Abstract
Human studies suggest that high-fat diets (HFD) increase the risk of breast cancer. The 7,12 dimethylbenz[a]anthracene (DMBA)-induced mammary carcinogenesis rat model is commonly used to evaluate the effects of lifestyle factors such as HFD on mammary-tumor risk. Past studies focused primarily on the effects of continuous maternal exposure on the risk of offspring at the end of puberty (PND50). We assessed the effects of prenatal HFD exposure on cancer susceptibility in prepubertal mammary glands and identified key gene networks associated with such disruption. During pregnancy, dams were fed AIN93G-based diets with isocaloric high olive oil, butterfat, or safflower oil. The control group received AIN-93G. Female offspring were treated with DMBA on PND21. However, a significant increase in tumor volume and a trend of shortened tumor latency were observed in rats with HFD exposure against the controls (p=0.048 and p=0.067 respectively). Large-volume tumors harbored carcinoma in situ. Transcriptome profiling identified 43 differentially expressed genes in the mammary glands of the HFBUTTER group as compared with control. Rapid hormone signaling was the most dysregulated pathway. The diet also induced aberrant expression of Dnmt3a, Mbd1, and Mbd3, consistent with potential epigenetic disruption. Collectively, these findings provide the first evidence supporting susceptibility of prepubertal mammary glands to DMBA-induced tumorigenesis that can be modulated by dietary fat that involves aberrant gene expression and likely epigenetic dysregulation.
Keywords: Breast cancer; epigenetics; developmental origin of health and disease; DNA methylation; epigenetics, DNA methyl transferase; methylated DNA binding domain
1. Introduction
Exposure to environmental factors, including lifestyle choices, is important in the etiology of breast cancer. The risk of breast cancer among Asian women born in their homeland is lower than that among those born in the United States (USA) [1]. Moreover, the risk of breast cancer is higher among Asian immigrants who have lived longer in the USA than among their more recently immigrated counterparts [2]. With every subsequent generation living in USA, the breast cancer risk increases progressively [1, 2]. These observations suggest strong environmental influences on breast cancer risk. One such influence is the Western diet, which is typically high in fat. Epidemiologic observations and intervention trials found positive associations between high-fat diets (HFD) (~40% total energy intake) and greater breast cancer risk in some [3-10] but not all studies [11-13]. Noteworthy is the focus of these human studies on the effects of high-fat consumption during adulthood; information on the impact of early-life (e.g., prenatal) exposure to HFD on human breast cancer risk is not yet available.
The most common model for studying the impact of lifestyle factors such as HFD on the risk of mammary carcinogenesis is the 7,12 dimethylbenz[a]anthracene (DMBA)-induced mammary tumor rat model. The polycyclic aromatic hydrocarbon, DMBA, which induces DNA damage via formation of epoxides, is routinely administered on postnatal day (PND) 50, a time close to the end of pubertal mammary-gland development [14]. The timing of HFD exposure was found to be an important determinant of mammary tumor risk in this model [15-17]. In contrast to HFD exposure during adulthood, exposure during the intrauterine period had the greatest impact on increasing the risk of mammary tumorigenesis later in life [15, 18-20], possibly because of the high degree of plasticity of the fetal mammary glands, rendering them more susceptible to reprogramming [21, 22]. Furthermore, a recent study reported that the effects could be observed in multiple generations (F1 and F2) [23], hence raising the question of whether such changes are “heritable.”
In addition to the timing of exposure, significant studies have been devoted to the differential effects of specific fatty acids. Mammary-tumor risk was higher among rats born to mothers exposed to a diet high in n-6 polyunsaturated fatty acids (PUFA) than in the offspring of mothers fed a diet low in n-6 PUFA [18]. Another study by the same group revealed that exposure to a diet high in n-3 PUFA in utero significantly reduced the risk of mammary cancer in offspring as compared with prenatal exposure to a diet high in n-6 PUFA [20]. We previously studied the effects of lifelong exposure to three different types of HFD on mammary cancer risk in the PND50 DMBA-induced mammary tumor model. We compared three HFD, 39% Kcal of olive oil (n-9 monounsaturated fatty acids), safflower oil (n-6 polyunsaturated fatty acids), and butterfat (saturated acids), with the reference AIN-93G diet containing 10% Kcal soy oil (n-6 mixed mono-, poly- and saturated fatty acids). HFD exposure induced marked increases in epithelial cell proliferation and a unique proliferation gene signature in PND21 and PND50 mammary glands, with no differences among the different types of fats [24]. These findings led us to re-examine whether prenatal exposure to these three HFDs has different effects and to ask the question of whether the susceptibility of the prepubertal mammary glands to DMBA can be modified by an HFD.
To our knowledge, only two studies have examined the prepubertal carcinogen window [25, 26]. Although, the prepubertal carcinogen window is a tentative model, understanding this window will generate a more precise understanding of the effects of diet in early life on breast cancer risk in later life, which has important ramifications on early prevention of breast cancer. To this end, this study provides the first evidence that the prepubertal mammary glands are susceptible to modulation of high-fat diets with regard to DMBA-induced tumorigenesis, possibly involving aberrant gene expression and epigenetic dysregulation.
2. Materials and Methods
2.1. Animals
Female, virgin Sprague-Dawley rats were obtained from Taconic Farms (Germantown, NY) at ~7 weeks of age. The rats were randomized into four groups (n=11) and placed on one of four diets: AIN-93G, AIN-based high-fat olive oil (HFOLIVE), high-fat butter (HFBUTTER) and high-fat safflower oil (HFSAFF) (Table 1) diets (Research Diets, New Brunswick, NJ), for 1 month. Animals were housed individually in a temperature- and humidity-controlled room in the AALAC-approved University of Cincinnati facility under a 12-h light-dark cycle. All rats were provided food and tap water ad libitum and were housed on Sani-chips bedding (P.J. Murphy Forest Products, Montville, NJ). All animal-care procedures were approved by the University of Cincinnati Institutional Animal Care and Use Committee, and the experiments were performed following the guidelines of the National Institutes of Health (NIH) for the proper and humane use of animals in biomedical research.
Table 1.
Modified AIN-93G rodent diets with 40% fat from olive oil, butter fat and safflower oil.
| Product | Modified AIN-93 G | Olive oil | Butter fat | Safflower oil | ||||
|---|---|---|---|---|---|---|---|---|
| g | kcal | g | kcal | g | kcal | g | kcal | |
| Protein | 19.7 | 20 | 23.4 | 20 | 23.4 | 20 | 23.4 | 20 |
| Carbohydrate | 67.6 | 70 | 46.4 | 40 | 46.4 | 40 | 46.4 | 40 |
| Fat | 4.2 | 10 | 20.1 | 39 | 20.1 | 39 | 20.1 | 39 |
| Total | 100 | 100 | 100 | 100 | ||||
| Kcal/g | 3.87 | 4.60 | 4.60 | 4.60 | ||||
| Ingredient | g | kcal | g | kcal | g | kcal | g | kcal |
| Casein,80 Mesh | 200 | 800 | 200 | 800 | 200 | 800 | 200 | 800 |
| L-Cystine | 3 | 12 | 3 | 12 | 3 | 12 | 3 | 12 |
| Corn starch | 463.5 | 1854 | 168 | 672 | 168 | 672 | 168 | 672 |
| Maltodextrin 10 | 125 | 500 | 125 | 500 | 125 | 500 | 125 | 500 |
| Sucrose | 100 | 400 | 100 | 400 | 100 | 400 | 100 | 400 |
| Cellulose, BW200 | 50 | 0 | 50 | 0 | 50 | 0 | 50 | 0 |
| Soybean oil | 43.8 | 394 | 15 | 135 | 15 | 135 | 15 | 135 |
| Olive oil | 0 | 0 | 160 | 1440 | 0 | 0 | 0 | 0 |
| Safflower oil | 0 | 0 | 0 | 0 | 0 | 0 | 160 | 1440 |
| Butter, anhydrous | 0 | 0 | 0 | 0 | 160 | 1440 | 0 | 0 |
| tBHQ | 0.014 | 0 | 0.0356 | 0 | 0.0356 | 0 | 0.0356 | 0 |
| S10012G, AIN-93G salts | 35 | 0 | 35 | 0 | 35 | 0 | 35 | 0 |
| Vitamin mix, V10037, AIN 93 vitamins | 10 | 40 | 10 | 40 | 10 | 40 | 10 | 40 |
| Choline bitartate | 2.5 | 0 | 2.5 | 0 | 2.5 | 0 | 2.5 | 0 |
| Total | 1032.814 | 4000 | 868.5856 | 3999 | 868.5856 | 3999 | 868.5856 | 3999 |
2.2. Diets
Diet formulations (Research Diets) are described in Table 1. Diets reflect a mixture of fatty acids as found in the typical diet. AIN-93G with a soy oil base, a widely used diet for mammary studies, was used here as the reference diet. The diets were controlled for caloric content, vitamins, salts, and protein but varied in fat and carbohydrate content and therefore in density [24].
