Abstract
A mapping population of 186 recombinant inbred lines developed from a cross between UC1110, an adapted California spring wheat, and PI610750, a synthetic derivative from CIMMYT’s wide-cross program, was evaluated for its response to current California races of stripe rust (Puccinia striiformis f. sp. tritici) in replicated field trials over four seasons (2007–2010) in the northern Sacramento Valley. A genetic map was constructed consisting of 1,493 polymorphic probes (SSRs, DArTs, and ESTs) mapped to 559 unique loci; and QTL analysis revealed the presence of four stripe rust resistance QTL segregating in this population, two from UC1110 (on chromosomes 3BS and 2BS) and two from PI610750 (5AL and 2AS). The two QTL of largest effects (on 3BS and 5AL) were validated in independent populations and their intervals narrowed to 2.5 cM and 4.7 cM, respectively. The 3BS QTL was shown, by allelism test and genotype, to carry a gene different from the Yr30/Sr2 complex. Mapped position also suggests that the 3BS QTL is associated with a gene different from either Yrns-B1 or YrRub, two stripe rust resistance genes mapped to this region in other studies. The 5AL QTL carries a previously unreported partial stripe rust resistance gene, designated here as Yr48. This paper discusses the individual contributions to resistance of these four QTL, their epistatic interactions, and their potential in durable resistance breeding strategies based on combinations of partial resistance genes.
Keywords: Puccinia striiformis, Triticum aestivum, DArT, Yr48, QYr.ucw-3BS, QYr.ucw-2BS
Introduction
Present throughout the world’s major wheat growing areas, the fungal disease stripe rust [causal organism Puccinia striiformis Westend. f. sp. tritici Eriks. (PST)] is an historical and continuing threat to wheat production, capable of significant reductions in both grain quality (Dimmock and Gooding 2002) and yield (Smith et al. 1986) in susceptible cultivars under conditions conducive to infection. The development and timely deployment of genetic resistance to this important disease is complicated by the fact that PST, like the causal agents of leaf rust (P. triticina Eriks.) and stem rust (P. graminis Pers. f. sp. tritici) of wheat, is a rapidly evolving population of distinct virulent races exhibiting a wide and complex range of interactions with host resistance genes.
In the United States, 59 races of PST, characterized by unique patterns of virulence on a standard set of 20 differential wheat lines, were identified during 35 years of race surveys conducted between 1963 and 1998. Then, in the year 2000 alone, 21 new races were detected; and since that year, nearly 60 more have been identified (Chen et al. 2010). As a class, these post-2000 races are noteworthy for their broad virulence profiles (including virulence on the previously effective major resistance genes Yr8 and Yr9), increased aggression (indicated by reduced latent periods and faster spore production), and extended environmental adaptation (specifically tolerance to higher temperatures) (Chen et al. 2010; Chen 2005, 2007; Hovmøller et al. 2008; Markell and Milus 2008; Milus et al. 2009). In California, the rise in frequency of these post-2000 races led to epidemic levels of stripe rust among previously-resistant varieties in the year 2003, with statewide yield losses estimated at more than 25% (Jackson et al. 2003).
This explosion of new races since the year 2000 has resulted in a situation in which deployed resistance in California now relies largely upon unknown genes and an increased presence of Yr5 and Yr15, two major resistance genes for which rapid deployment was possible due to the timely availability of molecular markers (Chen et al. 2003; Sun et al. 1997). Though these two genes thus far remain effective against races in the region, the regularly observed defeat of such race-specific genes (most recently Yr8, Yr9, Yr17, Yr19, and Yr27) calls into question the long-term effectiveness of strategies of resistance based upon major genes. Indeed, the race-specificity of such genes, coupled with the current dynamism observed within the global pathogen population, is a cause for concern (Hodson and Nazari 2010; Hovmøller et al. 2010) and underscores the urgent need for alternative defense strategies. Combinations of partial resistance genes represent one such alternative and thus are currently of great interest as sources of potentially more durable (i.e. race non-specific) resistance (see Lowe et al. 2010 for a recent discussion of the promise of partial resistance genes in achieving more durable rust resistance).
The mapping study presented here is intended as a contribution to the long-term, collective research effort to identify, characterize, and ultimately deploy genetic sources of partial resistance to stripe rust. The four years of phenotyping in this study were performed under field conditions in the Sacramento Valley of California, a region noteworthy for its diverse and burgeoning population of post-2000 races of PST (broadly speaking, races designated as PST-100 to PST-137 and above; see Chen et al. 2010). The consistent effectiveness under such conditions of the resistance QTL described here suggests their potential relevance to other regions, particularly since these post-2000 races, many of which were first detected in California, now predominate in the eastern United States (Markell and Milus 2008) and carry virulence profiles and AFLP patterns similar to those found among current races in Australia, Europe, China, and South Africa (Chen et al. 2009; Hovmøller et al. 2008). Donor lines and flanking molecular markers for all QTL described and validated in this paper are publicly available to facilitate their immediate use by interested wheat breeding and research programs (see Discussion).
Materials and Methods
Materials
The hexaploid wheat (Triticum aestivum L.) mapping population used in this study consists of 186 recombinant inbred lines (RILs) developed via single-seed descent from the cross UC1110 / PI610750. UC1110 (pedigree Chukar /// Yding // Bluebird / Chanate), an adapted California hard white spring wheat, is an elite breeding line from the UC Davis breeding program. PI610750 (CIMMYT number CYG90.248.1, pedigree Croc1 / Aegilops tauschii (Synthetic 205) // Kauz), also a spring wheat, is a synthetic derivative developed under the Wide Cross Program of the International Maize and Wheat Improvement Center (CIMMYT) in Mexico and was originally registered for its resistance to Septoria tritici leaf blotch (Mujeeb-Kazi et al. 2000).
A single spike from each F5 RIL was harvested during the 2007 field season and one F6 seed from each spike was grown in the greenhouse to provide F7 seed for headrow increases (Tulelake, CA, 2008). One F7 seed from each RIL was also sown in the greenhouse, providing tissue from which genomic DNA was extracted. These single, large-scale, high-quality DNA extractions per RIL were archived and used as the templates for all subsequent genotyping. F8 seed was used for the field trials in both 2009 and 2010 and was deposited in the National Small Grains Collection (see Discussion).
Field trial management
The RIL population was sown in replicated blocks during the 2007 (3 replications, F5 generation), 2008 (2 replications; F6), and 2009 (2 replications; F8) growing seasons in research fields located approximately two miles southwest of the town of Davis, CA (Yolo County; 38° 31’ 40” N, 121° 46’ 44” W, elevation 20 m; soil types: Yolo silt loam and Reiff very fine sandy loam). In 2010, the population was planted (2 replications; F8) in a commercial field approximately five miles southeast of Grimes, CA, along the Sacramento River (Colusa County; 39° 02’ 17” N, 121° 50’ 29” W, elevation 12 m; soil type: Grandbend loam). Within each block, approximately 30 seeds of each RIL were sown evenly across one-meter rows, with rows (i.e. RILs) spaced 30 cm apart to facilitate disease evaluation. Around and dividing each block, a double row of D6301, a dwarf variety highly susceptible to stripe rust, was planted as a spreader.
Trials were sown in November of each season, following a pre-plant fertilization of 100 kg N/ha. An additional 50 kg N/ha was applied in February, and the trials were flood-irrigated as needed, according to commercial production practices. In March of each season, chamber-grown plants (cv. Joaquin) infected with stripe rust race PST-100 were transplanted into the D6301 borders as a safeguard against the potential absence of naturally-occurring inoculum. Ultimately, such a safeguard proved unnecessary, as limited tissue samples taken each season revealed the presence of a representative diversity of post-2000 races at the time of phenotyping (see Results).
Disease evaluation
In the field, UC1110 exhibits a moderate level of resistance to current California races of PST, with an average flag leaf disease severity of 29% (range 0–85%) at grain fill over the four seasons of this study (2007–2010). While PI610750 exhibits a similar moderate level of resistance, with an average disease severity of 23% (range 5–60%), the population of RILs segregates clearly in its reaction to stripe rust. Images of typical reactions of the parents and the RILs are presented in Online Resource 1.
In response to the particular segregating characteristics of this population, an 11-point scale (Table 1 and Online Resource 1) was developed in an effort to capture in a single number two standard measures of disease response, namely the infection type (i.e. S, MS, MR, R) and the infection severity (i.e. % leaf area affected). In addition to this 11-point reaction type scale, used in all four seasons, all entries were also scored for standard disease severity (i.e. % area of the flag leaf infected by pustules) in 2008, 2009, and 2010. All phenotyping was performed between early-milk and hard-dough stages (Zadok’s scale: 7.3 – 8.5).
Table 1.
Brief descriptions of the levels of 11-point reaction type scale developed for and used to phenotype the population of RILs in this study
| Scalea | Description |
|---|---|
| 1 | No evidence of stripe rust |
| 2 | Flecking; no pustules |
| 3 | Trace necrotic striping (< 5%); 0-MR pustules |
| 4 | Trace necrotic striping (< 5 %); MS pustules |
| 5 | Evident necrotic striping (5–30%); 0-MR pustules |
| 6 | Evident necrotic striping (5–30%); MS pustules |
| 7 | Abundant necrotic striping (> 30%); 0-MR pustules |
| 8 | Abundant necrotic striping (> 30%); MS pustules |
| 9 | S pustules (< 30%), not preceded by necrosis |
| 10 | Abundant S pustules (30–70%); not preceded by necrosis |
| 11 | Very abundant S pustules (70–100%); not preceded by necrosis |
Generally speaking, “resistant” [R] reactions are indicated by scores 1–2, “moderately resistant” [MR] reactions by 3–6, “moderately susceptible” [MS] reactions by 7–8, and “susceptible” [S] reactions by 9–11. A more detailed discussion of this scale, including representative pictures for each level, can be found in Online Resource l.
