Abstract
We conducted a genome-wide linkage scan and positional association study to identify genes and variants influencing blood lipid levels among participants of the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study. The GenSalt study was conducted among 1906 participants from 633 Han Chinese families. Lipids were measured from overnight fasting blood samples using standard methods. Multipoint quantitative trait genome-wide linkage scans were performed on the high-density lipoprotein, low-density lipoprotein, and log-transformed triglyceride phenotypes. Using dense panels of single nucleotide polymorphisms (SNPs), single-marker and gene-based association analyses were conducted to follow-up on promising linkage signals. Additive associations between each SNP and lipid phenotypes were tested using mixed linear regression models. Gene-based analyses were performed by combining P-values from single-marker analyses within each gene using the truncated product method (TPM). Significant associations were assessed for replication among 777 Asian participants of the Multi-ethnic Study of Atherosclerosis (MESA). Bonferroni correction was used to adjust for multiple testing. In the GenSalt study, suggestive linkage signals were identified at 2p11.2–2q12.1 [maximum multipoint LOD score (MML) = 2.18 at 2q11.2] and 11q24.3–11q25 (MML = 2.29 at 11q25) for the log-transformed triglyceride phenotype. Follow-up analyses of these two regions revealed gene-based associations of charged multivesicular body protein 3 (CHMP3), ring finger protein 103 (RNF103), AF4/FMR2 family, member 3 (AFF3), and neurotrimin (NTM ) with triglycerides (P = 4 × 10−4, 1.00 × 10−5, 2.00 × 10−5, and 1.00 × 10−7, respectively). Both the AFF3 and NTM triglyceride associations were replicated among MESA study participants (P = 1.00 × 10−7 and 8.00 × 10−5, respectively). Furthermore, NTM explained the linkage signal on chromosome 11. In conclusion, we identified novel genes associated with lipid phenotypes in linkage regions on chromosomes 2 and 11.
Keywords: Lipids, Linkage analysis, Positional association analysis, Gene-based analysis
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide (He et al., 2005; Lozano et al., 2013; Murray et al., 2013). Suboptimal lipid levels contribute to the atherosclerotic process, with clinical trials and observational studies demonstrating a strong relation between blood lipid concentrations and CVD (Hokanson and Austin, 1996; LaRosa et al., 1999; Di Angelantonio et al., 2009; Huxley et al., 2011). The heritabilities of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglyceride concentrations have long been established (Friedlander et al., 1997; Malhotra and Wolford, 2005; Luo et al., 2010; Zhang et al., 2010). More recently, genome-wide association studies (GWASs) have made important strides in identifying single nucleotide polymorphisms (SNPs) that contribute to the inter-individual variability in these complex phenotypes (Saxena et al., 2007; Kathiresan et al., 2008; Kooner et al., 2008; Wallace et al., 2008; Willer et al., 2008; Aulchenko et al., 2009; Kathiresan et al., 2009; Teslovich et al., 2010; Waterworth et al., 2010; Kim et al., 2011; Tan et al., 2012). Despite such progress, up to 75% of the variance in lipid levels due to genetic factors remains unexplained (Teslovich et al., 2010). Further research is needed to identify novel variants, genes, and biological pathways with important influences on lipid phenotypes.
We conducted genome-wide linkage analyses to identify chromosomal regions harboring quantitative trait loci (QTLs) for LDL-C, HDL-C, and triglyceride phenotypes among Han Chinese participants of the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study. This unique analysis takes advantage of the large, multi-generational family-based design of the GenSalt study, which included very few participants taking lipid-lowering medications. To localize promising linkage signals, genome-wide scans were followed-up with positional association analyses leveraging dense panels of SNP markers. The association study included not only single-marker testing but also gene-based analyses, which may increase power to detect the modest effects of common SNPs by examining their joint genic contributions (Ma et al., 2013).
RESULTS
Characteristics of participants
The baseline characteristics of the 1865 GenSalt participants, including 661 probands, 68 spouses, 936 siblings, and 200 offspring from 633 families, are presented in Table 1. The average LDL-C level ranged from 78.4 mg/dL among offspring to 101.6 mg/dL among spouses, HDL-C ranged from 48.0 mg/dL among offspring to 51.8 mg/dL among siblings, and triglycerides ranged from 108.6 mg/dL among offspring to 138.7 mg/dL among probands.
Table 1.
Characteristics of 1865 GenSalt participants from 633 Han Chinese families
| Proband (n = 661) | Spouse (n = 68) | Sibling (n = 936) | Offspring (n = 200) | |
|---|---|---|---|---|
| Male (%) | 60.5 | 32.4 | 50.9 | 43.5 |
| Age (y, mean ± SD) | 41.0 ± 8.3 | 49.0 ± 6.7 | 39.5 ± 7.7 | 23.5 ± 6.5 |
| BMI (kg/m2, mean ± SD) | 24.2 ± 3.2 | 23.4 ± 3.6 | 23.0 ± 2.8 | 21.6 ± 3.3 |
| Blood lipids (mg/dL, mean ± SD) | ||||
| LDL-C | 99.2 ± 28.0 | 101.6 ± 27.9 | 94.9 ± 26.2 | 78.4 ± 22.7 |
| HDL-C | 50.8 ± 12.2 | 50.3 ± 11.7 | 51.8 ± 10.8 | 48.1 ± 9.3 |
| Triglycerides* | 138.7 ± 91.8 | 124.7 ± 65.7 | 114.8 ± 69.0 | 108.6 ± 59.8 |
BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SD, standard deviation.
Geometric mean ± SD.
