Abstract
Stem cell-derived cardiomyocytes have the potential to be used to study heart disease and maturation, screen drug treatments, and restore heart function. Here, we discuss the procedures involved in using micropost arrays to measure the contractile forces generated by stem cell-derived cardiomyocytes. Cardiomyocyte contractility is needed for the heart to pump blood, so measuring the contractile forces of cardiomyocytes is a straightforward way to assess their function. Microfabrication and soft lithography techniques are utilized to create identical arrays of flexible, silicone microposts from a common master. Micropost arrays are functionalized with extracellular matrix protein to allow cardiomyocytes to adhere to the tips of the microposts. Live imaging is used to capture videos of the deflection of microposts caused by the contraction of the cardiomyocytes. Image analysis code provides an accurate means to quantify these deflections. The contractile forces produced by a beating cardiomyocyte are calculated by modeling the microposts as cantilever beams. We have used this assay to assess techniques for improving the maturation and contractile function of stem cell-derived cardiomyocytes.
Keywords: Microposts, Induced Pluripotent Stem Cells, Cardiomyocytes, Soft Lithography, Cell Mechanics
1 Introduction
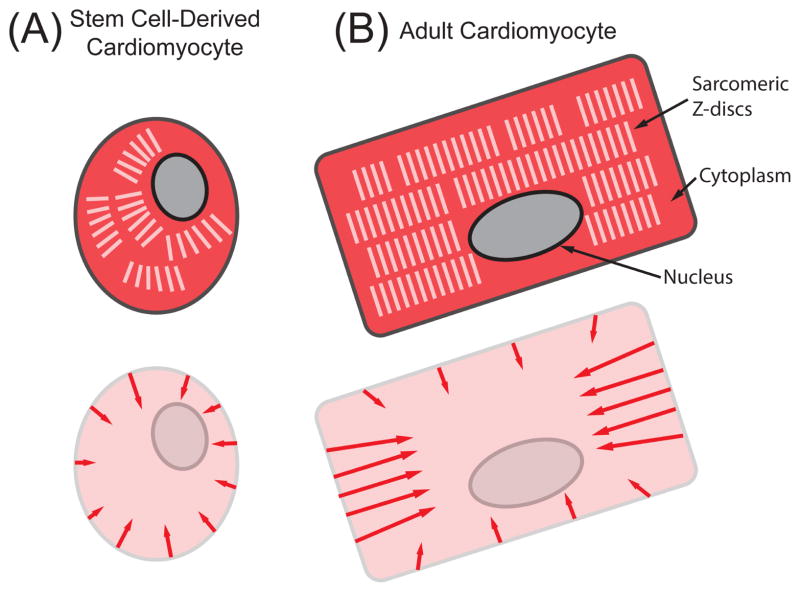
One of the most important functions of cardiomyocytes in the heart is to forcibly contract in order to pump blood through the body. Cardiomyocyte death and/or dysfunction are the underlying causes of many cardiac diseases [1]. Since cardiac tissue has a low capacity to regenerate or repair itself, cardiomyocytes derived from stem cells (SC-CMs) have the promise to restore function to the damaged regions of cardiac tissue. In addition to being used as a model to study heart maturation and disease [2–6], SC-CMs have also been used to screen potential drug treatments [5–7] and more recently to restore heart function [2, 8–13]. However, these studies have shown that the contractile output of SC-CMs is far lower than their adult counterparts, which is due in part to their small, round morphology and sarcomeric structure (Fig. 1A) [14]. Thus, it is important to characterize and study the contractile dynamics of SC-CMs to achieve better treatments for heart disease.
Figure 1.
Structural and force differences in (A) immature SC-CMs and (B) adult cardiomyocytes. In the top schematics, the cytoplasm is colored dark red, the sarcomeric Z-discs are white, and the nucleus is gray. The arrows in the bottom schematics indicate the primary directions of contractile forces. Stem cell-derived cardiomyocytes have a smaller, more round morphology, with unaligned myofibrils, resulting in less force generation than their adult counterparts.
Adult cardiomyocytes have a structure of actin and myosin filaments that is highly ordered (Fig. 1B). The alignment of these contractile filaments allows cardiomyocytes to produce strong forces by concentrating the generation of force along one direction. Many cellular assays have been used to measure the contractility of single cardiomyocytes, including: magnetic beads [15], atomic force microscopy (AFM) [16–19], traction force microscopy (TFM) [20–22], optical edge detection [23, 24], flexible cantilevers [25–27], and strain gauges [28]. These approaches have been used to study the biomechanics and mechanobiology of adult and neonatal cardiomyocytes. To the best of our knowledge, out of these techniques only AFM and TFM have been used successfully to measure forces from single human SC-CMs [19, 21].
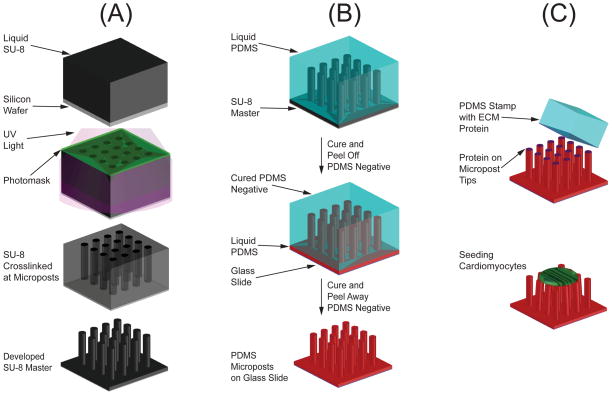
Microposts consist of arrays of vertical cantilever beams made from flexible silicone (Fig. 2). This technology has been widely used to measure contractile forces produced by cells [29–32], including cardiomyocytes [33–39]. Cells adhere to the tips of the microposts and are allowed to spread naturally across the tips of multiple microposts. When a cell contracts, it generates forces which cause the microposts to deflect. Our group has successfully used micropost arrays to assess the contractile maturation of SC-CMs [39–41].
Figure 2.
