Summary
Sample size and power calculations are an important part of designing new sequence-based association studies. In this paper, we explore an efficient and accurate approach to computing sample size and power (particularly at small significance level, e.g., 10−6) for the sequence kernel association test (SKAT), which is a powerful and widely used approach for testing variant set association. The recently developed SEQPower (Wang et al., 2014) and SPS programs (Li et al., 2015) adopted random Monte Carlo simulations to empirically estimate power for a series of variant set association test methods including the SKAT, which could be very computing intensive and time consuming. It is desirable to develop methods that can quickly and accurately compute power without intensive Monte Carlo simulations. To our knowledge, the only analytical approach to computing power for SKAT was proposed by at Lee et al. (2012), who used an approximate non-central χ2 distribution to efficiently compute sample size and power for SKAT and related methods. However we will show that the computed power based on the analytical approach of Lee et al. (2012) could be inflated especially for a small significance level, which is often of primary interest for large-scale whole genome and exome sequencing projects. We propose a new non-central χ2 approximation based approach to accurately and efficiently compute sample size and power. In addition we study and implement a more accurate “exact” method to compute power, which is more efficient than the Monte Carlo approach though generally involves more computations than the χ2 approximation method. The exact approach could produce very accurate results and be used to verify alternative approximation approaches. We implement the proposed methods in publicly available R programs that can be readily adapted when planning sequencing projects.
Keywords: Sample size, sequencing study, sequence kernel association test
Introduction
Sample size and power calculations are a crucial step for designing sequence-based association studies. Software programs exist for single SNP based association tests in the genome-wide association studies (GWAS) (see, for example, Purcell et al., 2003; Skol et al., 2006). Single variant based association tests have proven useful in discovery of hundreds of disease-associated common variants (Welter et al., 2014). However these identified common variants only explain a small proportion of most human trait variance and disease heritability (Manolio et al., 2009), which indicates more variants with small to moderate effects or rare variants with large effects may yet to be discovered. Recently there is growing interest in detecting the joint association of a set of variants in a gene or region, which can aggregate multiple weak effects to boost detection power. Many methods have been proposed for detecting variant set association (VSA). Among them, the two widely used approaches are the burden test (BT) and sequence kernel association test (SKAT) (see, for example, Morgenthaler and Thilly, 2007; Li and Leal, 2008; Kwee et al., 2008; Madsen and Browning, 2009; Morris and Zeggini, 2010; Price et al., 2010; Liu and Leal, 2010; Lin and Tang, 2011; Wu et al., 2010; Neale et al., 2011; Wu et al., 2011; Lin et al., 2011; Lee et al., 2012; Schaid et al., 2013; Wang et al., 2013; Zhang et al., 2014; Lee et al., 2014; Wu et al., 2015; Wang et al., 2015).
Compared to the well developed VSA test methods, sample size and power calculation methodology for VSA tests is greatly lacking in the literature. This is partly due to two challenges: the complicated population dependent genetic variant distribution and the mathematical intractability of most VSA test methods. A common strategy to overcome the first challenge is to simulate sequence data based on, for example, the Wright-Fisher formula, forward-time simulation, or coalescent theory (Hudson, 2002; Schaffner et al., 2005; Hellenthal and Stephens, 2007; Peng and Liu, 2010). With the availability of well characterized reference panels of outside samples (for example, the HapMap and 1000 Genome Projects) and more and more real-word sequencing datasets, for example, the National Heart, Lung, and Blood Institute Exome Sequencing Project (Tennessen et al., 2012), we can also efficiently simulate sequence data by resampling from the real data (Li and Li, 2008). As for the second challenge, a straightforward solution is the Monte Carlo approach. For example, the recently developed SEQPower (Wang et al., 2014) and SPS programs (Li et al., 2015) adopted the random Monte Carlo simulations to empirically estimate power for a series of rare variant set association test methods including the SKAT. In general, these simulations are very computing intensive and time consuming. It is desirable to develop methods that can quickly and accurately compute power without intensive Monte Carlo simulations. To our knowledge, the only analytical approach to computing power for SKAT is studied at Lee et al. (2012), who used an approximate non-central χ2 distribution to efficiently compute sample size and power for SKAT and related methods. However we will show that the computed power based on the analytical approach of Lee et al. (2012) could be inflated especially for a small significance level, which is of primary interest for large-scale whole genome and exome sequencing projects. We study a new non-central χ2 approximation based approach to accurately and efficiently compute sample size and power. In addition we also study and implement a more accurate “exact” method to compute power, which is more efficient than the Monte Carlo approach though generally involves more computations than the χ2 approximation method. The exact approach could produce very accurate results and be used to verify alternative approximation approaches. We implement the proposed methods in R programs that can be readily adapted when planning sequencing projects.
