Abstract
The brain endothelium is an important therapeutic target for the inhibition of cerebrovascular dysfunction in ischemic stroke. Previously, we documented the important regulatory roles of microRNAs in the cerebral vasculature, in particular the cerebral vascular endothelium. However, the functional significance and molecular mechanisms of other classes of non-coding RNAs in the regulation of cerebrovascular endothelial pathophysiology after stroke are completely unknown.
Using RNA sequencing (RNA-seq) technology, we profiled long non-coding RNA (lncRNA) expressional signatures in primary brain microvascular endothelial cells (BMECs) after oxygen-glucose deprivation (OGD), an in vitro mimic of ischemic stroke conditions. After 16h of OGD exposure, the expression levels for 362 of the 10,677 lncRNAs analyzed changed significantly, including a total of 147 lncRNAs increased and 70 lncRNAs decreased by more than 2-fold. Among them, the most highly upregulated lncRNAs include Snhg12, Malat1, and lnc-OGD 1006, whereas the most highly downregulated lncRNAs include 281008D09Rik, Peg13, and lnc-OGD 3916. Alteration of the most highly upregulated/downregulated ODG-responsive lncRNAs was further confirmed in cultured BMECs after OGD as well as isolated cerebral microvessels in mice following transient middle cerebral artery occlusion (MCAO) and 24h reperfusion by the quantitative real-time PCR approach. Moreover, promoter analysis of altered ODG-responsive endothelial lncRNA genes by bioinformatics showed substantial transcription factor binding sites on lncRNAs, implying potential transcriptional regulation of those lncRNAs. These findings are the first to identify OGD-responsive brain endothelial lncRNAs, which suggest potential pathological roles for these lncRNAs in mediating endothelial responses to ischemic stimuli. Endothelial-selective lncRNAs may function as a class of novel master regulators in cerebrovascular endothelial pathologies after ischemic stroke.
Keywords: RNA sequencing, long non-coding RNAs, transcriptome, brain microvascular endothelial cells, oxygen-glucose deprivation, ischemic stroke
INTRODUCTION
Stroke is one of the severest health issues in the United States (Moskowitz et al., 2010). Currently, thrombolytic therapy is not enough for the acute intervention of ischemic stroke and only small percentage of patients can receive it because of its narrow therapeutic time window (Schellinger et al., 2004; Stapf and Mohr, 2002). Thus, development of effective therapies is urgently required. During the past two decades, extensive studies showing the effectiveness of neuroprotectants in animal stroke models, but not in stroke clinical trials, imply that solely focusing on neuroprotection is not sufficient (Ginsberg, 2009; Iadecola and Anrather, 2011). Thus, greater attention has been paid to the local environment of the surviving neuron, such as the cerebral microvasculature, neurovascular units, and vascular neural network (Barone, 2009; del Zoppo, 2006; Ginsberg, 2009; Lok et al., 2007; Yin et al., 2010a; Yin et al., 2011; Yuan, 2009; Zhang et al., 2012). As a major structural and functional element of the brain microvasculature, the vascular endothelium plays a dominant role in maintaining its normal physiological functions. There is increasing evidence showing that ischemia-induced cerebral endothelial injury, endothelial inflammation, and subsequent impairment of endothelial function increase cerebrovascular permeability and BBB leakage, contributing to ischemic brain injury (del Zoppo and Hallenbeck, 2000; Ishikawa et al., 2004; Sandoval and Witt, 2008). Thus, it is important to identify the insightful mechanisms by which cerebrovascular disorders can be effectively inhibited or reduced through protection of the brain endothelium under ischemic stroke conditions (Fagan et al., 2004; Fisher, 2008; Rodriguez-Yanez et al., 2006). Understanding the critical mediators in regulating cerebrovascular endothelial dysfunction may eventually lead us to discover novel targets for the treatment of stroke.
Recently, accumulating studies have shown that non-coding RNAs (ncRNAs) are functional RNA molecules that are generally not translated into proteins but can actively regulate the expression and function of protein-coding genes by different mechanisms. It is worth noting that only approximately 1.5% of DNA sequences in the human genome are responsible for protein coding whereas at least 98% of the genome does not contain protein-coding DNA sequences but transcribed into various ncRNAs (Derrien et al., 2012; Qureshi and Mehler, 2012). In addition to well-known ncRNAs such as rRNAs and tRNAs, ncRNAs can be broadly classified as small (<200 nt) and long (>200 nt) ncRNAs (lncRNAs) (Bartel, 2004; Kim, 2005; Qureshi and Mehler, 2012; Schonrock et al., 2012; Taft et al., 2010; Vemuganti, 2013; Yin et al., 2014). NcRNAs have become a focus in biomedical research in the last half decade and are now regarded as important and essential mediators in major biological/physiological processes impacting development, differentiation, and metabolism, as well as pathologies in a variety of human diseases (Qureshi and Mehler, 2012; Schonrock et al., 2012; Suarez and Sessa, 2009; Taft et al., 2010; Vemuganti, 2013; Wienholds and Plasterk, 2005; Yin et al., 2014). So far in the stroke field, the role of a specific class of small ncRNAs, miRNAs, was mainly investigated in the pathogenesis of stroke (Eacker et al., 2013; Liu et al., 2013; Ouyang et al., 2013; Rink and Khanna, 2010; Saugstad, 2010; Tan et al., 2011; Vemuganti, 2010; Yin et al., 2013b, 2014). Notably, our research team has focused on studying the functional significance of stroke-associated endothelial ncRNAs with a special focus on miRNAs in cerebral endothelial pathophysiology after stroke (Yin et al., 2010a; Yin et al., 2013a; Yin et al., 2013b, 2014; Yin et al., 2012).
Non-coding transcripts >200 nucleotides up to ~100kb are defined as lncRNAs (Mattick, 2009; Mercer et al., 2009; Ponting et al., 2009; Vemuganti, 2013; Yin et al., 2014). Recently, whole transcriptome sequencing (e.g. RNA-seq) of human and other genomes has revealed significant numbers of lncRNAs which are long transcripts similar to mRNAs but lack protein coding (translation) potential (Derrien et al., 2012). LncRNAs can control gene expression by various mechanisms that include recruitment of histone modification complexes, modulating transcription factor activity and splicing machinery, increasing mRNA stability, and acting as transcription enhancers. LncRNAs function in various biological processes including gene transcription, cell differentiation, imprinting, immune responses, and stem cell pluripotency, and have cell-specific expression patterns that respond to external stimuli (Mattick, 2009; Mercer et al., 2009; Ponting et al., 2009). Involvement of lncRNA in human diseases, including stroke, have begun to appear and are expected to escalate. For example, recent microarray profiling studies identifying stroke-responsive alterations of lncRNAs (Dharap et al., 2012; Dharap et al., 2013; Vemuganti, 2013) underscore the importance of lncRNAs in the pathogenesis of ischemic stroke. However, the essential role and molecular mechanisms of lncRNA in ischemic stroke still remain poorly understood. In particular, nothing is known to date about the functional significance of lncRNAs in cerebral vascular endothelial biology and pathophysiology in ischemic stroke.
