Abstract
F-box proteins are substrate receptors of the SCF (SKP1-Cullin 1-F-box protein) E3 ubiquitin ligase that play important roles in a number of physiological processes and activities. Through their ability to assemble distinct E3 ubiquitin ligases and target key regulators of cellular activities for ubiquitylation and degradation, this versatile group of proteins is able to regulate the abundance of cellular proteins whose deregulated expression or activity contributes to disease. In this review, we describe the important roles of select F-box proteins in regulating cellular activities, the perturbation of which contributes to the initiation and progression of a number of human malignancies.
Keywords: F-box, SCF, Ubiquitin, E3 ubiquitin ligases, Cancer
1. The ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) regulates numerous biological processes including cell cycle progression, cell growth, transcription and apoptosis [1]. The UPS directs target proteins to the 26S proteasome, where they are digested into small peptides. Approximately 80% of intracellular proteins are targeted for proteasomal degradation through the UPS [2]. The ubiquitin-dependent proteolysis of proteins is a highly coordinated process that ensures the timely down-regulation of proteins, thereby controlling cellular activity and maintaining cell and tissue homeostasis [3-6]. It is frequently triggered by posttranslational modifications (e.g. phosphorylation) of the target substrate. Proteasomal degradation is preceded by the covalent attachment of multiple ubiquitin molecules linked together through lysine 48 (K48) to substrate proteins, a process known as polyubiquitylation (Figure 1A) [7, 8]. This form of polyubiquitylation is in contrast to the polyubiquitylation of protein substrates on lysine 63 (K63; involved in molecular assembly) or other residues (K6, K11, K27, K29 and the C-terminal methionine), and the covalent conjugation of a single ubiquitin moiety to the substrate protein (mono-ubiquitylation). These latter modifications have non-proteolytic functional consequences such as kinase activation, DNA repair, and protein trafficking [9, 10].
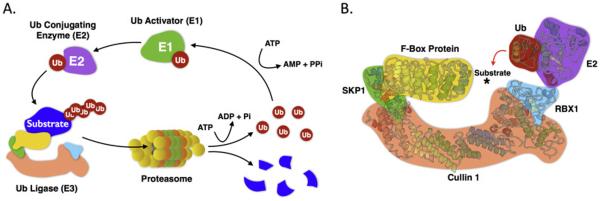
Figure 1.
(A) Schematic of the various steps involved in the ubiquitin-proteasome degradation pathway. Ubiquitin molecules (red circles) are covalently linked to the substrate in a three-step enzymatic process, resulting in protein degradation by the 26S proteasome. The final step is mediated by an E3 ubiquitin ligase. (B) Structural architecture of the CRL1 (SCF) ubiquitin ligase. Cullin 1 (orange) serves as a scaffold on which, the rest of the complex is built. The F-box protein (yellow; SKP2 in this illustration) confers substrate specificity by recruiting target substrates to the SCF core. 3D protein structures were obtained from RCSB Protein Data Base: 1FQV (SKP1-SKP2 complex), 1LDK (Cullin 1-RBX1-SKP1-F-box SKP2), 1TTE (UBC1), and 1FXT (UBC1 catalytic domain-Ubiquitin complex).
Polyubiquitylation involves three distinct and consecutive enzymatic steps (Figure 1A): Ubiquitin activation by an E1 ubiquitin-activating enzyme (UAE), the transfer of the AMP-charged, activated ubiquitin to an E2 ubiquitin-conjugating enzyme (UBC), and the transfer of ubiquitin to the substrate through the activity of an E3 ubiquitin ligase [3-5, 8]. E3 ubiquitin ligases are responsible for the selective recognition of the substrate protein prior to its ubiquitylation.
2. The SCF E3 ubiquitin ligases and F-box proteins
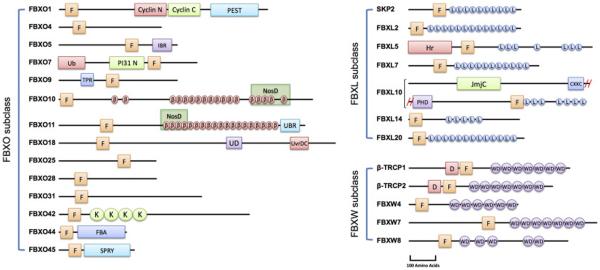
The human genome encodes more than 700 E3 ubiquitin ligases, classified into two main families: the RING (Really Interesting New Gene) and the HECT (Homologous to the E6-AP Carboxyl Terminus) domain containing E3 ubiquitin ligases [2, 11, 12]. Cullin-RING E3 ubiquitin Ligases (CRLs) constitute the largest family of E3 ligases and play significant roles in various physiological and pathological processes including tumorigenesis [13-15]. Family members include cullin 1, 2, 3, 4A, 4B, 5, and cullin 7 as well as the cullin-like proteins PARC and APC2. The general description of the structure and function of CRLs has been described in several excellent reviews [12, 14, 16-21]. The SCF (SKP1-Cullin1-F-Box protein; also known as CRL1) ubiquitin ligase is the prototype and most characterized member of this family of E3 ligases. The structure of the SCF ubiquitin ligase consists of a scaffold protein (cullin 1), which interacts via its C-terminus with RBX1, a RING-domain protein essential for the recruitment of E2s, and via its N-terminus with the SKP1 adaptor protein (Figure 1B). SKP1 in turn interacts with a number of proteins collectively called F-box proteins, which selectively recognize and recruit the substrate proteins for polyubiquitylation by the E2 enzymes. In mammalian cells, the SCF ligase associates with 69 unique substrate receptors, collectively known as F-box proteins, thus constituting a large family of distinct SCF ligases with varying specificity [22-24]. SCF ligases are best known for their roles in the regulation of cellular proliferation, apoptosis and differentiation. F-box proteins are classified into three subfamilies (FBXW, FBXL and FBXO) depending on the presence of specific domains other than the F-box motif, which is important for binding to the SKP1 adaptor (Figure 2). The FBXW subfamily contains WD40 repeats, which are known for their ability to mediate protein-protein interactions; the FBXL subfamily is characterized by the presence of leucine-rich repeats; and the FBXO (F-box only) subfamily is more or less a “catch-all” group with several members containing various other domains [2].
Figure 2.
Schematic of human F-box proteins with functional domains, arranged into subclasses FBXL, FBXO, and FBXW series. F: F-box motif; L: Leucine-rich repeats; Hr: Hemerythrin-like domain; JmjC: Jomonji C domain; CXXC: CXXC-type Zing finger; PHD: PHD-type Zing finger; Cyclin N: Cyclin N-terminal domain; Cyclin C: Cyclin C-terminal domain; PEST: proline (P), glutamic acid (E), serine (S), and threonine (T) sequence; IBR: In between RING fingers domain; Ub: Ubiquitin-like domain; PI31 N: Proteasome regulator, N-terminal domain; TPR: Tetratricopeptide repeat β: Parallel β-helix repeat; NosD: Parallel β-helix domain found in Periplasmic copper-binding protein; UBR: Ubiquitin protein ligase E component n-recognin; UD: UvrD-like helicase N-terminal domain; UvrDC: UvrD-like helicase C-terminal domain; K: Kelch repeat; FBA: F-box associated domain; SPRY: SPLA and the ryanodine receptor domain; D: D Domain of β-TRCP; WD: WD40 repeat.
A number of F-box proteins exhibit oncogenic or tumor-suppressive activities. Some of these, such as FBXW7, are mutated or exhibit deregulated expression at high frequencies in a large number of human malignancies, suggesting a prominent role in the development or progression of these cancers (Tables 1, 2). Others, such as β-TRCP1/2, exhibit context-dependent oncogenic or tumor-suppressive properties. Additional F-box proteins are emerging as critical regulators of cellular proliferation, metastasis, or cell death and are likely to be involved in tumorigenesis given their deregulated expression in cancer. Here we highlight the roles of some of the F-box proteins with cancer-related activities. In particular, we focus on F-box proteins that regulate the abundance of key proteins associated with diverse processes of particular relevance to the initiation and progression of human cancer, such as cell proliferation and cell death as well as invasion and metastasis (Figure 3). Although some of these F-box proteins lack definitive evidence supporting their classification as bona fide tumor suppressor or oncogenic proteins, they are described below on the basis of the available data supporting such roles for these proteins.
Table 1.
F-box proteins with established or potential oncogenic functions
| F-box protein |
Biological processes |
Deregulation in human malignancies |
Genetic evidence (mouse models of cancer) |
Ubiquitylation substrates (biochemical) |
|---|---|---|---|---|
|
SKP2
(FBXL1) |
Proliferation/ Metastasis/ Survival |
Overexpression - breast cancer, prostate cancer, colorectal cancer, pancreatic cancers, lymphoma, melanoma, and nasopharyngeal carcinoma [25-27]. |
Transgenic - enhanced tumorigenesis [28-30]. Knockout mice - impaired or delayed tumorigenesis [31- 35]. |
p27 [36-38], p21 [39, 40], p57 [41], p130 [42], Cyclin A [43], Cyclin D1 [40], Cyclin E [38], E2F-1 [44], ORC1 [45], CDT1 [46], CDK9 [47], c-MYC [48], TOB1 [49], RASSF1A [50], SMAD4 [51], RAG2 [52], UBP43 [53], FOXO1 [54], BRCA1 [55], BRCA2 [56], TRUSS [57], mH2A1 [58], MEF2C/D [59]. |
|
β-TRCP1/2
(FBXW1/11) |
Proliferation/ Genome stability |
Overexpression - colorectal [60], pancreatic [61], and liver cancers [62]. β-TRCP2 is overexpressed in breast, prostate and gastric cancers [63]. Somatic mutations in gastric cancer [64, 65], suggestive of tumor- suppressive role. |
Transgenic - mammary, ovarian and uterine cancers [66]. Dominant negative expression suppresses UVB-induced hyperplasia in the skin [67]. Knockout mice - non-tumorigenic [68, 69]. |
CDC25A [70, 71], β-Catenin [72], SNAIL [73], ATF4 [74], p105 [75], MDM2 [76], XLF [77], IkBα [78], Pro-caspase-3 [79], PDCD4 [80], p100 [81], DEPTOR [82], WEE1 [83], CReP [84], TIAM1 [85], EMI1 [68]. |
|
FBXO42
(JFK) |
Proliferation/ Survival/ Metastasis |
Overexpression - breast cancer [86]. | None | p53 [87], ING4 [86]. |
|
EMI1
(FBXO5) |
Proliferation | Overexpression - ovarian and other tumors [88-90]. |
Knockout mice - embryonic lethal [91]. |
Endogenous inhibitor of the APC/CCDH1 and APC/CCDC20 ubiquitin ligase [92]. No known ubiquitylation substrates. |
| FBXO44 | Genome stability |
Expression correlates negatively with BRCA1 expression in sporadic breast cancer [93]. |
None | BRCA1 [93], RGS2 (substrate for CRL4FBXO44) [94]. |
| FBXO9 | Survival | Overexpression - multiple myeloma [95]. |
None | TEL2 [95] TTI1 [95]. |
| FBXO28 | Proliferation | Overexpression - breast cancer [96]. | None | c-MYC (non-proteolytic ubiquitylation) [96]. |
|
FBXL10
(KDM2B) |
Proliferation/ Survival |
Overexpression - pancreatic cancer [97]. |
Transgenic - cooperates with KrasG12D to promote pancreatic ductal adenocarcinoma [97]. |
H2A (non-proteolytic ubiquitylation) [98]. |
| FBXO7 | Proliferation/ Survival |
Overexpressed in epithelial tumors [99]. |
Overexpression in transformed murine fibroblasts renders them tumorigenic in nude mice [99]. Promotes T cell lymphomagenesis [100]. |
CD43 [101], cIAP1 [102], HURP [103]. |
Table 2.