2.3. Experimental design
Female SD rats were randomized into four groups. After 1 month of exposure to the experimental diet, female rats were bred with male Sprague-Dawley rats ~3 months old in stainless steel hanging cages. Mating plugs were noted and recorded as gestational day 1, and females were returned to single housing during the remainder of the gestational period. During mating and gestation, the rats were fed with respective experimental diets. After the pups were born, dams and offspring were switched to the control AIN93G diet until sacrifice. Litters were culled to eight pups on the second day after birth (PND1), keeping all female pups and sufficient males to balance the number. Litters were weighed at PND21 and PND140 before sacrifice. Pups had access to dam milk, as well as tap water and food during lactation. Pups were weaned at PND21, housed on Sani-chips bedding with tap water and the reference diet (AIN-93G) ad libitum. One female pup from each litter group was sacrificed on PND21. One female pup from each litter group was selected and given a single gavage of DMBA (Fisher Acros, Fisher Scientific) (20 mg/kg/body weight) at PND21. Animals were palpated weekly for tumor formation. Tumor growth was measured by recording tumor diameter with a caliper and determining the length of the longest axis and the width perpendicular to the longest axis. The end points for data analysis were (a) latency to tumor appearance, (b) number of animals with tumors (tumor incidence), (c) number of tumors/rat (tumor multiplicity), and (c) the tumor volume. Three months (PND140) after DMBA exposure, all animals were sacrificed. At this time, all tumor tissues were fixed in formalin and imbedded in paraffin for future histologic characterization.
2.4. Histopathology
Formalin-fixed samples were processed for hematoxylin/eosin staining and scored for hyperplasia (intraductal/lobular), carcinoma in situ (CIS, ductal/lobular), microinvasion, and invasion as previously described [27, 28], and according to the Russos’ criteria [29]. Invasive breast cancers are believed to originate from abnormal growth of epithelial cells (i.e., hyperplasia). The expanded epithelial cells occupy the duct or lobule and become CIS. Over time, some malignant cells may invade beyond the basement membrane of the duct/lobule to form microinvasions and ultimately to progress to massive stromal invasion. All samples were graded by a certified pathologist (Dr. Yan Mei) on the basis of representative hematoxylin and eosin (H&E)-stained sections. The incidence and mean score per treatment group were determined and analyzed by Fisher's exact test.
2.5. RNA-Seq
Mammary glands from rats prenatally exposed with AIN-93G control (CTL) or high fat butter (HFBUTTER) were collected at PND21 for RNA-Seq analyses. One snap frozen mammary gland from each litter and six samples from each group (CTL vs HFBUTTER) were selected for the experiment. Total RNA was extracted using the Qiagen RNeasy Lipid kit (Qiagen, Sample & Assay Technologies). The RNA quality was checked using an Agilent Bioanalyzer, and the quantity determined with Nanodrop (Thermo Scientific). RNA libraries were prepared according to manufacturer's protocol (Illumina, San Diego, CA; TruSeq RNA sample preparation kit). RNA sequencing was performed using the Genome Analyzer II sequencing system in the Genomics, Epigenomics and Sequencing Core at University of Cincinnati.
Bioinformatics RNA-Seq data analysis
Sequence reads were aligned to the reference genome using the TopHat aligner [30], and reads aligning to each known transcript were counted using Bioconductor packages for next-generation sequencing data analysis [31]. The differential expression analysis between HFBUTTER and Control samples was performed separately based on the negative-binomial statistical model of read counts as implemented in the DESeq Bioconductor package [32]. We used differential expression p-values in LRpath (http://lrpath.ncibi.org/) gene set enrichment analyses [33] to identify the top 100 gene ontology (GO)–affected categories in each group. These gene ontologies (Gene Ontology; http://www.geneontology.org/) were hierarchically clustered on the basis of the LRpath enrichment z-score, with positive values denoting upregulation and negative values denoting downregulation. Clustering was performed with the GENE-E algorithm (http://www.broadinstitute.org/cancer/software/GENE-E/). The gene expression data and results [34] have been deposited in the Gene Expression Omnibus (GEO, GSE73604). The expression level of each gene was determined, and the differences of gene expression between two groups were determined by t-test, and the false discovery rate was based on previously published protocols [34]. Genes differentially expressed in HFBUTTER in mammary transcriptomes were analyzed further by knowledge-based Ingenuity Pathways (IPA) network online software (www.ingenuity.com).
2.6. Real-time PCR
Total RNAs were converted into cDNA by Superscript III (Life Technologies, Carlsbad, CA). Real-time PCR was performed with the ABI7900 system (Life Technologies) with SYBR Greener (Life Technologies) and gene-specific primers (Table 2). Previously published primers [35] for methylation-related genes, including DNA methyltransferases (DNA (cytosine-5-)-methyltransferase 1 (Dnmt1), DNA (cytosine-5-)-methyltransferase 3 alpha (Dnmt3a), and DNA (cytosine-5-)-methyltransferase 3 beta (Dnmt3b) and methyl-CpG binding domain proteins (methyl CpG binding protein 2 (Mecp2), methyl-CpG binding domain protein 1 (Mbd1), methyl-CpG binding domain protein 2 (Mbd2), methyl-CpG binding domain protein 3 (Mbd3), and methyl-CpG binding domain protein 4 (Mbd4), were applied to this study.
Table 2.
Primers sequence for this study.
| rBtn1a1 | ACGTTGGATTCAGCAGCTCCCT |
| AGTACCGCTGGCGACCTCGT | |
| rCadm4 | GCCAGGGACGGCACAGGAAG |
| TCGTCCTTCAGGGCTCGGGTG | |
| rCsn1s1 | AGCAACGGCAAGTGCTCAGGA |
| GCTGTTCCAGGGTGCATCGGT | |
| rDbf4 | CCCTGAAACTCGGCCAGCGG |
| ATGCCACCAGGGAGAGGTGCT | |
| rLrrn1 | AGTGGACCGCTATGCCCTGGA |
| GCCGGGACACTCCGGAAAGC | |
| rNf1 | CCTGACACCCACATCTCCTT |
| GGGGGAGAGTTCAACGTTCT | |
| rPla2g2a | GCCACAGATTGGTGCTGTGTGA |
| TGGCCCCCTCGGTAGGAGAAC | |
| rSlc6a14 | GTCGCTCTCGGATGGTGTAT |
| GTCTCCTTTGCAGGTTCAGC | |
| rTmem45b | CCACTCACTCCTGCTGTTCA |
| CATGTCCTTCTGGTCCCACT |
2.7. Statistical Analysis
Binary variables such as tumor incidence were compared between groups using Chi-square tests. Numerical variables were compared of means using t tests. They were inspected of empirical distributions to determine if the variables need to be transformed before performing the t tests. For counting data, a Poisson model was used in analysis to compare mean between groups. All statistical tests were repeated in HFBUTTER, HFOLIVE, HFSAFF subgroup analyses too. All statistical analyses were performed using SAS 9.4 software (SAS, Cary, NC). P-values<0.05 were considered statistically significant.
3. Results
3.1. High fat diet significantly increases tumor volume with no changes in tumor latency, incidence and multiplicity at 21 day DMBA-induced mammary carcinogenesis
A total of 122 rats were exposed to HFD, with the numbers of animals being 43, 36 and 43 in HFOLIVE, HFBUTTER and HFSAFF subgroups respectively. There were 44 rats in the control group. No significant difference was observed in terms of body weight at PND21 and PND140 in any of the high fat groups when compared with the control group (data not shown). Tumor incidence was 34 out of 44 or 77.3% in the control group, lower than that of 104 out 122 or 85.2% in the HFD group (Table 3), yet the difference was not statistically significant (p=0.226). Likewise, survival analysis suggested that tumor incidence for individual high fat diet was not significantly different from the control (data not shown). When we combined HFBUTTER and HFSAFF groups, the tumor incidence was 73 out of 86 or 84.88%, yet the difference was not statistically significant when compared with the control. The mean ± standard error (SE) of tumor latency (i.e. days to develop tumor) was 58.1 ± 1.7 days in the HFD group, about 6 days shorter than the control group (64.1 ± 2.8 days, p=0.067). Combined HFBUTTER and HFSAFF group showed the shorter latency (57.4 ± 2.1, p=0.06) compared with the control group. The HFSAFF subgroup by itself showed the shortest latency (56.2 ± 2.9 days) as compared with the control group (p=0.05).
Table 3.