(A) SU-8 lithography is used to create an SU-8 master of the microposts. (B) Manufacturing of the PDMS microposts uses a double-casting process with a PDMS negative mold. (C) Microcontact printing is used to coat the tips of microposts with extracellular matrix (ECM) proteins before seeding cells.
Adult cardiomyocytes contract along a principle axis and for this reason, several methods measure force in a single direction [16, 18, 28, 34, 37]. However, immature cardiomyocytes like SC-CMs do not have uniaxial contractions [42]. Microposts are advantageous in this regard, as they are capable of measuring cellular forces in two-dimensions and with subcellular spatial resolution. For some methods, cardiomyocytes must be affixed to the force transducer via clamping or gluing; this artificial adhesion may alter the behavior of cells. Additionally, some of these other methods, such as those using large-scale force sensing cantilevers and commercial force transducers, are unable to measure single-cell forces and can only report a bulk tissue values. In contrast, microposts make it possible to quantify the multi-directional forces of individual cardiomyocytes at each adhesion that forms on the tips of the microposts. Here, we describe the processes involved in using this assay, including microcontact printing, cell culturing, and the analysis steps to quantify the contractile forces and myofibril structure of SC-CMs.
2 Experimental Methods
2.1 Micropost Fabrication
Soft lithography techniques are used to create polydimethylsiloxane (PDMS) micropost arrays [29, 43]. First, a master structure must be fabricated from a photoresist via photolithography (Fig. 2A). We choose to use SU-8 due to its ability to produce high-aspect-ratio structures [44]. SU-8 is an epoxy-based negative photoresist that crosslinks when exposed to UV light and heat, causing the highly viscous liquid to solidify. Prior to fabrication of the SU-8 master, an opaque photomask is created with circular windows; the size and configuration of the windows defines the diameter and spacing of the microposts. Next, a silicon wafer (Silicon Quest International, Santa Clara, CA) undergoes dehydration at 200°C for 30 minutes on a hot plate. A 2 μm thick layer of SU-8 2005 (MicroChem, Newton, MA) is spin-coated onto the wafer to serve as a strong base for the microposts. The wafer is soft-baked at 65 °C for one minute followed by 95 °C for two minutes. After exposing the base layer of SU-8 to UV light (365 nm), the wafer undergoes a post-exposure bake at 65 °C for 1 minute followed by 95 °C for 1 minute, and allowed to cool to room temperature. The appropriate SU-8 manufacturer data sheet should be consulted for operational parameters when spin-coating and exposing [45].
For the microposts, a film of SU-8 is spin-coated on top of the cross-linked SU-8 base layer. We find that a film thickness of 10 μm yields appropriately stiff and tall microposts. Shorter microposts are generally stiffer, while taller posts may collapse during fabrication. A mask aligner (AB-M, San Jose, CA) is used to bring the photomask into contact with the wafer. It is important for there to be good contact between the mask and the wafer; any gap between the mask and the SU-8 will cause the UV light to diffract, resulting in misshapen microposts. We use an edge-bead removal step, where a cleanroom swab soaked with SU-8 developer is dragged along the edge of the wafer, and strong vacuum contact in the mask aligner to ensure that there is not a gap between the mask and the wafer. Next, the wafer is exposed to UV light and subsequently baked at 65 °C for 1 minute followed by 95 °C for 1 minute. To develop the pattern, the unexposed SU-8 is dissolved with SU-8 developer followed by a rinse in isopropyl alcohol to remove the developer solution. The wafer is dried with nitrogen gas before a hard-bake step at 150 °C for 5 minutes, which strengthens the SU-8 microposts. To prevent PDMS from adhering strongly to the SU-8 master, the arrays are plasma treated for 90 seconds (SPI Supplies, West Chester, PA) and silanized overnight with (tridecafluoro-1,1,2,2-tetrahyrooctyl)- 1-trichlorosilane (UCT, Horsham, PA) inside a desiccator.
With the SU-8 master fabricated, PDMS negative molds can be created (Fig. 2B). First, the PDMS base and crosslinker (Sylgard 184, Dow Corning, Midland, MI) are mixed at a 10:1 ratio. The liquid mixture of PDMS is poured over the SU-8 master in an aluminum dish before baking at 110 °C for 10 minutes to cure. The negative molds are plasma treated for 90 seconds to activate the surface, followed by silanization for 2 hours in a desiccator. This silanization step will allow the negative mold to cleanly separate from the microposts in a later step. Next, we recommend that the microposts be cast from the negative molds onto a hard surface to facilitate substrate handling. We find that glass microscope coverslips work well as a stiff, transparent substrate to cast on; specifically, we use No. 1 round glass coverslips, which fit directly into Attofluor® viewing chambers (Life Technologies, Grand Island, NY) discussed at the end of section 2.3. To begin the positive casting, a new batch of liquid PDMS is mixed. The glass coverslips are then plasma treated for 90 seconds to promote adhesion with the liquid PDMS and help secure the microposts to the glass coverslip. Next, a small droplet of the liquid PDMS is placed onto the coverslip, followed quickly by placing the silanized negative mold on top. The negative mold is allowed to settle on the liquid PDMS for 10 minutes to ensure that the micropost tips are not skewed, and all lie in the same focal plane. The assembly is baked at 110 °C for around 20 hours to ensure that the PDMS is fully cured. After curing, the negative mold is gently peeled from the PDMS on the glass coverslip, leaving behind PDMS microposts. Care should be taken during the peeling process to avoid large deformations of the microposts, which will cause the microposts to collapse, i.e. stick to each other or to the surface between the microposts. By changing the design of the photomask and thickness of SU-8, we have found it possible to fabricate micropost arrays without collapsed posts with heights between 6–10 μm, diameters between 2–3 μm, and center-to-center spacing between 6–9 μm. Different combinations of these dimensions dictate the bending stiffness of the microposts. These ranges of dimensions yield a substrate stiffness between 5–100 kPa, which is comparable to the stiffness of cardiac tissue (20–30 kPa for healthy human cardiac tissue, 50–70 kPa for infarcted tissue) [17]. The double-casting process with a negative mold and a common SU-8 master ensures that the dimensions of the microposts, and hence the stiffness of the microposts, are consistent between experiments.