F-box proteins with established or potential tumor suppressor activities
| F-box protein |
Biological processes |
Deregulation in human malignancies | Genetic evidence (mouse models of cancer) |
Ubiquitylation substrates (biochemical) |
|---|---|---|---|---|
|
FBXW7
(FBW7) |
Proliferation | Frequent deletion of chromosome 4q32 - in lung, head and neck, testicular, breast and endometrial cancers [138]. Mutations in T-ALL [140-143]. Mutations in multiple tumors [141, 154, 155]. |
Transgenic - Tumorigenic upon the expression of mutant FBXW7 [147, 148]. Knockout - Tumorigenic [156]. Deletion contributes to T-ALL [19, 157-160]. Deletion in the gut cooperates with APC or p53 loss to cause adenomas [148, 161, 162]. Deletion in mice suppresses chronic myeloid leukemia (CML) - oncogenic activity [163, 164]. |
Cyclin E [165], c-MYC [152], c-JUN [166], TGIF1 [167], NOTCH [168], MCL1 [169], KLF5 [170], PLK1 [171]. |
| FBXL2 | Proliferation | None | Transgenic mice [172]. Overexpression in lung cancer cells inhibited tumors in nude mice [173]. |
Cyclin D2 [174], Cyclin D3 [175], Aurora B [173], p85β [176], APP [172]. |
| FBXO4 | Proliferation/ Genome stability |
Low expression in prostate, thyroid, and breast adenocarcinomas and lymphoma [177]. Inactivating mutations in a subset of human esophageal cancers [178]. |
Knockout mice - develop lymphomas, histiocytic sarcomas, and mammary and hepatocellular carcinomas, and loss of Fbxo4 facilitates carcinogen-induced papilloma formation [179, 180]. Knockout mice - no tumorigenesis [181]. |
Cyclin D1 [177, 182], TRF1 [183]. |
| FBXO31 | Proliferation | Frequent LOH in 16q24.3 region in breast [184, 185], ovarian [186], and prostate [187] cancers. Low expression in breast [185], hepatocellular [188], and gastric cancers [189]. |
None | Cyclin D1 [190], CDT1 [191], MDM2 [192], PAR6C [193]. |
| FBXL7 | Proliferation/ Survival |
Increased ovarian and breast cancer risk [194, 195]. |
None | Aurora A [196], Survivin [197]. |
|
FBXL14
(PPA) |
EMT | None | None | SNAIL [198], SLUG [199], MKP3 [200], TWIST1 [201], SIP1 [201]. |
| FBXL20 | Genome stability |
Overexpressed in human colorectal adenocarcinoma [202]. |
Knockout mice - none tumorigenic [203]. | RIM1 [204], VPS34 [205]. |
| FBXL5 | Invasion and metastasis/ Genome stability |
Downregulation in metastatic gastric cancer [206]. Upregulated in lung cancer - oncogenic function [207]. |
Knockout mice - embryonic lethal [208, 209]. |
IRP2 [210, 211], p150 (Glued) [212], SNAIL [213], hSSB1 [207]. |
| FBXW8 | Proliferation | None | Knockout mice - none tumorigenic, embryonic lethal) [181, 214, 215]. |
IRS-1 [216], TBC1D3 [217], Cyclin D1 [218], IGFBP2 [214], HPK1 [219]. |
|
Cyclin F
(FBXO1) |
Genome stability |
Downregulated in hepatocellular carcinoma [220], and altered in lung cancer [221]. |
Knockout mice - embryonic lethal [222]. | CP110 [223], NUSAP [224], RRM2 [225], B-MYB [226]. |
|
Cyclin F
(FBXO1) |
Genome stability |
Downregulated in hepatocellular carcinoma [220], and altered in lung cancer [221]. |
Knockout mice - embryonic lethal [222]. | CP110 [223], NUSAP [224], RRM2 [225], B-MYB [226]. |
| FBXO45 | Survival/EMT | None | None | p73 [227], PAR4 [228], EMT factors: ZEB1/2, SNAIL, SLUG and TWIST1 [229]. |
| FBXO10 | Survival | Downregulated or inactivated in diffuse large B cell lymphomas (DLBCLs) [230], and associated with increased susceptibility of breast cancer [231, 232]. |
None | BCL2 [230]. |
| FBXO11 | Proliferation/ Invasion and metastasis |
Deleted or mutated in primary diffuse large B cell lymphomas (DLBCLs) [233]. |
Mouse mutant Jeff (JF) - none tumorigenic [234]. |
BCL6 [233], CDT2 [235, 236], p53 [237], SNAIL [238, 239], SLUG and SCRATCH [239]. |
|
FBXO18
(FBH1) |
Genome stability |
Frequently deleted in melanoma and lung cancer cell lines [240], and associated with breast cancer risk [241]. |
Knock out in mouse ES cell lines - impaired mitotic progression after decatenation stress [242]. |
ATF1 (in S. pombe) [243]. |
| FBXW4 | Unknown | Low expression, mutated, or deleted in several human cancers and correlates with poor patient survival in lung adenocarcinoma [244]. |
None | No known substrates |
| FBXO25 | Survival | Deleted in mantle cell lymphoma (MCL) [245]. |
Knockdown - accelerated lymphoma in Eμ-Myc mice and in a human MCL xenotransplant animal model [245]. |
HAX-1 [245], NKX2-5 [246], ISL1 [246], HAND1 [246], ELK-1 [247]. |
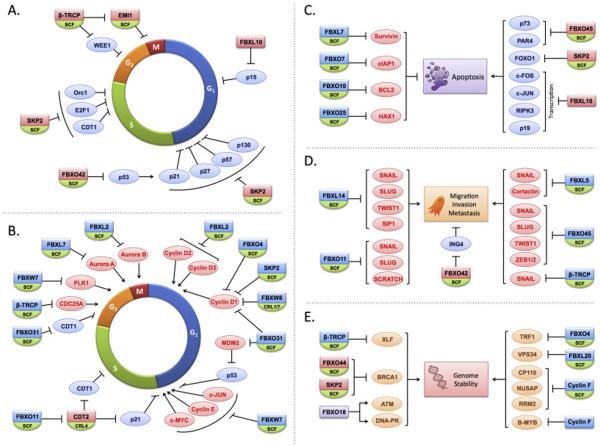
Figure 3.
The regulation of biological activities by F-box proteins implicated in human cancer. The schematics depict the impact of various F-box proteins that are known to be deregulated in human cancer on cellular processes relevant to the tumorigenic phenotype, including the cell cycle (A and B; with A: positive role in proliferation, with potential oncogenic F-box proteins, and B: with inhibitory role on proliferation, with potential tumor-suppressing role), cell survival (C), invasion and metastasis (D) as well as those that regulate the integrity of the genome (E). Positive regulators of cell proliferation, survival and metastasis are depicted in light red ovals, and those that negatively impact these processes in light blue. Some of these F-box proteins are represented more than once to highlight their involvement in the various biological activities. Factors which impact genomic stability are shown in orange ovals. The SCF ligase with the various F-box protein substrate receptors directs the ubiquitin-dependent proteasomal degradation of substrates to regulate these processes. Although they assemble SCF ligases, FBXL10, FBXO18 and cyclin F exhibit functions that are yet to be shown to require their assembly into SCF ligases, and as such are depicted as single subunits.
3. Oncogenic F-box proteins
A number of F-box proteins exhibit oncogenic activities, with SKP2 representing the most studied F-box oncoprotein. Others, such as β-TRCP1/2, have less established roles as bona fide oncogenes, but their deregulated expression in human cancer and evidence from experimental animal tumor models are consistent with their activities as potential oncogenes (Table 1, Figure 3). In this section, we describe the oncogenic activities of SKP2 and β-TRCP in detail, and highlight the emerging oncogenic activities associated with a number of additional F-box proteins.
3.1. SKP2 (FBXL1)
SKP2 (S-phase kinase-associated protein 2) was first identified as an interacting protein of cyclin A in transformed cells [43], and subsequently identified as the substrate recruiting subunit of the SCFSKP2 ubiquitin ligase [104]. SKP2 regulates a number of cellular activities including cell cycle regulation, metastasis, tumor differentiation, and apoptosis [27, 105-108]. Substantial evidence supports the conclusion that SKP2 is oncogenic. First, SKP2 is overexpressed in a large number of human tumors including breast, prostate, colorectal and pancreatic cancers as well as in lymphoma, melanoma, and nasopharyngeal carcinoma [25-27]. Second, studies with transgenic mice demonstrate that overexpressing Skp2 is sufficient to promote malignancy and that Skp2 cooperates with other oncogenes to drive malignancy. For example, Skp2 overexpression in the prostate gland induces hyperplasia, dysplasia and low-grade carcinoma [28], whereas its overexpression in the T-lymphoid lineage promotes Nras–induced T-cell lymphoma and decreased survival [29]. Furthermore, knockout studies demonstrated that SKP2 is critical for the initiation and/or progression of several tumors in a number of mouse tumor models, in large through the stabilization of the cyclin-dependent kinase (CDK) and cell cycle inhibitor p27, a well-established substrate of the SCFSKP2 ligase. For example, Skp2 deficiency suppressed spontaneous pituitary tumors in the Rb+/− mice [31] and delayed breast cancer development in MMTV-Neu mice [32]. In addition, Skp2 deficiency rendered mice resistant to the development of lymphomas and sarcomas that develop spontaneously in the Arf−/− background, and significantly suppressed adrenal and prostate tumors with inactivated tumor suppressor Pten [33]. It is noteworthy that the role of SKP2 in restraining p27 in the absence of functional PTEN seems to be important for the survival, growth and/or migration of a number of tumors of various origins. This is supported by several studies demonstrating a relationship between PTEN deletion or downregulation and SKP2 overexpression and p27 reduction in a number of cancer cell lines. For example, PTEN overexpression in the glioblastoma cells down-regulated SKP2 and increased the stability of p27 resulting in G1/S cell cycle arrest, which can be inhibited by SKP2 overexpression [109]. Similarly, thrombin-induced growth and migration of lung cancer cells is dependent on the downregulation of PTEN and concomitant increase in SKP2 and the resulting reduction of p27 [58]. Interestingly, Skp2 deficiency can also suppress tumors induced by chemical carcinogens such as the DMBA (7,12-dimethylbenz(a)anthracene)/TPA (12-O-tetradecanoylphorbol-13-acetate)-induced skin tumors, but this is independent of its ability to downregulate p27 [34].
The best understood mechanistic basis for the oncogenic functions of SKP2 stems from its role in promoting cell cycle progression via its ability to promote the ubiquitin-dependent proteolysis of the CDK inhibitors p21CIP1, p27KIP1 and p57KIP2. In addition, the SCFSKP2 ligase promotes the cell cycle-dependent degradation of cyclins D1, E and A, which are necessary activators of CDKs in G1, S and early G2 phase of the cell cycle. This latter activity ensures the availability of CDK molecules for assembling distinct cyclin-CDK complexes with varying specificity necessary for the irreversible progression of the cell cycle from one phase to the next. SKP2 also targets the retinoblastoma-like protein 2 (RBL2), also known as p130, for degradation, and this is likely to contribute to its oncogenic activity [42].
Paradoxically, several other SKP2 substrates, such as E2F1, ORC1, CDT1, and c-MYC are positive regulators of the cell cycle and thus, their degradation through the SCFSKP2 ubiquitin ligase may not directly contribute to its oncogenic activity, but may be important for terminating their aberrant activity during the wrong cell cycle stage. This is certainly the case for CDT1, which is a replication initiation factor necessary for the establishment of pre-replication initiation complexes (Pre-RC) from late mitosis until early entry into S-phase when replication initiation begins [110]. During S-phase, CDT1 must be inactivated or eliminated to prevent further re-initiation of DNA replication from the same origins of replication, a phenomenon referred to as re-replication. Re-replication is deleterious to cells owing to the accumulation of replication intermediates and collapsed replication forks, leading to check point activation and cell cycle arrest [110]. SCFSKP2 cooperates with the cullin 4-based E3 ligase CRL4CDT2 to ensure that CDT1 is effectively eliminated during S-phase of the cell cycle [111]. SKP2-mediated degradation of CDT1 is preceded by the phosphorylation CDT1 on Thr-29 by cyclin A-CDK2, which is necessary for recognition by SKP2 [46, 112, 113].
Similar to CDT1, the destabilization of E2F1 by the SCFSKP2 ligase may be important to limit its activity in S and G2 phases of the cell cycle [44]. The role of SKP2 in mediating the degradation of c-MYC however, is far less clear. On one hand, although SKP2 was shown to promote the degradation of c-MYC, SKP2 is transcriptionally activated by c-MYC and is critical for the induction of c-MYC-dependent genes [48, 114]. On the other hand, SKP2 is involved in the targeted proteolysis of another E3 ligase, TRPC4AP (transient receptor potential cation channel, subfamily C, member 4-associated protein)/TRUSS (tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein) [57], which is a CRL4 substrate receptor that is shown to promote the proteolysis of c-MYC and is downregulated in most cancer cell lines [115]. Thus, SKP2 also stabilizes c-MYC by downregulating an E3 ligase, which degrades c-MYC. Interestingly, SKP2 is also a direct transcriptional target of the v-MYC myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) [116, 117], and the amplification of MYCN is found in 25% of neuroblastoma resulting in increased proliferation and decreased apoptosis [118]. Whether SKP2 is critical for the development of these malignancies remains to be determined.
The pleiotropic transcription factors MEF2C and MEF2D are shown to be substrates of SKP2 in late G1 for degradation following their phosphorylation by cyclin D1/CDK4 [59]. Since MEF2C and MEF2D induce the expression of p21, the ubiquitylation and degradation of MEF2C/D by SKP2 ensures S-phase entry through reduction of p21 protein.
In addition to its role in promoting G1/S transition through the targeted proteolysis of p27 and p21, SKP2 has recently been shown to also promote G2/M transition. This is mediated through its ability to promote the degradation of macroH2A1 (mH2A1, also known as H2AFY), an epigenetic factor that helps HP1 promote transcriptional repression [119, 120]. This results in the induction of oncogenic CDK8 [58], which is frequently amplified in colorectal cancer [121]. Importantly, deregulation of the SKP2/mH2A1/CDK8 pathway is associated with human breast cancer progression and correlates with poor patient survival [58]. Consistently, mH2A1 knockdown or CDK8 expression was sufficient to restore tumorigenicity in breast tumors deficient of SKP2 in a mouse tumor model, demonstrating the importance of targeted proteolysis of mH2A1 by SKP2 in breast cancer. Interestingly, CDK8 was shown to promote p27 degradation by phosphorylation and subsequent ubiquitylation and degradation via the SCFSKP2 ligase [58]. This pathway may also be relevant in other tumors that are dependent on CDK8 expression. For example, mH2A1, through inhibiting CDK8, can suppress melanoma progression through direct transcriptional regulation of CDK8 [122].