In this article, we utilized RNA-seq technology to transcriptome profiling lncRNAs in brain microvascular endothelial cells (BMECs) after exposure to Oxygen-Glucose Deprivation (OGD), an in vitro mimic of stroke conditions. For the first time, we have identified substantial OGD-responsive lncRNAs in BMECs, and further confirmed the altered pathological lncRNA expression in the cerebral vascular endothelium in a mouse ischemic stroke model. Moreover, we also found a number of conserved transcription factor binding sites in the promoter region of these OGD-responsive endothelial lncRNAs. Therefore, our studies could uncover lncRNAs associated with brain endothelial dysfunction in ischemic stroke, potentially advancing the field by linking lncRNA biology to stroke-induced cerebral vasculature pathologies.
MATERIALS AND METHODS
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO), while cell culture supplies were purchased from Invitrogen Corporation (Carlsbad, CA) unless specified.
BMEC cultures and oxygen-glucose deprivation (OGD)
Mouse BMECs were prepared as previously described (Yin et al., 2002b). Briefly, mouse cerebral cortex from adult male C57BL/6J mice (male, body weight 25-30g, 3-4 months-old) (Jackson Laboratory, Bar Harbor, ME) was homogenized, filtered and sequentially digested with collagenase B, then collagenase/dispase (Roche Molecular Biochemicals, Indianapolis, IN), followed by centrifugation in 40% Percoll solution. The second band containing microvessels was collected and plated onto collagen-coated dishes. Mouse BMECs (1-5 passages, > 95% purity based on expression of factor VIII and exhibiting bradykinin receptor function) were grown to 85-95% confluency before use (Yin et al., 2002b).
To mimic ischemic-like conditions in vitro, cell cultures were exposed to OGD for a fixed time (Yin et al., 2002a). Briefly, mouse BMEC cultures were transferred into a temperature controlled (37±1°C) anaerobic chamber (Forma Scientific, Marietta, OH) containing a gas mixture composed of 5% CO2 and 95% N2. The culture medium was replaced with deoxygenated glucose-free Hanks’ Balanced Salt Solution (Invitrogen, Carlsbad, CA) and cells were maintained in the hypoxic chamber for 16 h. Control mouse BMECs were not exposed to OGD.
RNA sequencing and Bioinformatics analysis
RNA was extracted from cultured BMECs by using a miRNeasy Mini Kit (Qiagen, Valencia, CA). Eluted RNA was prepared for sequencing using Illumina protocols, and then sequenced on the HiSeq 2000 (Illumina) to generate 50 bp paired-end reads. We obtained about 41.4~59.1M reads for each sample (four independent biological replicates per group). The reads were aligned to the mouse genome (from UCSC Genome Browser) using Bowtie2 (2.1.0) (Langmead et al., 2009) with Tophat (2.0.8b) (Trapnell et al., 2009). We used Cufflinks to first assemble transcripts from all datasets using the NCBI Reference Sequence (RefSeq) mouse gene annotation as a reference guide. All mouse XenoRefSeq annotations that overlapped with the unannotated transcripts were identified. Nonoverlapping transcripts were assessed for protein-coding potential using PhyloCSF (Lin et al., 2011). Using a threshold previously used to identify non-coding RNAs, all multi-exonic transcripts with a PhyloCSF score >20 were called novel protein-coding transcripts and <20 as lncRNAs (Leung et al., 2013; Lin et al., 2011; Pauli et al., 2012). FPKM values were calculated for expression levels of each transcript. To assess the biological reproducibility between the replicates for each condition, Pearson's Correlation value was also determined for each pair of replicates. Cuffdiff (Trapnell et al., 2010) was used to identify differentially expressed transcripts in two conditions with the reference annotation containing mouse RefSeq genes and lncRNAs with an FPKM >0.5.
Hierarchical cluster analysis was performed on normalized array values with Cluster 3.0 with Euclidean distance and single linkage for >10,000 genes and average linkage for smaller gene sets. Heatmaps were generated with Java TreeView. Gene ontology (GO) analysis was completed using GO Elite with default settings.
To assess the significance of the proximity of co-regulated lncRNAs and protein-coding genes, we carried out 100,000 simulations of proximity measurements on randomly placed co-regulated lncRNAs and protein-coding genes in the mouse genome (Leung et al., 2013; Lin et al., 2011; Pauli et al., 2012). We analyzed 4,301 coding genes and 362 lncRNAs to be co-regulated and then counted the number of simulated OGD-responsive lncRNAs within 500kb of a simulated OGD-responsive gene.
Promoter analysis of transcription factor binding sites
As transcription factors are crucial in the process of transcription of non-coding RNAs, we checked and analyzed the promoter region of OGD-responsive lncRNA genes for the identification of conserved transcription factor binding sites. For each lncRNA, a 2-kb promoter sequence upstream to the transcription start site was analyzed for conserved TF binding sites by using the Genomatix Software. All TF matrices with a Z-score of >2 (representing P>0.05) were considered statistically significant (Dharap et al., 2011).
Mouse model of transient focal cerebral ischemia
Male C57BL/6J mice (male, 8- to 10-weeks-old) were purchased from Jackson Laboratory. Focal cerebral ischemia was induced by intraluminal middle cerebral artery occlusion (MCAO) using a nylon monofilament suture as described previously (Yin et al., 2010a; Yin et al., 2011; Yin et al., 2010b; Yin et al., 2013a). Briefly, mice were anesthetized with ketamine (100mg/kg) and xylazine (10mg/kg). After a midline skin incision, the left common carotid artery was exposed and then its branches were electrocoagulated. A 2-cm length of 6-0 rounded tip nylon suture was gently advanced from the external carotid artery up to the internal carotid artery until regional cerebral blood flow (rCBF) was reduced to less than 16% of baseline. After 60 minutes of proximal MCA occlusion, blood flow was restored by removing the suture. Changes in cerebral blood flow (CBF) at the surface of the cortex were recorded using a laser-Doppler flowmetry monitor (BPM2 System, Vasamedic, St. Paul, MN). Sham control animals were subjected to similar operations to expose the carotid arteries without occlusion of the middle cerebral artery. After 60 minutes of MCAO, the mice were allowed to recover for 24 hours. Arterial blood gases, mean arterial pressure, and heart rate were also monitored in selected animals 30 min before, during, and 30 min after MCAO. The rectal temperature was controlled at 37.0 ± 0.5°C during surgery with a feedback-regulated heating pad (Harvard, Holliston, MA). After the ischemic insult, mice were kept in an air-ventilated incubator at 24.0 ± 0.5°C. The animals were sacrificed at 24 h of reperfusion, and the brains were quickly removed for biochemical assays as well as infarct determination. All procedures using laboratory animals were approved by the University of Michigan Animal Care and Use Committee.