2.2 Micropost Stiffness Calibration
Cardiomyocyte forces are calculated based on the deflection of the microposts. To calculate these forces, the spring constant of the microposts first needs to be determined, which is a linear approximation for the relationship between the applied force and the deflection at the tip of each micropost. Two methods may be used to determine the spring constant: slender-beam theory and experimental calibration.
Using slender-beam theory, the spring constant is:
| (1) |
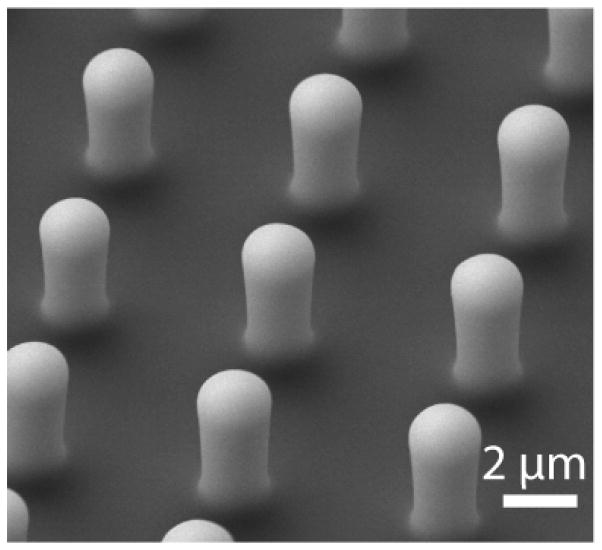
where E is the Young’s modulus of elasticity for PDMS, which is approximately 3.8 MPa for a 10:1 mixture of PDMS and cross-linker baked at 100 °C for a sufficiently long time (at least 20 hours) [46]. We note that this stiffness is highly dependent upon baking time, baking temperature, and mixing ratio. The diameter D and length L of the PDMS microposts are measured from side-view images obtained by scanning electron microscopy (Fig. 3). For large deformations, slender-beam theory may not be appropriate and thus, empirical fitting of experimental force data is preferred [43].
Figure 3.
SEM image of PDMS microposts used in estimating the spring constant. Length and diameter are measured from the image, accounting for the rounded edges, and used in Eq. 1 to determine the stiffness of the microposts.
For experimental determination of the spring constant, AFM or a calibrated piezoresistive cantilever may be used [36, 47]. For example, the tip of the piezoresistive cantilever is brought into contact with a single micropost, and force is measured simultaneously with the micropost tip deflection. Repeating this process for a number of different tip deflections generates an empirical curve that calibrates force and micropost tip deflection. Alternatively, micropost calibration can be achieved by deflecting the tip of a micropost with a glass micropipette of known stiffness kp [48, 49]. The deflections of both the micropipette and micropost are measured using a microscope. For a displacement of the micropipette xp, the tip of the micropost will deflect a distance x. With this information, the spring constant can be calculated by modeling the interaction of these structures as two springs in series:
| (2) |
These calibration methods assume that the micropost is in pure bending. Since the base layer to which the micropost is attached is also PDMS, elastic compliance of the base layer should be considered in this calibration. Based on the aspect ratio of the micropost (length-to-diameter) and the Poisson ratio for PDMS, a correction factor ranging between 0.5 and 0.9 accounting for the elastic base layer can be used in calculating the spring constant of a micropost [36, 50].
2.3 Microcontact Printing
Microcontact printing is used to coat the tips of the microposts with extracellular matrix (ECM) proteins (Fig. 2C). Microcontact printing utilizes the surface properties of PDMS to transfer proteins onto the tips of the microposts [51]. Other methods of securing cells to the force measurement device, such as gluing or clamping, may adversely interact with normal cellular function. By allowing cells to naturally adhere to ECM proteins, the physiological function of the cells is thought to be better preserved.
Microcontact printing may be used to confine the shape or spreading of cells [52]. Here, we describe a protocol for microcontact printing using stamps with no topological features. The stamps are created by pouring a 30:1 mixture of PDMS on a silanized flat silicon wafer and curing at 110 °C for 20 minutes. The PDMS is peeled from the wafer and cut into blocks the size of the micropost array to form the stamps. Under sterile conditions, a solution of ECM protein is aliquoted on top of the flat surface of each PDMS stamp. To ensure adequate coating of the ECM protein, it is helpful to begin by depositing small droplets near the edges of the stamp, connecting these to form an outer frame, and then successively dragging the droplets towards the middle of the stamp. This ECM protein is then allowed to adsorb onto the surface of the stamp for 1 hour. The stamp is then rinsed in sterile DI water and dried with nitrogen gas, leaving behind the deposited ECM proteins on the stamp surface.
Next, the micropost substrates are treated with UV/ozone (Jelight, Irvine, CA), causing the PDMS surface of the micropost to become hydrophilic and thereby allowing the ECM proteins to transfer from the stamp to the microposts [53]. The stamp is gently lowered perpendicular to the microposts, eventually forming contact with the tips of microposts. To ensure adequate transfer of the ECM protein, the stamp is left in this configuration for 30 seconds. The microposts then undergo a series of sterilizing and washing steps. First, the micropost substrate is submerged in 100% ethanol to sterilize and fully wet the spaces between the microposts. Next, the substrate is submerged in 70% ethanol. Finally, the substrate is rinsed by successively submerging in dishes of sterile DI water. The 70% ethanol washing step serves as a transition between 100% ethanol and sterile DI water, preventing post collapse due to the high surface tension forces of water. Additionally, the ethanol washes may also help raise any posts that collapsed during the stamping process.
After washing, the sides of the posts are fluorescently tagged, which enables the deflection of the microposts to be captured with fluorescence microscopy. We find that bovine serum albumin (BSA) conjugated with Alexa Fluor 594 (Life Technologies, Grand Island, NY) provides the best fluorescence for our image analysis. We dilute the BSA in DI water to 5μg/ml and submerge the microposts in the BSA for 1 hour. Next, the substrates are briefly submerged in DI water to rinse, followed by submerging in 0.2% Pluronic F-127 (BASF, Ludwigshafen, Germany) for 30 minutes. The hydrophobicity of PDMS allows Pluronic F-127 to adsorb to its surface where its co-block polymer chains of polyethylene glycol prevent the adsorption of additional proteins to the base or sidewalls of the microposts, thus confining cell adhesion to only the tips of the microposts [54–56]. Finally, the substrates are rinsed in phosphate buffered saline (PBS) before securing in an Attofluor® viewing chamber (Life Technologies, Grand Island, NY).
We place substrates in the Attofluor® viewing chambers for two primary reasons: 1) the chamber keeps the substrate securely in place, which is vital to the capture of video, and 2) the chamber allows for imaging from above the cardiomyocytes, improving video and image quality. The microposts may be cast onto other substrates and placed into a tissue culture dish or even cast directly onto a tissue culture dish itself, as long as the substrate does not move during video capturing.
2.4 Cell Culture and Preparation
The protocol that we employ for cell seeding, feeding, and maintenance has been previously reported [39], but will be summarized here. Prior to substrate manufacturing, we obtain differentiated human induced pluripotent SC-CMs. The SC-CMs are initially cultured on Matrigel®-coated plates (Corning Life Sciences, Corning, NY) in RPMI medium containing RPMI 1640 with L-glutamine (Life Technologies, Grand Island, NY), supplemented with B-27® plus insulin (Life Technologies, Grand Island, NY) and 1% penicillin/streptomyosin (Mediatech, Manassas, VA).
To seed the SC-CMs onto the microposts, they must first be dissociated from their initial culture substrate. First, the SC-CMs are washed in PBS. Next, the PBS is aspirated and Versene (Life Technologies, Grand Island, NY) is added to the culture dish. After incubation at 37 °C for 3 minutes, the Versene is aspirated and replaced with a solution of 0.25% trypsin ethylenediaminetetra-acetic acid (Mediatech, Manassas, VA) in Versene. After adding the trypsin-EDTA solution, the SC-CMs are placed in an incubator for approximately 5 minutes, observing the dissociation every minute. Once fully dissociated, the SC-CMs are pelleted and resuspended in RPMI medium supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY). The cell suspension is repeatedly triturated by pipette to break up any large clumps and ensure the suspension contains individual cells and not clusters of cells. The SC-CMs are then seeded onto the micropost arrays at a density of approximately 500,000 cells per 500 mm2 of substrate. Following 24 hours of culturing after seeding, the media is gently removed and quickly replaced with serum-free RPMI medium (Life Technologies, Grand Island, NY). This serum-free media prevents any unwanted cardiac fibroblasts from replicating, while SC-CMs are provided nutrients by the B-27® supplement. A majority of SC-CMs are found beating within a few days after initial seeding, and they are typically studied within 7 days after seeding. Prior to video recording, the media is replaced with HEPES RPMI medium (Life Technologies, Grand Island, NY).
3 Imaging
3.1 Video Recording
High speed video microscopy is necessary to capture the muscle twitch dynamics of the SC-CMs. We use a Hamamatsu ORCA-Flash 2.8 Scientific CMOS camera because of its ability to capture images at up to 100 frames per second. The frame rate of the camera is inversely correlated with the size of the region-of-interest (ROI), so we restrict the ROI to encompass the cell of interest as closely as possible while maintaining complete view of the attached microposts. A high frame rate is essential because the twitch frequency of SCCMs is irregular and a waveform that describes the dynamics of the twitch force has not been established. As such, we find that the higher frame rates ensure that our image analysis more accurately identifies the characteristic features of twitch contraction.
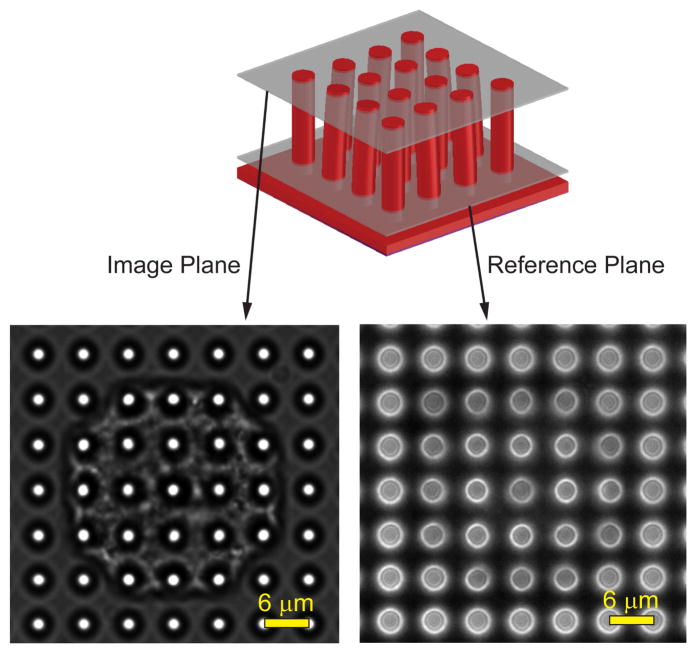
The camera is paired with a Nikon Eclipse Ti upright microscope which records images of individual SC-CMs through a 60× water immersion objective (NA 1.0). Before initializing a recording, the objective is focused on the bottom of the microposts and a fluorescent image is taken to determine the reference positions of each micropost (Fig. 4). Afterwards, the focus is shifted to an image plane containing the tips of the microposts and a phase contrast video is taken. As the SC-CMs beat, there are visible changes positions of the tips of the microposts in the phase contrast video. The change in position of the tips of microposts relative to their reference position in the fluorescent image is used to calculate the twitch force, which is described in Section 4.1.
Figure 4.
A phase contrast microscopy video is taken at an image plane that contains the tips of the microposts. A reference image is taken at an image plane near the base of the microposts. The video frames and reference image are analyzed with a series of image analysis steps using custom GUI. Adapted from [39].
