Summary
There are two ways to maintain fitness in the face of infection: resistance is a host’s ability to reduce microbe load and disease tolerance is the ability of the host to endure the negative health effects of infection. Resistance and disease tolerance should be applicable to any insult to the host and have been explored in depth with regards to infection, but have not been examined in the context of cancer. Here, we establish a framework for measuring and separating resistance and disease tolerance to cancer in Drosophila melanogaster. We plot a disease tolerance curve to cancer in wild-type flies and then compare this to natural variants, identifying a line with reduced cancer resistance. Quantitation of these two traits opens an additional dimension for analysis of cancer biology.
Results and Discussion
Host immune defense strategies can be separated into the ability to control pathogen burden, called resistance, and the ability of the host to endure the negative health effects of infection, called disease tolerance. Disease tolerance is the dose response curve relating host health to elicitor loads. While resistance is a heavily studied aspect of immune response, disease tolerance is less well understood. Originating in plant ecology studies (Caldwell et al., 1958; Schafer, 1971), the concept of disease tolerance was only recently introduced to animal immunity research (Ayres et al. 2008; Råberg et al., 2007). Distinguishing between resistance and disease tolerance is useful because they are fundamentally different strategies for surviving challenges. Applying the concepts of resistance and disease tolerance has improved our understanding of pathogenic infections (Iwasaki and Pillai, 2014; Medzhitov et al., 2012; Råberg, 2014; Vale et al. 2014) and should be applicable to any insult to host health, like cancer, not just infectious disease. We established a model to separate resistance and tolerance to cancer to understand the role of these immunological processes in cancer infections.
A system for separating resistance and tolerance to cancer
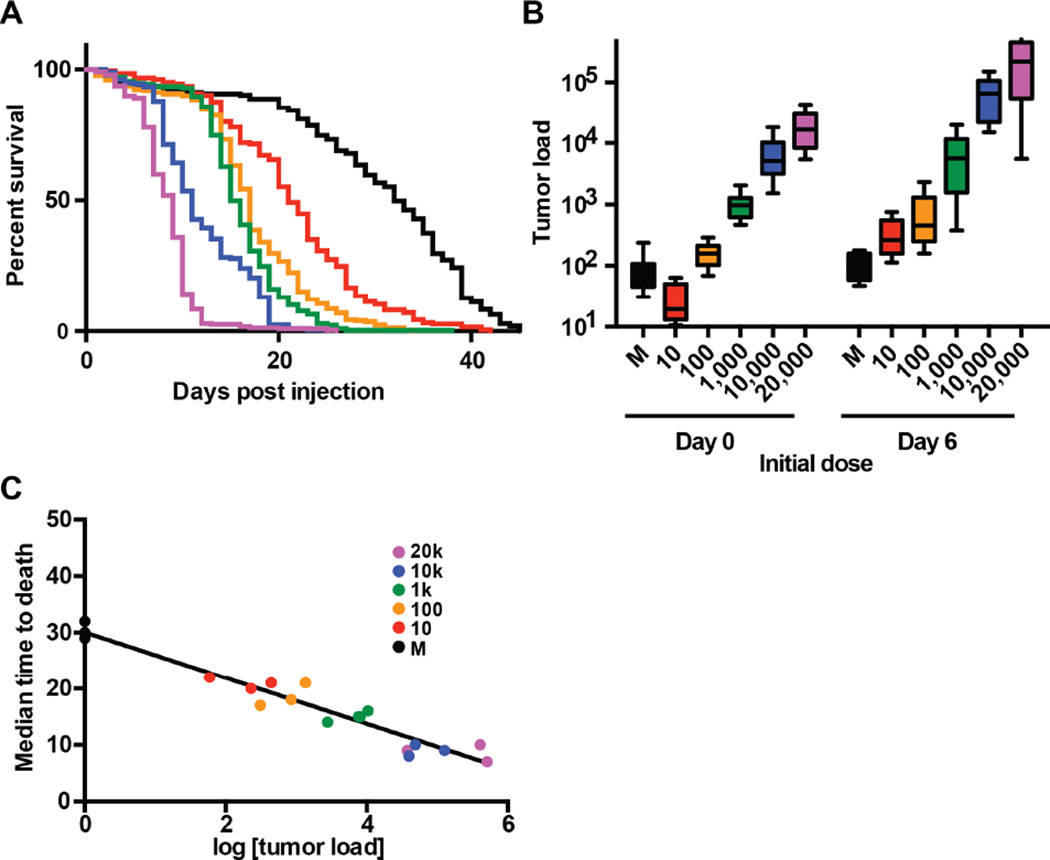
The model organism Drosophila melanogaster is useful for investigating both resistance and disease tolerance in infections because large numbers of animals can be infected with precise doses of pathogens and the growth of the pathogens and health of the host can be easily monitored (Ayres et al., 2008; Ayres and Schneider, 2009; Howick and Lazzaro, 2014; Rose et al., 2011; Rottschaefer and Lazzaro, 2012); we reasoned the fly would be suitable for studying resistance and tolerance to cancer. We used the Drosophila Oregon-R strain as an initial wild-type strain in our experiments. We chose to use a transplantable cancer model instead of an inducible one because it let us precisely regulate and measure input material (Ayres et al., 2008; Råberg et al., 2007; Regoes et al., 