Cerebral microvessel isolation
Cerebral microvessel isolation employed previously described methods with modifications (Pardridge et al., 1985; Zlokovic et al., 1993; (Yin et al., 2010a; Yin et al., 2006b). Briefly, mice were sacrificed by decapitation under anesthesia. The brains were immediately removed from the skull and immersed in ice-cold buffer A (103 mM NaCl, 4.7mM KC1, 2.5 mM CaC12, 1.2 mM KH2PO4, 1.2 mM MgSO4, 15 mM Hepes, pH 7.4). The brain was homogenized in a 5-fold volume excess of buffer B (103 mM NaCl, 4.7mM KC1, 2.5 mM CaC12, 1.2 mM KH2PO4, 1.2 mM MgSO4, 15 mM Hepes, 25 mM HCO3, 10 mM glucose, 1mM sodium pyruvate, and 1 g/100 ml dextran pH 7.4) with a Teflon homogenizer. The homogenate was suspended in an equal volume of 25% BSA and was centrifuged at 5,800 × g at 4°C for 30 min. The pellet was resuspended in 10 ml of buffer B and passed over an 85-μm nylon mesh (Tetko, Depu, NY). This filtrate was then passed over a 3 × 4-cm glass bead column (0.45-mm glass beads) with a 44-μm nylon mesh at the bottom, and then washed with 400 ml of buffer B. Microvessels adhered to the glass beads while contaminants passed unimpeded. Microvessels were recovered by repeated gentle agitation of the glass beads in buffer B. The supernatant with microvessels was decanted and spun at 500 × g for 5 min, and the final pellet was stored at −80°C until various biochemical assays were performed.
Quantitative real-time PCR
Total RNA was isolated from BMEC cultures or isolated cerebral microvessels by using a miRNeasy Mini Kit (Qiagen, Valencia, CA) or Trizol (Invitrogen). A quantitative real-time reverse-transcriptase polymerase chain reaction (RT-PCR) was carried out with a Bio-Rad thermocycler and an SYBR green kit (Bio-Rad, Hercules, CA) according to the manufacturer's recommendations. Specific lncRNA primers used for the reaction are listed in Supplemental Table 1. The relative lncRNA expression was normalized by 18S RNA levels. The PCR experiments were repeated 3 times, each using separate sets of cultures (Yin et al., 2006a; Yin et al., 2006b).
Immunofluorescence staining
Mouse BMECs grown on coverslips were then fixed with 4% paraformaldehyde for 30 min and washed 3 times with 0.1 M PBS (pH 7.4). The cells were incubated with a primary rat anti-CD31 antibody (1:50; BD Bioscience), mouse anti-Claudin 5 (1:50; Invitrogen), or mouse anti-Occludin antibody (1:50; Invitrogen) overnight at 4°C. On the following day, the cells were incubated with fluorescein-conjugated anti-rat or anti-mouse IgG (1:100; Vector Labs, Burlingame, CA) for 1 h. BMECs were counterstained with 1 μg/ml of DAPI (Molecular Probes; Eugene, OR) to visualize nuclear morphology. Slides were washed, wet mounted and examined with an Olympus fluorescence microscope (Yin et al., 2006a; Yin et al., 2002b).
Quantitative and Statistical analysis
Quantitative data are expressed as mean ± SD or SEM based on at least three independent experiments of triplicate samples. Differences among groups were statistically analyzed by oneway analysis of variance followed by Bonferroni's post-hoc test. Comparison between two experimental groups was based on a two-tailed t-test. A p-value less than 0.05 was considered significant.
RESULTS
RNA-seq identification of transcriptome profiling in mouse BMECs after OGD
To determine the transcriptomic effects of OGD, an in vitro mimic of ischemic stroke conditions, on the cerebral vascular endothelium, we treated mouse BMECs (Fig. 1) with OGD for 16 hours and then performed RNA sequencing (Fig. 2) of RNA isolated from non-OGD control and OGD-treated cells. RNA sequencing reads were generated and mapped to the mouse genome reference. A total of 79.68% of filtered reads could be aligned to the mouse reference genome. Four independent biological replicates were performed for each treatment and the transcript expression level (FPKM) was calculated as described in the Methods.
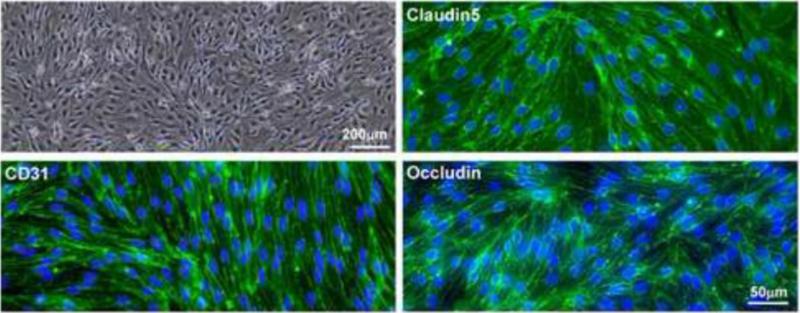
Figure 1.
Brain microvascular endothelial cell cultures. Phase-contrast light microscopy shows primary mouse BMEC cultures. Immunofluorescent stain using antibody against brain vascular endothelial cell marker, CD31 and BBB markers, Claudin 5 and Occludin reveals typical circumcellular signals of CD31, Claudin 5, and Occludin. Scale bars are indicated.
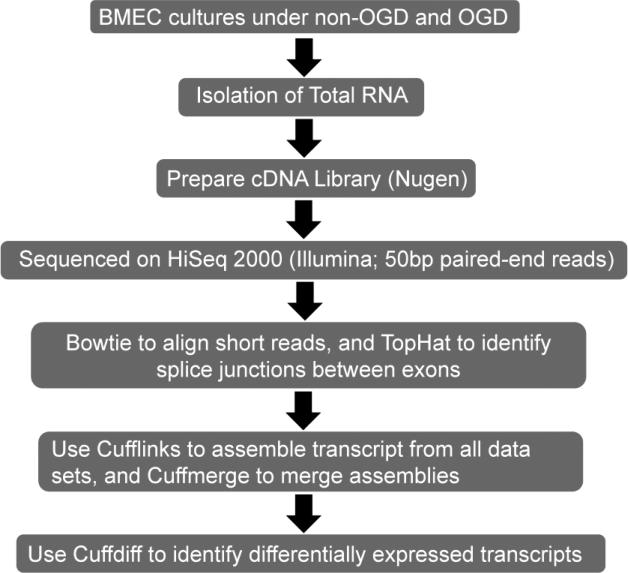
Figure 2.
Schematic representation of RNA-seq experiments and bioinformatics analysis.
