Abstract
Background
The genome of Schizophyllum commune encodes a diverse repertoire of degradative enzymes for plant cell wall breakdown. Recent comparative genomics study suggests that this wood decayer likely has a mode of biodegradation distinct from the well-established white-rot/brown-rot models. However, much about the extracellular enzyme system secreted by S. commune during lignocellulose deconstruction remains unknown and the underlying mechanism is poorly understood. In this study, extracellular proteins of S. commune colonizing Jerusalem artichoke stalk were analyzed and compared with those of two white-rot fungi Phanerochaete chrysosporium and Ceriporiopsis subvermispora and a brown-rot fungus Gloeophyllum trabeum.
Results
Under solid-state fermentation (SSF) conditions, S. commune displayed considerably higher levels of hydrolytic enzyme activities in comparison with those of P. chrysosporium, C. subvermispora and G. trabeum. During biodegradation process, this fungus modified the lignin polymer in a way which was consistent with a hydroxyl radical attack, similar to that of G. trabeum. The crude enzyme cocktail derived from S. commune demonstrated superior performance over a commercial enzyme preparation from Trichoderma longibrachiatum in the hydrolysis of pretreated lignocellulosic biomass at low enzyme loadings. Secretomic analysis revealed that compared with three other fungi, this species produced a higher diversity of carbohydrate-degrading enzymes, especially hemicellulases and pectinases acting on polysaccharide backbones and side chains, and a larger set of enzymes potentially supporting the generation of hydroxyl radicals. In addition, multiple non-hydrolytic proteins implicated in enhancing polysaccharide accessibility were identified in the S. commune secretome, including lytic polysaccharide monooxygenases (LPMOs) and expansin-like proteins.
Conclusions
Plant lignocellulose degradation by S. commune involves a hydroxyl radical-mediated mechanism for lignocellulose modification in parallel with the synergistic system of various polysaccharide-degrading enzymes. Furthermore, the complex enzyme system of S. commune holds significant potential for application in biomass saccharification. These discoveries will help unveil the diversity of natural lignocellulose-degrading mechanisms, and advance the design of more efficient enzyme mixtures for the deconstruction of lignocellulosic feedstocks.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0461-x) contains supplementary material, which is available to authorized users.
Keywords: Schizophyllum commune, Solid-state fermentation, White rot, Brown rot, Polysaccharide-degrading enzyme, Secretome, Hydroxyl radical, LPMO, Expansin
Background
Lignocellulosic feedstock from plant materials is considered to be an abundant renewable resource with increasing potential for the production of alternative liquid fuels. One of the main challenges for large-scale production of economically competitive bioethanol has been the high cost of enzymatic hydrolysis process [1]. Because conversion of biomass polysaccharides into fermentable sugars depends exclusively on the efficiency of enzyme mixtures [2], optimizing the mixture composition could contribute to the hydrolysis efficiency and reduction of enzyme loadings. Many fungi and bacteria are capable of utilizing cellulose and hemicellulose through the secretion of polysaccharide-degrading enzymes. The filamentous fungi Trichoderma reesei has been widely exploited in industrial applications for the production of commercial cellulases [3]. However, the enzyme pool produced by T. reesei via submerged fermentation is deficient on certain components and requires additional enzyme supplement for efficient and complete hydrolysis of complex lignocellulosic materials [4, 5]. For this reason, considerable research efforts have been devoted to exploring alternative fungi for the production of cost-effective enzymes and development of more efficient enzyme cocktails [6–8].
Schizophyllum commune is a widely distributed saprophytic basidiomycete around the world and its genome has been previously sequenced. The carbohydrate-active enzyme database (CAZy) annotation of S. commune identifies a total of 366 carbohydrate-active enzymes (CAZymes), of which 106 are predicted to be involved in plant polysaccharide degradation [9]. Compared to T. reesei, S. commune owns an equivalent number of members from GH families acting on cellulose, including GH5, 6, 7, 12, and 45. Analysis of the diversity of non-cellulosic polysaccharide-degrading enzymes showed that S. commune has more abundant xylan and pectin degradation-related glycoside hydrolases such as GH5, 10, 28, 43, 51, 53, 93, 105, and 115 families. In particular, the S. commune genome possesses a much larger number of GH43 proteins (19 in S. commune vs only 2 in T. reesei), indicating its great potential for hemicellulose and pectin deconstruction. In addition to hydrolytic enzymes, S. commune has an expanded complement of genes encoding lytic polysaccharide monooxygenases (LPMOs) of auxiliary activity (AA) family 9 (22 in S. commune vs 3 in T. reesei), which participate in oxidative cleavage of crystalline cellulose [10, 11]. Because the genome of S. commune encodes an extensive catalog of genes implicated in lignocellulose decomposition, its lignocellulolytic enzyme pool is expected to provide a prospective enzyme source for biotechnological applications. Several novel families of enzymes that remove xylan side chains, such as acetyl xylan esterases [12], glucuronoyl esterases [13], and α-glucuronidases [14, 15] have been identified in the cellulolytic system of S. commune. Among the few studies regarding its polysaccharide hydrolases, a recombinant endoxylanase from S. commune has been reported to exhibit very high level of activity against beechwood xylan [16], and crude enzymes containing mainly β-glucosidase from this fungal species could saccharify pretreated lignocellulose comparably well with a commercial β-glucosidase [17]. To date, studies of this species have focused mainly on the mating-type gene function [18, 19] and mushroom development [20, 21], whereas the lignocellulolytic enzymes produced by S. commune are far less characterized and a comprehensive study on its enzymatic system is still lacked.
Wood-degrading basidiomycetes have been typically classified as white-rot and brown-rot fungi according to the types of decay that they cause [22]. White-rot fungi such as Phanerochaete chrysosporium simultaneously break down all polymeric components of plant cell walls, including cellulose, hemicellulose, and lignin. Other white-rot species like Ceriporiopsis subvermispora selectively degrade lignin with little cellulose loss. In contrast, brown-rot fungi represented by Gloeophyllum trabeum, efficiently depolymerize the cellulose portion without substantial removal of lignin [23]. S. commune has been previously characterized as a white-rot species despite very limited lignin-degrading capacity [24]. With respect to lignin degradation, the genome of S. commune lacks genes encoding class II peroxidases of AA family 2, which is similar to brown-rot species [9]. Recent genomic comparisons of 33 basidiomycetes suggest that S. commune is an intermediate between white-rot and brown-rot species in terms of gene families encoding lignocellulose-degrading enzymes [25]. Given the limited reports characterizing its lignocellulolytic enzymes [26–28] and wood decay process [29], it is not clear what biodegradative strategy S. commune uses to decompose lignocellulosic biomass.
Jerusalem artichoke is one of the potential energy crops that can grow well on marginal lands in harsh environment [30]. This plant is considered as a promising candidate for consolidated bioprocessing (CBP), which enables the utilization of whole plant biomass (the tuber and stalk). Jerusalem artichoke tuber, with a high content of inulin, has been investigated as a sugar source for bioethanol production [31]. As for Jerusalem artichoke stalk, there is no report about using its lignocellulosic materials for fungal growth and induction of lignocellulolytic enzymes.
The aim of the present study is to identify potentially important enzymes and provide insight into the mechanism of plant cell wall deconstruction by S. commune. Here we report the degradation dynamics and lignocellulolytic enzyme pattern of S. commune cultivated on Jerusalem artichoke stalk during solid-state fermentation (SSF). This complex lignocellulosic substrate and the SSF culture conditions simulate more closely its decay process occurring in nature. The crude enzymes derived from S. commune were examined for the enzymatic saccharification of pretreated lignocellulosic biomass. The composition of extracellular protein profile of S. commune was identified by nano liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) and compared with those of representative white-rot and brown-rot basidiomycetes to gain a deeper understanding of the lignocellulose-degrading enzyme system of this fungus.
Results
Scanning electron microscopy (SEM)
Scanning electron microscopy was applied to investigate the micromorphological characteristics of degraded Jerusalem artichoke stalk after inoculation with different fungal species. Figure 1a and b depicted non-decayed stalk cell walls with an intact morphology, while in stalk sample inoculated with P. chrysosporium, hyphae and spores were visible growing in the cavities formed by extensive decay, implying the degradation of all cell wall components (Fig. 1c). Sample subjected to inoculation with C. subvermispora exhibited a similar eroded morphology, but to a lesser extent (Fig. 1d). In contrast to white rot, stalk sample inoculated with G. trabeum was not completely degraded but appeared to have a distorted and collapsed structure because of more brittle and weakened cell walls (Fig. 1e). In stalk sample colonized by S. commune, the plant cell walls remained relatively intact despite the fungal mycelia covering the stalk surface (Fig. 1f). However, adjacent cells appeared to be separated from each other, probably due to degraded middle lamellae. Physical invasion of cell walls by fungal mycelia was also seen through the fissures between cells.
Fig. 1.

Scanning electron micrographs of Jerusalem artichoke stalk. a, b SEM image of Jerusalem artichoke stalk without any fungal inoculation. c SEM image of stalk sample inoculated with P. chrysosporium for 30 days. d SEM image of sample inoculated with C. subvermispora for 30 days. e SEM image of sample inoculated with G. trabeum for 30 days. f SEM image of sample inoculated with S. commune for 30 days
Fungal degradation of Jerusalem artichoke stalk
Table 1 showed the degradation of the Jerusalem artichoke stalk components by four fungal species during SSF over an incubation period of 30 days. Incubation with S. commune for 30 days resulted in decrease in the cellulose and hemicellulose contents by 18.4 and 17.5 %, respectively, with a limited lignin degradation of 3.2 %. In comparison, C. subvermispora selectively degraded lignin as much as 45.4 %, while the cellulose content decreased by only 4.3 % in 30 days, indicating a preferential removal of lignin with respect to cellulose. And there was up to 22.9 % hemicellulose reduction along with lignin degradation, probably due to increasing accessibility of hemicellulose as lignin decomposed. At the end of cultivation, cellulose and hemicellulose loss caused by P. chrysosporium reached 33.3 and 31.6 %, respectively, the highest among four fungal species. A concomitant lignin degradation of 23.6 % was observed with the polysaccharide loss. Such simultaneous removal of all lignocellulosic components was consistent with the typical white rot. On the contrary, the brown-rot fungus G. trabeum caused a polysaccharide degradation of 12.2 % cellulose and 9.8 % hemicellulose, with a very minor lignin removal of 2.3 %.
Table 1.
Degradation of Jerusalem artichoke stalk components by four fungi during solid-state fermentation
| Days | S. commune | P. chrysosporium | C. subvermispora | G. trabeum | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loss (% initial content) | Loss (% initial content) | Loss (% initial content) | Loss (% initial content) | |||||||||
| Cel | Hem | Lig | Cel | Hem | Lig | Cel | Hem | Lig | Cel | Hem | Lig | |
| 5 | 2.1 ± 0.7 | 1.2 ± 0.1 | 0.6 ± 0.1 | 6.9 ± 0.3 | 4.9 ± 0.4 | 3.5 ± 0.2 | 1.5 ± 0.2 | 3.2 ± 0.6 | 7.7 ± 0.7 | 1.5 ± 0.2 | 1.7 ± 0.3 | 0.5 ± 0.1 |
| 10 | 3.7 ± 0.5 | 3.1 ± 0.2 | 1.1 ± 0.3 | 12.3 ± 0.7 | 9.3 ± 0.6 | 8.1 ± 0.2 | 1.9 ± 0.2 | 6.0 ± 0.2 | 15.9 ± 0.2 | 3.0 ± 0.3 | 3.0 ± 0.1 | 0.8 ± 0.1 |
| 15 | 8.7 ± 0.3 | 6.4 ± 0.2 | 1.4 ± 0.1 | 19.4 ± 0.3 | 17.2 ± 0.7 | 11.9 ± 0.7 | 2.5 ± 0.8 | 10.5 ± 0.7 | 25.4 ± 0.6 | 4.9 ± 0.5 | 3.8 ± 0.2 | 1.2 ± 0.1 |
| 20 | 11.3 ± 0.8 | 9.7 ± 0.8 | 2.1 ± 0.3 | 25.5 ± 0.6 | 22.0 ± 0.5 | 15.4 ± 0.5 | 3.1 ± 0.4 | 15.5 ± 0.6 | 33.4 ± 0.8 | 8.4 ± 0.6 | 4.6 ± 0.4 | 1.7 ± 0.2 |
| 25 | 15.9 ± 0.2 | 13.3 ± 0.9 | 2.7 ± 0.4 | 30.1 ± 0.6 | 27.5 ± 1.1 | 21.1 ± 0.6 | 3.8 ± 0.7 | 19.0 ± 1.2 | 38.8 ± 1.5 | 10.1 ± 0.8 | 7.7 ± 0.5 | 2.1 ± 0.3 |
| 30 | 18.4 ± 0.9 | 17.5 ± 1.2 | 3.2 ± 0.4 | 33.3 ± 1.2 | 31.6 ± 0.5 | 23.6 ± 0.2 | 4.3 ± 0.3 | 22.9 ± 0.6 | 45.4 ± 1.1 | 12.2 ± 1.1 | 9.8 ± 0.9 | 2.3 ± 0.3 |
Cellulose (Cel), hemicellulose (Hem), and lignin (Lig) losses of Jerusalem artichoke stalk are presented for each fungal species. The data indicate mean values ± standard deviations from three replicates
Pyrolysis gas chromatography-mass spectrometry (Py-GC/MS) analysis
Pyrolysis coupled with gas chromatography–mass spectrometry is a useful tool for characterizing decay patterns of wood-degrading fungi, especially chemical features of lignin structure [32]. During analysis of sound Jerusalem artichoke stalk and samples incubated with different fungal species, some differences were observed in the relative abundances of the released pyrolysis products (Table 2), indicating the divergent degradation patterns caused by the different fungi. The two white-rot fungi, C. subvermispora and P. chrysosporium preferentially degraded lignin moiety in stalk as evidenced by a decrease in the ratio of lignin- to carbohydrate-derived pyrolysates (lignin/carbohydrate ratio). G. trabeum and S. commune, on the other hand, caused an increased ratio between lignin and carbohydrate pyrolysis products, revealing a preferential consumption of polysaccharides relative to lignin. Regardless of the decay patterns and substrate preferences, all four fungi seemed to demethoxylate syringyl units of lignin, thus decreasing the ratio of syringyl- to guaiacyl-type pyrolysates (S/G ratio).