2014). We used the Rasv12-H7 line of Drosophila hyperplastic cancer cells, which expresses an oncogenic form of Ras, has a UAS-GFP reporter, and has previously been shown to metastasize throughout the fly and lead to premature death (Simcox et al., 2008) (see Experimental Procedures). The hyperplastic cells were delivered in a manner similar to microbial pathogens; the cells were cultured in vitro, quantified, diluted and injected into adult flies (Figure 1A–B). We used survival (median time to death) as a measure of disease progression and found that, similar to microbial infections, cancer kills in a dose-dependent manner, ranging from 8 to 21 days (Figure 1A) whereas wounding controls would live for 29 to 32 days. To measure tumor load we quantified the number of cancer cells on the day of infection (day 0) and six days post infection (PI) (day 6) by performing qPCR on DNA copies of the GFP gene, which was carried by the tumor cells but not the hosts (see Experimental Procedures). We chose to measure tumor load at 6 days PI to allow the cancer time to grow, but not so much time as to pass the median time to death for flies given high initial cancer doses. For each initial dose, cancer cells grew about tenfold by day 6 PI in OR flies (Figures 1B and S1).
Figure 1. A cancer disease tolerance curve establishes a framework for separating resistance and disease tolerance to cancer.
Wild-type (Oregon-R) adult male flies were injected with doses varying from 10–20,000 KRas hyperplastic cancer cells and were monitored for survival (disease progression) and cancer load (elicitor load). A. Survival curves were monitored for flies injected with 10–20,000 KRas hyperplastic cancer cells (n ≥ 180 flies per dose). The survival curves are significantly different (****, p<0.0001, Log-rank (Mantel-Cox) test). B. Rasv12-H7 fly cancer cells were injected to adult flies. The initial dose (day 0) and subsequent cancer growth (day 6) were quantitatively measured using a gfp marker present in the cancer cells but not the flies (n ≥ 150 flies per dose per day). C. A cancer disease tolerance curve was prepared by plotting pairs of cancer load and survival data for 18 cancer load/MTD pairs (n ≥ 110 flies per data point). This curve was fit with a linear regression model (r2>0.94) (Table S1).
We generated a cancer tolerance curve by plotting median time to death for a given dose of cells against the cancer growth (i.e. the number of cells measured 6 days post inoculation for that inoculation dose). (Figure 1C). These data were fit with a linear regression model (r2>0.94)(Table S1). This design allows the health of these flies to be described with two parameters: The first is vigor (the health of the animal in the absence of disease, which in this case is around 30 days, and the second is the slope of the curve, which for this curve is −4.080 days per log of tumor load (Figure 1C).