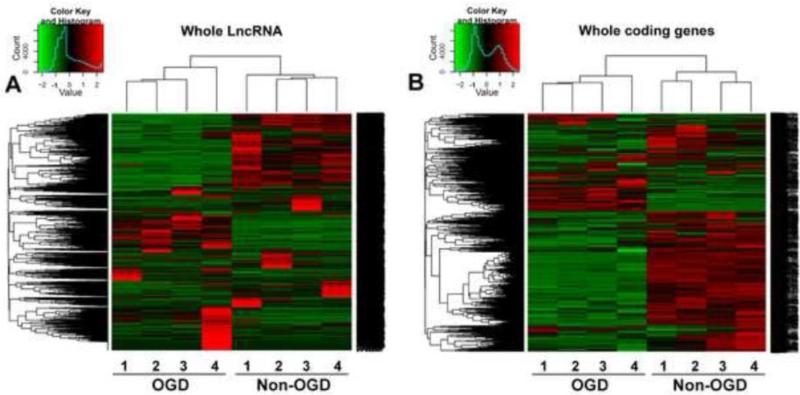
We discovered 55,832 transcripts using this approach, and further divided them into protein-coding or non-coding by assessing their protein-coding potential. We classified a total of 10,677 transcripts with limited protein-coding potential as lncRNAs and 43,705 as protein-coding transcripts (Table 1). Approximately 5,241 lncRNA transcripts that did not overlap with the known mouse genome were classified as novel (non-annotated), and named as lnc-OGD in numerical order (lnc-OGD 1-5,241) compared to 5,436 annotated lncRNAs (Table 1). Further bioinformatics analysis using Cuffdiff discovered 4,663 significantly differentially expressed transcripts (FDR<0.05), including 362 annotated or unannotated lncRNAs and 4,301 coding genes (Table 1). Hierarchical clustering showed systematic variations in the expression of lncRNAs and protein-coding RNAs in mouse BMECs after ischemic stimuli (Fig. 3). Among the 362 significantly changed lncRNAs analyzed, a total of 147 lncRNAs increased and 70 lncRNAs decreased by more than 2-fold in mouse BMECs after 16h of OGD exposure, and were classified as OGD-responsive endothelial lncRNAs (Table 1). The most highly-upregulated annotated lncRNAs are Snhg12, Malat1, and Srsf3, whereas the most highly upregulated non-annotated lncRNAs are lnc-OGD 1645, lnc-OGD 2327, and lnc-OGD 1006 (Table 2). Accordingly, the most highly downregulated annotated lncRNAs include 281008D09Rik, Peg13, and Pisd-ps1, whereas the most highly downregulated non-annotated lncRNAs include lnc-OGD 3916, lnc-OGD 2838, and lnc-OGD 301 (Table 3).
Table 1.
Overview of RNA-seq results In mouse BMECs after OGD
| Transcriptome | Classification | Number | Access ID |
|---|---|---|---|
| ncRNAs | Total | 12,127 | |
| LncRNAs | Total | 10,677 | |
| Annotated | 5,436 | NR_ or XR_ | |
| Non-annotated | 5,241 | Lnc-OGD | |
| Significant change | 362 | ||
| >2 fold increase | 147 | ||
| <2 fold decrease | 70 | ||
| mRNAs | Total | 43,705 | NM_ or XM_ |
| Significant change | 4,301 | ||
| >2 fold increase | 992 | ||
| <2 fold decrease | 748 |
Total 55.832 transcriptome isoform expression in mouse primary BMEC culture
Figure 3.
Altered lncRNA and mRNA profiles in mouse BMECs after OGD. Heatmap and hierarchical clustering analysis of 10,677 lncRNAs (A) and 43,705 protein-coding RNAs (B) that were differentially expressed (P<0.045 and FDR<0.05) in mouse BMECs 16 hours after OGD exposure. Four independent biological replicates were analyzed for non-OGD and OGD conditions. The clustering tree for lncRNAs and protein-coding RNAs is shown at the top. The expression values are represented in shades of red and green, indicating expression above and below the median value across all samples (log scale 2, from −2.00 to +2.00), respectively.
Table 2.
Most highly upregulated IncRNAs in BMECs after OGD
| LncRNAs | Ref. ID | Transcript length | Fold change | Chromosome | Start site | End site | p value |
|---|---|---|---|---|---|---|---|
| Lnc-OGD1645 | - | 268 | 27.13 | 5 | 113592196 | 113592464 | 5.00E-05 |
| Snhg12 | NR_029468.1 | 606 | 23.06 | 4 | 132308677 | 132311C33 | 5.00E-05 |
| Gm19660 | XR_104882.2 | 636 | 21.40 | 4 | 119108744 | 119139596 | 5.00E-06 |
| Lnc-OGD2327 | - | 237 | 12.93 | 7 | 126644602 | 126644839 | 0.0022 |
| LOC100861877 | XR_140599.1 | 554 | 12.05 | 3 | 65527482 | 65560603 | 5.00E-05 |
| Lnc-OGD1006 | - | 381 | 9.39 | 3 | 149326946 | 149327327 | 5.00E-05 |
| Lnc-OGD1784 | - | 367 | 9.00 | 6 | 28410358 | 26410725 | 5.00E-06 |
| Lnc-OGD3838 | - | 665 | 8.43 | 13 | 5860536 | 5851201 | 5.00E-05 |
| Lnc-OGD2085 | - | 445 | 7.66 | 7 | 25076565 | 25077010 | 5.00E-05 |
| Lnc-OGD2439 | - | 259 | 6.87 | 8 | 54441066 | 54441325 | 5.00E-05 |
| Gm3601 | XR_105655.1 | 219 | 6.63 | 14 | 49971362 | 49989708 | 5.00E-05 |
| Lnc-OGD1710 | - | 305 | 6.55 | 5 | 140403591 | 140403B95 | 5.00E-05 |
| Lnc-OGD3723 | - | 254 | 6.19 | 12 | 74803907 | 74804506 | 0.00825 |
| Malat1 | NR_002847.2 | 6424 | 6.05 | 19 | 5795687 | 5802950 | 5.00E-05 |
| Srsf3 | NR_036613.1 | 3041 | 6.05 | 17 | 29032659 | 29043735 | 5.00E-05 |
| LOC100861991 | XR_141282.1 | 535 | 5.88 | 15 | 80255183 | 80265023 | 5.00E-05 |
| Snhg1 | NR_002896.3 | 476 | 5.86 | 19 | 8723486 | 8726445 | 5.00E-05 |
| Lnc-OGD5236 | - | 1873 | 5.68 | Y | 90731450 | 90736460 | 5.00E-05 |
| Lnc-OGD3641 | - | 265 | 5.58 | 13 | 9834264 | 9834529 | 5.00E-05 |
| Gm13187 | XR_104650.1 | 866 | 5.46 | 2 | 4152862 | 4614043 | 0.0041 |
| Lnc-OGD3413 | - | 1160 | 5.34 | 11 | 67455361 | 67689220 | 0.00035 |
| Gm13351 | XR_140493.1 | 1957 | 5.31 | 2 | 18055236 | 18392830 | 5.00E-05 |
| Lnc-OGD456 | - | 476 | 5.25 | 2 | 98664878 | 98665371 | 5.00E-05 |
| Lnc-OGD2596 | - | 1689 | 5.24 | 9 | 13414327 | 13416072 | 0.00555 |
| Gm11974 | NR_045893.1 | 592 | 5.01 | 11 | 6525590 | 6528805 | 5.00E-05 |
| 1810026B05Rik | NR_037569.1 | 709 | 5.00 | 7 | 73426454 | 73558615 | 0.0017 |
| LOC100861894 | XR_140466.1 | 494 | 4.96 | 1 | 143999337 | 144016631 | 5.00E-05 |
| Gm20367 | XR_105161.1 | 315 | 4.67 | 7 | 126623245 | 126635195 | 5.00E-05 |
| 2410006H16Rik | NR_030738.1 | 461 | 4.39 | 11 | 62602876 | 62604806 | 5.00E-05 |
| Gm16702 | NR_045795.1 | 2384 | 4.35 | 17 | 8372395 | 8422728 | 5.00E-05 |
| Lnc-OGD3606 | - | 477 | 4.38 | 11 | 109011641 | 109012118 | 5.00E-06 |
| 4930470H14Rik | NR_045764.1 | 1734 | 4.12 | 17 | 4044657 | 40B2995 | 5.00E-05 |
| Lnc-OGD4481 | - | 204 | 4.10 | 16 | 17222307 | 17222511 | 5.00E-05 |
| Lnc-OGD2556 | - | 1703 | 4.09 | 8 | 125418062 | 125569817 | 5.00E-05 |
| 4930405P13Rik | NR_045955.1 | 811 | 4.03 | 11 | 88905927 | 88955442 | 0.01145 |
Table 3.