SKP2 may exert additional oncogenic activity through the ubiquitylation of several other proteins without a direct link to the cell cycle (Table 1). For example, SKP2 promotes the degradation of the transcription factor FOXO1 [54], which positively regulates apoptosis. In pituitary tumors deficient of Rb, SKP2 limits E2F1-dependent apoptosis primarily through the destabilization of p27 and the consequent increase in the binding of cyclin A to E2F1, which inhibits its activity [123]. Other substrates of SCFSKP2 without direct link to the cell cycle include SMAD4, RAG2, UBP43, BRCA2, and papillomavirus E7 [51-54, 56, 124].
3.2. β-TRCP1/2 (FBXW1 and FBXW11)
β-TRCP (β-transducin repeat-containing protein) proteins regulate multiple cellular processes by targeting various proteins for proteasomal degradation, including WEE1, claspin, CDC25A, β-catenin, IKBα, and EMI1 (Table 1). There are two paralogues of β-TRCP (β-TRCP1 and β-TRCP2) in mammals, but their functions with regards to substrate recognition and degradation are indistinguishable [63]. Several studies support an oncogenic role for these two proteins. For example, β-TRCP1 is overexpressed in colorectal [60] and pancreatic cancers [61] and in biopsy samples of hepatoblastoma [62]. Moreover, its expression in colorectal cancer is associated with poor patient outcome [60]. Similarly, β-TRCP2 is overexpressed in breast, prostate and gastric cancers [63]. Data in support of oncogenic β-TRCP proteins include the development of mammary, ovarian and uterine cancers in transgenic mice that overexpress β-TRCP1 in the mammary gland or other tissues [66]. Furthermore, suppression of β-TRCP2 activity by the expression of a dominant negative version of this protein in the epidermis of mice decreased UVB-induced hyperplasia in the skin, suggesting a tumor-promoting role in this tissue [67]. It is noteworthy that somatic mutations in β-TRCP1 and β-TRCP2 that disrupt the E3 ligase activity were found in a subset of gastric cancers and this was associated with the stabilization of β-catenin [64, 65], suggesting that these two proteins may also exhibit tumor-suppressive activities, at least in gastric cancer.
β-TRCP1/2 proteins are best known for their ability to regulate cell cycle progression primarily through regulating the activity of the CDK1 kinase [125]. These E3 ligases inhibit CDK1 both during S phase and in mitosis through the ubiquitylation and degradation of CDC25A and EMI1 (early mitotic inhibitor-1), respectively. CDC25A is a phosphatase that removes an inhibitory phosphorylation on CDK1, and its degradation ensures that CDK1 is not active until S-phase is completed. In mitosis, the activity of CDK1 is generally kept low by β-TRCP, except in early mitosis when the anaphase-promoting complex (APC) ubiquitin ligase APC/CCDC20 ubiquitylates and degrades the CDK1 inhibitor p21. β-TRCP additionally ubiquitylates and degrades EMI1, which is an F-box protein (see Section 3.3) and endogenous inhibitor of the APC ligase. This leads to APC/CCDC20 activation and the ubiquitylation and degradation of the CDK activators cyclins A and B. In G2 phase of the cell cycle, β-TRCP activates CDK1 through the ubiquitylation and degradation of WEE1, a tyrosine kinase that phosphorylates and inhibits CDK1, and whose degradation is essential for CDK1activity.
β-TRCP1/2 proteins are also crucial for regulating cellular responses to DNA damage, particularly during S and G2 phases of the cell cycle [125]. In response to DNA damage, CDC25A is degraded via the SCFβ-TRCP1/2 ubiquitin ligase following its phosphorylation by the DNA damage-activated CHK1 and CHK2 kinases. This results in CDK1 inhibition and cell cycle arrest [70, 71]. Following DNA repair, the SCFβ-TRCP1/2 ligase ubiquitylates and degrades claspin and WEE1 and restores CDK1 activity. It is tempting to speculate that the main oncogenic functions (and perhaps some of the tumor suppressor activity) of β-TRCP proteins may result from altered cell cycle progression and aberrant DNA damage control, although definitive evidence for this remains to be established.
β-TRCP1/2 proteins have been shown to play additional roles in the recovery of cells from DNA damage. Using the novel technique of Ubiquitin Ligase Substrate Trapping, wherein an E3 ligase is fused to a ubiquitin-associated (UBA) domain to trap and identify ubiquitylation substrates [126], Loveless, et al., identified the constitutive reverter of eIF2α phosphorylation (CReP) as a novel substrate of β-TRCP [84]. The phosphorylation of the eukaryotic transcription initiation factor eIF2α down-regulates protein synthesis under a variety of stress conditions [127]. CReP is a specificity subunit of the phosphatase Protein Phosphatase 1 (PP1), which targets eIF2α to promote the removal of a stress-induced inhibitory phosphorylation and increase cap-dependent translation. Importantly, the full downregulation of CReP by β-TRCP in response to DNA damage is required for the maximal induction of eIF2α phosphorylation, and is important for reducing translation as cells recover from DNA damage [84].
3.3. Other F-box proteins with oncogenic properties
The F-box protein JFK (Just one F-box and kelch domain-containing protein) has been implicated in the ubiquitylation and subsequent degradation of the p53 tumor suppressor [87]. RNAi-mediated knockdown of JFK stabilized p53 and promoted apoptosis, G1 cell cycle arrest, and sensitized cells to ionizing radiation (IR) [87]. More recently, ING4 (inhibitor of growth protein 4), a type II tumor suppressor protein, has been identified as a novel ubiquitylation substrate for JFK [86]. JFK-mediated downregulation of ING4 results in hyperactivation of the canonical NF-κB pathway. Consistent with its oncogenic role, JFK levels are upregulated in breast cancer, inversely correlating with ING4 levels. Moreover, the JFK-directed ubiquitylation of ING4 promoted angiogenesis and metastasis of breast cancer in vitro and in vivo [86].
Evidence pointing towards an oncogenic role for the APC inhibitor and F-box protein EMI1 (also known as FBXO5) comes primarily from its frequent overexpression in human malignancies. For example, EMI1 is significantly overexpressed in ovarian tumors and its overexpression correlates positively with high histological grade and poor patient survival [88, 89]. Similarly, EMI1 overexpression was noted in a number of malignant tumors when compared to benign tumors [90]. Experimental evidence supporting an oncogenic role for EMI1 comes from the observations that its overexpression enhances proliferation and genomic instability in p53-deficient cells [128], and enhances the proliferation of chronic myeloid leukemia cells expressing the BRC-ABL fusion oncoprotein [129]. Mechanistically, EMI1 regulates mitosis by inhibiting the APC/CCDC20 and APC/CCDH1 ubiquitin ligases through interaction with the APC activator CDC20 [92]. EMI1 controls the timing of APC ubiquitylation activity before the spindle checkpoint becomes active and stabilizes mitotic cyclins in early mitosis. However, it has also been shown that the APC ligase is activated on entry into mitosis regardless of the presence of non-degradable EMI1, and the timing of cyclin A degradation is not affected [130]. Di Fiore et al. showed that EMI1 regulates mitotic entry by promoting the stabilization of geminin and the mitotic cyclins A and B in G2 phase to prevent re-replication and allow cells to enter mitosis [130].
The F-box protein FBXO44 is another F-box protein with oncogenic properties. SCFFBXO44 has been reported to facilitate the ubiquitin-mediated degradation of BRCA1, which functions in DNA repair, cell cycle checkpoint regulation, apoptosis, and transcriptional regulation [93]. FBXO44 expression inversely correlated with BRCA1 expression in human breast cancer tumors, suggesting that FBXO44 may contribute to breast cancer progression [93]. More recently, the signaling protein RGS2 has been identified as a novel FBXO44 substrate, but this involves its assembly with the CRL4 ubiquitin ligase and not the SCF ligase [94]. RGS proteins regulate signaling via G-protein coupled receptors and low expression of RGS2 has been associated with prostate cancer [131], suggesting a possible link between FBXO44 and the progression of prostate cancer.
FBXL10 functions as a substrate receptor of the SCF complex, but also contains a JmjC domain for histone demethylase activity. FBXL10 is shown to be required for histone H2A monoubiquitylation on lysine 119, which contributes to the differentiation of embryonic stem cells (ESC) [98]. FBXL10 has also been shown to have oncogenic functions in pancreatic cancer as it is commonly overexpressed and its expression correlates with tumor progression and metastasis [97]. Tzatsos et al. showed that exogenous Fbxl10 cooperates with KrasG12D to promote pancreatic ductal adenocarcinoma (PDAC) formation in mouse models [97]. Notably, FBXL10 exhibits anti-apoptotic activity in vivo. Mice deficient in Fbxl10 exhibit neural tube defects associated with apoptosis in the neuroepithelium and mesenchyme of E9.5 embryos, and this was associated with increased expression of p19ARF, an inducer of apoptosis, in E8.5 embryos and mouse embryonic fibroblast cells [132]. FBXL10 may also suppress apoptosis via repression of c-FOS, c-JUN and RIPK3 promoters [133-135]. In addition, FBXL10 promotes cell proliferation via inhibiting the transcription of the CDK4 inhibitor p15 (INK4B) [136]. Paradoxically, FBXL10 was reported to function as a tumor suppressor protein, which inhibited cell growth and proliferation [137]. How FBXL10 impacts transcription from these genes, and whether these activities are dependent on the ubiquitylation of H2A or other substrates, or are dependent on its histone demethylase activity, remain to be determined.
The SCFFBXO9 ubiquitin ligase is reported to have oncogenic potential through its role in targeting TEL2 and TTI1 proteins for degradation [95]. Degradation of TEL2 and TTI1 within the mTORC1 complex was shown to inactivate mTORC1 signaling, thereby inhibiting cell growth, while concomitantly sustaining mTORC2 signaling through relief of negative feedback regulation in order to promote survival in human myeloma cells. FBXO9 is overexpressed in multiple myeloma, perhaps accounting for some of the anticancer activity of proteasome inhibitors in multiple myeloma [95].
Upon phosphorylation by CDK1/2, FBXO28 targets c-MYC for non-proteolytic ubiquitylation and promotes MYC-dependent transcription, proliferation and tumorigenesis in p53−/− immortalized mouse embryonic fibroblasts [96]. Accordingly, FBXO28 inactivation led to diminished MYC-induced transformation and tumorigenesis. Furthermore, enhanced expression and phosphorylation of FBXO28 correlate strongly with poor outcome and overall survival in breast cancer, corroborating the oncogenic role of FBXO28 in human cancer [96].
FBXO7 is another potential oncogene, which is highly expressed in epithelial tumors but not in normal tissues [99]. FBXO7 has been shown to directly bind to the cyclin D/CDK6 complex, promoting cell cycle progression. CDK6-dependent transformation of murine fibroblasts was observed with overexpression of FBXO7 in athymic nude mice [99]. Moreover, FBXO7 overexpression in hematopoietic stem and progenitor cells (HSPCs) reduces colony formation in a p53-dependent manner whereas in the absence of p53, FBXO7 promotes T cell lymphomagenesis in mice [100]. FBXO7 however, also exhibits tumor-suppressive properties via the ubiquitylation of certain oncogenes such as cIAP1 (Inhibitor of apoptosis protein 1) [102] and HURP (hepatoma upregulated protein), which requires cyclin B-CDK1-mediated phosphorylation for degradation [103]. Fbxo7 null mice showed increased populations of precursor pro-B cells and pro-erythroblasts although no functional information regarding tumorigenesis was observed [101].
4. F-box proteins with tumor-suppressive properties
The ability of cancer cells to sustain proliferation may be disrupted by F-box proteins that function to promote the degradation of key proteins essential for cell cycle progression or cell survival (Table 2, Figure 3). Several F-box proteins exhibit such activity and are described here as bona fide or candidate tumor suppressor proteins.
4.1. FBXW7
FBXW7 (also known as CDC4 and FBW7) is a well-established tumor suppressor gene, which resides on chromosome 4q32, a region deleted in 31% of all neoplasms, including 67% of lung cancers, 63% of head and neck cancers, 41% of testicular cancers, 27% of breast cancer and 17% of endometrial cancers [138]. In fact, FBXW7 is one of the most commonly mutated genes found in cancer [139]. Inactivating mutations of FBXW7 are also prevalent in a number of human malignancies, most noticeably in T cell acute lymphoblastic leukemia (T-ALL; approximately 30% of cases) [140-143]. The majority of these mutations are missense heterozygous mutations in three conserved arginines that confer high-affinity binding to the substrates [144-146], suggesting that these mutant FBXW7 proteins (FBXW7ARG) exhibit dominant negative activity [147, 148].
FBXW7 suppresses proliferation primarily through the targeted ubiquitylation and degradation of oncoproteins such as c-JUN, cyclin E, c-MYC, NOTCH1, MCL1, TGIF1 and KLF5 [139]. These and other FBXW7 substrates regulate a number of processes that are key to cancer initiation and progression, including proliferation, apoptosis, metabolism and differentiation. All of these substrates, except cyclin E, are oncogenic transcription factors, which regulate complex transcriptional programs culminating in enhanced cellular proliferation. There are three isoforms of FBXW7 protein, FBXW7α, FBXW7β and FBXW7γ, which exhibit differential localization to the nucleoplasm, cytoplasm, and nucleolus, respectively [149-153]. This ensures the targeted proteolysis of relevant substrates in the various cellular compartments. For example, nucleoplasmic c-MYC is ubiquitylated by FBXW7α, whereas c-MYC, which localizes in the nucleolus, is targeted for ubiquitylation by FBXW7γ, and both isoforms contribute to the suppression of the growth-promoting activity of c-MYC [149-153].