Genotyping
In a preliminary screen of 1,009 microsatellite markers [SSRs], the two parent lines were found to be polymorphic for 447 markers, a high rate of polymorphism (44%) reflecting the relatively exotic pedigree of PI610750. Of these 447 markers, 325 were considered reliable due to their codominant nature. The population was screened with this codominant subset of 325 putative polymorphic SSRs using dye-labeled primers on an ABI3130xl, with alleles scored using the software GeneMapper v3.7. Of these, usable results were obtained for 229 SSR markers. Concurrently, the population was screened with Diversity Array Technology (DArT) markers (Triticarte, Australia), 1,229 of which were found to be polymorphic among the RILs.
Map construction
Using the publicly-available software MAPMAKER/EXP 3.0 (Lincoln et al. 1993), an integrated SSR-DArT genetic map was developed for this population using the polymorphic SSRs and DArTs mentioned above. After reducing this dataset by merging perfectly-linked markers with one another, linkage groups were assigned to chromosomes based on published SSR maps; and marker orders within linkage groups were established based solely on linkage within this population.
SSR scores within subsequently detected stripe rust QTL were rechecked using unlabeled primers on vertical acrylamide gels; and the population was screened with a few additional targeted markers (barc133, barc147, Vrn2, BE495011, etc.) to provide greater genetic resolution of these regions. Of these additional markers, all primer sequences are readily available via GrainGenes (http://wheat.pw.usda.gov) except for the ones reported here: Vrn2 (F 5’– ttactggtactcataactgcctctt – 3’; R 5’ – agatgaagcttgggatggat – 3’) and BE495011 (F 5′– tgattactgtagctacctcctcct – 3’; R 5’ – ggtgcaagatgtgcctgtaa – 3’).
QTL analysis and model building
Stripe rust resistance QTL were detected in the population using the publicly-available software QTL Cartographer, version 1.17j (Basten et al. 2004). Specifically, composite interval mapping (CIM) analysis was performed on both reaction type (11-point scale) and disease severity (% infected flag leaf area) data for each season separately, after averaging the scores of each RIL across all replications within each season. Permutation analyses were conducted for each phenotypic dataset for each season and, for simplicity, the highest threshold LOD value (3.3) was chosen as a uniform threshold for all analyses.
For each QTL detected across multiple seasons in both phenotypic scales, the marker at the peak of that QTL was used as a class variable in a simple ANOVA model (SAS v9.1). Included in this model were all QTL main effects as well as all first-order interactions among these QTL; both seasons and blocks within seasons were included as class variables.
QTL validation
The map positions of the two largest QTL for stripe rust resistance detected in this population, namely the ones on chromosomes 3BS and 5AL, were validated via progeny tests in independent F2 populations. Supporting evidence for the map position of the comparatively minor stripe rust resistance QTL on chromosome 2BS is provided by four seasons of field data for two near isogenic lines (NILs), sister RILs found to be segregating for stripe rust susceptibility in 2007 (F5 segregants).
Results
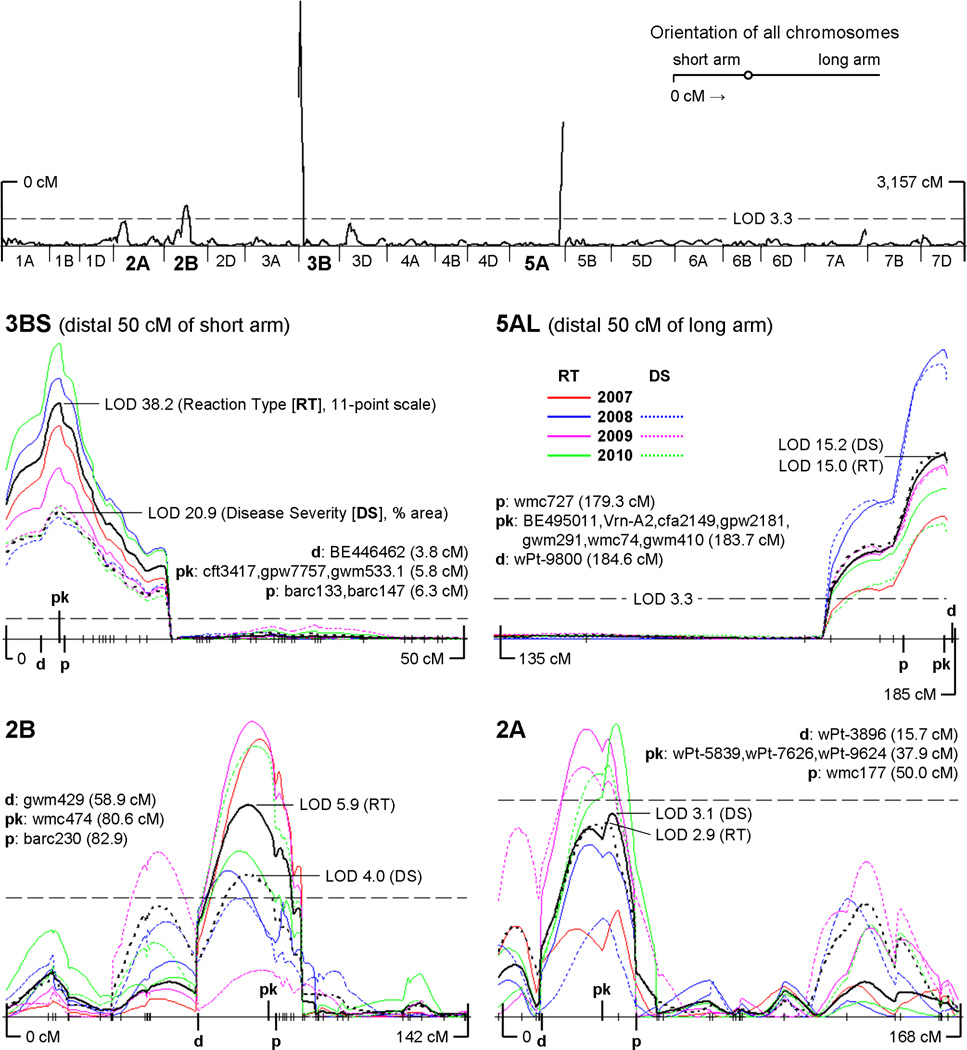
The complete genetic map developed for this population, with total coverage of 3,157 cM and an average marker spacing of 5.9 cM, is presented along with detailed notes in Online Resource 2; and a searchable spreadsheet of all mapped markers is provided in Online Resource 3 (Worksheet 2). Using this genetic map, CIM analysis of each phenotypic dataset (11-point reaction type and percent disease severity) for each season revealed the presence of four distinct stripe rust resistance QTL with significances exceeding the threshold level of LOD 3.3 over multiple years. In order of decreasing magnitude of effects, these QTL are located in the distal region of chromosome 3BS, in the distal region of 5AL, in centromeric 2BS, and in 2AS. The presence, relative sizes, and stability over multiple years of these four QTL can be seen in Fig. 1.
Fig. 1.
The top graph presents a whole-genome view of the results of the CIM analysis (global average of all LOD scores across all seasons and both phenotypic scales) with the threshold LOD of 3.3 indicated by a dashed horizontal line. All chromosomes in this whole-genome view are to scale and oriented with the telomere of the short arm to the left. As throughout this paper, distances are in Kosambi cM, with the telomere of the short arm positioned at 0 cM. Below is a panel of four LOD plots, one for each significant QTL region, with the threshold LOD of 3.3 included in each for scale. Within each plot, solid color lines indicate the CIM results for reaction type for each season (2007–2010, see color key in figure); dashed color lines indicate the CIM results for disease severity for each season (2008–2010). The solid black line is the average LOD plot of the four seasons of 11-point reaction type data. The dashed black line is the average LOD plot of three seasons of % disease severity data. “pk” = QTL peak marker (i.e. the locus associated with the highest overall average LOD score); “d” = distal flanking marker; “p” = proximal flanking marker. Hatch marks along the x-axes indicate the positions of mapped loci (see complete genetic map in Online Resource 2)
Table 2 presents the virulence profiles of PST races collected from the fields where the RIL population was evaluated for disease response. Each year, a small number of sporulating stripe rust-infected tissue samples were collected from throughout the population and submitted to the USDA-ARS Laboratory of the Wheat Genetics, Quality Physiology, and Disease Research Unit (Pullman, WA) for race identification, as part of that laboratory’s ongoing disease surveillance and monitoring work (see Chen et al. 2002 for a detailed description of methods). Field-collected spores were pathotyped using either the standard US set of differentials (years 2007–2009; Chen et al. 2002) or a new set of monogenic resistance differentials (2010; XM Chen and associates, unpublished data). These results show that the RIL population was under pressure from at least eight different broadly virulent post-2000 races over the course of this investigation. Given the season-to-season consistency of the resistance QTL detected in this study, it can be concluded that these QTL are effective against at least those races listed in Table 2.
Table 2.
Virulence profiles of field-collected spores confirm the presence of a diversity of broadly-virulent post-2000 races of PST during the course of this study. In light of the limited number (34) of samples analyzed, this list should not be understood as a comprehensive survey of all races present.
| Race | Year | Virulence profilea | |
|---|---|---|---|
| PST-100 | 2008 | 1,3,8,9,10,11,12,16,17,18,19,20 | |
| PST-111 | 2008 | 1,3,5,8,10,11,12,16,17,18,19,20 | |
| PST-113 | 2007,08 | 1,2,3,8,9,10,11,12,16,17,18,19,20 | |
| PST-115 | 2008 | 1,3,5,8,9,10,11,12,14,16,17,18,19,20 | |
| PST-122 | 2008 | 1,2,3,6,8,9,10,11,12,14,16,17,18,19,20 | |
| PST-127 | 2009 | 1,2,3,5,6,8,9,10,11,12,13,15,16,17,18,19,20 | |
| PST-128 | 2007 | 1,2,3,6,8,10,11,12,16,17,18,19,20 | |
| PST-129 | 2007,08 | 1,2,3,8,10,11,12,16,17,18,19,20 | |
| undesignated | 2010 | 2,6,7,8,9,26,32,43,44,Tr1,Exp2 | |
| undesignated | 2010 | 2,6,7,8,9,17,26,27,43,44,Tr1,Exp2 | |
| undesignated | 2010 | 2,6,7,8,9,17,26,27,43,44,Tr1,Exp2 | |
| undesignated | 2010 | 2,6,7,8,9,17,26,27,32,43,44,Tr1,Exp2 | |
| undesignated | 2010 | 1,2,6,7,8,9,10,24,27,32,43,44,Tr1,Exp2 | |
Note that while the virulence profiles for samples taken in 2007–2009 refer to a set of 20 historic differential lines (see Chen et al. 2002), those for samples taken in 2010 refer to specific resistance genes (e.g. Yr2, Yr6, Yr7, etc.). Due to this recent transition to a set of monogenic differentials, the samples collected in 2010 have yet to receive official race designations.
The reliable field evaluation of adult plant resistance to stripe rust can be complicated by seasonal shifts in the pathogen population, spatially non-uniform disease pressure, environmental differences, and high inoculum loads. Such factors can introduce substantial noise into an experiment, thereby preventing the detection of sources of resistance even if they exist. In this particular study, however, four significant stripe rust resistance QTL were detected consistently across four seasons and two locations in spite of these variables. The strong correlation between population replications within each season, with R2 values ranging from 0.83–0.94 (11-point reaction type) and 0.80–0.96 (percent disease severity), indicates limited spatial non-uniformity of disease pressure within seasons (Online Resource 4, Fig. S1). Across seasons, shifts in the pathogen population and differences in environmental conditions and overall inoculum load appear to have introduced slightly more variability into the study, though the correlations remain high, with R2 values ranging from 0.76–0.86 (reaction type) and 0.63–0.84 (disease severity) (Online Resource 4, Fig. S2).
The fact that roughly 1/16 of the population (12 RILs) exhibits complete susceptibility (average disease severity ≥ 90%) is consistent with the identification of four significant resistance QTL. And the fact that two of the resistance QTL (on 3BS and 2BS) are contributed by UC1110 and the other two (on 5AL and 2AS) are contributed by PI610750 provides some insight into the moderately resistant phenotypes observed for the two parents (see Online Resource 1). Before proceeding to discuss these four QTL in more detail, it is worth mentioning that two other stripe rust resistance QTL of very consistent but sub-threshold effect were also detected on chromosomes 3DS and 7AL. While these QTL are presented in Online Resource 4 (Fig. S3) as a point of interest, they are not included in any of the subsequent analyses presented here.
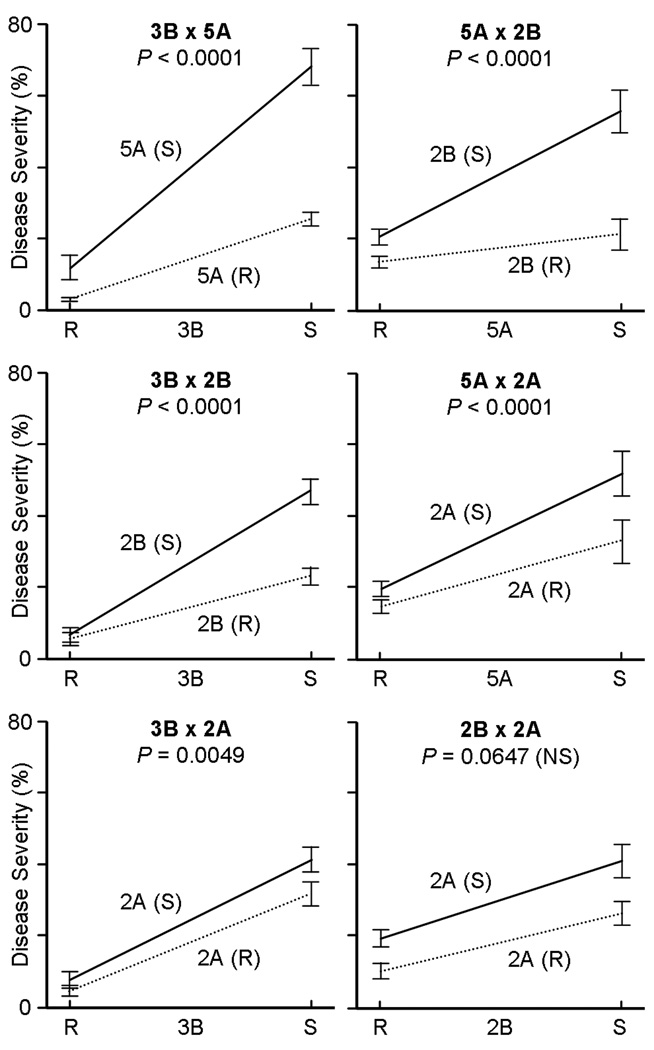
The mapped locus associated with the highest overall average LOD score within each of the QTL regions was designated the “peak” marker for that region (see Fig. 1). These four peak markers (gwm533.1 for 3BS, cfa2149 for 5AL, wmc474 for 2BS, and wPt-5839 for 2AS) were then used as class variables, along with years (2007–2010) and blocks in the field (nested within years), in an analysis of variance (ANOVA). The results of this analysis (P-values and % variation explained), including the main effects of these four QTL and their first-order interactions, are presented in Table 3. For the 11-point reaction type data, this ANOVA model accounts for 71.8% of the phenotypic variation observed in the field; for the percent disease severity data, this model accounts for 64.5% of the observed variation. In general, the analyses using the two different phenotypic scales are in close agreement (see Discussion). Interaction plots (% disease severity) for the four QTL are presented in Fig. 2, and equivalent plots for reaction type can be found in Online Resource 4 (Fig. S4).
Table 3.
Analysis of variance (ANOVA) results from a simple model in which each of the four detected QTL are represented by their peak markers
| Reaction Type (11-point scale) | Disease Severity (% leaf area) | |||
|---|---|---|---|---|
| Effecta | R2= 71.8% | R2= 64.5% | ||
| P | % Variation | P | % Variation | |
| 3B | < 0.0001 | 36.4% | < 0.0001 | 22.2% |
| 5A | < 0.0001 | 6.5% | < 0.0001 | 10.2% |
| 2B | < 0.0001 | 4.2% | < 0.0001 | 4.5% |
| 2A | < 0.0001 | 1.1% | < 0.0001 | 2.3% |
| 3B × 2B | < 0.0001 | 2.3% | < 0.0001 | 3.0% |
| 5A × 2B | < 0.0001 | 1.9% | < 0.0001 | 2.7% |
| 3B × 5A | < 0.0001 | 1.8% | < 0.0001 | 2.4% |
| 5A × 2A | 0.0137 | 0.1% | < 0.0001 | 0.7% |
| 3B × 2A | 0.0087 | 0.1% | 0.0049 | 0.3% |
| 2B × 2A | 0.1060 (NS) | 0.0% | 0.0647 (NS) | 0.1% |
The peak markers used to represent the four QTL in this model are gwm533.1 (3B), cfa2149 (5A), wmc474 (2B), and wPt-5839 (2A). Included in the model are the main effects of the four QTL, the first-order interactions among these QTL, years (not shown), and blocks within the field (nested within years, also not shown). P-values are presented for all main and first-order QTL effects. The overall model R2 values for the two phenotypic datasets are provided, and the percents of total phenotypic variation explained by each effect are also given.
Fig. 2.
Two-way interaction plots for the four detected QTL. In each plot, the QTL region of larger magnitude is shown on the x-axis (“R” = resistant; “S” = susceptible). The two phases of the smaller QTL are indicated by the two lines in each plot (“R” = dotted line; “S” = solid line). Error bars are ± 2 standard errors, and associated p-values are indicated
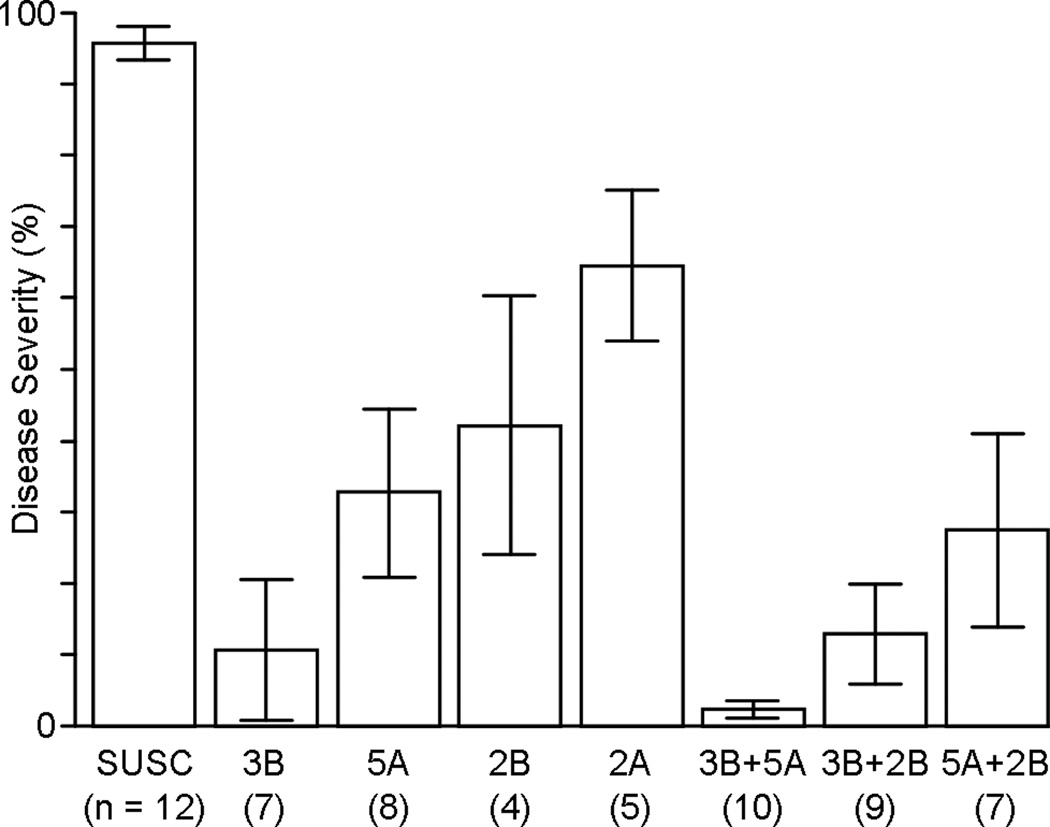
The general trend illustrated in the interaction plots (Fig. 2) is that the observed effects of each of the four QTL are greater in the absence of resistant alleles from the other QTL than in their presence (except for the 2BS + 2AS combination). The relative slopes of the lines in the interaction plots in Fig. 2 are indeed informative in terms of QTL combinability, but the values of their endpoints do not accurately represent the individual effects of the four QTL. The reason for this is that the endpoints of these lines are the averages of all 186 RILs, classified according to the two QTL under consideration but making no attempt to control for the effects of the other two QTL. For example, the values presented in the 2B × 2A interaction plot are biased by the effects of the 3B and 5A QTL. To obtain a more accurate sense of the individual and combined effects of these QTL, the flanking markers for the four QTL regions were used to select subsets of RILs in which these effects are controlled (e.g. RILs possessing only the 2BS resistance were selected to represent the effect of the 2BS QTL, etc.). The results of this more controlled comparison are presented in Fig. 3, along with a more detailed description of these subsets of RILs. An equivalent chart for reaction type data can be found in Online Resource 4 (Fig. S5).