Table 2.
Relative peak areas (%) of lignin-derived pyrolysates identified after Py–GC/MS of fungi-rotting stalk
| Compound | Control | S. commune | P. chrysosporium | C. subvermispora | G. trabeum |
|---|---|---|---|---|---|
| Guaiacol (G) | 0.29 | 0.42 | 0.54 | 0.47 | 0.39 |
| 3-Methoxycatechol | ndb | 0.32 | nd | nd | 0.43 |
| 4-vinylguaiacol | 0.45 | 0.63 | 0.32 | 0.20 | 0.69 |
| Syringol(S) | 0.42 | 0.52 | 0.30 | 0.23 | 0.34 |
| 4-Methylsyringol | nd | 0.15 | nd | 0.08 | 0.16 |
| Trans-Isoeugenol | 0.10 | 0.18 | 0.11 | nd | 0.09 |
| 4-Vinylsyringol | nd | 0.17 | 0.12 | nd | 0.14 |
| 4-Allylsyringol | 0.28 | 0.21 | 0.12 | 0.10 | 0.23 |
| Carbohydrate | 41.37 | 38.17 | 46.30 | 50.45 | 35.42 |
| Syringyl/guaiacyl ratio | 0.83 | 0.68 | 0.56 | 0.61 | 0.74 |
| Pch-C1,2/Pch-C3 ratioa | 1.18 | 3.26 | 1.91 | 2.80 | 3.09 |
| Lignin/carbohydrate ratio | 0.04 | 0.07 | 0.03 | 0.02 | 0.07 |
Jerusalem artichoke stalk without fungal inoculation is used as control
a Ratio of phenylmethane and phenylethane to phenylpropane-type compounds
b not detected
Although G. trabeum and S. commune preferentially degraded the carbohydrate polymers, both fungal species caused a significant increase in the ratio of phenylmethane and phenylethane units to phenylpropane units (Ph-C1,C2/Ph-C3 ratio), suggesting modification on the lignin moiety through extensive cleavage of side-chain linkages. A closer inspection of lignin-derived compounds showed that methoxyl groups linked to syringyl subunits were demethylated, as evidenced by the identification of 3-methoxycatechol pyrolysate. Such pyrolysis product was undetectable in the spectrum of sound stalk and samples decayed by C. subvermispora and P. chrysosporium.
Determination of lignocellulolytic enzyme activities and iron-reducing capacity during SSF
The present study found that the four fungal species showed different time courses of lignocellulolytic enzyme activities in the extracellular extracts during growth on Jerusalem artichoke stalk (Fig. 2). In general, S. commune produced significantly higher levels of cellulolytic, xylanolytic, and pectinolytic activities compared to the other three fungi.
Fig. 2.
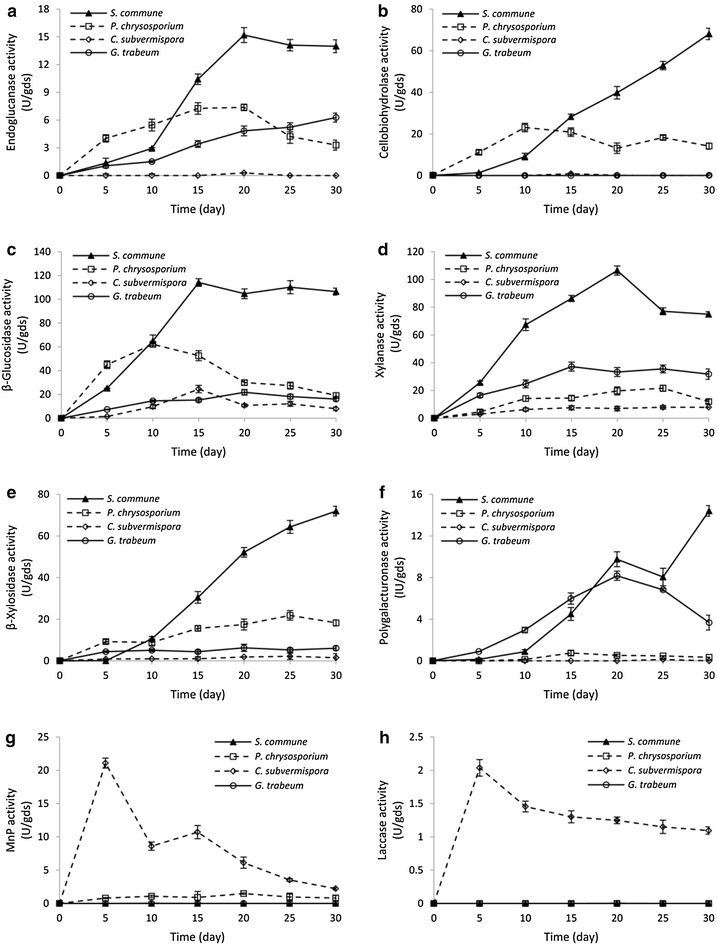
Lignocellulolytic enzyme activities in the secretomes of four fungi during SSF on Jerusalem artichoke stalk. The activities of cellulases (endoglucanase, cellobiohydrolase, and β-glucosidase) are shown in a, b, and c, respectively. The activities of hemicellulases (endoxylanase and β-xylosidase) are shown in d and e, respectively. The polygalacturonase activities are shown in f. The activities of ligninolytic enzymes (manganese peroxidase and laccase) are shown in g and h, respectively. The values shown are the mean of three replicates and the error bars indicate standard deviations from the mean values
When comparing the activities of cellulases produced by the four fungal species, endoglucanase activity in S. commune extract increased gradually over time and reached a fairly constant level of around 15.2 ± 1.1 U/gds after day 20 (Fig. 2a). As for P. chrysosporium, the activity increased slowly to achieve maximum on day 20 (7.4 ± 1.2 U/gds), and declined afterward. Endoglucanase activity in G. trabeum extract increased over time (a maximum of 6.3 ± 1.5 U/gds on day 30), while it was negligible in C. subvermispora. Cellobiohydrolase activity in S. commune extract increased over the incubation time, and the highest activity (68.1 ± 2.2 U/gds) was obtained at the end of cultivation (Fig. 2b). In contrast, the maximum exoglucanase activity in P. chrysosporium extract (23.1 ± 3.0 U/gds) was obtained on day 10, and the activity was not detected or too low in C. subvermispora and G. trabeum(Fig. 1b). Beta-glucosidase activities were detected in all four fungi, but with different activity levels (Fig. 2c). In S. commune extract, the activity increased gradually to reach its highest level (114.1 ± 7.2 U/gds), which was more than twofold, fourfold, and fivefold as high as those of P. chrysosporium (62.2 ± 3.2 U/gds), C. subvermispora (24.5 ± 1.5 U/gds), and G. trabeum (21.8 ± 1.1 U/gds), respectively. Beta-glucosidase activities in the latter three fungal extracts increased at first and decreased subsequently.
Among hemicellulases, the xylanase activity of S. commune achieved a maximum of 106.5 ± 3.3 U/gds on day 20, after which it decreased to 44.9 ± 1.8 U/gds at the end of cultivation (Fig. 2d). The corresponding activity in P. chrysosporium increased over time and a peak (21.6 ± 1.4 U/gds) appeared on day 25. For G. trabeum, the activity increased to its highest value on day 15 (37.2 ± 2.5 U/gds) and then became relatively constant. Xylanase activity in C. subvermispora remained at a low level during the incubation. From the fifth day, β-xylosidase activity of S. commune increased gradually until the end of the incubation (72.0 ± 2.3 U/gds), while the corresponding activity produced by P. chrysosporium achieved a maximum of 21.8 ± 1.7 U/gds on day 25 (Fig. 2e). The activity in G. trabeum fluctuated at a level between 4.4 ± 0.3 and 6.3 ± 0.9 U/gds during the incubation period. C. subvermispora extract contained only a minor β-xylosidase activity.
As for pectinases, the maximum polygalacturonase activity of S. commune (14.4 ± 2.3 U/gds) was obtained at the end of incubation (Fig. 2f). As for G. trabeum, the polygalacturonase activity increased to a maximum of 8.2 ± 0.4 U/gds on day 20 and decreased markedly afterward. For comparison, the activity remained at a low level in P. chrysosporium extract in the incubation period and was not detected in C. subvermispora extract. Pectin lyase activity was below detectable level in all four fungal extracts (see Additional file 1: Figure S1).
In terms of ligninolytic enzymes, manganese peroxidase (MnP) from C. subvermispora showed high activity (21.1 ± 1.6 U/gds) on day 5 and the activity declined quickly with the incubation time (Fig. 2g). The MnP activity in P. chrysosporium extract maintained at a low level with a maximum activity of 1.5 ± 0.1 U/gds. Laccase (Lac) activity was detected only in C. subvermispora and showed a similar trend to MnP, but the peak activity was merely 2.0 ± 0.1 U/gds on the fifth day (Fig. 2g). A very low lignin peroxidase (LiP) activity was observed in P. chrysosporium extract, while no LiP activity was detected in C. subvermispora (see Additional file 1: Figure S1). In the case of S. commune and G. trabeum, no ligninolytic activities were detected in their culture extracts during the overall incubation period. For the selective ligninolytic fungus C. subvermispora, the ligninolytic enzymes (Lac and MnP) were produced from the beginning of cultivation and their activities decreased over time, while the activities of cellulases and hemicellulases were detected after 5 days of cultivation. Our results with enzyme activities are in accordance with recent secretomic analysis showing the temporal expression patterns of lignocellulytic enzymes of C. subvermispora in aspen-containing liquid medium [33].
For typical Fenton reactions, a system with the capability to potentiate the reduction of Fe3+ to Fe2+ is required. A remarkable difference was observed regarding the Fe3+-reducing activities in the fungal culture extracts (Fig. 3). Analysis of extracellular extracts recovered from S. commune showed that its Fe3+-reducing activity was constantly increased during the incubation period, suggesting the production of extracellular metabolites or enzymes capable of reducing iron ions. As for G. trabeum, the Fe3+-reducing activity increased rapidly in the first 10 days and then decreased slightly. It has been shown that G. trabeum can produce iron-reducing hydroquinones [34, 35] and low-molecular weight glycopeptides [36] for Fenton chemistry. C. subvermispora, on the other hand, showed a rapid decline of Fe3+-reducing activity in its extracellular extracts in the first 5 days of biodegradation, which was in agreement with previous report that hexadecylitaconic acids (known as ceriporic acid B) produced by C. subvermispora could inhibit the iron reduction [37]. P. chrysosporium also showed a similar trend of decreased Fe3+-reducing activity. This decrease of the Fe3+-reducing activity could be related to the consumption of extractives with Fe3+-reducing activity.
Fig. 3.