Summary of Tumor latency, incidence and multiplicity
| Groups/Sub Groups | Number of animals | Animals with tumors | % of tumors incidence | Mean ± SE of Latency (in Days) | Mean (95% CI) of multiplicity of tumors |
|---|---|---|---|---|---|
| AIN | 44 | 34 | 77.30% | 64.1 ± 2.8 | 2.7 (2.2, 3.4) |
| All HFD | 122 | 104 | 85.20% | 58.1± 1.7* | 3.1 (2.7, 3.5) |
| HFBUTTER/HFSAFF | 86 | 73 | 84.88% | 57.4 ± 2.1Ψ | 3.0 (2.6, 3.4) |
| HFBUTTER | 43 | 37 | 86.10% | 58.5 ± 2.8 | 2.9 (2.3, 3.5) |
| HFOLIVE | 36 | 31 | 86.10% | 59.8 ± 3.1 | 3.3 (2.6, 4.1) |
| HFSAFF | 43 | 36 | 83.70% | 56.2 ± 2.9† | 3.1 (2.5, 3.8) |
p=0.067 as compared to the AIN group.
p=0.06 as compared to the AIN Group.
p=0.050 as compared to the AIN group.
Tumor volume was 8.5 (6.2, 11.6) cm3 for the HFD group, almost double the size of the control group (4.5 (2.6, 7.8) cm3, p=0.048) (Table 4). In combined HFBUTTER and HFSAFF group, the tumor volume was (9.6 (6.7, 13.8) cm3, p=0.02) compared with the control. The HFSAFF subgroup showed the largest tumor size on average (9.7 (5.8, 16.5) cm3) and significant as against the control group (p=0.048). However, in combined HFBUTTER and HFSAFF, the occurrence of CIS (90%) was significantly higher than in the control (79%).
Table 4.
Tumor volume of individual HFDs and combined analysis of all HFD groups
| Variable | Groups | Occurrence of tumors | Mean (95% CI) | p |
|---|---|---|---|---|
| Volume | AIN | 34 | 4.5 (2.6, 7.8) | |
| All HFD | 104 | 8.5 (6.2, 11.6) | 0.048* | |
| HFBUTTER/HFSAFF | 73 | 9.6 (6.7,13.8) | 0.025† | |
| HFBUTTER | 37 | 9.4 (5.6, 15.8) | 0.056 | |
| HFOLIVE | 31 | 6.4 (3.6, 11.3) | 0.384 | |
| HFSAFF | 36 | 9.7 (5.8, 16.5) | 0.048* |
p<0.05, when compared with control AIN diet.
p<0.02, when compared with control AIN diet.
3.2. Significant increase in number of tumors with greater tumor volume with CIS and microinvasion
We further evaluated the pathohistology of tumors collected from HFD-exposed vs. control groups. Histology data showed no statistical differences among the three HFD groups in CIS (ductal/lobular) and microinvasion when individually compared with the control group (Table 5). However, when all HFD groups were pooled, the occurrence of CIS (90%) was higher than in the control group (79%) (Table 5). We also compared the multiplicity of tumors with CIS in the pooled HFD group (2.5) with the control group (1.8) and found a significant difference (p<0.01). We next calculated the tumor volume of CIS-containing tumors in the HFD group (4.0 cm3) and compared it with the control group (2.2 cm3) and observed a trend of increase (p = 0.08). Interestingly, the multiplicity of tumors with CIS in combined HFBUTTER and HFSAFF group was significantly higher than the one in control group (p<0.034). With regards to the tumor volume, the CIS-containing tumors in this combined group seems to be larger but did not reach statistical significant when compared with the control group (p<0.078).
Table 5.
Histological types showing percentage of occurrence and volume in combined analysis of all HFD groups
| Tumor type | Groups | Total number of tumors | Occurrence of CIS and Microivasion | Occurrence rate (%) | Number of CIS/microinvasive tumors /animal (95% CI) | Tumor volume (cm3) mean (95% CI) |
|---|---|---|---|---|---|---|
| CIS | AIN | 34 | 27 | 79 | 1.8 (1.4, 2.2) | 2.2 (1.17, 3.99) |
| All HFD | 104 | 94 | 90 | 2.4 (2.1, 2.7)* | 4.0 (2.88, 5.47)## | |
| HFBUTTER/HFSAFF | 73 | 66 | 90 | 2.3 (2.0, 2.7)& | 4.2 (2.85,6.11)$ | |
| HFBUTTER | 37 | 33 | 89 | 2.2 (1.8, 2.7) | 3.63 (2.10, 6.26) | |
| HFOLIVE | 31 | 28 | 90 | 2.5 (2.0, 3.1) | 3.53 (1.95, 6.38) | |
| HFSAFF | 36 | 33 | 91 | 2.4 (2.0, 3.0) | 4.78 (2.79, 8.18) | |
| Microinvasive | AIN | 34 | 20 | 59 | 1.6 (1.3, 2.0) | 6.3 (3.90, 10.22) |
| All HFD | 104 | 51 | 49 | 1.3 (1.2, 1.5)# | 12.8 (9.45, 17.26)++ | |
| HFBUTTER/HFSAFF | 73 | 37 | 51 | 1.3 (1.1, 1.5)&& | 13.8 (10.0, 19.9)$$ | |
| HFBUTTER | 37 | 22 | 59 | 1.3 (1.1, 1.5) | 12.64 (7.88, 20.29) | |
| HFOLIVE | 31 | 14 | 45 | 1.4 (1.1, 1.8) | 10.34 (5.79, 18.46) | |
| HFSAFF | 36 | 15 | 41 | 1.3 (1.0, 1.6) | 15.57 (9.05, 26.77) |
p=0.01, when all HFD compared with control AIN diet.
p=0.087, when all HFD compared with control AIN diet.
p=0.034, when HFBUTTER /HBSAFF compared with control AIN diet.
p=0.078, when HFBUTTER /HBSAFF compared with control AIN diet.
p=0.074, when all HFD compared with control AIN diet.
p=0.018, when all HFD compared with control AIN diet.
p=0.068, when HFBUTTER /HBSAFF compared with control AIN diet.
p=0.016, when HFBUTTER /HBSAFF compared with control AIN diet.
On the other hand, no statistically significant difference in the occurrence and frequency of tumor with microinvasion was noted between the HFD pooled groups and in the combined HFBUTTER and HFSAFF groups when compared with the control group (Table 5). However, when we calculated the volume of tumors with microinvasion, we noted significantly larger tumors in the HFD group (12.8 cm3) and in the combined HFBUTTER and HFSAFF groups (13.8 cm3) as compared with the control group (6.3 cm3) (p=0.018, 0.016) (Table 5).
3.3. Identification of the gene signature associated with in utero exposure to HFBUTTER in PND21 mammary gland (without DMBA treatment)
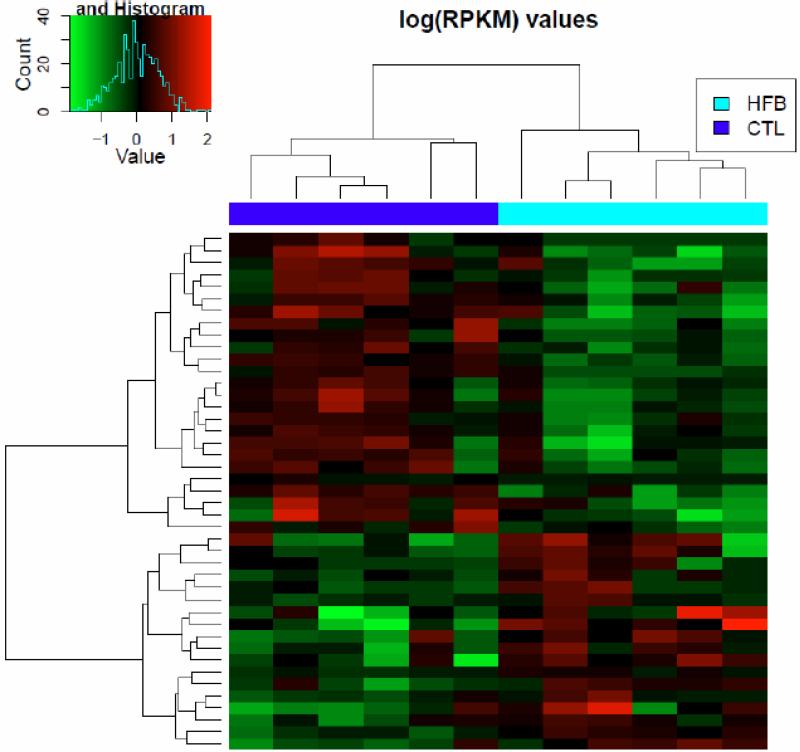
In our laboratory, we chose butter as a source of fat because the fact that butter-based diet is a popular diet in western countries. Due to the health issues associated with trans fats, consumption of margarine declined rapidly but butter consumption in the US in 2013 reached its highest levels in four decades according to recent statistics from American Butter Institute. Because of this reason, we prioritize our effort to understand how butter impacts mammary gland development with RNA sequencing experiments. We used transcriptome profiling with RNA-Seq to identify differentially expressed gene patterns in PND21 mammary glands before treatment with DMBA in the HFBUTTER-treated and the control group. Two-way unsupervised clustering analysis revealed distinct dysregulation of gene expression patterns in the two groups (Figure 1). These early changes in gene expression may be related to the differential susceptibility of PND21 mammary glands to DMBA-induced mammary tumorigenesis.