FBXW7 recognizes phosphorylated peptides, known as CDC4-phosphodegrons (CPD) motifs, within the substrate proteins. These motifs are commonly phosphorylated by glycogen synthase kinase 3 (GSK3) and antagonized by mitogenic signals through the PI3K-AKT pathway, which inhibit GSK3 activity. Thus, in addition to inactivating mutations in FBXW7, the abnormal AKT and PTEN activity, which is commonly found in human cancers, may additionally and indirectly inactivate the tumor-suppressive functions of FBXW7 through inhibiting the GSK3-dependent phosphorylation of FBXW7 oncogenic substrates. Importantly, some of the FBXW7 oncogenic substrates are stabilized (activated) in human cancers thorough mutations in their CPD motifs rendering them insensitive to FBXW7. For example, the degradation of c-MYC is enhanced by phosphorylation at Thr-56 [248, 249], which is located within the c-MYC CPD, and missense mutations in this region are found in lymphomas [250, 251]. Similarly, activating mutations in NOTCH1, which occurs in approximately in 50% of T-ALL, often target the PEST domain [252] containing the CPD motif, and these mutations are mutually exclusive with FBXW7 mutations found in these malignancies [140-143].
Experimental evidence for the potent tumor suppressor activity of FBXW7, particularly in hematologic malignancies, comes from data showing that Fbxw7 deletion in T cells or in hematopoietic stem cells (HSCs) was sufficient to cause T-ALL, which can be accelerated by the activation of oncogenes such as Notch1, or by the loss of additional tumor suppressor genes, such as Pten or p53 [147, 157-160]. Fbxw7ARG/+ mice however, did not develop T-ALL, but the Fbxw7ARG allele cooperated with deregulated Notch1 to drive T-ALL, and this was largely dependent on increased stability of c-Myc [147]. In addition to NOTCH1 and c-MYC, deregulated stability of cyclin E is also a key player in FBXW7-driven T-ALL. This is demonstrated in mice expressing FBXW7-resistant allele of cyclin E1 (Cyclin EΔCPD), which developed T cell malignancies, but additionally exhibited a chromosomal instability phenotype not seen in Fbw7ARG/+ T-ALL [253].
Deletion of FBXW7 in the mouse gut on the other hand, is not sufficient to cause colorectal tumors, but cooperates with other mutations commonly seen in human colorectal cancers, such as APC or p53 inactivation, to accelerate adenoma formation and increase tumor burden without progressing to more advanced stages [139, 161, 162]. This finding supports a functional role for FBXW7 mutations that are found in early stage human colon adenomas [254].
It is noteworthy that FBXW7 has been shown to exhibit oncogenic activity in chronic myeloid leukemia (CML). Deletion of Fbxw7 in mice resulted in the exhaustion/eradication of leukemia-initiating cells (LICs) by allowing them to exit quiescence or undergo apoptotic cell death. In one study, deletion of Fbxw7 resulted in c-Myc overexpression and p53-dependent apoptosis in LICs that inhibited tumor progression [163]. In another, the upregulation of c-Myc following Fbxw7 deletion abrogated quiescence in LICs, and rendered them sensitive to imatinib treatment in mice [164]. These results demonstrate that in CML, the main function of FBXW7 is to maintain the quiescence state of stem cell population. In principle, this is an anti-proliferative activity, but its abrogation results in tumor suppression due to the exhaustion of cancer initiating cells. Whether this role of FBXW7 is conserved in other cancer stem cells is yet to be determined.
4.2. FBXL2
Recent work from the Mallampalli laboratory identified potential tumor suppressor activities for the F-box protein FBXL2 by facilitating the ubiquitin-mediated degradation of crucial cell cycle regulators including cyclin D2, cyclin D3 and Aurora B. D-type cyclins partner with CDK4 and CDK6 to drive G1-to-S cell-cycle progression. Consequently, failure of D-type cyclin degradation may lead to premature entry into S-phase, whereas their accelerated degradation may arrest cells in G1. Indeed, Chen et al. found that ectopic expression of FBXL2 induces G0 cell cycle arrest and apoptosis in B-lymphoblastoid and leukemic cell lines. Importantly, the levels of FBXL2 are reduced in tissue samples from acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL) patients, and were accompanied by increases in cyclin D2 protein levels. FBXL2-mediated degradation of cyclin D2 is inhibited by calmodulin, which binds to the same site required for FBXL2-cyclin D2 interaction and suppressed FBXL2-mediated degradation of cyclin D2 [174].
In lung epithelial cells, ectopic expression of FBXL2 induced mitotic arrest, which was accompanied by the appearance of supernumerary centrosomes and tetraploidy and inhibited the growth and migration of tumorigenic cells in vitro, suppressing tumor formation in athymic nude mice [175]. This was likely mediated through its ability to ubiquitylate and promote the degradation of cyclin D3, which localizes to centrosomes [255], as well as to the accelerated degradation of Aurora B (see below). FBXL2-induced degradation of cyclin D3 in lung carcinoma cells was also stimulated by the chemotherapeutic agent vinorelbine, which increases FBXL2 protein, resulting in enhanced cyclin D3 degradation and apoptosis [175]. Similar to cyclin D2, FBXL2-mediated degradation of cyclin D3 is inhibited by calmodulin, which binds to cyclin D3 and prevents FBXL2-cyclin D3 binding and whose depletion accentuates vinorelbine-induced apoptosis [175].
The induction of mitotic arrest and apoptosis following FBXL2 expression in human lung cancer cells is aided through its ability to ubiquitylate and target for degradation Aurora B [173], a kinase involved in spindle assembly, chromosomal cohesion, alignment and segregation and in cytokinesis. Importantly, the stable expression of FBXL2 in A549 lung cancer cells inhibited tumor formation of athymic nude mice [173]. Similar results were obtained following the stabilization of FBXL2 in the human leukemic monocyte lymphoma cell line by BC-1258, a small molecule inhibitor of the SCFFBXO3 ubiquitin ligase, that targets FBXL2 for ubiquitin-mediated proteolysis [256], thus providing a therapeutic tool for lung and hematologic malignancies [173].
4.3. FBXO4
Unlike cyclin D2 and cyclin D3, which are ubiquitylated and degraded via a seemingly phosphorylation-independent recognition of cyclin D2 and D3 proteins, the degradation of cyclin D1 is mediated through a phosphorylation-dependent recognition of cyclin D1 by a number of F-box proteins (Tables 1, 2). Cyclin D1 is a key regulator of G1/S transition and its expression is often elevated in human malignancies such as lymphoma, breast cancer and esophageal cancer [181]. The FBXO4 protein has long been implicated in the regulation of cyclin D1 turnover in normal cycling cells, and this was shown to be dependent on the chaperone protein αβ-crystallin, which recognizes cyclin D1 following its phosphorylation on Thr-286 by GSK3β [177]. Consequently, the loss of FBXO4 in cancer cell lines and in a subset of primary cancers was associated with cyclin D1 accumulation [177], suggesting a possible tumor-suppressive function for this F-box protein. Importantly, inactivating mutations in FBXO4 are found in a subset of human esophageal cancers and these mutations prevented the dimerization of FBXO4, resulting in impaired cyclin D1 degradation and oncogenic transformation in colony formation assays [178]. Of note, dimerization and subsequent activity of the SCFFBXO4 E3 ligase are dependent upon GSK3β-mediated phosphorylation during G1/S transition [178, 257], and on the interaction between SCFFBXO4 and the 14-3-3ε protein [258]. The FBXO4-dependent degradation of cyclin D1 is antagonized by the mTOR kinase mTORC2 in non-small cell lung carcinoma cells [259]. Additional studies in mouse cells demonstrated that FBXO4 also promotes the ubiquitylation and degradation of cyclin D1 following DNA damage, and this too, was mediated by GSK3β-dependent phosphorylation of cyclin D1 on Thr-286, which in turn was dependent upon ATM signaling [260]. The loss of FBXO4-mediated degradation of cyclin D1 stimulated radio-resistant DNA synthesis and impaired the intra-S-phase checkpoint response in NIH3T3 cells, resulting in the accumulation of chromatid breaks and S-phase sensitivity to chemotherapeutic agent camptothecin (CPT) [260].
Consistent with a tumor-suppressive role for FBXO4, Vaites et. al., demonstrated that Fbxo4+/− and Fbxo4−/− mice developed lymphomas, histiocytic sarcomas, and mammary and hepatocellular carcinomas [179]. Furthermore, tumor and tissue samples extracted from these mice displayed elevated cyclin D1 levels [179]. The same group also demonstrated that Fbxo4 deficiency induces Braf-driven melanoma and this was dependent on cyclin D1 [261]. In addition, papilloma growth in Fbxo4+/− and Fbxo4−/− mice was reported following treatment with the esophageal carcinogen, N-nitrosomethylbenzylamine [180]. Furthermore, treatment of mice harboring these tumors with the CDK4/6 specific inhibitor PD0332991 reduced the number and size of these tumors, suggesting that sustained tumorigenesis is dependent on cyclin D1/CDK4 activity [180].
4.4. FBXO31
In addition to FBXO4 and unlike the case in the NIH3T3 mouse fibroblasts, the degradation of human cyclin D1 following DNA damage was reported to be dependent on the FBXO31 protein [190]. The FBXO31 gene resides on 16q24.3 and is a candidate tumor suppressor gene that exhibits loss of heterozygozity (LOH) in breast [184, 185], ovarian [186] and prostate [187] cancers. FBXO31 expression is also down-regulated in breast [185], hepatocellular [188] and gastric cancers [189]. In response to DNA damage, the ATM kinase was shown to phosphorylate FBXO31 on Ser-278, resulting in increased FBXO31 protein level and the degradation of cyclin D1, which was also phosphorylated by ATM on Thr-286, leading to cell cycle arrest in the G1 [190]. Conversely, RNAi-mediated knockdown of FBXO31 prevented DNA damage-induced cyclin D1 degradation and increased the sensitivity of melanoma cells to IR. This suggests that therapeutic targeting of FBXO31 could induce radiosensitization in melanoma. It is important to note that this study was conducted in a single melanoma cancer cell line, SK-MEL-28, and thus whether FBXO31 has similar activities in other cell types remains to be seen.
A recent study has challenged the role of FBXO4, FBXO31, SKP2, and FBXW8 (see below) in regulating cyclin D1 stability [181]. Kanie et al. developed a mouse knockout of Fbxo4 and found that these Fbxo4−/− mice developed normally and did not develop neoplasms up to 1 year of age, and additionally exhibited normal cyclin D1 levels. Furthermore, the apparent normal stability of cyclin D1 in Fbox4−/− mouse embryo fibroblasts (MEFs) cannot be accounted for through redundant or compensatory ubiquitylation and degradation of cyclin D1 by the other F-box proteins SKP2, FBXW8 or FBXO31, since the simultaneous deletion/depletion of these proteins did not impact cyclin D1 stability. This important finding casts doubt on the identity of the E3 ubiquitin ligase(s) that regulate cyclin D1 stability in cancer cells and may suggest that the aforementioned F-box proteins may only regulate cyclin D1 stability in a cell type-specific manner [181]. Clearly, additional studies are required to understand the mechanism underlying the rapid turnover of cyclin D1 protein, the consequences of its deregulated proteolysis, and whether it contributes to cyclin D1-induced tumorigenicity.
More recent findings suggested that FBXO31 may suppress tumor formation through the targeted proteolysis of two additional ubiquitylation substrates, CDT1 [191] and MDM2 [192]. The CDT1 protein is a chromatin licensing and DNA replication factor that associates with CDC6 to form the pre-RCs and is critical for the recruitment of the DNA replicative helicase MCM2-7 [191]. FBXO31-mediated ubiquitylation and degradation of CDT1 occurs in G2 and early M phase to prevent re-replication that occurs as a consequence of CDT1 accumulation [191]. Consistent with this observation, the depletion of FBXO31 by siRNA was shown to induce re-replication albeit at low levels [191]. CDT1 however, and as mentioned above, is degraded primarily during S-phase and this is mediated by the SCFSKP2 and CRL4CDT2 ubiquitin ligases. SCFSKP2 ubiquitylates CDT1 following its phosphorylation by cyclin A-CDK2 and degrades soluble CDT1, whereas CRL4CDT2-dependent ubiquitylation and degradation of CDT1 requires the interaction between CDT1 and chromatin-bound PCNA [110, 262]. We have recently demonstrated that CDT1, similar to p21 and SET8, two other substrates of the CRL4CDT2 [110, 262], begins to re-accumulate as cells complete S-phase and enter G2 [263]. Consistently, it was found that in G2 cells, CDT1 is prevented from being ubiquitylated and degraded by the CRL4CDT2 through two independent mechanisms. The first mechanism is mediated through the phosphorylation of CDT1 by the JNK and p38 kinases, thus preventing CDT1 recognition by the CRL4CDT2 ligase [264]. In addition, a second mechanism ensures that CDT2 is prevented from being recruited to chromatin through its phosphorylation by CDK1 [265]. Notably, the re-accumulation of CDT1, similar to SET8, in G2 was shown to be important for cell cycle progression [265]. Because extensive DNA re-replication is deleterious to cells, the re-replication resulting from failure to degrade CDT1 by FBXO31 in tumors with low or absent FBXO31 is likely to be only minor. Consistently, CDT1 increase or activation in response to CRL4CDT2 inactivation or the depletion of geminin (an endogenous inhibitor of CDT1) results in robust re-replication and is associated with cell cycle arrest and severe growth inhibition [110, 262]. On the other hand, inactivation of FBXO31 only resulted in the subtle levels of re-replication (7.6% vs. 4.6% in control cells), and does not inhibit growth [191]. Thus, the ability of FBXO31 to degrade CDT1 in G2 is limited and may be required only to dampen the level of CDT1 rather than obliterate its activity. It is tempting to speculate that in the absence of FBXO31, higher than necessary levels of CDT1 may result in minute levels of re-replication that may contribute to gene amplifications and/or genome instability associated with the tumorigenic phenotype.