Iron-reducing capacities of the extracellular extracts produced by four fungi grown on Jerusalem artichoke stalk. Extracellular extracts are sampled after inoculation for 5, 10, 20, and 30 days, respectively. Extracts of Jerusalem artichoke stalk before fungal inoculation are used as reference. The data represent the mean of three replicates and the error bars indicate standard deviations from the mean values
Saccharification experiment
The extracellular enzyme system of S. commune was selected for the hydrolysis of lignocellulosic biomass since it produced the highest hydrolytic enzyme activities among four fungi in the study. Specific activities of the crude enzyme cocktail derived from S. commune were determined using model substrates and compared with those of a commercially available enzyme preparation from Trichoderma longibrachiatum (Table 3). The S. commune enzyme cocktail displayed comparable levels of cellulase activities to the commercial preparation. Specifically, enzymes from S. commune were slightly higher in endoglucanase and exoglucanase activities but lower in β-glucosidase activity. For hemicellulosic substrates, much higher levels of endoxylanase and β-xylosidase activities were present in the S. commune cocktail than the T. longibrachiatum preparation, indicating its high hydrolytic activity toward xylan. The two enzyme mixtures also showed difference in their specific activities against pectin. The S. commune enzymes exhibited more than twofold higher level of polygalacturonase activity relative to T. longibrachiatum enzymes.
Table 3.
Comparison of specific activities (U/mg protein) of the two enzyme cocktails on model substrates
| Enzyme source | CMC | PNPC | PNPG | Beechwood xylan | pNPX | Pectin |
|---|---|---|---|---|---|---|
| S. commune | 21.5 ± 1.6 | 60.7 ± 2.8 | 108.8 ± 3.9 | 112.5 ± 2.3 | 55.6 ± 3.0 | 17.3 ± 1.6 |
| T. longibrachiatum | 19.7 ± 2.9 | 52.2 ± 1.9 | 129.0 ± 2.7 | 26.6 ± 3.2 | 34.8 ± 1.4 | 5.2 ± 1.3 |
The S. commune enzyme cocktail was then examined for the saccharification capacity on pretreated lignocellulosic substrates. After 24 h of hydrolysis at an enzyme loading of 20 mg protein/g glucan, a significantly higher amount of reducing sugars was released by the S. commune cocktail from all substrates tested compared to the commercial enzymes (Fig. 4). In particular, reducing sugar yield produced by the S. commune cocktail from partially delignified Jerusalem artichoke stalk was approximately fivefold as high as that released by the T. longibrachiatum preparation. For the S. commune enzyme cocktail, switchgrass was the most digestible of all four substrates in the enzymatic hydrolysis, followed by Jerusalem artichoke stalk, Miscanthus, and corn stover, respectively. The observed difference in the hydrolysis yields of pretreated lignocellulosic materials could be related to differing effects of the sodium chlorite treatment on the chemical composition of the resulting biomass residue (see Additional file 2: Table S1).
Fig. 4.

Release of reducing sugars from pretreated lignocellulosic biomass by S. commune enzymes and commercial enzymes. The hydrolysis reactions were performed at 37 °C for 24 h incubation with 2 % (w/v) biomass loading. Enzyme loading was set at 20 mg protein/g glucan. The amount of reducing sugars released was determined using the DNS method. Se S. commune enzyme cocktail, Ce commercial enzyme preparation. The mean values of three replicates and standard deviations are presented
The saccharification performance of the S. commune cocktail was further investigated at a relatively low enzyme loading of 5 mg protein/g glucan. After 72 h of hydrolysis, the highest glucan conversion (about 95 %) was achieved by the S. commune enzymes from pretreated switchgrass, 2.5-fold as high as that (about 38 %) by the commercial T. longibrachiatum preparation (Fig. 5a). Such remarkable conversion difference was also observed when saccharification was performed with the other pretreated substrates. In fact, the hydrolysis extents of glucan by the S. commune enzymes within 9 h already exceeded those by the commercial enzymes after 72 h. While conducting these saccharification experiments, we also found that the S. commune cocktail could achieve much higher xylan hydrolysis yields from pretreated biomass in comparison with the commercial preparation (Fig. 5b). For example, after 72 h the xylan conversion in the pretreated switchgrass by the S. commune enzymes was 75.9 %, while the commercial T. longibrachiatum preparation hydrolyzed only 19.1 % of the xylan in the same substrate. Time course of saccharification demonstrated that the enzyme cocktail prepared from S. commune was capable of higher levels of overall hydrolysis of pretreated biomass at a faster rate than the T. longibrachiatum preparation.
Fig. 5.
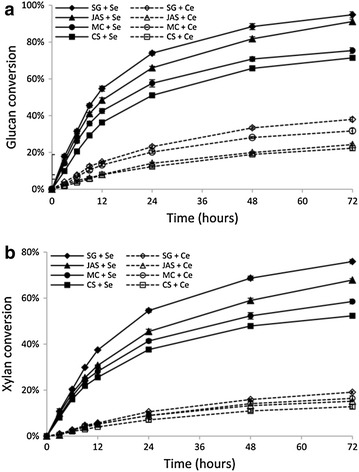
Conversion of (a) glucan and (b) xylan during hydrolysis of pretreated lignocellulosic substrates. The hydrolysis reactions were performed at 37 °C for 72 h incubation with 2 % (w/v) pretreated biomass. S. commune enzyme cocktail and commercial enzyme preparation were both dosed at 5 mg enzyme/g glucan. SG switchgrass, JAS Jerusalem artichoke stalk, MC Miscanthus, CS corn stover. Se S. commune enzyme cocktail, Ce commercial enzyme preparation. Each test was conducted in duplicate
Extracellular proteomic analysis
Proteomic analysis using nanoLC-MS/MS was performed to compare the lignocellulolytic enzyme profile of the S. commune secretome with those of P. chrysosporium, C. subvermispora and G. trabeum during solid-state fermentation on Jerusalem artichoke stalk. A total of 229 proteins were identified in the S. commune secretome, 112 proteins were identified in the P. chrysosporium secretome, 95 proteins for C. subvermispora, and 109 for G. trabeum (see Additional file 3: Table S2). Four fungal species were noticeably different in the number of proteins from glycoside hydrolase (GH) and family distribution (Fig. 6). Enzymes produced by the four fungi can be classified into 38, 26, 19, and 30 GH families, respectively. Proteins from some GH families, such as GH3 (β-glucosidase), GH5 (endoglucanase), GH10 (endoxylanase), GH15 (glucoamylase), GH18 (chitinase), GH20 (N-acetylhexosaminidase), GH27 (α-galactosidase), GH28 (polygalacturonase), GH47 (1,2-α-mannosidase), GH51 (α-l-arabinofuranosidase), GH55 (exo-β-1,3-glucanase), GH88 (d-4,5-unsaturated glucuronyl hydrolase), and GH92 (α-mannosidase) were represented in all four species. A number of other GH families, including GH11 (endo-1,4-β-xylanase), GH17 (glucan endo-1,3-β-glucosidase), GH32 (arabinosidase), GH38 (1,2-α-mannosidase), GH45 (β-1,4-glucanase), GH53 (endo-β-1,4-galactanase), GH62 (α-l-arabinofuranosidase), GH81 (β-1,3-glucanase), and GH93 (exo-1,5-α-l-arabinanase), were detected exclusively in the secretome of S. commune, while hydrolases from GH29 (α-l-fucosidase), GH78 (α-l-rhamnosidase), and GH79 (β-glucuronidase) were represented in the G. trabeum secretome only. GH131 (exo-β-1,3/1,6-glucanase) was detected only in the P. chrysosporium secretome and GH95 (α-l-fucosidase) only in C. subvermispora. The classification results based on GH families revealed that S. commune produced a much larger battery of GH family representatives than those of the other three species.
Fig. 6.

Distribution of glycosyl hydrolase (GH) family proteins identified in the secretomes of four fungi. GH family annotations of the secreted proteins are based on the carbohydrate-active enzyme database (CAZy)
Functional annotation analysis of the secreted proteins indicated that among the four fungal species, S. commune released the highest amount of extracellular enzymes involved in the degradation of all carbohydrate components of plant cell walls, including cellulose, hemicellulose, pectin, starch, and other polysaccharides (Fig. 7), which indicated its high capacity for enzyme secretion. The complete hydrolysis of cellulose depends on the complementary interaction of three different classes of core cellulases: endo-1,4-β-glucanases, exo-1,4-β-glucanases, and β-glucosidases [38, 39]. Both P. chrysosporium and S. commune produced a full enzymatic suite necessary for cellulose degradation (Table 4). Three endoglucanases (GH5), three exoglucanases (GH6 and GH7), and six β-glucosidases (GH1 and GH3) were detected in the S. commune secretome, and two endoglucanases (GH5), three exoglucanases (GH6 and GH7), and three β-glucosidases (GH3) were detected in P. chrysosporium. In contrast, identified cellulolytic enzymes of C. subvermispora comprised only two endoglucanases and two β-glucosidases and no exo-acting cellobiohydrolase was detected. As for G. trabeum, no GH6 or GH7 cellobiohydrolases were detected although its secretome contained three endoglucanases (GH5 and GH12) and four β-glucosidases (GH3 and GH30).
Fig. 7.

Functional classification of the proteins released by four fungi grown on Jerusalem artichoke stalk. Functional predictions of (a) S. commune proteins, (b) P. chrysosporium proteins, (c) C. subvermispora proteins, and (d) G. trabeum proteins are based on the Joint Genome Institute (JGI) databases for Schizophyllum commune H4-8 v3.0, Phanerochaete chrysosporium RP-78 v2.2, Ceriporiopsis (Gelatoporia) subvermispora B, and Gloeophyllum trabeum v1.0, respectively
Table 4.
Summary of cellulose-degrading proteins identified in the secretomes of four fungi
| Species | Accession no.a | Familyb | Predicted proteina | Upc | SignalPd |
|---|---|---|---|---|---|
| S. commune | Endoglucanase | ||||
| Schco3|2602020 | GH5 | endo-β-1,4-glucanase | 18 | N | |
| Schco3|2151684 | GH5 | endo-β-1,4-glucanase | 7 | Y | |
| Schco3|17204 | GH45 | endo-β-1,4-glucanase | 5 | Y | |
| Exoglucanase | |||||
| Schco3|17408 | GH6 | cellobiohydrolase | 11 | Y | |
| Schco3|13879 | GH7 | reducing end-acting cellobiohydrolase | 17 | Y | |
| Schco3|2361770 | GH7 | reducing end-acting cellobiohydrolase | 12 | Y | |
| Β-glucosidase | |||||
| Schco3|2450343 | GH1 | β-glucosidase | 9 | N | |
| Schco3|105341 | GH1 | β-glucosidase | 2 | Y | |
| Schco3|2266284 | GH3 | β-glucosidase | 16 | N | |
| Schco3|2003814 | GH3 | β-glucosidase | 13 | Y | |
| Schco3|1099292 | GH3 | β-glucosidase | 4 | N | |
| Schco3|2026622 | GH3 | β-glucosidase | 20 | N | |
| P. chrysosporium | Endoglucanase | ||||
| Phchr2|2536644 | GH5,CBM1 | endoglucanase | 2 | Y | |
| Phchr2|2864492 | GH5,CBM1 | endoglucanase | 2 | Y | |
| Exoglucanase | |||||
| Phchr2|133052 | GH6,CBM1 | exocellobiohydrolase | 5 | Y | |
| Phchr2|129072 | GH7,CBM1 | 1,4-β-cellobiohydrolase | 8 | Y | |
| Phchr2|137372 | GH7,CBM1 | cellobiohydrolase | 5 | Y | |
| Β-glucosidase | |||||
| Phchr2|139063 | GH3 | β-glucosidase | 3 | Y | |
| Phchr2|1854790 | GH3 | β-glucosidase | 3 | N | |
| Phchr2|2894234 | GH3,CBM1 | β-glucosidase | 2 | Y | |
| C. subvermispora | Endoglucanase | ||||
| Cersu1|106487 | GH5 | endoglucanase | 7 | N | |
| Cersu1|102329 | GH5,CBM1 | endoglucanase | 2 | Y | |
| Β-glucosidase | |||||
| Cersu1|108602 | GH3 | β-glucosidase | 13 | Y | |
| Cersu1|103439 | GH3 | β-glucosidase | 8 | Y | |
| G. trabeum | Endoglucanase | ||||
| Glotr1_1|57704 | GH5 | endoglucanase | 9 | Y | |
| Glotr1_1|63180 | GH5 | endoglucanase | 7 | Y | |
| Glotr1_1|138821 | GH12 | endo-1,4-β-glucanase | 4 | Y | |
| Β-glucosidase | |||||
| Glotr1_1|71534 | GH3 | β-glucosidase | 14 | Y | |
| Glotr1_1|75899 | GH3 | β-glucosidase | 18 | N | |
| Glotr1_1|46629 | GH30 | β-glucosidase | 5 | Y | |
| Glotr1_1|75778 | GH30 | β-glucosidase | 7 | Y | |
aAccession numbers and predicted proteins were obtained from the corresponding Joint Genome Institute (JGI) databases
bFamily information was obtained from the carbohydrate-active enzyme database (CAZy)
cUnique peptides
dPrediction of signal peptides was based on SignalP analysis
Heteroxylan has a backbone of β-(1,4)-linked xylopyranosyl units that can be substituted with side groups. Heteroxylan degradation requires the concerted action of both main chain and side-group hemicellulases. All four fungi in this study produced a variety of enzymes involved in the xylan degradation, but differed in both the number and family distribution (Table 5). Present in all four secretomes were endo-1,4-β-xylanase from GH10, α-galactosidase from GH27, α-l-arabinofuranosidase from GH51, and acetyl xylan esterase from carbohydrate esterase (CE) family 1. In addition, P. chrysosporium secreted two endo-1,4-β-mannanases (GH5), one galactan 1,3-β-galactosidase (GH43), and one oligoxyloglucan cellobiohydrolase (GH74). C. subvermispora secretome contained two beta-mannosidases (GH2) and one beta-galactosidase (GH35) besides an α-l-fucosidase (GH95). Compared with the white-rot species, S. commune and G. trabeum expressed more hemicellulases cleaving xylan side chains (20 and 13, respectively). Specifically, side-chain-degrading hemicellulases of S. commune included three acetyl xylan esterases (CE1), one feruloyl esterase (CE1), one glucuronoyl esterase (CE15), one α-xylosidase (GH31), five β-galactosidases (GH35), six α-l-arabinofuranosidases (GH43, GH51 and GH62), and one α-glucuronidase (GH115).