Sample size and power calculation for sequence-based association study
For illustration we consider a continuous trait with no covariates. We note that the following results can be easily extended to accommodate covariates and various outcomes (for example, disease status) following the approach of Lee et al. (2012). Consider n unrelated individuals. Denote the n outcomes as Y = (y1, …, yn)T, and the m-vector of genotype scores as Gi for individual i = 1, …, n. We assume the linear regression model, yi = β0 + μi + εi, where εi is a zero mean normal random variable with variance σ2, is the contribution from the variant set, and β is a m-vector of regression coefficients. Without loss of generality, we assume the genotype scores have been centered and the outcome is standardized with unit variance σ = 1.
We can write the SKAT test statistic as Q = (Y − Ȳ)TGWWGT (Y − Ȳ)/n, where , G = (G1, …, Gn)T, and W = diag(w1, …,wm) is a diagonal matrix of weights pre-specified for each variant (typically determined by the variant MAF). Here we have scaled the typical SKAT statistic by n for ease of derivation. Let . We have Q = ZTZ. Our sample size and power calculation will be based on the m-vector Z (typically m ≪ n).
Note that G is centered, therefore , and hence Z follows a multivariate normal distribution with mean and covariance matrix Σ = WGTGW/n. Note that GTG/n is the pairwise covariance matrix of the m genotype scores. Σ can be readily computed from simulated sequencing data or existing real-world sequencing data. Denote the eigen decomposition Σ = UDUT, where U is an orthogonal matrix and D = diag(λ1, …, λm) consists of the eigen values. Note that UUT equals to the identity matrix, hence we can write Q = (UTZ)T (UTZ). Here UTZ follows a multivariate normal distribution with mean UTη and covariance matrix UTΣU = D. So Q is essentially the sum of squares of m independent normal random variables. Therefore Q asymptotically follows a mixture of one degree of freedom (DF) non-central χ2 distributions, , where the vector of non-centrality parameter (δ1, …, δm) equal to the square of .
Conditional on the genotype scores G, Lee et al. (2012) approximated the distribution of Q using the non-central χ2 distribution under the null and alternative derived by matching the first four moments of Q based on the approach of Liu et al. (2009). This moment-matching based non-central χ2 approximation enables us to analytically and quickly compute the sample size and power. As shown in Duchesne and Lafaye De Micheaux (2010), the computed probabilities from the non-central χ2 approximation of Liu et al. (2009) could be very far from the true values. For small significance level in the magnitude of 10−6, which is typically of our main interest in whole genome and exome sequencing studies, the approach of Lee et al. (2012) can lead to over-estimated power as we show in the following. To account for the randomness of G, we can easily average over randomly generated sequence data, or approximately replace η and Σ with their expected values following Lee et al. (2012), which often leads to accurate results for relatively large sample size.
In the following we first derive a general formula to calculate power for SKAT and then discuss various computation approaches.
Power calculation
Denote the p-value function P(Y ; n,β,G) which computes the significance p-value for n individuals with observed outcome Y and genotype G. Here β is the postulated standardized genotype regression coefficients. For a given significance level α and genotype G, the power can be computed as S(α; n,β,G) = EY {I[P(Y; n,β,G) ≤ α]}, where the expectation is with respect to the outcome distribution conditional on G. Then we can compute the power as θ = EG[S(α; n,β,G)], where the expectation is with respect to the genotype distribution.
The SEQPower (Wang et al., 2014) and SPS programs (Li et al., 2015) computed θ by Monte Carlo simulation of both sequence data and outcomes to approximate EY and EG. Lee et al. (2012) computed S(α; b,β,G) analytically based on the non-central χ2 distributions, and then approximately computed EG(S) by evaluating S at the expected genotype covariance matrix and non-centrality parameters based on simulated sequence data. We can invert the power formula to compute the sample size.
Exact and approximation approaches
We can analytically compute S(α; n,β,G) for given genotype using the Davies’ method (Davies, 1980), which is based on the numerical integration to invert the characteristic function of Q. The Davies’ method can achieve very high accuracy but generally requires very large number of integration terms. It has been implemented in the R package CompQuadForm (Lafaye De Micheaux, 2013). To accurately compute small p-values using the Davies’ method, we set the error bound as 10−12 and maximum number of integration terms as 108. For a given significance level α, sample size n, and G, we first numerically solve the quantile under the null hypothesis (setting β = 0) based on the Davies’ method, which is then used to compute the corresponding power for the given β. Specifically we use the R ‘uniroot()’ function to numerically solve the null quantile. We need to calculate the expectation EG to compute the exact power, which can be approached by averaging over randomly simulated G to estimate the power. We will also evaluate a more computationally efficient approach where we use EG(Σ) and EG(η) to compute the corresponding power to approximate the exact average power.
For fast and accurate computation of more extreme significance p-values, we use the the non-central χ2 approximation approach of Wu and Pankow (2015), which is based on matching the higher moments of Q to achieve more accuracy, while Lee et al. (2012) matched the first four moments as in Liu et al. (2009). Specifically we approximate the distribution of Q using a k-DF non-central χ2 distribution with non-centrality parameter γ ≥ 0. Following the approach of Liu et al. (2009), we compute
where , μχ = k + γ, and . Here k and γ can be estimated by moment-matching. Specifically Liu et al. (2009) matched the skewness and minimized the kurtosis difference. Lee et al. (2012) proposed to match the kurtosis to improve the tail probability estimation. Both approaches lead to analytical solutions for k and γ. We can check that the jth cumulant of Q is (see, for example, Liu et al., 2009), and the jth cumulant of is κ̃j = 2j−1(j − 1)!(k + jγ). From the cumulants, we can easily compute their central moments denoted as νj and ν̃j for Q and respectively (see, for example, Lange, 2010). Let and . When , let , both the Liu and Lee method have γ = s1a3 − a2 and k = a2 − 2λ. When , the Liu method has γ = 0 and , and the Lee method has γ = 0 and k = 1/s2. Both methods provide poor approximation to small tail probabilities (Wu and Pankow, 2015). To improve the tail probability approximation accuracy, we can match higher moments. Specifically we follow the approach of Wu and Pankow (2015) to minimize the standardized (J − 1)th and Jth moments differences
We set J = 12 to accurately compute small tail probabilities under the null hypothesis following Wu and Pankow (2015). For approximating relatively large tail probabilities under alternative, setting J = 6 leads to an overall good performance in our numerical studies. Note that both k and γ just need to be solved once and can be stored for power calculation for any significance level.
For a given sample size n and genotype G, denote the estimated non-central χ2 distribution parameters as k0, γ0 under the null hypothesis (β = 0), and k1, γ1 under the alternative hypothesis. Under significance level α, the power can be computed as , where is the cumulative distribution function of . To compute the average power over genotype distribution EG, we can average over randomly simulated genotype G to compute the power. Generally we need relatively small number of simulations (for example, 100) to obtain accurate power estimate. Instead of computing the average of power over random sequence data, we can estimate EG(Σ) and EG(η) and use them to compute the corresponding power to approximate the exact average power.
In the following we evaluate the numerical accuracy of various approaches for computing power.
Numerical example
We compared the performance of the the analytical approach of Lee et al. (2012), the proposed non-central χ2 approximation and Davies’ method. We compared their estimated power over 1000 simulated sequencing datasets, and the approximate power based on the average genotype covariance matrix. For a benchmark, we also included the Monte Carlo approach of estimating power over 108 simulations. Specifically each time we simulate both outcomes and genotypes, and compute the SKAT p-values. The collection of 108 p-values are then used to estimate power at a given significance level α.
For illustration we analyze common and rare variant sets in the gene G6PC2 using data from the Atherosclerosis Risk in Communities (ARIC) Study (The ARIC Investigators, 1989). The ARIC study is a prospective investigation of atherosclerotic disease with a total of 15792 individuals recruited from four U.S. communities participating in the baseline examination in 1987–1989. G6PC2 is a glucose-6-phosphatase gene. Both common and rare G6PC2 variants have shown significant associations with fasting glucose in large scale GWAS and sequencing studies (see, for example, Dupuis et al., 2010; Service et al., 2014; Mahajan et al., 2015; Wessel et al., 2015). First, we study a selected set of eighteen common G6PC2 variants, which have squared pairwise correlation smaller than 0.8 based on the GWAS data from 5947 non-diabetic white ARIC participants (Dupuis et al., 2010). Second, we study a set of nine rare G6PC2 variants measured using the illumina exome chip in 5866 non-diabetic white ARIC participants (Wessel et al., 2015). We treat the subset of measured ARIC genotypes G as the true population and use Bootstrap resampling to generate random sequencing data for any given sample size. We compute the standardized residuals from regressing the measured fasting glucose levels on the covariates (age, gender, and study center). We then regress the standardized residuals on genotypes to estimate β, which is treated as the true values.
We set weight for common variants and wi = (1 − pi)24 for rare variants, where pi is the MAF of the ith variant. We plug in the population genotype MAF and covariance matrix to approximate the average of Σ and η for a finite sample. Generally there is large variation associated with rare variants, and it is possible that we do not observe a rare variant in a finite sample. Following Lee et al. (2012), we compute an “observation probability” ri = 1 − (1 − pi)2n for the ith rare variant in a finite sample of n individuals. We then adjust the average of Σ and η as follows. Denote the population genotype covariance matrix as R with its (i, j)th element being σij. Define a matrix R̃ with its (i, j)th element being . We approximate the average of Σ with WR̃W, and the average of η with .
Table 1 and 2 summarize the estimated power at significance level 2.5 × 10−6 for the common and rare variant sets respectively. Overall we can see that the Lee et al. (2012) approach over-estimated power. The proposed non-central χ2 approximation and Davies’ method lead to very similar results and both agree with the Monte Carlo approach very well. In general we need a very large number of simulations for the Monte Carlo approach to obtain stable estimates especially for the rare variant set. For the common variant set, the approximate powers computed at the expected genotype covariance matrix are very close to the average power computed from 1000 simulations. Due to the large variation of sampling the rare variant set, the approximate approach generally under-estimated the power for a relatively small sample size, but the approximation improved with increased sample size.
Table 1.
Estimated power (%) at significance level 2.5 × 10−6 for the set of eighteen common G6PC2 variants: listed within parenthesis are the standard errors (%) of the estimated average powers over 1000 simulations for Davies, Lee and WP methods, and 108 simulations for the Monte Carlo (MC) approach. The Davies1, Lee1, and WP1 are the approximate powers computed at the expected genotype covariance matrix.
| n | MC | Davies | Davies1 | WP | WP1 | Lee | Lee1 |
|---|---|---|---|---|---|---|---|
| 1000 | 1.09 (0.001) | 1.09 (0.02) | 1.00 | 1.07 (0.02) | 0.99 | 1.49 (0.02) | 1.40 |
| 2000 | 15.8 (0.004) | 15.8 (0.11) | 15.5 | 15.6 (0.11) | 15.3 | 19.0 (0.12) | 18.9 |
| 3000 | 51.2 (0.005) | 51.1 (0.16) | 51.2 | 50.8 (0.16) | 50.8 | 56.3 (0.16) | 56.4 |
| 4000 | 82.0 (0.004) | 82.0 (0.10) | 82.3 | 81.8 (0.10) | 82.0 | 85.3 (0.09) | 85.5 |
| 5000 | 95.6 (0.002) | 95.7 (0.03) | 95.8 | 95.6 (0.04) | 95.7 | 96.8 (0.03) | 96.8 |
| 6000 | 99.2 (0.001) | 99.3 (0.01) | 99.3 | 99.2 (0.01) | 99.3 | 99.5 (0.01) | 99.5 |
Table 2.
Estimated power (%) at significance level 2.5 × 10−6 for the set of nine rare G6PC2 variants: listed within parenthesis are the standard errors (%) of the estimated average powers over 1000 simulations for Davies, Lee and WP methods, and 108 simulations for the Monte Carlo (MC) approach. The Davies1, Lee1, and WP1 are the approximate powers computed at the expected genotype covariance matrix.
| n | MC | Davies | Davies1 | WP | WP1 | Lee | Lee1 |
|---|---|---|---|---|---|---|---|
| 4000 | 6.70 (0.003) | 6.58 (0.12) | 5.44 | 6.49 (0.11) | 5.35 | 8.20 (0.13) | 7.11 |
| 6000 | 25.7 (0.004) | 25.4 (0.27) | 23.8 | 25.2 (0.27) | 23.5 | 29.6 (0.28) | 28.3 |
| 8000 | 53.2 (0.005) | 52.9 (0.35) | 52.4 | 52.6 (0.35) | 52.0 | 58.1 (0.34) | 58.0 |
| 10000 | 77.1 (0.004) | 77.1 (0.26) | 77.4 | 76.8 (0.26) | 77.1 | 81.0 (0.23) | 81.5 |
| 12000 | 91.1 (0.003) | 91.1 (0.14) | 91.7 | 90.9 (0.15) | 91.6 | 93.1 (0.12) | 93.7 |
| 15000 | 98.5 (0.001) | 98.5 (0.03) | 98.8 | 98.5 (0.04) | 98.8 | 99.0 (0.03) | 99.2 |
Effect of variant weight on rare variant set association test power
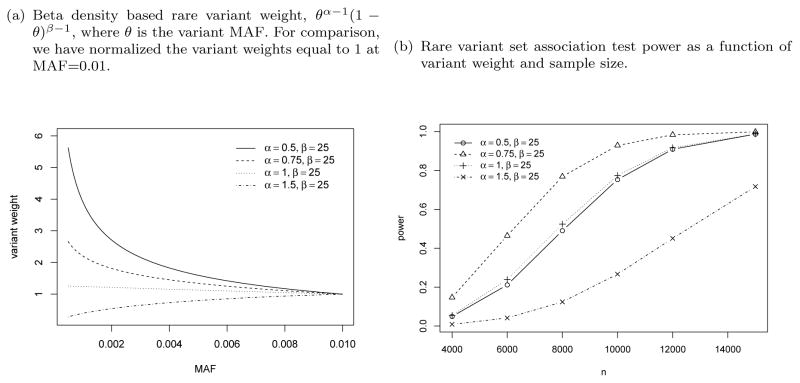
For SKAT of rare variant set, rarer variants are typically up-weighted to increase the detection power, which is based on the assumption that rarer variants tend to have larger effect sizes. Here we empirically investigate the SKAT power as a function of variant weight. Figure 1 compares four choices of variant weights based on the Beta density function, θα−1(1−θ)β−1, where θ is the variant MAF. For comparison, we have normalized the variant weights equal to 1 at MAF=0.01. The default weight of SKAT is (α = 1, β = 25) following Wu et al. (2011). Here we fix β = 25 and set α at 0.5, 0.75, 1 and 1.5. In general rarer variants get more weights with smaller α. For the first three sets of weights with α ≤ 1, the weight is decreasing with variant MAF, hence we are assigning more weights to rarer variants. For the last set of weights with α = 1.5, the weight is increasing with variant MAF, hence we are down-weighting rarer variants. Figure 1(b) compares the empirical power (computed using the Davies method) using previous G6PC2 rare variant set under different sample size and variant weights. Overall we can see that up-weighting rarer variants (α ≤ 1) does improve the SKAT power compared to down-weighting rarer variants (α = 1.5). And there are appreciable power difference between the first three sets of weights: α = 0.75 has the overall best power, and α = (0.5, 1) have similar performance. We expect that correct rare variant weighting can boost power, but generally the correct weights depend on the data and are unknown. It is worthwhile to study methods that can adaptively assign variant weight based on the data.
Figure 1.
Effects of rare variant weight and sample size on the variant set association test power.
Discussion
In this paper, we studied sample size and power calculations for designing sequencing studies for variant set association tests. Methods to perform these calculations are widely available for single SNP tests, but have been less well developed for variant set association tests, which are likely to increase in importance with the advent of whole exome or whole genome sequencing projects. Our numerical studies have suggested that care should be taken when computing and reporting power at a small significance level especially for rare variant sets due to their inherent large variations. The non-central χ2 distribution based analytical approximation method of Lee et al. (2012) in general over-estimated the power. We developed alternative approaches based on a new non-central χ2 approximation and Davies’ method to efficiently and accurately estimate sample size and power. We recommend computing/verifying sample size and power at those small significance level by using alternative methods, e.g., computationally intensive Monte Carlo simulation or the proposed methods.
We have implemented the proposed methods in R programs posted at http://www.umn.edu/~baolin/research/katsp_Rcode.html. They can be readily adapted to help investigators optimally design sequencing studies. Our proposed methods and results will offer important insights into the design of appropriately powered sequencing studies to maintain study power and reduce costs.
Acknowledgments
This research was supported in part by NIH grants GM083345 and CA134848. We are grateful to the University of Minnesota Supercomputing Institute for assistance with the computations. We want to thank the reviewer for the constructive comments which have greatly improved the presentation of the paper.
The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN-268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN-268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Support for exome chip genotyping in the ARIC Study was provided by the National Institutes of Health (NIH) American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419).
References
- Barnett IJ, Lee S, Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet Epidemiol. 2013;37(2):142–151. doi: 10.1002/gepi.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RB. Algorithm AS 155: the distribution of a linear combination of χ2 random variables. J R Stat Soc Ser C Appl Stat. 1980;29(3):323–333. [Google Scholar]
- Duchesne P, Lafaye De Micheaux P. Computing the distribution of quadratic forms: Further comparisons between the Liu–Tang–Zhang approximation and exact methods. Comput Stat Data Anal. 2010;54(4):858–862. [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JRB, Egan JM, Lajunen T, Grarup N, Spars T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proena C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YDI, Chines P, Clarke R, Coin LJM, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day INM, Geus EJCd, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PRV, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martnez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CNA, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AFH, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurosson G, Sijbrands EJG, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JCM, Yarnell JWG, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJF, Meneton P, Magnusson PKE, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ros M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WHL, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF Diagram Consortium, Giant Consortium, Global BPgen Consortium, Anders Hamsten on behalf of Procardis Consortium the MAGIC Investigators. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farebrother RW. Algorithm AS 204: The Distribution of a Positive Linear Combination of χ2 Random Variables. J R Stat Soc Ser C Appl Stat. 1984;33(3):332–339. [Google Scholar]
- Hellenthal G, Stephens M. msHOT: modifying Hudson’s ms simulator to incorporate crossover and gene conversion hotspots. Bioinformatics. 2007;23(4):520–521. doi: 10.1093/bioinformatics/btl622. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright?Fisher neutral model of genetic variation. Bioinformatics. 2002;18(2):337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Imhof JP. Computing the distribution of quadratic forms in normal variables. Biometrika. 1961;48(3–4):419–426. [Google Scholar]
- Kuonen D. Saddlepoint approximations for distributions of quadratic forms in normal variables. Biometrika. 1999;86(4):929–935. [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A Powerful and Flexible Multilocus Association Test for Quantitative Traits. Am J Hum Genet. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaye De Micheaux P. CompQuadForm: Distribution function of quadratic forms in normal variables. R package version 1.4.1. 2013 http://cran.r-project.org/web/packages/CompQuadForm/index.html.
- Lange K. Numerical Analysis for Statisticians. 2. Springer; New York: 2010. [Google Scholar]
- Lee S, Abecasis G, Boehnke M, Lin X. Rare-variant association analysis: Study designs and statistical tests. Am J Hum Genet. 2014;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13(4):762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li M. GWAsimulator: a rapid whole-genome simulation program. Bioinformatics. 2008;24(1):140–142. doi: 10.1093/bioinformatics/btm549. [DOI] [PubMed] [Google Scholar]
- Li J, Sham PC, Song Y, Li M. SPS: A Simulation Tool for Calculating Power of Set-Based Genetic Association Tests. Genet Epidemiol. 2015;39(5):395–397. doi: 10.1002/gepi.21898. [DOI] [PubMed] [Google Scholar]
- Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. Am J Hum Genet. 2011;89(3):354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Cai T, Wu MC, Zhou Q, Liu G, Christiani DC, Lin X. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genet Epidemiol. 2010;35(7):620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM. A Novel Adaptive Method for the Analysis of Next-Generation Sequencing Data to Detect Complex Trait Associations with Rare Variants Due to Gene Main Effects and Interactions. PLoS Genet. 2010;6(10):e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Tang Y, Zhang HH. A new chi-square approximation to the distribution of non-negative definite quadratic forms in non-central normal variables. Comput Stat Data Anal. 2009;53(4):853–856. [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM, Locke AE, Grarup N, Im HK, Cingolani P, Flannick J, Fontanillas P, Fuchsberger C, Gaulton KJ, Teslovich TM, Rayner NW, Robertson NR, Beer NL, Rundle JK, Bork-Jensen J, Ladenvall C, Blancher C, Buck D, Buck G, Burtt NP, Gabriel S, Gjesing AP, Groves CJ, Hollensted M, Huyghe JR, Jackson AU, Jun G, Justesen JM, Mangino M, Murphy J, Neville M, Onofrio R, Small KS, Stringham HM, Syvanen AC, Trakalo J, Abecasis G, Bell GI, Blangero J, Cox NJ, Duggirala R, Hanis CL, Seielstad M, Wilson JG, Christensen C, Brandslund I, Rauramaa R, Surdulescu GL, Doney ASF, Lannfelt L, Linneberg A, Isomaa B, Tuomi T, Jorgensen ME, Jorgensen T, Kuusisto J, Uusitupa M, Salomaa V, Spector TD, Morris AD, Palmer CNA, Collins FS, Mohlke KL, Bergman RN, Ingelsson E, Lind L, Tuomilehto J, Hansen T, Watanabe RM, Prokopenko I, Dupuis J, Karpe F, Groop L, Laakso M, Pedersen O, Florez JC, Morris AP, Altshuler D, Meigs JB, Boehnke M, McCarthy MI, Lindgren CM, Gloyn AL On Behalf of the T2D-GENES consortium, GoT2D consortium. Identification and functional characterization of G6PC2 coding variants in uencing glycemic traits define an effector transcript at the g6pc2-ABCB11 locus. PLoS Genet. 2015;11:e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat Res. 2007;615(1–2):28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oualkacha K, Dastani Z, Li R, Cingolani PE, Spector TD, Hammond CJ, Richards JB, Ciampi A, Greenwood CMT. Adjusted sequence kernel association test for rare variants controlling for cryptic and family relatedness. Genet Epidemiol. 2013;37(4):366–376. doi: 10.1002/gepi.21725. [DOI] [PubMed] [Google Scholar]
- Peng B, Liu X. Simulating Sequences of the Human Genome with Rare Variants. Hum Hered. 2010;70(4):287–291. doi: 10.1159/000323316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PIW, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86(6):832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15(11):1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Sinnwell JP, Thibodeau SN. Multiple Genetic Variant Association Testing by Collapsing and Kernel Methods With Pedigree or Population Structured Data. Genet Epidemiol. 2013;37(5):409–418. doi: 10.1002/gepi.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service SK, Teslovich TM, Fuchsberger C, Ramensky V, Yajnik P, Koboldt DC, Larson DE, Zhang Q, Lin L, Welch R, Ding L, McLellan MD, O’Laughlin M, Fronick C, Fulton LL, Magrini V, Swift A, Elliott P, Jarvelin MR, Kaakinen M, McCarthy MI, Peltonen L, Pouta A, Bonnycastle LL, Collins FS, Narisu N, Stringham HM, Tuomilehto J, Ripatti S, Fulton RS, Sabatti C, Wilson RK, Boehnke M, Freimer NB. Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci. PLoS Genet. 2014;10:e1004147. doi: 10.1371/journal.pgen.1004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, OConnor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ARIC Investigators. The atherosclerosis risk in communities (aric) study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- Wang GT, Li B, Santos-Cortez RPL, Peng B, Leal SM. Power analysis and sample size estimation for sequence-based association studies. Bioinformatics. 2014;30(16):2377–2378. doi: 10.1093/bioinformatics/btu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee S, Zhu X, Redline S, Lin X. GEE-Based SNP Set Association Test for Continuous and Discrete Traits in Family-Based Association Studies. Genet Epidemiol. 2013;37(8):778–786. doi: 10.1002/gepi.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xing EP, Schaid DJ. Kernel methods for large-scale genomic data analysis. Brief Bioinform. 2015;16(2):183–192. doi: 10.1093/bib/bbu024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich L, Hidalgo B, Fox K, Huffman JE, An P, Lu Y, Rasmussen-Torvik LJ, Grarup N, Ehm MG, Li L, Baldridge AS, Stancakova A, Abrol R, Besse C, Boland A, Bork-Jensen J, Fornage M, Freitag DF, Garcia ME, Guo X, Hara K, Isaacs A, Jakobsdottir J, Lange LA, Layton JC, Li M, Hua Zhao J, Meidtner K, Morrison AC, Nalls MA, Peters MJ, Sabater-Lleal M, Schurmann C, Silveira A, Smith AV, Southam L, Stoiber MH, Strawbridge RJ, Taylor KD, Varga TV, Allin KH, Amin N, Aponte JL, Aung T, Barbieri C, Bihlmeyer NA, Boehnke M, Bombieri C, Bowden DW, Burns SM, Chen Y, Chen YD, Cheng CY, Correa A, Czajkowski J, Dehghan A, Ehret GB, Eiriksdottir G, Escher SA, Farmaki AE, Franberg M, Gambaro G, Giulianini F, Goddard WA, Goel A, Gottesman O, Grove ML, Gustafsson S, Hai Y, Hallmans G, Heo J, Hoffmann P, Ikram MK, Jensen RA, Jorgensen ME, Jorgensen T, Karaleftheri M, Khor CC, Kirkpatrick A, Kraja AT, Kuusisto J, Lange EM, Lee IT, Lee WJ, Leong A, Liao J, Liu C, Liu Y, Lindgren CM, Linneberg A, Malerba G, Mamakou V, Marouli E, Maruthur NM, Matchan A, McKean-Cowdin R, McLeod O, Metcalf GA, Mohlke KL, Muzny DM, Ntalla I, Palmer ND, Pasko D, Peter A, Rayner NW, Renstrom F, Rice K, Sala CF, Sennblad B, Serafetinidis I, Smith JA, Soranzo N, Speliotes EK, Stahl EA, Stirrups K, Tentolouris N, Thanopoulou A, Torres M, Traglia M, Tsafantakis E, Javad S, Yanek LR, Zengini E, Becker DM, Bis JC, Brown JB, Adrienne Cupples L, Hansen T, Ingelsson E, Karter AJ, Lorenzo C, Mathias RA, Norris JM, Peloso GM, Sheu WHH, Toniolo D, Vaidya D, Varma R, Wagenknecht LE, Boeing H, Bottinger EP, Dedoussis G, Deloukas P, Ferrannini E, Franco OH, Franks PW, Gibbs RA, Gudnason V, Hamsten A, Harris TB, Hattersley AT, Hayward C, Hofman A, Jansson JH, Langenberg C, Launer LJ, Levy D, Oostra BA, O’Donnell CJ, O’Rahilly S, Padmanabhan S, Pankow JS, Polasek O, Province MA, Rich SS, Ridker PM, Rudan I, Schulze MB, Smith BH, Uitterlinden AG, Walker M, Watkins H, Wong TY, Zeggini E, Laakso M, Borecki IB, Chasman DI, Pedersen O, Psaty BM, Shyong Tai E, van Duijn CM, Wareham NJ, Waterworth DM, Boerwinkle E, Linda Kao WH, Florez JC, Loos RJF, Wilson JG, Frayling TM, Siscovick DS, Dupuis J, Rotter JI, Meigs JB, Scott RA, Goodarzi MO The EPIC-InterAct Consortium. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Pankow JS, Guan W. Sequence kernel association analysis of rare variant set based on the marginal regression model for binary traits. Genet Epidemiol. 2015;39(6):399–405. doi: 10.1002/gepi.21913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Pankow JS. On computing the tail probability of non-negative definite quadratic forms in central normal variables. 2015 Submitted. tech report http://www.umn.edu/~baolin/research/katpval_WuPankow.pdf.
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-Set Analysis for Case-Control Genome-wide Association Studies. Am J Hum Genet. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang L, Koboldt D, Boreki IB, Province MA. Adjusting family relatedness in data-driven burden test of rare variants. Genet Epidemiol. 2014;38(8):722–727. doi: 10.1002/gepi.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]