performed as described in Methods section. Data are based on the indicated number (n) of independent experiments.
The mutants affected the allosteric potency of S‐CIT to a degree comparable with previous results (Plenge et al., 2012) showing significant changes for all mutants (one‐way anova with Bonferroni's multiple comparisons test). For the R104K mutant, which displays unaffected S1 binding (Plenge et al., 2012), the effect on S‐CIT was so dramatic that it was not possible to assess the allosteric potency (Figure 5). For compound 3, we also observed a dramatic effect of the R104K mutation resulting in an immeasurable allosteric potency. For the other mutations, we observed significant effects of the W103H and A486E mutations, whereas L99H and I179H showed no significant change relative to WT. For the W103H and I179H mutations, the changes in allosteric potency for compound 3 were significantly less than for S‐CIT (Figure 5). Together, these data suggest that S‐CIT and compound 3 are probably both accommodated in the S2 binding pocket but that the orientation and precise set of interactions might differ between the two compounds.
Discussion and conclusions
The allosteric binding site in SERT has been investigated for more than three decades (Wennogle and Meyerson, 1982). Only recently, due to high‐resolution X‐ray crystal structures of LeuT (Singh et al., 2007) and dDAT (Wang et al., 2015), has it been possible to obtain reliable molecular models of SERT and, in conjunction with site‐directed mutagenesis, to provide evidence for localization of the site to the extracellular vestibule of the transporter (Plenge et al., 2012). Structures of dDAT (Penmatsa et al., 2013; Penmatsa et al., 2015) and LeuT (Wang et al., 2013) have also together with mutagenesis studies (Henry et al., 2006; Andersen et al., 2009; Sinning et al., 2010; Andersen et al., 2014) provided convincing evidence that the high‐affinity binding site for TCAs and SSRIs are all overlapping the S1 site. However, despite the fact that evidence has been provided for the location of the allosteric binding site, the question still remains whether allosteric binding to the site has any specific pharmacological implications. Indeed, it has been suggested that the faster onset of action and higher efficacy of S‐CIT compared with the racemic mixture is due to its allosteric action at the transporter (Mørk et al., 2003; Sanchez et al., 2004; Storustovu et al., 2004; Sanchez, 2006; Mansari et al., 2007). Other compounds with no apparent similarity to citalopram also possess allosteric properties for SERT (Rothman et al., 2015; Nightingale et al., 2005; Nandi et al., 2004). But rather than inhibiting dissociation, a common characteristic for these compounds is that they only show a 40–60% inhibition of [3H]‐5‐HT uptake and [125I]‐RTI‐55 binding. Accordingly, those compounds could possess a different allosteric mechanism than S‐CIT. Further analysis must elucidate these differences.
A way to increase our understanding of the allosteric site versus the orthosteric site would be to develop tool compounds possessing high selectivity towards either site. Already, compounds are available showing selectivity for the orthosteric S1 site (Plenge et al., 2007; Chen et al., 2005b). To date, the compound reported with the highest allosteric potency is, to our knowledge, S‐CIT. However, its potency is still low (micromolar range), and together with its high affinity (nanomolar range) for the S1 site, S‐CIT is not a useful tool for elucidating the specific role of the allosteric site. Here, we present a systematic SAR study of a series of compounds derived from citalopram with the purpose of identifying the molecular determinants for its allosteric potency. As previously found for the S1 binding site (Andersen et al., 2011), we find that the cyano‐group is absolutely essential for the present series of compounds to carry an allosteric potential. In addition to the cyano‐group, the fluorine substituent increases the allosteric potency the most, followed by the phthalane dimethyl groups, whereas the number of N‐methyl substituents is a minor determinant for the allosteric potency (Figure 6).
Figure 6.