Fig. 1.
Clustering analysis of genes with p<0.05 in control and treatment groups. Heat map shows the two signature panels of differentially expressed genes in response to an HFBUTTER (HFB) diet and a control AIN diet (CTL). Green represents low expression; red represents high expression.
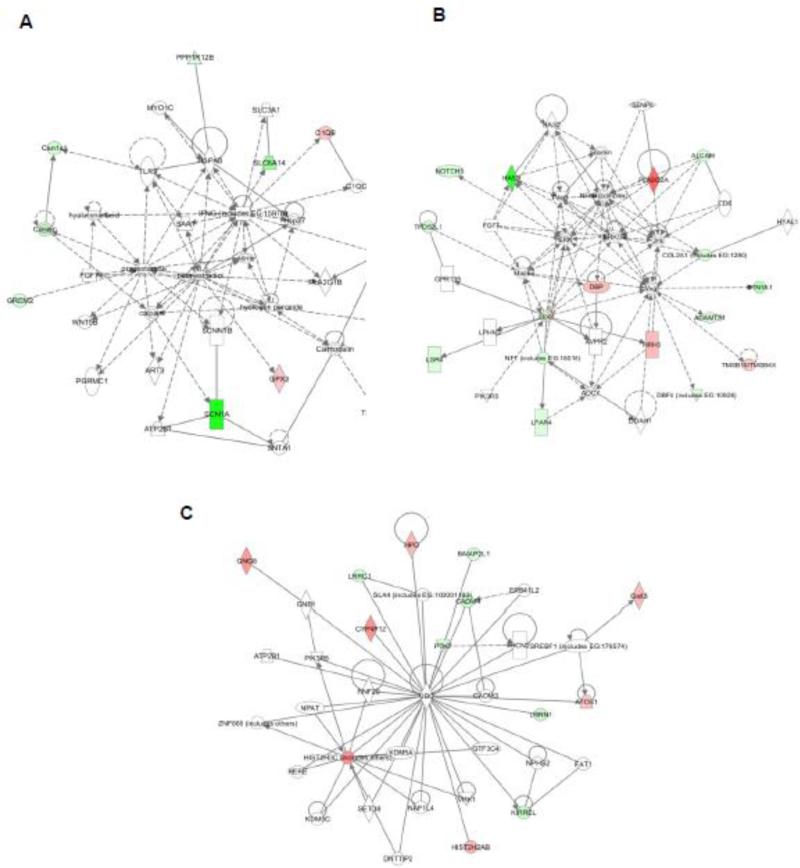
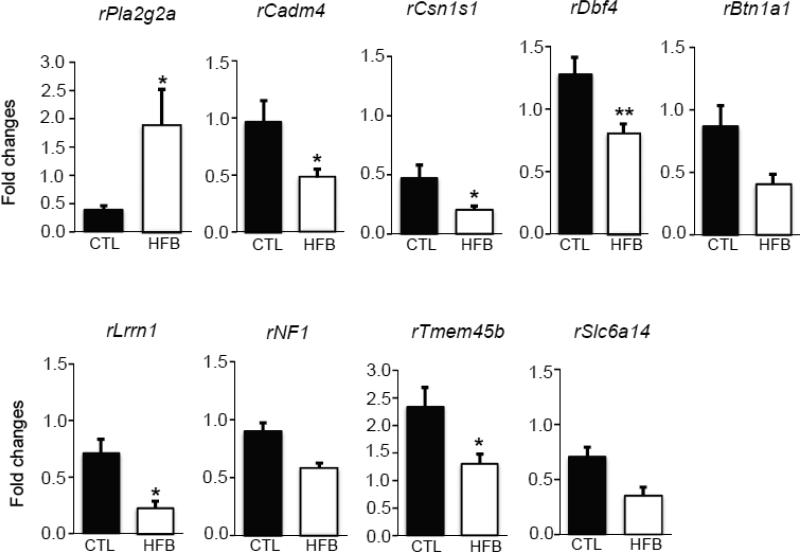
In this RNA-Seq study, we found 43 genes that were differentially expressed in two groups (HFBUTTER vs. CTL) (Table 6). To gain insights into their biological relationship, we used the Ingenuity Pathway Analysis (IPA, http://analysis.ingenuity.com) to map these genes into knowledge networks (Figure 2). We identified three major networks: 1) carbohydrate metabolism, drug metabolism, and small-mole biochemistry; 2) cell morphology, cell death, and renal necrosis/cell death; and 3) lipid metabolism, small-molecule biochemistry, and vitamin and mineral metabolism, which involve estradiol, progesterone, ERK/MAPK, NFk-B, VEGF, and ubiquitin C signaling. Using qPCR, we confirmed the differential expression of nine genes selected from the top pathways in HFBUTTER compared with the control (Figure 3). These include phospholipase A2, group IIA (Pla2g2a), most upregulated gene and cell adhesion molecule 4 (Cadm4), casein alpha s1 (Csn1s), DBF4 zinc finger (Dbf4), leucine-rich repeat neuronal 1 (Lrrn1), butyrophilin, subfamily 1, member A1 (Btn1a1), neurofibromin 1 (Nf1), transmembrane protein 45B (Tmem45b), and solute carrier family 6 (amino acid transporter), member 14 (Slc6a14) the most downregulated genes as compared with expression levels in the control group (Figure 3).
Table 6.
Differential expressed genes in HFBUTTER vs CTL (n=6), p<0.05
| SYMBOL | GENENAME | Fold | pval |
|---|---|---|---|
| Pla2g2a | phospholipase A2, group IIA (platelets, synovial fluid) | 4.720 | 0.0130 |
| Add2 | adducin 2 (beta) | 3.345 | 0.0212 |
| Gng8 | guanine nucleotide binding protein (G protein), gamma 8 | 3.289 | 0.0397 |
| Hist2h3c2 | histone cluster 2, H3c2 | 2.705 | 0.0497 |
| Scand1 | SCAN domain-containing 1 | 2.319 | 0.0011 |
| Cyp2u1 | cytochrome P450, family 2, subfamily u, polypeptide 1 | 2.035 | 0.0479 |
| Gstt3 | glutathione S-transferase, theta 3 | 1.939 | 0.044 |
| Hist2h2ab | histone cluster 2, H2ab | 1.922 | 0.016 |
| LOC691921 | hypothetical protein LOC691921 | 1.909 | 0.047 |
| Dbp | D site of albumin promoter (albumin D-box) binding protein | 1.833 | 0.009 |
| Fxyd2 | FXYD domain-containing ion transport regulator 2 | 1.690 | 0.042 |
| Sepw1 | selenoprotein W, 1 | 1.609 | 0.011 |
| Rps2 | ribosomal protein S2 | 1.596 | 0.015 |
| Plac9 | placenta-specific 9 | 1.594 | 0.041 |
| Selm | selenoprotein M | 1.581 | 0.030 |
| C1qb | complement component 1, q subcomponent, B chain | 1.555 | 0.011 |
| Tmsb10 | thymosin, beta 10 | 1.546 | 0.037 |
| Atp6v0b | ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b | 1.539 | 0.027 |
| Polr2l | polymerase (RNA) II (DNA directed) polypeptide L | 1.531 | 0.028 |
| Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 1.523 | 0.034 |
| Gpx3 | glutathione peroxidase 3 | 1.517 | 0.013 |
| Rpl39 | ribosomal protein L39 | 1.507 | 0.048 |
| Atox1 | ATX1 antioxidant protein 1 homolog (yeast) | 1.498 | 0.031 |
| Rpl41 | ribosomal protein L41 | 1.475 | 0.048 |
| Ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 | 1.451 | 0.044 |
| Bcat2 | branched chain aminotransferase 2, mitochondrial | 1.446 | 0.033 |
| C1qa | complement component 1, q subcomponent, A chain | 1.438 | 0.048 |
| Atp5i | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit E | 1.431 | 0.042 |
| Aldoa | aldolase A, fructose-bisphosphate | 1.429 | 0.033 |
| Ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | 1.410 | 0.049 |
| Mylk | myosin light chain kinase | −1.496 | 0.046 |
| Kitlg | KIT ligand | −1.503 | 0.045 |
| Adamts9 | a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 9 | −1.557 | 0.042 |
| Adamts1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | −1.654 | 0.048 |
| Cdh1 | cadherin 1 | −1.719 | 0.047 |
| Ptpn13 | protein tyrosine phosphatase, non-receptor type 13 | −1.901 | 0.042 |
| Col2a1 | collagen, type II, alpha 1 | −1.956 | 0.027 |
| Lpar4 | lysophosphatidic acid receptor 4 | −2.115 | 0.0244 |
| Notch3 | Notch homolog 3 (Drosophila) | −2.186 | 0.0091 |
| Csn1s1 | casein alpha s1 | −2.410 | 0.0012 |
| Mpz | myelin protein zero | −2.686 | 0.0063 |
| Nf1 | neurofibromin 1 | −2.691 | 0.0072 |
| Dll1 | delta-like 1 (Drosophila) | −2.790 | 0.0170 |
| Cadm4 | cell adhesion molecule 4 | −3.225 | 0.0421 |
| Cd244 | Cd244 molecule, natural killer cell receptor 2B4 | −3.726 | 0.0342 |
| Btn1a1 | butyrophilin, subfamily 1, member A1 | −4.114 | 0.0070 |
| Lrrn1 | leucine rich repeat neuronal 1 | −4.170 | 0.0167 |
| Svs4 | seminal vesicle secretory protein 4 | Infinitive | 0.0016 |
Fig. 2.