Table 5.
Summary of hemicellulose-degrading proteins identified in the secretomes of four fungi
| Species | Accession no.a | Familyb | Predicted proteina | Upc | SignalPd |
|---|---|---|---|---|---|
| S. commune | Main chain | ||||
| Schco3|2357273 | GH2 | β-mannosidase | 2 | Y | |
| Schco3|2174600 | GH2 | β-mannosidase | 18 | Y | |
| Schco3|2573073 | GH10 | β-1,4-xylanase | 4 | N | |
| Schco3|2252725 | GH10 | β-1,4-xylanase | 2 | Y | |
| Schco3|2599608 | GH10, CBM1 | β-1,4-xylanase | 12 | Y | |
| Schco3|2170032 | GH11 | endo-1,4-β-xylanase A | 7 | Y | |
| Schco3|103086 | GH43 | β-xylosidase | 2 | Y | |
| Schco3|2147860 | GH74 | xyloglucanase | 5 | Y | |
| Side group | |||||
| Schco3|2159785 | GH27 | α-galactosidase | 7 | N | |
| Schco3|2310213 | GH31 | α-xylosidase | 1 | Y | |
| Schco3|2304995 | GH35 | β-galactosidase | 15 | Y | |
| Schco3|2669535 | GH35 | β-galactosidase | 1 | Y | |
| Schco3|110618 | GH35 | β-galactosidase | 1 | Y | |
| Schco3|2138893 | GH35 | β-galactosidase | 13 | Y | |
| Schco3|13761 | GH35 | β-galactosidase | 5 | N | |
| Schco3|2347041 | GH43 | α-l-arabinofuranosidase | 9 | Y | |
| Schco3|15661 | GH43 | α-l-arabinofuranosidase | 4 | N | |
| Schco3|2226774 | GH43 | α-l-arabinofuranosidase | 3 | N | |
| Schco3|2085606 | GH51 | α-l-arabinofuranosidase | 3 | N | |
| Schco3|108819 | GH51, CBM4 | α-l-arabinofuranosidase | 4 | Y | |
| Schco3|2167580 | GH62 | α-l-arabinofuranosidase | 5 | Y | |
| Schco3|1339154 | GH115 | α-glucuronidase | 8 | Y | |
| Schco3|2275054 | CE1 | acetyl xylan esterase | 3 | Y | |
| Schco3|2061511 | CE1, CBM1 | acetyl xylan esterase | 2 | N | |
| Schco3|2061509 | CE1, CBM1 | acetyl xylan esterase | 2 | Y | |
| Schco3|1161522 | CE1, CBM17 | feruloyl esterase | 4 | Y | |
| Schco3|2151545 | CE1 | esterase | 2 | N | |
| Schco3|2138510 | CE15 | glucuronoyl esterase | 6 | Y | |
| P. chrysosporium | Main chain | ||||
| Phchr2|1969118 | GH3 | β-xylosidase | 5 | Y | |
| Phchr2|2686185 | GH5, CBM1 | endo-1,4-β-mannanase | 2 | Y | |
| Phchr2|2972357 | GH5, CBM1 | endo-1,4-β-mannanase | 2 | Y | |
| Phchr2|2970250 | GH10 | endo-β-1,4-xylanase | 6 | Y | |
| Phchr2|138715 | GH10, CBM1 | endo-1,4-β-xylanase C | 1 | Y | |
| Phchr2|2970992 | GH10, CBM1 | endo-1,4-β-xylanase | 4 | Y | |
| Phchr2|134556 | GH74, CBM1 | oligoxyloglucan cellobiohydrolase | 8 | Y | |
| Side group | |||||
| Phchr2|125033 | GH27 | α-galactosidase | 5 | Y | |
| Phchr2|2973689 | GH43, CBM35 | galactan 1,3-β-galactosidase | 2 | Y | |
| Phchr2|2305691 | GH51, CBM4 | α-l-arabinofuranosidase | 4 | N | |
| Phchr2|126075 | CE1, CBM1 | acetyl xylan esterase | 1 | Y | |
| C. subvermispora | Main chain | ||||
| Cersu1|107994 | GH2 | β-mannosidase | 5 | Y | |
| Cersu1|103229 | GH2 | β-mannosidase | 2 | N | |
| Cersu1|157432 | GH10 | endo-1,4-β-xylanase | 5 | Y | |
| Cersu1|116326 | GH10 | endo-1,4-β-xylanase A | 4 | Y | |
| Cersu1|67561 | GH10, CBM1 | endo-1,4-β-xylanase A | 3 | Y | |
| Side group | |||||
| Cersu1|102094 | GH27 | α-galactosidase | 4 | N | |
| Cersu1|124631 | GH27 | α-galactosidase | 2 | Y | |
| Cersu1|110070 | GH35 | β-galactosidase | 18 | Y | |
| Cersu1|99896 | GH51 | α-l-arabinofuranosidase A | 4 | Y | |
| Cersu1|106199 | GH95 | α-l-fucosidase | 3 | Y | |
| Cersu1|106142 | CE1, CBM1 | acetyl xylan esterase | 2 | Y | |
| G. trabeum | Main chain | ||||
| Glotr1_1|115191 | GH2 | β-mannosidase | 13 | Y | |
| Glotr1_1|122002 | GH3 | β-xylosidase | 16 | Y | |
| Glotr1_1|114574 | GH5 | endo-β-mannanase | 10 | Y | |
| Glotr1_1|46499 | GH10 | endo-1,4-β-xylanase | 19 | N | |
| Glotr1_1|138785 | GH10 | β-1,4-xylanase | 5 | Y | |
| Glotr1_1|140289 | GH10, CBM1 | endo-1,4-β-xylanase | 12 | Y | |
| Glotr1_1|68887 | GH74 | oligoxyloglucan cellobiohydrolase | 21 | Y | |
| Side group | |||||
| Glotr1_1|81012 | GH1 | β-galactosidase-like protein | 23 | Y | |
| Glotr1_1|117566 | GH27 | α-galactosidase | 12 | Y | |
| Glotr1_1|61165 | GH29 | α-l-fucosidase | 10 | N | |
| Glotr1_1|141329 | GH31 | α-xylosidase | 29 | Y | |
| Glotr1_1|119185 | GH31 | α-xylosidase | 19 | Y | |
| Glotr1_1|81512 | GH35 | β-galactosidase | 24 | N | |
| Glotr1_1|111095 | GH35 | β-galactosidase | 37 | Y | |
| Glotr1_1|69366 | GH43, CBM6 | galactan 1,3-β-galactosidase | 10 | Y | |
| Glotr1_1|134804 | GH51 | α-l-arabinofuranosidase | 14 | Y | |
| Glotr1_1|111463 | GH51, CBM4 | α-l-arabinofuranosidase | 12 | Y | |
| Glotr1_1|121308 | GH115 | α-glucuronidase | 24 | Y | |
| Glotr1_1|117128 | CE1 | acetyl xylan esterase | 10 | Y | |
| Glotr1_1|46545 | CE15 | glucuronoyl esterase | 7 | Y | |
a Accession numbers and predicted proteins were obtained from the corresponding Joint Genome Institute (JGI) databases
b Family information was obtained from the carbohydrate-active enzyme database (CAZy)
c Unique peptides
d Prediction of signal peptides was based on SignalP analysis
Pectin is a highly branched structural heteropolysaccharide contained in the primary cell walls of terrestrial plants [40]. Because of the complexity of its structural and chemical composition, a broad range of enzymes are required for the degradation of pectin. Polygalacturonases from GH28 and pectin methylesterases from CE8 were represented in all four species (Table 6). G. trabeum produced the most endo- and exo-acting polygalacturonases and it also secreted an endo-1,5-α-l-arabinanase (GH43), an α-l-rhamnosidase (GH78), and a β-glucuronidase (GH79). As for S. commune, it produced a higher diversity of pectinolytic enzymes targeting side chains, including one rhamnogalacturonan lyase (PL4), four pectate lyases (PL1 and PL3), one endo-β-1,4-galactanase (GH53), one exo-1,5-α-l-arabinanase (GH93), and one rhamnogalacturonan acetylesterase (CE12).
Table 6.
Summary of pectin-degrading proteins identified in the secretomes of four fungi
| Species | Accession no.a | Familyb | Predicted proteina | Upc | SignalPd |
|---|---|---|---|---|---|
| S. commune | Main chain | ||||
| Schco3|110819 | GH28 | polygalacturonase | 3 | Y | |
| Schco3|2141311 | PL1 | pectate lyase | 9 | Y | |
| Schco3|2264630 | PL1 | pectate lyase | 9 | Y | |
| Schco3|2164161 | PL3 | pectate lyase | 3 | Y | |
| Schco3|2130270 | PL3 | pectate lyase | 3 | Y | |
| Schco3|2063343 | PL4 | rhamnogalacturonan lyase | 12 | Y | |
| Side group | |||||
| Schco3|2287708 | GH16 | endo-β-1,3-galactanase | 4 | Y | |
| Schco3|2601007 | GH16 | endo-β-1,3-galactanase | 4 | Y | |
| Schco3|2189132 | GH43 | endo-1,5-α-l-arabinosidase | 5 | Y | |
| Schco3|2011638 | GH43 | α-l-arabinosidase | 7 | Y | |
| Schco3|2601132 | GH43 | α-l-arabinosidase | 6 | Y | |
| Schco3|2187351 | GH43 | exo-β-1,3-galactanase | 1 | Y | |
| Schco3|2086209 | GH43 | arabinosidase | 3 | Y | |
| Schco3|13837 | GH43,CBM35 | exo-β-1,3-galactanase | 9 | Y | |
| Schco3|15841 | GH53 | endo-β-1,4-galactanase | 3 | Y | |
| Schco3|15206 | GH93 | exo-1,5-α-l-arabinanase | 4 | Y | |
| Schco3|2183364 | CE8 | pectinesterase | 7 | N | |
| Schco3|231570 | CE12 | rhamnogalacturonan acetylesterase | 6 | Y | |
| P. chrysosporium | Main chain | ||||
| Phchr2|2865709 | GH28 | endo-polygalacturonase | 7 | Y | |
| Side group | |||||
| Phchr2|132137 | CE8 | pectin methylesterase | 3 | Y | |
| C. subvermispora | Main chain | ||||
| Cersu1|105970 | GH28 | endo-polygalacturonase | 7 | Y | |
| Cersu1|124872 | GH28 | exo-polygalacturonase | 2 | Y | |
| Side group | |||||
| Cersu1|112103 | GH43 | endo-1,5-α-l-arabinase | 3 | Y | |
| Cersu1|103663 | CE8 | pectin methylesterase | 4 | Y | |
| G. trabeum | Main chain | ||||
| Glotr1_1|6650 | GH28 | exo-polygalacturonase | 7 | N | |
| Glotr1_1|110574 | GH28 | endo-polygalacturonase | 4 | Y | |
| Glotr1_1|117232 | GH28 | exo-polygalacturonase | 7 | Y | |
| Glotr1_1|120615 | GH28 | endo-polygalacturonase | 7 | Y | |
| Glotr1_1|138836 | GH28 | exo-polygalacturonase | 7 | Y | |
| Side group | |||||
| Glotr1_1|58475 | GH43 | endo-1,5-α-l-arabinanase | 4 | Y | |
| Glotr1_1|136552 | GH78 | α-l-rhamnosidase | 12 | Y | |
| Glotr1_1|116837 | GH79 | β-glucuronidase | 10 | N | |
| Glotr1_1|77537 | CE8 | pectin methylesterase | 3 | Y | |
| Glotr1_1|112531 | CE8 | pectin methylesterase | 8 | Y | |
a Accession numbers and predicted proteins were obtained from the corresponding Joint Genome Institute (JGI) databases
b Family information was obtained from the carbohydrate-active enzyme database (CAZy)
c Unique peptides
d Prediction of signal peptides was based on SignalP analysis
A clear difference between four fungi was observed in the number of non-hydrolytic proteins involved in polysaccharide depolymerization. Eight and three members of AA9 family were identified in the S. commune and P. chrysosporium secretomes, respectively, while only one was detected in G. trabeum (Table 7). No AA9 proteins were found in the C. subvermispora secretome. The S. commune secretome also contained three expansin-like proteins, as opposed to no such proteins found in the secretomes of the other three species.