Effect of citalopram substituents on allosteric potency. Schematic representation of how the different substituents change allosteric potency. Firstly, the eight compounds depicted all contain the cyano‐group, which is required for possessing an allosteric effect significantly different from control dissociation. Secondly, the N‐methyl substituent did not contribute to allosteric potency, so the two otherwise identical compounds are grouped together. The arrows denote when the added substituent increases the allosteric potency. Grey arrows: smaller (mean < 2.5‐fold) increase in potency. Black arrows: larger (mean > 2.5‐fold) increase in potency.
Interestingly, the phthalane dimethyl substituent contributes to a marked decrease in S1 affinity, as assessed previously with [125I]‐RTI‐55 (Andersen et al., 2011) and herein using [3H]‐S‐CIT competition (Table 2). In contrast to the decreased effect on S1 affinity, the phthalane dimethyl substituent are accommodated by the S2 site. This structural feature is essential for creating analogues with similar or even minor preference for the S2 site over the S1 site. Accordingly, the compound with the highest allosteric potency and lowest orthosteric affinity reported here is compound 3. In fact, it showed a twofold increase in allosteric potency over the orthosteric affinity. Accordingly, the SAR study performed here revealed that dimethyl citalopram possesses the highest affinity for the allosteric site relative to the S1 site in SERT. To the best of our knowledge, this is the first compound reported with a preference towards the allosteric site. However, the twofold selectivity is modest and for practical purposes not useful for investigating the putative potential of the allosteric site. In addition, there are at least two uncertainties discrediting the reported affinities: firstly, the allosteric potency is a dose–response measurement and does not reflect an actual binding event in equilibrium conditions. Due to the low‐affinity nature of allosteric ligands, the fast off rate would hamper determination of specific binding of a radioligand to the allosteric site. Therefore, it must be determined as the ability of the allosteric compound to impede dissociation of a high‐affinity radioligand bound to the orthosteric site. Thus, the actual affinity could differ from the allosteric potency measured. Secondly, as both S1 inhibition binding and allosteric potency are measured as an impairment or displacement of S1‐bound [3H]‐S‐CIT, an S2 bound compound 3 might also contribute to [3H]‐S‐CIT displacement in the binding assay. Thus, the S1 affinity for compound 3 could be lower than the reported IC50 value, which would contribute to a higher selectivity to the allosteric site. Further studies are needed to elucidate the specific contribution from the two binding sites to the measured affinity of compound 3. Due to the lack of consensus for quantitative determination of the allosteric potency, it is difficult to directly relate our findings to previous reports on SERT binding compounds possessing allosteric properties (Nandi et al., 2004; Nightingale et al., 2005; Kortagere et al., 2013; Rothman et al., 2015). We have previously attempted to produce high‐affinity allosteric ligands for SERT (Banala et al., 2013), but none of them showed a notable loss in S1 affinity and, thus, did not show an S2: S1 affinity ratio comparable with the compounds reported herein.
All the tested compounds were racemic structures. Accordingly, the two enantiomers might behave differently at the two binding sites. It is well established that it is S‐CIT that possesses the allosteric properties. It is therefore possible that the (S)‐enantiomer of the tested citalopram analogues would possess higher allosteric affinity than their corresponding racemates and therefore be more potent S2 inhibitors.
Based on the results presented, it could be argued that citalopram itself is not the best starting point or lead compound for developing higher affinity allosteric SERT modulators. On the other hand, the data do provide suggestions for which substituents on citalopram it is possible to modify with a potential gain of allosteric potency. For example, the N‐methyl substituent did not contribute to gain or loss of allosteric potency but might enable other substitutions in this position for gain of potency. Indeed, we have previously shown that specific substituents at this position do confer an allosteric potency (Banala et al., 2013). It is possible that proper substitutions here will lead to increased S2 affinities. Further experiments are necessary to clarify this issue.