Pathway analysis of 43 genes differentially expressed in HFBUTTER (HFB) vs. CTL groups. The major networks involved are estradiol, progesterone, ERK/MAPK, NFk-B, VEGF, and ubiquitin (Ub) C signaling. Ingenuity pathway analysis shows the most significant gene networks of the HFB group. A, estradiol and progesterone network: 11 genes were mapped principally to this network as distinct node. B, ERK, NFk-B, and VEGF network: 16 genes were mapped to these networks as a distinct node. C, Ub network: 13 genes were mapped to a network with Ub as a central node. The intensity of the node color indicates the degree of up- or downregulation. Genes in uncolored nodes were not identified as differentially expressed in our array experiments and were incorporated into individual networks on the basis of the IPA knowledge database, indicating a relevance to this network.
Fig. 3.
Real-time PCR analyses of differentially expressed genes in this study. RNA sequencing shows transcripts levels of 9 genes. Relative levels of transcript expression of rPla2g2a (upregulated), rCadm4, rCsn1s1, rDbf4, rLrrn1, rBtn1a1, rNf1, rTmem45b, and rSlc6a14 (downregulated) in day 21 rat mammary gland of HFBUTTER (HFB) groups (n=5) compared with control (CTL) (n=5). Comparison with CTL used Student's t-test. p<0.05 compared with control.
3.4. Expression profiles of specific genes encoding DNA methylation–modifying proteins are altered by in utero exposure to HFBUTTER
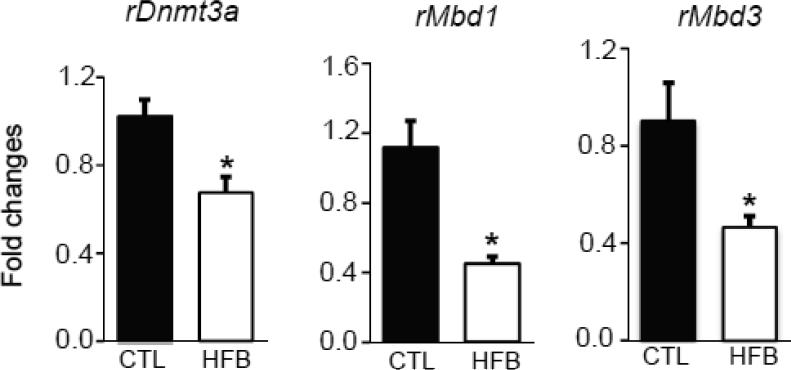
Early gene perturbation may be caused by aberrant expression of DNA methylation–related enzymes or proteins. We therefore analyzed the expression of genes encoding the most well-known enzymes/proteins involved in cytosine methylation: DNA methyltransferases (Dnmt1, Dnmt3a, and Dnmt3b) and methyl-CpG binding domain proteins (Mecp2, Mbd1, Mbd2, Mbd3, and Mbd4). We observed significant repression of Dnmt3a, Mbd1, and Mbd3 (Figure 4) but not Dnmt1, Dnmt3b, Mecp2, Mbd2, and Mbd4 in the HFBUTTER group as compared with the control.
Fig. 4.
Expression of rDNMT3a, rMBD1, and rMBD3 in HFBUTTER (HFB) vs. CTL. Relative levels of transcript expression of rDnmt3a, rMbd1 and Mbd3 in day 21 rat mammary gland of HFB (n=5) groups compared with control (CTL) (n=5). Comparison was performed with CTL using Student's t-test. p<0.01 compared with control.
3.5. Differential expression profiles of 9 genes in top pathways and genes encoding DNA methylation-modifying proteins in HFBUTTER, HFSAFF and HFOLVE groups
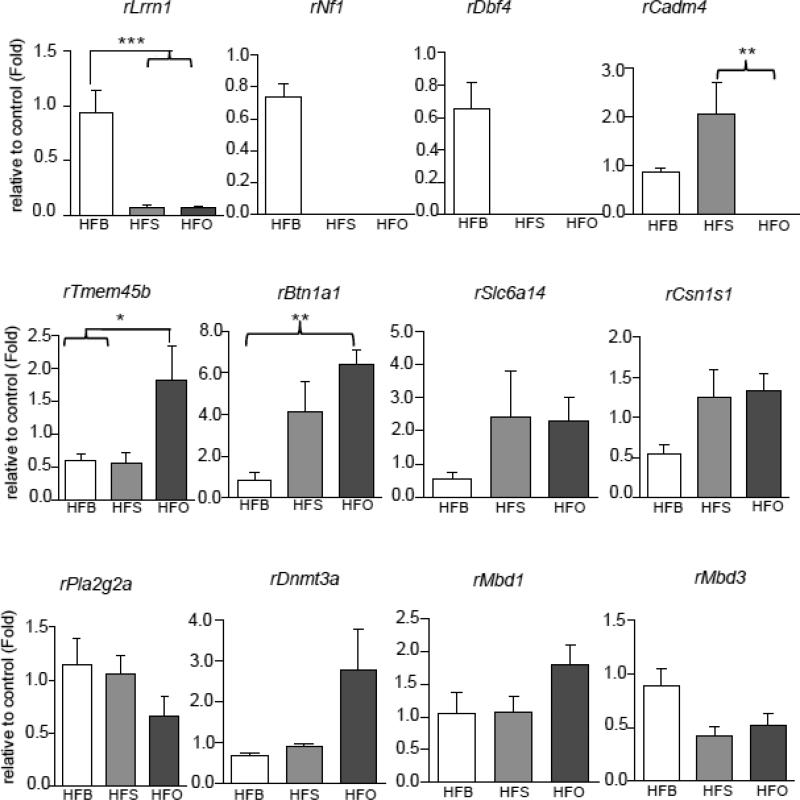
We next determined whether those genes investigated in HFBUTTER can also be altered by other two fat diets (e.g. HFSAFF and HFOLIVE) (Figure 5). Interestingly, we found that Lrrn1, Nf1 and Dbf4, which were down-regulated in HFBUTTER group, were almost totally shut down in both HFSAFF and HFOLIVE groups. Cadm4 seems to be induced in HFSAFF group but repressed in both HFBUTTER and HFOLIVE groups. Tmem45b and Btn1a1 were significantly augmented in HFOLIVE group when compared with HFBUTTER group. On the other hand, those three methylation-related genes found significantly down-regulated in HFBUTTER group did not show any statistically significant change in either HFSAFF or HFOLIVE group although HFOLIVE showed higher level expression of Dnmt3a and Mbd1 among other fat diets.
Fig. 5.
Real-time PCR analyses of differentially expressed genes in HFBUTTER (HFB), HFSAFF (HFS) and HFOLIVE (HFO) groups. Significant difference in the gene expression patterns of Lrrn1, Nf1, Dbf4, Cadm4, Tmem45b, and Btn1a1 genes among HFB, HFS and HFO groups relative to control (CTL). Comparison was performed using one way ANOVA. *p<0.05, **p<0.01, ***p<0.001 compared among three different fats relative to control.
4. Discussion
The primary aim of this study was to determine if prenatal exposure to different high-fat diets would modify prepubertal mammary glands in a fat-specific manner and alter their susceptibility to the development of mammary tumors accordingly. However, our data did not show any marked difference among the three high-fat-diet groups except for a marginal shortening of tumor latency. In contrast, when we considered all three HFD groups as a single group, we found that prenatal HFD exposure did promote the development of larger tumors, most notably in groups exposed to HFSAFF. Of interest, the exposure appeared to promote the development of more tumors harboring CIS that were twice the size of those developed in the control group. We also combined HFBUTTER and HFSAFF groups to identify, whether we see a trend in these two combined groups. However, the results indicated HFBUTTER and HFSAFF not always have the same effect. Despite the modest changes in tumorigenic outcome in the HFD group, we were able to identify a significant number of differentially expressed genes in the prepubertal glands. The gene ontology data suggest that the rapid signaling networks involving estradiol, progesterone, ERK/MAPK, NFk-β, VEGF, and ubiquitin C were the major regulatory pathways involved. Methylation-associated genes, including Dnmt3a, Mbd1, and Mbd3, were found to be disrupted. Collectively, these data are consistent with that HFD may alter mammary tumor risk via early epigenetic reprogramming in prepubertal mammary glands.
We chose PND21 to determine whether cancer susceptibility could be attained even at this early time point (PND21), as opposed to a classical cancer window (PND50). Mammary gland development at this earlier stage relies on the regulation of paracrine communication between neighboring epithelial and mesenchymal cells and is likely independent of ovarian hormones [36, 37]. The use of this model can avoid the impact of ovarian hormones and extensive mammary gland development during puberty that might confound the transcriptome analyses and the cancer-induction study. Moreover, the mammary glands of day 21 offspring have more of the undifferentiated structures called terminal end buds (TEBs), where it is generally accepted that stem-cell activity is found. These structures form the growing tips of the extending ducts and consist of a mass of “body cells” that are surrounded by a layer of “cap cells.” It has been speculated that cap cells are stem cells [38]. Terminal end buds harboring stem cells have a long life because of their resistance to apoptosis and thus can accumulate DNA damage and mutations, making them ideal candidates for the initiation of DMBA-induced mammary tumors [39, 40].
After DMBA induction, rats exposed prenatally to different HFDs showed a general trend toward increasing tumor volume, indicating that fatty acids influence mammary tumorigenesis by promoting the growth of tumors. Our findings are consistent with the report of Hilakivi-Clarke et al, [41] stating that the adipose cells of pregnant rats fed high-fat diets will produce and release estrogen in the body, promoting tumor growth. Our histopathologic analysis indicated that carcinomas from HFD animals displayed increased tumor volume and exhibited CIS. In contrast, most carcinomas from the control group were well circumscribed lobular and intraductal hyperplasia, showing the least infiltrative pattern with less morphologic aggressiveness. Our results are in concordance with those in the existing literature demonstrating that exposure to a high-fat diet may lead to the development of more aggressive mammary tumors [42, 43].
From the tumor data and histology pattern it appears that in utero exposure to HFD may change the course of mammary-gland development even at an early age. To understand the molecular changes, we analyzed the gene expression of the early prepubertal mammary glands. Using Ingenuity Pathway analysis, we found that the top signaling pathways include estradiol, progesterone, Erk/Mapk, Nfkb, Vegf, and ubiquitin C signaling. Steroid hormones exert profound effects on cell growth, development, differentiation, and homeostasis [24]. Breast development is stimulated by 17β-estradiol (E2) and progesterone, the predominant steroids and the most biologically active hormones in breast tissue. Besides the classical genomic mechanism, estradiol modulates gene expression by an indirect mechanism that involves the interaction of ER with other transcriptional factors and activation of a variety of signal transduction pathways such as Erk/Mapk, p38/Mapk, PI3K/AKT, and PLC/PKC [44]. Aberrant activation of these signaling pathways plays a key role in cell proliferation and malignant transformation in mammary tissues [45-47]. Our network- mapping analysis suggests a link between several transcripts related to inflammation, cell-cell adhesion, calcium transport, DNA replication, cell proliferation, and tumor suppressor, and other processes [48-51]. This finding contrasts with the findings of our previous study [23], demonstrating that the lifelong exposure to dietary fatty acids substantially altered the proliferation pathways in the mammary epithelial cells in similar rodent model. This suggests that rapid hormone signaling may be disturbed first during the prepubertal period and followed by changing cellular proliferation pathways after prolonged HFD treatment.
The five differentially expressed genes in our study; Lrrn1, NF1, DBF4, Cadm4, Tmem45b, Btn1a1 has been discussed in various study associated with breast and different forms of cancers [52-61]. In particular, few studies showed NF1 as tumor suppressors and Btn1a1 as a potential biomarker in breast cancer [53-56, 61]. The report by Pomp et al [61] revealed Btn1a1 is candidate biomarkers of breast cancer metastasis and it is significantly altered by dietary high fat in metastatic breast cancer. Significant higher expression of Btn1a1 in HFOLIVE group in our study supports, HFOLIVE differentially modified Btn1a1 gene expression with no or lesser effect in mammary tumorigenesis compared with HFBUTTER and HFSAFF. Difference in expression pattern of these five genes in HFBUTTER, HFSAFF and HFOLIVE in early developmental gland supports the existence of certain diet-dependent and independent cancer modifier networks underlying differential susceptibility to mammary cancer risk in adult life.
Exposure to HFD in utero can also modify mammary gland development through epigenetic mechanisms critical for gene expression. Recent work by Hilakivi-Clarke and associates [23] states that dietary and estrogenic exposures during pregnancy increase breast cancer risk in multiple generations of offspring, possibly through epigenetic mechanisms. The studies by Andrade et al [62, 63] showed exposure to a lard-based HF diet during early life changes the fatty acid profile and transcriptional network in mammary gland in young adult rats, which supports our findings that saturated butter fat, modifies the transcriptome and subsequent gene expression patterns. Another study by Pan et al [64] revealed maternal high fat exposure represses p16 (INK4a) gene expression in the mammary gland of offspring through changes of histone modifications and HDAC3 binding activity within the regulatory regions of the p16 (INK4a) gene. DNA methylation is one of the major epigenetic events that regulate gene expression. Specific DNA methylation marks, 5′-methylated cytosine (mC) in CG dinucleotides, can be maintained throughout life and transmitted for many generations [65, 66]. This is one of the key mechanisms that explain development-based adult disease caused by environmental exposure in many environmental models. Several studies state that alterations in the fetal estrogenic environment result in epigenetic modifications and increased breast cancer risk [23, 67-69].
DNA methylation, an integral part of epigenetic reprogramming, involves at least two classes of proteins, DNMTs and MBDs [70]. Here we studied the expression of eight genes encoding these proteins after in utero exposure to HFBUTTER, HFSAFF and HFOLIVE. Of the eight genes analyzed, the levels of expression of Dnmt3a, Mbd1, and Mbd3 were those most affected by in utero exposure to HFBUTTER. Since Dnmt1 expression was unchanged by HFBUTTER exposure, altered DNA methylation in the affected genes after prenatal HFBUTTER exposure must have relied on de novo methylation via Dnmt3a. Although all members of the MBD family of proteins share a highly conserved methylated DNA- binding domain and function to establish a locally compact chromatin and transcriptional repression [71], each member has been shown to play a distinct role in epigenetic regulation [72]. Specifically, Mbd1 preferentially binds to methylated CpG(s), and mediated gene silencing and absence of Mbd1 results in loss of heterochromatin formation. In this study, exposure to HFBUTTER in utero elicited significant downregulation of Mbd1 and Mbd3, suggesting that they may promote aberrant promoter hypomethylation of target genes. Collectively, Dnmt3a, Mbd1, and Mbd3 may be involved in early-life reprogramming in this rat model.
These results, taken together, underscore the complexity of gene reprogramming by early-life factors and consistent with that DNA methylation may act as a type of epigenetic memory for early insult. There are different studies which has proved maternal high fat diet alters the epigenetic histone codes resulting in adverse health outcome using different in vivo models [73-76]. However, delineating the precise effect of HFD on each epigenetic modulation, their associations with mammary gland development, and subsequent mammary tumor risk is a challenging task. Our knowledge of the association between HFD exposure in utero and epigenetics in mammary tumor model is still limited. In particular, the effects of HFD on histone methylation and chromatin remodeling complexes are largely unknown.
In conclusion, our present study suggests that exposure to HFD during pregnancy may modulate rapid hormone signaling in the prepubertal mammary gland and increase the susceptibility of female offspring to more aggressive mammary tumors. It is apparent from our findings that the complex interplay of diet and timing of dietary exposure may significantly reprogram the developing mammary gland, facilitating a permissive environment for breast cancer development.
Acknowledgements
We thank Dr. Mei Yan for pathology support and Ms. Nancy K. Voynow for her professional editing of this manuscript. This work was supported by NIH grants ES020988, ES019480, and ES006096 to SMH, ES006096 to SMH, MM, YKL and VA grant BX000675 to SMH, and HL111638 and HL127624 to MM, and an internal funding source from the University of Cincinnati Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
We declare that there are no conflicts of interest.
References
- 1.Buell P. Changing incidence of breast cancer in Japanese-American women. J Natl Cancer Inst. 1973;51:1479–83. doi: 10.1093/jnci/51.5.1479. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler RG, Hoover RN, Pike MC, Hildesheim A, Nomura AM, West DW, Wu-Williams AH, Kolonel LN, Horn-Ross PL, Rosenthal JF, Hyer MB. Migration patterns and breast cancer risk in Asian-American women. J Natl Cancer Inst. 1993;85:1819–27. doi: 10.1093/jnci/85.22.1819. [DOI] [PubMed] [Google Scholar]
- 3.Kushi L, Giovannucci E. Dietary fat and cancer. Am J Med. 2002;113:63S–70S. doi: 10.1016/s0002-9343(01)00994-9. [DOI] [PubMed] [Google Scholar]
- 4.Freedman LS, Clifford C, Messina M. Analysis of dietary fat, calories, body weight, and the development of mammary tumors in rats and mice: a review. Cancer Res. 1990;50:5710–19. [PubMed] [Google Scholar]
- 5.Prentice RL, Caan B, Chlebowski RT, Patterson R, Kuller LH, Ockene JK, et al. Low-fat dietary pattern and risk of invasive breast cancer: the Women's Health Initiative Randomized Controlled Dietary Modification Trial. JAMA. 2006;295:629–42. doi: 10.1001/jama.295.6.629. [DOI] [PubMed] [Google Scholar]
- 6.Prentice RL, Shaw PA, Bingham SA, Beresford SA, Caan B, Neuhouser ML, et al. Biomarker-calibrated energy and protein consumption and increased cancer risk among postmenopausal women. Am J Epidemiol. 2009;169:977–89. doi: 10.1093/aje/kwp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tinker LF, Rosal MC, Young AF, Perri MG, Patterson RE, Van Horn L, et al. Predictors of dietary change and maintenance in the Women's Health Initiative Dietary Modification Trial. J Am Diet Assoc. 2007;107:1155–66. doi: 10.1016/j.jada.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman M, The second World Cancer Research Fund/American Institute for Cancer Research expert report Food, nutrition, physical activity, and the prevention of cancer: a global perspective. Proc Nutr Soc. 2008;67:253–6. doi: 10.1017/S002966510800712X. [DOI] [PubMed] [Google Scholar]
- 9.Turner LB. A meta-analysis of fat intake, reproduction, and breast cancer risk: an evolutionary perspective. Am J Hum Biol. 2011;23:601–8. doi: 10.1002/ajhb.21176. [DOI] [PubMed] [Google Scholar]
- 10.Ritenbaugh C, 1, Patterson RE, Chlebowski RT, Caan B, Fels-Tinker L, Howard B, et al. The Women's Health Initiative Dietary Modification trial: overview and baseline characteristics of participants. Ann Epidemiol. 2003;13:S87–97. doi: 10.1016/s1047-2797(03)00044-9. [DOI] [PubMed] [Google Scholar]
- 11.Martin LJ, Li Q, Melnichouk O, Greenberg C, Minkin S, Hislop G, et al. A randomized trial of dietary intervention for breast cancer prevention. Cancer Res. 2011;71:123–33. doi: 10.1158/0008-5472.CAN-10-1436. [DOI] [PubMed] [Google Scholar]
- 12.Hunter DJ, Spiegelman D, Adami HO, Beeson L, van den Brandt PA, Folsom AR, et al. Cohort studies of fat intake and the risk of breast cancer—a pooled analysis. N Engl J Med. 1996;334:356–61. doi: 10.1056/NEJM199602083340603. [DOI] [PubMed] [Google Scholar]
- 13.Willett WC, Hunter DJ, Stampfer MJ, Colditz G, Manson JE, Spiegelman D, et al. Dietary fat and fiber in relation to risk of breast cancer. An 8-year follow-up. JAMA. 1992;268:2037–44. [PubMed] [Google Scholar]
- 14.Russo IH, Russo J. Mammary gland neoplasia in long-term rodent studies. Environ Health Perspect. 1996;104:938–67. doi: 10.1289/ehp.96104938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo CY, Hsieh PH, Chen HF, Su HM. A maternal high-fat diet during pregnancy in rats results in a greater risk of carcinogen-induced mammary tumors in the female offspring than exposure to a high-fat diet in postnatal life. Int J Cancer. 2009;125:767–73. doi: 10.1002/ijc.24464. [DOI] [PubMed] [Google Scholar]
- 16.Welsch CW. Relationship between dietary fat and experimental mammary tumorigenesis: a review and critique. Cancer Res. 1992;52:2040s–48s. [PubMed] [Google Scholar]
- 17.Boyd NF, Stone J, Vogt KN, Connelly BS, Martin LJ, Minkin S. Dietary fat and breast cancer risk revisited: a meta-analysis of the published literature. Br J Cancer. 2003;89:1672–85. doi: 10.1038/sj.bjc.6601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilakivi-Clarke L, Onojafe I, Raygada M, Cho E, Clarke R, Lippman ME. Breast cancer risk in rats fed a diet high in n-6 polyunsaturated fatty acids during pregnancy. J Natl Cancer Inst. 1996;88:1821–27. doi: 10.1093/jnci/88.24.1821. [DOI] [PubMed] [Google Scholar]
- 19.Hilakivi-Clarke L, Clarke R, Lippman M. The influence of maternal diet on breast cancer risk among female offspring. Nutrition. 1999;15:392–401. doi: 10.1016/s0899-9007(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 20.Hilakivi-Clarke L, Cho E, Cabanes A, DeAssis S, Olivo S, Helferich W, et al. Dietary modulation of pregnancy estrogen levels and breast cancer risk among female rat offspring. Clin Cancer Res. 2002;8:3601–10. [PubMed] [Google Scholar]
- 21.Barker DJ. In utero programming of chronic disease. Clin Sci. 1998;95:115–28. [PubMed] [Google Scholar]
- 22.de Assis S, Khan G, Hilakivi-Clarke L. High birth weight increases mammary tumorigenesis in rats. Int J Cancer. 2006;119:1537–46. doi: 10.1002/ijc.21936. [DOI] [PubMed] [Google Scholar]
- 23.de Assis S, Warri A, Cruz MI, Laja O, Tian Y, Zhang B, et al. High-fat or ethinyl-oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring. Nat Commun. 2012;3:1053. doi: 10.1038/ncomms2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvedovic M, Gear R, Freudenberg JM, Schneider J, Bornschein R, Yan M, et al. Influence of fatty acid diets on gene expression in rat mammary epithelial cells. Physiol Genomics. 2009;38:80–8. doi: 10.1152/physiolgenomics.00007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimano N, Uehara N, Kiyozuka Y, Shikata N, Tsubura A. Effects of prepubertal indole-3-carbinol treatment on development of N-methyl-N-nitrosourea-induced mammary carcinomas in female Sprague-Dawley rats. In Vivo. 2007;21:983–8. [PubMed] [Google Scholar]
- 26.Zhu Z, Jiang W, McGinley JN, Prokopczyk B, Richie JP, Jr, El Bayoumy K, et al. Mammary gland density predicts the cancer inhibitory activity of the N-3 to N-6 ratio of dietary fat. Cancer Prev Res (Phila) 2011;4:1675–85. doi: 10.1158/1940-6207.CAPR-11-0175. [DOI] [PubMed] [Google Scholar]
- 27.Heffelfinger S, Gear R, Schneider J, LaDow K, Yan M, Lu F, et al. TNP-470 Inhibits DMBA-Induced Mammary Tumor Formation When Administered Prior to the Formation of CIS but is not Additive with Tamoxifen. Lab Invest. 2003;83:1001–1011. doi: 10.1097/01.lab.0000075641.27128.67. [DOI] [PubMed] [Google Scholar]
- 28.Heffelfinger S, Yan M, Gear R, Schneider J, LaDow K, Warshawsky D. Inhibition of VEGFR2 Prevents DMBA-Induced Mammary Tumor Formation. Lab Invest. 2004;84:989–998. doi: 10.1038/labinvest.3700128. [DOI] [PubMed] [Google Scholar]
- 29.Russo J, Russo I. Atlas and histologic classification of tumors of the rat mammary gland. J Mammary Gland Biol Neoplasia. 2000;5:187–200. doi: 10.1023/a:1026443305758. [DOI] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;259:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Meth. 2015;2:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protocols. 2013;9:1765–86. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 33.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Chen J, Ko CI, Fan Y, Carreira V, Chen Y, et al. Disruption of aryl hydrocarbon receptor homeostatic levels during embryonic stem cell differentiation alters expression of homeobox transcription factors that control cardiomyogenesis. Environ Health Perspect. 2013;121:1334–43. doi: 10.1289/ehp.1307297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, Ho SM. Neonatal exposure to estradiol/bisphenol A alters promoter methylation and expression of Nsbp1 and Hpcal1 genes and transcriptional programs of Dnmt3a/b and Mbd2/4 in the rat prostate gland throughout life. Endocrinology. 2012;153:42–55. doi: 10.1210/en.2011-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 37.Wysolmerski JJ, Philbrick WM, Dunbar ME, Lanske B, Kronenberg H, Broadus AE. Rescue of the parathyroid hormone-related protein knockout mouse demonstrates that parathyroid hormone-related protein is essential for mammary gland development. Development. 1998;125:1285–1294. doi: 10.1242/dev.125.7.1285. [DOI] [PubMed] [Google Scholar]
- 38.Williams JM, Daniel CW. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- 39.Russo J, Russo IH. Influence of differentiation and cell kinetics on the susceptibility of the rat mammary gland to carcinogenesis. Cancer Res. 1980;40:2677–2687. [PubMed] [Google Scholar]
- 40.Russo J, Russo IH. Biological and molecular bases of mammary carcinogenesis. Lab Invest. 1987;57:112–137. [PubMed] [Google Scholar]
- 41.Hilakivi-Clarke L, Clarke R, Onojafe I, Raygada M, Cho E, Lippman M. A maternal diet high in n - 6 polyunsaturated fats alters mammary gland development, puberty onset, and breast cancer risk among female rat offspring. Proc Natl Acad Sci U S A. 1997;94:9372–7. doi: 10.1073/pnas.94.17.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon RR, La Merrill M, Hunter KW, Sørensen P, Threadgill DW, Pomp D. Dietary fat-dependent transcriptional architecture and copy number alterations associated with modifiers of mammary cancer metastasis. Clin Exp Metastasis. 2010;5:279–93. doi: 10.1007/s10585-010-9326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose DP, Connolly JM, Meschter CL. Effect of dietary fat on human breast cancer growth and lung metastasis in nude mice. J Natl Cancer Inst. 1991;83:1491–5. doi: 10.1093/jnci/83.20.1491. [DOI] [PubMed] [Google Scholar]
- 44.Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics. 2006;7:497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy KB, Glaros S. Inhibition of the MAP kinase activity suppresses estrogen-induced breast tumor growth both in vitro and in vivo. Int J Oncol. 2007;30:971–5. [PubMed] [Google Scholar]
- 46.Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, et al. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137–44. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 47.Lønne GK, Cornmark L, Zahirovic IO, Landberg G, Jirström K, Larsson C. PKC alpha expression is a marker for breast cancer aggressiveness. Mol Cancer. 2010;14:9–76. doi: 10.1186/1476-4598-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawa M, Yamashita S, Sakamoto K, Ikei S. Elevation of serum group II phospholipase A2 in patients with cancers of digestive organs. Res Commun Chem Pathol Pharmacol. 1991;74:241–244. [PubMed] [Google Scholar]
- 49.Williams YN, Masuda M, Sakurai-Yageta M, Maruyama T, Shibuya M, Murakami Y. Cell adhesion and prostate tumor suppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4. Oncogene. 2006;25:1446–53. doi: 10.1038/sj.onc.1209192. [DOI] [PubMed] [Google Scholar]
- 50.Abbas T, Keaton MA, Dutta A. Genomic instability in cancer. Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a012914. doi: 10.1101/cshperspect.a012914. doi: 10.1101/cshperspect.a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace MD, Pfefferle AD, Shen L, McNairn AJ, Cerami EG, Fallon BL, et al. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192:385–96. doi: 10.1534/genetics.112.142802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dmitriev AA, Rudenko EE, Kudryavtseva AV, Krasnov GS, Gordiyuk VV, Melnikova NV, et al. Epigenetic alterations of chromosome 3 revealed by NotI-microarrays in clear cell renal cell carcinoma. Biomed Res Int. 2014;2014:735292. doi: 10.1155/2014/735292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross JS, Badve S, Wang K, Sheehan CE, Boguniewicz AB, Otto GA, et al. Genomic profiling of advanced-stage, metaplastic breast carcinoma by next-generation sequencing reveals frequent, targetable genomic abnormalities and potential new treatment options. Arch Pathol Lab Med. 2015;5:642–9. doi: 10.5858/arpa.2014-0200-OA. [DOI] [PubMed] [Google Scholar]
- 54.Vasan N, Yelensky R, Wang K, Moulder S, Dzimitrowicz H, Avritscher R, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist. 2014;5:453–8. doi: 10.1634/theoncologist.2013-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, Yelensky R, Pérez-Fidalgo JA, Wang Y, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;5:1382–9. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A Molecular Portrait of High-Grade Ductal Carcinoma In Situ. Cancer Res. 2015;18:3980–90. doi: 10.1158/0008-5472.CAN-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansouri M, Mirzaei SA, Lage H, Mousavi SS, Elahian F. The cell cycle arrest and the anti-invasive effects of nitrogen-containing bisphosphonates are not mediated by DBF4 in breast cancer cells. Biochimie. 2014;99:71–6. doi: 10.1016/j.biochi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Bonte D, Lindvall C, Liu H, Dykema K, Furge K, Weinreich M. Cdc7-Dbf4 kinase overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia. 2008;9:920–31. doi: 10.1593/neo.08216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuppuswamy U, Ananthasubramanian S, Wang Y, Balakrishnan N, Ganapathiraju MK. Predicting gene ontology annotations of orphan GWAS genes using protein-protein interactions. Algorithms Mol Biol. 2014;1:10. doi: 10.1186/1748-7188-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaRocca J, Pietruska J, Hixon M. Akt1 is essential for postnatal mammary gland development, function, and the expression of Btn1a1. PLoS One. 2011;9:e24432. doi: 10.1371/journal.pone.0024432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.La Merrill M, Gordon RR, Hunter KW, Threadgill DW, Pomp D. Dietary fat alters pulmonary metastasis of mammary cancers through cancer autonomous and non-autonomous changes in gene expression. Clin Exp Metastasis. 2010;2:107–16. doi: 10.1007/s10585-009-9302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Oliveira Andrade F, Fontelles CC, Rosim MP, de Oliveira TF, de Melo Loureiro AP, Mancini-Filho J, et al. Exposure to lard-based high-fat diet during fetal and lactation periods modifies breast cancer susceptibility in adulthood in rats. J Nutr Biochem. 2014;6:613–22. doi: 10.1016/j.jnutbio.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrade Fde O, de Assis S, Jin L, Fontelles CC, Barbisan LF, Purgatto E, et al. Lipidomic fatty acid profile and global gene expression pattern in mammary gland of rats that were exposed to lard-based high fat diet during fetal and lactation periods associated to breast cancer risk in adulthood. Chem Biol Interact. 2015;239:118–28. doi: 10.1016/j.cbi.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 64.Zheng S, Li Q, Zhang Y, Balluff Z, Pan YX. Histone deacetylase 3 (HDAC3) participates in the transcriptional repression of the p16 (INK4a) gene in mammary gland of the female rat offspring exposed to an early-life high-fat diet. Epigenetics. 2012;2:183–90. doi: 10.4161/epi.7.2.18972. [DOI] [PMC free article] [PubMed] [Google Scholar]; 57 Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res C Embryo Today. 2011;93:51–55. doi: 10.1002/bdrc.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmer JR, Wise LA, Hatch EE, Troisi R, Titus-Ernstoff L, Strohsnitter W, et al. Prenatal diethylstilbestrol exposure and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1509–14. doi: 10.1158/1055-9965.EPI-06-0109. [DOI] [PubMed] [Google Scholar]
- 67.Troisi R, Hatch EE, Titus-Ernstoff L, Hyer M, Palmer JR, Robboy SJ, et al. Cancer risk in women prenatally exposed to diethylstilbestrol. Int J Cancer. 2007;121:356–60. doi: 10.1002/ijc.22631. [DOI] [PubMed] [Google Scholar]
- 68.Hoover RN, Hyer M, Pfeiffer RM, Adam E, Bond B, Cheville AL, et al. Adverse health outcomes in women exposed in utero to diethylstilbestrol. N Engl J Med. 2011;365:1304–14. doi: 10.1056/NEJMoa1013961. [DOI] [PubMed] [Google Scholar]
- 69.Akhavan-Niaki H, 1, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67:501–13. doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- 70.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 71.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–65. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borengasser SJ, Kang P, Faske J, Gomez-Acevedo H, Blackburn ML, Badger TM, et al. High fat diet and in utero exposure to maternal obesity disrupts circadian rhythm and leads to metabolic programming of liver in rat offspring. PLoS One. 2014;1:e84209. doi: 10.1371/journal.pone.0084209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suter MA, Chen A, Burdine MS, Choudhury M, Harris RA, Lane RH, et al. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB J. 2012;12:5106–14. doi: 10.1096/fj.12-212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masuyama H, Hiramatsu Y. Effects of a high-fat diet exposure in utero on the metabolic syndrome-like phenomenon in mouse offspring through epigenetic changes in adipocytokine gene expression. Endocrinology. 2012;6:2823–30. doi: 10.1210/en.2011-2161. [DOI] [PubMed] [Google Scholar]
- 75.Strakovsky RS, Zhang X, Zhou D, Pan YX. Gestational high fat diet programs hepatic phosphoenolpyruvate carboxykinase gene expression and histone modification in neonatal offspring rats. J Physiol. 2011;11:2707–17. doi: 10.1113/jphysiol.2010.203950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;2:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]