Abstract
Feline calicivirus depends on host-cell proteins for its replication. We previously showed that knockdown of nucleolin (NCL), a phosphoprotein involved in ribosome biogenesis, resulted in the reduction of FCV protein synthesis and virus yield. Here, we found that NCL may not be involved in FCV binding and entry into cells, but it binds to both ends of the FCV genomic RNA, and stimulates its translation in vitro. AGRO100, an aptamer that specifically binds and inactivates NCL, caused a strong reduction in FCV protein synthesis. This effect could be reversed by the addition of full-length NCL but not by a ΔrNCL, lacking the N-terminal domain. Consistent with this, FCV infection of CrFK cells stably expressing ΔrNCL led to a reduction in virus protein translation. These results suggest that NCL is part of the FCV RNA translational complex, and that the N-terminal part of the protein is required for efficient FCV replication.
Keywords: FCV, translation, nucleolin, AGRO100, aptamer, RNA virus, UTRs
Introduction
Human caliciviruses (HuCVs), from the Norovirus and Sapovirus genera in the Caliciviridae family, are a major cause of epidemic gastroenteritis that affects people of all ages (Widdowson et al., 2005). Despite their impact on public health, information regarding the HuCVs replicative cycle has been limited due to the difficulty to grow them in cultured cells (Duizer et al., 2004; Jones et al., 2014; Taube et al., 2013). Feline calicivirus (FCV), a member of the genus Vesivirus, can be propagated efficiently in conventional cell culture (Kreutz et al., 1994), and has served as an important model for the study of calicivirus molecular biology (Papafragkou et al., 2013; Vashist et al., 2009). Similar to other positive strand RNA viruses, FCV replication involves interactions of the virus proteins with a number of host cellular factors (Li and Nagy, 2011; Liu et al., 2009). The junctional adhesion molecule 1 protein (JAM-1) is a functional receptor for FCV virions (Makino et al., 2006); some eukaryotic translation initiation factors, such as eIF4E, eIF4A, and eIF4G, promote translation of the viral RNA (Chaudhry et al., 2006; Goodfellow et al., 2005). Moreover, the polypyrimidine tract-binding protein (PTB) can bind to the 5′ terminal end of the FCV genomic and subgenomic RNAs, and is required for viral replication (Karakasiliotis et al., 2010). It was suggested that PTB functions as a negative regulator of FCV translation, promoting the synthesis of RNA (Karakasiliotis et al., 2010).
Several more studies have reported the identification of a number of host-cell proteins that interact with the 5′- and 3′-ends of calicivirus genomes (Gutierrez-Escolano et al., 2000; Gutierrez-Escolano et al., 2003; Vashist et al., 2012). However, the functional role of these interactions remains poorly understood. Our previous work showed that nucleolin (NCL) in vitro associates with the 3′ end from the FCV genomic RNA. The NCL association with the genomic RNA was further confirmed in infected cells, and a reduction of its expression by siRNA decreased protein synthesis and viral yield, confirming its role in the FCV life cycle (Cancio-Lonches et al., 2011).
NCL is a multifunctional phosphoprotein with a molecular weight of 100–110 kDa (Tajrishi et al., 2011). This protein resides primarily in the cell nucleolus; however it can also be found in the nucleus, cytoplasm and on the surface of some cells (Borer et al., 1989; Hovanessian et al., 2000; Losfeld et al., 2011; Tajrishi et al., 2011). NCL has three well-defined domains. The N-terminal domain is involved in nuclear localization and responsible for binding rDNA and histone H1, and some ribosomal proteins (Erard et al., 1988; Ginisty et al., 1998). The central region contains an RNA-binding domain with four RNA recognition motifs (RRM), and is responsible for interactions with p53, beta-globin, Hsp90, and Bcl-XL mRNAs (Abdelmohsen et al., 2011; Jiang et al., 2006; Serin et al., 1996; Takagi et al., 2005; Wang et al., 2011; Zhang et al., 2008). The NCL C-terminal region contains the glycine-arginine-rich (GAR) domain that is essential for binding of ribosomal proteins and the telomerase RNA subunit hTERT (Bouvet et al., 1998; Khurts et al., 2004).
NCL controls a wide range of fundamental cellular processes such as ribosome biogenesis, proliferation and cellular cycle regulation (Cong et al., 2012; Ginisty et al., 1998; Ugrinova et al., 2007). On the cell surface, NCL serves as a receptor for ligands such as lactoferrin (Legrand et al., 2004), midkine protein (MK) (Take et al., 1994), pleiotrophin (PTN) (Said et al., 2005), and AGRO100, an oligonucleotide aptamer that has been shown to bind with high specificity and affinity to the NCL RRMs (Bates et al., 2009; Dahl et al., 1987; Mongelard and Bouvet, 2010; Reyes-Reyes et al., 2010; Soundararajan et al., 2009; Xu et al., 2001). The cell surface-expressed NCL is required for the efficient entry of human parainfluenza virus type 3 (HPV 3) into human lung epithelial A549 cells (Bose et al., 2004). NCL has been also identified as a receptor for the human respiratory syncytial virus (RSV) (Tayyari et al., 2011), and has been implicated as a low affinity receptor for the human immunodeficiency virus (HIV) (Said et al., 2002). The cytoplasmic NCL has been reported to interact with several viral proteins, such as hepatitis C virus (HCV) nonstructural protein 5B (NS5B), (Bouvet et al., 1998; Hirano et al., 2003; Kusakawa et al., 2007). Recently, it was shown that NCL could interact with dengue virus (DV) capsid protein, suggesting its role in viral morphogenesis (Balinsky et al., 2013).
In addition to protein-protein interactions, NCL has been shown to bind viral RNAs, such as the 5′ end of the HCV, poliovirus (PV), and rhinovirus (HRV) genomic RNAs (Lu et al., 2004). NCL interaction with the untranslated regions (UTRs) of the PV and HRV genomic RNAs has been found to stimulate translation of viral proteins (Izumi et al., 2001; Waggoner and Sarnow, 1998). On the other hand, NCL (Nsr1p in yeast), bound to the 3′ UTR of the tombusvirus (TBVS) RNA has been shown to inhibit its replication by interfering with recruitment of the viral RNA for replication (Jiang et al., 2010).
Due to the different processes that NCL modulates during viral replication, and because in our previous studies we demonstrated the requirement of NCL for an efficient FCV viral production (Cancio-Lonches et al., 2011), we wanted to investigate the specific function of this cellular protein during the different steps of the FCV replicative cycle. In the present report, we examined a role of NCL in binding and entry of FCV into the CrFK cells. We demonstrated that NCL specifically binds to the 5′ and 3′ end of the FCV genomic RNA, and stimulates its translation in vitro. We also showed that NCL mutants lacking the amino terminal domain (but containing RRMs) inhibit FCV RNA translation in vitro, and reduce viral protein synthesis and viral production during infection.
Results
Detection of NCL on the surface of CrFK cells
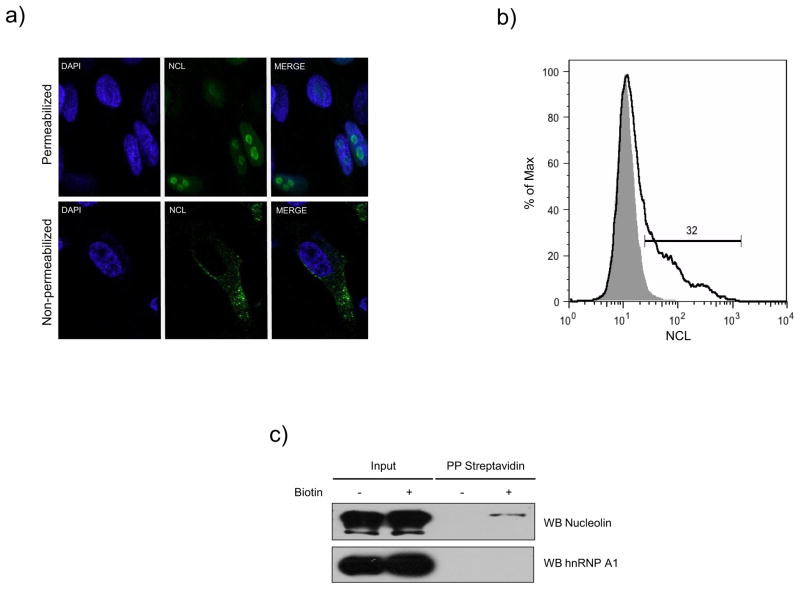
We previously reported that NCL associates with the FCV 3′ UTR and is required for an efficient viral replication cycle (Cancio-Lonches et al., 2011), since knockdown in CrFK cells by siRNAs caused a reduction in FCV production. To establish the specific role of NCL in the FCV replicative cycle, we analyzed first its possible role in virus binding and entry. It has been reported that JAM-1 serves as a functional receptor for some strains of FCV (Makino et al., 2006). The expression of this protein is not sufficient for productive infection by some FCV isolates (Ossiboff and Parker, 2007), suggesting that additional receptors or co-receptors may be present on permissive cells. To determine if NCL could have a role in FCV binding and entry, it was necessary to determine if NCL was present on the surface of CrFK cells. Non-permeabilized and permeabilized CrFK cells were incubated with specific anti-NCL antibody (H-250, Santa Cruz Biotechnology) and NCL subcellular localization was detected by immunofluorescence. NCL was predominantly localized in the nucleoli of permeabilized CrFK cells (Fig. 1A, upper panel), while, in non-permeabilized cells, NCL was detected predominantly on the surface (Fig. 1A, lower panel). The presence of NCL was also detected on the surface of non-permeabilized CrFK cells by flow cytometry using the same antibody described above (Fig. 1B). Non-permeabilized CrFK cells were incubated with an anti-NCL (black line) or an anti-rabbit IgG antibody (gray shaded area) as an isotype control. In the representative histogram, the fluorescence intensity of CrFK cells labeled with anti-NCL antibody (black line) shifted from the basal fluorescence of cells incubated with the isotype control antibody (grey). Approximately 32% of the analyzed cells showed the presence of the NCL protein on their surface (Fig. 2B). To detect the presence of cell-surface NCL biochemically, the surface proteins of CrFK cells were biotinylated (Fig. 1C, +) or not (Fig. 1C, −) with a cell-impermeable biotinylation reagent. The biotinylated proteins were isolated by absorption to streptavidin-agarose beads and subjected to immunoblotting by anti-NCL and anti-heterogeneous nuclear ribonucleoprotein (hnRNP) A1 antibodies. Detection of the 110-kDa NCL-specific protein band among the streptavidin-precipitated biotinylated proteins was consistent with the presence of NCL on the surface of CrFK cells (Fig. 1C). hnRNP A1, a protein present in the cytoplasm and predominantly in the nucleus, served as a control for the cell surface labeling: it was not detected in the streptavidin-bound plasma membrane fraction (Fig. 1C).