Most highly downregulated IncRNAs in BMECs after OGD
| LncRNAs | Ref. ID | Transcript length | Fold change | Chromosome | Start site | End site | p value |
|---|---|---|---|---|---|---|---|
| LHC-OGD1996 | - | 408 | 0.07 | 6 | 128843185 | 128843593 | 5.00E-05 |
| Lnc-OGD3916 | - | 988 | 0.08 | 13 | 34875479 | 34906067 | 0.0011 |
| Lnc-OGD2838 | - | 280 | 0.17 | 13 | 3198305 | 3198585 | 5.00E-05 |
| 2810008D09Rik | NR_027059.1 | 600 | 0.17 | 11 | 117076719 | 117079086 | 5.00E-05 |
| LHC-OGD301 | - | 660 | 0.18 | 2 | 13011326 | 13014685 | 5.00E-05 |
| Gm19767 | XR_105707.2 | 1138 | 0.21 | 15 | 99029890 | 99038110 | 5.00E-05 |
| LOC100882008 | XR_140736.1 | 399 | 0.22 | 5 | 110135761 | 110176504 | 5.00E-05 |
| Lnc-OGD4126 | - | 1184 | 0.23 | 14 | 54431485 | 54443711 | 0.0012 |
| Lnc-OGD3114 | - | 1938 | 0.23 | 10 | 61428297 | 61430235 | 5.00E-05 |
| Lnc-OGD124 | - | 273 | 0.24 | 1 | 93918070 | 93918343 | 0.00605 |
| Peg 13 | NR_002864.1 | 4721 | 0.25 | 15 | 72589619 | 73061204 | 5.00E-05 |
| Pisd-ps1 | NR_003517.1 | 606 | 0.25 | 11 | 3124020 | 3194157 | 5.00E-05 |
| C920009B18Rik | NR_015465.1 | 336 | 0.25 | 10 | 22173444 | 22374139 | 0.001 |
| Gm10374 | XR_106218.1 | 584 | 0.25 | 11 | 84870922 | 84871631 | 5.00E-05 |
| 2610203C20Rik | NR_015483.2 | 2578 | 0.25 | 9 | 41474936 | 41615185 | 5.00E-05 |
| Lnc-OGD4922 | - | 1866 | 0.28 | 18 | 60593012 | 60649002 | 0.00125 |
| LOC100861838 | XR_140809.1 | 540 | 0.33 | 7 | 35772345 | 35838074 | 0.0089 |
| 2700023E23Rik | NR_015531.1 | 798 | 0.30 | 5 | 74093064 | 74094336 | 0.0117 |
| LOC100861735 | XR_140828.1 | 611 | 0.31 | 7 | 55794147 | 55833964 | 5.00E-05 |
| LOC100861846 | XR_140548.1 | 364 | 0.31 | 2 | 72476218 | 72499861 | 5.00E-05 |
| LOC100861633 | XR_140781.1 | 593 | 0.32 | 6 | 127116352 | 127212419 | 5.00E-05 |
| LOC100861797 | XR_140919.1 | 722 | 0.32 | 8 | 106150398 | 106198704 | 0.00075 |
| Lnc-OGD5097 | - | 1675 | 0.33 | 19 | 56601953 | 56603628 | 5.00E-05 |
| 2700038G22Rik | NR_045040.1 | 1370 | 0.33 | 5 | 23850512 | 23855109 | 5.00E-05 |
| LHC-OGD2057 | - | 941 | 0.34 | 7 | 16348401 | 16049342 | 5.00E-05 |
| Lnc-OGD3950 | - | 1881 | 0.34 | 13 | 57999967 | 58001848 | 5.00E-05 |
| Gm19841 | XR_104928.1 | 713 | 0.35 | 5 | 35697179 | 35729383 | 0.00265 |
| Lnc-OGD1718 | - | 582 | 0.35 | 5 | 142866909 | 142867491 | 5.00E-05 |
| LOC100862093 | XR_140579.1 | 478 | 0.35 | 3 | 108591237 | 108722299 | 0.00035 |
| LOC100862295 | XR_141255.1 | 541 | 0.35 | 15 | 10486033 | 10493909 | 5.00E-05 |
| Gm14966 | XR_106193.1 | 2631 | 0.35 | 19 | 6341160 | 6378158 | 0.00445 |
Interestingly, we also found that a total of 992 protein-coding RNAs increased and 748 decreased by more than 2-fold in mouse BMECs after the same ischemic exposure (Table 1). Within the listed differentially expressed genes (Supplementary Table 2-3), GO analysis results were determined and KEGG pathway analysis (Supplementary Fig.1) revealed substantial enrichment-related pathways, including cell degradation, cell adhesion, cell cycle, p53, MAPK, VEGF, and other significant signaling pathways in this profile, many of which are associated closely with cell death, abnormal cell metabolism, ER stress, oxidative stress, inflammation, or cerebral endothelial pathologies under ischemic stroke.
It has been reported that lncRNAs may prefer to co-regulate with neighboring genes and exert enhancer-like activities (Cabili et al., 2011; Leung et al., 2013; Orom et al., 2010a; Orom et al., 2010b). In order to determine the significance of the proximity of co-regulated OGD-responsive endothelial lncRNAs and protein-coding genes, we carried out 100,000 simulations of proximity measurement on randomly placed co-regulated lncRNAs and protein-coding genes in the mouse genome. In particular, we focused on 4,301 significantly changed genes and the most highly altered lncRNAs (Table 2-3) to be co-regulated. We counted the number of simulated OGD-responsive lncRNAs that were within 500kb of a simulated OGD-responsive gene. We found that most highly upregulated or downregulated lncRNAs are proximal (< 500kb) to differentially expressed OGD-responsive genes (Supplementary Table 4-5). Further functional association or regulation between OGD-responsive lncRNAs and their neighboring/adjacent protein-coding genes needs to be further verified in the future.