Table 7.
Summary of non-hydrolytic proteins involved in polysaccharide deconstruction identified in the secretomes of four fungi
| Species | Accession no.a | Familyb | Predicted proteina | Upc | SignalPd |
|---|---|---|---|---|---|
| S. commune | Schco3|1244931 | AA9 | lytic polysaccharide monooxygenase | 4 | Y |
| Schco3|1324169 | AA9 | lytic polysaccharide monooxygenase | 7 | Y | |
| Schco3|1280218 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| Schco3|2601330 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| Schco3|1219608 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| Schco3|1105422 | AA9 | lytic polysaccharide monooxygenase | 4 | Y | |
| Schco3|1144769 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| Schco3|1192808 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| Schco3|2146679 | CBM1 | carbohydrate-binding module family 1 protein | 11 | Y | |
| Schco3|2246934 | CBM13 | carbohydrate-binding module family 13 protein | 2 | Y | |
| Schco3|2034642 | CBM13 | carbohydrate-binding module family 13 protein | 2 | Y | |
| Schco3|2195917 | CBM63 | expansin-like protein | 3 | Y | |
| Schco3|2195885 | CBM63 | expansin-like protein | 5 | Y | |
| Schco3|2437508 | CBM63 | expansin-like protein | 2 | Y | |
| P. chrysosporium | Phchr2|2609667 | AA9 | lytic polysaccharide monooxygenase | 3 | Y |
| Phchr2|122129 | AA9 | lytic polysaccharide monooxygenase | 2 | Y | |
| Phchr2|1841316 | AA9 | lytic polysaccharide monooxygenase | 3 | Y | |
| C. subvermispora | Cersu1|100632 | CBM13 | carbohydrate-binding module family 13 protein | 4 | N |
| G. trabeum | Glotr1_1|63531 | AA9 | lytic polysaccharide monooxygenase | 7 | Y |
a Accession numbers and predicted proteins were obtained from the corresponding Joint Genome Institute (JGI) databases
b Family information was obtained from the carbohydrate-active enzyme database (CAZy)
c Unique peptides
d Prediction of signal peptides was based on SignalP analysis
With regard to lignin degradation, the secretion patterns of oxidoreductases differed substantially between the four fungi (Table 8). Extracellular protein profile produced by P. chrysosporium included three lignin peroxidases and one manganese peroxidases, while two manganese peroxidases and one Lac were detected in the C. subvermispora secretome. In contrast, neither of the S. commune and G. trabeum secretomes contained ligninolytic enzymes such as manganese peroxidases, lignin peroxidases, or laccases. All fungal species secreted enzymes involved in peroxide generation, but were different in the family distribution and amount of members.
Table 8.
Summary of oxidoreductases involved in lignin degradation identified in the secretomes of four fungi
| Species | Accession no.a | Familyb | Predicted proteina | Upc | SignalPd |
|---|---|---|---|---|---|
| S. commune | Peroxide generation | ||||
| Schco3|2106675 | AA3 | aryl-alcohol oxidase | 11 | Y | |
| Schco3|2186806 | AA3 | glucose oxidase | 14 | Y | |
| Schco3|13888 | AA3 | glucose oxidase | 14 | Y | |
| Schco3|2443737 | AA3 | alcohol oxidase | 9 | N | |
| Schco3|2110714 | AA3 | alcohol oxidase | 3 | Y | |
| Schco3|1099997 | AA3 | alcohol oxidase | 4 | N | |
| Schco3|1110525 | AA5 | copper radical oxidase | 2 | Y | |
| Schco3|1133515 | AA7 | glucooligosaccharide oxidase | 9 | Y | |
| Schco3|1149633 | AA7 | glucooligosaccharide oxidase | 2 | Y | |
| Iron reduction | |||||
| Schco3|114791 | AA3 | cellobiose dehydrogenase | 14 | Y | |
| Schco3|13527 | AA6 | 1,4-benzoquinone reductase | 5 | Y | |
| Schco3|2599696 | ferric reductase | 4 | Y | ||
| P. chrysosporium | Ligninolysis | ||||
| Phchr2|1719525 | AA2 | Ligninase B | 4 | Y | |
| Phchr2|122202 | AA2 | Ligninase LG6 | 3 | Y | |
| Phchr2|1385954 | AA2 | Ligninase precursor | 2 | Y | |
| Phchr2|1179466 | AA2 | Manganese peroxidase H3 | 5 | Y | |
| Peroxide generation | |||||
| Phchr2|11068 | AA5 | glyoxal oxidase precursor | 4 | Y | |
| Phchr2|134241 | AA5 | copper radical oxidase variant A | 3 | Y | |
| Phchr2|124009 | AA5 | copper radical oxidase | 8 | Y | |
| Iron reduction | |||||
| Phchr2|11098 | AA3 | Cellobiose dehydrogenase | 12 | Y | |
| C. subvermispora | Ligninolysis | ||||
| Cersu1|108863 | AA1 | Laccase | 3 | Y | |
| Cersu1|107141 | AA2 | manganese-dependent peroxidase | 7 | Y | |
| Cersu1|22243 | AA2 | manganase peroxidase | 3 | Y | |
| Peroxide generation | |||||
| Cersu1|107672 | AA3 | aryl-alcohol oxidase precursor | 6 | Y | |
| Cersu1|121682 | AA3 | alcohol oxidase | 5 | N | |
| Cersu1|106232 | AA5 | copper radical oxidase | 1 | Y | |
| Cersu1|106640 | AA5 | copper radical oxidase | 3 | Y | |
| Iron reduction | |||||
| Cersu1|69058 | AA6 | 1,4-benzoquinone reductase | 5 | N | |
| G. trabeum | Peroxide generation | ||||
| Glotr1_1|81501 | AA3 | aryl-alcohol oxidase | 13 | Y | |
| Glotr1_1|82487 | AA3 | aryl-alcohol oxidase-like protein | 13 | Y | |
| Glotr1_1|74773 | AA3 | alcohol oxidase | 11 | Y | |
| Glotr1_1|65654 | AA5 | glyoxal oxidase | 8 | Y | |
| Iron reduction | |||||
| Glotr1_1|113732 | AA3 | cellobiose dehydrogenase | 3 | N | |
a Accession numbers and predicted proteins were obtained from the corresponding Joint Genome Institute (JGI) databases
b Family information was obtained from the carbohydrate-active enzyme database (CAZy)
c Unique peptides
d Prediction of signal peptides was based on SignalP analysis
Discussion
During SEM and stalk component degradation analysis, it is interesting to note that S. commune, with the highest degradative enzyme activities after 15 days, did not produce strong polysaccharide degradation in Jerusalem artichoke stalk. P. chrysosporium, on the other hand, showed the highest consumption of cellulose and hemicellulose despite moderate enzyme activities. The inconsistency in the enzyme activities and degradation kinetics can be explained by the different lignin-degrading strategies employed by different fungi. In plant cell walls, lignin is intimately associated with structural polysaccharides. The complex hydrophobic network of lignin acts as a form of protection for the interior carbohydrate components, thus making them recalcitrant to hydrolytic attack [41]. Although they cannot utilize lignin as a carbon source, both P. chrysosporium and C. subvermispora secreted a wide array of extracellular peroxidases and oxidases to efficiently depolymerize and mineralize this formidable substrate. The interaction of ligninolytic enzymes resulted in the degradation of lignin and consequently, the exposure of the polysaccharide constituents of plant cell walls, rendering them more available to classical cellulases and hemicellulases. In contrast, the brown-rot fungus G. trabeum has been suggested to employ hydroquinone-driven Fenton chemistry as a degradative mechanism for initiating polysaccharide depolymerization [23]. Similar to G. trabeum, the S. commune secretome did not contain the core components of ligninolytic enzyme system. It caused a very limited lignin removal from the untreated stalk due to the lack of extracellular class II peroxidases like MnPs and LiPs. The huge bulk of lignin not only impedes enzymatic access to cell wall polysaccharides but also causes non-productive adsorption of hydrolytic enzymes [42]. Despite the incomplete ligninolysis, S. commune still caused an appreciable degradation of polysaccharides in Jerusalem artichoke stalk compared with G. trabeum.
During the lignocellulose biodegradation S. commune demonstrated a preferential degradation of polysaccharides with respect to lignin. The pyrolysis analysis provided evidence that attack of plant cell wall constituents by S. commune also resulted in some modifications of lignin substructure, including demethoxylation, demethylation, and side-chain oxidation. Such oxidative alterations of lignin moieties were very similar to that by the brown-rot fungus G. trabeum. Although the lignin residues after hydroxyl radical attack remain in situ, the original inter-monomer side-chain linkages within lignin have been disrupted and the resulting aromatic polymer is no longer recognizable as lignin [43]. It is probable that S. commune utilized an oxidative mechanism to enable the diffusion of carbohydrate-degrading enzymes through the lignocellulosic matrix to interior polysaccharides.
In the lignin degradation systems of white-rot fungi, an array of redox enzymes, such as copper radical oxidases and glucose-methanol-choline (GMC) oxidoreductases, are physiologically coupled to peroxidases via H2O2 generation. In view of Fenton chemistry employed by brown-rot fungi, H2O2 production by copper radical oxidases and GMC oxidoreductases potentially supports hydroxyl radical generation. In the absence of class II peroxidases, it is interesting to note that S. commune released a more diverse assortment of oxidoreductases than P. chrysosporium and C. subvermispora. Specifically, we observed a high number of GMC oxidoreductases in the protein profile of the S. commune secretome relative to the other two species, including an aryl-alcohol oxidase (>jgi|Schco3|2106675), two glucose oxidases (>jgi|Schco3|2186806 and >jgi|Schco3|13888), and three alcohol oxidase (>jgi|Schco3|2443737, >jgi|Schco3|2110714, and >jgi|Schco3|1099997). It has been found that alcohol oxidases could use methanol, the primary product of lignin demethylation by brown-rot fungi, as a substrate to generate H2O2 [44]. Likewise, copper radical oxidase (>jgi|Schco3|1110525) and glucooligosaccharide oxidase (>jgi|Schco3|1133515) played a part in extracellular peroxide generation. Given the decay pattern of S. commune observed during stalk degradation, there remains the possibility that these seemingly redundant oxidoreductases serve as a source of H2O2 to support Fenton chemistry.
This proposed non-enzymatic oxidative mechanism was further supported by the fact that S. commune secreted multiple reductive enzymes involved in the Fe2+ production required for Fenton chemistry. In this connection, cellobiose dehydrogenase (CDH) (>jgi|Schco3|114791) and ferric reductase (>jgi|Schco3|2599696) may be of relevance to Fenton chemistry by their role in the Fe3+-reduction [45, 46]. Additionally, benzoquinone reductase (>jgi|Schco3|13527) and phenylalanine ammonia lyase (>jgi|Schco3|1191717) may participate in iron reduction system driven by quinone redox cycling [46]. These observations indicated that S. commune possessed the enzymatic apparatus to carry out Fenton reactions during lignocellulose degradation. The identification of these proteins in the S. commune secretome, viewed together with the production of iron-reducing agents and oxidative modification of lignin structure, supported a biodegradative role for extracellular Fenton system in plant cell wall degradation by this fungus.