Figure 1.
Nucleolin is expressed in the surface of CrFK cells. A) Non-permeabilized and permeabilized CrFK cells with acetone, were stained with an anti-NCL antibody (green), and visualized by indirect immunofluorescence using Alexa 488 as secondary antibody. DAPI was used for nuclei staining (blue). B) Non-permeabilized CrFK cells were incubated with anti-NCL (black line) or anti-rabbit IgG antibodies (gray shaded area), used as an isotype control. C) CrFK cells were treated with sulfosuccinimidyl-6-(biotin-amide) hexanoate (+) or vehicle (−) and biotinylated proteins were precipitated with streptavidin-agarose, and separated by SDS-PAGE. NCL and hnRNP A1 proteins were detected by immunoblotting using specific anti-NCL and anti-hnRNP A1 antibodies respectively.
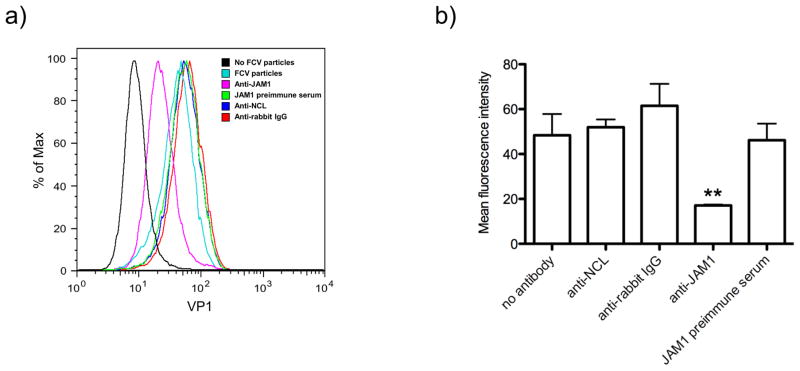
Figure 2.
Anti-NCL antibody does not inhibit FCV binding to CrFK cells. A) Non treated (black and blue light lanes), and CrFK cells treated with anti-JAM-1 (pink line), anti-NCL (blue line), and anti-rabbit IgG (red line) antibodies, or with the JAM-1 preimmune serum (green lanes), were incubated or no (black line) with FCV particles. FCV particles present on the cell surface were detected by flow cytometry using a commercial anti-FCV antibody (Santa Cruz Biotechnology). B) The bar chart illustrates the amount of binding inhibition as measured by mean fluorescence intensity. The error bars represent the standard deviations from three independent experiments. Values of P < 0.001 (**) are indicated.
Antibodies against NCL did not inhibit FCV-binding to CrFK cells surface
To investigate whether NCL on the CrFK cells surface is involved in FCV binding, NCL was blocked with anti-NCL antibody prior to virus infection, and FCV presence on the cell surface was tested by flow cytometry (Fig. 2). Pre-incubation of cells with anti-NCL antibody (40 μg/μl) did not inhibit FCV binding to the cell surface (Fig. 2A, blue line), in contrast, pre-incubation of cells with a 1:5 dilution of an anti-JAM1 guinea antibody partially (~50%) inhibited FCV binding to CrFK cells (Fig. 2A, pink line). Untreated cells or cells that were pre-incubated with JAM1 pre-immune serum (1:5 dilution) or with anti-rabbit IgG antibodies (40 μg/μl), did not show inhibition of FCV binding (Fig. 2A, blue light, green and red lines respectively). The data from three independent binding experiments are summarized in Fig. 2B.
Antibodies against NCL and FL rNCL soluble protein did not inhibit FCV plaque formation in CrFK cells
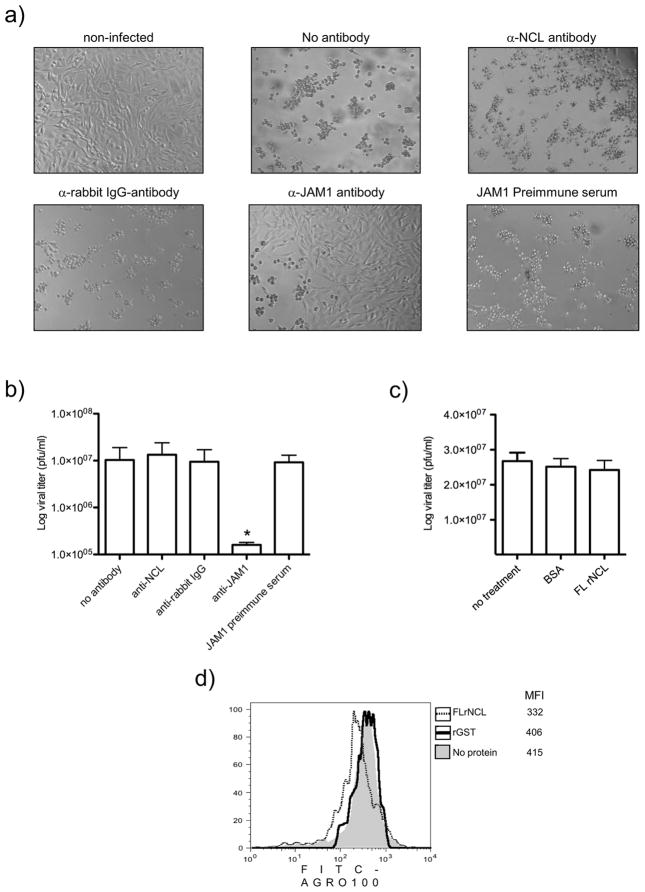
Even though anti-NCL antibody did not interfere with FCV binding to CrFK cells, it was important to test whether it has any capacity to offset the development of the FCV CPE and infection (Fig. 3). Pre-incubation of CrFK cells with a 1:5 dilution of JAM-1 antibody, significantly reduced the ability of FCV to produce CPE (Fig 3A). In contrast, pre-incubation of cells with a 40 μg/μl dilution of anti-NCL antibody (Fig. 3A) did not inhibit CPE produced by FCV infection. As expected, anti-rabbit IgG (40 μg/μl) and JAM1 pre-immune serum (1:5 dilution) used as negative controls, had no substantial effect on the CPE produced by FCV. Virus titration analysis of collected cell supernatants showed that only CrFK cells pretreated with anti-JAM 1 antiserum had a reduced level of virus production (Fig. 3B). Virus infection of cells pretreated with anti-NCL antibody resulted in a level of virus amplification similar to that in untreated cells or in cells pretreated with anti-rabbit IgG antibody, or JAM1 pre-immune serum (Fig. 3B).
Figure 3.
Anti-NCL antibody an FL NCL soluble protein does not have an effect on FCV infection in CrFK cells. A) Non treated and CrFK cells treated for 1 h at 37°C with anti-NCL, anti-rabbit IgG, anti-JAM-1 antibodies, and JAM-1 preimmune serum, were non infected or infected with FCV at an MOI of 0.01 for 24h. Cytopathic effect was observed by phase contrast microscopy. B) The bar chart illustrates the virus yield in supernatants determined by plaque assay. Error bars represent the standard deviations from three independent experiments. Value of P ≤ 0.05 (*) is indicated. C) Pre-incubated FCV (0.01 MOI) with BSA or FL rNCL proteins were added or not (no treatment) to CrFK cells for 24 h, and virus yield was determined in the supernatants by plaque assay. D) CrFK cells treated for 45 min on ice with AGRO-100 (gray shaded area) or a mix of 1μM FL rNCL (dashed line) or rGST (black line) proteins pre-incubated with 5μM FITC-AGRO for 2 h at 4°C. Mean fluorescence intensity (MFI) of FITC-AGRO100 was analyzed by flow cytometry.