Another FBXO31 ubiquitylation substrate, MDM2, is an E3 ligase, which ubiquitylates and promotes the degradation of p53. In response to DNA damage, MDM2 is targeted for degradation by the SCFFBXO31 ligase and this requires phosphorylation of both MDM2 and FBXO31 by ATM [192]. The proteasomal degradation of MDM2 results in the stabilization of p53 and growth arrest. It remains unclear however, the individual contribution of cyclin D1, CDT1 and MDM2 to the tumor suppressor functions of FBXO31 or whether FBXO31 exhibits additional tumor-suppressive activities independent of its ability to downregulate these proteins.
4.5. Other F-box proteins with tumor-suppressive functions
The gene encoding FBXO11 has been shown to be frequently and monoallelically deleted in diffuse large B-cell lymphoma cell lines (DLBCL) and in primary B cell-lymphomas, suggesting that it functions as a haploinsufficient tumor suppressor protein [233]. The tumor-suppressive function of FBXO11 in DLBCL is attributed to its ability to promote the ubiquitylation and degradation of BCL6, a key oncoprotein involved in the development of these malignancies. Restoration of FBXO11 expression in FBXO11-deficient cells promoted BCL6 degradation and suppressed proliferation and tumorigenesis in a xenograft mouse animal model of DLBCL [233]. In addition, CDT2, a substrate receptor for the CRL4CDT2 ubiquitin ligase, which is overexpressed in multiple human cancers [262] has been identified as a novel substrate of FBXO11, and its degradation is important for regulating cellular responses to TGF-β, cellular migration and exit from the cell cycle [235, 236, 266]. SCFFBXO11-mediated degradation of CDT2 stabilizes the CRL4CDT2 substrates p21 and SET8, and this is important for the response of epithelial cells to TGF-β stimulation and for cellular migration. Importantly, the migration defect of epithelial cells depleted of FBXO11 could be corrected by the reconstitution of catalytically active SET8 protein [235].
The F-box protein FBXW8 exhibits characteristics of tumor suppressor proteins. Unlike other F-box proteins, FBXW8 is unique in its ability to associate both with cullin 1 and with cullin 7 ligases [215]. FBXW8 has been shown to regulate cellular proliferation via ubiquitin-dependent degradation of proteins that promote cellular proliferation, such as IRS-1 [216, 267], TBC1D3 [217], cyclin D1 [218], and HPK1 [219]. The Cul7FBXW8 complex mediates the degradation of IRS-1, a crucial regulator of the IGF-1 signaling pathway [267]. Ectopic expression of FBXW8 in breast cancer cells led to increased IRS-1 degradation, a process dependent on mTOR/S6K activity [267]. More recently, the mTORC2 complex was shown to phosphorylate and stabilize FBXW8, directing it to the cytosol where it interacts with and facilitates the destruction of IRS-1 [216]. Cul7FBXW8 has also been reported to mediate the proteasomal degradation of the TBC1D3 oncoprotein in response to serum stimulation and growth factor signaling [217].
Contrary to the growth suppressing activity of FBXW8 described above, recent findings suggest that this protein may also be required for cell proliferation. For example, depletion of FBXW8 in human colon carcinoma cells significantly inhibited proliferation and this was attributed to the accumulation of its substrate cyclin D1 in the cytoplasm and sequestration of CDK1 [218]. This is the only example where failure to degrade cyclin D1 was shown to increase its abundance, and was associated with growth inhibition instead of growth stimulation. Suppressed growth following FBXW8 depletion was also seen in the human choriocarcinoma JEG-3 cells, and this was associated with cell cycle arrest in G2/M phase with concurrent upregulation of the p27 protein [268]. Whether cyclin D1 plays a role in this growth suppression remains to be investigated. Finally, knockdown of FBXW8 in pancreatic cancer cells decreased cellular proliferation via stabilization of HPK1, a serine/threonine kinase depleted by proteasomal degradation in over 95% of pancreatic cancer [219].
The gene encoding FBXL7 has been identified as a discriminative gene for ovarian cancer [194] and variants of FBXL7 have been associated with increased breast cancer risk [195], suggesting that this F-box protein may have a tumor-suppressive function. Although its specific role in tumorigenesis has not been elucidated, FBXL7 has been shown to target proteins involved in mitosis, cellular proliferation, and mitochondrial function [196, 197]. SCFFBXL7 mediates the proteasomal degradation of Aurora A, responsible for regulating mitotic spindle formation and chromosome alignment and segregation [196]. Interaction between FBXL7 and Aurora A occurred at the centrosome, specifically during mitosis, suggesting that FBXL7-directed destruction of Aurora A may be a crucial regulator of mitosis and cell proliferation. Consistently, overexpression of FBXL7 in transformed lung epithelial cells was sufficient to induce G2/M arrest and polyploidy, as well as an increase in apoptosis [196].
Recently, FBXW4 has also been implicated as a potential tumor suppressor protein, given that FBXW4 is under-expressed, mutated, or deleted in several human cancers and in clinical samples [244]. Furthermore, decreased expression of FBXW4 correlated with poor patient survival in lung adenocarcinoma [244]. The characterization of FBXW4 ubiquitylation substrates will further elucidate its mechanism of tumor-suppression in cancer.
5. Regulation of cell survival and apoptosis by F-box proteins
An important characteristic of cancer cells is their ability to evade apoptotic signals and survive in a growth-suppressing environment. This often results through the upregulation of anti-apoptotic or pro-survival genes or reciprocally, through the downregulation of pro-apoptotic proteins. The ability to initiate apoptosis following cellular stress or DNA damage is an important characteristic of some tumor suppressor proteins. Alternatively, oncogenic proteins may promote tumorigenesis by disabling their apoptosis-inducing machinery. In this section, we highlight the roles of some of the F-box proteins, which have been shown to regulate apoptosis (Figure 3).
5.1. F-box proteins with pro-apoptotic activities
In a recent study, the gene encoding FBXO25 was shown to function as a haploinsufficient tumor suppressor gene, which is heterozygously deleted in a subset of mantel cell lymphoma (MCL) [245]. In these cancers, the loss of one FBXO25 allele was sufficient to promote the survival of MCL cells through failure to ubiquitylate and degrade the anti-apoptotic and pro-survival protein HCLS1 (hematopoietic cell-specific Lyn substrate 1)-associated protein X-1, HAX-1 [245]. Both HAX-1 and FBXO25 are phosphorylated by protein kinase C delta (PRKCD) to initiate the FBXO25-mediated degradation of HAX-1 following apoptotic stress, such as that induced by the chemotherapeutic and DNA damaging agent etoposide. In addition, and consistent with a tumor-suppressive role for this gene, FBXO25 has also been found to interact with and facilitate the degradation of the proto-oncogene ELK-1 to suppress ELK-1-dependent transcriptional activation of c-FOS and EGR-1 in response to tumor promoter TPA [247]. Together, these studies highlight the role of FBOX25 in suppressing tumorigenesis.
FBXO10 has also been found to initiate apoptotic cell death in human lymphoma by targeting the anti-apoptotic protein BCL2 for proteasomal degradation [230]. Consistent with its putative tumor-suppressive role, FBXO10 is inactivated or its expression is reduced in DLBCL [230]. Similarly, downregulation of FBXO10 transcript levels in T cells has been correlated with increased breast cancer susceptibility [231, 232]. Whether FBXO10 exhibits tumor-suppressive functions independent of BCL2 downregulation remains to be seen.
As mentioned in Section 4.5, the ectopic expression of FBXL7 in transformed lung epithelial cells induces apoptosis through the degradation of Aurora A [196]. A recent study reports that SCFFBXL7 also targets the anti-apoptotic protein survivin for ubiquitin-mediated degradation, thereby regulating mitochondrial activity [197]. Survivin is a member of the inhibitor of apoptosis (IAP) family that prevents caspase activation, resulting in the induction of apoptosis. Overexpression of FBXL7 in lung epithelia was shown to induce mitochondrial dysfunction, while efficient knockdown of FBXL7 protected the mitochondria from damage caused by depolarization [197].
Finally, and as described in Section 3.3, the FBXO7 protein has some pro-apoptotic activity via the ubiquitylation of cIAP1 (Inhibitor of apoptosis protein 1) [102], although this protein exhibits mostly oncogenic activity.
5.2. F-box proteins with anti-apoptotic activities
FBXO45 mediates the proteasomal degradation of the pro-apoptotic tumor suppressor protein p73, a member of the p53 family of transcription factors [227]. Furthermore, siRNA-mediated silencing of FBXO45 was sufficient to stabilize p73, thereby sensitizing breast cancer cells to chemotherapeutic agent-induced death, suggesting a possible oncogenic function for FBXO45 [227]. Another study implicated FBXO45 in the degradation of tumor suppressor protein PAR4 (Prostate apoptosis response protein 4), which promotes apoptosis in cancer cells [228]. The depletion of FBXO45 was shown to stabilize Par4 in mouse embryonic stem cells, resulting in enhanced apoptosis [228].
As stated in Section 3.3, FBXL10 exhibits anti-apoptotic function via the regulation of apoptosis inducer p19 (ARF) [132]. Furthermore, FBXL10 has been shown to suppress apoptosis via direct binding and repression of c-FOS, c-JUN and RIPK3 promoters [133-135]. FBXL10 also promotes cell proliferation and inhibits senescence through inhibiting the transcription of the CDK4 inhibitor p15 (INK4B) [136]. In addition, SKP2 suppresses apoptosis via degradation of the transcription factor FOXO1 [54], which positively regulates apoptosis.
6. Regulation of invasion and metastasis by F-box proteins
Another hallmark of cancer cells is their ability to invade local tissue and vasculature and establish colonies at a distal site. This process is often driven by activation of epithelial-mesenchymal transition (EMT), which enables epithelial cells to lose cell-cell adhesion and acquire the migratory and invasive capacity of mesenchymal stem cells. Here we highlight the roles of several F-box proteins in regulating EMT and tumor metastasis (Figure 3).
As mentioned in Section 4.5, the tumor-suppressive function of FBXO11 has been attributed to its ability to downregulate the oncoprotein BCL6 [233] and potentially CDT2 [235, 236]. More recently, FBXO11 has also been identified as a crucial inhibitor of metastasis in breast cancer. This is mediated through the targeted ubiquitylation and degradation of SNAIL [238], a transcription factor that represses the CDH1 gene encoding E-cadherin to promote EMT in human epithelial cells. FBXO11 recognizes and targets SNAIL protein for degradation following its phosphorylation on Ser-11 in the SNAG domain by protein kinase D1 (PKD1). FBXO11-mediated downregulation of SNAIL inhibited tumorigenesis of the Neu-transformed human mammary epithelial cells HMLEN in nude mice. In addition, FBXO11 overexpression inhibited the metastasis of the mouse mammary tumor cell line 4T1 to the lungs, and this was largely attributed to its ability to suppress SNAIL expression and inhibit EMT. Reciprocally, depletion of endogenous FBXO11 promoted lung metastasis in this breast cancer model system. Importantly, immunohistochemistry staining of breast cancer tissue samples demonstrated a significant correlation between low FBXO11 expression and high SNAIL protein, which correlated with the invasive properties of these breast cancers [238]. These results demonstrate a critical role for the FBXO11-PDK1-SNAIL degradation axis in suppressing tumorigenesis and metastasis in breast cancer. This mechanism of downregulation of SNAIL is distinct from the mechanism mediated by β-TRCP1, which is dependent on SNAIL phosphorylation at two separate motifs by GSK3β and regulates SNAIL’s subcellular localization and ubiquitylation [73].
The ubiquitylation and degradation of SNAIL, as well as two other SNAIL family members, SLUG and SCRATCH, by FBXO11 was confirmed in a second study [239]. However, this study demonstrated that neither the SNAG domain, nor SNAIL phosphorylation were required for recognition by FBXO11. Nevertheless, both studies demonstrated clearly that the FBXO11 acts as a master regulator of EMT and metastasis, at least in the context of breast tumorigenesis.
In addition to FBXO11 and β-TRCP1, FBXL5 has also been shown to mediate the turnover of SNAIL [206, 213]. The downregulation of SNAIL by FBXL5 was shown to suppress metastasis in gastric cancer cells, and FBXL5 protein levels were significantly reduced in metastatic gastric cancer tissue samples, and negatively correlated with SNAIL protein levels [206]. Another study reported that the downregulation of the actin-interacting protein cortactin by FBXL5 inhibits gastric cancer cell migration and invasion, whereas the expression of non-degradable cortactin protein promoted cell migration in gastric cancer cells [269]. Notably, FBXL5 protein levels are downregulated following IR, resulting in the stabilization of SNAIL [213] and providing a mechanistic basis for the induction of EMT following radiotherapy.