Natural variation of cancer resistance
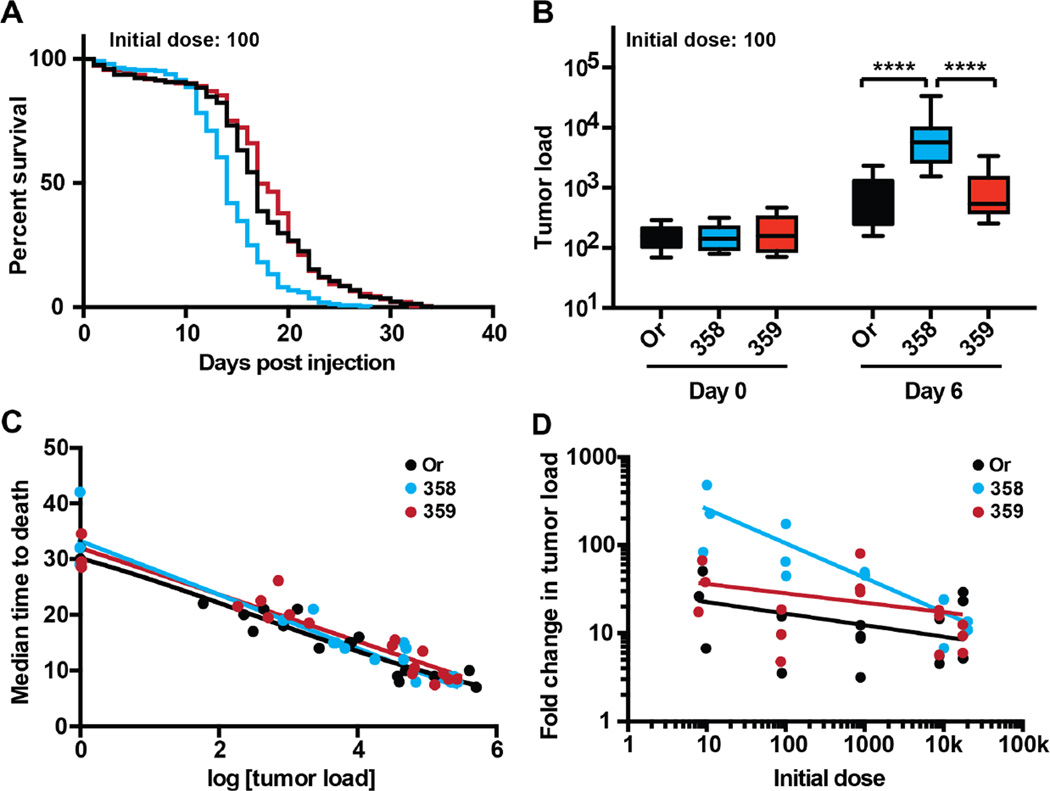
To investigate how genetic variation might influence resistance and/or tolerance to cancer we used two natural variant fly lines from the Drosophila Genetics Reference Panel (DGRP; lines RAL-358 and RAL-359) (Mackay et al., 2012). We selected RAL-358 and RAL-359 based on data from a pilot screen (Figure S2A). We generated a survival dose response curve for these fly lines as described above (Figure S3B–G). RAL-358 died significantly faster than both wild-type and RAL-359 at all cell doses other than the highest dose of 20,000 cells (Figures 2A and S3B–G). To determine the role resistance played in these survival changes, we measured the tumor load of these lines at 6 days PI. RAL-358 had consistently higher loads than either wild-type or RAL-359 when injected with an initial dose of 10, 100, or 1,000 cells (Figures 2B and S1). All three fly lines had equal tumor loads 6 days PI when injected with high initial doses (10,000 or 20,000 cells) (Figure S1A–B). These data demonstrate that RAL-358 has a resistance defect, experiencing more than 100-fold tumor growth when given low initial doses of tumor cells (Figures 2B–D and S1).
Figure 2. Genetic variation alters resistance to cancer.