On the other hand, FCV pretreated with FL rNCL (OriGene) or BSA (bovine serum albumin) did not affect virus infection (Fig. 3C). However, CrFK cells treated with the mix of FL rNCL and the NCL ligand AGRO100 coupled to a FITC molecule (FITC-AGRO100) reduced its ability to bind to CrFK cells (Fig. 3D). Taken together, these results strongly suggest that FL rNCL soluble protein does not inhibit FCV infection of CrFK cells.
ΔrNCL binds in vitro to both terminal ends of the FCV genomic RNA
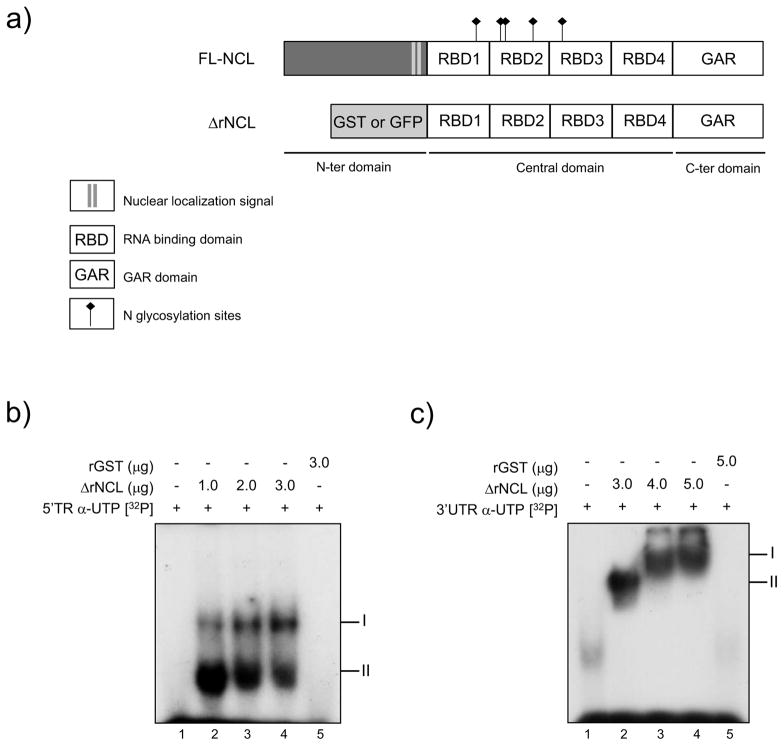
Since antibody-blocking experiments showed that administration of anti-NCL-specific antibody prior to FCV infection had no effect on the virus binding and entry, we examined a possible role of NCL in subsequent steps of the virus replication cycle such as translation and replication of the virus RNA. The terminal ends of viral genomes contain important sequences and structural elements that are recognized by cellular proteins and are critical for translation and RNA replication (Ahlquist et al., 2003). NCL is known to play a regulatory role during translation and replication of several viruses (Izumi et al., 2001; Jiang et al., 2010). Interestingly, NCL forms an RNP complex with the 3′UTR of the FCV RNA (Cancio-Lonches et al., 2011); then, to determine if NCL can directly bind to the 5′ end and the 3′ UTR from the FCV genomic RNA, a recombinant NCL was produced. To date it has not been possible to express full-length NCL, most probably due to the propensity of the N-terminal highly acidic domain to cause NCL auto-proteolysis (Chen et al., 1991; Izumi et al., 2001; Srivastava et al., 1989; Valdez et al., 1995); thus, a truncated form of NCL, ΔrNCL, that contain the protein central and C-terminal parts with four RNA recognition motifs (RRM) was expressed and purified (Fig. 4A). When in vitro synthesized RNAs containing the first 260 nt from the 5′ TR and 50 nt from the 3′ UTR of FCV genomic RNA were incubated with the ΔrNCL, the protein formed RNA-protein complexes that could be visualized by EMSA. ΔrNCL was able to specifically bind to the 5′ TR of the FCV RNA, forming two complexes (Fig. 4A). Moreover, ΔrNCL was also able to interact with the FCV 3′ UTR, and form a complex (II), which is enriched with increasing amounts of ΔrNCL (I) (Fig. 4B). No complex formation was observed when rGST was incubated with any of the FCV RNAs (Fig. 4A and B). These results indicate that ΔrNCL binds directly to the 5′ TR and the 3′ UTR of the FCV RNA.
Figure 4.
NCL recombinant protein binds to 5′ terminal region and 3′ UTR from the FCV genomic RNA. A) Schematic representation of NCL and ΔrNCL proteins. The RNA binding domains (RBD), nuclear localization signal (NLS), and the GAR domain are highlighted. N-glycosylation sites, located at positions 317–319, 399–401, 403–405, 477–479, and 491–493 are indicated. GST or GFP are located in the N-terminal end of the ΔrNCL protein. B) The [α-32P] UTP-labeled RNAs corresponding to the 5′ terminal region or C) the 3′ UTR of the FCV genomic RNA, were interacted with increasing amounts of ΔrNCL, (lanes 2, 3 and 4 respectively), or with the rGST protein (lanes 5). Lanes 1: free RNA. Migration of complexes is indicated.
NCL is required for an efficient FCV in vitro translation
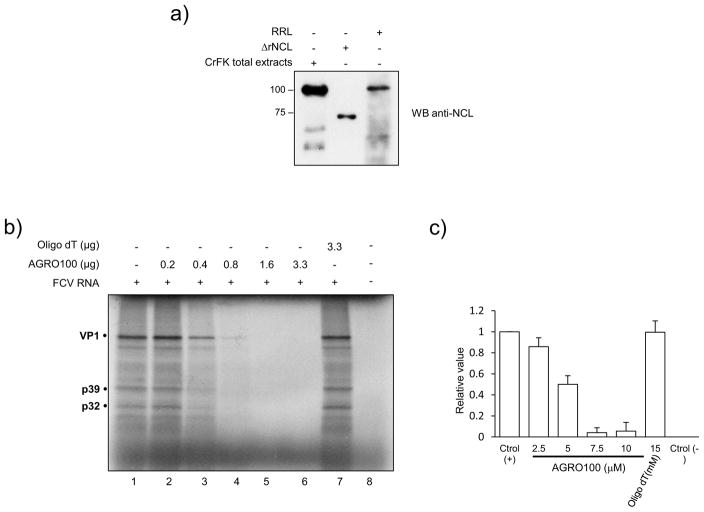
Because NCL could specifically bind both ends of the FCV genomic RNA, we investigated a possible role of this protein in regulating viral RNA translation. The effect of NCL on translation was analyzed using a rabbit reticulocyte lysate (RRL) system, which has been reported to support FCV RNA translation (Goodfellow et al., 2005; Karakasiliotis et al., 2010). The presence of NCL in these lysates could be readily detected by Western blot, using anti-NCL antibody (kindly donated by Kurt Gustin, University of Arizona, Phoenix) (Fig. 5A). The virus RNA was translated in vitro in the presence of the NCL functional inhibitor AGRO100. The in vitro translation assay showed that increasing amounts of AGRO100 included in the RRL resulted in an inhibition of FCV RNA translation that was dose-dependent (Fig. 5B and Fig. 5C). In contrast, the inclusion of a non-specific oligonucleotide (oligo-dT) in the RRL, did not affect FCV translation (Fig. 5B, lane 7 and Fig. 5C), suggesting that inactivation of NCL by AGRO100 was responsible for the inhibition of viral protein synthesis.
Figure 5.
AGRO100 inhibits FCV in vitro translation. A) Total extracts from CrFK cells, ΔrNCL and extracts from RRL (lanes 1, 2 and 3 respectively) were analyzed by SDS-PAGE and NCL was detected by western blot using a specific monoclonal antibody. B) FCV in vitro translation was performed in RRL extracts and the 35S methionine labeled viral proteins were analyzed by SDS-PAGE in the absence (lane 2) and the presence of increasing concentrations of AGRO100 (lanes 3–7), or oligo dT (lane 8) used as a non-specific control. C) The bar chart illustrates the amount of FCV protein synthesis as percentage respect of the control (0 μg AGRO100) as measured by ImageJ quantification. Error bars represent the standard deviation from three different experiments.
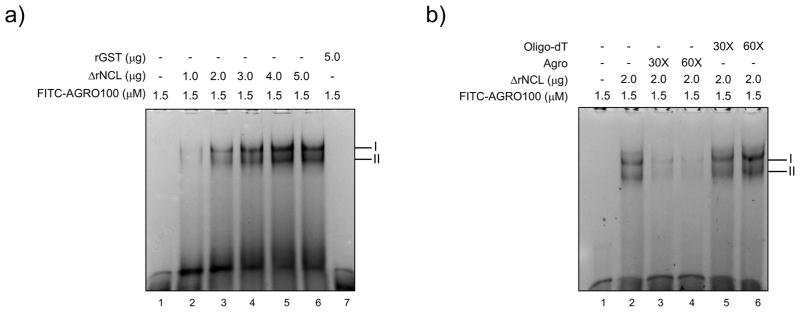
ΔrNCL binds specifically to AGRO100
To determine if AGRO100 binds to NCL, FITC-AGRO100 was incubated with the ΔrNCL, and the complex formation was analyzed by EMSA (Fig. 6). Two complexes were formed in the presence of 1 μg of ΔrNCL, and gradual increase in intensity of both bands was observed when FITC-AGRO100 was incubated with increasing amounts of ΔrNCL (Fig. 6A, lanes 2–6). In contrast, no complex formation was observed when rGST was added to the reaction (Fig. 6A, lane 7), suggesting that the interaction between AGRO100 and ΔrNCL was specific. To further confirm the specificity of the interaction between AGRO-FITC and ΔrNCL, the interaction was performed in the presence of unlabeled AGRO100 or oligo-dT molecules (Fig. 6B). The addition of 30x and 60x of AGRO100 (non coupled to FITC), strongly reduced complex formation with ΔrNCL, while the addition of the same amount of oligo-dT did not affect complex formation (Fig. 6B, lanes 3–4 and 5–6, respectively), indicating that AGRO100 specifically binds to ΔrNCL.
Figure 6.
AGRO100 specifically interacts with the NCL recombinant protein. A) FITC-labeled AGRO100 (1.5 μg) were interacted with increasing amounts of ΔrNCL, (lanes 2–6), or with the rGST protein (lane 7). B) Two μg of ΔrNCL (lanes 2–6), were interacted with 30X and 60X unlabeled AGRO100 (lanes 3 and 4 respectively), or unlabeled oligo dT (5, and 6 respectively), previous to the addition of 1.5 μg of FITC-labeled AGRO100. Lanes 1: free RNA. Migration of complexes is indicated.
NCL enhances FCV RNA in vitro translation and blocks the AGRO100 inhibitory effect
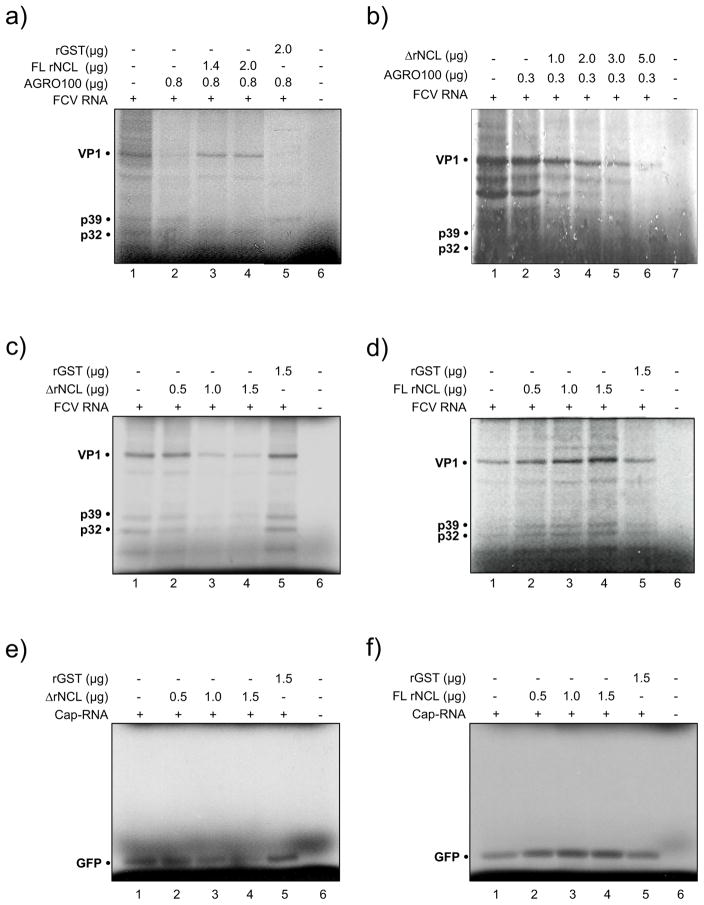
To determine if FL rNCL and ΔrNCL have the ability to rescue FCV protein synthesis lost by the presence of AGRO100, these proteins were included into the reactions containing inhibitory levels of AGRO100 (Fig. 7A and 7B). The inclusion of 1.4 and 2.0 μg of FL rNCL restored in vitro translation of FCV inhibited by AGRO100 (Fig. 7A, lanes 3 and 4 respectively), however, the same amount of ΔrNCL (Fig. 7B, lanes 3 and 4) or 2.0 μg rGST (Fig. 7A, lane 5) failed to restore FCV in vitro translation. A possible explanation for this might be that the N-terminal end of the NCL protein is required for the stimulatory/translational activity of this protein. Consistent with that, deletion of the N-terminus in ΔrNCL resulted in failure of the truncated version of NCL to reduce the AGRO100 inhibitory effect.
Figure 7.
Full length NCL but not the recombinant NCL positively regulates FCV in vitro translation. FCV in vitro translation was performed in RRL extracts and the 35S methionine labeled viral proteins were analyzed by SDS-PAGE. A) in the presence of 0.8 μg of AGRO100 (lanes 2–5), and increasing concentrations of FL rNCL (lanes 3 and 4); B) in the presence of 0.3 μg of AGRO100 (lanes 2–6) and increasing concentrations of ΔrNCL (lanes 3 and 6); C) in the presence of increasing amounts of ΔrNCL (lanes 2–4); and D) in the presence of increasing amounts of FL rNCL (lanes 2–4). E) non-viral capped RNA (GFP) in the presence of increasing concentrations of ΔrNCL (lanes 2–4); and F) in the presence of increasing amounts of FL rNCL (lanes 2–4). Lanes 1: FCV or GFP in vitro translation. Reactions performed in the presence of rGST or in the absence of FCV or Cap-RNAs are indicated.
The addition of increasing amounts of ΔrNCL to the FCV in vitro translation reaction strongly reduced FCV viral protein synthesis (Fig. 7C, lanes 2–4). In contrast, when increasing amounts of FL rNCL were added, FCV protein synthesis was enhanced in a dose-dependent manner (Fig. 7D, lanes 2–4), confirming that FL rNCL is a positive regulator of FCV in vitro translation. Parallel experiments were carried out to determine the effect of ΔrNCL and FL rNCL in cap-dependent translation. The addition of increasing amounts of ΔrNCL to the capped-GFP RNA in vitro translation reaction caused a reduction of the protein synthesis (Fig. 7E, lanes 2–4), while increasing amounts of FL rNCL promoted protein synthesis in a dose-dependent manner (Fig. 7E, lanes 2–4). Similar results were obtained with capped-Hsp70 RNA (data not shown). Since NCL is also required for PV and human rhinovirus (HRV) IRES dependent translation (Izumi et al., 2001), it is possible that this protein could be a general positive regulator in the RRL in vitro translation system.
The inability of the ΔrNCL to restore FCV in vitro translation inhibition by AGRO100, as well as its capacity to inhibit FCV viral protein synthesis suggests that this protein could have a negative regulatory effect in FCV translation.
NCL N-terminal end is required for the FCV protein synthesis and an efficient viral production
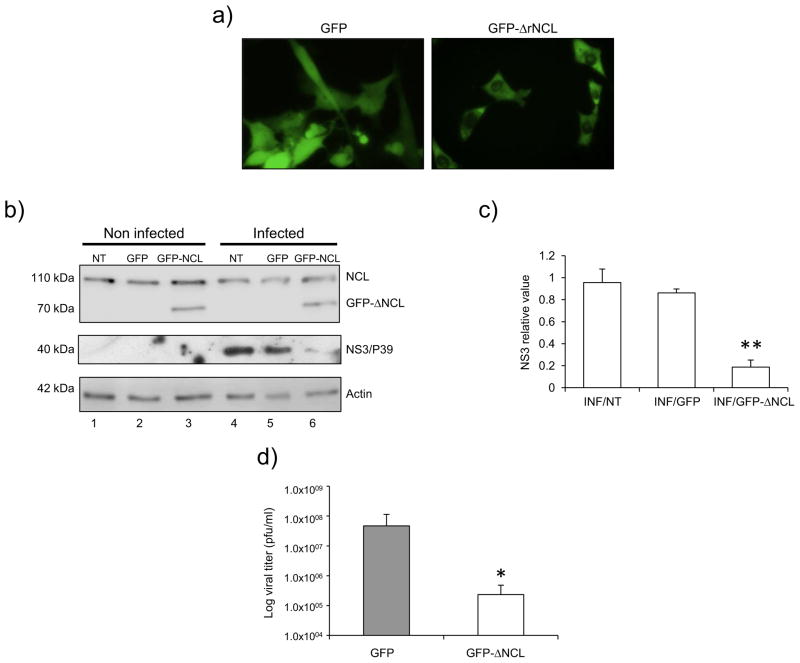
To further verify the dominant-negative effect of the ΔNCL on FCV infection, a CrFK cell line that stably expresses GFP-ΔrNCL was generated (Fig. 8A). Analysis of epifluorescence images of CrFK-GFP-ΔrNCL cells revealed a cytoplasmic and nucleolar presence of the GFP-ΔrNCL (Fig. 8, right panel). In contrast, GFP transfected cells showed a uniform GFP distribution throughout the cell (Fig. 8A, left panel). To verify the GFP-ΔrNCL expression, total cell lysates derived from non-transfected (NT), GFP- and GFP-ΔrNCL-transfected cells were analyzed by western blot using the specific anti-NCL antibody (Fig. 8B). The presence of the 110-kDa-protein band corresponding to the endogenous NCL was observed in all samples tested (Fig. 8B, lanes 1–6). However, the presence of a 70 kDa protein corresponding to the GFP-ΔrNCL fusion protein was only observed in the GFP-ΔrNCL transfected cells (Fig. 8B, lane 3 and 6), as expected.
Figure 8.
FCV protein synthesis and virus production in CrFK cells expressing ΔrNCL. A) CrFK cells were transfected with the empty vector (GFP) or pEGFP–C1-ΔNCL (GFP-ΔrNCL 302-707 aa) plasmids. GFP or GFP-ΔrNCL G418-resistant cells were selected using fluorescence microscopy. (B) Non transfected (NT), and GFP and GFP-ΔrNCL transfected CrFK cells were mock-infected (NI) (lanes 1, 2, and 3) or infected (INF) (lanes 4, 5, and 6) with FCV at MOI of 5. Protein lysates were obtained after 5 hpi and the presence of NCL and NS3 was determined by Western blot using specific anti-NCL and anti-NS3 antibodies. Actin was used as the loading control. C) NS3 band intensities were quantified by densitometry analysis using ImageJ software and are expressed as the relative value in relation to the actin expression level. Error bars represent the standard deviation from three independent experiments. Value of P < 0.01 (**) is indicated. D) The bar chart illustrates the virus yield in supernatants of CrFK cells transfected with GFP and GFP-ΔrNCL, quantified by plaque assay. Error bars represent the standard deviations from three independent experiments.
After having determined the stable expression of GFP-ΔrNCL by microscopy and western blot (Fig. 8A and B), non-transfected as well as transfected cell lines were infected with FCV at an MOI of 5, and the production of NS3 protein was analyzed at 5 hpi (Fig. 8B and 8C). The non-transfected as well as the GFP-transfected cells infected with FCV, showed equivalent amounts of NS3 expression (Fig. 8B, lanes 4 and 5 respectively); however, the cells expressing GFP-ΔrNCL showed a reduced amount of NS3 protein (Fig. 8B, lane 6, and 8C), suggesting that the amino-terminal end of the NCL was required for an efficient production of viral proteins in culture cells. It was also possible that GFP-ΔrNCL behaves as a dominant-negative mutant for NCL, and thus block or reduce the synthesis of NCL-dependent cellular proteins required for FCV RNA translation. Then, in vitro translation assays of CrFK cells transfected with GFP or GFP-ΔrNCL in the presence of 35S-Met were performed, and the protein synthesis profile was compared (Supplementary Fig. 1). The amount of the labeled bands was similar in both conditions, suggesting that GFP-ΔrNCL does not affect the synthesis of other cellular proteins.
Finally, the titers of virus produced from the cells transfected with GFP or GFP-ΔrNCL were determined (Fig 8D); Significantly lower (two-log reduction) virus titers than those obtained from cells transfected with GFP were observed for CrFK-GFP-ΔNCL cells (Fig. 8D), suggesting that the expression of the NCL deletion mutant was responsible for the reduction of viral production. The result could be the consequence of a direct interaction of the non-functional ΔNCL bound to the FCV RNA, acting as a negative dominant and thus, interfering with the interaction of the complete and functional NCL.
Discussion
As obligate intracellular parasites, viruses rely on cellular pathways and processes for almost all phases of their life cycle (Ahlquist et al., 2003; Lin et al., 2009). In our previous report (Cancio-Lonches et al., 2011), we identified NCL as a host cell factor present in RNP complexes formed with both Norwalk virus (NV) and FCV RNAs. Moreover, we showed that siRNA knockdown of NCL in CrFK cells, caused a reduction in cytopathic effect and in FCV production, confirming a requirement for NCL in some aspect(s) of the FCV life cycle (Cancio-Lonches et al., 2011). The purpose of our present study was to investigate a potential mechanism for the function of NCL in FCV replication.
We first examined a possible role for NCL as a receptor or attachment ligand during virus binding and entry. The JAM-1 protein (Makino et al., 2006) and sialic acid α2,6 (Stuart and Brown, 2007) have been implicated in early entry events for a number of FCV strains, but NCL had not been investigated. We demonstrated for the first time that NCL is expressed on permissive CrFK cells. However, pretreatment of CrFK cells with anti-NCL specific antibody, did not affect FCV binding and infection. Moreover, FL rNCL prevented the binding of the NCL ligand FITC-AGRO to CrFK cells, but not the binding of FCV. These findings suggest that NCL may not participate in early stages of the FCV life cycle, but rather is involved in post-entry stages of infection.
Our previous studies showed that NCL was part of an RNP complex formed with the FCV genomic RNA (Cancio-Lonches et al., 2011), however, a direct interaction with either the 5′ TR or the 3′ UTR had not been demonstrated. The binding of NCL to both terminal regions of the FCV genome, and the correlation of its knockdown by siRNA to a reduction in NS viral protein synthesis (Cancio-Lonches et al., 2011), suggest a role in viral RNA translation and/or replication. It is well known that positive-strand RNA virus genomes have processes inherently linked, both as a template for replication by the RNA polymerase and as a template for translation; thus, we studied the role of NCL in viral protein synthesis in the absence of viral RNA replication using RRL (Karakasiliotis et al., 2010). Calicivirus translation in RRL has been previously demonstrated (Goodfellow et al., 2005; Karakasiliotis et al., 2010), thus, the presence of NCL in these lysates, reported here and by others (Chen et al., 2012a), suggested that this protein might have a role in FCV translation. Three observations supported a role for NCL in translation. First, the NCL-specific ligand AGRO100 inhibited FCV RNA translation in a dose-dependent manner. Second, translation efficiency could be rescued by the addition of intact FL rNCL. Finally, FL rNCL could enhance translation when added to RRL in a dose dependent manner. Taken together, intact FL rNCL was linked to efficient translation in this in vitro translation assay.
Our next series of experiments examined the location of the domain in the NCL protein responsible for the enhancement of translation. We engineered a truncated protein construct designated ΔrNCL that lacked the N-terminal domain involved in nuclear localization and binding to a number of ribosomal proteins, but that retained the central RNA binding domains and the C-terminal ribosome binding domain. This truncated protein was unable to restore translation efficiency in the presence of AGRO100. Furthermore, its presence inhibited the translation of viral RNA in RRLs. Cells stably expressing this truncated form of NCL showed reduced viral protein synthesis and decreased virus yields (2 log reduction in titer). Taken together, these results strongly suggest that the ΔrNCL acts as a negative regulator, possibly competing out binding of endogenous NCL to FCV RNA. In support of this hypothesis, the FL NCL has been reported to stimulate the PV IRES-mediated translation in vitro, while NCL mutants containing the C-terminal RNA binding domains but lacking the N-terminal domain were found to inhibit in vitro translation of the same RNA. The translation inhibitory activity of these mutants correlates with their ability to bind the 5′ UTR sequence (Izumi et al., 2001). On the other hand, the lack of the N-terminal domain of ΔrNCL may prevent protein interactions with NCL that might be required for its function (Bouvet et al., 1998; Erard et al., 1988; Ginisty et al., 1998; Sicard et al., 1998). Here, we demonstrate that NCL has a role in FCV VPg-mediated translation in vitro; however, due to the fact that ΔrNCL also inhibited cap-dependent synthesis of GFP and Hsp70 in RRL, and that the addition of FL rNCL enhances their translation in a dose dependent manner, it is quite possible that NCL could act as a general translation factor, by its ability to bind a variety of mRNAs, as previously suggested (Izumi et al., 2001).
NCL has the capacity to bind G-rich elements located in both coding and noncoding sequences of several cellular mRNAs that encode cell growth- and cancer related proteins, and enhances their translation (Abdelmohsen et al., 2011). In some cases, NCL promotes mRNA translation without influencing mRNA stability (Abdelmohsen et al., 2011), while in others, it can differentially regulate both RNA and protein levels (Woo et al., 2013). NCL has the ability to bind to the 5′ and 3′ UTRs of several cellular mRNAs; when binding to the 3′UTR of the cytokine CSF-1 mRNA, promotes deadenylation and decay of CSF-1. On the other hand, its interaction with the poly A binding protein (PABP) C, promotes the formation of the mRNP closed loop, and enhances CSF-1 protein levels (Woo et al., 2013). NCL is one of the protein factors which participates in the circularization of the α1 (I) collagen mRNA in a 5′ stem-loop dependent manner; it has been proposed that NCL stabilizes the mRNA and relaxes the secondary structure of the 5′ stem-loop, through its RNA helicase activity, thus, making the start codon accessible to ribosomes. The ribosomal protein L26 (RPL26) and NCL are involved in the translational control of p53. In unstressed cells, NCL repression of p53 translation occurs by its binding to an intrinsic double-stranded RNA formed by base pairing between the 5′-3′-UTR interaction region of human p53 mRNA. NCL RNA binding domains (RBD) are responsible for their own dimerization/oligomerization, and for the stabilization of the nucleic acid hybrid structure within the p53 mRNA. After DNA damage, the RBD of NCL recruits RPL26 to the same interaction regions of the human p53 mRNA, disrupting the NCL homodimer by forming a heterodimer that facilitates translation of p53 mRNA. Thus, disruption of NCL-NCL homodimer by RPL26 may be the switch between translational repression and activation after stress (Chen et al., 2012b).
Then, the regulation of translation by NCL in several cases is determined by the protein ability to interact with both ends of the mRNA, and is often involved in the circularization of its target RNA, alone, or as a part of a protein complex. Circularization of the cellular mRNAs has been involved as a strategy for a more efficient protein synthesis, however, for viral genomic RNAs, the 5′-3′ end interactions are also implicated in the regulation of RNA replication (Alvarez et al., 2005; Herold and Andino, 2001; Nicholson and White, 2014). Since our previously published data report that NCL forms a RNP complex with the 3′UTR and with NS6/7, and here, we demonstrated the direct interaction of NCL with both the 5′ and the 3′ UTR from the FCV genomic RNA, it is possible that its role as a positive regulator of FCV translation, may involve the circularization of the FCV genomic RNA. In this regard, evidence of the 5′-3′ end interactions in other caliciviruses such as NV and murine norovirus (MNV) (Lopez-Manriquez et al., 2013; Sandoval-Jaime and Gutierrez-Escolano, 2009), require an RNA-RNA interaction that is further stabilized by cellular proteins, and NCL could be one of the candidates for this function. On the other hand, we cannot rule out the possibility that NCL may also be involved in the regulation of other steps during the FCV replicative cycle, such as RNA replication.
Materials and methods
Cell culture and virus
CrFK cells obtained from the American Type Culture Collection (ATCC) (Rockville, MD) were grown in Eagle’s minimal essential medium with Earle’s balanced salt solution and 2 mM l-glutamine that was modified by the ATCC to contain 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, 1.5 g/liter sodium bicarbonate. The medium was supplemented with 10% bovine fetal serum, 5,000 U of penicillin and 5 μg/ml of streptomycin. Cells were grown in a 5% CO2 incubator at 37°C. In this study, CrFK cells were infected with the FCV F9 strain American Type Culture Collection (ATCC) (Rockville, MD). Virus titer was determined by plaque assay as reported (Escobar-Herrera et al., 2007).
Immunofluorescence assays
CrFK cells (1.5 × 105) were seeded in a 6-well plate containing glass coverslips pretreated with poly-L-lysine (0.1%) and grown overnight. The cells were washed once with cold phosphate buffer (PBS) and treated for 15 min with 2% paraformaldehyde in PBS at 4°C. Samples were permeabilized with acetone 100%, washed three times with cold PBS for 5 min and blocked with anti-goat sera at 5%, for 30 min, at 4°C. Then, cells were washed one time with cold PBS for 1 min and incubated with 8 μg/ml of an anti-NCL antibody (H-250, Santa Cruz Biotechnology) diluted in 5% goat sera at 4°C overnight. The samples were washed three times with cold PBS for 10 min and incubated with the corresponding secondary antibody (Invitrogen) diluted in 5% goat serum for 2 h at room temperature (RT). The samples were washed three times with PBS and incubated with 1 μg/ml of 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) for 10 min. Following the DAPI staining, the coverslips were washed two times with cold PBS, and mounted onto glass slides using Vecta-Shield liquid mounting media (Vector Laboratories A. C.). The images were captured using a TCS SP8 Leica confocal microscope.
Cell Surface Biotinylation
Biotinylation of the CrFK cells surface proteins was performed as described previously with some modifications (Hirano et al., 2005; Soundararajan et al., 2009). A monolayer of CrFK cells (1 × 107 cells) were washed with cold PBS, that contained 0.1 mM Ca2+ and 1 mM Mg2+(PBS+), and incubated with 1 mg/ml sulfosuccinimidyl-6-(biotin-amide) hexanoate in PBS (Pierce) at 4°C for 2 h with gentle agitation. Then, unreacted biotin was quenched by incubating the treated cells in PBS-100 mM glycine for 2 min at 4°C. Cells treated without the biotinylation reagent served as a negative control. After the cells were washed with PBS+, they were lysed with a buffer containing 1% Triton X-100, 0.05% sodium deoxycholate, 5 mm EDTA, 30 mm Tris-HCl (pH 7.4), 150 mm NaCl, protease inhibitor (complete mini Roche) and 10% glycerol. The lysates were centrifuged for 5 min at 15,000 x g. To isolate biotinylated proteins, the supernatants containing 3 mg of a protein were applied to 1 ml of streptavidin-agarose (Pierce) pre-equilibrated with lysis buffer and were incubated overnight at 4°C with gentle agitation. The matrices were thoroughly washed with lysis buffer, and bound proteins were solubilized with SDS-PAGE sample buffer (50 mM Tris-HCL (pH 6.8), 2% SDS, 6% glycerol, 0.1 M dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.015% bromophenol blue) at 95°C for 5 min and subjected to SDS-PAGE (10% gel). Immunoblotting for the detection of NCL was performed using anti-NCL, and anti-hnRNP A1 antibodies (sc-32301, Santa Cruz Biotechnology) were used as a control of non-membranous fraction. Proteins were visualized by chemiluminescence using SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Scientific™).
Detection of NCL on CrFK cells surface by flow cytometry
CrFK cells were dispersed with 0.02% EDTA in PBS. 1 × 106 cells were washed once in flow cytometry buffer (FCB) containing PBS-BSA 2% and 0.1% NaN3 and incubated with anti-NCL antibody for 1 h at 4°C. The cells were washed three times with FCB and incubated with the corresponding secondary antibody (Invitrogen) for 1 h at 4°C. The cells were washed three times with FCB and resuspended in 4% formaldehyde-FCB. The samples were analyzed with a FACSCalibur (Becton Dickinson, Franklin Lakes, MN).
Virus-binding assay by flow cytometry
Virus-binding assay was performed using a method previously described (Makino et al., 2006). CrFK cells (1 × 106) were washed once in FCB and incubated with 3 × 105 FCV particles, for 30 min at 4°C. For blocking experiments, cells were incubated with anti-JAM-1 pre- and post-immune sera (1:1), anti-NCL (40 μg/μl), or anti-rabbit IgG (40 μg/μl) antibodies for 1 h at 4°C, before the virus was added. After that, the samples were washed twice with FCB and incubated with anti-FCV antibody (FCV1-43, Santa Cruz Biotechnology), that recognizes an epitope on the capsid protein, for 1 h at 4°C. Then, the samples were washed three times with FCB and incubated with the corresponding secondary antibody for 1 h at 4°C. The cells were washed three times with FCB and analyzed with a FACSCalibur (Becton Dickinson, Franklin Lakes, MN). For binding assays, CrFK cells were grown in 175-cm2 flasks and infected with FCV virus; the supernatant was harvested at 24-h post infection (hpi), layered onto a 25% sucrose cushion, and centrifuged at 120,000 × g for 2 h at 4°C. The pellet was resuspended in PBS. The suspension was centrifuged at 650 × g to remove insoluble matter and used for binding assays.
Inhibition experiments
CrFK cells were plated in 96-well plates (1.5 × 104 cells per well) and allowed to grow for 24 h at 37°C before being washed once with cold PBS. Washed cells were incubated with anti-JAM-1 preimmune serum (2.4 μg/μl) (1:5), anti-JAM-1 (2.4 μg/μl) (1:5), anti-NCL (40 μg/μl) or anti-rabbit IgG (40 μg/μl) antibodies for 1 h at 37°C. After antibodies were removed, the samples were inoculated with FCV at a multiplicity of infection of 0.01 and incubated for 1 h at 37°C. The cytopathic effect (CPE) was monitored at 24 hpi by evaluating cell morphology using a phase-contrast microscope. For competition experiments, FL rNCL or BSA (120 nM each) were incubated with FCV (MOI= 0.01) for 3h, at RT, and protein pre-incubated viruses were added to ~ 80% confluent CrFK cells monolayers in 96-well plates and kept them at 37°C for 1 h. After removal of unbounded viruses, cells were kept for an additional 24 h at 37°C. Supernatants were collected and virus titration was performed by plaque assay. For competition experiments using the NCL ligand FITC-AGRO, 5 X105 cells were incubated 45 min on ice with a mix of 1μM FL rNCL or rGST proteins pre-incubated with 5μM FITC-AGRO for 2 h at 4°C. The samples were analyzed with a FACS Calibur (Becton Dickinson, Franklin Lakes, MN).
Cloning, expression, and purification of the recombinant ΔrNCL protein
DNA fragment corresponding to amino acids 301-710 from the human NCL (ΔrNCL) was amplified from HeLa cells (ATCC, Rockville, MD) cDNA, by using Pfu DNA polymerase (Thermo Fisher Scientific) and pair of primers: FW5′-ACCATGGAATTCACAGAACCGACTACGGCTTTC-3′, and RV5′-GCCTTTCTCGAGCTATTCAAACTTCGTCTTCTTTCC-3′. The sequences of primers included EcoRI and XhoI recognition sites (underlined). The amplicon was cloned into the cloning vector pJET1.2 (Thermo Scientific), and sub-cloned into the EcoRI and XhoI enzyme sites of the prokaryotic expression vector pGEX5X.1. The resulting plasmid was named pGEX5X.1-ΔrNCL and contained a truncated version of the NCL gene. To express ΔrNCL, E. coli BL21 cells were transformed with pGEX5X.1-ΔrNCL, transformed cells were grown to A600 and protein expression was induced by 0.1 mM IPTG for 4 h at 37 °C. Recombinant protein purification was performed as follows: the cells were pelleted by centrifugation (JA-10 Beckman Coulter 30 min at 11,435 x g) and resuspended in a lysis buffer that contained PBS-TritonX-100 1%, lysozyme (100 mg/ml) and protease inhibitor (Complete Mini, Roche). The cells were incubated 30 min at 4 °C and then sonicated. After the lysate was centrifuged (JA-20 Beckman Coulter 30 min at 17,250 x g), the supernatant was incubated with glutathione-agarose (G4510 Sigma-Aldrich) for 1 h, at 4°C, the beads were washed three times with PBS-Triton X100-1% and the bound protein was eluted with 500 μl of elution buffer (10 mM L-glutathione reduced G4251 (Sigma-Aldrich), 15 mM Tris-HCl pH 8.0). The eluted protein was dialyzed against buffer containing 20 mM MOPS pH 7.2, 10 mM NaCl, 50 mM KCl, 1.1 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol (DTT). The purified ΔrNCL was stored at −80 °C.
For the expression of GFP-ΔrNCL in CrFK cells, the amplicon corresponding to amino acids 301-710 from the human NCL (ΔrNCL), was amplified from HeLa cells cDNA by using Pfu DNA polymerase (Thermo Fisher Scientific) and a pair of primers with EcoRI and ApaI sites (underlined): FW5′-ACCATGGAATTCTACAGAACCGACTACGGCTTTC-3′, and RV5′-GCCTTTGGGCCCCTATTCAAACTTCGTCTTCTTTCC. The amplicon was cloned into the cloning vector pJET1.2, and subcloned into the EcoRI and ApaI enzyme sites of the eukaryotic expression vector pEGFP-C1. The resulting plasmid was named pEGFP-C1-ΔrNCL and contained a truncated version of the NCL gene under control of cytomegalovirus (CMV) promoter. All plasmids containing the NCL amplicons were sequenced to verify the inserts.
Stable transfection of GFP-ΔrNCL into CrFK cells
Transfections of CrFK cells with one μg of pEGFP-C1 (Clontech laboratories, Inc.) or pEGFP-C1-ΔrNCL plasmids were performed by electroporation using Nucleofector™ Technology (T-020). For stable transfection, the electroporated cells were selected using growth medium that contained 2.5 mg/ml G418 (Sigma-Aldrich). Stably transfected cells were maintained as populations in the presence of 0.5 mg/mL G418.
In vitro transcription assays
Two amplicons that corresponded to the 5′ terminal region (TR) (nts 1 to 182) and the complete 3′UTR (nts 7646 to 7690) from the FCV genomic RNA were obtained by PCR from FCV-infected CrFK cells cDNA (M-MLV Reverse Transcriptase, Invitrogene) using a sense primer that contained the bacteriophage T7 promoter sequence (5′RT-FW 5′-TAATACGACTCACTATAGGGGTAAAAGAAATTTGAGAC-3′, 5′RT-RV5′-CATTGTCGAAGACCCGTC-3′) and (3′UTR-FW-5′ TAATACGACTCACTATAGGGTCATATATCCCTTTGGG, 3′UTR-RV-5′ CCCTGGGGTTAGGCGCAGG-3′). The PCR reactions were performed with a Pfu DNA Polymerase (Thermo Fisher Scientific) at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C 30 s, using a Perkin-Elmer Cetus DNA thermocycler. RNA transcripts corresponding to the 5′TR and 3′UTR FCV regions were synthesized from the purified amplicons using In vitro MEGAscript® T7 Transcription Kit (Life Technologies) according to the manufacturer’s protocol. Following the synthesis, the reaction mixtures were treated with DNAse 1 at 37°C for 30 min to remove the template DNAs. Unincorporated nts were removed by phenol:chloroform extraction and isopropanol precipitation. For the synthesis of radiolabeled transcripts, [α32P]-UTP (Perkin Elmer) was included in the transcription reaction mixture.
Electrophoretic mobility shift assays (EMSA)
EMSA was performed using a method described previously (Cancio-Lonches et al., 2011). Briefly, increasing concentrations of ΔrNCL were pre-incubated for 15 min at 4°C with the same amount of tRNA in a buffer containing 10 mM HEPES (pH 7.4), 0.1 mM EDTA, 0.2 mM DTT, 8 mM MgCl2 and 10% (v/v) glycerol in a final volume of 15 μl. The reaction mixture was incubated for 30 min at 4°C. The [α32P]-UTP-labeled (4 × 105 cpm) RNA transcripts corresponding to the FCV 5′TR and 3′UTR were added to each reaction mixture and incubated for 15 min at 4°C. Then, the samples were incubated with 20 units of RNase A and 20 μg of RNase T1 for 15 min at RT before loading the gels. The RNA-protein complexes were analyzed in a 4% (5′ TR) and 8% (3′UTR) native gels. The gels were dried and autoradiographed. For EMSA performed with FITC-labeled-AGRO-100 (Sigma-Aldrich), the complexes were observed in a Chemidoc™ MP Imagine system with Image lab software (Bio-Rad).
In Vitro Translation assays
FCV VPg-linked RNA was purified from FCV-infected (MOI=40) CrFK cells at 8 hpi. In vitro translation assays were performed in Flexi rabbit reticulocyte lysates (RRLs, Promega). Each 12.5 μl translation reaction included 6.25 μl of RRL, 20 μM amino acid mixture minus methionine, 100 mM KCl, 2 mM DTT, 0.5 mM MgOAc, 5 μCi35S methionine (1000 Ci/mmol) (GE Healthcare) and 400 ng of FCV VPg-linked RNA. The reactions were incubated at 30°C for 1.5 h. The gels were dried and autoradiographed. Different amounts of ΔrNCL, FL rNCL (OriGene) dialyzed in (20 mM MOPS pH 7.2, 10 mM NaCl, 50 mM KCl, 1.1 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT) and AGRO100 (Sigma-Aldrich) were added into translation reactions as indicated. When FL rNCL was used, the translation reaction was performed in a final volume of 37 μl.
[35S]-methionine experiments
CrFK cells (1.25 × 105), expressing GFP or GFP-ΔrNCL were incubated with methionine-free MEM for 1 h, at 37 °C, and pulsed with 50 μCi/ml)of [35S]-methionine for 1h at 37 °C. Cells were solubilized with SDS-PAGE sample buffer, and subjected to SDS-PAGE and autoradiography.
Supplementary Material
Supplementary Figure 1. GFP-Δ-rNCL transfected in CrFK cells does not affect de novo protein synthesis. CrFK cells transfected with GFP (lane 1) or GFP-ΔrNCL (lane 2) were pulsed with [35S]-methionine (50 μCi/ml) for 1 h. Cells were solubilized with SDS-PAGE sample buffer, and subjected to SDS-PAGE and autoradiography.
Highlights.
NCL is present in CrFK cells surface but it may not be involved in FCV binding or entry
NCL directly binds to the 5′ and the 3′ ends of the FCV genomic RNA
AGRO100 binds and inactivates NCL causing a reduction in FCV protein synthesis
AGRO100 effect can be reversed by full length recombinant NCL
NCL N-terminal end is involved in an efficient FCV translation and virus production
Acknowledgments
We thank Rosa del Angel for helpful suggestions during the development of this research. We also thank Beatriz Gómez and Carlos A. Santiago for technical assistance. This work was supported by grants 154767 from CONACyT and ICYTDF/247/12 from ICyTDF and in part, from the Division of Intramural Research, NIAID, National Institutes of Health, U. S. Department of Health and Human Services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, Selimyan R, Martindale JL, Yang X, Carrier F, Zhan M, Becker KG, Gorospe M. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic acids research. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. Journal of virology. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Bose S, Basu M, Banerjee AK. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. Journal of virology. 2004;78:8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- Cancio-Lonches C, Yocupicio-Monroy M, Sandoval-Jaime C, Galvan-Mendoza I, Urena L, Vashist S, Goodfellow I, Salas-Benito J, Gutierrez-Escolano AL. Nucleolin interacts with the feline calicivirus 3′ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication. Journal of virology. 2011;85:8056–8068. doi: 10.1128/JVI.01878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eIF4F components. Journal of Biological Chemistry. 2006;281:25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- Chen CM, Chiang SY, Yeh NH. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J Biol Chem. 1991;266:7754–7758. [PubMed] [Google Scholar]
- Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem. 2012a;287:16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem. 2012b;287:16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong R, Das S, Ugrinova I, Kumar S, Mongelard F, Wong J, Bouvet P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic acids research. 2012;40:9441–9454. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl BH, Nielsen J, Dahl O. Mechanistic studies on the phosphoramidite coupling reaction in oligonucleotide synthesis. I. Evidence for nucleophilic catalysis by tetrazole and rate variations with the phosphorus substituents. Nucleic acids research. 1987;15:1729–1743. doi: 10.1093/nar/15.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. J Gen Virol. 2004;85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem. 1988;175:525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x. [DOI] [PubMed] [Google Scholar]
- Escobar-Herrera J, Medina-Ramirez FJ, Gutierrez-Escolano AL. A carboxymethyl-cellulose plaque assay for feline calicivirus. Journal of Virological Methods. 2007;146:393–396. doi: 10.1016/j.jviromet.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. Embo J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO reports. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Escolano AL, Brito ZU, del Angel RM, Jiang X. Interaction of cellular proteins with the 5′ end of Norwalk virus genomic RNA. Journal of Virology. 2000;74:8558–8562. doi: 10.1128/jvi.74.18.8558-8562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Escolano AL, Vazquez-Ochoa M, Escobar-Herrera J, Hernandez-Acosta J. La, PTB, and PAB proteins bind to the 3′ untranslated region of Norwalk virus genomic RNA. Biochemical and Biophysical Research Communications. 2003;311:759–766. doi: 10.1016/j.bbrc.2003.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Molecular Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Miki Y, Hirai Y, Sato R, Itoh T, Hayashi A, Yamanaka M, Eda S, Beppu M. A multifunctional shuttling protein nucleolin is a macrophage receptor for apoptotic cells. J Biol Chem. 2005;280:39284–39293. doi: 10.1074/jbc.M505275200. [DOI] [PubMed] [Google Scholar]
- Hirano M, Kaneko S, Yamashita T, Luo H, Qin W, Shirota Y, Nomura T, Kobayashi K, Murakami S. Direct interaction between nucleolin and hepatitis C virus NS5B. J Biol Chem. 2003;278:5109–5115. doi: 10.1074/jbc.M207629200. [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, Krust B. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261:312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus research. 2001;76:17–29. doi: 10.1016/s0168-1702(01)00240-4. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li Z, Nagy PD. Nucleolin/Nsr1p binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology. 2010;396:10–20. doi: 10.1016/j.virol.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xu XS, Russell JE. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Molecular and cellular biology. 2006;26:2419–2429. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasiliotis I, Vashist S, Bailey D, Abente EJ, Green KY, Roberts LO, Sosnovtsev SV, Goodfellow IG. Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PLoS One. 2010;5:e9562. doi: 10.1371/journal.pone.0009562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurts S, Masutomi K, Delgermaa L, Arai K, Oishi N, Mizuno H, Hayashi N, Hahn WC, Murakami S. Nucleolin interacts with telomerase. J Biol Chem. 2004;279:51508–51515. doi: 10.1074/jbc.M407643200. [DOI] [PubMed] [Google Scholar]
- Kreutz LC, Seal BS, Mengeling WL. Early interaction of feline calicivirus with cells in culture. Arch Virol. 1994;136:19–34. doi: 10.1007/BF01538814. [DOI] [PubMed] [Google Scholar]
- Kusakawa T, Shimakami T, Kaneko S, Yoshioka K, Murakami S. Functional interaction of hepatitis C Virus NS5B with Nucleolin GAR domain. J Biochem-Tokyo. 2007;141:917–927. doi: 10.1093/jb/mvm102. [DOI] [PubMed] [Google Scholar]
- Legrand D, Vigie K, Said EA, Elass E, Masson M, Slomianny MC, Carpentier M, Briand JP, Mazurier J, Hovanessian AG. Surface nucleolin participates in both the binding and endocytosis of lactoferrin in target cells. Eur J Biochem. 2004;271:303–317. doi: 10.1046/j.1432-1033.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Nagy PD. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8:305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Manriquez E, Vashist S, Urena L, Goodfellow I, Chavez P, Mora-Heredia JE, Cancio-Lonches C, Garrido E, Gutierrez-Escolano AL. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1. Journal of virology. 2013;87:11371–11387. doi: 10.1128/JVI.03433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losfeld ME, Leroy A, Coddeville B, Carpentier M, Mazurier J, Legrand D. N-Glycosylation influences the structure and self-association abilities of recombinant nucleolin. Febs J. 2011;278:2552–2564. doi: 10.1111/j.1742-4658.2011.08180.x. [DOI] [PubMed] [Google Scholar]
- Lu H, Li WQ, Noble WS, Payan D, Anderson DC. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J Proteome Res. 2004;3:949–957. doi: 10.1021/pr0499592. [DOI] [PubMed] [Google Scholar]
- Makino A, Shimojima M, Miyazawa T, Kato K, Tohya Y, Akashi H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J Virol. 2006;80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr Opin Mol Ther. 2010;12:107–114. [PubMed] [Google Scholar]
- Nicholson BL, White KA. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol. 2014;12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossiboff RJ, Parker JSL. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. Journal of Virology. 2007;81:13608–13621. doi: 10.1128/JVI.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papafragkou E, Hewitt J, Park GW, Greening G, Vinje J. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PloS one. 2013;8:e63485. doi: 10.1371/journal.pone.0063485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said EA, Courty J, Svab J, Delbe J, Krust B, Hovanessian AG. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. Febs J. 2005;272:4646–4659. doi: 10.1111/j.1742-4658.2005.04870.x. [DOI] [PubMed] [Google Scholar]
- Said EA, Krust B, Nisole S, Svab J, Briand JP, Hovanessian AG. The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J Biol Chem. 2002;277:37492–37502. doi: 10.1074/jbc.M201194200. [DOI] [PubMed] [Google Scholar]
- Sandoval-Jaime C, Gutierrez-Escolano AL. Cellular proteins mediate 5′-3′ end contacts of Norwalk virus genomic RNA. Virology. 2009;387:322–330. doi: 10.1016/j.virol.2009.02.041. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- Sicard H, Faubladier M, Noaillac-Depeyre J, Leger-Silvestre I, Gas N, Caizergues-Ferrer M. The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N terminus and its RNA-binding domains. Mol Biol Cell. 1998;9:2011–2023. doi: 10.1091/mbc.9.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan S, Wang L, Sridharan V, Chen W, Courtenay-Luck N, Jones D, Spicer EK, Fernandes DJ. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol Pharmacol. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Fleming PJ, Pollard HB, Burns AL. Cloning and Sequencing of the Human Nucleolin Cdna. Febs Letters. 1989;250:99–105. doi: 10.1016/0014-5793(89)80692-1. [DOI] [PubMed] [Google Scholar]
- Stuart AD, Brown TD. Alpha2,6-linked sialic acid acts as a receptor for Feline calicivirus. J Gen Virol. 2007;88:177–186. doi: 10.1099/vir.0.82158-0. [DOI] [PubMed] [Google Scholar]
- Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4:267–275. doi: 10.4161/cib.4.3.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Take M, Tsutsui J, Obama H, Ozawa M, Nakayama T, Maruyama I, Arima T, Muramatsu T. Identification of nucleolin as a binding protein for midkine (MK) and heparin-binding growth associated molecule (HB-GAM) J Biochem-Tokyo. 1994;116:1063–1068. doi: 10.1093/oxfordjournals.jbchem.a124628. [DOI] [PubMed] [Google Scholar]
- Taube S, Kolawole AO, Hohne M, Wilkinson JE, Handley SA, Perry JW, Thackray LB, Akkina R, Wobus CE. A Mouse Model for Human Norovirus. MBio. 2013;4 doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17:1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. Bmc Mol Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez BC, Henning D, Busch RK, Srivastava M, Busch H. Immunodominant RNA recognition motifs of human nucleolin/C23. Mol Immunol. 1995;32:1207–1213. doi: 10.1016/0161-5890(95)00093-3. [DOI] [PubMed] [Google Scholar]
- Vashist S, Bailey D, Putics A, Goodfellow I. Model systems for the study of human norovirus Biology. Future Virol. 2009;4:353–367. doi: 10.2217/fvl.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Urena L, Chaudhry Y, Goodfellow I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. Journal of Virology. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S, Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. Journal of virology. 1998;72:6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SA, Li HY, Hsu TI, Chen SH, Wu CJ, Chang WC, Hung JJ. Heat shock protein 90 stabilizes nucleolin to increase mRNA stability in mitosis. J Biol Chem. 2011;286:43816–43829. doi: 10.1074/jbc.M111.310979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdowson MA, Monroe SS, Glass RI. Are noroviruses emerging? Emerging infectious diseases. 2005;11:735–737. doi: 10.3201/eid1105.041090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HH, Baker T, Laszlo C, Chambers SK. Nucleolin mediates microRNA-directed CSF-1 mRNA deadenylation but increases translation of CSF-1 mRNA. Mol Cell Proteomics. 2013;12:1661–1677. doi: 10.1074/mcp.M112.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hamhouyia F, Thomas SD, Burke TJ, Girvan AC, McGregor WG, Trent JO, Miller DM, Bates PJ. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J Biol Chem. 2001;276:43221–43230. doi: 10.1074/jbc.M104446200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tsaprailis G, Bowden GT. Nucleolin stabilizes Bcl-X L messenger RNA in response to UVA irradiation. Cancer Res. 2008;68:1046–1054. doi: 10.1158/0008-5472.CAN-07-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. GFP-Δ-rNCL transfected in CrFK cells does not affect de novo protein synthesis. CrFK cells transfected with GFP (lane 1) or GFP-ΔrNCL (lane 2) were pulsed with [35S]-methionine (50 μCi/ml) for 1 h. Cells were solubilized with SDS-PAGE sample buffer, and subjected to SDS-PAGE and autoradiography.