3.2 Immunofluorescence Staining
After live experiments are completed, the myofibril structures of the SC-CMs are immunofluorescently stained. First, a Triton extraction procedure is used to permeabilize the samples. The samples are submerged for 10 seconds in buffer solution of 10 mM PIPES (Avantor, Center Valley, PA), 50 mM NaCl (EMD Chemicals, VWR Inc., Radnor, PA), 150 mM sucrose (Avantor, Center Valley, PA), 2 mM phenylmethylsulfonyl fluoride (Electron Microscopy Sciences, Hatfield, PA), 3 mM MgCl (BDH), 20 μg/ml aprotinin (G-Biosciences, St. Louis, MO), 1 μg/ml leupeptin (G-Biosciences, St. Louis, MO), and 1 μg/ml pepstatin (G-Biosciences, St. Louis, MO) at pH 6.5. Samples are then moved to a new buffer solution containing the same mixture listed before, but with the addition of 0.5% Triton X-100 (Sigma Aldrich, St. Louis, MO) for 2 minutes.
Samples are fixed with 4% paraformaldehyde (EMD Chemicals) in PBS. Next, the proteins of interest are stained; we find that staining cell nuclei and sarcomeric α-actinin are the most typical for analyzing internal structure of cardiomyocytes. Cell nuclei are stained with Hoechst 33342 (Life Technologies) and sarcomeric α-actinin is marked with monoclonal mouse anti-α-actinin (Sigma Aldrich) and stained with goat anti-mouse AlexaFluor 488 (Life Technologies, Grand Island, NY). A Nikon Ti Eclipse inverted microscope with a 40× oil immersion lens (NA 1.0) is used to image these stained substrates.
4 Image and Data Analysis
4.1 Video Analysis
Since contractility is a major function of cardiomyocytes, contractile force during a twitch contraction is an obvious choice as a marker of maturation. The contractile velocity is also an important factor; immature SC-CMs have lower levels of Na+ channel Nav1.5 and L-type calcium channels than adult cardiomyocytes, resulting in a slower upstroke velocity [42]. Since both force and velocity are important, the contractile power, which is the product of force and velocity, is arguably the most accurate and useful metric for assessing maturation. At high temporal resolution, the change in deflection of the microposts can be used to determine the twitch velocity. The twitch power is then calculated as a product of the force and velocity traces for each micropost.
A custom MATLAB (Mathworks, Natick, MA) code is used to obtain the contractile properties of individual cardiomyocytes from live images. The change in the position of each micropost is tracked by a series of image analysis steps. First, a fluorescent reference image and corresponding phase contrast video are selected (Fig. 4). The video is converted into an image stack using the function “videoreader” and the user is prompted to rotate the images such that the microposts appear as an array of rows and columns. At this step, a pixel-to-micrometer conversion value is established based on the known spacing between microposts. The user is then prompted to select a region of interest that encloses the cell: the exclusion of posts exterior to the cell allows for faster data analysis. Based on the selected region, the image is divided into a grid such that each box within the grid encloses an individual micropost (Fig. 6A). Each box is converted to a binary image with the function “im2bw” using a thresholding value that is unique to that micropost. Using a local threshold in this manner compensates for uneven illumination in the image and ensures that each micropost is binarized to a circular shape with similar diameters. Any noise in the binarized image is removed using “bwmorph” with the ‘clean’ and ‘majority’ options, and any holes are filled in using “bwfill” with the ‘holes’ option. Then, the centroid of the binarized image is calculated using “regionprops” and the pixel distance between its position in the phase image relative to the reference image is calculated (Figs. 6B,C). This process is repeated for each micropost and for each frame within the captured video.
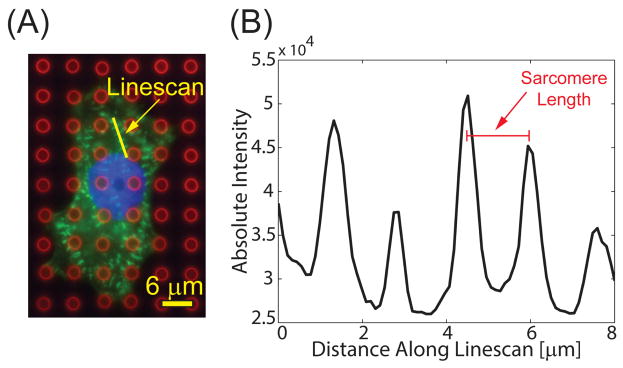
Figure 6.
(A) Composite immunofluorescent image showing microposts (red), sarcomeres (green), and nucleus (blue). Adapted from [40]. (B) A linescan (shown in yellow) is drawn across at least four sarcomeres, and the distance between the peaks is averaged to determine sarcomere length.
With a data array containing the change in position of a micropost for each frame, the pixel distance is then converted into a corresponding deflection based on the pixel-to-micrometer conversion value. With the deflection δi determined, the force Fi acting on the ith micropost is calculated by Hooke’s law:
| (3) |
where the stiffness, k, is calculated using slender-beam bending theory as seen in Eq. 1, in which the micropost is assumed to have one fixed end and undergoes small deflections. The total force generated by a cell is calculated by summation of the absolute magnitude of the forces measured at each micropost underneath the cell. The total passive tension and total active tension are on the order of 100 nN and 10 nN, respectively.