Wild-type (Oregon-R) and natural variant adult male flies were injected with doses varying from 10–20,000 KRas hyperplastic cancer cells and were monitored for survival (disease progression) and cancer load (pathogen load). Wild-type (Oregon-R) is in black, RAL-358 is in blue, and RAL-359 is shown in red. A. A survival curve of adult flies injected with 100 cancer cells, comparing wild-type and natural variant flies. RAL-358 dies significantly faster than wild-type or RAL-359 (****, p<0.0001, Log-rank (Mantel-Cox) test) (n ≥ 180 flies per line). Whereas there is no significant difference between wild-type and RAL-359. B. A cancer growth plot showing the initial dose of 100 cells and the cancer burden of flies 6 days post injection. RAL-358 has a significantly higher cancer load than either wild-type or RAL-359 (****, p<0.0001, two-way ANOVA Tukey’s multiple comparisons test) (n ≥ 150 flies per dose per day). C. A cancer disease tolerance curve was prepared for each of the three fly lines (wild-type, RAL-358, and RAL-359) by plotting pairs of cancer load and survival data for 18 cancer load/MTD pairs for each line (n ≥ 110 flies per data point). These curves were each fit with a linear regression model (r2>0.94 for wild-type, r2>0.91 for RAL-358, and r2>0.91 for RAL-359). The slope of these lines is similar (−4.1 for wildtype, −4.8 for RAL-358, and −4.2 for RAL-359) and the 95% confidence intervals overlap. D. The ratio of cancer growth over six days PI. Wild-type is in black, RAL-358 is in blue, and RAL-359 is shown in red (n ≥ 150 flies per line per day in all of these experiments). The data for each of these lines were fit with a log-log non-linear regression, 0.12, 0.57, and 0.13 R2 for Or, RAL-358, and RAL-359 respectively. The slope of the log-log nonlinear regression is −0.13, −0.4, and −0.11 for Or, RAL-358, and RAL-359 respectively. The 95% confidence interval for the slope of RAL-358 remains negative while the 95% confidence interval for the slope of wild-type and RAL-359 ranges from negative to positive. In a non-linear log-log line regression F test, one curve does not fit all the data (p < 0.0001).
Resistance and disease tolerance are not mutually exclusive defense strategies but can act in concert in host defense. We wondered if the decrease in survival in cancer-laden RAL-358 could also have a tolerance component. To investigate the differences in cancer disease tolerance, we plotted tolerance curves for all three fly lines (Figure 2C). We found that they had similar tolerance curves, suggesting that tolerance was not changing. Changes in resistance, however, are apparent in the growth plots and disease tolerance curves (Figures 2B–D and S1). For example, the data for RAL-358 in the tolerance curve are shifted toward the bottom right of the plot compared to wild-type and RAL-359 (Figure 2C). The resistance defect we found in RAL-358 is dose-dependent, manifesting only at low initial tumor inoculations (10–100 cells) (Figure 2D). This could be due to decreased immune surveillance sensitivity, where defenses, which can protect against tumor growth (Pastor-Pareja et al., 2008), turn on only at higher initial tumor loads. These experiments highlight the importance of performing a dose response curve when testing a health insult.
Applying disease tolerance to cancer
While the importance of resistance to cancer is well understood and has been well leveraged in current cancer treatment programs, cancer disease tolerance is not explicitly studied. There is evidence in some human cancers that health loss and survival rates do not always correlate with cancer burden, provocatively suggesting a role for cancer disease tolerance where hosts show different health effects given equal tumor loads (Heyneman et al., 2001; Patz et al., 2000). Our line of inquiry here provides a quantitative conceptual methodology for identifying unexplored aspects of host-cancer interactions that could be utilized to improve the treatment and outcomes for cancer patients, even when reduction or removal of cancer tissue is not possible.