Taken together, the pharmacological significance of the allosteric binding site in SERT has yet to be elucidated. A compound with high selectivity and affinity to the site is a prerequisite for such investigations. The investigations herein provide a novel tool compound with at least similar activity at S1 and S2 and demonstrate which substituents on the chemical skeleton of citalopram can confer affinity towards the allosteric binding site. The results could be useful to guide future synthesis of compounds bearing high selectivity and high affinity towards the allosteric binding site.
Author contributions
M. A. B. L. and P. P. performed the research. M. A. B. L. and C. J. L. designed the research study. J. A., J. N. N. E., A. S. K., K. P. B., K. S. and B. B‐A. contributed essential reagents and tools. M. A. B. L., P. P. and C. J. L. analysed the data. M. A. B. L. and C. J. L. wrote the first draft. All authors contributed in writing the paper.
Conflict of interest
K. B. and B. B‐A. are employees of H. Lundbeck A/S.
Acknowledgements
Lone Rosenquist and Bente Bennicke are thanked for excellent technical assistance. Assoc. Prof. Harrie C.M. Boonen (University of Copenhagen, DK) is thanked for extensive help with statistical analysis. The work was supported in part by the Danish Independent Research Council – Sapere Aude (0602‐02100B), the Lundbeck Foundation (R108‐A10755) and bioSYNergy – University of Copenhagen's Excellence Programme for Interdisciplinary Research.
Larsen, M. A. B. , Plenge, P. , Andersen, J. , Eildal, J. NN. , Kristensen, A. S. , Bøgesø, K. P. , Gether, U. , Strømgaard, K. , Bang‐Andersen, B. , and Loland, C. J. (2016) Structure–activity relationship studies of citalopram derivatives: examining substituents conferring selectivity for the allosteric site in the 5‐HT transporter. British Journal of Pharmacology, 173: 925–936. doi: 10.1111/bph.13411.
References
- Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, et al. (2015). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. Br J Pharmacol 172: 6110–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Taboureau O, Hansen KB, Olsen L, Egebjerg J, Stromgaard K, et al. (2009). Location of the antidepressant binding site in the serotonin transporter: importance of Ser‐438 in recognition of citalopram and tricyclic antidepressants. J Biol Chem 284: 10276–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Olsen L, Hansen KB, Taboureau O, Jorgensen FS, Jorgensen AM, et al. (2010). Mutational mapping and modeling of the binding site for (S)‐citalopram in the human serotonin transporter. J Biol Chem 285: 2051–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Stuhr‐Hansen N, Zachariassen L, Toubro S, Hansen SM, Eildal JN, et al. (2011). Molecular determinants for selective recognition of antidepressants in the human serotonin and norepinephrine transporters. Proc Natl Acad Sci U S A 108: 12137–12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Stuhr‐Hansen N, Zachariassen LG, Koldso H, Schiott B, Stromgaard K, et al. (2014). Molecular basis for selective serotonin reuptake inhibition by the antidepressant agent fluoxetine (Prozac). Mol Pharmacol 85: 703–714. [DOI] [PubMed] [Google Scholar]
- Banala AK, Zhang P, Plenge P, Cyriac G, Kopajtic T, Katz JL, et al. (2013). Design and synthesis of 1‐(3‐(dimethylamino)propyl)‐1‐(4‐fluorophenyl)‐1,3‐dihydroisobenzofuran‐5‐carboni trile (citalopram) analogues as novel probes for the serotonin transporter S1 and S2 binding sites. J Med Chem 56: 9709–9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, et al. (2008). The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci 11: 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik L, Sinning S, Severinsen K, Hansen CG, Moller MS, Bols M, et al. (2008). Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation J Am Chem Soc 130: 3853–3865. [DOI] [PubMed] [Google Scholar]
- Chen F, Larsen MB, Neubauer HA, Sanchez C, Plenge P, Wiborg O (2005a). Characterization of an allosteric citalopram‐binding site at the serotonin transporter. J Neurochem 92: 21–28. [DOI] [PubMed] [Google Scholar]