Contrary to the tumor-suppressive effects of FBXL5 described above, FBXL5 also appears to exhibit tumor-promoting activities. For example, depletion of FBXL5 in HeLa cells was reported to suppress anchorage-independent growth, presumably through E-cadherin upregulation at the post-transcriptional level [270]. Moreover, FBXL5 expression was upregulated in lung cancer samples, and this correlated with lower levels of its ubiquitylation substrate hSSB1, a crucial regulator of genome stability [207]. The FBXL5-directed ubiquitylation and degradation of hSSB1 was shown to abrogate the DNA damage response during genotoxic stress [207]. This latter result suggests that FBXL5 may also contribute to cancer development, at least in the context of lung cancer.
FBXL14 regulates key EMT-associated transcription factors, including SLUG [199], SNAIL [198], TWIST1 and SIP1 [201]. FBXL14 was shown to mediate the proteasomal degradation of SLUG during neural crest development in Xenopus embryos [199], and SNAIL in human tumor cells [198]. Under hypoxic conditions, FBXL14 is downregulated, which is accompanied by SNAIL stabilization [198]. TWIST1 and SIP1 were subsequently identified as additional FBXL14 substrates in Xenopus embryos [201]. The identification of multiple regulators of EMT as FBXL14 substrates underscores the importance of FBXL14 in regulating EMT, suggesting that it may represent yet another suppressor of metastasis.
In addition to its oncogenic properties (Section 5.2), FBXO45 also exhibits tumor-suppressive characteristics through regulation of EMT-inducing transcription factors ZEB1/2, SNAIL, SLUG and TWIST1 [229]. Accordingly, repression of FBXO45 by miR-27a resulted in enhanced EMT initiation and cancer progression [229].
In contrast to the F-box proteins which suppress EMT and metastasis, JFK (Section 3.3) has been shown to promote angiogenesis and metastasis of breast cancer via turnover of ING4 (inhibitor of growth protein 4) [86].
7. F-box proteins and the regulation of genome integrity
Genome instability is regarded as an enabling characteristic of tumorigenesis [271]. Some F-box proteins maintain genome integrity and prevent the accumulation of cancer-causing mutations by promoting DNA repair and ensuring mitotic fidelity and appropriate telomere function. Some, such as β-TRCP1/2, are critical regulators of the DNA damage response (Section 3.2) and they directly impact the integrity of the genome. Others impact the integrity of the genome, but only indirectly. In this section, we highlight the activity of some F-box proteins whose deregulated expression or activity may also contribute to tumorigenesis through impacting the integrity of the genome (Figure 3).
7.1. Cyclin F (FBXO1)
Cyclin F is the founding member of the F-box family of proteins, in which the F-box motif was first identified [104]. Cyclin F regulates cell cycle transitions and oscillates in a pattern similar to cyclin A2 and cyclin B1, although unlike most cyclins, it does not partner with any CDK [222, 272]. Mice deficient of Cyclin F exhibited placental development abnormalities and subsequently died at E10.5, whereas Cyclin F+/− mice were normal and fertile. Although cyclin F was not essential for MEF viability, MEFs devoid of Cyclin F had impaired cell cycle progression, with a delayed doubling time and postponed cell cycle reentry from quiescence [222].
Consistent with a tumor-suppressive role for this F-box protein, low expression levels of cyclin F were found in hepatocellular carcinoma and significantly correlated with tumor size, clinical stage, and tumor multiplicity [220]. Reduced expression of cyclin F was also associated with poor tumor differentiation, poor overall survival and poor progression-free survival, validating the prognostic impact of cyclin F in hepatocellular carcinoma [220]. In addition, cyclin F has been identified by expression profiling as a potential cell cycle gene that is differentially altered in lung cancer [221].
As a functional component of the SCF complex, cyclin F has been shown to prevent genome instability and centrosome duplication by facilitating the proteasomal degradation of CP110 [223], NUSAP [224] and RRM2 [225]. Cyclin F regulates CP110 levels during G2 to preserve genome integrity and mitotic fidelity [223]. CP110 is necessary for centrosome replication and its degradation ensures the precise duplication of centrosomes, preventing centrosomal and mitotic anomalies due to centrosome reduplication. Depletion of cyclin F resulted in centrosomal and mitotic defects, which could be rescued by co-silencing CP110 [223]. CP110 is also regulated by the deubiquitinase USP33, which antagonizes SCFCyclinF activity during S and G2/M to stabilize CP110, causing mitotic abnormalities [273].
Cyclin F also regulates chromosome stability via the downregulation of NUSAP, a microtubule binding protein that functions in chromosome alignment and segregation [224]. SCFCyclinF facilitates NUSAP degradation in response to UV-induced DNA damage during S and G2 phases of the cell cycle [224]. Like CP110, NUSAP is also involved in microtubule spindle assembly, highlighting the importance of Cyclin F in regulating chromosomal stability.
RRM2, the ribonucleotide reductase family member 2, is another ubiquitylation substrate for the SCFCyclinF ligase [225]. RRM2 is required for the cell cycle-dependent activity of ribonucleotide reductase (RNR), an enzyme essential for DNA synthesis. RNR is responsible for catalyzing the conversion of ribonucleotides (rNTPs) to deoxyribonucleotides (dNTPs) during DNA replicative and repair synthesis. RRM2 is ubiquitylated and subsequently degraded upon CDK-dependent phosphorylation of RRM2 on Thr-33 in G2 for the purpose of maintaining balanced dNTP pools [225]. Failure to degrade RRM2 either through the depletion of cyclin F by siRNA or following the expression of cyclin F-resistant and non-degradable RRM2 led to enhanced mutation frequency and genomic instability as a consequence of excessive dNTP accumulation. Furthermore, DNA damage elicited by different agents e.g. doxorubicin, CPT, UV, MMS, or γ-irradiation, stimulated ATR-dependent downregulation of cyclin F, leading to accumulation of RRM2. Importantly, defective elimination of cyclin F prevented DNA damage-induced accumulation of RRM2, delayed DNA repair, sensitized cells to DNA damage and promoted cell death, and these phenotypes were reverted by the expression of cyclin F-resistant mutant of RRM2 protein [225].
Cyclin F plays another role in cell cycle checkpoint control via the suppression of oncoprotein B-MYB [226], a transcriptional activator involved in cell cycle progression. Following IR, Cyclin F interacts with B-MYB through its cyclin box domain to prevent B-MYB phosphorylation by cyclin A-CDK [226]. Consequently, B-MYB is unable to promote G2 progression until DNA damage can be repaired. This ensures the preservation of the G2 checkpoint response following IR [226].
7.2. Other F-box proteins involved in the regulation of genome stability
FBXO4 has been implicated in telomere homeostasis through its role in the regulation of the telomere regulator TRF1 protein [183]. TRF1 is downregulated in many human cancers and plays a role in cell cycle checkpoint regulation and in telomere elongation [183]. Ectopic expression of FBXO4 was shown to reduce TRF1 protein stability, resulting in telomere elongation. Conversely, FBXO4 depletion increased TRF1 protein levels, resulting in telomere shortening and diminished cell growth [183]. This finding however, is more consistent with a tumor promoting, rather than a tumor-suppressing activity for FBXO4 (Section 4.3). Although some evidence suggests conditional knockout of TRF1 increases incidence of neoplastic lesions in mice, the extent to which TRF1 degradation contributes to tumorigenesis has yet to be fully clarified [274].
FBXO18 functions both as a DNA-helicase and as a component of the SCFFBXO18 E3 ubiquitin ligase [275], although no ubiquitylation substrates for human FBXO18 have been identified to date. The FBXO18 locus is frequently deleted in melanoma and lung cancer cell lines [240], and rare variants within the FBXO18 gene were reported to be strongly associated with breast cancer risk [241], suggesting that FBXO18 may function as a tumor suppressor. Consistent with this notion, depletion of FBXO18 in melanocytes promoted UV irradiation-induced transformation. The helicase activity of FBXO18 has been shown to regulate homologous recombination (HR) repair, thereby contributing to the maintenance of the genome [276]. In response to DNA replication stress induced by UV, FBXO18 promotes the helicase-dependent induction of DNA double-strand breakage and activation of ATM and DNA-PK [277]. Once activated, RPA2 and p53 are phosphorylated, resulting in apoptosis [277]. Likewise, another study reported that FBXO18 was able to induce double strand DNA breaks and cell death in response to replicative stress with the help of MUS81 nuclease in promoting endonucleolytic DNA cleavage [278]. It is conceivable that the tumor-suppressive role of FBXO18 may be attributed to its contribution to genomic maintenance, although the identification of FBXO18 substrates will shed further light on its role in tumorigenesis.
In response to DNA damage, the F-box protein FBXL20 targets Vps34 (Vacuolar protein-sorting 34) for proteasomal degradation [205]. Vps34 is the catalytic subunit in the class III PtdIns3 (phosphatidylinositol 3) kinase complex that is necessary for mTORC1 autophagy activity. In response to the DNA damaging agent CPT, cyclin B1-CDK1 phosphorylates Vps34 on Thr-159 to initiate its degradation, leading to inhibition of autophagy and receptor degradation. Furthermore, the transcription of FBXL20 is induced by p53, suggesting that FBXL20-mediated downregulation of Vps34 may represent a novel mediator of p53-checkpoint that regulates autophagy in response to DNA damage [205]. Additionally, FBXL20 may function as an oncogene, given that FBXL20 overexpression was detected in human colorectal adenocarcinoma [202]. Therefore, further investigation is required to elucidate the role of FBXL20 in cancer.
The ubiquitin-dependent degradation of BRCA1 may also be relevant to the oncogenicity of SKP2. BRCA1, a scaffold protein for the assembly of HR proteins at DNA double-strand breaks (DSBs) [279], was shown to be phosphorylated by CHK2 kinase upon γ-irradiation and ubiquitylated and degraded through the SCFSkp2 [55]. BRCA1 degradation begins in late G1 phase, abolishing its inhibition of the nuclease activity of the double-strand break repair protein MER11A, and ensuring the initiation of HR in S and G2 phases of the cell cycle.
β-TRCP1/2 play important roles not only in regulating cellular response to DNA damage and the recovery from DNA damage as discussed above (Section 3.2), but also directly impact the repair of damaged DNA. For example, in a recently published study, Liu et al, found that β-TRCP1/2 target XLF (XRCC4 like factor, also called NHEJ1), a component of the DNA ligase IV/XRCC4 complex involved in the repair of DSBs by non-homologous end-joining (NHEJ), for ubiquitin-mediated degradation [77]. This degradation is dependent on AKT-mediated phosphorylation of XLF on Thr-181, which triggers its dissociation from the XRCC4/DNA ligase IV complex and cytoplasmic retention, where it is polyubiquitylated and degraded via the SCFβ-TRCP ubiquitin ligase. This finding highlights the interplay between aberrant AKT hyperactivation commonly observed in cancer, the oncogenic activity of β-TRCP, and the deficiency in timely repair of double-strand breaks, leading to genome instability and tumorigenesis.
8. Conclusions and Future Perspectives
Significant advances have been made in our understanding of the roles of the F-box proteins in cancer. An abundance of literature has now shown clearly that deregulation of F-box proteins by means of mutations, deletions, or over- and under-expression plays critical roles in cancer development, progression and metastasis. Key molecular events that contribute to deregulated proliferation as a consequence of deregulated proteolysis by this versatile group of proteins have been identified, and many of these findings were verified in human cancer and in experimental animal models of tumorigenesis. For example, the well-documented tumor suppressor FBXW7 is mutated at high frequencies or deleted in a large number of human malignancies (Table 1) Other F-box proteins with emerging tumor-suppressive functions are also deleted, under-expressed, or mutated in a subset of human cancers (Table 2).
Gain of function activity of other F-box proteins is also abundant in human malignancies, although the vast majority of these are only overexpressed, without activating mutations or gene amplifications (Table 2). Yet, their frequent overexpression in human neoplasms (e.g. SKP2 and β-TRCP1/2), and its association with patient outcome, as well as the validation of their tumorigenic activity in experimental animals clearly point towards their oncogenic potential. Other F-box proteins with oncogenic potential are also overexpressed in several cancers (Table 1), but more validation of their tumorigenic roles is required.
Given their frequent deregulation in human cancer and the key roles they play in regulating various aspects of the tumorigenic phenotype (Figure 3), this class of proteins represents attractive molecular targets for therapeutic intervention [2]. The pharmacological targeting of the SCF ligases in particular or E3 ubiquitin ligases in general however, is not a simple task given that this class of proteins lack enzymatic activity, and inhibition of the E3 ligase-substrate interaction is necessary to achieve targeted inhibition. Although challenging, pharmacological inhibitors of few F-box proteins have been developed, and show significant promise [280-283]. For example, pharmacological inactivation of the SCFSKP2 ligase was recently shown to exhibit potent antitumor activities in multiple animal models and cooperates with chemotherapeutic agents to reduce cancer cell survival [283]. However, the complexity underlying the regulation of these E3 ligases, as well as the targeted proteolysis of many substrates involved in a number of physiological activities by the same SCF ligase (Figure 3), calls for caution regarding the potential side effects that may arise through disrupting their activities. Thus, a careful examination of the biochemical activities of these proteins with regards to the selective ubiquitylation and degradation of key substrates may represent ideal and suitable targets of intervention.
Although the pharmacological targeting of oncogenic F-box proteins is straight forward, reactivation of tumor suppressor F-box proteins is a more challenging task, especially for the proteins that are mutated in cancer, such as FBXW7. This requires a different mode of intervention that must be based either on restoring the wild type active form of the protein, or on the direct inactivation of the oncogenic protein that fails to be degraded by the mutant F-box protein. Because multiple oncogenic proteins may be stabilized in cancers with mutated F-box proteins, restoration of the wild type F-box gene may be the only viable option for these diseases. Here, the recent development of new tools for genome editing, such as the CRISPR/Cas9, combined with new advances in gene therapy approaches [284], may prove useful for achieving these goals.