Experimental Procedures
Flies, cells, and media
The Oregon-R strain was used as a wild-type control while Drosophila Genetic Reference Panel lines RAL-358 and RAL-359 were used as natural variants. Fly strains were obtained from the Bloomington Drosophila Stock Center (Mackay et al., 2012). Flies were kept in standard fly bottles containing dextrose medium (129.4g dextrose, 7.4g agar, 61.2g corn meal, 32.4g yeast, and 2.7g tegosept per liter; polypropylene round bottom 8oz bottles plugged with bonded dense weave cellulose acetate plugs, Genesee Scientific Cat #49–100) and were housed at 25°C with 65% relative humidity and a 12 hour light and 12 hour dark cycle. The hyperplastic cell line Rasv12-H7, marked with a UAS-GFP construct, was obtained from the Drosophila Genomics Resource Center (Simcox et al., 2008). Cells were cultured as previously described (Simcox et al., 2008; Simcox, 2013). Briefly, cells were cultured in Schneider’s media supplemented with 10% heat inactivated fetal calf serum (Sigma® cat # F4135-100ML) and 1% penicillin – streptomycin solution solution (HyClone® cat # SV30010). Rasv12-H7 cells are strongly adherent and were passaged by first rinsing the cells in chilled PBS, then incubated for 5 minutes in 0.05% trypsin (HyClone® cat # SH30236.01). Cells were collected and diluted into an equal volume of media, centrifuged for 2 minutes, and resuspended in new media. For infection experiments the cells were quantified using a hemocytometer and diluted to the desired doses. Frozen stocks of cells were produced as previously described (Simcox, 2013).
Cell injections into adults
Different doses of Rasv12-H7 cells (10, 100, 1,000, 10,000, and 20,000 cells) were injected into adult male flies aged 5–7 days old with control flies being injected with cell culture media. Flies received 50 nl injections in the anterior abdomen. Injections were performed using a Picospritzer® III (Parker Instrumentation) and a pulled glass needle as previously described (Ayres et al., 2008). Cancer cell doses were counted using a hemocytometer immediately prior to injection. Each dose was injected into ≥60 adult male flies and each experiment was replicated 3 times, thus n = ≥180 flies per dose, per fly line tested; resulting in ≥3,240 individual D. melanogaster being injected for the survival portion of these experiments. Survival of the cancer injected flies was counted every 24 hr post infection (PI) until all flies were dead. The median time to death (MTD), for each experiment was determined as used as a measure of disease progression.
Quantifying cancer cell growth
Rasv12-H7 cells carry an integrated UAS-GFP marker. Cancer load in flies was determined by quantitative-PCR, amplifying DNA copies of the gfp maker using previously published primers (Simcox et al., 2008; Portugal et al., 2011). Total DNA was extracted using phenol-chloroform. We quantified gfp by pooling five adult male flies to make one sample, and ten samples were taken per dose both on the day of infection (day 0) and six days PI (day 6). Thus n ≥ 50 flies per dose per day, for five doses plus media controls, and each experiment was replicated three times; resulting in the extraction of ≥ 1,800 flies in batches of 5. qPCR was performed on a StepOnePlus qRT-PCR system and analyzed using StepOne™ software v2.2.2 (Applied Biosystems®). Reactions were done using SYBR Green PCR Master Mix (Applied Biosystems®) using 15µl reactions, in 96 well plates, using a relative standard curve. Following a primary denaturation of 10 minutes at 95°C, the reactions were done for 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. We used gfp primers from Portugal et al. (2011) 5’- GTC AGT GGA GAG GGT GAA GG– 3’ and 5’ – ACT TCA GCA CGT GTC TTG TAG TTC – 3’. The standard curve was performed using a serial dilution series (1:10) of Rasv12-H7 cells, quantified using a hemocytometer. The gfp abundance relative to our standard curve provided a quantitative measure of initial cancer dose and cancer burden after 6 days post infection. Using MTD as a measure of disease progression and the cell growth at 6 days PI, we were able to plot disease tolerance curves as shown in Figures 1 and 2.
Statistical analyses
Statistical analyses were done using GraphPad Prism version 6 for Mac OS X. The log-rank (Mantel-Cox) test was used to evaluate the survival curves. Two-way ANOVA with Tukey’s multiple comparisons tests were used to analyze cancer growth among the fly lines. Disease tolerance curves were fit with a linear regression model after testing several other models (four-parameter sigmoid, three-parameter sigmoid, and quadratic).