- Chen F, Larsen MB, Sanchez C, Wiborg O (2005b). The S‐enantiomer of R,S‐citalopram, increases inhibitor binding to the human serotonin transporter by an allosteric mechanism. Comparison with other serotonin transporter inhibitors. Eur Neuropsychopharmacol 15: 193–198. [DOI] [PubMed] [Google Scholar]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander SPA, Giembycz MA, et al. (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172: 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eildal JN, Andersen J, Kristensen AS, Jorgensen AM, Bang‐Andersen B, Jorgensen M, et al. (2008). From the selective serotonin transporter inhibitor citalopram to the selective norepinephrine transporter inhibitor talopram: synthesis and structure–activity relationship studies. J Med Chem 51: 3045–3048. [DOI] [PubMed] [Google Scholar]
- Henry LK, Field JR, Adkins EM, Parnas ML, Vaughan RA, Zou MF, et al. (2006). Tyr‐95 and Ile‐172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J Biol Chem 281: 2012–2023. [DOI] [PubMed] [Google Scholar]
- Hill ER, Huang X, Zhan CG, Ivy Carroll F, Gu HH (2011). Interaction of tyrosine 151 in norepinephrine transporter with the 2beta group of cocaine analog RTI‐113. Neuropharmacology 61: 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen JP, Plenge P, Sachs BD, Pehrson AL, Cajina M, Du Y, et al. (2014). The interaction of escitalopram and R‐citalopram at the human serotonin transporter investigated in the mouse. Psychopharmacology (Berl) 231: 4527–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortagere S, Fontana AC, Rose DR, Mortensen OV (2013). Identification of an allosteric modulator of the serotonin transporter with novel mechanism of action. Neuropharmacology 72: 282–290. [DOI] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, et al. (2011). SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63: 585–640. [DOI] [PubMed] [Google Scholar]
- Malinauskaite L, Quick M, Reinhard L, Lyons JA, Yano H, Javitch JA, et al. (2014). A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat Struct Mol Biol 21: 1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansari ME, Wiborg O, Mnie‐Filali O, Benturquia N, Sanchez C, Haddjeri N (2007). Allosteric modulation of the effect of escitalopram, paroxetine and fluoxetine: in‐vitro and in‐vivo studies. Int J Neuropsychopharmacol 10: 31–40. [DOI] [PubMed] [Google Scholar]
- Mohammad‐Zadeh LF, Moses L, Gwaltney‐Brant SM (2008). Serotonin: a review. J Vet Pharmacol Ther 31: 187–199. [DOI] [PubMed] [Google Scholar]
- Mørk A, Kreilgaard M, Sanchez C (2003). The R‐enantiomer of citalopram counteracts escitalopram‐induced increase in extracellular 5‐HT in the frontal cortex of freely moving rats. Neuropharmacology 45: 167–173. [DOI] [PubMed] [Google Scholar]
- Nandi A, Dersch CM, Kulshrestha M, Ananthan S, Rothman RB (2004). Identification and characterization of a novel allosteric modulator (SoRI‐6238) of the serotonin transporter. Synapse 53: 176–183. [DOI] [PubMed] [Google Scholar]
- Nightingale B, Dersch CM, Boos TL, Greiner E, Calhoun WJ, Jacobson AE, et al. (2005). Studies of the biogenic amine transporters. XI. Identification of a 1‐[2‐[bis(4‐fluorophenyl)methoxy]ethyl]‐4‐(3‐phenylpropyl)piperazine (GBR12909) analog that allosterically modulates the serotonin transporter. J Pharmacol Exp Ther 314: 906–915. [DOI] [PubMed] [Google Scholar]
- Owens MJ, Knight DL, Nemeroff CB (2001). Second‐generation SSRIs: human monoamine transporter binding profile of escitalopram and R‐fluoxetine. Biol Psychiatry 50: 345–350. [DOI] [PubMed] [Google Scholar]
- Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SP, Buneman OP et al., NC‐IUPHAR (2014). The IUPHAR/BPS guide to PHARMACOLOGY: an expert‐driven knowledge base of drug targets and their ligands. Nucleic Acids Res 42: D1098–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E (2013). X‐ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E (2015). X‐ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat Struct Mol Biol . doi:10.1038/nsmb.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge P, Mellerup ET (1985). Antidepressive drugs can change the affinity of [3 H]imipramine and [3 H]paroxetine binding to platelet and neuronal membranes. Eur J Pharmacol 119: 1–8. [DOI] [PubMed] [Google Scholar]
- Plenge P, Gether U, Rasmussen SG (2007). Allosteric effects of R‐ and S‐citalopram on the human 5‐HT transporter: evidence for distinct high‐ and low‐affinity binding sites. Eur J Pharmacol 567: 1–9. [DOI] [PubMed] [Google Scholar]
- Plenge P, Shi L, Beuming T, Te J, Newman AH, Weinstein H, et al. (2012). Steric hindrance mutagenesis in the conserved extracellular vestibule impedes allosteric binding of antidepressants to the serotonin transporter. J Biol Chem 287: 39316–39326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman RB, Ananthan S, Partilla JS, Saini SK, Moukha‐Chafiq O, Pathak V, et al. (2015). Studies of the biogenic amine transporters 15. Identification of novel allosteric dopamine transporter ligands with nanomolar potency J Pharmacol Exp Ther 353: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res 34: D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C (2006). The pharmacology of citalopram enantiomers: the antagonism by R‐citalopram on the effect of S‐citalopram. Basic Clin Pharmacol Toxicol 99: 91–95. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Bogeso KP, Ebert B, Reines EH, Braestrup C (2004). Escitalopram versus citalopram: the surprising role of the R‐enantiomer. Psychopharmacology (Berl) 174: 163–176. [DOI] [PubMed] [Google Scholar]
- Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA (2008). The mechanism of a neurotransmitter: sodium symporter – inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell 30: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E (2007). Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448: 952–956. [DOI] [PubMed] [Google Scholar]
- Sinning S, Musgaard M, Jensen M, Severinsen K, Celik L, Koldso H, et al. (2010). Binding and orientation of tricyclic antidepressants within the central substrate site of the human serotonin transporter. J Biol Chem 285: 8363–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovstrup S, Taboureau O, Brauner‐Osborne H, Jorgensen FS (2010). Homology modelling of the GABA transporter and analysis of tiagabine binding. ChemMedChem 5: 986–1000. [DOI] [PubMed] [Google Scholar]
- Stolzenberg S, Quick M, Zhao C, Gotfryd K, Khelashvili G, Gether U, et al. (2015). Mechanism of the association between Na+ binding and conformations at the intracellular gate in neurotransmitter: sodium symporters. J Biol Chem 290: 13992–14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storustovu S, Sanchez C, Porzgen P, Brennum LT, Larsen AK, Pulis M, et al. (2004). R‐citalopram functionally antagonises escitalopram in vivo and in vitro: evidence for kinetic interaction at the serotonin transporter. Br J Pharmacol 142: 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Goehring A, Wang KH, Penmatsa A, Ressler R, Gouaux E (2013). Structural basis for action by diverse antidepressants on biogenic amine transporters. Nature 503: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E (2015). Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 521: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennogle LP, Meyerson LR (1982). Serotonin modulates the dissociation of [3 H]imipramine from human platelet recognition sites. Eur J Pharmacol 86: 303–307. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E (2005). Crystal structure of a bacterial homologue of Na+/Cl−‐dependent neurotransmitter transporters. Nature 437: 215–223. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith ME, et al. (2007). LeuT‐desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317: 1390–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Law CJ, Reith ME, Wang DN (2009). Antidepressant specificity of serotonin transporter suggested by three LeuT‐SSRI structures. Nat Struct Mol Biol 16: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]