Despite the significant advances made in understanding the biological consequences of deregulated proteolysis via the SCF ligases, several issues remain unanswered. For example, with few exceptions, the signaling cascades leading to the activation or inactivation of the various ligases remain poorly understood. Tissue-specific regulation of these ligases, interaction with various genetic backgrounds in certain tumor types, and the differential cell type-specific phenotypic outputs downstream of deregulated activity of the various ligases, are all issues that need careful examination. Another unaddressed challenge is the cross-regulation between these F-box proteins and other E3 ligases or other components of the UPS system. Finally, the majority of F-box proteins remain uncharacterized, without identifiable substrates, and are likely to play a role in tumorigenesis. Clearly, we are just beginning to understand the complexity underlying deregulated proteolysis by this class of E3 ubiquitin ligases and its contribution to cancer, and novel and more comprehensive approaches are needed to address these pressing challenges.
Acknowledgments
We apologize to authors whose primary work was not cited owing to space restrictions. Work in the Abbas laboratory is supported by a grant from the National Institutes of Health (NIH; CA009109) and a New Idea Award (NIA) from the leukemia and lymphoma society (LLS; P-NIA-0214-13) to TA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nature reviews. Drug discovery. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182:1–11. doi: 10.1002/(SICI)1097-4652(200001)182:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- [4].Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- [5].Ciechanover A, Schwartz AL. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35:3–6. doi: 10.1053/jhep.2002.30316. [DOI] [PubMed] [Google Scholar]
- [6].Amir R, Ciechanover A, Cohen S. [The ubiquitin-proteasome system: the relationship between protein degradation and human diseases] Harefuah. 2001;140:1172–1176. 1229. [PubMed] [Google Scholar]
- [7].Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annual review of biochemistry. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- [8].Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol. 2003;35:606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- [9].Yang WL, Zhang X, Lin HK. Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene. 2010;29:4493–4503. doi: 10.1038/onc.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- [11].Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- [13].Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- [14].Hotton SK, Callis J. Regulation of cullin RING ligases. Annual review of plant biology. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- [15].Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen Z, Sui J, Zhang F, Zhang C. Cullin family proteins and tumorigenesis: genetic association and molecular mechanisms. J Cancer. 2015;6:233–242. doi: 10.7150/jca.11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- [21].Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- [24].Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [26].Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci U S A. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am J Pathol. 2004;165:175–180. doi: 10.1016/S0002-9440(10)63286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, Zhang H. Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate. Cancer research. 2003;63:1583–1588. [PubMed] [Google Scholar]
- [29].Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci U S A. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Umanskaya K, Radke S, Chander H, Monardo R, Xu X, Pan ZQ, O'Connell MJ, Germain D. Skp2B stimulates mammary gland development by inhibiting REA, the repressor of the estrogen receptor. Mol Cell Biol. 2007;27:7615–7622. doi: 10.1128/MCB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, Zhu L. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nature genetics. 2010;42:83–88. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sistrunk C, Kim SH, Wang X, Lee SH, Kim Y, Macias E, Rodriguez-Puebla ML. Skp2 deficiency inhibits chemical skin tumorigenesis independent of p27(Kip1) accumulation. The American journal of pathology. 2013;182:1854–1864. doi: 10.1016/j.ajpath.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao H, Bauzon F, Fu H, Lu Z, Cui J, Nakayama K, Nakayama KI, Locker J, Zhu L. Skp2 deletion unmasks a p27 safeguard that blocks tumorigenesis in the absence of pRb and p53 tumor suppressors. Cancer Cell. 2013;24:645–659. doi: 10.1016/j.ccr.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- [37].Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- [38].Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, Kitagawa M, Nakayama K, Hatakeyama S. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. The EMBO journal. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- [40].Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A. 2003;100:10231–10236. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2) Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- [44].Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- [45].Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Molecular cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- [46].Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- [47].Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbe JC, Lorca T, Nakayama Ki K, Benkirane M. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol Cell Biol. 2001;21:7956–7970. doi: 10.1128/MCB.21.23.7956-7970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- [49].Hiramatsu Y, Kitagawa K, Suzuki T, Uchida C, Hattori T, Kikuchi H, Oda T, Hatakeyama S, Nakayama KI, Yamamoto T, Konno H, Kitagawa M. Degradation of Tob1 mediated by SCFSkp2-dependent ubiquitination. Cancer Res. 2006;66:8477–8483. doi: 10.1158/0008-5472.CAN-06-1603. [DOI] [PubMed] [Google Scholar]
- [50].Song MS, Song SJ, Kim SJ, Nakayama K, Nakayama KI, Lim DS. Skp2 regulates the antiproliferative function of the tumor suppressor RASSF1A via ubiquitin-mediated degradation at the G1-S transition. Oncogene. 2008;27:3176–3185. doi: 10.1038/sj.onc.1210971. [DOI] [PubMed] [Google Scholar]
- [51].Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng XH, Lin X. Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol. 2004;24:7524–7537. doi: 10.1128/MCB.24.17.7524-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- [53].Tokarz S, Berset C, La Rue J, Friedman K, Nakayama K, Nakayama K, Zhang DE, Lanker S. The ISG15 isopeptidase UBP43 is regulated by proteolysis via the SCFSkp2 ubiquitin ligase. J Biol Chem. 2004;279:46424–46430. doi: 10.1074/jbc.M403189200. [DOI] [PubMed] [Google Scholar]
- [54].Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R, Lin HK, Li R, Paull TT, Hu Y. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle. 2015;14:437–448. doi: 10.4161/15384101.2014.972901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Moro L, Arbini AA, Marra E, Greco M. Up-regulation of Skp2 after prostate cancer cell adhesion to basement membranes results in BRCA2 degradation and cell proliferation. J Biol Chem. 2006;281:22100–22107. doi: 10.1074/jbc.M604636200. [DOI] [PubMed] [Google Scholar]
- [57].Jamal A, Swarnalatha M, Sultana S, Joshi P, Panda SK, Kumar V. The G1 phase E3 ubiquitin ligase TRUSS that gets deregulated in human cancers is a novel substrate of the S-phase E3 ubiquitin ligase Skp2. Cell Cycle. 2015:0. doi: 10.1080/15384101.2015.1056946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xu D, Li CF, Zhang X, Gong Z, Chan CH, Lee SW, Jin G, Rezaeian AH, Han F, Wang J, Yang WL, Feng ZZ, Chen W, Wu CY, Wang YJ, Chow LP, Zhu XF, Zeng YX, Lin HK. Skp2-macroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis. Nat Commun. 2015;6:6641. doi: 10.1038/ncomms7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Di Giorgio E, Gagliostro E, Clocchiatti A, Brancolini C. The control operated by the cell cycle machinery on MEF2 stability contributes to the downregulation of CDKN1A and entry into S phase. Mol Cell Biol. 2015;35:1633–1647. doi: 10.1128/MCB.01461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, Minamoto T. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96:1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- [61].Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, Kalthoff H, Folsch UR, Schafer H. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer research. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- [62].Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Waha A, Wharton KA, Jr., Fuchs SY, von Schweinitz D, Pietsch T. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res. 2005;11:4295–4304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- [63].Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- [64].Saitoh T, Katoh M. Expression profiles of betaTRCP1 and betaTRCP2, and mutation analysis of betaTRCP2 in gastric cancer. Int J Oncol. 2001;18:959–964. [PubMed] [Google Scholar]
- [65].Kim CJ, Song JH, Cho YG, Kim YS, Kim SY, Nam SW, Yoo NJ, Lee JY, Park WS. Somatic mutations of the beta-TrCP gene in gastric cancer. APMIS. 2007;115:127–133. doi: 10.1111/j.1600-0463.2007.apm_562.x. [DOI] [PubMed] [Google Scholar]
- [66].Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–8194. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bhatia N, Demmer TA, Sharma AK, Elcheva I, Spiegelman VS. Role of beta-TrCP ubiquitin ligase receptor in UVB mediated responses in skin. Arch Biochem Biophys. 2011;508:178–184. doi: 10.1016/j.abb.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK, Yamasaki L, Pagano M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Developmental cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- [69].Kanarek N, Horwitz E, Mayan I, Leshets M, Cojocaru G, Davis M, Tsuberi BZ, Pikarsky E, Pagano M, Ben-Neriah Y. Spermatogenesis rescue in a mouse deficient for the ubiquitin ligase SCF{beta}-TrCP by single substrate depletion. Genes Dev. 2010;24:470–477. doi: 10.1101/gad.551610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- [71].Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- [74].Pons J, Evrard-Todeschi N, Bertho G, Gharbi-Benarous J, Tanchou V, Benarous R, Girault JP. Transfer-NMR and docking studies identify the binding of the peptide derived from activating transcription factor 4 to protein ubiquitin ligase beta-TrCP. Competition STD-NMR with beta-catenin. Biochemistry. 2008;47:14–29. doi: 10.1021/bi7014212. [DOI] [PubMed] [Google Scholar]
- [75].Ciechanover A, Gonen H, Bercovich B, Cohen S, Fajerman I, Israel A, Mercurio F, Kahana C, Schwartz AL, Iwai K, Orian A. Mechanisms of ubiquitin-mediated, limited processing of the NF-kappaB1 precursor protein p105. Biochimie. 2001;83:341–349. doi: 10.1016/s0300-9084(01)01239-1. [DOI] [PubMed] [Google Scholar]
- [76].Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, Stommel JM, Gygi S, Lahav G, Asara J, Xiao ZX, Kaelin WG, Harper Jr., J.W., Wei W. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu P, Gan W, Guo C, Xie A, Gao D, Guo J, Zhang J, Willis N, Su A, Asara JM, Scully R, Wei W. Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair. Mol Cell. 2015;57:648–661. doi: 10.1016/j.molcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Molecular cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- [79].Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, Lu H, Sun Y. SAG/ROC-SCF beta-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- [81].Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- [82].Wang Z, Zhong J, Gao D, Inuzuka H, Liu P, Wei W. DEPTOR ubiquitination and destruction by SCF(beta-TrCP), American journal of physiology. Endocrinology and metabolism. 2012;303:E163–169. doi: 10.1152/ajpendo.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Loveless TB, Topacio BR, Vashisht AA, Galaang S, Ulrich KM, Young BD, Wohlschlegel JA, Toczyski DP. DNA Damage Regulates Translation through beta-TRCP Targeting of CReP. PLoS Genet. 2015;11:e1005292. doi: 10.1371/journal.pgen.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhu G, Fan Z, Ding M, Mu L, Liang J, Ding Y, Fu Y, Huang B, Wu W. DNA damage induces the accumulation of Tiam1 by blocking beta-TrCP-dependent degradation. J Biol Chem. 2014;289:15482–15494. doi: 10.1074/jbc.M114.553388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yan R, He L, Li Z, Han X, Liang J, Si W, Chen Z, Li L, Xie G, Li W, Wang P, Lei L, Zhang H, Pei F, Cao D, Sun L, Shang Y. SCF(JFK) is a bona fide E3 ligase for ING4 and a potent promoter of the angiogenesis and metastasis of breast cancer. Genes & development. 2015;29:672–685. doi: 10.1101/gad.254292.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sun L, Shi L, Li W, Yu W, Liang J, Zhang H, Yang X, Wang Y, Li R, Yao X, Yi X, Shang Y. JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc Natl Acad Sci U S A. 2009;106:10195–10200. doi: 10.1073/pnas.0901864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gutgemann I, Lehman NL, Jackson PK, Longacre TA. Emi1 protein accumulation implicates misregulation of the anaphase promoting complex/cyclosome pathway in ovarian clear cell carcinoma. Mod Pathol. 2008;21:445–454. doi: 10.1038/modpathol.3801022. [DOI] [PubMed] [Google Scholar]
- [89].Min KW, Park MH, Hong SR, Lee H, Kwon SY, Hong SH, Joo HJ, Park IA, An HJ, Suh KS, Oh HK, Yoo CW, Kim MJ, Chang HK, Jun SY, Yoon HK, Chang ED, Kim DW, Kim I. P. Gynecologic Pathology Study Group of the Korean Society of, Clear cell carcinomas of the ovary: a multi-institutional study of 129 cases in Korea with prognostic significance of Emi1 and Galectin-3. Int J Gynecol Pathol. 2013;32:3–14. doi: 10.1097/PGP.0b013e31825554e9. [DOI] [PubMed] [Google Scholar]
- [90].Lehman NL, Tibshirani R, Hsu JY, Natkunam Y, Harris BT, West RB, Masek MA, Montgomery K, van de Rijn M, Jackson PK. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. The American journal of pathology. 2007;170:1793–1805. doi: 10.2353/ajpath.2007.060767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lee H, Lee DJ, Oh SP, Park HD, Nam HH, Kim JM, Lim DS. Mouse emi1 has an essential function in mitotic progression during early embryogenesis. Mol Cell Biol. 2006;26:5373–5381. doi: 10.1128/MCB.00043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- [93].Lu Y, Li J, Cheng D, Parameswaran B, Zhang S, Jiang Z, Yew PR, Peng J, Ye Q, Hu Y. The F-box protein FBXO44 mediates BRCA1 ubiquitination and degradation. The Journal of biological chemistry. 2012;287:41014–41022. doi: 10.1074/jbc.M112.407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sjögren B, Swaney S, Neubig RR. FBXO44-Mediated Degradation of RGS2 Protein Uniquely Depends on a Cullin 4B/DDB1 Complex. PloS one. 2015;10:e0123581. doi: 10.1371/journal.pone.0123581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fernandez-Saiz V, Targosz BS, Lemeer S, Eichner R, Langer C, Bullinger L, Reiter C, Slotta-Huspenina J, Schroeder S, Knorn AM, Kurutz J, Peschel C, Pagano M, Kuster B, Bassermann F. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol. 2013;15:72–81. doi: 10.1038/ncb2651. [DOI] [PubMed] [Google Scholar]