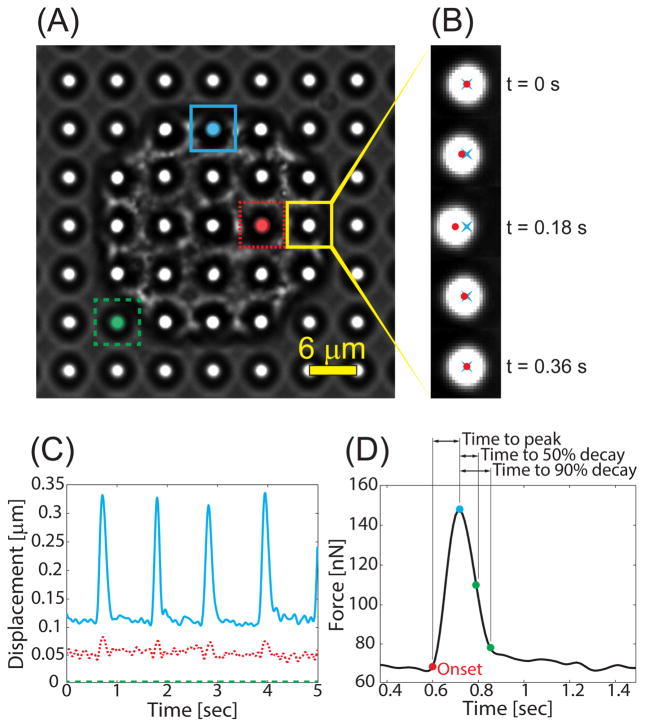
Individual contractile events are identified by drawing a user-selected box, which defines the baseline region before and after contraction. The peak force is determined by calculating the maximum force during each contraction (Fig. 5). The passive tension is determined by calculating the average resting force before and after a twitch. The twitch force is the difference between the passive tension and peak force for each contraction. The twitch frequency (in Hz) can be determined by calculating the inverse of the difference in time between adjacent peak forces. We define the onset and end of each pulse (with a user-defined threshold value) as the first and last times that the force is above 5% of the pulse amplitude, respectively. If the data is excessively noisy or otherwise irregular, the pulse onset and end points may be selected manually. The pulse onset and end times are then used to calculate the pulse duration and time to peak. For a study, we calculate the averages of each contractile property across multiple beats of a cell and for multiple cells within a population.
Figure 5.
(A) Phase contrast image of a cardiomyocyte on microposts. The image is segmented into a grid such that each grid-box contains a single micropost. Adapted from [39]. (B) The centroid of the micropost is measured for each video frame (red ●), and the distance from the reference position (blue ×) is used to measure its displacement. (C) For each micropost, the deflection is calculated at each frame to produce a waveform. Posts near the edge of the cell (blue) typically deform much more than posts near the middle (red). (D) The calculated force waveform from a single contraction from the cardiomyocyte and the characteristic times.
With the same deflection data, the instantaneous velocity of a micropost can be determined by the difference in its deflection before and after the time point of interest, expressed as a central difference:
| (4) |
The resulting velocity is on the order of 1 μm/s. With the values for twitch force and twitch velocity determined, the twitch power can be calculated as:
| (5) |
In similar fashion to force and velocity, the total power generated by the cell is calculated by summing the power at each micropost at each instance of time, yielding a twitch power produced by the cell on the order of 10 fW. Overall, the entire video analysis process takes between 5 to 10 minutes per cell for someone experienced with the code. Most of this analysis time is spent selecting and inputting settings, and only a short time (less than 2 minutes) is required to load the video and obtain the post displacements in MATLAB. A custom GUI was developed to make the analysis easier and faster. The MATLAB code and GUI are available on request by e-mailing the corresponding author.
4.2 Immunofluorescent Analysis
While contraction is a good means of assessing an SCCM’s functional maturation, its structural maturation can be assessed by analyzing the organization and size of its myofibrils. In particular, sarcomeres are known to be key in force generation and align along the major axis of a matured cardiomyocyte [1]. Maturity causes an increase in the Z-band distance between sarcomere units [42], indicating the formation of actin-myosin cross bridges and better coupling between the sarcomeres. Therefore, sarcomere length and Z-band width are good indicators of myofibril maturation.
For the immunofluorescently stained substrates, we conduct our image analysis with the NIH ImageJ software. Sarcomere length is measured by selecting a region of parallel sarcomeres within the α-actinin image and drawing a line across the center of four or more consecutive sarcomeres. The Plot Profile function reports the intensity profile along the line, which is used to determine the distance between the brightest points of each sarcomere (Fig. 6). We repeat this process for at least three different regions within each cell to account for spatial variability, and average these values to characterize the sarcomere length, which is on the order of 1 μm. To measure the average Z-band width, a “Fast Filter” function is implemented to obtain the outlines of the individual sarcomeres. The “Despeckle” and “Remove Outlier” functions remove background noise before the image is converted to a binary image. Once binarized, a region of interest enclosing at least four sarcomeres is specified. Each sarcomere is then fitted to an ellipse using the “Analyze Particles” function, and the major axis is measured for each ellipse to get the Z-band width. Similar to sarcomere length measurements, this process is repeated at least three times in different sections of the cell to obtain an average measurement.
5 Conclusions
Micropost arrays offer many advantages over other techniques for characterizing the contractility of cardiomyocytes, including the ability to measure multi-directional forces, twitch velocity, and twitch power. Additionally, the ordered arrays of microposts offer simple calculation of force from deflection data using small deformation cantilever beam theory. By changing the height, diameter, or spacing of the microposts, different substrate stiffnesses can be achieved. Soft lithography can produce a high yield of identical arrays of microposts from a common master, which reduces experiment to experiment heterogeneity. The use of single-cell assays also allows for more direct assessment of cardiomyocyte forces without the extracellular environment or multicellular interactions obscuring this measurement.
Our lab has utilized micropost arrays to assess the effect of ECM protein coatings [39], thyroid hormone (T3) treatment [40], and let-7 microRNA upregulation [41] on the contractile maturation of human SC-CM. Our ECM study showed that laminin provided increased cell spreading and attachment to the microposts when compared to fibronectin and collagen IV, but the generated force, velocity, and power were insignificant between the three ECM proteins. In the thyroid hormone study, we found that human induced pluripotent SC-CMs subjected to a treatment by thyroid hormone T3 showed significantly increased contractile force (from 7.5 nN in control cells to 12.3 nN in T3 treated cells), significantly decreased time to peak (from 0.25 s to 0.15 s) and time to 90% relaxation, and significantly increased sarcomere length (from 1.67 μm to 1.73 μm) all suggesting increased maturation. Finally, the overexpression of let-7, a microRNA family that is highly upregulated during cardiac maturation, showed increased contractile force generation from 7.77 nN to 11.32 and 9.28 nN when let-7i and let-7g, respectively, were overexpressed. Additionally, overexpression of let-7i lowered the beat frequency from 1.57 Hz to 1.05 Hz, and overexpression of let- 7g reduced it to 0.92 Hz. Increased force generation and decreased beat frequency both suggest increased contractile maturation. The combination of these techniques, as well as other mechanical or electrical stimuli, may lead to improved pro-maturation culture conditions for SC-CMs, potentially leading to more effective therapies for cardiac disease.
Micropost arrays are limited to measuring forces in a two-dimensional environment, which is an environment cells rarely experience in vivo, so some physiological context is lost. Additionally, analysis of the microposts requires optical measurement, so the spatial and temporal resolutions can be limited. However, comparisons can be between SC-CMs on microposts and engineered constructs on larger-sized microposts to understand cardiac contraction from the cellular scale to the tissue-level scale [57–60]. In the future, microposts may be simultaneously combined with electrical stimulation and calcium sensing, providing answers about electromechanical coupling during maturation in cardiomyocytes. Additionally, the effect of mechanical stimulation on maturation and contractility may be studied by stretching the micropost substrate (e.g. FlexCell®) while simultaneously measuring force and velocity. Finally, magnetic wires or magnetic nanoparticles may be mixed into the PDMS to potentially be used for mechanical stimulation by deflecting posts via magnetic fields or as a non-optical measurement system.
Highlights.
We manufacture microposts to measure forces from stem cellderived cardiomyocytes.
Soft lithography creates identical micropost arrays from a common master.
Microcontact printing functionalizes micropost tips with extracellular matrix.
Live imaging and image analysis quantify the deflections of micropost tips.
Contractile forces are calculated by modeling microposts as cantilever beams.
Acknowledgments
The authors wish to acknowledge support from the following sources: an NIH T32 fellowship award to KMB (EB001650), an NSF GRFP fellowship awarded to MLR, a DoEd GAANN fellowship awarded to NT (P200A130025), an NIH F32 fellowship award to AL (HL126332), and an NSF CAREER grant awarded to NJS. Part of this work was conducted at the University of Washington microfabrication/Nanotechnology User Facility, a member of the NSF National Nanotechnology Infrastructure Network.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braunwald E, Bonow RO. Braunwald's heart disease : a textbook of cardiovascular medicine. 9. Saunders; Philadelphia: 2012. [Google Scholar]
- 2.Freund C, Mummery CL. J Cell Biochem. 2009;107:592–599. doi: 10.1002/jcb.22164. [DOI] [PubMed] [Google Scholar]
- 3.Das AK, Pal R. J Tissue Eng Regen Med. 2010;4:413–421. doi: 10.1002/term.258. [DOI] [PubMed] [Google Scholar]
- 4.Dambrot C, Passier R, Atsma D, Mummery CL. Biochem J. 2011;434:25–35. doi: 10.1042/BJ20101707. [DOI] [PubMed] [Google Scholar]
- 5.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 6.Bellin M, Marchetto MC, Gage FH, Mummery CL. Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 7.Mercola M, Colas A, Willems E. Circ Res. 2013;112:534–548. doi: 10.1161/CIRCRESAHA.111.250266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosna F, Annunziato F, Pizzolo G, Krampera M. Cardiovasc Hematol Agents Med Chem. 2010;8:227–243. doi: 10.2174/187152510792481216. [DOI] [PubMed] [Google Scholar]
- 9.Mauritz C, Martens A, Rojas SV, Schnick T, Rathert C, Schecker N, Menke S, Glage S, Zweigerdt R, Haverich A, Martin U, Kutschka I. Eur Heart J. 2011;32:2634–2641. doi: 10.1093/eurheartj/ehr166. [DOI] [PubMed] [Google Scholar]
- 10.Singla DK, Long X, Glass C, Singla RD, Yan B. Mol Pharm. 2011;8:1573–1581. doi: 10.1021/mp2001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamura M, Miyagawa S, Miki K, Saito A, Fukushima S, Higuchi T, Kawamura T, Kuratani T, Daimon T, Shimizu T, Okano T, Sawa Y. Circulation. 2012;126:S29–37. doi: 10.1161/CIRCULATIONAHA.111.084343. [DOI] [PubMed] [Google Scholar]
- 12.Masumoto H, Matsuo T, Yamamizu K, Uosaki H, Narazaki G, Katayama S, Marui A, Shimizu T, Ikeda T, Okano T, Sakata R, Yamashita JK. Stem Cells. 2012;30:1196–1205. doi: 10.1002/stem.1089. [DOI] [PubMed] [Google Scholar]
- 13.Miki K, Uenaka H, Saito A, Miyagawa S, Sakaguchi T, Higuchi T, Shimizu T, Okano T, Yamanaka S, Sawa Y. Stem Cells Transl Med. 2012;1:430–437. doi: 10.5966/sctm.2011-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson C, Tran DD, George SC. Stem Cells. 2013;31:829–837. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin S, Zhang X, Zhan C, Wu J, Xu J, Cheung J. Biophys J. 2005;88:1489–1495. doi: 10.1529/biophysj.104.048157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domke J, Parak WJ, George M, Gaub HE, Radmacher M. Eur Biophys J. 1999;28:179–186. doi: 10.1007/s002490050198. [DOI] [PubMed] [Google Scholar]
- 17.Jacot JG, Martin JC, Hunt DL. J Biomech. 2010;43:93–98. doi: 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang WT, Yu D, Lai YC, Lin KY, Liau I. Anal Chem. 2012;85:1395–1400. doi: 10.1021/ac3022532. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Sun N, Bruce MA, Wu JC, Butte MJ. PLoS One. 2012;7:e37559. doi: 10.1371/journal.pone.0037559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacot JG, McCulloch AD, Omens JH. Biophys J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, Delgado SM, Wakatsuki T, Crone WC, Pruitt BL, Palecek SP. Int J Cell Biol. 2012;2012:508294. doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hersch N, Wolters B, Dreissen G, Springer R, Kirchgebner N, Merkel R, Hoffmann B. Biology Open. 2013;2:351–361. doi: 10.1242/bio.20133830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brixius K, Hoischen S, Reuter H, Lasek K, Schwinger RH. J Card Fail. 2001;7:335–341. doi: 10.1054/jcaf.2001.29902. [DOI] [PubMed] [Google Scholar]
- 24.Edes IF, Czuriga D, Csanyi G, Chlopicki S, Recchia FA, Borbely A, Galajda Z, Edes I, van der Velden J, Stienen GJM, Papp Z. Am J Physio Reg Int Comp Physio. 2007;293:R20–R29. doi: 10.1152/ajpregu.00537.2006. [DOI] [PubMed] [Google Scholar]
- 25.Park J, Ryu J, Choi SK, Seo E, Cha JM, Ryu S, Kim J, Kim B, Lee SH. Analytical Chemistry. 2005;77:6571–6580. doi: 10.1021/ac0507800. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Park J, Na K, Yang S, Baek J, Yoon E, Choi S, Lee S, Chun K, Park J, Park S. Journal of Biomechanics. 2008;41:2396–2401. doi: 10.1016/j.jbiomech.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 27.You J, Moon H, Lee BY, Jin JY, Chang ZE, Kim SY, Park J, Hwang YS, Kim J. Journal of Biomechanics. 2014;47:400–409. doi: 10.1016/j.jbiomech.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Vannier C, Chevassus H, Vassort G. Cardiovasc Res. 1996;32:580–586. [PubMed] [Google Scholar]
- 29.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saez A, Ghibaudo M, Buguin A, Silberzan P, Ladoux B. Proc Natl Acad Sci U S A. 2007;104:8281–8286. doi: 10.1073/pnas.0702259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saez A, Anon E, Ghibaudo M, du Roure O, Di Meglio JM, Hersen P, Silberzan P, Buguin A, Ladoux B. Journal of Physics- Condensed Matter. 2010;22 doi: 10.1088/0953-8984/22/19/194119. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Zhang X. Sensors and Actuators a-Physical. 2006;125:398–404. [Google Scholar]
- 34.Tanaka Y, Morishima K, Shimizu T, Kikuchi A, Yamato M, Okano T, Kitamori T. Lab on a Chip. 2006;6:230–235. doi: 10.1039/b512099c. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Lim CC, Sawyer DB, Liao RL, Zhang X. Cell Motility and the Cytoskeleton. 2007;64:718–725. doi: 10.1002/cm.20218. [DOI] [PubMed] [Google Scholar]
- 36.Kim K, Taylor R, Sim JY, Park SJ, Norman J, Fajardo G, Bernstein D, Pruitt BL. Micro & Nano Letters. 2011;6:317–322. [Google Scholar]
- 37.Taylor RE, Kim K, Sun N, Park SJ, Sim JY, Fajardo G, Bernstein D, Wu JC, Pruitt BL. Biomedical Microdevices. 2013;15:171–181. doi: 10.1007/s10544-012-9710-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribeiro AJS, Zaleta-Rivera K, Ashley EA, Pruitt BL. Acs Applied Materials & Interfaces. 2014;6:15516–15526. doi: 10.1021/am5042324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ. J Biomech Eng. 2014;136:051005-051001–051005-051010. doi: 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang XL, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, Sniadecki NJ, Ruohola-Baker H, Murry CE. Journal of Molecular and Cellular Cardiology. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, Sniadecki NJ, Laflamme MA, Ruzzo WL, Murry CE, Ruohola-Baker H. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XL, Pabon L, Murry CE. Circulation Research. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sniadecki NJ, Chen CS. Cell Mechanics. 2007;83:313–328. doi: 10.1016/S0091-679X(07)83013-5. [DOI] [PubMed] [Google Scholar]
- 44.del Campo A, Greiner C. Journal of Micromechanics and Microengineering. 2007;17:R81–R95. [Google Scholar]
- 45.MicroChem. Processing guidelines for SU-8 2000 permanent epoxy negative photoresist. Newton, MA, USA: [Google Scholar]
- 46.Fuard D, Tzvetkova-Chevolleau T, Decossas S, Tracqui P, Schiavone P. Microelectronic Engineering. 2008;85:1289–1293. [Google Scholar]
- 47.Schoen I, Pruitt B, Vogel V, Clarke D. Annual Review of Materials Research. 2013;43:589–618. [Google Scholar]
- 48.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sniadecki NJ, Han SJ, Ting LH, Feghhi S. Methods Cell Biol. 2014;121:61–73. doi: 10.1016/B978-0-12-800281-0.00005-1. [DOI] [PubMed] [Google Scholar]
- 50.Schoen I, Hu W, Klotzsch E, Vogel V. Nano Lett. 2010;10:1823–1830. doi: 10.1021/nl100533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia YN, Whitesides GM. Annual Review of Materials Science. 1998;28:153–184. [Google Scholar]
- 52.Ruiz SA, Chen CS. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 53.Efimenko K, Wallace WE, Genzer J. Journal of Colloid and Interface Science. 2002;254:306–315. doi: 10.1006/jcis.2002.8594. [DOI] [PubMed] [Google Scholar]
- 54.Amiji M, Park K. Biomaterials. 1992;13:682–692. doi: 10.1016/0142-9612(92)90128-b. [DOI] [PubMed] [Google Scholar]
- 55.Liu VA, Jastromb WE, Bhatia SN. Journal of Biomedical Materials Research. 2002;60:126–134. doi: 10.1002/jbm.10005. [DOI] [PubMed] [Google Scholar]
- 56.Boxshall K, Wu MH, Cui Z, Cui ZF, Watts JF, Baker MA. Surface and Interface Analysis. 2006;38:198–201. [Google Scholar]
- 57.Eschenhagen T, Fink C, Remmers U, Scholz H, Wattchow J, Weil J, Zimmerman W, Dohmen HH, Schafer H, Bishopric N, Wakatsuki T, Elson EL. Faseb Journal. 1997;11:683–694. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- 58.Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boudou T, Legant WR, Mu AB, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS. Tissue Engineering Part A. 2012;18:910–919. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Spreeuwel AC, Bax NA, Bastiaens AJ, Foolen J, Loerakker S, Borochin M, van der Schaft DW, Chen CS, Baaijens FP, Bouten CV. Integr Biol (Camb) 2014;6:422–429. doi: 10.1039/c3ib40219c. [DOI] [PubMed] [Google Scholar]