In the conversion process of lignocellulosic feedstocks to bioethanol, the complete hydrolysis of structural polysaccharides into fermentable sugars requires the cooperative action of various degradative enzymes with complementary activities. In this context, S. commune is a promising enzyme source as its enzyme system favored a more complex inventory of polysaccharide-degrading enzymes with enhanced enzyme activities. Comparison of the lignocellulolytic enzyme profiles of S. commune and other fungi in the study showed that S. commune had significantly higher levels of hydrolytic enzyme activities, including cellulolytic, xylanolytic, and pectinolytic activities. Enzyme cocktail from S. commune showed more than threefold higher xylanase activity and twofold higher polygalacturonase activity than the T. longibrachiatum enzyme preparation with comparable levels of cellulolytic activities. Proteomic analysis revealed that its secretome contained a more diverse repertoire of GH families and more members in each family than the other fungi. Biological function analysis of extracellular proteins indicated that S. commune produced a larger variety of accessory enzymes that facilitate the complete hydrolysis of non-cellulosic polysaccharides, such as pectin and heteroxylan.
The present study showed that S. commune secreted a wide variety of hemicellulases and pectinases acting not only on main chains (xylanase, polygalacturonase, and pectate lyase), but also side groups, such as acetyl xylan esterase, α-l-arabinofuranosidase, α-glucuronidase, feruloyl esterase, endo-β-1,3-galactanase, and α-l-arabinosidase. It is noteworthy that the complementary action of cellulases and accessory enzymes such as hemicellulases, pectinases, and LPMOs, plays a key role in the hydrolysis of lignocellulosic materials as observed in previous studies. Xylanase supplementation to cellulase enzyme mixtures has been shown to improve the overall hydrolysis of pretreated lignocellulosic substrates due to increased cellulose accessibility to cellulases through solubilization of xylan and alteration of fiber features [47]. A notable improvement in cellulose hydrolysis was also observed when a pectinase-enriched complex was added to cellulase preparations [4]. Side-chain-degrading enzymes may also contribute to the conversion yields of lignocellulosic materials containing branched polysaccharides by enhancing the hydrolysis effectiveness of main chain-cleaving enzymes. It has been reported that acetyl xylan esterases present in enzyme mixtures could promote xylan solubilization, and thus the subsequent hydrolysis of xylan and cellulose in the biomass by removing acetyl groups from xylan, as demonstrated in the saccharification of wheat straw and giant weed [48]. When a very low amount (1.2 mg/g glucan) of bacterial α-arabinofuranosidase (GH51) and α-glucuronidase (GH67) was added to a mixture of cellulase and xylanase, the total enzyme loading required for a 80 % sugar yield from AFEX-pretreated corn stover can be decreased by about 33 % [49]. As most commercial cellulase preparations contain relatively low levels of hemicellulases and pectinases, the enzyme pool of S. commune via solid-state fermentation offers a preferable alternative as it favors a complex cocktail of synergistically acting accessory enzymes, which promote enzyme loading reduction without decreasing hydrolysis yields.
Comparison with the other wood-rotting fungi showed that in addition to glycoside hydrolases, S. commune produced higher amounts of LPMOs, expansin-like proteins together with carbohydrate-binding modules. These non-hydrolytic proteins are suggested to reduce the recalcitrance of the lignocellulosic matrix, thereby enhancing the efficiency of enzymatic hydrolysis by hydrolases. AA9 family is a recently discovered class of fungal copper-dependent polysaccharide monooxygenases that oxidatively cleave cellulose [39]. By introducing chain breaks in the crystalline cellulose microfibrils, LPMOs from AA9 family enhance cellulosic digestibility by conventional hydrolytic enzymes. Intensive studies have shown that LPMO addition can significantly boost the hydrolysis efficiency of cellulase mixtures [50, 51]. Several fungal and bacterial expansins and expansin-like proteins have been reported to improve the enzymatic hydrolysis of crystalline cellulose by cellulases [52–54] and xylan by xylanases [55] without detectable hydrolytic activity. More recently, a novel expansin from S. commune, ScExlx1 has been found to act on both cellulose and chitin [56]. It is suggested that expansins are able to loosen the tightly packed architecture of plant cell wall matrix through the disruption of hydrogen bonds in polysaccharide networks. The high abundance of non-hydrolytic proteins such as LPMOs and expansin-like proteins in the S. commune secretome suggested that they may contribute to the high saccharification capacity of the S. commune enzyme cocktail observed in this work.
The commercial T. longibrachiatum preparation used for comparison in our study contained a variety of hydrolytic enzymes, including cellulase, xylanase, pectinase, mannanase, xyloglucanase, laminarase, β-glucosidase, β-xylosidase, α-l-arabinofuranosidase, and amylase. The enzyme preparation has been used in the hydrolysis of alkaline-pretreated Miscanthus giganteus and achieved higher hydrolysis yield of polysaccharides than another commonly used cellulase preparation Celluclast 1.5L [57]. During the saccharification experiments, the S. commune cocktail exhibited high activity compared to the T. longibrachiatum preparation against all substrates tested, converting 71–95 % of glucan compared to 22–38 % glucan conversion by the commercial preparation. It should be noted that all the comparative hydrolysis in this study were carried out at the optimal temperature and pH of the commercial preparation. One would expect that when the optimal conditions for the S. commune enzymes were used, the effectiveness of enzymatic hydrolysis by the S. commune cocktail can be further improved. Additionally, most commercial cellulase mixtures need the supplementation of exogenous β-glucosidase for efficient hydrolysis due to end-product inhibition. The β-glucosidase activity in the S. commune enzyme mixture was sufficiently high to ensure a complete hydrolysis of cellobiose to glucose.
When using LC-MS/MS to analyze the extracellular proteome, we noticed that the expression patterns of some homologous proteins differed among fungi. For instance, both P. chrysosporium and S. commune encode two glucuronoyl esterases (CE15) in their genomes [58, 59], and we identified one CE15 protein (Schco3|2138510) in the S. commune secretome. However, no peptides matching CE15 proteins of P. chrysosporium were detected in our study, even though an earlier study detected such protein in extracellular liquid cultures [60]. It should be noted that the LC-MS/MS approach reported here favored identification of soluble proteins, whereas proteins bound to the solid substrate may be overlooked. As shown by SEM, the hyphae of P. chrysosporium penetrated through the stalk cell walls, while the mycelia of S. commune mostly covered the stalk surface over the cultivation period. As the growth of P. chrysosporium on stalk was characterized by invasive mycelia, we assumed that some extracellular proteins were adsorbed on the substrate colonized by fungi. Consequently, protein recovery from such closely intertwined substrate/mycelia mixture might be restricted by the extraction method. Therefore, growth conditions should be taken into account when it comes to the interpretation of the absence of detectable extracellular proteins.
Conclusions
The current work described for the first time the decay pattern and the composition of the secretome of S. commune growing on complex lignocellulosic substrates. Comparison with the representatives of white-rot and brown-rot fungi revealed that S. commune employed a Fenton chemistry-based oxidative mechanism for lignocellulose modification, while possessing a composite polysaccharide-degrading enzyme system. Furthermore, the enzymatic cocktail derived from S. commune displayed high hydrolysis effectiveness of polysaccharide in lignocellulose materials at low protein loadings. Proteomic analysis of the S. commune extracellular enzyme profile indicated that the significant saccharification efficiency could be attributed to the synergistic cooperation of cellulases and hemicellulases, pectinases, and non-hydrolytic proteins like LPMOs and expansins. These findings presented here shed light on the complex degradative enzyme system of S. commune, and highlight the important role of accessory enzymes in lignocellulose hydrolysis.
Methods
Fungal strains and culture conditions
Fungal strain S. commune SH12 was isolated from forest soil (Shaanxi province, China) and preserved on potato dextrose agar (PDA) plates at 4 °C. P. chrysosporium PC2 were obtained from China General Microbiological Culture Collection Center (CGMCC) and C. subvermispora CBS 347.63 were obtained from the fungal culture collections of the Centraalbureau voor Schimmelcultures (CBS, Baarn, The Netherlands). These strains were maintained following suppliers’ protocol at 4 °C.Prior to solid-state fermentation, the fungi were cultivated at 28 °C for 7 days on PDA-medium plates.
Jerusalem artichoke stalk, mainly composed of 38.64 % cellulose, 22.58 % arabinoxylan (consisted of 18.05 % xylose and 4.53 % arabinose), 4.97 % pectin and 18.10 % lignin, was chopped into small chips of approximately 1 cm squares. The solid-state fermentation was carried out in 150 mL Erlenmeyer flasks containing 3 g of sterilized Jerusalem artichoke stalk and 10 ml of Mandels’ salts solution (pH 6.5, 95 ml of water, 5 ml 20X nitrate salts, and 0.1 ml 1000X trace elements). Each flask was inoculated with four mycelium agar plugs of 1 cm in diameter from fungal species and incubated at 28 °C without humidity control. Non-inoculated Jerusalem artichoke stalk incubated under the same conditions was used as control. All treatments were performed in triplicate.
Scanning electron microscopy (SEM)
Scanning electron microscopy (SEM) was carried out to observe the morphological changes on Jerusalem artichoke stalk after 30 days of incubation with respective fungal species. Samples were prepared for scanning electron microscopy as described by [61]. Micrographs of non-inoculated and fungi-inoculated samples were taken using a Hitachi S3400N scanning electron microscope (Hitachi) at an acceleration voltage of 15.0 kV.
Pyrolysis gas chromatography–mass spectrometry
Pyrolysis gas chromatography–mass spectrometry (Py-GC/MS) of sound and degraded stalk was performed using a EGA/PY-3030D pyrolyzer (Frontier Laboratories, Japan) coupled to a Shimadzu single quadrupole GCMS-QP2010 Ultra gas chromatograph–mass spectrometer (Shimadzu Scientific, Japan) with a Phenomenex Zebron™ ZB-5HT GC capillary column (30 m × 0.25 mm × 0.25 μm). The pyrolysis was conducted at 500 °C. The chromatography temperature was programmed from 45 °C for 3 min to 300 °C at 15 °C min−1 and then held for 5 min. Helium was used as carrier gas at a rate of 1 ml min−1. The pyrolysis products were identified by comparing the mass spectra with those of the Wiley and NIST libraries.
Extracellular protein extraction
The cultures of each fungus were harvested every 5 days until the end of incubation. The fungal cultures were incubated with 30 mL Milli-Q water at 4 °C with shaking at a rate of 200 rpm for 2 h. The water-soluble phases containing the total proteins were collected by centrifugation at 10,000× g for 15 min at 4 °C and then passed through a 0.25-μm filter. The filtrates were assayed directly for enzyme activities before further concentration and desalting with 3-kDa cutoff Amicon Ultra-15 Centrifugal Filter Unit (Merck Millipore) and lyophilized for secretome analysis. The fungal mycelia were removed from the solid residues by incubating in a mixture of acetic acid and nitric acid (8:1, v/v) on ice with agitation for 1 h, and washed with deionized water until it was pH-neutral. The mycelium-free residues were oven-dried at 60 °C to constant weight for the subsequent determination of chemical composition. The structural carbohydrate and lignin contents were determined according to the NREL laboratory analytical procedure (version 08-03-2012). In brief, 0.5 g of sample (dry weight) was extracted with 200 mL of ethanol at 95 °C and hydrolyzed at 30 °C with 3.0 mL H2SO4 (72 %) for 60 min. Then 84 mL of water was added and a second hydrolysis was carried out in the autoclave at 121 °C for 1 h. The mixture was then filtered by porcelain filter crucibles with glass filters. The amounts of sugars in the filtrates were determined by HPLC. The content of Klason lignin was determined by subtracting the ash content from the solid residue dried at 105 °C overnight. Ash content was determined by heating the solid residue at 575 °C for 3 h.
Enzyme assays
Activities of endoglucanase, endoxylanase, and polygalacturonase were determined by the dinitrosalicylic acid (DNS) assay [62], with 1.0 % low-viscosity carboxymethylcellulose (CMC), beechwood xylan, and polygalacturonic acid as substrates, respectively. 50 μL crude enzyme was incubated with 150 μL of 1.0 % (w/v) substrate in 50 mM sodium acetate buffer (pH 5.0) at 50 °C for 10 min, and the reaction was stopped with 50 μL of 1M NaOH. Calculation of enzyme activities was based on corresponding standards containing glucose, xylose, and galacturonic acid. Enzyme activities are represented in units per gram of dry substrate (U/gds), with one unit of enzyme activity defined as the amount of enzyme needed to liberate 1 μmol of reducing sugars in 1 min from the substrate under given assay conditions.
Cellobiohydrolase, β-glucosidase, and β-xylosidase activities were quantified using the respective substrates p-nitrophenyl-β-d-cellobioside (pNPC), p-nitrophenyl-β-d-glucopyranoside (pNPG), and p-nitrophenyl-β-d-xylopyranoside (pNPX) [63]. The reactions were carried out in 1.5 ml centrifuge tubes containing 50 μL crude enzyme, 50 μL of 200 mM sodium acetate buffer (pH 5.0), and 100 μL of 5 mM substrate. After 10 min incubation at 50 °C, the reaction was terminated by adding 100 μL 1M Na2CO3, and the color development was recorded at 405 nm. Enzyme activities were calculated using p-nitrophenol standard. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of pNP in 1 min under the assay conditions.
MnP activity was measured by the oxidative dimerization of 2,6-dimethoxyphenol (2,6-DMP) in the presence of H2O2 [64]. The reaction mixture contained 1 mM 2,6-dimethoxyphenol (2,6-DMP), 1 mM MnSO4, 50 mM sodium tartrate (pH 4.5), 100 μM H2O2, and crude enzyme in a total volume of 1000 μL. LiP activity was assayed with veratryl alcohol as a substrate in 100 mM sodium tartrate (pH 3.0) [65]. The crude enzyme was mixed with 40 mM veratryl alcohol and 10 mM H2O2 in a total volume of 1000 μL at room temperature. Laccase (Lac) activity was determined by monitoring the oxidation of 2,2′-azonodi-3-ethylbenzothiazoline-6-sulfuric acid (ABTS) at room temperature [66]. The reaction mixture contained 1 mM ABTS in 100 mM sodium acetate buffer (pH 4.5) and crude enzyme in a total volume of 1000 μL. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of corresponding products in one min under the assay conditions.
Reducing capacity analysis
Assessment of the iron-reducing ability of extracellular soluble extracts from fungal species was based on the formation of the Fe2+-ferrozine complex [67]. The reaction mixture contained 200 μL of 20 mM sodium acetate buffer (pH 5.0), 60 μL of 40 mM ferrozine, 100 μL sample, and 40 μL of 10 mM freshly prepared FeCl3. The absorbance change at 562 nm was recorded after 10 min of incubation at room temperature.
Biomass saccharification
All lignocellulosic substrates were delignified with sodium chlorite before enzymatic hydrolysis [68]. Enzyme preparations used in the saccharification assay were crude enzyme cocktail from S. commune and a cellulase cocktail from Trichoderma longibrachiatum (C9748, Sigma). The hydrolysis reactions were carried out with 2 % (w/v) pretreated biomass in 50 mM sodium acetate buffer (pH 5.0) in a final volume of 1 ml. Saccharifications were performed at two enzyme loadings (20 and 5 mg protein/g delignified biomass) in an orbital shaker incubator at 37 °C. Control assays including enzyme without substrate and substrate without enzyme are conducted under the same conditions. Protein concentration was determined using the Bradford Protein Assay Kit (GenStar, China) according to the manufacturer’s instructions.
The hydrolysis was terminated by heating the reaction mixture at 100 °C for 10 min to inactivate the enzymes. The supernatants were collected by centrifugation at 12,000 rpm for 10 min and further clarified by filtration through a 0.25-μm filter. The total contents of reducing sugars were quantified by the DNS method. The individual concentration of glucose, xylose, and arabinose in the supernatants was determined by Essentia LC-15C high performance liquid chromatography (Shimadzu, Japan) equipped with an Rezex ROA-Organic acid H+ (8 %) column (Phenomenex, USA) and a RID-10A refractive index detector (Shimadzu, Japan). The glucan and xylan conversions were calculated based on the initial contents of polysaccharides in the pretreated substrates. All hydrolysis experiments were conducted in duplicate.
Protein identification by nanoLC-MS/MS
50 μg of total protein was separated by SDS-PAGE in triplicate using 12.5 % polyacrylamide at 100 V, stained with Coomassie Blue G-250 overnight. Each lane was divided into six fractions, and each fraction was cut into small pieces (approximately 1 mm2). The gel pieces were washed with pure water for 3 times, destained with 10 % acetic acid, and washed again with pure water. The gel pieces were immerged in acetonitrile and then vacuum-dried. The dried gel pieces were incubated in 10 mM dithiothreitol (DTT) solution at 56 °C for 45 min, after which they were alkylated with 55 mM iodoacetamide (IAM) solution in the dark at room temperature for 30 min. The gel pieces were washed with acetonitrile again before vacuum-dried. Then the gel pieces were subject to trypsin digestion at 37 °C overnight. The peptides were extracted with 30 % acetonitrile in 0.1 % formic acid for 30 min followed by 60 % acetonitrile for 30 min, and dried in vacuum concentrator. The residue was reconstituted with 0.1 % formic acid for nanoLC-MS/MS analysis.
NanoLC separation was achieved with a Waters (Milford, MA, USA) nanoACQUITY nano-HPLC. Both trap column and analytical column were home made. The trap column was made with 100 μm I.D. fused silica capillary (Polymicro, Phoenix, AZ, USA) filled with 2 cm of C18 stationary phase (Phenomenex, Torrance, CA, USA). The analytical column was made with 50 μm I.D. fused silica capillary (Polymicro) filled with 10 cm of C18 stationary phase. A 3-μm-diameter spray tip was pulled at the end of the analytical column with a Sutter P-2000 laser micropipette puller (Sutter Instrument, Novato, CA, USA). A gradient elution program was used. Gradients of different lengths were used for samples of different complexities. Following LC nanospray, ESI-MS was performed on a Thermo Q Exactive high-resolution mass spectrometer (Thermo Scientific, Waltham, MA, USA). The 10 most intensive peptide signals from the full scan were selected for MS/MS scans.
Raw data from the mass spectrometer were preprocessed with Mascot Distiller 2.5 for peak picking. The resulting peak lists were searched against the Joint Genome Institute (JGI) databases for S. commune H4-8 v3.0, P. chrysosporium RP-78 v2.2, and C. (Gelatoporia) subvermispora B using Mascot search engine (version 2.5.1, Matrix Science). The search parameters included carbamidomethyl cysteine as fixed modification and oxidized methionine as the variable modification. A maximum of two missed tryptic cleavages was allowed. The peptide mass tolerance was set at 15 ppm, and MS/MS fragment mass tolerance was set at 0.02 Da. For protein identification, only proteins containing a minimum of two significant peptides were considered and at least one unique peptide needed to be identified in the protein. The significance threshold p is set to be less than 0.01, thus bringing peptide and protein false discovery rate (FDR) below 1 %. For each identified protein, the theoretical isoelectric point (pI) and molecular weight (Mw) was calculated using the Compute pI/Mw tool in ExPASy (http://web.expasy.org/compute_pi). SignalP (http://www.cbs.dtu.dk/services/SignalP) was used to predict possible secretion signals in the identified proteins. A schematic representation of the nanoLC-MS/MS procedure is shown in Additional file 4: Figure S2.
Authors’ contributions
NZ and HY conceived and designed the experiments. NZ performed the majority of the laboratory work, analyzed the results, and drafted the manuscript. JL participated in the determination of enzyme activities and SEM experiments, and performed the enzymatic hydrolysis. JY contributed to the interpretation of results and helped to revise the manuscript. YL assisted in the statistical analysis of LC-MS/MS data. YY, LJ, and ML carried out the material pretreatment and determination of chemical compositions. HY supervised the overall work, discussed the results, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors acknowledge the financial support for this work from the National High Technology Research and Development Program of China (863 Program, number: 2011AA10A206).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- SSF
solid-state fermentation
- LPMO
lytic polysaccharide monooxygenase
- CAZy
carbohydrate-active enzyme database
- AA
auxiliary activity
- NanoLC-MS/MS
nano liquid chromatography-tandem mass spectrometry
- Py-GC/MS
pyrolysis coupled with gas chromatography-mass spectrometry
- MnP
manganese peroxidase
- LiP
lignin peroxidase
- Lac
laccase
- GH
glycoside hydrolase
- CE
carbohydrate esterase
- PL
polysaccharide lyase
- CDH
cellobiose dehydrogenase
- GMC oxidoreductase
glucose-methanol-choline oxidoreductase
- CMC
carboxymethylcellulose
- 2,6-DMP
2,6-dimethoxyphenol
- ABTS
2,2′-azonodi-3-ethylbenzothiazoline-6-sulfuric acid
- DNS
3,5-Dinitrosalicylic acid
- pNPC
p-nitrophenyl-β-d-cellobioside
- pNPG
p-nitrophenyl-β-d-glucopyranoside
- pNPX
p-nitrophenyl-β-d-xylopyranoside
- DTT
dithiothreitol
- IAM
iodoacetamide
- FDR
false discovery rate
Additional files
10.1186/s13104-016-1932-7 Pectin lyase and lignin peroxidase activities of four fungi during SSF on Jerusalem artichoke stalk. Pectin lyase activities are shown in A and lignin peroxidase activities are shown in B. The values shown are the mean of three replicates and the error bars indicate standard deviations from the mean values.
10.1186/s13104-016-1932-7 Chemical composition of lignocellulosic substrates before and after sodium chlorite delignification. The main components of lignocellulosic substrates include anhydroglucose (Glu), anhydroxylose (Xyl) and anhydroarabinose (Ara). The data represent the mean values of three replicates.
10.1186/s13104-016-1932-7 Identified proteins in the secretomes of four fungi during SSF on Jerusalem artichoke stalk.
10.1186/s13104-016-1932-7 Schematic representation of nanoLC-MS/MS analysis of the extracellular proteins.
Contributor Information
Ning Zhu, Email: joylign@yeah.net.
Jiawen Liu, Email: 1070184179@qq.com.
Jinshui Yang, Email: yangjsh1999@163.com.
Yujian Lin, Email: 15210562063@163.com.
Yi Yang, Email: yang11yi@aliyun.com.
Lei Ji, Email: jilei.1010@163.com.
Meng Li, Email: 421719421@qq.com.
Hongli Yuan, Email: hlyuan@cau.edu.cn.
References
- 1.Margeot A, Hahn-Hagerdal B, Edlund M, Slade R, Monot F. New improvements for lignocellulosic ethanol. Curr Opin Biotechnol. 2009;20:372–380. doi: 10.1016/j.copbio.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Van Dyk JS, Pletschke BI. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-Factors affecting enzymes, conversion and synergy. Biotechnol Adv. 2012;30:1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Singhania RR, Sukumaran RK, Patel AK, Larroche C, Pandey A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb Technol. 2010;46:541–549. doi: 10.1016/j.enzmictec.2010.03.010. [DOI] [Google Scholar]
- 4.Berlin A, Maximenko V, Gilkes N, Saddler J. Optimization of enzyme complexes for lignocellulose hydrolysis. Biotechnol Bioeng. 2007;97:287–296. doi: 10.1002/bit.21238. [DOI] [PubMed] [Google Scholar]
- 5.Ahamed A, Vermette P. Enhanced enzyme production from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger LMA grown as fed batch in a stirred tank bioreactor. Biochem Eng J. 2008;42:41–46. doi: 10.1016/j.bej.2008.05.007. [DOI] [Google Scholar]
- 6.Fujii T, Fang X, Inoue H, Murakami K, Sawayama S. Enzymatic hydrolyzing performance of Acremonium cellulolyticus and Trichoderma reesei against three lignocellulosic materials. Biotechnol Biofuels. 2009;2:24. doi: 10.1186/1754-6834-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusakov AV. Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 2011;29:419–425. doi: 10.1016/j.tibtech.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Marx IJ, van Wyk N, Smit S, Jacobson D, Viljoen-Bloom M, Volschenk H. Comparative secretome analysis of Trichoderma asperellum S4F8 and Trichoderma reesei Rut C30 during solid-state fermentation on sugarcane bagasse. Biotechnol Biofuels. 2013;6:172. doi: 10.1186/1754-6834-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 10.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bey M, Zhou SM, Poidevin L, Henrissat B, Coutinho PM, Berrin JG, et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl Environ Microb. 2013;79:488–496. doi: 10.1128/AEM.02942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biely P, Puls J, Schneider H. Acetyl xylan esterases in fungal cellulolytic systems. FEBS Lett. 1985;186:80–84. doi: 10.1016/0014-5793(85)81343-0. [DOI] [Google Scholar]
- 13.Spanikova S, Biely P. Glucuronoyl esterase—Novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 2006;580:4597–4601. doi: 10.1016/j.febslet.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 14.Tenkanen M, Siika-aho M. An alpha-glucuronidase of Schizophyllum commune acting on polymeric xylan. J Biotechnol. 2000;78:149–161. doi: 10.1016/S0168-1656(99)00240-0. [DOI] [PubMed] [Google Scholar]
- 15.Chong SL, Battaglia E, Coutinho PM, Henrissat B, Tenkanen M, de Vries RP. The alpha-glucuronidase Agu1 from Schizophyllum commune is a member of a novel glycoside hydrolase family (GH115) Appl Microbiol Biot. 2011;90:1323–1332. doi: 10.1007/s00253-011-3157-y. [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Lee YG, Choi IS, Lee KH, Cho EJ, Bae HJ. Heterologous expression of endo-1,4-beta-xylanase A from Schizophyllum commune in Pichia pastoris and functional characterization of the recombinant enzyme. Enzyme Microb Technol. 2013;52:170–176. doi: 10.1016/j.enzmictec.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Lee YM, Lee H, Kim JS, Lee J, Ahn BJ, Kim GH, et al. Optimization of medium components for beta-glucosidase production in Schizophyllum commune KUC9397 and enzymatic hydrolysis of lignocellulosic biomass. Bioresources. 2014;9:4358–4368. [Google Scholar]
- 18.Fowler TJ, Mitton MF, Rees EI, Raper CA. Crossing the boundary between the B alpha and B beta mating-type loci in Schizophyllum commune. Fungal Genet Biol. 2004;41:89–101. doi: 10.1016/j.fgb.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann S, Freihorst D, Raudaskoski M, Schmidt-Heck W, Jung EM, Senftleben D, et al. Transcriptome and functional analysis of mating in the basidiomycete Schizophyllum commune. Eukaryot Cell. 2012;11:571–589. doi: 10.1128/EC.05214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer GE, Horton JS. Mushrooms by magic: making connections between signal transduction and fruiting body development in the basidiomycete fungus Schizophyllum commune. FEMS Microbiol Lett. 2006;262:1–8. doi: 10.1111/j.1574-6968.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohm RA, de Jong JF, de Bekker C, Wosten HAB, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol Microbiol. 2011;81:1433–1445. doi: 10.1111/j.1365-2958.2011.07776.x. [DOI] [PubMed] [Google Scholar]
- 22.Daniel G. Use Of electron-microscopy for aiding our understanding of wood biodegradation. FEMS Microbiol Rev. 1994;13:199–233. doi: 10.1111/j.1574-6976.1994.tb00043.x. [DOI] [Google Scholar]
- 23.Jensen KA, Houtman CJ, Ryan ZC, Hammel KE. Pathways for extracellular fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microb. 2001;67:2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt O, Liese W. Variability Of Wood Degrading Enzymes of Schizophyllum commune. Holzforschung. 1980;34:67–72. doi: 10.1515/hfsg.1980.34.2.67. [DOI] [Google Scholar]
- 25.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A. 2014;111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willick GE, Morosoli R, Seligy VL, Yaguchi M, Desrochers M. Extracellular proteins secreted by the basidiomycete Schizophyllum commune in response to carbon source. J Bacteriol. 1984;159:294–299. doi: 10.1128/jb.159.1.294-299.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xavier-Santos S, Carvalho CC, Bonfa M, Silva R, Capelari M, Gomes E. Screening for pectinolytic activity of wood-rotting Basidiomycetes and characterization of the enzymes. Folia Microbiol. 2004;49:46–52. doi: 10.1007/BF02931645. [DOI] [PubMed] [Google Scholar]
- 28.Doria E, Altobelli E, Girometta C, Nielsen E, Zhang T, Savino E. Evaluation of lignocellulolytic activities of ten fungal species able to degrade poplar wood. Int Biodeter Biodegr. 2014;94:160–166. doi: 10.1016/j.ibiod.2014.07.016. [DOI] [Google Scholar]
- 29.Padhiar A, Albert S, Nagadesi PK, Arya A. Lignin degradation by Flavodon flavus (Klotzsch.) Ryv. and Schizophyllum commune Fr. on Mangifera indica and Syzygium cumini woods. J Wood Chem Technol. 2010;30:129–139. doi: 10.1080/02773810903207770. [DOI] [Google Scholar]
- 30.Yuan WJ, Chang BL, Ren JG, Liu JP, Bai FW, Li YY. Consolidated bioprocessing strategy for ethanol production from Jerusalem artichoke tubers by Kluyveromyces marxianus under high gravity conditions. J Appl Microbiol. 2012;112:38–44. doi: 10.1111/j.1365-2672.2011.05171.x. [DOI] [PubMed] [Google Scholar]
- 31.Matias J, Encinar JM, Gonzalez J, Gonzalez JF. Optimisation of ethanol fermentation of Jerusalem artichoke tuber juice using simple technology for a decentralised and sustainable ethanol production. Energy Sustain Dev. 2015;25:34–39. doi: 10.1016/j.esd.2014.12.009. [DOI] [Google Scholar]
- 32.Laskar DD, Ke J, Zeng JJ, Gao X, Chen SL. Py-GC/MS as a powerful and rapid tool for determining lignin compositional and structural changes in biological processes. Curr Anal Chem. 2013;9:335–351. doi: 10.2174/1573411011309030003. [DOI] [Google Scholar]
- 33.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, et al. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism’s strategy for degrading lignocellulose. Appl Environ Microb. 2014;80:2062–2070. doi: 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerem Z, Jensen KA, Hammel KE. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven fenton reaction. FEBS Lett. 1999;446:49–54. doi: 10.1016/S0014-5793(99)00180-5. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol. 2006;8:2214–2223. doi: 10.1111/j.1462-2920.2006.01160.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Gao PJ. Function and mechanism of a low-molecular-weight peptide produced by Gloeophyllum trabeum in biodegradation of cellulose. J Biotechnol. 2003;101:119–130. doi: 10.1016/S0168-1656(02)00321-8. [DOI] [PubMed] [Google Scholar]
- 37.Rahmawati N, Ohashi Y, Watanabe T, Honda Y, Watanabe T. Ceriporic acid B, an extracellular metabolite of Ceriporiopsis subvermispora, suppresses the depolymerization of cellulose by the Fenton reaction. Biomacromolecules. 2005;6:2851–2856. doi: 10.1021/bm050358t. [DOI] [PubMed] [Google Scholar]
- 38.Kostylev M, Wilson D. Synergistic interactions in cellulose hydrolysis. Biofuels. 2012;3:61–70. doi: 10.4155/bfs.11.150. [DOI] [Google Scholar]
- 39.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohyd Res. 2009;344:1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen H, Kristensen JB, Felby C. Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuels Bioproducts & Biorefining-Biofpr. 2007;1:119–134. doi: 10.1002/bbb.4. [DOI] [Google Scholar]
- 42.Zeng Y, Zhao S, Yang S, Ding SY. Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr Opin Biotechnol. 2014;27:38–45. doi: 10.1016/j.copbio.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Yelle DJ, Ralph J, Lu FC, Hammel KE. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ Microbiol. 2008;10:1844–1849. doi: 10.1111/j.1462-2920.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- 44.Daniel G, Volc J, Filonova L, Plihal O, Kubatova E, Halada P. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microb. 2007;73:6241–6253. doi: 10.1128/AEM.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldrian P, Valaskova V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 46.Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu JG, Arantes V, Saddler JN. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol Biofuels. 2011;4:36. doi: 10.1186/1754-6834-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang JH, Siika-aho M, Tenkanen M, Viikari L. The role of acetyl xylan esterase in the solubilization of xylan and enzymatic hydrolysis of wheat straw and giant reed. Biotechnol Biofuels. 2011;4:60. doi: 10.1186/1754-6834-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao DH, Uppugundla N, Chundawat SPS, Yu XR, Hermanson S, Gowda K, et al. Hemicellulases and auxiliary enzymes for improved conversion of lignocellulosic biomass to monosaccharides. Biotechnol Biofuels. 2011;4:5. doi: 10.1186/1754-6834-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris PV, Welner D, McFarland KC, Re E, Poulsen JCN, Brown K, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 51.Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VGH, Igarashi K, et al. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS One. 2011;6:e27807. doi: 10.1371/journal.pone.0027807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen XA, Ishida N, Todaka N, Nakamura R, Maruyama JI, Takahashi H, et al. Promotion of efficient saccharification of crystalline cellulose by Aspergillus fumigatus Swo1. Appl Environ Microb. 2010;76:2556–2561. doi: 10.1128/AEM.02499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quiroz-Castaneda RE, Martinez-Anaya C, Cuervo-Soto LI, Segovia L, Folch-Mallol JL. Loosenin, a novel protein with cellulose-disrupting activity from Bjerkandera adusta. Microb Cell Fact. 2011;10:8. doi: 10.1186/1475-2859-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Liu C, Ma Y, Liu X, Zhang K, Zhang M. Improved production of two expansin-like proteins in Pichia pastoris and investigation of their functional properties. Biochem Eng J. 2014;84:16–27. doi: 10.1016/j.bej.2013.12.018. [DOI] [Google Scholar]
- 55.Lee HJ, Kim IJ, Kim JF, Choi IG, Kim KH. An expansin from the marine bacterium Hahella chejuensis acts synergistically with xylanase and enhances xylan hydrolysis. Bioresour Technol. 2013;149:516–519. doi: 10.1016/j.biortech.2013.09.086. [DOI] [PubMed] [Google Scholar]
- 56.Tovar-Herrera OE, Batista-Garcia RA, Sanchez-Carbente MD, Iracheta-Cardenas MM, Arevalo-Nino K, Folch-Mallol JL. A novel expansin protein from the white-rot fungus Schizophyllum commune. Plos One. 2015;10:e0122296. doi: 10.1371/journal.pone.0122296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swiatek K, Lewandowska M, Swiatek M, Bednarski W, Brzozowski B. The improvement of enzymatic hydrolysis efficiency of rape straw and Miscanthus giganteus polysaccharides. Bioresource Technol. 2014;151:323–331. doi: 10.1016/j.biortech.2013.10.090. [DOI] [PubMed] [Google Scholar]
- 58.Duranova M, Spanikova S, Wösten HAB, Biely P, Vries RP. Two glucuronoyl esterases of Phanerochaete chrysosporium. Arch Microbiol. 2008;191:133–140. doi: 10.1007/s00203-008-0434-y. [DOI] [PubMed] [Google Scholar]
- 59.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 60.Wymelenberg AV, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J, et al. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl Environ Microb. 2011;77:4499–4507. doi: 10.1128/AEM.00508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saykhedkar S, Ray A, Ayoubi-Canaan P, Hartson SD, Prade R, Mort AJ. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol Biofuels. 2012;5:52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller GL. Use Of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 63.Saha BC, Bothast RJ. Production, purification, and characterization of a highly glucose-tolerant novel beta-glucosidase from Candida peltata. Appl Environ Microb. 1996;62:3165–3170. doi: 10.1128/aem.62.9.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinfling A, Ruiz-Duenas FJ, Martinez MJ, Bergbauer M, Szewzyk U, Martinez AT. A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett. 1998;428:141–146. doi: 10.1016/S0014-5793(98)00512-2. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka H, Koike K, Itakura S, Enoki A. Degradation of wood and enzyme production by Ceriporiopsis subvermispora. Enzyme Microb Technol. 2009;45:384–390. doi: 10.1016/j.enzmictec.2009.06.003. [DOI] [Google Scholar]
- 66.Pickard MA, Roman R, Tinoco R, Vazquez-Duhalt R. Polycyclic aromatic hydrocarbon metabolism by white rot fungi and oxidation by Coriolopsis gallica UAMH 8260 laccase. Appl Environ Microb. 1999;65:3805–3809. doi: 10.1128/aem.65.9.3805-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodell B, Daniel G, Jellison J, Qian YH. Iron-reducing capacity of low-molecular-weight compounds produced in wood by fungi. Holzforschung. 2006;60:630–636. doi: 10.1515/HF.2006.106. [DOI] [Google Scholar]
- 68.Park J, Shin H, Yoo S, Zoppe JO, Park S. Delignification of lignocellulosic biomass and its effect on subsequent enzymatic hydrolysis. Bioresources. 2015;10:2732–2743. [Google Scholar]