- [96].Cepeda D, Ng HF, Sharifi HR, Mahmoudi S, Cerrato VS, Fredlund E, Magnusson K, Nilsson H, Malyukova A, Rantala J, Klevebring D, Vinals F, Bhaskaran N, Zakaria SM, Rahmanto AS, Grotegut S, Nielsen ML, Szigyarto CA, Sun D, Lerner M, Navani S, Widschwendter M, Uhlen M, Jirstrom K, Ponten F, Wohlschlegel J, Grander D, Spruck C, Larsson LG, Sangfelt O. CDK-mediated activation of the SCF(FBXO) (28) ubiquitin ligase promotes MYC-driven transcription and tumourigenesis and predicts poor survival in breast cancer. EMBO Mol Med. 2013;5:999–1018. doi: 10.1002/emmm.201202341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park PJ, Bardeesy N. KDM2B promotes pancreatic cancer via Polycomb-dependent and - independent transcriptional programs. J Clin Invest. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Molecular cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- [99].Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E, Knowles P, McDonald N, Boshoff C. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. The EMBO journal. 2005;24:3104–3116. doi: 10.1038/sj.emboj.7600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lomonosov M, Meziane el K, Ye H, Nelson DE, Randle SJ, Laman H. Expression of Fbxo7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. PLoS One. 2011;6:e21165. doi: 10.1371/journal.pone.0021165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Meziane el K, Randle SJ, Nelson DE, Lomonosov M, Laman H. Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. Journal of cell science. 2011;124:2175–2186. doi: 10.1242/jcs.080465. [DOI] [PubMed] [Google Scholar]
- [102].Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022–1026. doi: 10.1016/j.bbrc.2006.02.061. [DOI] [PubMed] [Google Scholar]
- [103].Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–32602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- [104].Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- [105].Yokoi S, Yasui K, Iizasa T, Takahashi T, Fujisawa T, Inazawa J. Down-regulation of SKP2 induces apoptosis in lung-cancer cells. Cancer Sci. 2003;94:344–349. doi: 10.1111/j.1349-7006.2003.tb01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- [107].Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- [108].Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1424. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- [109].Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2) Curr Biol. 2001;11:263–267. doi: 10.1016/s0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- [110].Abbas T, Keaton MA, Dutta A. Genomic instability in cancer. Cold Spring Harb Perspect Biol. 2013;5:a012914. doi: 10.1101/cshperspect.a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, Lygerou Z, Nishimoto T. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem. 2004;279:17283–17288. doi: 10.1074/jbc.C300549200. [DOI] [PubMed] [Google Scholar]
- [113].Takeda DY, Parvin JD, Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem. 2005;280:23416–23423. doi: 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- [114].Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- [115].Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Evans L, Chen L, Milazzo G, Gherardi S, Perini G, Willmore E, Newell DR, Tweddle DA. SKP2 is a direct transcriptional target of MYCN and a potential therapeutic target in neuroblastoma. Cancer Lett. 2015;363:37–45. doi: 10.1016/j.canlet.2015.03.044. [DOI] [PubMed] [Google Scholar]
- [117].Bretones G, Acosta JC, Caraballo JM, Ferrandiz N, Gomez-Casares MT, Albajar M, Blanco R, Ruiz P, Hung WC, Albero MP, Perez-Roger I, Leon J. SKP2 oncogene is a direct MYC target gene and MYC down-regulates p27(KIP1) through SKP2 in human leukemia cells. J Biol Chem. 2011;286:9815–9825. doi: 10.1074/jbc.M110.165977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- [120].Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- [121].Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lu Z, Bauzon F, Fu H, Cui J, Zhao H, Nakayama K, Nakayama KI, Zhu L. Skp2 suppresses apoptosis in Rb1-deficient tumours by limiting E2F1 activity. Nature communications. 2014;5:3463. doi: 10.1038/ncomms4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Oh KJ, Kalinina A, Wang J, Nakayama K, Nakayama KI, Bagchi S. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J Virol. 2004;78:5338–5346. doi: 10.1128/JVI.78.10.5338-5346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Mark KG, Simonetta M, Maiolica A, Seller CA, Toczyski DP. Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell. 2014;53:148–161. doi: 10.1016/j.molcel.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- [128].Lehman NL, Verschuren EW, Hsu JY, Cherry AM, Jackson PK. Overexpression of the anaphase promoting complex/cyclosome inhibitor Emi1 leads to tetraploidy and genomic instability of p53-deficient cells. Cell cycle. 2006;5:1569–1573. doi: 10.4161/cc.5.14.2925. [DOI] [PubMed] [Google Scholar]
- [129].Chen JY, Wang MC, Hung WC. Bcr-Abl-induced tyrosine phosphorylation of Emi1 to stabilize Skp2 protein via inhibition of ubiquitination in chronic myeloid leukemia cells. Journal of cellular physiology. 2011;226:407–413. doi: 10.1002/jcp.22346. [DOI] [PubMed] [Google Scholar]
- [130].Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Cao X, Qin J, Xie Y, Khan O, Dowd F, Scofield M, Lin MF, Tu Y. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–3734. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- [132].Fukuda T, Tokunaga A, Sakamoto R, Yoshida N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Molecular and cellular neurosciences. 2011;46:614–624. doi: 10.1016/j.mcn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- [133].Ge R, Wang Z, Zeng Q, Xu X, Olumi AF. F-box protein 10, an NF-kappaB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 2011;18:1184–1195. doi: 10.1038/cdd.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Koyama-Nasu R, David G, Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol. 2007;9:1074–1080. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- [135].Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J. The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology, and the metabolome of fibroblasts. J Biol Chem. 2012;287:30984–30992. doi: 10.1074/jbc.M112.341040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- [138].Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. The American journal of pathology. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer research. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- [141].Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- [145].Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- [146].Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- [147].King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, Sandy P, Shen SS, Ferrando A, Aifantis I. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Davis H, Lewis A, Behrens A, Tomlinson I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut. 2014;63:792–799. doi: 10.1136/gutjnl-2013-304719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. The Journal of cell biology. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Grim JE, Gustafson MP, Hirata RK, Hagar AC, Swanger J, Welcker M, Hwang HC, Ericsson J, Russell DW, Clurman BE. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. The Journal of cell biology. 2008;181:913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Current biology : CB. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- [152].Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, Sgroi DC, Program NIHISCCS, Hieter P, Mullikin JC, Merino MJ, Bell DW. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nature genetics. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- [157].Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr., Liu Y, Huang Y, Kang HC, DelRosario R, Jen KY, Perez-Losada J, Wei G, Balmain A, Mao JH. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, Miyamoto K, Yoshiwara H, Hosokawa K, Nakamura Y, Gomei Y, Iwasaki H, Hayashi Y, Matsuzaki Y, Nakayama K, Ikeda Y, Hata A, Chiba S, Nakayama KI, Suda T. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22:986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, Lewis A, Watson S, Behrens A, Tomlinson I, Nateri AS. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- [163].Reavie L, Buckley SM, Loizou E, Takeishi S, Aranda-Orgilles B, Ndiaye-Lobry D, Abdel-Wahab O, Ibrahim S, Nakayama KI, Aifantis I. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell. 2013;23:362–375. doi: 10.1016/j.ccr.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell. 2013;23:347–361. doi: 10.1016/j.ccr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- [165].Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- [166].Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- [167].Bengoechea-Alonso MT, Ericsson J. Tumor suppressor Fbxw7 regulates TGFbeta signaling by targeting TGIF1 for degradation. Oncogene. 2010;29:5322–5328. doi: 10.1038/onc.2010.278. [DOI] [PubMed] [Google Scholar]
- [168].Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, Wang W, Wang R, Wang Q, Sun J, Wang P. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Giraldez S, Herrero-Ruiz J, Mora-Santos M, Japon MA, Tortolero M, Romero F. SCF(FBXW7alpha) modulates the intra-S-phase DNA-damage checkpoint by regulating Polo like kinase-1 stability. Oncotarget. 2014;5:4370–4383. doi: 10.18632/oncotarget.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Watanabe T, Hikichi Y, Willuweit A, Shintani Y, Horiguchi T. FBL2 regulates amyloid precursor protein (APP) metabolism by promoting ubiquitination-dependent APP degradation and inhibition of APP endocytosis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:3352–3365. doi: 10.1523/JNEUROSCI.5659-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Chen BB, Glasser JR, Coon TA, Mallampalli RK. Skp-cullin-F box E3 ligase component FBXL2 ubiquitinates Aurora B to inhibit tumorigenesis. Cell death & disease. 2013;4:e759. doi: 10.1038/cddis.2013.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, McDyer JF, Boyiadzis M, Mallampalli RK. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132–3141. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012;31:2566–2579. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, Washburn MP, Pagano M. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–480. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Molecular cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Vaites LP, Lee EK, Lian Z, Barbash O, Roy D, Wasik M, Klein-Szanto AJP, Rustgi AK, Diehl JA. The Fbx4 tumor suppressor regulates cyclin D1 accumulation and prevents neoplastic transformation. Molecular and cellular biology. 2011;31:4513–4523. doi: 10.1128/MCB.05733-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lian Z, Lee EK, Bass AJ, Wong KK, Klein-Szanto AJ, Rustgi AK, Diehl JA. FBXO4 loss facilitates carcinogen induced papilloma development in mice. Cancer biology & therapy. 2015;16:750–755. doi: 10.1080/15384047.2015.1026512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Kanie T, Onoyama I, Matsumoto A, Yamada M, Nakatsumi H, Tateishi Y, Yamamura S, Tsunematsu R, Matsumoto M, Nakayama KI. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Molecular and cellular biology. 2012;32:590–605. doi: 10.1128/MCB.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Jia L, Sun Y. F-box proteins FBXO31 and FBX4 in regulation of cyclin D1 degradation upon DNA damage. Pigment Cell Melanoma Res. 2009;22:518–519. doi: 10.1111/j.1755-148X.2009.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- [184].Miller BJ, Wang D, Krahe R, Wright FA. Pooled analysis of loss of heterozygosity in breast cancer: a genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. American journal of human genetics. 2003;73:748–767. doi: 10.1086/378522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Cornelisse CJ, Cleton-Jansen A-M, Callen DF. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer research. 2005;65:11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- [186].Launonen V, Mannermaa A, Stenbäck F, Kosma VM, Puistola U, Huusko P, Anttila M, Bloigu R, Saarikoski S, Kauppila A, Winqvist R. Loss of heterozygosity at chromosomes 3, 6, 8, 11, 16, and 17 in ovarian cancer: correlation to clinicopathological variables. Cancer genetics and cytogenetics. 2000;122:49–54. doi: 10.1016/s0165-4608(00)00279-x. [DOI] [PubMed] [Google Scholar]
- [187].Härkönen P, Kyllönen AP, Nordling S, Vihko P. Loss of heterozygosity in chromosomal region 16q24.3 associated with progression of prostate cancer. The Prostate. 2005;62:267–274. doi: 10.1002/pros.20147. [DOI] [PubMed] [Google Scholar]
- [188].Huang H-L, Zheng W-L, Zhao R, Zhang B, Ma W-L. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncology reports. 2010;24:715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- [189].Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F, Li W, Yang Q, Zeng J, Jia J, Liu Z. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178–6190. doi: 10.18632/oncotarget.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Johansson P, Jeffery J, Al-Ejeh F, Schulz RB, Callen DF, Kumar R, Khanna KK. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J Biol Chem. 2014;289:18514–18525. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci U S A. 2015;112:8632–8637. doi: 10.1073/pnas.1510929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmuller J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One. 2013;8:e57530. doi: 10.1371/journal.pone.0057530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Kim S, Kon M, DeLisi C. Pathway-based classification of cancer subtypes. Biology direct. 2012;7:21. doi: 10.1186/1745-6150-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wang X, Pankratz VS, Fredericksen Z, Tarrell R, Karaus M, McGuffog L, Pharaoh PDP, Ponder BAJ, Dunning AM, Peock S, Cook M, Oliver C, Frost D, Sinilnikova OM, Stoppa-Lyonnet D, Mazoyer S, Houdayer C, Hogervorst FBL, Hooning MJ, Ligtenberg MJ, Spurdle A, Chenevix-Trench G, Schmutzler RK, Wappenschmidt B, Engel C, Meindl A, Domchek SM, Nathanson KL, Rebbeck TR, Singer CF, Gschwantler-Kaulich D, Dressler C, Fink A, Szabo CI, Zikan M, Foretova L, Claes K, Thomas G, Hoover RN, Hunter DJ, Chanock SJ, Easton DF, Antoniou AC, Couch FJ. Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Human Molecular Genetics. 2010;19:2886–2897. doi: 10.1093/hmg/ddq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora A, causing mitotic arrest. Cell cycle. 2012;11:721–729. doi: 10.4161/cc.11.4.19171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Liu Y, