Abstract
Excess synapses formed during early postnatal development are pruned over an extended period, while the remaining synapses mature. Synapse pruning is critical for activity-dependent refinement of neuronal connections and its dysregulation has been found in neurodevelopmental disorders such as autism spectrum disorders; however, the mechanism underlying synapse pruning remains largely unknown. As dendritic spines are the postsynaptic sites for the vast majority of excitatory synapses, spine maturation and pruning are indicators for maturation and elimination of these synapses. Our previous studies have found that dendritically localized mRNA for brain-derived neurotrophic factor (BDNF) regulates spine maturation and pruning. Here we investigated the mechanism by which dendritic Bdnf mRNA, but not somatically restricted Bdnf mRNA, promotes spine maturation and pruning. We found that neuronal activity stimulates both translation of dendritic Bdnf mRNA and secretion of its translation product mainly as proBDNF. The secreted proBDNF promotes spine maturation and pruning, and its effect on spine pruning is in part mediated by the p75NTR receptor via RhoA activation. Furthermore, some proBDNF is extracellularly converted to mature BDNF and then promotes maturation of stimulated spines by activating Rac1 through the TrkB receptor. In contrast, translation of somatic Bdnf mRNA and the release of its translation product mainly as mature BDNF are independent of action potentials. These results not only reveal a biochemical pathway regulating synapse pruning, but also suggest that BDNF synthesized in the soma and dendrites is released through distinct secretory pathways.
Keywords: Brain-derived neurotrophic factor, local protein synthesis, spine maturation, spine pruning, TrkB, p75NTR
Introduction
Dendritic spines are the postsynaptic sites for the vast majority of excitatory synapses (Harris, 1999). In many cortical areas of humans and other mammals, spine density increases over a short period in early postnatal life, followed by an extended period when spine numbers are reduced to reach mature levels. During this pruning phase, up to 40% of spines are selectively eliminated while the remaining spines mature and change in morphology from long and thin to short and stubby (Grutzendler et al., 2002; Huttenlocher, 1979; Marin-Padilla, 1967; Rakic et al., 1986; Zuo et al., 2005). Spine maturation and spine pruning are dependent upon neuronal activity and are required for the refinement of neuronal connections in the developing brain (Churchill et al., 2002; Ethell and Pasquale, 2005; Mataga et al., 2004; Zuo et al., 2005). However, the mechanism governing spine maturation is not completely understood and very little is known about the molecular mechanism underlying spine pruning.
One protein known to be important for the control of spine maturation and pruning is fragile X mental retardation protein (FMRP). Its loss, due to transcriptional silencing, results from the expansion of CGG repeats in the 5′ untranslated region (UTR) of the FMR1 gene. This expansion causes fragile X syndrome, the most common form of inherited mental retardation (Bagni and Greenough, 2005). Neurons in patients with fragile X syndrome have a higher density of dendritic spines and their dendritic spines are often longer and thinner, compared to neurons in control subjects (Hinton et al., 1991; Irwin et al., 2001). The same spine dysmorphogenesis has been observed in Fmr1 knockout mice (Comery et al., 1997; Grossman et al., 2006; McKinney et al., 2005). Since FMRP is localized to synapses and is associated with numerous mRNAs, it is thought that FMRP affects the structure and function of postsynaptic sites by regulating dendritic protein synthesis (Weiler et al., 1997; Zalfa et al., 2003).
We have found that brain-derived neurotrophic factor (BDNF) synthesized in dendrites is a key regulator of spine pruning and maturation (An et al., 2008; Kaneko et al., 2012; Orefice et al., 2013). BDNF is synthesized as a precursor (proBDNF), which is cleaved to yield mature BDNF (mBDNF). Cleavage occurs either intracellularly by proconvertases such as furin or extracellularly by the serine protease plasmin and specific matrix metalloproteinases (Lee et al., 2001; Pang et al., 2004). Neurons release both mBDNF and proBDNF (Nagappan et al., 2009; Yang et al., 2009), which interact with the TrkB receptor and the sortilin-p75NTR receptor complex, respectively (Reichardt, 2006; Teng et al., 2005). The rodent and human Bdnf genes produce two populations of mRNA species, one with a short 3′ UTR (~0.35 kb) and the other with a long 3′ UTR (~2.85 kb) (Timmusk et al., 1993). Our previous studies have showed that short 3′ UTR Bdnf mRNA is restricted to cell bodies in cortical and hippocampal neurons, whereas long 3′ UTR Bdnf mRNA is also transported to dendrites for local translation (An et al., 2008). Mice lacking long 3′ UTR Bdnf mRNA display thinner and denser spines on dendrites of CA1 pyramidal neurons in the hippocampus and L2/3 pyramidal neurons in the visual cortex (An et al., 2008; Kaneko et al., 2012). Furthermore, knocking down long 3′ UTR Bdnf mRNA or blocking transport of long 3′ UTR Bdnf mRNA to dendrites inhibits spine maturation and pruning, whereas overexpressing long 3′ UTR Bdnf mRNA enhances spine maturation and pruning in cultured hippocampal neurons (Orefice et al., 2013). Here, we investigated the molecular mechanisms by which dendritically synthesized BDNF regulates pruning and maturation of dendritic spines.
Materials and Methods
Animals and DNA constructs
Pregnant female Sprague Dawley rats were purchased from the Charles River Laboratories. The p75NTR +/− mouse strain was obtained from the Jackson Laboratory (stock #: 002213) and maintained in the C57BL6/J genetic background. Both genders of mice were used in these experiments. The animal care and use committees at the Scripps Research Institute Florida and Georgetown University approved all animal procedures used in this study.
The pActin-GFP construct was previously described (Fischer et al., 1998). The pRK5-myc-Rac1-T17N plasmid (Rac1 dominant-negative construct) was produced by Gary Bokoch and purchased from Addgene (plasmid #12984). The mouse full-length TrkB plasmid was previously described (Fu et al., 2010). The human p75NTR construct was a gift from Dr. Sung Ok Yoon at Ohio State University and was previously described (Yoon et al., 1996). The constructs, pBDNF-A and pBDNF-A*B, were previously described (Orefice et al., 2013). In pBDNF-A*B, the first polyadenylation site is mutated such that this construct only produces long 3′ UTR Bdnf mRNA. To generate the pBDNFAAA-A*B construct, three arginine residues at the proBDNF cleavage site were mutated to three alanine residues in pBDNF-A*B, using the Quickchange Site Directed Mutagenesis Kit (Stratagene).
To generate a construct expressing either shRNA or a scrambled derivative of the shRNA (control shRNA), two pairs of oligonucleotides were designed to produce DNA duplexes and subsequently cloned into the vector pSUPER (Brummelkamp et al., 2002). The following oligonucleotides were used to generate shRNAs.
p75NTR shRNA:
5′-gatccccctcctgccaggacaaacagttcaagagactgtttgtcctggcaggagtttttggaaa-3′
5′-agcttttccaaaaactcctgccaggacaaacagtctcttgaactgtttgtcctggcaggagggg-3′
tPA shRNA:
5′-cgcgtccccctggatccaagacaacatgttcaagagacatgttgtcttggatccagtttttggaaat-3′
5′-cgatttccaaaaactggatccaagacaacatgtctcttgaacatgttgtcttggatccaggggga-3′
TrkB shRNA:
5′-gatcccccggcaacctgcggcacatcttcaagagagatgtgccgcaggttgccgtttttggaaa-3′
5′-agcttttccaaaaacggcaacctgcggcacatctctcttgaagatgtgccgcaggttgccgggg-3′ Scrambled shRNA #1:
5′-gatccccgccggacgcaccaggtagattcaagagatctacctggtgcgtccggctttttggaaa-3′
5′ agcttttccaaaaagccggacgcaccaggtagatctcttgaatctacctggtgcgtccggcggg-3′ Scrambled shRNA #2:
5′-gatccccgaccgccaccggctcatgattcaagagatcatgagccggtggcggtctttttggaaa-3′
5′-agcttttccaaaaagaccgccaccggctcatgatctcttgaatcatgagccggtggcggtcggg-3′
Culture and transfection of primary neurons
Rat hippocampal neurons were cultured and transfected as previously described (Orefice et al., 2013). For the experiments involving the TrkB shRNA, a caspase inhibitor (Boc-Asp(OMe)-fluoromethyl ketone, Sigma Aldrich) was added to cultures every 2–3 days at a final concentration of 25 μM. TrkB-Fc chimera (Sigma Aldrich) was used at a final concentration of 2.5 μg/ml. Fourteen days after transfection (DIV21), TrkB-Fc or vehicle was added to 200 μl of media and applied to cultured neurons in a drop-wise manner. At DIV25 the procedure was repeated. Following 7 days of treatment, cultures were fixed and stained.
Time-lapse imaging of dendritic spines
Cultured rat hippocampal neurons were transfected with pActin-GFP at DIV7 and imaged at DIV17 and DIV23. Z-series time-lapse imaging was performed on a Nikon C2+ confocal microscope (Nikon Instruments Inc.) equipped with a stage-top incubator (INUG2A-TIZ, Tokai Hit Co., LTD, Japan). The chamber was humidified and maintained at 37 ºC with 5% CO2. Images were acquired using a 60X oil immersion objective (NA = 1.4, CFI Plan Apo, Nikon) with Nikon Type A immersion oil under 488 nm of excitation laser and a 525/50 nm emission filter. We applied Nyquist resolution to all time-lapse images, taken at 512 x 512 image size, 0.5 μm step at Z axis, and under a 40-μm pinhole size. Images were collected at 30-minute intervals for ten hours. Each imaged dendritic segment was 50 to 100 μm away from the soma. Z-series images was deconvoluted in DeconvolutionLab (Vonesch and Unser, 2008), an ImageJ plugin, with Tikhonov-Miller algorithm and a Born & Wolf point spread function (PSF) model generated by PSF Generator (Kirshner et al., 2013). Events of spine formation and spine elimination were identified manually.
Local protein synthesis assay
Local protein synthesis constructs were previously described (Liao et al., 2012). A sequence encoding the Src myristoylation peptide (MGSSKSKPK) and a sequence encoding the SV40 large T antigen nuclear localization sequence (PKKKRKV) were added to the 5′ and 3′ ends of a PCR-amplified d1EGFP insert from plasmid pd1EGFP-N1, respectively. The PCR product was inserted into a plasmid downstream of the human synapsin promoter, generating phSYN-myr-d1GFP. The mouse sequences for the Bdnf short (A) and long (A*B) (where the first polyadenylation signal AATAAA was mutated to TTTTTT) 3’ UTRs were cloned into phSYN-myr-d1GFP, generating phSYN-myr-d1GFP-A (myr-d1GFP-A) and phSYN-myr-d1GFP-A*B (myr-d1GFP-A*B). These two constructs were transfected into cultured rat hippocampal neurons at DIV14. One day after transfection, the cultured neurons were treated with 50 μM NMDA or vehicle for 1 hr and then fixed. The longest dendrites of transfected neurons were analyzed by quantification of the fluorescent intensity of a line drawn through the center of the dendrite using NIH ImageJ software, and mean intensity values for each condition were calculated in 50 μm bins.
Production of adeno-associated virus (AAV)
To generate BDNF-expressing AAV constructs, we digested the pAAV-MCS vector (Stratagene) with NotI to remove the 1.7-kb fragment containing the CMV promoter, the β-globin intron, the multiple cloning sites (MCS), and the human growth hormone polyadenylation signal between the two AAV2 inverted terminal repeats. We then subcloned the NotI fragments from pBDNF-A, pBDNF-A*B and pBDNFAAAA*B into the NotI-digested pAAV-MCS vector, generating pAAV-BDNF-A, pAAV-BDNF-A*B and pAAV-BDNFAAA-A*B, respectively. HEK-293T cells were cultured in DMEM growth media (containing 4.5 g/L glucose, 110 mg/L sodium pyruvate, and 4 mM L-glutamine; Life Technologies) supplemented with 10% (v/v) heat-inactivated fetal bovine serum. Plasmid pAAV-BDNF-A or pAAV-BDNF-A*B was co-transfected with helper plasmid pDC2 into HEK-293T cells at >90% confluence. Media were refreshed 24 h post transfection. Cells and media were harvested 48 h post transfection. Viral particles were then purified through 4 consecutive freeze/thaw cycles, followed by centrifugation. For all Western blot experiments, cultures were infected with viral particles at DIV21; cell lysates and/or media were harvested and analyzed at DIV35.
KCl treatment of cultured hippocampal neurons
Fourteen days post AAV infection (DIV35), KCl or vehicle was added to 200 μl of media in the presence of a protease inhibitor cocktail for use in tissue culture media (Sigma Aldrich). This solution was added to cultured neurons in a drop-wise manner, and cultures were incubated in this solution for 30 minutes at 37 °C. Following treatment, media and cell lysates from each well were collected and stored at −80 °C for Western blotting.
Immunocytochemistry
Immunocytochemistry of cultured neurons was performed as previously described (An et al., 2008). The following primary antibodies were used: rabbit anti-GFP (Clontech Laboratories), 1:5,000 dilution; mouse anti-GFP (BD Bioscience), 1:1000 dilution; mouse anti-Myc (Sigma Aldrich), 1:5000 dilution; rabbit anti-Myc (Cell Signaling Technology) 1:1,000 dilution; mouse anti-Flag (Sigma Aldrich); mouse anti-p75 (R&D Systems) 1:1,000 dilution. For protein distribution, fluorescent immunocytochemistry was performed. Appropriate DyLight-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove) and used at a dilution of 1:500. For spine density and morphology analyses, immunocytochemistry was performed using a biotinylated secondary antibody and an avidin-biotin complex solution (Vector Labs). Immunoreactivity was visualized using a solution containing 0.05% DAB and 0.003% H2O2 in 100 mM Tris pH7.5. After colorimetric reaction was complete, cells were rinsed with 100 mM Tris and dehydrated, then coverslips were mounted onto glass slides using DPX mounting medium.
Spine analysis
Density and head width of dendritic spines on cultured hippocampal neurons were analyzed as previously described (Orefice et al., 2013). In brief, treatment groups were assigned in a pseudo-random order, such that at least one well for each condition was represented per culture plate. Spine densities of at least 10 neurons from multiple coverslips were analyzed for each experimental condition, with at least 200 μm of continuous dendrite analyzed per neuron. The majority of analyzed neurons were CA1 pyramidal neurons, as pyramidal neurons with 3–5 main dendritic branch points were selected for analysis. To analyze the morphology of spines, multiple high-magnification images using an oil-immersion lens (60X, NA1.40) were taken along a segment of one main dendrite that is 50–100 μm away from the soma for each neuron. Spine head width was measured using NIH ImageJ software. Spine head width was defined as the maximum width of the spine head. At least 10 neurons from multiple coverslips and approximately 50 spines per neuron were analyzed for each experimental condition. Analyses of spine density and spine head width were performed blindly.
Western blotting
Western blots of the media and cell lysates from cultured neurons were performed using the Odyssey Infrared Imaging System (LI-COR Biosciences). Cell lysates were collected using a lysis buffer containing protease inhibitors for mammalian cell/tissue extracts (Sigma Aldrich). Media samples were immunoprecipitated with Myc antibodies bound to protein G Sepharose beads (Thermo Scientific). The following primary antibodies were used: mouse anti-Myc (Sigma Aldrich), 1:500 dilution; rabbit anti-Myc (Cell Signaling Technology), 1:1,000 dilution; mouse anti-Flag (Sigma Aldrich), 1:1,000 dilution; mouse anti-alpha tubulin (Sigma Aldrich), 1:1,000 dilution; chicken anti-TrkB (Abcam) 1:15,000 dilution; rabbit anti-TrkB (Cell Signaling Technology) 1:1,000 dilution; rabbit anti-phospho TrkB (Cell Signaling Technology) 1:1,000 dilution. Appropriate IRDYE infrared secondary antibodies (LI-COR Biosciences) or biotinylated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were purchased and used at a dilution of 1:10,000.
Golgi impregnation
Golgi staining was performed using the FD Rapid GolgiStain Kit (FD NeuroTechnologies) according to the manufacturer’s protocol. Sections (180 μm thick) were taken throughout the hippocampus of each animal. For spine density, we used Neurolucida software to trace spines along dendrites of granule cells in the anterior hippocampus at 60X magnification and calculated spine density as described above. Spine density for each animal was obtained from at least five neurons in multiple sections, with a total length of 1000 2000 μm. To measure spine morphology, we took images of distal dendrites with similar widths in the anterior hippocampus at 60X magnification, and measured the length and width of dendritic spines located at 50–100 μm away from the soma, as described above using NIH ImageJ Software. At least 10 neurons from multiple sections and approximately 50 spines per neuron were analyzed for each animal.
Active Rho GTPase pull down assays
Active Cdc42, Rac1 and RhoA pull down experiments were performed using the ThermoScientific Active GTPase Pull Down and Detection Kits, according to the manufacturer’s protocol. Cultured hippocampal neurons were lysed and harvested in Lysis/Binding/Wash Buffer. A specific GST-fusion protein that contains a binding domain from a downstream effector protein of each GTPase, along with glutathione agarose resin, was used to specifically pull down active Cdc42, Rac1, and RhoA. Following pull-down assays, Western blotting was performed and samples were probed using an antibody to each GTPase for detection (anti-Cdc42, 1:167 dilution; anti-Rac1, 1:1,000 dilution; anti-RhoA, 1:666 dilution). For each experiment, samples were separated into two aliquots: 100 μg of each sample was used for the pull down assay and 20 μg of each sample was used directly for Western blotting, to determine total levels (active plus non-active) of each GTPase.
Surface biotinylation assay
Surface TrkB experiments were performed using DIV35 hippocampal cultures that had been treated with 50 μM APV for 48 hours, according to a previously described protocol (Elia, 2012). Briefly, on the day of harvest, cells were washed and surface proteins were labeled with Sulfo-NHS-SS-Biotin at 500 μg/ml PBS (Thermo Scientific) on ice with gentle rocking for 10 minutes. After quenching, cells were lysed and centrifuged for 2 minutes to collect proteins. To isolate biotin-labeled (surface) proteins, lysate was added to pre-washed Streptavidin SepharoseTM High Performance resin (GE Healthcare) and incubated for 2 hours at room temperature with occasional mixing. Resin was washed and then incubated with elution buffer at 96 °C for 15 minutes with occasional agitation. Eluted proteins were analyzed for TrkB by Western blotting. Prior to isolation of biotin-labeled proteins, 20 μg of each sample was saved to determine total levels of TrkB using immunoblotting analyses.
Statistical analysis
All data are expressed as mean ± SEM. The significance of differences was tested using unpaired Student’s t test, one-way or two-way ANOVA with post-hoc Bonferroni test. Differences are considered significant for p values <0.05. One or two-way ANOVAs were performed using GraphPad Prism (GraphPad Software).
Results
Activity-dependent maturation and pruning of dendritic spines in vitro
We previously showed that in dissociated rat embryonic hippocampal cultures dendritic spines mature during the 3rd and 4th weeks in vitro, as evidenced by an increase in spine head width and a reduction in spine length. Furthermore, these cultured neurons exhibit a reduction in spine density during the 4th week in vitro (Orefice et al., 2013). To confirm that the reduction in spine density reflects a pruning process, we monitored GFP-labeled dendritic spines via time-lapse imaging at the 17th or 23rd day in vitro (DIV17 or DIV23) (Figures 1A and 1B). Over a 6-hour period, the spine elimination rate was not significantly different from the spine formation rate at DIV17 (Figure 1C), but was higher than the spine formation rate at DIV23 (Figure 1D), indicating that the density reduction observed during the 4th week in this culture system is a result of spine pruning.
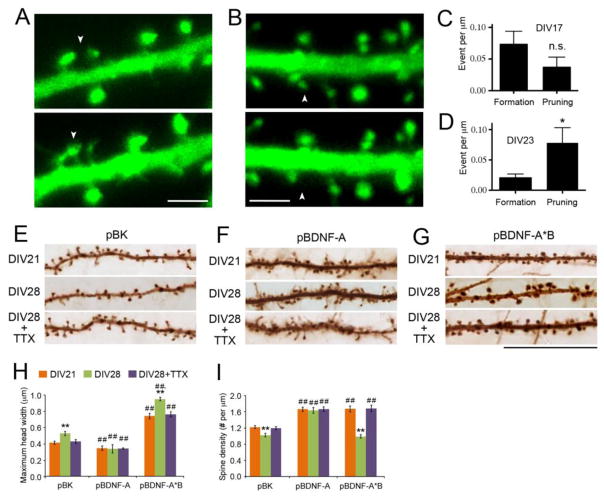
Figure 1. Activity-dependent maturation and elimination of dendritic spines in neuronal culture.
(A and B) Time-lapse imaging of dendritic spines at DIV23. Neurons were transfected at DIV7 with pActin-GFP. The arrows denote one new spine in (A) and one eliminated spine in (B). Scale bars represent 5 μm.
(C and D) The number of spine formation and spine pruning events over a 6-hour period in cultured rat hippocampal neurons at DIV17 and DIV23. Student’s t test: *p < 0.05; n.s., not significant.
(E G) Representative images of dendrites, showing morphology of dendritic spines at DIV21 and DIV28. Neurons were transfected at DIV7 with pActin-GFP and either pBK (empty vector), pBDNF-A or pBDNF-A*B, following one week of 1 μM TTX treatment from DIV21 to 28. Scale bar, 25 μm.
(H and I) Average head width and density of dendritic spines in neurons treated with 1 μM TTX from DIV21 to DIV28 (n=10 neurons/condition). Two-way ANOVA with post-hoc Bonferroni test: **p < 0.01 when compared to DIV21 neurons transfected with the same constructs; ##p < 0.01 when compared to pBK-transfected neurons at the same time point.
Data are reported as mean ± SEM. See also Figure S1.
Neuronal activity plays a key role in maturation and pruning of dendritic spines in the brain (Ethell and Pasquale, 2005; Zuo et al., 2005). To determine whether in vitro development of dendritic spines is also dependent on neuronal activity, we transfected hippocampal neurons at DIV7 with an empty vector (pBK) and a construct expressing actin-GFP to label dendritic spines. We applied 1 μM tetrodotoxin (TTX, an action potential blocker) to cultures at DIV21 and then fixed and analyzed the number and morphology of dendritic spines at DIV28 (Figure 1E). TTX prevented spine head enlargement and spine pruning that normally occurs from DIV21 to DIV28 (Figures 1H and 1I). Thus, cultured hippocampal neurons could be used to study activity-dependent maturation and pruning of dendritic spines.
Our previous work shows that BDNF translated from long 3′ UTR Bdnf mRNA stimulates maturation and pruning of dendritic spines (Orefice et al., 2013). We asked whether this stimulation was also dependent upon neuronal activity, by transfecting neurons at DIV7 with a construct expressing long 3′ UTR Bdnf mRNA (pBDNF-A*B) and treating neurons with TTX from DIV21 to DIV28 (Figure 1G). In agreement with our previous observation (Orefice et al., 2013), neurons overexpressing long 3′ UTR Bdnf mRNA had larger spine heads at DIV21 and DIV28 (Figure 1H) and greater spine pruning from DIV21 to DIV28 (Figure 1I), compared with pBK-transfected neurons. TTX blocked both head enlargement and pruning of dendritic spines from DIV21 to DIV28 in these neurons (Figures 1G–I). In contrast, TTX did not affect the size or density of dendritic spines in pBDNF-A-transfected neurons that overexpress short 3′ UTR Bdnf mRNA (Figures 1F, 1H, and 1I), which impairs trafficking of long 3′ UTR Bdnf mRNA to dendrites and therefore blocks maturation and pruning of dendritic spines (Orefice et al., 2013). In excitatory hippocampal neurons, action potentials are initiated through activation of the NMDA receptor (NMDAR). As expected, the selective NMDAR antagonist APV had the same effect as TTX on maturation and pruning of dendritic spines in both control neurons and BDNF-overexpressing neurons (Figure S1). These results, along with our previous observation that long 3′ UTR Bdnf mRNA is essential for head enlargement and pruning of dendritic spines in vivo and in vitro (An et al., 2008; Kaneko et al., 2012; Orefice et al., 2013), suggest that BDNF translated from the long form of Bdnf mRNA regulates maturation and pruning of dendritic spines in an NMDAR-dependent manner.
NMDAR-dependent synthesis and release of BDNF in neurons
We hypothesized that NMDAR activation might regulate spine maturation and pruning by stimulating translation of long 3′ UTR Bdnf mRNA in dendrites and then secretion of its translation product. To test this, we utilized an in vitro local protein synthesis reporter assay based on GFP expression. Myr-d1GFP-nls reporter constructs used in this study contain four key elements: (1) a sequence encoding a myristoylation peptide (myr) that brings the GFP fusion protein to the plasma membrane; (2) a nuclear localization signal (nls) at the C-terminus that further restricts protein diffusion; (3) a sequence encoding a destabilized GFP with a half life of 1 hour; and, (4) a genomic sequence encoding either the Bdnf long 3′ UTR (myr-d1GFP-A*B) or short 3′ UTR (myr-d1GFP-A). Once this construct is expressed, GFP will attach to the membrane or move into the nucleus to restrict movement, while destabilized GFP will be degraded quickly, thus reporting the site at which mRNA from the construct is translated. This strategy has been successfully employed to examine local synthesis of CaMKIIα and LIMK1 in dendrites (Aakalu et al., 2001; Schratt et al., 2006). Cultured hippocampal neurons at DIV14 were transfected with either myr-d1GFP-A*B or myr-d1GFP-A. One day later, neurons were treated with either vehicle or 50 μM NMDA for 1 hour before fixation, and then analyzed for GFP signal intensity along a main dendrite. We had previously established that long 3′ UTR Bdnf mRNA is trafficked to the dendrites and short 3′ UTR Bdnf mRNA is restricted to the soma (An et al., 2008), and thus we expected to see dendritic changes in local synthesis only in neurons transfected with the long 3′ UTR construct. We found that NMDA significantly increased GFP signal intensity along the dendrites of neurons transfected with myr-d1GFP-A*B, but not myr-d1GFP-A (Figures 2A and 2B). Thus, NMDA stimulates dendritic synthesis of BDNF in hippocampal neurons.
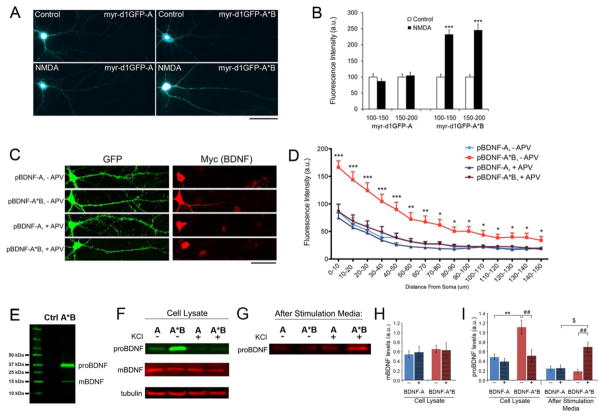
Figure 2. Activity-dependent translation of Bdnf mRNA and release of proBDNF.
(A) Representative whole cell images of cultured rat hippocampal neurons expressing either myr-d1GFP-A or myr-d1GFP-A*B. Neurons were transfected at DIV14 and treated with either vehicle (control) or 50 μm NMDA (NMDA) at DIV15 for one hour and then fixed for analysis. Scale bar, 50 μm.
(B) Quantification of myr-d1GFP fluorescence in dendrites. Fluorescence intensities on distal dendrites (100–150 μm and 150–200 μm away from somata) were measured and normalized to control levels (n=22–25).
(C) Representative whole cell images of cultured rat hippocampal neurons transfected at DIV7 with pActin-GFP and either pBDNF-A or pBDNF-A*B, and treated with either vehicle (− APV) or 50 μm APV (+ APV) from DIV26 to 28. Scale bar, 50 μm.
(D) Quantification of Myc immunoreactivity as an indicator of BDNF levels in dendrites (n=23–29).
(E) Immunoblot analysis of Myc-tagged BDNF in cell lysates from neurons infected with AAV-BDNF-A*B. The Myc tag is at the C-terminus of BDNF. The lysate of un-transduced neurons was used as a negative control (Ctrl).
(F and G) Western blot analysis of Myc-tagged BDNF in cell lysates or media from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B), that were either unstimulated (−) or stimulated with 50 mM KCl (+) for 30 minutes at 37°C.
(H and I) Quantification of mBDNF and proBDNF levels from Western blots represented in F and G (n=8 for cell lysate and n=3 for after-stimulation media).
Data are reported as mean ± SEM. ANOVA with post-hoc Bonferroni test: *p < 0.05, **p < 0.01 and ***p < 0.001 when compared to control; $p < 0.05 and ##p < 0.01 when comparisons were done as indicated. See also Figure S2.
We further confirmed that NMDA receptor activation is required for local synthesis of BDNF in dendrites using BDNF-expressing constructs. We transfected hippocampal neurons at DIV7 with pBK, pBDNF-A or pBDNF-A*B, in combination with pActin-GFP. At DIV26, cultures were treated with either 50 μM APV or vehicle for 48 hours and then immediately fixed and stained with antibodies to GFP (to mark transfected neurons) and Myc (for exogenous BDNF expression) (Figure 2C). APV treatment significantly reduced dendritic levels of BDNF in neurons transfected with pBDNF-A*B, but not neurons transfected with pBDNF-A (Figures 2C and 2D). APV treatment did not significantly change levels of Myc-tagged BDNF in the cell bodies of neurons transfected with either pBDNF-A (217 ± 9 with vehicle vs. 211 ± 6 with APV in arbitrary units, p = 0.5678) or pBDNF-A*B (172 ± 12 with vehicle vs.164 ± 9 with APV in arbitrary units, p = 0.5966). These results suggest that NMDAR activation is necessary for synthesis of BDNF in dendrites, but not in cell bodies of hippocampal neurons.
We then asked whether BDNF translated from long 3′ UTR Bdnf mRNA is released in an activity-dependent manner. A high KCl concentration in the culture medium induces membrane depolarization of cultured neurons, leading to NMDAR activation (Ghosh et al., 1994). Treatment with 50 mM KCl has been shown to induce nerve growth factor release from cultured hippocampal neurons, which is dependent on action potentials (Blochl and Thoenen, 1995). Thus, we employed the KCl treatment to examine activity-dependent BDNF release from cultured hippocampal neurons. We infected cultured hippocampal neurons at DIV21 with AAV-BDNF-A expressing short 3′ UTR Bdnf mRNA or AAV-BDNF-A*B expressing long 3′ UTR Bdnf mRNA. At DIV35, we collected media (termed “conditioned media”) and added neurobasal media with or without 50 mM KCl to the cultures for 30 minutes at 37 °C, in the presence of protease inhibitors. Following the KCl treatment, media (termed “after-stimulation media”) were collected and cell lysates were prepared from the cultures. BDNF in the media was immunoprecipitated with a mouse anti-Myc antibody. We employed immunoblotting to detect BDNF in the cell lysates (Figure 2E) and immunoprecipitates, using a rabbit anti-Myc antibody. Analyses revealed that without stimulation, cell lysates of neurons infected with AAV-BDNF-A contained similar amounts of mBDNF to, but significantly less proBDNF than, neurons infected with AAV-BDNF-A*B (Figures 2F, 2H, and 2I). KCl treatment significantly reduced the amount of proBDNF in cell lysates of neurons infected with AAV-BDNF-A*B, while increasing the amount of proBDNF in the after-stimulation media, compared to unstimulated neurons infected with AAV-BDNF-A*B (Figures 2F, 2G, and 2I). This treatment, however, did not affect levels of mBDNF in lysates from neurons infected with either virus (Figures 2F and 2H). We were unable to detect mBDNF in the media following the 30-min KCl stimulation, likely due to binding of released mBDNF to the cultured neurons (Balkowiec and Katz, 2000). KCl stimulation did not alter levels of proBDNF in either the cell lysates or after-stimulation media from neurons infected with AAV-BDNF-A (Figures 2F, 2G, and 2I). These results indicate that the translation product of long 3′ UTR Bdnf mRNA remains in the proBDNF form inside neurons and its release is dependent upon action potentials.
We previously reported that conditioned media from neurons infected with AAV-BDNF-A had greater amounts of mBDNF than media from neurons infected with AAV-BDNF-A*B (Orefice et al., 2013). To determine whether any synaptic activity is involved in secretion of mBDNF, we treated hippocampal neuron cultures infected with AAV-BDNF-A with TTX or TTX + APV + CNQX (an AMPA/kainate receptor antagonist) for 2 days and then measured the amount of mBDNF in the conditioned media. The released mBDNF accumulated over 2 days to reach a detectable level (Figure S2A). We found that TTX had no effect on secretion of mBDNF (Figure S2); however, blockade of both action potentials and miniature excitatory postsynaptic potentials (mEPSPs) using TTX + APV + CNQX significantly reduced the amount of mBDNF in the conditioned media (Figure S2). This observation indicates that mEPSPs are critical for secretion of mBDNF.
In summary, our results indicate that NMDAR activation is essential for translation of long 3′ UTR Bdnf mRNA in dendrites and release of the translation product in the proBDNF form. In contrast, somatic short 3′ UTR Bdnf mRNA is translated independent of action potentials, and the translation product is mainly released in the form of mature BDNF in a mEPSP-dependent manner. These findings indicate that BDNF translated in dendrites and cell bodies is released from neurons via distinct secretory pathways.
Promotion of spine pruning and maturation by proBDNF via p75NTR
As p75NTR is a component of the high affinity receptor complex for proBDNF (Teng et al., 2005), we reasoned that dendritically synthesized and released proBDNF might interact with p75NTR to mediate spine pruning. We mutated the pBDNF-A*B construct to produce a protease-resistant proBDNF, in which the 3 arginine residues at the cleavage site between the BDNF propeptide and mBDNF were mutated to 3 alanine residues (termed pBDNFAAA-A*B). Therefore, the cleavage site is not recognized by proteases and only proBDNF is produced from this construct (Figure 3A). Indeed, we detected proBDNF, but no mature BDNF, in the cell lysates of HEK293T cells transfected with pBDNFAAA-A*B (Figure 3B).
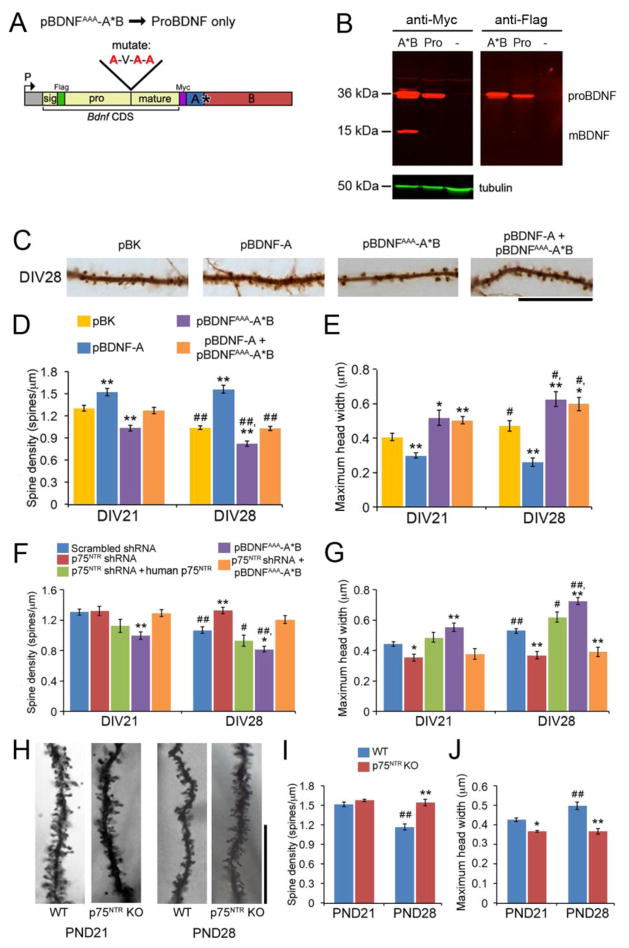
Figure 3. proBDNF derived from long 3′ UTR Bdnf mRNA mediates pruning and maturation of dendritic spines through the p75NTR receptor.
(A) Diagram of the pBDNFAAA-A*B construct. Three arginine residues at the proBDNF cleavage site were changed to three alanine residues.
(B) Western blot analysis of lysates from non-transfected HEK293T cells (-) and HEK293T cells transfected with either pBDNF-A*B (A*B) or pBDNFAAA-A*B (Pro), indicating that the three Arg→Ala changes prevent cleavage of proBDNF (~36 kDa) into mBDNF (~15 kDa).
(C) Representative images of dendrites, showing morphology of dendritic spines at DIV28. Neurons were transfected at DIV7 with pActin-GFP and either pBK (empty vector), pBDNF-A, pBDNFAAA-A*B, or pBDNF-A plus pBDNFAAA-A*B at DIV7. Scale bar, 20 μm.
(D and E) Overexpressing proBDNF promotes pruning and maturation of dendritic spines in cultured hippocampal neurons. Neurons were transfected at DIV7, and their dendritic spines were analyzed at DIV21 and DIV28 (n=10 neurons/condition).
(F and G) p75NTR is required for proBDNF to mediate pruning of dendritic spines. Neurons were transfected at DIV7, and their dendritic spines were analyzed at DIV21 and DIV28 (n=10 neurons/condition).
(H) Representative dendrites of Golgi-stained hippocampal granule cells from WT and p75NTR KO mice at PND21 and PND28. Scale bar, 25 μm.
(I and J) Spine density and spine head width of Golgi-stained hippocampal granule cells from WT or p75NTR KO mice at PND21 and PND28 (n=4 WT and 2 KO animals at PND21; n=4 WT and 3 KO animals at PND28).
Data are reported as mean ± SEM. Two-way ANOVA with post-hoc Bonferroni test: *p < 0.05 and **p < 0.01 when compared to control neurons at the same time point; #p < 0.05 and ##p < 0.01 when compared to an earlier time point with the same treatment or genotype. See also Figure S3.
We transfected cultured hippocampal neurons at DIV7 with either pBK, pBDNF-A, pBDNFAAA-A*B, or pBDNF-A plus pBDNFAAA-A*B, in combination with pActin-GFP. Overexpression of proBDNF from long 3′ UTR Bdnf mRNA (pBDNFAAA-A*B) decreased spine density at both DIV21 and DIV28, compared to control neurons transfected with pBK (Figures 3C and 3D). Co-transfection of pBDNFAAA-A*B with pBDNF-A was able to correct the increased spine density phenotype observed in neurons transfected with pBDNF-A alone (Figures 3C and 3D). Interestingly, overexpression of dendritic proBDNF alone or dendritic proBDNF plus short 3′ UTR Bdnf mRNA (pBDNF-A) caused a small but significant increase in spine head width, compared to control neurons at both DIV21 and DIV28 (Figure 3E). These results suggest that proBDNF plays key roles in spine pruning as well as spine head enlargement.
We further examined the role of proBDNF in spine pruning and maturation by knocking down the expression of its receptor, p75NTR, using shRNA (Figure S3). Expressing p75NTR shRNA, starting at DIV7, increased spine density at DIV28 and decreased spine head width at both DIV21 and DIV28, compared to neurons expressing a scrambled shRNA (Figures 3F and 3G). These effects were reversed by co-expression of human p75NTR that is resistant to p75NTR shRNA (Figures 3F and 3G), indicating that the shRNA specifically targets p75NTR. In the presence of p75NTR shRNA, proBDNF lost its stimulatory effect on spine pruning and spine head enlargement, such that neurons transfected with p75NTR shRNA and pBDNFAAA-A*B had a higher spine density and smaller spine heads at DIV28, compared to neurons transfected with the scrambled shRNA construct or pBDNFAAA-A*B alone (Figures 3F and 3G). These results further support the view that proBDNF stimulates spine pruning and maturation via the p75NTR receptor.
To corroborate our in vitro evidence that proBDNF binds to p75NTR to mediate spine pruning and spine maturation, we performed Golgi staining on brains from wild-type (WT) and p75NTR knockout (KO) mice at postnatal day 21 (PND21) and 28 (PND28) (Figure 3H). At PND21, hippocampal granule cells from WT and p75NTR KO mice did not differ in spine density (Figure 3I), suggesting that spine formation is normal in p75NTR KO mice. From PND21 to PND28, spine density of WT granule cells was significantly reduced, indicating that spine pruning occurs from PND21 to PND28. However, the spine density of p75NTR KO granule cells remained largely unchanged, thus leading to a significantly higher spine density in p75NTR KO granule cells at PND28, compared to WT granule cells (Figure 3I). Granule cells in p75NTR KO mice had significantly smaller spine heads at both PND21 and PND28, compared to WT mice (Figure 3J). These data indicate that p75NTR plays key roles in spine pruning and spine maturation in vivo.
Stimulatory effects of TrkB on spine formation
We previously reported that somatically synthesized BDNF promotes spine formation (Orefice et al., 2013). To determine whether this effect is mediated by the TrkB receptor, we generated an shRNA construct directed at TrkB. The TrkB shRNA reduced endogenous TrkB levels by more than 50% in somata and 70% in dendrites of cultured hippocampal neurons (Figure S4). Because TrkB is important for cell survival, a pan caspase inhibitor was added to cultures every 2–3 days for the duration of these experiments. Expressing TrkB shRNA in cultured hippocampal neurons, beginning at DIV7, decreased spine density at DIV28, compared to expressing a scrambled shRNA (Figure 4A). Co-expressing mouse TrkB that is resistant to the shRNA reversed this effect on spine density (Figure 4A). This reduction in spine density is probably a result of impaired spine formation, because expressing TrkB shRNA, starting at DIV14 when spine density reaches a plateau, did not affect spine density of hippocampal neurons at DIV28, even with overexpression of short or long 3′ UTR Bdnf mRNA (Figure 4C). In support of this view, neurons expressing TrkB shRNA beginning at DIV7 showed a significant reduction in spine density at DIV21, compared to neurons expressing a scrambled control (Figure 4E). Because at DIV21 the majority of spines have not yet been pruned, this result indicates that the decreased spine density of neurons expressing TrkB shRNA is indeed due to deficits in spine formation rather than enhanced spine pruning. Furthermore, expressing TrkB shRNA beginning at DIV7 also significantly reduced spine density of neurons overexpressing long 3′ UTR Bdnf mRNA at DIV21 (Figure 4E). These results indicate that TrkB mediates the stimulatory effect of somatically synthesized BDNF on spine formation.
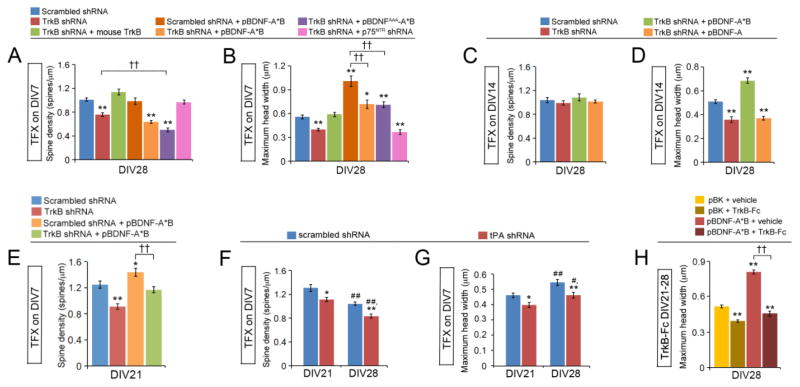
Figure 4. BDNF promotes spine head enlargement through TrkB.
(A and B) TrkB knockdown at DIV7 reduces density and head width of dendritic spines. Neurons were transfected at DIV7, and their dendritic spines were analyzed at DIV28 (n=10 neurons/condition). One-way ANOVA with post-hoc Bonferroni test: *p < 0.05 and **p < 0.01 when compared to neurons expressing scrambled shRNA; ††p < 0.01 when two indicated groups were compared.
(C and D) TrkB knockdown at DIV14 reduces head width of dendritic spines without affecting spine density. Neurons were transfected at DIV14, and their dendritic spines were analyzed at DIV28 (n=10 neurons/condition). One-way ANOVA with post-hoc Bonferroni test: **p < 0.01 when compared to neurons expressing scrambled shRNA.
(E) TrkB knockdown at DIV7 reduces density of dendritic spines at DIV21. Neurons were transfected at DIV7, and their dendritic spines were analyzed at DIV21 (n=10 neurons/condition). One-way ANOVA with post-hoc Bonferroni test: *p < 0.05 and **p < 0.01 when compared to neurons expressing scrambled shRNA; ††p < 0.01 when two indicated groups were compared.
(F and G) Knockdown of tPA with shRNA reduces density and head width of dendritic spines. Neurons were transfected at DIV7, and their dendritic spines were analyzed at DIV21 and 28 (n=10 neurons/condition). Two-way ANOVA with post-hoc Bonferroni test: *p < 0.05 and **p < 0.01 when compared to neurons expressing scrambled shRNA; #p < 0.05 and ##p < 0.01 when compared to DIV21 neurons in same condition.
(H) Removal of extracellular BDNF with TrkB-Fc inhibits spine head enlargement. Neurons were treated with either vehicle or TrkB-Fc (2.5 μg/ml) from DIV21 to 28, and their dendritic spines were analyzed at DIV28 (n=10 neurons/condition). One-way ANOVA with post-hoc Bonferroni test: **p < 0.01 when compared to the non-treated control within the same condition; ††p < 0.01 when indicated groups are compared.
Data are reported as mean ± SEM. See also Figure S4.
Two lines of evidence indicate that TrkB is not essential for dendritically synthesized proBDNF to mediate spine pruning that occurs after DIV21 in our culture system. First, neurons co-transfected with pBDNFAAA-A*B and TrkB shRNA had significantly lower spine density than neurons expressing TrkB shRNA alone at DIV28 (Figure 4A). This indicates that proBDNF mediates spine pruning in the absence of TrkB receptors. Second, expression of TrkB shRNA, starting at DIV14, did not affect spine density of hippocampal neurons at DIV28 (Figure 4C), indicating that pruning occurs normally in the absence of TrkB. Because TrkB and p75NTR have distinct roles in spine formation and spine pruning, expression of both TrkB shRNA and p75NTR shRNA likely cancelled out the opposing effects of each receptor on spine density, leading to a normal spine density in neurons at DIV28 (Figure 4A).
Stimulatory effects of TrkB on spine maturation
Hippocampal neurons transfected with pBDNF-A*B had larger spine heads than neurons transfected with pBDNFAAA-A*B (Figures 1H and 3E; 0.895 ± 0.030 μm for pBDNF-A*B vs. 0.649 ± 0.045 μm for pBDNFAAA-A*B at DIV28, p < 0.001), suggesting that dendritically synthesized BDNF also stimulates spine maturation through TrkB, in addition to p75NTR. Indeed, hippocampal neurons transfected with TrkB shRNA at DIV7 exhibited decreased spine head width in hippocampal neurons at DIV28, as compared to neurons transfected with a scrambled shRNA (Figure 4B). This effect from knocking down TrkB could be reversed with mouse TrkB overexpression (Figure 4B). Knocking down TrkB after DIV14 still produced a similar effect on spine head width (Figure 4D), which is in agreement with the observation that spine heads grow during the 3rd and 4th weeks in our culture system (Orefice et al., 2013). Notably, hippocampal neurons overexpressing BDNF from either pBDNF-A*B or pBDNFAAA-A*B, but not pBDNF-A, in the presence of TrkB shRNA had significantly larger spine heads than control neurons expressing the scrambled shRNA alone (Figure 4B). However, the spine heads of these neurons were still much smaller than those of neurons overexpressing BDNF from pBDNF-A*B in the presence of the scrambled shRNA (Figure 4B). These results indicate that TrkB and p75NTR have non-redundant and additive roles in mediating the effect of dendritically synthesized BDNF on spine head enlargement.
Our results indicate that the translation product of long 3′ UTR Bdnf mRNA is mostly secreted in the form of proBDNF (Figures 2F-I). In order for proBDNF to interact with TrkB to promote spine maturation, it has to be converted to mature BDNF by the extracellular protease plasmin, which is derived from inactive plasminogen. If this hypothesis is true, knocking down tissue plasminogen activator (tPA), which converts plasminogen to plasmin, should increase proBDNF levels and decrease mBDNF levels, leading to fewer and less mature dendritic spines due to increased spine pruning and impaired spine maturation. Indeed, hippocampal neurons expressing a tPA shRNA, which was reported to knock down endogenous tPA by 80% (Wang et al., 2009), had fewer and thinner dendritic spines at DIV21 and DIV28 than neurons expressing a scrambled shRNA (Figures 4F and 4G). To further demonstrate that extracellular mBDNF activates TrkB on the cell surface to promote spine maturation, we added the TrkB-Fc chimera protein to neuronal cultures to remove extracellular mBDNF. Addition of the protein from DIV21 to DIV28 inhibited spine head enlargement in control neurons and neurons overexpressing long 3′ UTR Bdnf mRNA (Figure 4H), indicating that BDNF must be secreted from cells to regulate spine maturation. Taken together, these experiments indicate that the translation product of long 3′ UTR Bdnf mRNA is secreted as proBDNF and extracellularly converted to mBDNF, which binds to the surface TrkB receptors to promote spine maturation.
Activation of Cdc43, RhoA, and Rac1 by BDNF
Three small Rho GTPases (Cdc42, Rac1 and RhoA) have previously been shown to regulate spine morphology and are expressed at high levels in the developing hippocampus (Nakayama et al., 2000). A loss of function mutation in Cdc42 lead to a significant reduction in spine density (Scott et al., 2003); inhibition of Rac1 signaling prevented spine head growth and increased the proportion of long, thin spines (Tashiro and Yuste, 2004); overexpression of RhoA caused a reduction in spine density (Nakayama et al., 2000). We therefore investigated the possibility that TrkB and p75NTR regulate spine formation, spine maturation, and spine pruning by activating these GTPases.
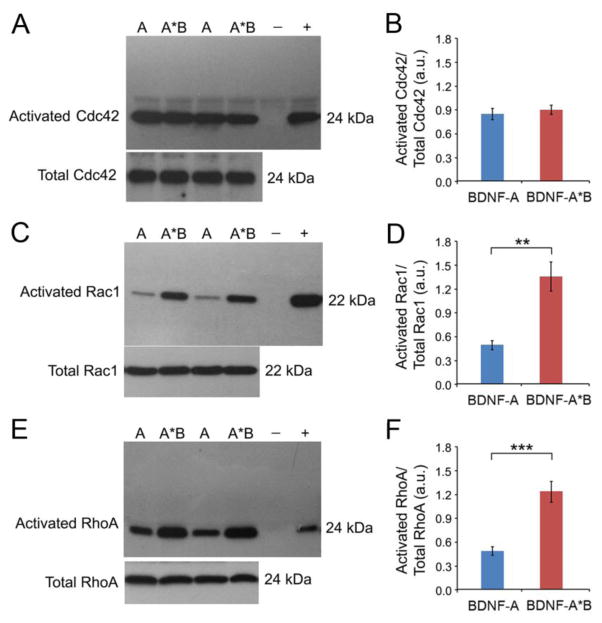
We infected cultured hippocampal neurons at DIV21 with AAV-BDNF-A or AAV-BDNF-A*B and harvested cell lysates at DIV35 to probe for levels of activated Cdc42, RhoA, and Rac1 using pull-down assays. Analyses revealed that Cdc42 was similarly activated in the two BDNF-expressing cultures, compared to a control culture (Figures 5A, 5B, S5A, and S5B). Because overexpressing either short or long 3′ UTR Bdnf mRNA increases spine formation (Orefice et al., 2013), this observation suggests that somatically synthesized BDNF promotes spine formation in part by activating Cdc42. To further support this view, we found that activated Cdc42 was at a high level at DIV14 and decreased significantly by DIV28 and DIV35 in cultured hippocampal neurons (Figures S5E and S5F), which is in accordance with our finding that spine density reaches a plateaus at DIV14.
Figure 5. BDNF-mediated activation of Cdc42, RhoA, and Rac1.
(A and B) Western blot analysis and quantification of activated Cdc42 and total Cdc42 in cell lysates from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B) (n=6 samples/condition). Negative (-) and positive (+) Cdc42-GTP controls were included.
(C and D) Western blot analysis and quantification of activated Rac1 and total Rac1 in cell lysates from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B) (n=6 samples/condition). Negative (−) and positive (+) Rac1-GTP controls were included.
(E and F) Western blot analysis and quantification of activated RhoA and total RhoA in cell lysates from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B) (n=6 samples/condition). Negative (−) and positive (+) RhoA-GTP controls were included.
Data are reported as mean ± SEM. Student’s t test: **p < 0.01 and ***p < 0.001. See also Figure S5.
In contrast to Cdc42 activation, overexpressing long 3′ UTR Bdnf mRNA activated Rac1 much more strongly than overexpressing short 3′ UTR Bdnf mRNA in cultured hippocampal neurons (Figures 5C and 5D). This was due to a combination of an inhibitory effect of short 3′ UTR Bdnf mRNA and a stimulatory effect of long 3′ UTR Bdnf mRNA on Rac1 activation (Figures S5C and S5D). Similarly, overexpressing long 3′ UTR Bdnf mRNA activated RhoA much more strongly than overexpressing short 3′ UTR Bdnf mRNA in cultured hippocampal neurons (Figures 5E and 5F). As overexpressing long 3′ UTR Bdnf mRNA promotes spine maturation and pruning while overexpressing short 3′ UTR Bdnf mRNA does the opposite (Figure 1), these results suggest that the translation product of long 3′ UTR Bdnf mRNA exerts this effect by activating Rac1 and RhoA.
An essential role of activated RhoA in proBDNF-dependent spine pruning
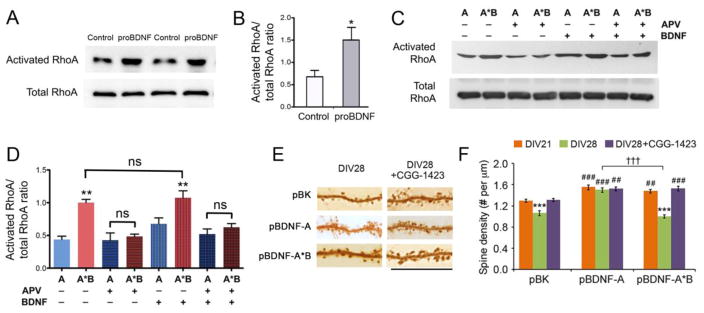
It has been shown that p75NTR activates RhoA by interacting with Kalirin 9, a guanidine nucleotide exchange factor (Harrington et al., 2008). Furthermore, overexpression of RhoA causes a reduction in spine density (Nakayama et al., 2000). We have shown that the translation product of long 3′ UTR Bdnf mRNA is largely secreted as proBDNF (Figure 2G), a high-affinity ligand for p75NTR. These observations raised the possibility that proBDNF derived from long 3′ UTR Bdnf mRNA mediates spine pruning by activating RhoA through p75NTR.
To examine the activity of dendritically synthesized proBDNF in activating RhoA, we infected cultured hippocampal neurons at DIV21 with AAV-BDNFAAA-A*B and harvested cell lysates at DIV35 to probe for levels of activated RhoA. Compared to non-infected control neurons, neurons infected with AAV-BDNFAAA-A*B showed significantly increased levels of activated RhoA (Figures 6A and 6B). As shown earlier, overexpressing long 3′ UTR Bdnf mRNA was much more potent in activating RhoA than overexpressing short 3′ UTR Bdnf mRNA (Figures 6C and 6D). We found that addition of the NMDAR antagonist, APV, completely abolished RhoA activation in neurons overexpressing long 3′ UTR Bdnf mRNA (Figures 6C and 6D). Application of mature BDNF had no effect on RhoA activation in neuronal cultures, either in the absence or in the presence of APV (Figures 6C and 6D). These results indicate that proBDNF translated from long 3′ UTR Bdnf mRNA activates RhoA in an NMDAR-dependent manner.
Figure 6. The proBDNF derived from long 3′ UTR Bdnf mRNA promotes spine pruning by activating RhoA.
(A and B) Representative images and quantification of RhoA Western blots of cell lysates from non-infected neurons (control) or neurons infected with AAV-BDNFAAA-A*B (proBDNF) at DIV35 (n=4 samples/condition). Student’s t-test: *p < 0.05.
(C and D) Representative images and quantification of RhoA Western blots of cell lysates from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B), and treated with either vehicle, 50 μM APV, 100 ng/ml BDNF, or both APV and BDNF for 48 hours (n=3 samples/condition). One-way ANOVA with post-hoc Bonferroni test: **p < 0.01 when compared to the AAV-BDNF-A cultures without APV and BDNF treatments; ns, not significant; P > 0.05 when a comparison was made between A+APV and A+APV+BDNF or between A*B+APV and A*B+APV+BDNF.
(E) Representative images of dendrites, showing morphology of dendritic spines at DIV28. Neurons were transfected at DIV7 with pBK, pBDNF-A or pBDNF-A*B, in combination with pActin-GFP. Vehicle or 1 μM CGG-1423 was applied to neuronal cultures from DIV21 to DIV28. Scale bar, 20 μm.
(F) Average spine density of neurons described in (E). Two-way ANOVA with post-hoc Bonferroni test (n=10 neurons/condition): ***p < 0.001 when compared to DIV21 neurons transfected with the same constructs; ##p < 0.01 and ###p < 0.001 when compared to pBK-transfected neurons at the same time point; †††p < 0.001 when indicated groups are compared.
Data are reported as mean ± SEM. See also Figure S6.
To determine the role of activated RhoA in spine pruning, we added a RhoA inhibitor, CGG-1423 to hippocampal neuron cultures from DIV21 to DIV28. CGG-1423 completely blocked spine pruning from DIV21 to DIV28 in control neurons (pBK) and neurons overexpressing long 3′ UTR Bdnf mRNA (pBDNF-A*B) without affecting spine density in neurons overexpressing short 3′ UTR Bdnf mRNA (pBDNF-A) (Figures 6E and 6F). These results indicate that active RhoA is required for spine pruning in cultured hippocampal neurons. In support of this claim, we found that levels of activated RhoA were increased at DIV28 and DIV35 when rates of spine pruning are highest (Figures S6A and S6B).
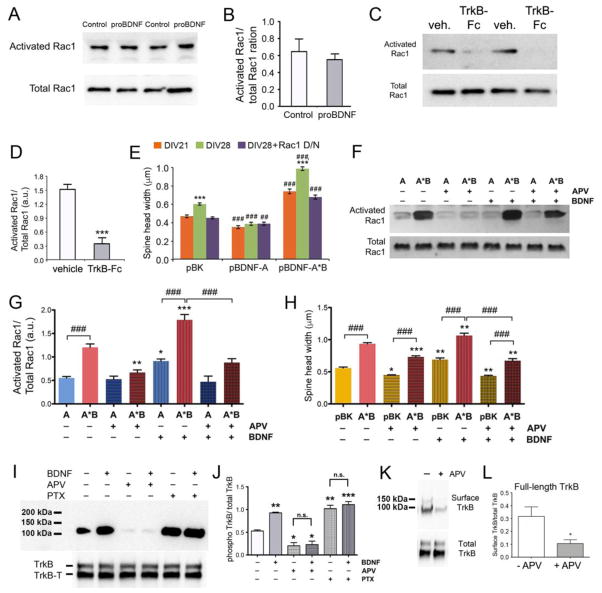
Requirement of activated Rac1 for mBDNF-dependent spine maturation
There are two proteins, proBDNF and mBDNF, derived from the translation product of long 3′ UTR Bdnf mRNA. We found that overexpressing proBDNF from long 3′ UTR Bdnf mRNA did not activate Rac1 in neurons (Figures 7A and 7B), although it did activate RhoA (Figures 6A and 6B). Furthermore, addition of TrkB-Fc to neuronal cultures to scavenge extracellular mBDNF blocked Rac1 activation (Figures 7C and 7D). These results indicate that mBDNF derived from long 3′ UTR Bdnf mRNA mediates Rac1 activation through TrkB.
Figure 7. Dendritically synthesized BDNF promotes spine maturation by activating Rac1 through TrkB.
(A and B) Representative images and quantification of Rac1 Western blots of cell lysates from non-infected neurons (control) or neurons infected with AAV-BDNFAAA-A*B (proBDNF) at DIV35 (n=4 samples/condition). Student’s t test: p > 0.05.
(C and D) Representative images and quantification of Western blots of activated Rac1 and total Rac1 in cell lysates from DIV35 neurons treated with either vehicle or TrkB-Fc (2.5 μg/ml) from DIV33 to DIV35 (n=4 samples/condition). Student’s t-test: ***p < 0.001.
(E) Average spine head width of neurons transfected at DIV7 with pBK, pBDNF-A or pBDNF-A*B in combination with either empty vector control or a Rac1 dominant/negative construct (Rac1 D/N). Two-way ANOVA with post-hoc Bonferroni test (n=10 neurons/condition): ***p < 0.001 when compared to DIV21 neurons transfected with the same constructs; ###p < 0.001 when compared to pBK-transfected neurons at the same time point.
(F and G) Representative images and quantification of Rac1 Western blots of cell lysates from DIV35 neurons infected with either AAV-BDNF-A (A) or AAV-BDNF-A*B (A*B), and treated with either vehicle, 50 μM APV, 100 ng/ml BDNF, or both APV and BDNF for 48 hours (n=3 samples/condition). One-way ANOVA with post-hoc Bonferroni test: *p < 0.05, **p < 0.01 and ***p < 0.001 when compared to the non-treated control neurons overexpressing the same Bdnf mRNA; ###p < 0.001 when indicated groups are compared.
(H) Average spine head width of neurons transfected at DIV7 with pBK or pBDNF-A*B and treated with either vehicle, 50 μM APV, 100 ng/ml BDNF, or both APV and BDNF from DIV26 to DIV28. One-way ANOVA with post-hoc Bonferroni test (n=10 neurons/condition): *p < 0.05, **p < 0.01 and ***p < 0.001 when compared to the non-treated control neurons transfected with the same constructs; ###p < 0.001 when indicated groups are compared.
(I) Western blot analyses of phosphorylated TrkB (p-TrkB), total full length TrkB (TrkB), and total truncated TrkB (TrkB-T) in lysates from cultured DIV33-35 neurons treated with BDNF, APV, and/or PTX.
(J) Quantification of p-TrkB and total TrkB levels from Western blots represented in panel I. One-way ANOVA with post-hoc Bonferroni test (n=3): *p < 0.05, **p < 0.01 and ***p < 0.001 when compared to the non-treated control neurons.
(K) Western blot analysis of surface TrkB and total TrkB in lysates from cultured DIV33-35 neurons treated with APV. Surface TrkB was labeled with biotin and pull-downed with streptavidin agarose beads.
(L) Quantification of surface full-length TrkB/total TrkB ratios from Western blots represented in panel K (n=4). *p < 0.05 by Student’s t test.
Data are reported as mean ± SEM. See also Figure S7.
We next determined whether activation of Rac1 is required for long 3′ UTR Bdnf mRNA to stimulate spine head enlargement. We employed a Rac1 dominant-negative mutant (Rac1 D/N) to block Rac1 activity. Expression of Rac1 D/N abolished spine head growth from DIV21 to DIV28 in control neurons and neurons overexpressing long 3′ UTR Bdnf mRNA, without affecting spine head width in neurons overexpressing short 3′ UTR Bdnf mRNA (Figure 7E). Control neurons are dependent upon endogenous long 3′ UTR Bdnf mRNA for spine head growth, and overexpressing short 3′ UTR Bdnf mRNA inhibits spine head growth by blocking trafficking of long 3′ UTR Bdnf mRNA to dendrites (Orefice et al., 2013). Therefore, these results indicate that dendritically synthesized BDNF mediates spine head growth through a pathway that requires activation of Rac1. In support of this claim, we found that the level of activated Rac1 was increased from DIV21-DIV35, when spine heads are growing in cultured hippocampal neurons (Figures S7A and S7B).
Finally, we investigated why mBDNF derived from short 3′ UTR Bdnf mRNA does not promote spine maturation, and hypothesized that it could be related to neuronal activity. To this end, we examined the ability of BDNF derived from overexpressed Bdnf mRNA, as well as recombinant mBDNF, to activate Rac1 in the absence or presence of APV. As shown above, levels of activated Rac1 were much higher in neurons overexpressing long 3′ UTR Bdnf mRNA than in neurons overexpressing short 3′ UTR Bdnf mRNA (Figures 7F and 7G). Addition of recombinant mBDNF further increased levels of activated Rac1 in neurons overexpressing either short or long 3′ UTR Bdnf mRNA. Blockade of NMDAR activity with APV abolished the ability of recombinant mBDNF to activate Rac1 (Figures 7F and 7G). This abolishment was correlated with the inability of recombinant mBDNF to activate TrkB (Figures 7I and 7J), due to the failure in transport of TrkB to the cell surface (Figures 7K, 7L, and S7C), in the presence of APV. In agreement with the Rac1 activation data, application of APV also blocked the stimulatory effect of recombinant mBDNF on spine head growth (Figure 7H). These observations suggest that activation of the NMDAR induces coupling of mBDNF production and secretion with TrkB responsiveness, so that local mBDNF concentration at stimulated synapses is high enough to activate TrkB and promote spine maturation. In contrast, mBDNF derived from short 3′ UTR Bdnf mRNA is released independent of action potentials (Figure S2), and its concentration may not be high enough to activate TrkB when it diffuses to stimulated synapses.
Discussion
Our previous studies indicate that long 3′ UTR Bdnf mRNA, but not short 3′ UTR Bdnf mRNA, promotes spine maturation and pruning, although the two forms of transcripts encode the same protein (An et al., 2008; Kaneko et al., 2012; Orefice et al., 2013). In this study we uncovered that the reason for distinct roles of the two forms of Bdnf mRNA in spine development is due to release of their translation products through different secretory pathways. Our results indicate that the translation product of long 3′ UTR Bdnf mRNA is released as proBDNF and converted to mBDNF at a stimulated synapse, which then activates TrkB to promote spine head growth of the same synapse. NMDAR activation is key to this process, by inducing release of proBDNF and by priming the same synapse to respond to mBDNF via insertion of TrkB onto the plasma membrane. Furthermore, the non-converted proBDNF will bind to p75NTR, likely at nearby silent synapses to induce spine pruning. In contrast, the translation product of short 3′ UTR Bdnf mRNA is mainly released as mBDNF independent of action potentials. When the released mBDNF reaches TrkB at a stimulated synapse through diffusion, its concentration would be too low to activate TrkB to stimulate spine growth. In addition, the affinity of mBDNF for p75NTR is low; mBDNF would not be able to activate p75NTR to induce spine pruning.
Secretory pathway of dendritically synthesized BDNF
Most of studies on BDNF secretion have been done in cultured neurons that overexpress a BDNF-GFP fusion protein from a mammalian expression vector containing a 3′ UTR that restricts transcripts to cell bodies (Egan et al., 2003; Hartmann et al., 2001; Kojima et al., 2001; Matsuda et al., 2009). Similar to short 3′ UTR Bdnf mRNA (Orefice et al., 2013), the majority of the translation products from these somatically-restricted Bdnf transcripts are present as mBDNF inside neurons (Egan et al., 2003). KCl-induced depolarization stimulates release of mBDNF from cultured neurons infected with BDNF-expressing sindbis virus (Egan et al., 2003). We were not able to detect mBDNF in the media of cultured neurons infected with BDNF-expressing AAV after 30-min KCl treatment, likely due to the fact that secreted BDNF tends to stick to cultured cells and that sindbis virus expresses protein at a much higher level than AAV does. Microscope imaging detected release of BDNF-GFP from dendrites in response to KCl treatment or electrical stimulation (Hartmann et al., 2001; Kojima et al., 2001; Matsuda et al., 2009). We found that 30-min KCl treatment did not significantly alter levels of intracellular mBDNF in neurons infected with either AAV-BDNF-A or AAV-BDNF-A*B. This observation suggests that action-potential-dependent mBDNF release may only account for a small fraction of BDNF secretion.
We have found that mEPSPs play an important role in release of mBDNF from neurons. Blockade of mEPSPs significantly reduced the amount of mBDNF secreted from neurons over a 2-day period. This result indicates that mBDNF is released from neurons in a mEPSP-dependent manner, in addition to the well-documented constitutive and action-potential-dependent pathways.
We previously reported that the translation product of long 3′ UTR Bdnf mRNA largely remains as proBDNF in neurons (Orefice et al., 2013). In this study, we found that addition of KCl to neuronal cultures induced release of approximately 50% of proBDNF from neurons infected with AAV-BDNF-A*B. This is in sharp contrast with the observation that KCl treatment did not significantly reduce levels of proBDNF in neurons overexpressing short 3′ UTR Bdnf mRNA or levels of mBDNF in neurons overexpressing either short or long 3′ UTR Bdnf mRNA. We believe that these results indicate that dendritically synthesized BDNF is packaged into vesicles that are highly sensitive to action potentials and that activity-dependent release of endogenous BDNF is mainly from local translation of long 3′ UTR Bdnf mRNA and is in the form of proBDNF, which is then extracellularly converted to mBDNF. Thus, we speculate that somatically and dendritically synthesized BDNF may be sorted into different types of vesicles. In agreement with this view, the typical Golgi complex, which is required for processing of precursor proteins in the cytoplasm, is sparse in dendrites (Horton et al., 2005). Further studies are needed to determine how dendritically synthesized BDNF is packaged into vesicles and released.
Molecular control of spine pruning
Protein synthesis regulators such as FMRP, TSC2, and mGluR5, are needed for spine pruning (Comery et al., 1997; Grossman et al., 2006; Hinton et al., 1991; Irwin et al., 2001; McKinney et al., 2005; Tang et al., 2014; Wilkerson et al., 2014). Furthermore, our previous studies have shown that translation of long 3′ UTR Bdnf mRNA in dendrites is essential for spine pruning (An et al., 2008; Kaneko et al., 2012; Orefice et al., 2013). These findings indicate that some proteins synthesized in dendrites control spine pruning and that one of the proteins is BDNF. Because the current study shows that both translation of dendritic long 3′ UTR Bdnf mRNA and release of the translation product are NMDAR-dependent, it may explain the reason why spine pruning is activity-dependent. It remains to be determined if FMRP, TSC2 and mGluR5 regulate local BDNF synthesis in dendrites.
In this study we were able to employ cultured hippocampal neurons and p75NTR knockout mice to demonstrate that proBDNF derived from long 3′ UTR Bdnf mRNA in dendrites controls spine pruning by activating RhoA through the p75NTR receptor. In support of this finding, increasing levels of endogenous proBDNF has been found to decrease spine density of hippocampal neurons in mice (Yang et al., 2014). It has been shown that proBDNF can induce neuronal apoptosis through the p75NTR-sortilin receptor complex (Teng et al., 2005), likely by activating proapoptotic caspases. Interestingly, a recent study found that caspase-3 played an active role in spine pruning (Erturk et al., 2014). It would be intriguing to test whether dendritically synthesized proBDNF promotes spine pruning by activating caspase-3 in addition to RhoA.
Spine maturation
Because somatically and dendritically synthesized BDNF have opposing effects on spine density by stimulating spine formation and spine pruning, respectively, hippocampal neurons may have a normal number of spines when the Bdnf gene is deleted. However, these spines do not go through the enlargement and pruning process mediated by BDNF derived from long 3′ UTR Bdnf mRNA and would not function properly. Indeed, we found that neurons expressing both TrkB shRNA and p75NTR shRNA exhibited normal spine density at DIV28, but decreased spine head width, compared to control neurons. This might explain the observation that while hippocampal neurons of Bdnf knockout mice display a normal number of spines, these spines are thinner (Rauskolb et al., 2010) and exhibit impaired long-term potentiation (Korte et al., 1995; Korte et al., 1996; Patterson et al., 1996).
This study uncovers a biochemical pathway underlying the role of BDNF in spine maturation. Our results suggest that proBDNF translated from long 3′ UTR Bdnf mRNA in dendrites is extracellularly converted to mature BDNF by the tPA/plasmin system, which activates Rac1 through TrkB to promote spine maturation. We found that dendritically synthesized BDNF also stimulated spine maturation through p75NTR, as overexpressing proBDNF from long 3′ UTR Bdnf mRNA promoted spine head enlargement and knocking down p75NTR inhibited spine head enlargement. Of note, the effect of p75NTR on spine enlargement is much smaller than that of TrkB. It is possible that materials from eliminated spines are recycled to activated spines and thus facilitate their growth. However, this view does not explain our observation that granule cells in p75NTR KO mice had smaller spine heads than WT mice at PND21 when significant spine loss has not occurred, suggesting that a distinct signaling cascade downstream of p75NTR may also stimulate spine maturation. If this speculation were true, it would be interesting to understand how proBDNF can coincidently mediate spine pruning and spine maturation.
Supplementary Material
HIGHLIGHTS.
The translation product of dendritic Bdnf mRNA is largely released as proBDNF
NMDA receptor activation stimulates dendritic BDNF synthesis and proBDNF release
ProBDNF promotes spine pruning by activating RhoA through the p75NTR receptor
Mature BDNF stimulates spine growth at active synapses by activating Rac1 via TrkB
Acknowledgments
We thank Dr. Maria Donoghue for allowing us to conduct some experiments in her laboratory and Dr. Sung Ok Yoon for the human p75NTR construct. This work was supported by the grants from the National Institutes of Health to BX (R01 NS073930) and LLO (F31 NS074840).
Abbreviations
- BDNF
brain-derived neurotrophic factor
- DIV
day in vitro
- FMRP
fragile X mental retardation protein
- MHCI
major histocompatibility class I
- mEPSPs
miniature excitatory postsynaptic potentials
- myr
myristoylation peptide
- nls
nuclear localization signal
- TTX
tetrodotoxin
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Balkowiec A, Katz DM. Activity-dependent release of endogenous brain-derived neurotrophic factor from primary sensory neurons detected by ELISA in situ. J Neurosci. 2000;20:7417–7423. doi: 10.1523/JNEUROSCI.20-19-07417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochl A, Thoenen H. Characterization of nerve growth factor (NGF) release from hippocampal neurons: evidence for a constitutive and an unconventional sodium-dependent regulated pathway. Eur J Neurosci. 1995;7:1220–1228. doi: 10.1111/j.1460-9568.1995.tb01112.x. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Churchill JD, Grossman AW, Irwin SA, Galvez R, Klintsova AY, Weiler IJ, Greenough WT. A converging-methods approach to fragile X syndrome. Dev Psychobiol. 2002;40:323–338. doi: 10.1002/dev.10036. [DOI] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Elia G. Cell surface protein biotinylation for SDS-PAGE analysis. Methods Mol Biol. 2012;869:361–372. doi: 10.1007/978-1-61779-821-4_29. [DOI] [PubMed] [Google Scholar]
- Erturk A, Wang Y, Sheng M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J Neurosci. 2014;34:1672–1688. doi: 10.1523/JNEUROSCI.3121-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Fu X, Zang K, Zhou Z, Reichardt LF, Xu B. Retrograde neurotrophic signaling requires a protein interacting with receptor tyrosine kinases via C2H2 zinc fingers. Mol Biol Cell. 2010;21:36–49. doi: 10.1091/mbc.E09-04-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Grossman AW, Elisseou NM, McKinney BC, Greenough WT. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006 doi: 10.1016/j.brainres.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Harrington AW, Li QM, Tep C, Park JB, He Z, Yoon SO. The role of Kalirin9 in p75/nogo receptor-mediated RhoA activation in cerebellar granule neurons. J Biol Chem. 2008;283:24690–24697. doi: 10.1074/jbc.M802188200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM. Structure, development, and plasticity of dendritic spines. Curr Opin Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Heumann R, Lessmann V. Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. Embo J. 2001;20:5887–5897. doi: 10.1093/emboj/20.21.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR. Synaptic density in human frontal cortex - developmental changes and effects of aging. Brain Res. 1979;163:195–205. doi: 10.1016/0006-8993(79)90349-4. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Xie Y, An JJ, Stryker MP, Xu B. Dendritic BDNF synthesis is required for late-phase spine maturation and recovery of cortical responses following sensory deprivation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:4790–4802. doi: 10.1523/JNEUROSCI.4462-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner H, Aguet F, Sage D, Unser M. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc. 2013;249:13–25. doi: 10.1111/j.1365-2818.2012.03675.x. [DOI] [PubMed] [Google Scholar]
- Kojima M, Takei N, Numakawa T, Ishikawa Y, Suzuki S, Matsumoto T, Katoh-Semba R, Nawa H, Hatanaka H. Biological characterization and optical imaging of brain-derived neurotrophic factor-green fluorescent protein suggest an activity-dependent local release of brain-derived neurotrophic factor in neurites of cultured hippocampal neurons. J Neurosci Res. 2001;64:1–10. doi: 10.1002/jnr.1080. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M, Griesbeck O, Gravel C, Carroll P, Staiger V, Thoenen H, Bonhoeffer T. Virus-mediated gene transfer into hippocampal CA1 region restores long-term potentiation in brain-derived neurotrophic factor mutant mice. Proc Natl Acad Sci U S A. 1996;93:12547–12552. doi: 10.1073/pnas.93.22.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- Liao GY, An JJ, Gharami K, Waterhouse EG, Vanevski F, Jones KR, Xu B. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat Med. 2012;18:564–571. doi: 10.1038/nm.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M. Number and distribution of the apical dendritic spines of the layer V pyramidal cells in man. J Comp Neurol. 1967;131:475–490. doi: 10.1002/cne.901310407. [DOI] [PubMed] [Google Scholar]
- Mataga N, Mizuguchi Y, Hensch TK. Experience-dependent pruning of dendritic spines in visual cortex by tissue plasminogen activator. Neuron. 2004;44:1031–1041. doi: 10.1016/j.neuron.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Lu H, Fukata Y, Noritake J, Gao H, Mukherjee S, Nemoto T, Fukata M, Poo MM. Differential activity-dependent secretion of brain-derived neurotrophic factor from axon and dendrite. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:14185–14198. doi: 10.1523/JNEUROSCI.1863-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Grossman AW, Elisseou NM, Greenough WT. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am J Med Genet B Neuropsychiatr Genet. 2005;136:98–102. doi: 10.1002/ajmg.b.30183. [DOI] [PubMed] [Google Scholar]
- Nagappan G, Zaitsev E, Senatorov VV, Jr, Yang J, Hempstead BL, Lu B. Control of extracellular cleavage of ProBDNF by high frequency neuronal activity. Proc Natl Acad Sci U S A. 2009;106:1267–1272. doi: 10.1073/pnas.0807322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orefice LL, Waterhouse EG, Partridge JG, Lalchandani RR, Vicini S, Xu B. Distinct roles for somatically and dendritically synthesized brain-derived neurotrophic factor in morphogenesis of dendritic spines. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:11618–11632. doi: 10.1523/JNEUROSCI.0012-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, Lu B. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Rauskolb S, Zagrebelsky M, Dreznjak A, Deogracias R, Matsumoto T, Wiese S, Erne B, Sendtner M, Schaeren-Wiemers N, Korte M, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L. Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J Neurosci. 2003;23:3118–3123. doi: 10.1523/JNEUROSCI.23-08-03118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol Cell Neurosci. 2004;26:429–440. doi: 10.1016/j.mcn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, Kermani P, Torkin R, Chen ZY, Lee FS, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Vonesch C, Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE transactions on image processing : a publication of the IEEE Signal Processing Society. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- Wang WY, Xu GZ, Tian J, Sprecher AJ. Inhibitory effect on LPS-induced retinal microglial activation of downregulation of t-PA expression by siRNA interference. Current eye research. 2009;34:476–484. doi: 10.1080/02713680902916108. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson JR, Tsai NP, Maksimova MA, Wu H, Cabalo NP, Loerwald KW, Dictenberg JB, Gibson JR, Huber KM. A role for dendritic mGluR5-mediated local translation of Arc/Arg3.1 in MEF2-dependent synapse elimination. Cell reports. 2014;7:1589–1600. doi: 10.1016/j.celrep.2014.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Harte-Hargrove LC, Siao CJ, Marinic T, Clarke R, Ma Q, Jing D, Lafrancois JJ, Bath KG, Mark W, et al. proBDNF negatively regulates neuronal remodeling, synaptic transmission, and synaptic plasticity in hippocampus. Cell Rep. 2014;7:796–806. doi: 10.1016/j.celrep.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Siao CJ, Nagappan G, Marinic T, Jing D, McGrath K, Chen ZY, Mark W, Tessarollo L, Lee FS, et al. Neuronal release of proBDNF. Nat Neurosci. 2009;12:113–115. doi: 10.1038/nn.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Lois C, Alvirez M, Alvarez-Buylla A, Falck-Pedersen E, Chao MV. Adenovirus-mediated gene delivery into neuronal precursors of the adult mouse brain. Proc Natl Acad Sci U S A. 1996;93:11974–11979. doi: 10.1073/pnas.93.21.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005;436:261–265. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.