Linkage analysis
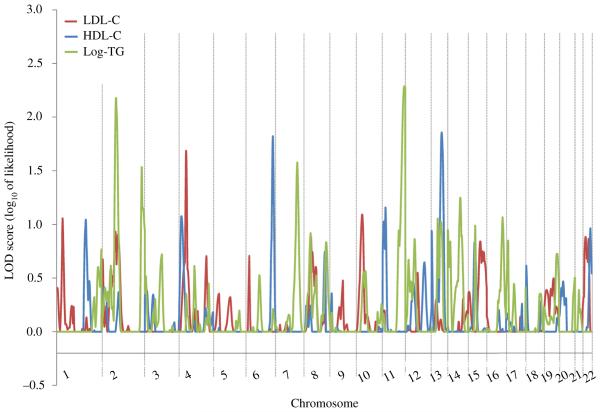
The heritabilities of LDL-C, HDL-C and triglycerides were 51.9%, 42.7% and 30.8%, respectively, in the GenSalt sample (all P < 0.001). Genome-wide linkage scan results for LDL-C, HDL-C, and triglycerides are shown in Fig. 1. We observed suggestive linkage (LOD > 2) of chromosomal regions 2p11.2–2q12.1 and 11q24.3–11q25 to the triglycerides phenotype. For triglycerides, maximum multipoint LOD scores of 2.177 and 2.288 were achieved at 2q11.2 and 11q25, respectively (Table 2).
Fig. 1. Genome-wide linkage scan results for lipid phenotypes.
Genome-wide linkage scan results for HDL-C (blue), LDL-C (red), and log-TG (green). HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; log-TG, log-transformed triglycerides.
Table 2.
Chromosome regions harboring LOD scores >2 for the blood lipid phenotypes
| Chromosomal location | Physical distance (kb) | Map distance (cM) | Maximum multipoint LOD score |
||
|---|---|---|---|---|---|
| HDL-C | LDL-C* | Log-TG | |||
| 2p11.2–2q12.1 | 84,800–103,700 | 106.00–115.00 | 0.132 | 0.927 | 2.177 |
| 11q24.3–11q25 | 128,826–134,170 | 141.00–158.00 | 0 | 0 | 2.288 |
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; Log-TG, log-transformed triglycerides.
Kurtosis adjusted LOD score.
Positional association analysis
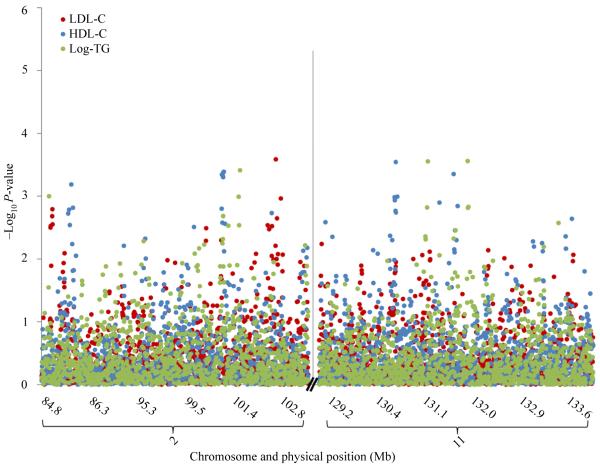
Fig. 2 shows the associations between 1929 tag-SNPs within the two suggestive linkage regions and LDL-C, HDL-C and triglyceride concentrations. No variants were significantly associated with the lipid phenotypes after adjustment for multiple testing.
Fig. 2. −Log10 P-values for the association between 1929 tag-SNPs in suggestive linkage regions and lipid phenotypes.
−Log10 P-values for the association between 1929 SNPs in suggestive linkage regions (LOD > 2) and HDL-C (blue), LDL-C (red), and log-TG (green). HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; log-TG, log-transformed triglycerides.
Gene-based analysis
A total of eight genes [charged multivesicular body protein 3 (CHMP3), ring finger protein 103 (RNF103), AF4/FMR2 family, member 3 (AFF3), mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), type II interleukin 1 receptor (IL1R2), type I interleukin 1 receptor (IL1R1), trans-membrane protein 182 (TMEM182), and neurotrimin (NTM )] from the identified linkage regions were significantly associated with lipid phenotypes in the GenSalt study (Table 3). Four of the identified genes were associated specifically with triglyceride levels, which corresponded to the identified linkage signals. These genes included CHMP3, RNF103 and AFF3 at 2p11.2–2q12.3 and NTM at 11q24.3–11q25 (P = 4.00 × 10−4, 1.00 × 10−5, 2.00 × 10−5, and 1.00 × 10−7, respectively). AFF3 and NTM were successfully replicated among the Asian participants of the Multi-ethnic Study of Atherosclerosis (MESA) (Table 3). P-values for all gene-based association tests are shown in the Table S1.
Table 3.
Significant gene-based findings (P-value) under linkage regions of log-transformed triglycerides
| Gene | Chr. | Start | End | GenSalt |
MESA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of SNPs |
LDL-C | HDL-C | Log-TG | Top SNP P* |
Number of SNPs |
LDL-C | HDL-C | Log-TG | Top SNP P* | ||||
| CHMP3 | 2 | 86730554 | 86948245 | 15 | 0.3851 | 0.5142 | 0.0004 | 0.0178 | 13 | 0.4394 | 0.4963 | 0.4778 | |
| RNF103 | 2 | 86830516 | 86850989 | 7 | 0.3597 | 0.5712 | 1.00E-5 | 0.0178 | 6 | 0.5538 | 0.5391 | 0.5649 | |
| AFF3 | 2 | 100162323 | 100759201 | 59 | 0.8594 | 0.2188 | 2.00E-5 | 0.0059 | 58 | <1.00E-7 | 0.1659 | <1.00E-7 | 0.0008†/0.0014€ |
| MAP4K4 | 2 | 102313312 | 102511149 | 9 | 0.3259 | 2.00E-7 | 0.5073 | 0.0023 | 9 | 0.4639 | 0.5065 | 3.00E-5 | 0.0040 |
| IL1R2 | 2 | 102608306 | 102645006 | 8 | 0.1400 | 1.00E-5 | 0.0996 | 0.0030 | 8 | 0.4639 | 0.3207 | 0.2333 | |
| IL1R1 | 2 | 102681004 | 102796334 | 34 | 0.7557 | 1.00E-7 | 0.7905 | 0.0003 | 34 | 0.1106 | 0.8421 | 0.4863 | |
| TMEM182 | 2 | 103353367 | 103460352 | 6 | 0.0002 | 0.4977 | 0.3901 | 0.0181 | 6 | 0.4670 | 0.2300 | 0.1708 | |
| NTM | 11 | 131240373 | 132206716 | 123 | 0.9623 | 0.6806 | 1.00E-7 | 0.0003 | 119 | 0.7680 | 0.2313 | 8.00E-5 | 0.0009 |
Chr., chromosome; SNP, single nucleotide polymorphism; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TG, triglyceride.
P-value of the most significant SNP in each significant gene-lipid association.
P-value of the most significant SNP for the AFF3 and LDL-C association.
P-value of the most significant SNP for the AFF3 and log-TG association. Bolded are significant findings after adjustment for multiple testing.
Sensitivity analysis
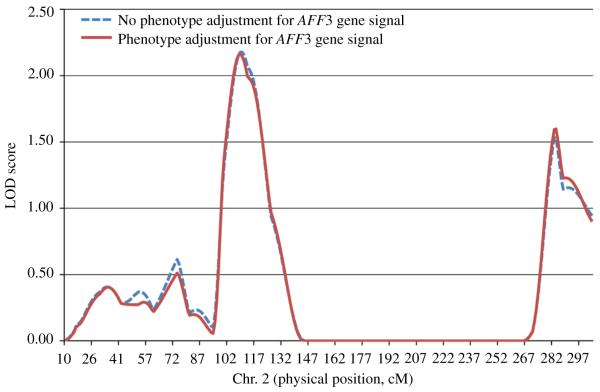
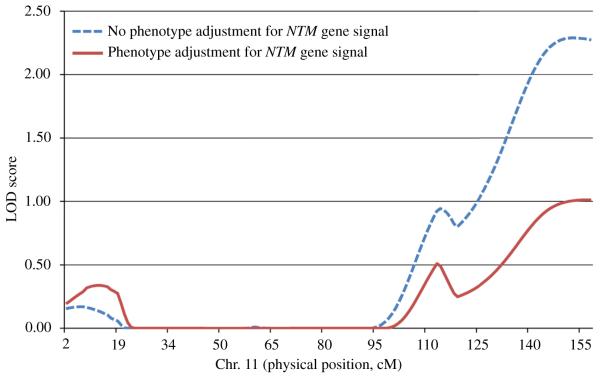
Linkage analyses before and after phenotype adjustment for the identified AFF3 and NTM genes are shown in Figs. 3 and 4, respectively. The NTM gene appeared to explain the linkage signal for log-transformed triglycerides on chromosome 11, with the maximum LOD score on chromosome 11 dropping from 2.29 to 1.01 after phenotype adjustment for the identified NTM gene signal (Fig. 4). The linkage signal on chromosome 2 only slightly changed after controlling for the AFF3 gene signal (Fig. 3).
Fig. 3. Sensitivity analysis for linkage signal on chromosome 2.
Linkage scan results on chromosome 2 for log-transformed triglyceride before and after controlling for the AFF3 gene signal among 1906 GenSalt participants.
Fig. 4. Sensitivity analysis for linkage signal on chromosome 11.
Linkage scan results on chromosome 11 for log-transformed triglyceride before and after controlling for the NTM gene signal among 1906 GenSalt participants.
DISCUSSION
The current analysis identified QTLs at 2p11.2–2q12.1 and 11q24.3–11q25 which may influence lipid phenotypes. We observed maximum multipoint LOD scores of 2.177 and 2.288 at 2q11.2 and 11q25, respectively, for triglycerides. Follow-up analyses of the 2p11.2–2q12.1 linkage signal revealed significant associations of seven protein-coding genes (RNF103, AFF3, MAP4K4, ILIR2, ILIR1, CHMP3, and TMEM182) with the lipid phenotypes. Under 11q24.3–11q25, follow-up gene-based analyses identified one protein-coding gene, NTM, significantly associated with triglycerides. The associations of both the AFF3 and NTM genes with triglycerides were successfully replicated among Asian MESA participants. Of particular interest, the NTM gene appeared to explain the observed linkage signal for triglycerides on chromosome 11. Given the lack of single-marker associations, these findings highlight the relevance of joint SNP analyses for detecting potentially important genes and biological pathways influencing serum lipid concentrations.
Chromosomal region 2p11.2–2q12.1 showed the first evidence of suggestive linkage to triglyceride levels in the current study. Follow-up gene-based analyses of protein-coding genes under this region revealed seven genes which were significantly associated with lipid phenotypes. Three of these genes, CHMP3, RNF103, and AFF3 were associated with triglycerides, corresponding to the observed linkage signal. Particularly noteworthy was the strong association of the AFF3 gene with triglycerides in GenSalt participants, which was robustly replicated among Asian MESA participants. AFF3 represents a biologically plausible candidate gene, having been associated previously with conditions which commonly include serum lipid alterations (Moschovi et al., 2004; Liu and Rosner, 2006; Guy et al., 2009; Steiner and Urowitz, 2009), such as acute lymphoblastic leukemia (von Bergh et al., 2002), end stage renal disease (Sandholm et al., 2012), type I diabetes (Barrett et al., 2009; Wallace et al., 2012), and rheumatoid arthritis (Barton et al., 2009; Plant et al., 2010). Since the AFF3 gene could not explain the observed linkage signal, it is likely that other genes and variants in this regions influence triglyceride levels. While CHMP3 and RNF103 showed an association that was consistent with the linkage signal among GenSalt participants, these findings could not be replicated in the smaller replication study. Future work in larger sample sizes will be needed to determine the relevance of these genes in serum lipid concentrations. Among the remaining four genes (MAP4K4, ILR2, ILIR1, and TMEM182), none were associated with triglycerides (the linked phenotype) nor were they replicated among MESA participants. While MAP4K4 significantly associated with HDL-C in the GenSalt study, it is interesting to note an association of this gene with triglycerides in MESA. Previous studies showed that MAP4K4 is involved in the suppression of lipid synthesis (Puri et al., 2008; Danai et al., 2013). Furthermore, Danai and colleagues (2013) showed that silencing the MAP4K4 gene in adipocytes enhanced the expression of lipogenic enzymes, and increased the level of triglycerides and fatty acids. Future studies focusing on this gene may be warranted.
Region 11q24.3–11q25 also showed suggestive evidence of linkage to triglyceride levels in the current study. This finding is similar to that of Bosse and colleagues who reported linkage of the 11q24 region to triglycerides among participants of the Quebec Family Study (Bosse et al., 2004). Under the 11q24.3–11q25 signal, the NTM gene was strongly associated with the triglyceride levels among GenSalt study participants. Interestingly, this finding appeared to explain the linkage signal for triglycerides in this region. Furthermore, NTM was successfully replicated among Asian MESA participants. Our findings are bolstered by previous functional and epidemiological studies demonstrating that the NTM gene may be involved in cholesterol homeostasis (Ramirez et al., 2011). Ramirez and colleagues (2011) reported that cellular cholesterol content in macrophages and livers of mice fed with high-fat diet can down-regulate miR-758, a microRNA involved in the down-regulation of NTM expression. In addition, Lukkonen and colleagues (2012) identified an association of NTM with intracranial and thoracic aortic aneurysm, an important dyslipidemia related disease. In aggregate, these findings provide strong evidence for a role of NTM in the regulation of serum lipid concentrations.
Our study has several important strengths. The large sample size and homogeneity of the GenSalt study population with respect to lifestyle and environmental factors should provide increased power to detect both linkage and association signals. In addition, study attributes, including the recruitment of only Han Chinese individuals, should make the association analysis robust to population stratification. Furthermore, stringent quality control procedures were employed during phenotype measurement, genotyping, and data cleaning. Certain limitations should also be addressed. The linkage signal on chromosome 2 could not be explained by the identified AFF3 gene, suggesting that other genomic factors in this region may influence triglycerides. While coverage of common genetic variation should be excellent in the current study (Nishida et al., 2008), further examination of any untagged common variants, structural variation, and low-frequency or rare variants in this region may be warranted to explain the linkage signal. Furthermore, due to the limited number of SNPs, gene-based analyses for 56 genes could not be conducted. Further research will be needed to explore any associations of these genes with lipid levels.
The current study described two chromosomal regions, including 2p11.2–2q12.1 and 11q24.3–11q25, which may harbor important susceptibility loci for blood lipid levels. Follow-up gene-based analysis of GenSalt participants identified eight protein-coding genes associated with lipid phenotypes in these regions. Two of these genes, AFF3 at 2p11.2 and NTM at 11q25, demonstrated robust replication of the observed triglyceride associations among Asian participants of the MESA study. These findings highlight the utility of gene-based analyses in helping to elucidate the biological pathways underlying serum lipid concentration. Furthermore, this research contributes additional information to our growing understanding of the genomic mechanisms underlying lipoprotein metabolism.
MATERIALS AND METHODS
Study population
The GenSalt study is a unique dietary feeding study designed to examine gene-dietary sodium and potassium interactions on blood pressure (BP). The GenSalt dietary intervention included 1906 Han Chinese participants from 633 families recruited from six field centers located in rural areas of northern China. A detailed description of the study design and participants has been presented elsewhere (Group, 2007). In brief, probands and their families were identified through a community-based BP screening conducted among persons aged 18–60 years in the study villages. Probands with a mean systolic BP of 130–160 mmHg and/or a mean diastolic BP between 85–100 mmHg and no use of antihypertensive medications were recruited for the study, along with their siblings, spouses, and offspring. Individuals who had stage-2 hypertension, secondary hypertension, clinical cardiovascular disease, chronic kidney disease, or diabetes, who used antihypertensive medications, or who were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study. After additional exclusion of 16 participants taking lipid-lowering medications and 25 participants missing genotype data, phenotype data or information on important covariables, a total of 1,865 GenSalt participants were included in the current analysis (97.8%).
Phenotype measurement
During the 3-day GenSalt baseline examination period, a standard questionnaire was administered by trained staff to collect information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors. Body weight and height were measured twice with participants in light indoor clothing and no shoes. Body mass index (BMI) was calculated as weight in kilograms divided by the square of height in meters. Overnight fasting blood samples were drawn by venipuncture on day one of the baseline examination. Blood specimens were processed at the field center and immediately frozen until they were shipped to the central clinical laboratory in Beijing. Total cholesterol, HDL-C and triglycerides were analyzed enzymatically using commercially available reagents (Allain et al., 1974). LDL-C was calculated using the Friedewald equation for participants with triglyceride levels <400 mg/dL: LDL-C = total cholesterol – HDL-C – triglyceride/5 (Friedewald et al., 1972).
Microsatellite marker and SNP genotyping
Lymphocytic DNA was extracted from whole blood samples and used for genotyping microsatellite markers spaced at approximately 9 cM intervals (407 markers, Marshfield Screening Set 12). Microsatellite genotyping used fluorescently labeled PCR primers for marker amplification followed by capillary electrophoresis on automated DNA sequencers (ABI 3730xl DNA Analyzer; Applied Biosystems, USA). Quality control samples included blind duplicates, no DNA controls, and Centre d’Etudes du Polymorphisme Humain (CEPH) DNA standards (mother, father, offspring with known genotypes). GeneMapper software (Applied Biosystems) was used to assign genotypes. ASPEX and GRR were used to check for potential misreported relationships in the GenSalt family pedigrees (Hinds and Risch, 1996; Abecasis et al., 2001). MapMaker/Sibs and PedCheck were used to check for Mendelian inconsistencies within families for each marker (O’Connell and Weeks, 1998; Pratt et al., 2000). A total of 359 microsatellite markers passed quality control and were included in the analysis.
SNPs located in promising linkage regions were genotyped using chip based hybridization assays (Affymetrix 6.0, Santa Clara, CA, USA). SNPs were excluded if they had a call rate less than 95%, were not in Hardy Weinberg Equilibrium, or had a minor allele frequency (MAF) less than 1%. Among 3999 SNPs in the linkage regions, 3284 SNPs met the quality control criteria. From these SNPs, 1929 were tagged (r2 < 0.9) using Haploview software, and were selected for inclusion in the single-marker analysis. Among the 116 protein-coding genes located in the promising linkage regions (Flicek et al., 2014), 60 genes with genotype data for at least three SNPs were included in the gene-based analysis (Table S1).
Statistical analysis
Triglyceride values were logarithmically transformed in order to normalize their distribution for all analyses. The means, geometric means, or percent of important covariables and lipid phenotypes were calculated for GenSalt probands, as well as their siblings, spouses and offspring.
Prior to linkage analysis, multipoint identity by descent estimates were calculated using Merlin software. In addition, HDL-C, LDL-C, and log-transformed triglyceride phenotypes were adjusted for the effects of age, BMI, gender and field center. In brief, each phenotype was regressed on the covariates in a stepwise manner, and only significant terms (P < 0.05) were retained. The residual variance was also examined (i.e., heteroscedasticity) by regressing the squared residual from the first regression on the same covariates (stepwise) and retaining significant terms. The final adjusted indicator was computed as the residual from the first regression, divided by the square root of the predicted score from the second regression. Heritability and multipoint genome-wide linkage scans of the adjusted blood lipid phenotypes were performed with SOLAR software (Almasy and Blangero, 1998). For linkage analyses, we used a multipoint linkage scan interval of 1 cM. Due to high residual kurtosis of the LDL-C phenotype (kurtosis = 2.60), we use a LOD score adjusted method implemented in SOLAR to ensure reliable results for this phenotype.
Additive single-marker associations between each SNP located in promising linkage regions (LOD > 2) and the lipid phenotypes (HDL-C, LDL-C, and log-transformed triglycerides) were examined using a mixed linear regression model to account for familial correlations. The same covariates used in the linkage analysis, including age, BMI, gender, and field center, were adjusted in multivariable analysis. We used a Bonferroni correction to adjust for the multiple testing (α-threshold = 0.05/1929 = 2.59 × 10−5). Significant SNPs were examined for replication among 777 Asian participants of MESA with GWAS and phenotype data available in the database of Genotypes and Phenotypes (dbGaP accession phs000209v11.p3.c2 and phs00420v4.p3.c2) (Mailman et al., 2007). The association analyses were conducted using SAS software (Version 9.3; SAS Institute, Inc., Cary, North Carolina, USA).
Gene-based associations with each lipid phenotype were tested by combining P-values from single SNP association analyses within each gene using TPM (Sheng and Yang, 2013; Yang et al., 2012). This method accommodates the correlations between SNPs through simulation, and has high power to detect gene-based associations compared to other meta-analysis techniques (Yang et al., 2012; Ma et al., 2013; Sheng and Yang, 2013). TPM has been evaluated extensively through simulation and applied to several gene-based studies of cardiometabolic phenotypes (Yang et al., 2012, 2013; Li et al., 2014; Zhu et al., 2014). For the current analysis, the truncation point was set as τ = 0.10, and the P-values for genes were estimated by 10,000 simulations. The simulations were increased up to 10,000,000 if 10,000 simulations failed to generate P-values. Genes significantly associated with lipid phenotypes in GenSalt were further tested for replication among the Asian participants of the MESA study. Bonferroni correction was applied to account for testing of 60 genes in the GenSalt study (α-threshold = 0.05/60 = 8.33 × 10−4) and eight genes in MESA replication study (α-threshold = 0.05/8 = 6.25 × 10−3). Gene-based analyses were performed using R software (Version 2.15.2, http://www.r-project.org).
Sensitivity analyses were conducted to determine whether those genes and variants identified in GenSalt and successfully replicated in MESA could explain the observed linkage signals. For this analysis, linkage scans were reconducted after phenotype adjustment for significant genes variants (along with adjustment for all previously described covariables) (Almasy and Blangero, 1998). Dampening of the LOD score after phenotype adjustment for identified association signals would provide evidence that such signals were responsible for the observed linkage result.
Ethnics statement
Institutional review boards at the Tulane University Health Sciences Center, Washington University School of Medicine, University of Texas School of Public Health, Fuwai Hospital and Chinese National Human Genome Center at Beijing, and Chinese Academy of Medical Sciences approved the GenSalt study. Written informed consents for the baseline observation and for the intervention program were obtained from each participant. The institutional review board at Tulane University approved of the use of publicly available genotype and phenotype data from MESA participants.
Supplementary Material
ACKNOWLEDGMENTS
The GenSalt is supported by a cooperative agreement project grant (Nos. U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health, Bethesda, MD. Dr. Bazzano was supported by a career development award (No. K08HL091108) from NHLBI. MESA and the MESA SHARe project were conducted and supported by NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and CTSA UL1-RR-024156. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetric Genome-Wide Human SNP Array 6.0.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgg.2015.02.003.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Allain CC, Poon LS, Chan CS, Richmond W, Fu PC. Enzymatic determination of total serum cholesterol. Clin. Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BWJH, Janssens ACJW, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin M-R, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga J-J, de Geus EJC, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJG, Uitterlinden AG, Witteman JCM, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Doring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L, Consortium, E Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS, Consortium, T.D.G Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A, Eyre S, Ke X, Hinks A, Bowes J, Flynn E, Martin P, Wilson AG, Morgan AW, Emery P, Steer S, Hocking LJ, Reid DM, Harrison P, Wordsworth P, Thomson W, Worthington J, Consortium, Y. Consortium, B Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum. Mol. Genet. 2009;18:2518–2522. doi: 10.1093/hmg/ddp177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J. Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- Danai LV, Guilherme A, Guntur KV, Straubhaar J, Nicoloro SM, Czech MP. Map4k4 suppresses Srebp-1 and adipocyte lipogenesis independent of JNK signaling. J. Lipid Res. 2013;54:2697–2707. doi: 10.1194/jlr.M038802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, Packard CJ, Collins R, Thompson SG, Danesh J, Collaboration ERF. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt S, Johnson N, Juettemann T, Kähäri AK, Keenan S, Kulesha E, Martin FJ, Maurel T, McLaren WM, Murphy DN, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ruffier M, Sheppard D, Taylor K, Thormann A, Trevanion SJ, Vullo A, Wilder SP, Wilson M, Zadissa A, Aken BL, Birney E, Cunningham F, Harrow J, Herrero J, Hubbard TJ, Kinsella R, Muffato M, Parker A, Spudich G, Yates A, Zerbino DR, Searle SM. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- Friedlander Y, Austin MA, Newman B, Edwards K, Mayer-Davis EI, King MC. Heritability of longitudinal changes in coronary-heart-disease risk factors in women twins. Am. J. Hum. Genet. 1997;60:1502–1512. doi: 10.1086/515462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group, G.C.R. GenSalt: rationale, design, methods and baseline characteristics of study participants. J. Hum. Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Ogden L, Wadwa RP, Hamman RF, Mayer-Davis EJ, Liese AD, D’Agostino R, Marcovina S, Dabelea D. Lipid and lipoprotein profiles in youth with and without type 1 diabetes: the SEARCH for Diabetes in Youth case-control study. Diabetes Care. 2009;32:416–420. doi: 10.2337/dc08-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Gu D, Wu X, Reynolds K, Duan X, Yao C, Wang J, Chen C-S, Chen J, Wildman RP, Klag MJ, Whelton PK. Major causes of death among men and women in China. N. Engl. J. Med. 2005;353:1124–1134. doi: 10.1056/NEJMsa050467. [DOI] [PubMed] [Google Scholar]
- Hinds D, Risch N. The ASPEX package: affected sib-pair exclusion mapping. 1996 http://aspex.sourceforge.net/
- Hokanson JE, Austin MA. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J. Cardiovasc. Risk. 1996;3:213–219. [PubMed] [Google Scholar]
- Huxley RR, Barzi F, Lam TH, Czernichow S, Fang X, Welborn T, Shaw J, Ueshima H, Zimmet P, Jee SH, Patel JV, Caterson I, Perkovic V, Woodward M, Asia Pacific Cohort Studies Collaboration and the Obesity in Asia Collaboration Isolated low levels of high-density lipoprotein cholesterol are associated with an increased risk of coronary heart disease: an individual participant data meta-analysis of 23 studies in the Asia-Pacific region. Circulation. 2011;124:2056–2064. doi: 10.1161/CIRCULATIONAHA.111.028373. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen M-R, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans [Erratum appears in Nat. Genet. 2008 Nov, 40, 1384] Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PIW, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang J-Y, Oh JH, Kim D-J, Kim NH, Kim S, Hong EJ, Kim J-H, Min H, Kim Y, Zhang R, Jia W, Okada Y, Takahashi A, Kubo M, Tanaka T, Kamatani N, Matsuda K, consortium, M. Park T, Oh B, Kimm K, Kang D, Shin C, Cho NH, Kim H-L, Han B-G, Lee J-Y, Cho YS. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- LaRosa JC, He J, Vupputuri S. Effect of statins on risk of coronary disease: a meta-analysis of randomized controlled trials [Summary for patients in J. Am. Geriatr. Soc. 2002 Feb, 50, 391–393] JAMA. 1999;282:2340–2346. doi: 10.1001/jama.282.24.2340. [DOI] [PubMed] [Google Scholar]
- Li C, Yang X, He J, Hixson JE, Gu D, Rao DC, Shimmin LC, Huang J, Gu CC, Chen J, Li J, Kelly TN. A gene-based analysis of variants in the serum/glucocorticoid regulated kinase (SGK ) genes with blood pressure responses to sodium intake: the GenSalt Study. PLoS One. 2014;9:e98432. doi: 10.1371/journal.pone.0098432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rosner MH. Lipid abnormalities associated with end-stage renal disease. Semin. Dial. 2006;19:32–40. doi: 10.1111/j.1525-139X.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FGR, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo J-P, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KMV, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh P-H, Yip P, Zabetian A, Zheng Z-J, Lopez AD, Murray CJL. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BF, Du L, Li JX, Pan BY, Xu JM, Chen J, Yin XY, Ren Y, Zhang F. Heritability of metabolic syndrome traits among healthy younger adults: a population based study in China. J. Med. Genet. 2010;47:415–420. doi: 10.1136/jmg.2009.068932. [DOI] [PubMed] [Google Scholar]
- Luukkonen TM, Pöyhönen M, Palotie A, Ellonen P, Lagström S, Lee JH, Terwilliger JD, Salonen R, Varilo T. A balanced translocation truncates Neurotrimin in a family with intracranial and thoracic aortic aneurysm. J. Med. Genet. 2012;49:621–629. doi: 10.1136/jmedgenet-2012-100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Clark AG, Keinan A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 2013;9:e1003321. doi: 10.1371/journal.pgen.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Wolford JK. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 2005;54:3007–3014. doi: 10.2337/diabetes.54.10.3007. [DOI] [PubMed] [Google Scholar]
- Moschovi M, Trimis G, Apostolakou F, Papassotiriou I, Tzortzatou-Stathopoulou F. Serum lipid alterations in acute lymphoblastic leukemia of childhood. J. Pediatr. Hematol. Oncol. 2004;26:289–293. doi: 10.1097/00043426-200405000-00006. [DOI] [PubMed] [Google Scholar]
- Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, Abraham J, Ackerman I, Aggarwal R, Ahn SY, Ali MK, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Bahalim AN, Barker-Collo S, Barrero LH, Bartels DH, Basanez M-G, Baxter A, Bell ML, Benjamin EJ, Bennett D, Bernabe E, Bhalla K, Bhandari B, Bikbov B, Bin Abdulhak A, Birbeck G, Black JA, Blencowe H, Blore JD, Blyth F, Bolliger I, Bonaventure A, Boufous S, Bourne R, Boussinesq M, Braithwaite T, Brayne C, Bridgett L, Brooker S, Brooks P, Brugha TS, Bryan-Hancock C, Bucello C, Buchbinder R, Buckle G, Budke CM, Burch M, Burney P, Burstein R, Calabria B, Campbell B, Canter CE, Carabin H, Carapetis J, Carmona L, Cella C, Charlson F, Chen H, Cheng AT-A, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahiya M, Dahodwala N, Damsere-Derry J, Danaei G, Davis A, De Leo D, Degenhardt L, Dellavalle R, Delossantos A, Denenberg J, Derrett S, Des Jarlais DC, Dharmaratne SD, Dherani M, Diaz-Torne C, Dolk H, Dorsey ER, Driscoll T, Duber H, Ebel B, Edmond K, Elbaz A, Ali SE, Erskine H, Erwin PJ, Espindola P, Ewoigbokhan SE, Farzadfar F, Feigin V, Felson DT, Ferrari A, Ferri CP, Fevre EM, Finucane MM, Flaxman S, Flood L, Foreman K, Forouzanfar MH, Fowkes FGR, Fransen M, Freeman MK, Gabbe BJ, Gabriel SE, Gakidou E, Ganatra HA, Garcia B, Gaspari F, Gillum RF, Gmel G, Gonzalez-Medina D, Gosselin R, Grainger R, Grant B, Groeger J, Guillemin F, Gunnell D, Gupta R, Haagsma J, Hagan H, Halasa YA, Hall W, Haring D, Haro JM, Harrison JE, Havmoeller R, Hay RJ, Higashi H, Hill C, Hoen B, Hoffman H, Hotez PJ, Hoy D, Huang JJ, Ibeanusi SE, Jacobsen KH, James SL, Jarvis D, Jasrasaria R, Jayaraman S, Johns N, Jonas JB, Karthikeyan G, Kassebaum N, Kawakami N, Keren A, Khoo J-P, King CH, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Laden F, Lalloo R, Laslett LL, Lathlean T, Leasher JL, Lee YY, Leigh J, Levinson D, Lim SS, Limb E, Lin JK, Lipnick M, Lipshultz SE, Liu W, Loane M, Ohno SL, Lyons R, Mabweijano J, MacIntyre MF, Malekzadeh R, Mallinger L, Manivannan S, Marcenes W, March L, Margolis DJ, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGill N, McGrath J, Medina-Mora ME, Meltzer M, Mensah GA, Merriman TR, Meyer A-C, Miglioli V, Miller M, Miller TR, Mitchell PB, Mock C, Mocumbi AO, Moffitt TE, Mokdad AA, Monasta L, Montico M, Moradi-Lakeh M, Moran A, Morawska L, Mori R, Murdoch ME, Mwaniki MK, Naidoo K, Nair MN, Naldi L, Narayan KMV, Nelson PK, Nelson RG, Nevitt MC, Newton CR, Nolte S, Norman P, Norman R, O’Donnell M, O’Hanlon S, Olives C, Omer SB, Ortblad K, Osborne R, Ozgediz D, Page A, Pahari B, Pandian JD, Rivero AP, Patten SB, Pearce N, Padilla RP, Perez-Ruiz F, Perico N, Pesudovs K, Phillips D, Phillips MR, Pierce K, Pion S, Polanczyk GV, Polinder S, Pope CA, 3rd, Popova S, Porrini E, Pourmalek F, Prince M, Pullan RL, Ramaiah KD, Ranganathan D, Razavi H, Regan M, Rehm JT, Rein DB, Remuzzi G, Richardson K, Rivara FP, Roberts T, Robinson C, De Leon FR, Ronfani L, Room R, Rosenfeld LC, Rushton L, Sacco RL, Saha S, Sampson U, Sanchez-Riera L, Sanman E, Schwebel DC, Scott JG, Segui-Gomez M, Shahraz S, Shepard DS, Shin H, Shivakoti R, Singh D, Singh GM, Singh JA, Singleton J, Sleet DA, Sliwa K, Smith E, Smith JL, Stapelberg NJC, Steer A, Steiner T, Stolk WA, Stovner LJ, Sudfeld C, Syed S, Tamburlini G, Tavakkoli M, Taylor HR, Taylor JA, Taylor WJ, Thomas B, Thomson WM, Thurston GD, Tleyjeh IM, Tonelli M, Towbin JA, Truelsen T, Tsilimbaris MK, Ubeda C, Undurraga EA, van der Werf MJ, van Os J, Vavilala MS, Venketasubramanian N, Wang M, Wang W, Watt K, Weatherall DJ, Weinstock MA, Weintraub R, Weisskopf MG, Weissman MM, White RA, Whiteford H, Wiebe N, Wiersma ST, Wilkinson JD, Williams HC, Williams SRM, Witt E, Wolfe F, Woolf AD, Wulf S, Yeh P-H, Zaidi AKM, Zheng Z-J, Zonies D, Lopez AD. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, Inoue I, Tokunaga K. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics. 2008;9:431. doi: 10.1186/1471-2164-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant D, Flynn E, Mbarek H, Dieudé P, Cornelis F, Arlestig L, Dahlqvist SR, Goulielmos G, Boumpas DT, Sidiropoulos P, Johansen JS, Ørnbjerg LM, Hetland ML, Klareskog L, Filer A, Buckley CD, Raza K, Witte T, Schmidt RE, Worthington J. Investigation of potential non-HLA rheumatoid arthritis susceptibility loci in a European cohort increases the evidence for nine markers. Ann. Rheum. Dis. 2010;69:1548–1553. doi: 10.1136/ard.2009.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L. Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am. J. Hum. Genet. 2000;66:1153–1157. doi: 10.1086/302830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V, Virbasius JV, Guilherme A, Czech MP. RNAi screens reveal novel metabolic regulators: RIP140, MAP4k4 and the lipid droplet associated fat specific protein (FSP) 27. Acta Physiol. (Oxf) 2008;192:103–115. doi: 10.1111/j.1748-1716.2007.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CM, Dávalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suárez Y, Fernández-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Mäkinen VP, Swan EJ, Palmer C, Boright AP, Ahlqvist E, Deshmukh HA, Keller BJ, Huang H, Ahola AJ, Fagerholm E, Gordin D, Harjutsalo V, He B, Heikkilä O, Hietala K, Kytö J, Lahermo P, Lehto M, Lithovius R, Osterholm AM, Parkkonen M, Pitkäniemi J, Rosengård-Bärlund M, Saraheimo M, Sarti C, Söderlund J, Soro-Paavonen A, Syreeni A, Thorn LM, Tikkanen H, Tolonen N, Tryggvason K, Tuomilehto J, Wadén J, Gill GV, Prior S, Guiducci C, Mirel DB, Taylor A, Hosseini SM, Parving HH, Rossing P, Tarnow L, Ladenvall C, Alhenc-Gelas F, Lefebvre P, Rigalleau V, Roussel R, Tregouet DA, Maestroni A, Maestroni S, Falhammar H, Gu T, Möllsten A, Cimponeriu D, Ioana M, Mota M, Mota E, Serafinceanu C, Stavarachi M, Hanson RL, Nelson RG, Kretzler M, Colhoun HM, Panduru NM, Gu HF, Brismar K, Zerbini G, Hadjadj S, Marre M, Groop L, Lajer M, Bull SB, Waggott D, Paterson AD, Savage DA, Bain SC, Martin F, Hirschhorn JN, Godson C, Florez JC, Groop PH, Maxwell AP, Group, D.E.R. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Sheng X, Yang J. Truncated product methods for panel unit root tests. Oxf. Bull. Econ. Stat. 2013;75:624–636. doi: 10.1111/j.1468-0084.2012.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner G, Urowitz MB. Lipid profiles in patients with rheumatoid arthritis: mechanisms and the impact of treatment. Semin. Arthritis Rheum. 2009;38:372–381. doi: 10.1016/j.semarthrit.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Tan A, Sun J, Xia N, Qin X, Hu Y, Zhang S, Tao S, Gao Y, Yang X, Zhang H, Kim S-T, Peng T, Lin X, Li L, Mo L, Liang Z, Shi D, Huang Z, Huang X, Liu M, Ding Q, Trent JM, Zheng SL, Mo Z, Xu J. A genome-wide association and gene-environment interaction study for serum triglycerides levels in a healthy Chinese male population. Hum. Mol. Genet. 2012;21:1658–1664. doi: 10.1093/hmg/ddr587. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee J-Y, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J.a., Lamina C, Ziegler A, Zhang W, Zee RYL, Wright AF, Witteman JCM, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJG, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BWJH, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PKE, Lucas G, Luben R, Loos RJF, Lokki M-L, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw K-T, Kaprio J, Kaplan LM, Johansson A, Jarvelin M-R, Janssens ACJW, Ingelsson E, Igl W, Kees Hovingh G, Hottenga J-J, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJC, de Faire U, Crawford G, Collins FS, Chen Y.-d.I., Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr., Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJP, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergh AR, Beverloo HB, Rombout P, van Wering ER, van Weel MH, Beverstock GC, Kluin PM, Slater RM, Schuuring E. LAF4, an AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2002;35:92–96. doi: 10.1002/gcc.10091. [DOI] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C, Rotival M, Cooper JD, Rice CM, Yang JH, McNeill M, Smyth DJ, Niblett D, Cambien F, Tiret L, Todd JA, Clayton DG, Blankenberg S, Consortium, C Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum. Mol. Genet. 2012;21:2815–2824. doi: 10.1093/hmg/dds098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, Luan JA, Ashford S, Wheeler E, Young EH, Hadley D, Thompson JR, Braund PS, Johnson T, Struchalin M, Surakka I, Luben R, Khaw K-T, Rodwell SA, Loos RJF, Boekholdt SM, Inouye M, Deloukas P, Elliott P, Schlessinger D, Sanna S, Scuteri A, Jackson A, Mohlke KL, Tuomilehto J, Roberts R, Stewart A, Kesaniemi YA, Mahley RW, Grundy SM, Wellcome Trust Case Control, C. McArdle W, Cardon L, Waeber G, Vollenweider P, Chambers JC, Boehnke M, Abecasis GR, Salomaa V, Jarvelin M-R, Ruokonen A, Barroso I, Epstein SE, Hakonarson HH, Rader DJ, Reilly MP, Witteman JCM, Hall AS, Samani NJ, Strachan DP, Barter P, van Duijn CM, Kooner JS, Peltonen L, Wareham NJ, McPherson R, Mooser V, Sandhu MS. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen W-M, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhu Y, Cole SA, Haack K, Zhang Y, Beebe LA, Howard BV, Best LG, Devereux RB, Henderson JA, Henderson P, Lee ET, Zhao J. A gene-family analysis of 61 genetic variants in the nicotinic acetylcholine receptor genes for insulin resistance and type 2 diabetes in American Indians. Diabetes. 2012;61:1888–1894. doi: 10.2337/db11-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhu Y, Lee ET, Zhang Y, Cole SA, Haack K, Best LG, Devereux RB, Roman MJ, Howard BV, Zhao J. Joint associations of 61 genetic variants in the nicotinic acetylcholine receptor genes with subclinical atherosclerosis in American Indians: a gene-family analysis. Circ. Cardiovasc. Genet. 2013;6:89–96. doi: 10.1161/CIRCGENETICS.112.963967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu X, Necheles J, Tsai HJ, Wang G, Wang B, Xing H, Li Z, Zang T, Xu X, Wang X. Genetic and environmental influences on serum lipid tracking: a population-based, longitudinal Chinese twin study. Pediatr. Res. 2010;68:316–322. doi: 10.1203/PDR.0b013e3181eeded6. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yang J, Yeh F, Cole SA, Haack K, Lee ET, Howard BV, Zhao J. Joint association of nicotinic acetylcholine receptor variants with abdominal obesity in American Indians: the Strong Heart Family Study. PLoS One. 2014;9:e102220. doi: 10.1371/journal.pone.0102220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.