Fig. 3.
The effects of different QTL (and pairs of QTL) on disease severity in otherwise susceptible backgrounds. “SUSC” = The subset of fully susceptible RILs (i.e. lines with susceptible haplotypes at all four QTL regions); “3B” = The subset of RILs possessing only the 3B resistance; “5A” = The subset of RILs possessing only the 5A resistance; etc. “3B+5A” = The subset of RILs possessing 3B and 5A resistance only (i.e. no 2B or 2A resistance); etc. The numbers in parentheses under each label indicate the numbers of RILs in each class. Bar heights are averages over all seasons; error bars are ± 2 standard errors. For a detailed list of the RILs in each class, please refer to Online Resource 4 (Figure S3)
Seeds of the mapping population parents and all RILs were deposited in the National Small Grain Collection and are available to interested breeding and research programs (GSTR numbers 13501–13687; for a table of ID’s, see Online Resource 3, Worksheet 4) through the USDA Germplasm Resources Information Network (www.ars-grin.gov). To facilitate the use of this mapping population for other QTL studies, full genotypic data for all RILs are available both through the GrainGenes database (http://wheat.pw.usda.gov/report?class=mapdata;name=Wheat,+UC1110+x+PI610750) and in Online Resource 4 (Worksheet 2). Complete phenotypic data for this study is also available in Online Resource 4 (Worksheet 3).
3BS QTL (henceforth QYr.ucw-3BS)
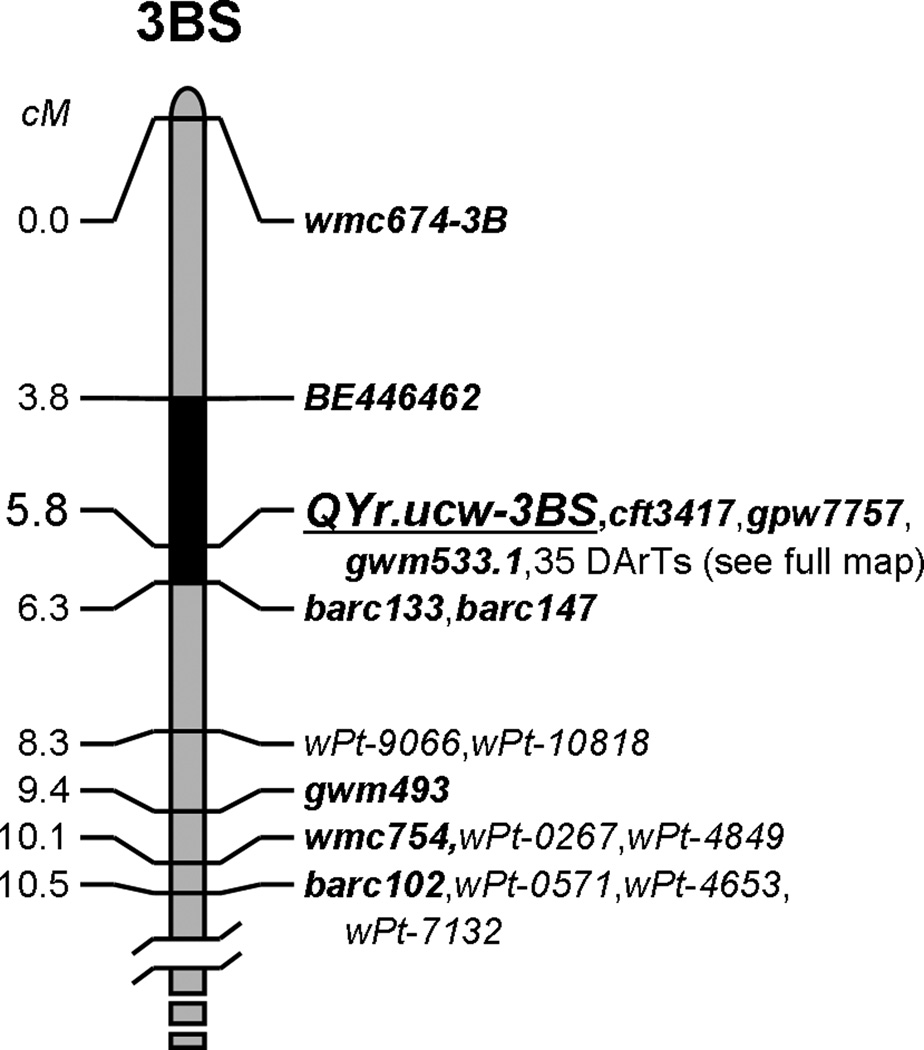
The QTL of largest effect in this population, contributed by UC1110, maps to the distal region of the short arm of chromosome 3B and is linked to locus gwm533.1 (Fig. 4). Some caution is necessary with regard to this marker because two different gwm533 loci have been mapped to chromosome 3B (Röder et al. 1998; Suenaga et al. 2003; Xue et al. 2008): gwm533.1, located 5–12 cM distal of gwm493 and perfectly linked to QYr.ucw-3BS in the present mapping population (Figure 4); and gwm533.2, located approximately 30–40 cM proximal of gwm493 (Röder et al. 1998; Xue et al. 2008). It is important to note that Suenaga et al. (2003) also mapped the same two gwm533 loci but inverted their names (gwm533.1 and gwm533.2) relative to the original map from Röder et al. (1998). The nomenclature adopted in this paper follows the precedent of Röder et al. (1998).
Fig. 4.
Mapped and validated position of stripe rust resistance QTL QYr.ucw-3BS on the short arm of chromosome 3B (source UC1110). The black region demarcates the current size of the QTL region (2.5 cM). This resistance is perfectly linked with gwm533.1 as well as two other SSR’s and 35 DArT markers. See Online Resource 2 for the full genetic map
In terms of disease severity, the main effect of this QTL alone accounts for 22% of the observed variation in the field and is associated with an average reduction in disease severity of 85%, compared to fully susceptible lines. As indicated by Fig. 2 and Fig. 3, the effects of the more minor 2BS and 2AS QTL are essentially nullified in the presence of the stronger 3BS resistance. Such is not the case, however, with the QTL on 5AL; though their effects are not additive, some additional protection is achieved through the combination of these two QTL (an average reduction in disease severity of 93%) compared to either alone.
QYr.ucw-3BS validation
To validate the presence and mapped position of the 3BS QTL, an independent F2 population was developed from the cross RIL149 / RIL233. Since RIL149 is a completely susceptible line from the original mapping population (i.e. it lacks all four resistance QTL) and RIL233 carries only the 3BS resistance (i.e. it lacks the 5AL and 2BS resistances), this F2 population segregates only for the 3BS resistance, thus Mendelizing the trait. Out of 90 F2 plants from this cross, 18 were found to be homozygous for the resistant RIL233 haplotype (based on both flanking markers and the peak marker gwm533.1) and 19 were found to be homozygous for the susceptible RIL149 haplotype. These 37 plants were inoculated in the greenhouse with spores collected during the 2010 field season and their reaction types (resistant or susceptible) were found to co-segregate perfectly with the two gwm533.1 alleles (Fisher’s Exact Probability Test P < 0.0001). The details of this screen, including images of typical reactions, are presented in Online Resource 5 (Table S1).
QYr.ucw-3BS dissection
The flanking markers for this 2.5 cM region (see Fig. 4) were determined by means of a detailed characterization of RILs with recombination events close to the QTL peak (Online Resource 5, Table S2). The distal flanking marker (EST-based marker BE446462) is set by the phenotype of RIL199, a 5AL- and 2BS-resistant line with a recombination event between this EST marker and peak marker gwm533.1 and a clear absence of the 3BS resistance (48% average disease severity, 7.3 average reaction type; refer to Table S2). The phenotypes of other RILs with recombination events in this area (e.g. RILs 178 and 204, Table S2) further confirm BE446462 as a reliable distal flanking marker. The phenotype of RIL160, a line with a recombination event between gwm533.1 and barc133, suggests barc133 as a reliable proximal flanking marker (Figure 4, Table S2). Due to the very close proximity (0.5 cM) of this marker to the peak of QYr.ucw-3BS, two independent F2 populations were developed to validate this boundary: 1) RIL160 / RIL149 (fully susceptible line) and 2) RIL160 / RIL233 (RIL carrying only the 3BS resistance). For each population, adult F2 plants lacking the 5AL resistance but homozygous for either of the parental 3BS haplotypes were inoculated in the greenhouse with field spores collected during the 2009 field season. The resistant and susceptible reaction types showed perfect linkage with gwm533.1 (Freeman-Halton extension of Fisher’s Exact Probability Test P = 0.007), indicating that RIL160 carries the QYr.ucw-3BS resistant allele and confirming barc133 as a reliable proximal marker (Figure 4, Table S2). Based on these flanking markers, QYr.ucw-3BS can be assigned to the Chinese Spring deletion bin 3BS8-0.78–1.00 and appears to span contigs 986, 412, 960, 1, 541, 11, 344, 6, 287, and 774 of the 3B physical map (Paux et al. 2008).
Allelism test between QYr.ucw-3BS and Yr30
A number of rust resistance genes and QTL have been mapped to the distal region of the short arm of chromosome 3B, including Yr30 (Singh et al. 2000; Suenaga et al. 2003), Yrns-B1 (Börner et al. 2000; Khlestkina et al. 2007), and YrRub (Bansal et al. 2010). Given this fact, substantial work beyond the scope of this current study is needed to determine if the QTL detected in this population is a novel gene, a novel allele of a previously mapped gene, or simply a rediscovery of an already known allele. Until such comprehensive work is done, no formal gene designation for this detected QTL is suggested. That being said, an allelism test was conducted to investigate the relationship between QYr.ucw.3BS and the adult plant resistance gene Yr30.
For this test, a population of 300 F2 plants from the cross RIL233 (donor of QYr.ucw-3BS) / Opata 85 (donor of Yr30; Suenaga et al. 2003) was planted in the field in Davis in 2010. Of these 300 plants, 7 were found to be completely susceptible (reaction type 11, disease severity > 90%). This result is compatible with the association of the peak of the QTL for Yr30 with gwm389 (Suenaga et al. 2003), a marker which maps in 4–12 cM distal of wms533.1 in other populations (Kota et al. 2006; Sherman et al. 2010). Based on this allelism test, it appears that the 3BS QTL detected in this population, being completely linked to gwm533.1, carries a gene for stripe rust resistance that is different from Yr30. This conclusion is strengthened by the fact that UC1110 does not possess Sr2, the slow-rusting stem rust resistance complex associated with Yr30 (Singh et al. 2000). The absence of Sr2 in UC1110 is supported by a recently developed diagnostic marker for Sr2 (Mago et al. 2010), by the 142 bp length of the amplified fragment of gwm533.1 in UC1110 (Sr2 is associated with a 120 bp fragment; Spielmeyer et al. 2003), as well as the lack of the Sr2-associated morphological trait pseudo-black chaff in both UC1110 and the mapping population.
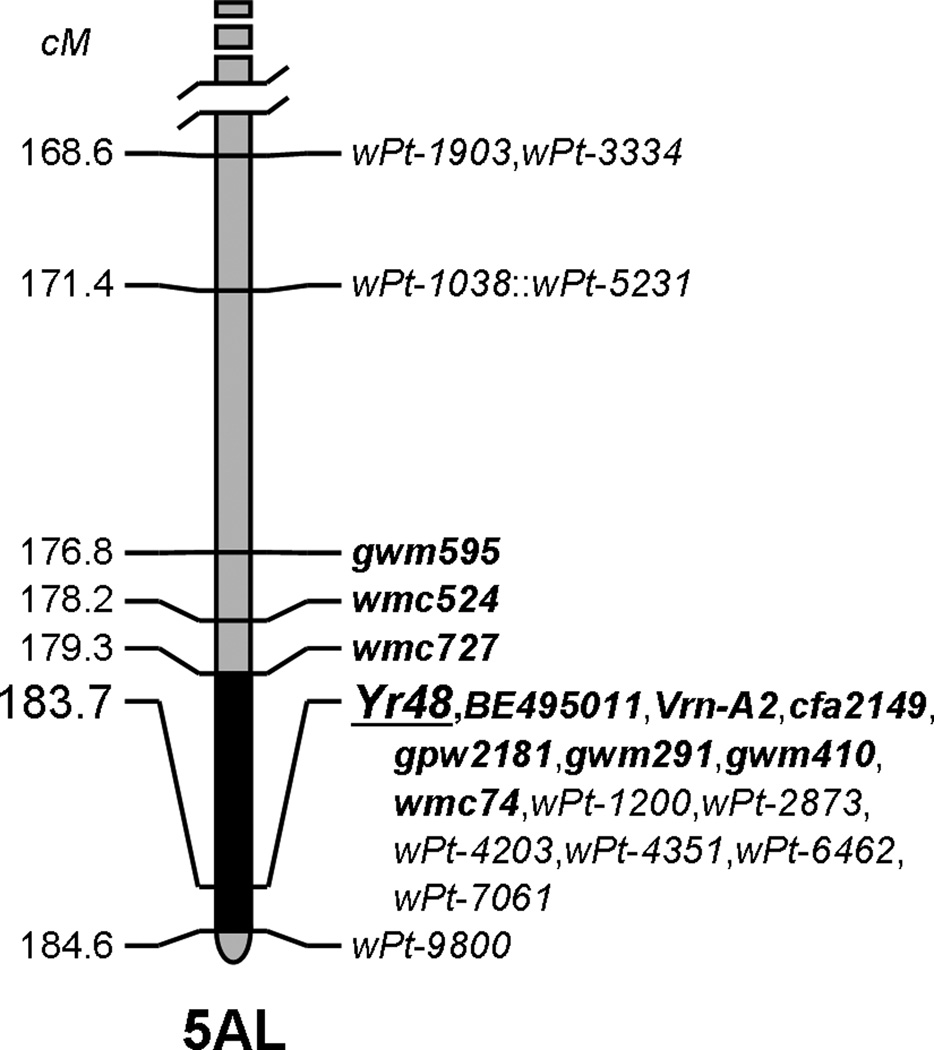
5AL QTL (henceforth Yr48, see Discussion)
The second most significant QTL in this population, contributed by the synthetic derivative PI610750, is located in the distal region of the long arm of chromosome 5A and is completely linked to the vernalization locus Vrn2 (Yan et al. 2004) (Figure 5). In terms of disease severity, the main effect of this QTL alone accounts for 10% of the observed variation in the field and is associated with an average reduction in disease severity of 63%, compared to fully susceptible lines. As indicated by Fig. 2 and Fig. 3 (and Online Resource 4, Fig. S4 and Fig. S5), this QTL combines well with the one on 3BS to enhance the level of protection in the field. Slight gains in protection were also observed through its combination with the QTL on 2BS and, to a lesser degree, the QTL on 2AS.
Fig. 5.
Mapped and validated position of the partial stripe rust resistance QTL on the long arm of chromosome 5A (source PI610750). The gene underlying this QTL is designated Yr48. The black region demarcates the current size of the QTL region (4.7 cM). This resistance is perfectly linked with the EST-derived marker BE495011, as well as vernalization gene Vrn2. See Online Resource 2 for the full genetic map
Yr48 validation
To validate the presence and position of this QTL, an independent F2 population was developed from the cross RIL149 / RIL167. Since RIL149 is a completely susceptible line from the original mapping population (i.e. it lacks all four resistance QTL) and RIL167 carries only the 5AL resistance (i.e. it lacks the 3BS and 2BS resistances), this F2 population segregates clearly for the 5AL resistance. Out of 150 F2 plants, 34 were found to be homozygous for the resistant RIL167 haplotype (based on both flanking markers and the peak marker cfa2149) and 32 were found to be homozygous for the susceptible RIL149 haplotype. These 66 plants were transplanted as seedlings into the field at Davis in 2010 and scored at booting stage under natural infection pressure as either resistant or susceptible. This binary reaction type was found to segregate perfectly with the two haplotypes (Fisher’s Exact Probability Test P < 0.0001); and the details of this validation trial, including images of typical reactions, can be found in Online Resource 5 (Table S1).
Yr48 dissection
The flanking markers for this 4.7 cM QTL region (see Fig. 5) were determined by means of a detailed characterization of RILs with recombination events close to the QTL peak. The distal flanking marker (gwm291) is set by the phenotype of RIL223, a line with a recombination event between gwm291 and peak marker cfa2149. Because this line lacks the 3BS and 2BS QTL, its 5.9 average reaction type and 20% average disease severity clearly indicates that it carries Yr48. The phenotypes of RIL149, RIL150, and RIL243, three lines carrying independent recombination events between cfa2149 at the peak of the QTL and wmc727, suggest wmc727 as a reliable proximal flanking marker. RILs 149 and 150 are completely susceptible lines (10.9 and 10.9 average reaction types, 87% and 91% average disease severities) that clearly lack the 3BS, 5AL, and 2BS resistances. RIL243, lacking the 3BS and 2BS resistances, is shown by its phenotype to carry the 5AL resistance (7.0 average reaction type, 38% average disease severity). Based on these flanking markers, Yr48 can be assigned to the Chinese Spring deletion bin 5AL23-0.87–1.00. Dissection data for this QTL is summarized in Online Resource 5 (Table S4).
Effects of development and temperature on QYr.ucw-3BS and Yr48
Given the sheer magnitude of its average effect (> 80% reduction in disease severity, Fig. 3), the possibility exists that QYr.ucw-3BS is a major resistance gene. But controlled chamber inoculations of seedling and adult plants at low- and high-temperatures with PST-100, a broadly virulent race of PST representative of the post-2000 group of races, suggest that QYr.ucw-3BS behaves instead like a high-temperature adult-plant (HTAP) partial resistance gene (see Table 4). At the seedling stage, the 3BS QTL (see RIL233) offers no significant protection against PST-100 but does provide effective resistance at the adult plant stage under high temperatures. Both the lack of resistance at the seedling stage and the temperature dependence of the resistance at the adult plant stage suggest that the 3BS QTL carries a partial HTAP resistance gene rather than a major gene conferring race-specific protection against PST-100. At the adult stage, the 3BS QTL combines with the one on 5AL (Yr48, see RIL8) to offer effective levels of resistance even at low temperatures. This combination of resistance QTL also confers partial resistance at the seedling stage, a developmental stage at which neither QYr.ucw-3BS nor Yr48 alone exert any discernable effect.
Table 4.
Summary of results of controlled inoculations using post-2000 stripe rust race PST-100
| Linea | QTL | Seedling (IT)b | Adult (% Severity)c | ||
|---|---|---|---|---|---|
| Low Td | High T | Low T | High T | ||
| RIL149 | SUSC | 8 | 8 | 30–80 | 20–50 |
| RIL233 | 3BS | 8 | 7 | 30–60 | 1–5 |
| RIL167 | 5AL | 8 | 7–8 | 40–80 | 50–80 |
| RIL8 | 3BS+5AL | 3–5 | 2 | 1–10 | 1–5 |
| Lemhi | - | 9 | 9 | 95–100 | 95–100 |
Based on haplotypes, RIL149 is a fully susceptible RIL (no 3BS, 5AL, 2BS, or 2AS resistance), RIL233 carries only the 3BS resistance, RIL167 carries only the 5AL resistance, and RIL8 carries the 3BS and the 5AL resistances in combination. Lemhi was included as a susceptible control.
Indicated seedling infection types (typical 1–9 scale) are the averages of 5–7 plants.
Indicated adult disease severities (% affected area of flag leaf) are the full ranges observed for four plants.
Low temperature tests were conducted using a 4–20°C cycle, and high temperature tests were conducted using a 10–30°C cycle.
2BS QTL (henceforth QYr.ucw-2BS)
A minor but still significant stripe rust resistance QTL in this population, contributed by UC1110, is located in the centromeric region of the short arm of chromosome 2B (see Fig. 1 and Online Resource 2). Assignment to the short arm is based on the mapping of flanking markers gwm429 and barc230 to 2BS using nullitetrasomic (N2AT2B, N2BT2D) and ditelosomic (Dt2BL) cytogenetic stocks (Sears 1954; Sears and Steinitz-Sears 1978). In terms of disease severity, the main effect of this QTL accounts for 4.5% of the observed variation in the field and is associated with an average reduction in disease severity of 53.6%, compared to fully susceptible lines. As indicated by Fig. 2 and Fig. 3 (and Online Resource 4, Fig. S4 and Fig. S5), this QTL combines well with the ones on 5AL and 2AS to enhance the level of protection in the field. In the presence of the relatively stronger 3BS resistance, however, the contribution of the 2BS QTL is marginal for this population over the years of this study.
QYr.ucw-2BS validation
While no independent populations were developed to validate the presence and boundaries of this QTL, a late-generation (F5) RIL found to be segregating for reaction type provides support for the role played by this region of chromosome 2BS in stripe rust resistance. Specifically, in 2007, one single-seed descent line in the mapping population was observed to be segregating for stripe rust resistance (reaction type 5–6 and reaction type 10–11, respectively). Seed from two plants within this line, representing the two reaction types observed, were harvested and advanced as two separate RILs (RIL193 and RIL194). Subsequent genotyping revealed these two RILs to be near-isogenic lines (NILs), monomorphic at 94.1% of the 559 mapped loci screened in this population. The remaining 5.9% polymorphic loci are concentrated in a 24.5 cM region on chromosome 2B (including the centromeric 2BS QTL region) and a small 3.7 cM region on chromosome 7AS (see Online Resource 3, Worksheet 2). Since these two NILs, based on their haplotypes, lack the 3BS, 5AL, and 2AS resistances, their relative performances over the three years of this study provide information on the putative role of this region on chromosome 2BS. An ANOVA comparing disease severity between these two NILs, using year and block as classification variables, lends strong support to the existence of a stripe rust QTL on 2BS (R2 = 99.8%, P < 0.0001) and provides a more accurate estimate of its effect in a susceptible UC1110 / PI610750 background (average RIL193 severity = 99.0%, average RIL194 severity = 68.3%). Assuming the small region on 7AS to play no role (an assumption backed by the four years of data, see Fig. 1), comparison of these NILs supports both the presence of a stripe rust QTL on 2BS as well as the reliability of flanking marker gwm429 (see Online Resource 5, Table S2).
QYr.ucw-2BS dissection
Like the 3BS and 5AL QTL described above, the flanking markers for this relatively large 24.0 cM QTL region (see Fig. 1 and Online Resource 2) are based on the characterization of RILs carrying recombination events close to its peak. The distal flanking marker (gwm429) is set by the phenotype of RIL150, a fully susceptible line with a recombination event between gwm429 and peak marker wmc474 and a clear absence of the 2BS resistance (91% average disease severity, 10.9 average reaction type). Further evidence for gwm429 as a distal flanking marker is provided by RILs 193 and 194 (see previous section). Similarly, the proximal flanking marker (barc230) is set by the phenotype of RIL72, a fully susceptible line with a recombination event between barc230 and peak marker wmc474 and a clear absence of the 2BS resistance (98% average disease severity, 10.8 average reaction type). Dissection data for this QTL is summarized in Online Resource 5 (Table S2).
2AS QTL (henceforth QYr.ucw-2AS)
A very minor but still marginally significant stripe rust resistance QTL in this population, contributed by the synthetic derivative PI610750, is located in the short arm of chromosome 2A (see Fig. 1 and Online Resource 2). Assignment to the short arm is based on the mapping of cfa2201, a marker proximal to the defined QTL region, to 2AS using nullitetrasomic (N2AT2B, N2BT2D) and ditelosomic (Dt2AS) cytogenetic stocks (Sears 1954; Sears and Steinitz-Sears 1978). In terms of disease severity, the main effect of this QTL accounts for a very small 2.3% of the observed variation in the field and an average reduction in disease severity of 31.2%, compared to fully susceptible lines. As indicated by Fig. 2 and Fig. 3 (and Online Resource 4, Fig. S4 and Fig. S5), this QTL combines well with the ones on 5AL and 2BS to enhance the level of protection in the field. In the presence of the relatively stronger 3BS resistance, however, the contribution of QYr.ucw-2AS is undetectable for this population over the years of this study.
Peak LOD scores which surpassed the threshold (LOD 3.3) over multiple seasons and in both phenotypic datasets (reaction type and disease severity) dictated the inclusion of this QTL in the ANOVA model used here, but no subsequent attempt was made in this study to validate its presence or boundaries in independent populations. The flanking markers for this very large 34.3 cM QTL region (see Figure 1 and Online Resource 2) are based on drops in two LOD units from the peaks of the averaged curves for both phenotypic datasets and therefore do not demarcate this QTL region with the certainty of the flanking markers for the 3BS, 5AL, or 2BS QTL regions.
Discussion
Through this mapping study of the sources of genetic resistance to broadly virulent post-2000 races of PST segregating in the population UC1110/PI610750, a resistance QTL of large effect (QYr.ucw-3BS) was identified and validated on the short arm of chromosome 3B. Preliminary dissection of this QTL narrowed its size to less than 4 cM. Despite its large average effect (> 80% reduction in disease severity, Fig. 3), QYr.ucw-3BS is indicated by its lack of effect against PST-100 at the seedling stage and its combinability with Yr48 to be a partial adult plant resistance gene. As such, QYr.ucw-3BS may prove valuable in breeding strategies for resistance based on combinations of partial resistance genes, particularly if it can be incorporated into a cassette of such genes in the 3BS region (see next section). As the possible race-specificity of QYr.ucw-3BS has consequences in terms of its usefulness in providing durable resistance to stripe rust, further characterization of this QTL is certainly needed. Toward this end, efforts are underway to dissect this QTL and eventually clone the underlying gene.
This mapping study also identified and validated the presence of Yr48, a gene underlying the partial stripe rust resistance QTL on the long arm of chromosome 5A. Preliminary dissection of this QTL narrowed its size to less than 5 cM. Considering the average levels of reaction type and disease severity observed in the field among RILs carrying only the Yr48 resistance (6.2 and 33%, respectively) and the good combinability of this gene with the other three detected QTL in this population, Yr48 also appears to be a partial rather than a major, race-specific resistance gene. In further support of this claim, controlled inoculations with PST-100 showed a lack of resistance at the seedling stage but a significant contribution toward effective resistance at the adult plant stage; in combination with QYr.ucw-3BS, it also provided resistance at the seedling stage (Table 3). Given the stability of the partial resistance provided by this gene across the years of this study, its relatively exotic source (synthetic derivative PI610750), and the effective role it plays in the gene combinations investigated in this study, Yr48 represents a new line of defense that may be of interest to wheat breeders. As with QYr.ucw-3BS, further characterization of Yr48 is certainly needed and efforts to clone the underlying gene are underway.
Though it has been validated and is also likely, based on its small effect and its good combinability with other QTL, to be a partial resistance gene, the smaller QTL on 2BS (QYr.ucw-2BS) requires further dissection (i.e. finer mapping) and characterization to be of practical use to breeding programs. The very small QTL on 2AS (QYr.ucw-2AS), potentially of little interest to breeding programs due to the magnitude of its effect, must be validated and more precisely mapped before its usefulness can be ascertained. Due to its relatively small effect on stripe rust reaction and severity, the development of near-isogenic lines will likely be required for validating and dissecting this QTL, an endeavor beyond the scope of this initial mapping study and perhaps not worth the investment, at least in terms of developing easily deployable packages of durable resistance.
With regard to methodology, this study illustrates the essential equivalence, in the context of QTL mapping, of scoring adult plant reactions to PST in the field using a canopy-wide reaction type scale versus the more common leaf-specific disease severity scale. In this population, the reaction type scale captured a higher proportion of the variation in stripe rust resistance (71.8%) than the disease severity scale (64.5%); and the QTL analyses resulted in equal or larger LOD scores (Fig. 1), suggesting a slight improvement in scoring phenotype. Moreover, the season-to-season correlations of RIL phenotypes were found to be consistently higher using the reaction type scale (Online Resource 4, Figure S2), further suggesting that such a scale, by integrating both infection severity and infection type in a single number, may be more robust under environmental variation and thus more suited to a multi-year, multi-location field evaluation of adult plant resistance.
The coincidence of the QTL peaks of both scoring methods is worth mentioning due to the fact that, in our experience, disease evaluation using a reaction type scale is much less time intensive, requiring less than half the amount of time per line, than the finer-scale disease severity evaluation based on percent leaf area. As the cost and speed of high-throughput genotyping continue to improve, any potential gains in phenotyping efficiency should be given consideration in order to accelerate the gene discovery process. While a crude reaction type scale is likely not appropriate for subsequent gene characterization work, the nuances of which may be best captured by disease severity scales, areas under the disease progress curve, histological studies, or other phenotypes, this study indicates that it may be sufficient for initial QTL detection and mapping work in some cases. It is possible, too, that the relative performance of the scales may offer some insight into the character of resistance genes. For example, the fact that QYr-ucw.3BS explains over twice as much of the total phenotypic variation in the reaction type scale than the disease severity scale, while Yr48 explains over twice as much in the disease severity scale than the reaction type scale, may be an indication of the differences in mechanism underlying these two sources of resistance.
Gene postulation
QYr.ucw-3BS
Based on an allelism test and the absence of Sr2 in UC1110 (donor of QYr.ucw-3BS), the gene underlying the 3BS QTL appears to be different from Yr30/Sr2. Several pieces of evidence also suggest that the detected QTL on 3BS carries a different gene than Yrns-B1. To begin, the adult plant resistance gene Yrns-B1 has been mapped 2.5 cM proximal of gwm493 (Börner et al. 2000; Khlestkina et al. 2007); in this study, in contrast, QYr.ucw-3BS was mapped 3.6 cM distal of gwm493. Second, in a screen of 23 German and UK wheat lines, the gwm533.1 allele fragment associated with Yrns-B1 resistance was found to be 117 bp in size while the resistance on 3BS from UC1110 is associated with a PCR fragment size of 142 bp. In terms of both mapped location and associated allele size, it appears that the 3BS QTL detected in this population may correspond to a stripe rust resistance gene that is different from Yrns-B1.
In another recent paper (Bansal et al. 2010), a QTL for stripe rust resistance carried by cv. Rubric (designated YrRub and likely allelic with Yr4) was also mapped to the distal region of chromosome 3BS. However, YrRub was mapped 10.1 ± 4.2 cM distal of gwm533.1, a distance that suggests the location of YrRub to be outside of the validated QYr.ucw-3BS region described here. Further support for the hypothesis that YrRub and QYr.ucw-3BS are different resistance genes is provided by the fact that YrRub is effective against post-2002 Australian PST races at the seedling stage (Bansal et al. 2010), a characteristic not observed for QYr.ucw-3BS (Table 3).
Despite the evidence presented here, the high density of rust resistance genes in the distal 3BS region means significant work remains to determine the potential novelty of QYr.ucw-3BS. Nevertheless, these results raise the distinct possibility of engineering, through simple recombination, a deployable cassette of stripe rust resistance genes in this region of the short arm of chromosome 3B. Specifically, in terms of packages of resistance based on stacked adult plant resistance genes, the combination of QYr.ucw-3BS and Yrns-B1, along with the Yr30/Sr2 complex, presents a promising subject of investigation.
Yr48
As for the 5AL QTL identified in this study, the only other stripe rust resistance gene reported in the long arm of chromosome 5A is Yr34 (Bariana et al. 2006). But because this gene maps 27.9 cM distal of the distal flanking marker in this study (gwm291), the QTL in this study appears to contain a different gene, hereby designated Yr48 (McIntosh et al. 2010). The additional fact that the adult plant reaction associated with Yr34 is comparatively quite resistant (10% R-MR, compare with Fig. 3) also supports this conclusion, though in and of itself this is not sufficient evidence due to the presence of different races and environmental conditions in the two studies.
QYr.ucw-2BS
Numerous stripe rust resistance genes have been mapped to chromosome 2B (e.g. Yr5, Yr7, Yr27, Yr31, Yr41, Yr43, Yr44, YrP81, YrSP, YrSte, YrV23, and QYrlo.wpg-2BS), and no attempt was made in this initial mapping study to establish their relationships with QYr.ucw-2BS. It is worth noting, however, that all but one (QYrlo.wpg-2BS) of these previously reported genes are known race-specific, all-stage resistance genes (Chen 2005; Cheng and Chen 2008; Luo et al. 2005; Pu et al. 2010; Sui et al. 2009). As with QYr.ucw-3BS and Yr48, the partial resistance provided by QYr.ucw-2BS to a mixture of broadly-virulent PST races (Table 1) suggests that it may not be a major gene, and seedling inoculations with appropriate races should prove useful in distinguishing it from the many all-stage resistance genes in the region. The one previously mapped and validated QTL for adult plant resistance to stripe rust is QYrlo.wpg-2BS, a QTL found to be most strongly associated with wmc474 (135 bp fragment1) from cv. Louise (Carter et al. 2009). The peak association of QYr-ucw-2BS with this same marker (wmc474, 136 bp fragment) suggests that it may be allelic to QYrlo.wpg-2BS; certainly further study is needed to determine the relationship between these two QTL. A small but consistent secondary QTL peak on 2BS, distal of flanking marker gwm429 (see Fig. 1), suggests that yet another gene may be present and segregating in the mapping population described in this study; but in terms of effect sizes, this study indicates that future investigation should prioritize QYr.ucw-3BS, Yr48, and possibly QYr.ucw-2BS over this other small QTL on 2BS or the minor QTL on 2AS, 3DS, or 7DL.
Breeding and applications
To be durable, a stripe rust resistance gene, or more likely a suite of resistance genes deployed in combination, must offer protection not only against current PST races but also against future races as they arise and gain in frequency. Durability, then, is intimately related to the concept of race-specificity; and until the mechanisms of resistance are better understood, the durability of any gene or combination of genes will elude prediction and remain a simple matter of “time will tell.” Nevertheless, the consistent detection of the resistance QTL identified in this study, particularly QYr.ucw-3BS, Yr48, and QYr.ucw-2BS, is an indication of their promise. Importantly, for four years, these QTL provided significant and consistent levels of protection against a diverse and evolving natural population of broadly virulent post-2000 PST races (see Table 1), a fact which recommends them for further characterization and utilization.
With narrow QTL regions well-defined by publicly available flanking molecular markers, both QYr.ucw-3BS and Yr48 are suited for immediate use by interested marker-assisted breeding programs. In terms of donor lines, RIL148 (GSTR 13606) and RIL191 (GSTR 13634) are recommended as adapted spring habit breeding lines carrying all three of the larger resistance QTL described in this paper (on 3BS, 5AL, and 2BS). For research programs interested in further basic characterization of these resistances, RIL140 (GSTR 13600) and RIL233 (GSTR 13664) are recommended as sources of the 3BS resistance alone (i.e. they are susceptible for 5AL and 2BS). RIL4 (GSTR 13504) and RIL167 (GSTR 13618) are recommended as sources of the 5AL resistance (Yr48) alone.
Despite the relatively exotic origin of the synthetic derivative PI610750, the chromosomal lengths in the developed genetic map indicate no significant barriers to recombination in this population. In fact, the only large collapse of genetic distance was found in the short arm of chromosome 1B, a suppression of recombination due to the presence of the 1RS translocation in PI610750, not its synthetic pedigree. While it is possible that the wide-cross origin of PI610750 could affect recombination rates at a finer scale in certain regions of the genome, the overall size of this genetic map and the high rate of marker polymorphism (44% of the initial 1,009 SSR’s screened on the parents) suggest not only that this population is well-suited for QTL mapping and trait dissection and that the RILs listed above may be utilized by breeding programs as QTL donor lines without restrictions.
Finally, the co-location of resistance QTL with major genes controlling development can present problems to breeders interested in their introgression into adapted germplasm. With this in mind, it is important to point out that, although Yr48 is completely linked to the vernalization locus Vrn2, this particular locus (VRN-A2) has been demonstrated to be non-functional in cultivated tetraploid wheat (Distelfeld et al. 2009), meaning that the resistance associated with Yr48 is most likely not an effect of development and that its deployment should have no significant impact on vernalization requirement. In similar vein, although QYr.ucw-2BS maps to the same chromosome arm as the photoperiod gene Ppd-B1, the mapped position of Ppd-B1 is 5.4 cM proximal of gwm257 (Mohler et al. 2004), a marker which is over 10 cM distal of gwm429 (the distal flanking marker for the QYr-ucw.2BS region) and over 30 cM distal of wmc474 (the peak marker for the QYr-ucw.2BS region). These genetic distances indicate that there is no causal link between the QYr-ucw.2BS resistance and Ppd-B1 and no obstacle to recombination between the two loci. Due to the large size and unvalidated boundaries of QYr-ucw.2AS region, the relationship of this minor source of resistance to the Ppd-A1 photoperiod gene (mapped within 3 cM of wmc177; Wilhelm et al. 2009) is less certain.
Supplementary Material
Acknowledgments
This project was supported by the National Research Initiative Competitive Grants no. 2009-65300-05640 and the Triticeae-CAP proposal 2010-04348 from the USDA National Institute of Food and Agriculture. I. Lowe received valuable support through the Graduate Student Research Fellowship program of the UC Davis Department of Plant Sciences. The authors thank O. Chicaiza, Z. Abate, A. Distelfeld, D. Fu, F. Paraiso, C. Uauy, A. Wan, D. Feltus, M. Osenga, V. Talbott, and especially X. Zhang for excellent technical assistance.
Abbreviations
- AFLP
Amplified fragment length polymorphism
- ANOVA
Analysis of Variance
- cM
centiMorgan (in this population, Kosambi)
- DArT
Diversity array technology
- EST
Expressed sequence tag
- HTAP
High-temperature adult plant
- LOD
Log of the odds
- NIL
Near-isogenic line
- PST
Puccinia striiformis Westend. f. sp. tritici Eriks
- QTL
Quantitative trait locus or quantitative trait loci, depending on context
- RIL
Recombinant inbred line
- SSR
Simple sequence repeat (microsatellite)
- 3BS
The short arm of chromosome 3B
- 5AL
The long arm of chromosome 5A
- 2BS
The short arm of chromosome 2B
- 2AS
The short arm of chromosome 2A;
Footnotes
The reported fragment length of 154 bp has been adjusted here for the 19-bp M13 tail for the sake of comparison.
Concise Descriptions of Online Resources
Online Resource 1. Detailed description, with images, of the parental phenotypes and the 11-point reaction type disease evaluation scale used in this study
Online Resource 2. Complete genetic map for the UC1110 / PI610750 mapping population, including detailed notes and summary QTL information
Online Resource 3. Searchable spreadsheet of the complete UC1110 / PI610750 genetic map, including all genotypic and phenotypic data used in this study; also GSTR assignments of deposited lines
Online Resource 4. Supplementary figures (S1-S6): Correlation scatterplots among replications within each year, correlation scatterplots between years, LOD plots of the sub-threshold 3D and 7AL QTL regions, QTL interaction plots (11-point reaction type), bar charts of effects of isolated QTL regions (reaction type), and bar chart of effects of 3BS and 5AL QTL regions by year
Online Resource 5. Supplementary tables (S1-S3) summarizing validation and dissection data for the 3BS, 5AL, and 2BS QTL regions
References
- Bansal U, Hayden M, Gill M, Bariana H. Chromosomal location of an uncharacterised stripe rust resistance gene in wheat. Euphytica. 2010;171:121–127. [Google Scholar]
- Bariana H, Parry N, Barclay I, Loughman R, McLean R, Shankar M, Wilson R, Willey N, Francki M. Identification and characterization of stripe rust resistance gene Yr34 in common wheat. Theoretical and Applied Genetics. 2006;112:1143–1148. doi: 10.1007/s00122-006-0216-3. [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B. QTL Cartographer Version 1.17. 2004 http://statgen.ncsu.edu/qtlcart.
- Börner A, Röder MS, Unger O, Meinel A. The detection and molecular mapping of a major gene for non-specific adult-plant disease resistance against stripe rust (Puccinia striiformis) in wheat. Theor Appl Genet. 2000;100:1095–1099. [Google Scholar]
- Carter AH, Chen XM, Garland-Campbell K, Kidwell KK. Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar ‘Louise’. Theor Appl Genet. 2009;119:1119–1128. doi: 10.1007/s00122-009-1114-2. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Wu LR, Liu TG, Xu SC, Jin SL, Peng YL, Wang BT. Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 2009;93:1093–1101. doi: 10.1094/PDIS-93-11-1093. [DOI] [PubMed] [Google Scholar]
- Chen X, Penman L, Wan A, Cheng P. Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Canadian Journal of Plant Pathology. 2010 [Google Scholar]
- Chen X, Soria MA, Yan G, Sun J, Dubcovsky J. Development of user-friendly PCR markers for wheat stripe rust resistance gene Yr5. Crop Sci. 2003;43:2058–2064. [Google Scholar]
- Chen XM, Moore M, Milus EA, Long DL, Line RF, Marshall D, Jackson L. Wheat stripe rust epidemics and races of Puccinia striiformis f. sp tritici in the United States in 2000. Plant Dis. 2002;86:39–46. doi: 10.1094/PDIS.2002.86.1.39. [DOI] [PubMed] [Google Scholar]
- Chen XM. Epidemiology and control of stripe rust (Puccinia striiformis f. sp tritici) on wheat. Canadian Journal of Plant Pathology. 2005;27:314–337. [Google Scholar]
- Chen XM. Challenges and solutions for stripe rust control in the United States. Aust J Agr Res. 2007;58:648–655. [Google Scholar]
- Cheng P, Chen X. Molecular mapping of a gene for resistance to stripe rust in spring wheat cultivar IDO377s. APS Annual Meeting. Phyto. 2008;98:S38. doi: 10.1007/s00122-010-1302-0. [DOI] [PubMed] [Google Scholar]
- Dimmock JPRE, Gooding MJ. The influence of foliar diseases, and their control by fungicides, on the protein concentration in wheat grain: a review. Journal of Agricultural Science. 2002;138:349–366. [Google Scholar]
- Hodson D, Nazari K. Serious outbreaks of wheat stripe or yellow rust in Central and West Asia and North Africa. [Accessed June 2010];Borlaug Global Rust Initiative. 2010 http://www.globalrust.org/traction?type=single&proj=Pathogen&sort=2&rec=206.
- Hovmøller MS, Walter S, Justesen AF. Escalating threat of wheat rusts. Science. 2010;329:369. doi: 10.1126/science.1194925. [DOI] [PubMed] [Google Scholar]
- Hovmøller MS, Yahyaoui AH, Milus EA, Justesen AF. Rapid global spread of two aggressive strains of a wheat rust fungus. Mol Ecol. 2008;17:3818–3826. doi: 10.1111/j.1365-294X.2008.03886.x. [DOI] [PubMed] [Google Scholar]
- Jackson LF, Dubcovksy J, Gallagher LW, Chicaiza O, Stewart D, Gibbs LK, Prato-Mayo D, Kirby D, Carlson H, Canevari M, Marsh B, Meister H, Munier D, Orloff S, Roberts B, Schmierer J, Vargas R, Wilson R, Wright S. 2003 Regional barley, common and durum wheat, triticale, and oat performance tests in California. [Accessed June 2010];Cal Coop Ext Agr Prog Rep. 2003 :286. http://agric.ucdavis.edu/crops/cereals/2003/oct2003_apr286.htm.
- Khlestkina E, Röder M, Unger O, Meinel A, Börner A. More precise map position and origin of a durable non-specific adult plant disease resistance against stripe rust (Puccinia striiformis) in wheat. Euphytica. 2007;153:1–10. [Google Scholar]
- Kota R, Spielmeyer W, McIntosh RA, Lagudah ES. Fine genetic mapping fails to dissociate durable stem rust resistance gene Sr2 from pseudo-black chaff in common wheat (Triticum aestivum L.) Theor Appl Genet. 2006;112:492–499. doi: 10.1007/s00122-005-0151-8. [DOI] [PubMed] [Google Scholar]
- Lincoln SE, Daly MJ, Lander ES. MAPMAKER/EXP Version 3.0. 1993 http://linkage.rockefeller.edu/soft/mapmaker.
- Lowe I, Cantu D, Dubcovsky J. Durable resistance to the wheat rusts: Integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica. 2010 doi: 10.1007/s10681-010-0311-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo PG, Ren ZL, Zhang HQ, Zhang HY. Identification, chromosome location, and diagnostic markers for a new gene (YrCN19) for resistance to wheat stripe rust. Phytopathology. 2005;95:1266–1270. doi: 10.1094/PHYTO-95-1266. [DOI] [PubMed] [Google Scholar]
- Markell SG, Milus EA. Emergence of a novel population of Puccinia striiformis f. sp. tritici in eastern United States. Phytopathology. 2008;98:632–639. doi: 10.1094/PHYTO-98-6-0632. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC. [Accessed June 2010];Catalogue Of Gene Symbols For Wheat: 2010 Supplement. 2010 http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2010.pdf.
- Milus EA, Kristensen K, Hovmøller MS. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp tritici causing stripe rust of wheat. Phytopathology. 2009;99:89–94. doi: 10.1094/PHYTO-99-1-0089. [DOI] [PubMed] [Google Scholar]
- Mohler V, Lukman R, Ortiz-Islas S, William M, Worland A, van Beem J, Wenzel G. Genetic and physical mapping of photoperiod insensitive gene Ppd-B1 in common wheat. Euphytica. 2004;138:33–40. [Google Scholar]
- Mujeeb-Kazi A, Gilchrist LI, Villareal RL, Delgado R. Registration of ten wheat germplasm lines resistant to Septoria tritici leaf blotch. Crop Sci. 2000;40:590–591. [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, Lagudah E, Somers D, Kilian A, Alaux M, Vautrin S, Berges H, Eversole K, Appels R, Safar J, Simkova H, Dolezel J, Bernard M, Feuillet C. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Pu ZJ, Chen GY, Wei YM, Yang WY, Yan ZH, Zheng YL. Identification and molecular tagging of a stripe rust resistance gene in wheat line P81. Plant Breeding. 2010;129:53–57. [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. The aneuploids of common wheat. Univ Mo Agric Exp Stn Res Bull. 1954;572:1–58. [Google Scholar]
- Sears ER, Steinitz-Sears LM. The telocentric chromosomes of common wheat. In: Ramanujam S, editor. Proceedings of the 5th International Wheat Genetics Symposium, New Delhi, 23–28 February 1988; New Delhi, India. Indian Society of Genetics and Plant Breeding; 1978. pp. 389–407. [Google Scholar]
- Sherman JD, Weaver DK, Hofland ML, Sing SE, Buteler M, Lanning SP, Naruoka Y, Crutcher F, Blake NK, Martin JM, Lamb PF, Carlson GR, Talbert LE. Identification of novel QTL for sawfly resistance in wheat. Crop Sci. 2010;50:73–86. [Google Scholar]
- Singh RP, Nelson JC, Sorrells ME. Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 2000;40:1148–1155. [Google Scholar]
- Smith RCG, Heritage AD, Stapper M, Barrs HD. Effect of stripe rust (Puccinia striiformis West.) and irrigation on the yield and foliage temperature of wheat. Field Crop Res. 1986;14:39–51. [Google Scholar]
- Spielmeyer W, Sharp PJ, Lagudah ES. Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.) Crop Sci. 2003;43:333–336. [Google Scholar]
- Suenaga K, Singh RP, Huerta-Espino J, William HM. Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology. 2003;93:881–890. doi: 10.1094/PHYTO.2003.93.7.881. [DOI] [PubMed] [Google Scholar]
- Sui XX, Wang MN, Chen XM. Molecular mapping of a stripe rust resistance gene in spring wheat cultivar Zak. Phytopathology. 2009;99:1209–1215. doi: 10.1094/PHYTO-99-10-1209. [DOI] [PubMed] [Google Scholar]
- Sun G, Fahima T, Korol A, Turpeinen T, Grama A, Ronin Y, Nevo E. Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat Triticum dicoccoides. Theor Appl Genet. 1997;94:622–628. [Google Scholar]
- Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor Appl Genet. 2009;118:285–294. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- Xue S, Zhang Z, Lin F, Kong Z, Cao Y, Li C, Yi H, Mei M, Zhu H, Wu J, Xu H, Zhao D, Tian D, Zhang C, Ma Z. A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor Appl Genet. 2008;117:181–189. doi: 10.1007/s00122-008-0764-9. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.