Supplementary Material
Acknowledgements
We would like to thank lab members and colleagues for critical reading of the manuscript (Alejandra Hotson, Alexander Louie, Brenda Torres, Damian Trujillo, Jose Oliveira, Katherine Cumnock, and Poonam Rath). We also thank Alejandra Hotson, Alexander Louie, Jose Oliveira, Katherine Cumnock, and Poonam Rath for their help in counting fly survival. We thank Amanda Simcox, the Bloomington Drosophila Stock Center (NIH P40OD018537), and the Drosophila Genomics Resource Center (NIH 2P40OD010949-10A1) for fly stocks and cell cultures. A.R.D. is a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation. This work was supported by an NIH Pioneer Award (DP1 AT007753) to D.S.S. and an NIH training grant (5T32HG000044-17) to A.R.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ARD and DSS conceived and designed the experiments. ARD performed the experiments. ARD and DSS analyzed and interpreted the data and wrote and revised the manuscript.
References
- Ayres JS, Freitag N, Schneider DS. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics. 2008;178:1807–1815. doi: 10.1534/genetics.107.083782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biology. 2009;7:e1000150. doi: 10.1371/journal.pbio.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RM, Schafer JF, Compton LE, Patterson FL. Tolerance to cereal leaf rusts. Science. 1958;128:714–715. doi: 10.1126/science.128.3326.714. [DOI] [PubMed] [Google Scholar]
- Heyneman LE, Herndon JE, Goodman PC, Patz EF., Jr Stage distribution in patients with a small (< or = 3 cm) primary nonsmall cell lung carcinoma. Implication for lung carcinoma screening. Cancer. 2001;92:3051–3055. doi: 10.1002/1097-0142(20011215)92:12<3051::aid-cncr10106>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Howick VM, Lazzaro BP. Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evolutionary Biology. 2014;14:56. doi: 10.1186/1471-2148-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature Reviews Immunology. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Disease Models & Mechanisms. 2008;1:144–154. doi: 10.1242/dmm.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz EF, Jr, Rossi S, Harpole DH, Jr, Herndon JE, Goodman PC. Correlation of tumor size and survival in patients with stage IA non-small cell lung cancer. Chest. 2000;117:1568–1571. doi: 10.1378/chest.117.6.1568. [DOI] [PubMed] [Google Scholar]
- Portugal S, Carret C, Recker M, Armitage AE, Goncalves LA, Epiphanio S, Sullivan D, Roy C, Newbold CI, Drakesmith H, et al. Host-mediated regulation of superinfection in malaria. Nature Medicine. 2011;17:732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- Råberg L. How to live with the enemy: Understanding tolerance to parasites. PLoS Biology. 2014;12:e1001989. doi: 10.1371/journal.pbio.1001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes RR, McLaren PJ, Battegay M, Bernasconi E, Calmy A, Gunthard HF, Hoffmann M, Rauch A, Telenti A, Fellay J, et al. Disentangling human tolerance and resistance against HIV. PLoS Biology. 2014;12:e1001951. doi: 10.1371/journal.pbio.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, Cherry S. Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host & Microbe. 2011;10:97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottschaefer SM, Lazzaro BP. No effect of Wolbachia on resistance to intracellular infection by pathogenic bacteria in Drosophila melanogaster. PloS One. 2012;7(7):e40500. doi: 10.1371/journal.pone.0040500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JF. Tolerance to plant disease. Annual Review of Phytopathology. 1971;9:235–252. [Google Scholar]
- Simcox A, Mitra S, Truesdell S, Paul L, Chen T, Butchar JP, Justiniano S. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PloS Genetics. 2008;4:e1000142. doi: 10.1371/journal.pgen.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox A. Progress towards Drosophila epithelial cell culture. In: Randell SH, Fulcher ML, editors. In Epithelial Cell Culture Protocols. Second Edition. Humana Press; 2013. pp. 1–11. [Google Scholar]
- Vale P, Fenton A, SP B. Limiting damage during infections: Lessons from infection tolerance for novel therapeutics. PLoS Biology. 2014;12:31001769. doi: 10.1371/journal.pbio.1001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.