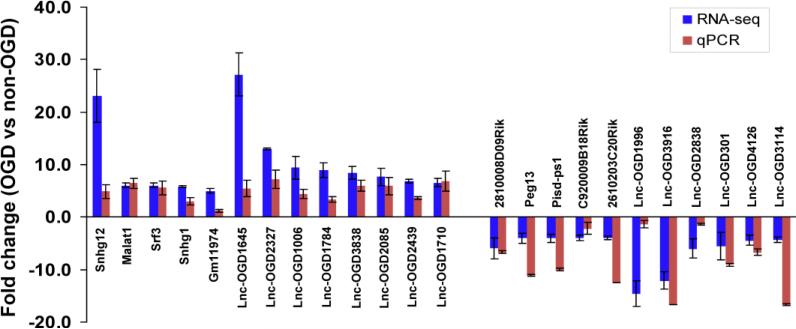
QPCR verification of RNA-seq identified OGD-responsive brain endothelial lncRNAs
In order to validate the expression profiles generated from RNA-seq, we performed quantitative real-time PCR to identify the correlation between the two datasets. As shown in Figure 4, the most highly upregulated or downregulated brain endothelial lncRNAs found in RNA-seq profiling experiments were further verified and showed consistent alteration tendencies in our qPCR study. These findings are the first to identify OGD-responsive brain endothelial lncRNAs, which suggest potential functional roles for these lncRNAs in mediating vascular endothelial responses to ischemic stimuli.
Figure 4.
PCR verification of expression profiles of lncRNA in mouse BMECs after OGD. The relative expression levels of most highly changed lncRNAs as indicated were examined using qRT-PCR in mouse BMECs that were treated with OGD for 16 hours. Data from RNA-seq were well-matched to quantitative PCR results. Data are expressed as mean ± SEM from 3 separate experiments.
The expression of OGD-responsive endothelial lncRNAs in cerebral microvasculature in mice after transient focal cerebral ischemia
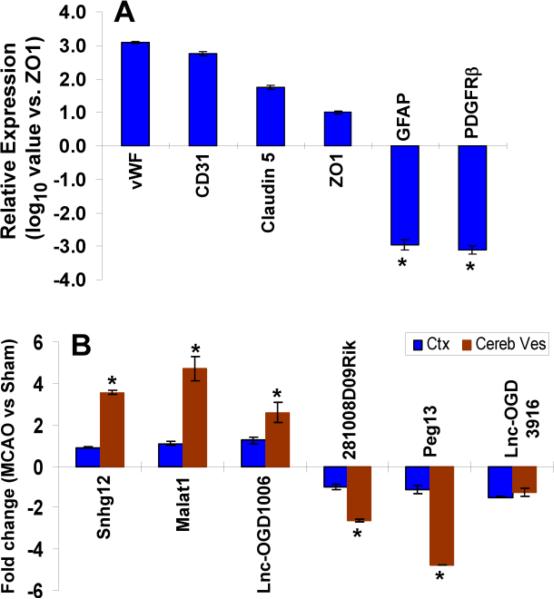
Extending our in vitro observations into in vivo studies, we further examined if OGD-responsive endothelial lncRNAs (upregulated and downregulated) have similar pathological expression patterns in mouse cerebral vasculature after ischemic stroke. To quantitatively determine expression of OGD-responsible lncRNAs in the cerebral vasculature, whole microvessel isolates from mouse brains after MCAO (n=6) were prepared by vessel-enriched isolation as described in our previous publications (Yin et al., 2010a; Yin et al., 2006b). Total RNA was isolated and subjected to quantitative PCR for the detection of lncRNA levels. As demonstrated in Figure 5, microvessels were isolated from mouse brains and qPCR analysis was characterized for high expression of brain EC markers ZO-1, claudin-5, CD31, and vWF with much lower levels of the astrocyte marker, GFAP and pericyte marker, platelet-derived growth factor receptor-β, indicating a relative high EC abundance (Fig. 5A). Of note, three most highly upregulated (Snhg12, Malat1, lnc-OGD1006) or downregulated (281008D09Rik, Peg13, and lnc-OGD3916) OGD-responsive endothelial lncRNA transcriptswere also significantly increased or reduced in isolated mouse cerebral microvessels in response to ischemic stroke (Fig. 5B), respectively. These data suggest that the expression of OGD-responsive endothelial lncRNAs will also be significantly altered in the cerebral vascular endothelium after in vivo ischemic insults, suggesting that these OGD-responsive lncRNA transcripts are associated with cerebrovascular pathogenesis after ischemic stroke, and warrant further investigation.
Figure 5.
Altered lncRNA expression profiles in cerebral microvasculature and brain in mice after focal cerebral ischemia. (A) Brain microvessels were isolated from mouse cerebral cortex and the expression of multiple neurovascular cell markers was analyzed by qPCR. Characterized high expression of brain EC markers, ZO-1, claudin-5, CD31, and vWF with much lower levels of the astrocyte marker, GFAP and pericyte marker, platelet-derived growth factor receptor-β, indicates a relative high EC abundance. (B) QPCR data demonstrated that three most highly upregulated and downregulated OGD-responsive lncRNAs as indicated are also significantly increased and reduced respectively in cerebral microvessels (Cereb Ves) but not in brain parenchyma (cerebral cortex, Ctx) in mice after 1h MCAO followed by 24h reperfusion. Data are expressed as mean + SEM. *p<0.05 vs. Sham or ZO-1 group.
Identification of transcription factors in OGD-responsive brain endothelial lncRNAs
The function of non-coding RNAs appears to be under the control of many transcription factors (TFs) (Bethke et al., 2009; Schanen and Li, 2010; Yin et al., 2010a). We analyzed a total of 128 transcription factors with FPKM>1 in all conditions of our RNA-seq data. To explore the possibility that TFs regulate OGD-responsive lncRNA expression in a transcriptional manner, we employed Genomatix software to screen for TF binding sites in the promoter region of 362 OGD-responsive endothelial lncRNAs (Table 4). Our bioinformatics analysis has identified that Sp1 and Krüppel-like transcription factors (KLFs) are the two highest overrepresented TFs in the promoter region of OGD-responsive lncRNA genes, whereas LOC100861682 and 2610019E17Rik have the highest number of TFs bound in their promoters (Table 4). These first-step results suggest that TFs may specifically affect the activity of OGD-responsive endothelial lncRNAs under ischemic stimuli.
Table 4.
Top transcription factors in the promoter of OGD-responsible IncRNAs
| Name of TFs | # of IncRNA promoters bound | Name of IncRNAs | # of TF bound in the promoter |
|---|---|---|---|
| SP1 | 141 | LOC100861682 | 137 |
| KLF4 | 137 | 2610019E17Rik | 127 |
| MAZ | 123 | Msx1as | 119 |
| PATZ1 | 119 | D330041H03Rik | 111 |
| TFAP2C | 112 | Lnc-OGD4751 | 103 |
| MZF1 | 107 | LOC100862031 | 102 |
| TFDP1 | 107 | Lnc-OGD3606 | 101 |
| YY1 | 86 | LOC100861830 | 93 |
| ELF2 | 85 | Prr3 | 89 |
| TFAP2B | 83 | LOC100861838 | 89 |
| SPI1 | 79 | Malat1 | 76 |
| SPIB | 79 | Srsf7 | 76 |
| GTF2I | 78 | LOC100861979 | 74 |
| PAX5 | 78 | LOC100862175 | 74 |
| SPZ1 | 76 | LOC100862295 | 74 |
| TCF4 | 75 | Gm10374 | 72 |
| KLF12 | 74 | 1110038B12Rik | 70 |
| MTF1 | 71 | Lnc-OGD3898 | 70 |
| MAFB | 70 | Lnc-OGD648 | 68 |
| MYOD1 | 70 | Gm17690 | 67 |
| HIC1 | 69 | Lnc-OGD3838 | 65 |
| GABPA | 68 | LOC100862023 | 65 |
| HOXB6 | 66 | Gm19767 | 65 |
| EGR3 | 65 | LOC100862184 | 64 |
| GATA1 | 65 | Gm20184 | 63 |
| FEV | 64 | Lnc-OGD2327 | 62 |
| RREB1 | 63 | Lnc-OGD2789 | 62 |
| GFI1 | 62 | Snhg5 | 61 |
| NHLH1 | 62 | Gm5853 | 61 |
| NKX3-2 | 61 | A030001D20Rik | 61 |
| ZNF148 | 60 | Lnc-OGD4044 | 60 |
| ARNT | 56 | LOC100861862 | 60 |
| NFE2L1 | 56 | LOC100861829 | 60 |
| BPTF | 55 | LOC100882158 | 59 |
DISCUSSION
The major finding of our study is the identification of OGD-responsive brain endothelial lncRNAs by a state-of-the-art RNA-seq profiling approach. For the first time, our data have shown that brain vascular endothelial cells respond to ischemic stimuli by dramatically altering hundreds of lncRNA transcriptomic profiles, suggesting potential and uncovered functional roles for these differentially expressed lncRNAs in the brain vascular pathogenesis of ischemic stroke.
During the past few years, our understanding of the complex aspects of vascular endothelial biology dramatically increased by the discovery of ncRNAs (Bartel, 2004; Kim, 2005; Qureshi and Mehler, 2012; Schonrock et al., 2012; Taft et al., 2010; Vemuganti, 2013; Yin et al., 2014). In that context, ncRNAs such as miRNAs have been shown to play crucial roles in the control of endothelial cell biology, including cerebral endothelial biology (Yin et al., 2013b, 2014). Except for endothelial miRNAs, only a few reports have demonstrated the participation of specific lncRNAs in the regulation of endothelial biology and function during physiological and disease conditions (Gordon et al., 2010; Li et al., 2010; Yuan et al., 2012). A recent study has shown that genetic inactivation of Meg3 in mice leads to a significant increase in the expression of pro-angiogenic genes that promote new microvessel formation in the brain cortex (Gordon et al., 2010; Li et al., 2010; Yuan et al., 2012). Up to now, the expression and function of lncRNAs in cerebral vascular endothelial biology and stroke-induced cerebral vascular pathologies are totally unknown (Yin et al., 2014).
In this study, we employed RNA-seq approaches to provide definitive evidence that cerebral vascular endothelial cells express high levels of lncRNA transcripts. In particular, we identified a total of 362 annotated and unannotated OGD-responsive endothelial lncRNAs in cultured BMECs, including 147 OGD-upregulated and 70 OGD-downregulated lncRNA transcripts. These most highly altered expression profiles of cerebral endothelial lncRNAs were further confirmed by our quantitative PCR analysis in cultured BMECs after in vitro stimuli. Importantly, the expression levels of the most highly upregulated lncRNAs (Snhg12, Malat1, and lnc-OGD 1006) as well as the most highly downregulated lncRNAs (281008D09Rik, Peg13, and lnc-OGD 3916) have also been found to increase or decrease respectively in cerebral microvasculature after in vivo ischemic stimuli (mouse ischemic stroke model). These findings strongly suggest that OGD-responsive brain endothelial lncRNAs may have functional roles in mediating endothelial responses to ischemic stimuli, and may serve as endogenous modulators to regulate endothelial cell biology and also pathophysiology after cerebral ischemia. Further studies are needed in the future to show the significance of individual lncRNA molecules in the regulation of post-stroke cerebrovascular pathophysiology and neurological outcomes. Also, the molecular/transcriptional regulation of selective lncRNAs in cerebral vascular endothelium was warranted for further investigation. These results will provide novel insights into a better understanding of lncRNA-based molecular regulatory mechanisms in the control of impaired cerebral vascular endothelial structure and function after cerebral ischemia.
LncRNAs are known to recruit chromatin-modifying proteins such as Polycomb Repressive Complex 2 (PRC2) to specific sites of genome and affect gene expression through regulating chromatin states (Rinn and Chang, 2012). Human lncRNA HOTAIR is the first such RNAs recognized: HOTAIR physically associates with PRC2, and modulates PRC2 and H3K27me3 localization of hundreds of sites throughout the genome (Rinn et al., 2007; Tsai et al., 2010). Also, a number of studies indicated that many TFs are able to transactivate or transrepress the transcription of lncRNAs and thus affect their functions (Bethke et al., 2009; Huang et al., 2015; Schanen and Li, 2010; Yin et al., 2010a). For example lincRNA-p21 expression can be activated by p53 through physical interaction with p53, lincRNA-p21 then serves as a transcriptional repressor in the p53 pathway and plays a role in triggering apoptosis (Huarte et al., 2010). In another study, lncRNA TUG1 expression was found induced by nuclear transcription factor SP1 and TUG1 further repressed Krüppel-like factor 2 transcription in the epigenetic level (Xu et al., 2015).
Consistent with the previous studies, our bioinformatics analysis has shown that there are a substantial number of TF binding sites in the promoter regions of the 362 OGD-responsive lncRNAs, including Krüppel-like transcription factors (KLFs) and Sp1 as the two highest overrepresented TFs in the promoter regions of most OGD-responsive lncRNA genes. These data imply the possibility that TFs may regulate OGD-responsive lncRNA expression in a transcriptional manner. In order to further validate TF transcriptional regulation of OGD-responsive lncRNAs, a series of further experiments, including luciferase transcription activity assay, chromatin immunoprecipitation (ChIP) analysis, qPCR, and Western blotting, will need to be performed in the future as described in our previous publications (Yin et al., 2010a; Yin et al., 2013a).
In summary, this study is the first to profile lncRNA regulatory transcripts in cerebral vascular endothelium under ischemic conditions by using high-throughput deep sequencing (RNA-seq) technology. The functional importance and molecular regulatory mechanisms of lncRNAs in the cerebral vasculature, especially in the endothelium following ischemic stroke, are totally unknown. Identification of new lncRNA transcripts will provide a novel window of opportunity to study RNA-directed epigenetic regulators in cerebral endothelial biology and their essential role in stroke-induced vascular endothelium-dependent cerebrovascular pathologies, as well as likely reveal novel targets for a promising translational future of lncRNA-based diagnostics and therapeutics in ischemic stroke.
Supplementary Material
Highlights.
➢ A first study to profile ischemic-responsive brain endothelial long non-coding RNAs.
➢ Total 217 lncRNAs were classified as ischemic-responsive endothelial lncRNAs.
➢ The most highly altered lncRNAs were confirmed in brain microvessels after stroke.
➢ Identified key transcription factors that regulate ischemic-responsive lncRNAs.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grants: NS094930, NS091175, NS086820 (K.J. Yin); and NS066652 (Y.E. Chen).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barone FC. Ischemic stroke intervention requires mixed cellular protection of the penumbra. Curr Opin Investig Drugs. 2009;10:220–223. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science (New York, N.Y. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Zoppo GJ. Stroke and neurovascular protection. The New England journal of medicine. 2006;354:553–555. doi: 10.1056/NEJMp058312. [DOI] [PubMed] [Google Scholar]
- del Zoppo GJ, Hallenbeck JM. Advances in the vascular pathophysiology of ischemic stroke. Thrombosis research. 2000;98:73–81. doi: 10.1016/s0049-3848(00)00218-8. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43:2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Pokrzywa C, Vemuganti R. Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN neuro. 2013;5:283–289. doi: 10.1042/AN20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. The interplay of microRNA and neuronal activity in health and disease. Frontiers in cellular neuroscience. 2013;7:136. doi: 10.3389/fncel.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan SC, Hess DC, Hohnadel EJ, Pollock DM, Ergul A. Targets for vascular protection after acute ischemic stroke. Stroke. 2004;35:2220–2225. doi: 10.1161/01.STR.0000138023.60272.9e. [DOI] [PubMed] [Google Scholar]
- Fisher M. Injuries to the vascular endothelium: vascular wall and endothelial dysfunction. Reviews in neurological diseases. 2008;5(Suppl 1):S4–11. [PubMed] [Google Scholar]
- Ginsberg MD. Current status of neuroprotection for cerebral ischemia: synoptic overview. Stroke. 2009;40:S111–114. doi: 10.1161/STROKEAHA.108.528877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P, Shu YQ. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Molecular cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C, Anrather J. Stroke research at a crossroad: asking the brain for directions. Nature neuroscience. 2011;14:1363–1368. doi: 10.1038/nn.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Zhang JH, Nanda A, Granger DN. Inflammatory responses to ischemia and reperfusion in the cerebral microcirculation. Frontiers in bioscience : a journal and virtual library. 2004;9:1339–1347. doi: 10.2741/1330. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Zhang ZG. MicroRNAs in cerebral ischemia-induced neurogenesis. Journal of neuropathology and experimental neurology. 2013;72:718–722. doi: 10.1097/NEN.0b013e31829e4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok J, Gupta P, Guo S, Kim WJ, Whalen MJ, van Leyen K, Lo EH. Cell-cell signaling in the neurovascular unit. Neurochem Res. 2007;32:2032–2045. doi: 10.1007/s11064-007-9342-9. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS genetics. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Moskowitz MA, Lo EH, Iadecola C. The science of stroke: mechanisms in search of treatments. Neuron. 2010;67:181–198. doi: 10.1016/j.neuron.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010a;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harbor symposia on quantitative biology. 2010b;75:325–331. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Stary CM, Yang GY, Giffard R. microRNAs: innovative targets for cerebral ischemia and stroke. Current drug targets. 2013;14:90–101. doi: 10.2174/138945013804806424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink C, Khanna S. MicroRNA in Ischemic Stroke Etiology and Pathology. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00158.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Yanez M, Castellanos M, Blanco M, Mosquera E, Castillo J. Vascular protection in brain ischemia. Cerebrovasc Dis 21 Suppl. 2006;2:21–29. doi: 10.1159/000091700. [DOI] [PubMed] [Google Scholar]
- Sandoval KE, Witt KA. Blood-brain barrier tight junction permeability and ischemic stroke. Neurobiol Dis. 2008;32:200–219. doi: 10.1016/j.nbd.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanen BC, Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2010 doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellinger PD, Kaste M, Hacke W. An update on thrombolytic therapy for acute stroke. Curr Opin Neurol. 2004;17:69–77. doi: 10.1097/00019052-200402000-00012. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- Stapf C, Mohr JP. Ischemic stroke therapy. Annual review of medicine. 2002;53:453–475. doi: 10.1146/annurev.med.53.082901.104106. [DOI] [PubMed] [Google Scholar]
- Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Tan JR, Koo YX, Kaur P, Liu F, Armugam A, Wong PT, Jeyaseelan K. microRNAs in stroke pathogenesis. Curr Mol Med. 2011;11:76–92. doi: 10.2174/156652411794859232. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNASeq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science (New York, N.Y. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuganti R. The MicroRNAs and Stroke: No Need to be Coded to be Counted. Transl Stroke Res. 2010;1:158–160. doi: 10.1007/s12975-010-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuganti R. All's well that transcribes well: non-coding RNAs and post-stroke brain damage. Neurochemistry international. 2013;63:438–449. doi: 10.1016/j.neuint.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, Huang MD, Shu YQ. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- Yin KJ, Chen SD, Lee JM, Xu J, Hsu CY. ATM gene regulates oxygen-glucose deprivation-induced nuclear factor-kappaB DNA-binding activity and downstream apoptotic cascade in mouse cerebrovascular endothelial cells. Stroke. 2002a;33:2471–2477. doi: 10.1161/01.str.0000030316.79601.03. [DOI] [PubMed] [Google Scholar]
- Yin KJ, Cirrito JR, Yan P, Hu X, Xiao Q, Pan X, Bateman R, Song H, Hsu FF, Turk J, Xu J, Hsu CY, Mills JC, Holtzman DM, Lee JM. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006a;26:10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010a;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Hamblin M, Zhang J, Chen YE. Vascular PPARdelta protects against stroke-induced brain injury. Arterioscler Thromb Vasc Biol. 2011;31:574–581. doi: 10.1161/ATVBAHA.110.221267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010b;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Fan Y, Hamblin M, Zhang J, Zhu T, Li S, Hawse JR, Subramaniam M, Song CZ, Urrutia R, Lin JD, Chen YE. KLF11 mediates PPARgamma cerebrovascular protection in ischaemic stroke. Brain. 2013a;136:1274–1287. doi: 10.1093/brain/awt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Hamblin M, Chen YE. Angiogenesis-Regulating microRNAs and Ischemic Stroke. Current vascular pharmacology. 2013b doi: 10.2174/15701611113119990016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Hamblin M, Chen YE. Non-coding RNAs in cerebral endothelial pathophysiology: Emerging roles in stroke. Neurochemistry international. 2014 doi: 10.1016/j.neuint.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Hsu CY, Hu XY, Chen H, Chen SW, Xu J, Lee JM. Protein phosphatase 2A regulates bim expression via the Akt/FKHRL1 signaling pathway in amyloid-beta peptide-induced cerebrovascular endothelial cell death. J Neurosci. 2006b;26:2290–2299. doi: 10.1523/JNEUROSCI.5103-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Lee JM, Chen SD, Xu J, Hsu CY. Amyloid-beta induces Smac release via AP-1/Bim activation in cerebral endothelial cells. J Neurosci. 2002b;22:9764–9770. doi: 10.1523/JNEUROSCI.22-22-09764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Olsen K, Hamblin M, Zhang J, Schwendeman SP, Chen YE. Vascular Endothelial Cell-specific MicroRNA-15a Inhibits Angiogenesis in Hindlimb Ischemia. J Biol Chem. 2012;287:27055–27064. doi: 10.1074/jbc.M112.364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. Neuroprotective strategies targeting apoptotic and necrotic cell death for stroke. Apoptosis. 2009;14:469–477. doi: 10.1007/s10495-008-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, Zhang L, Wang F, Sun SH, Zhou WP. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Badaut J, Tang J, Obenaus A, Hartman R, Pearce WJ. The vascular neural network--a new paradigm in stroke pathophysiology. Nature reviews. Neurology. 2012;8:711–716. doi: 10.1038/nrneurol.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.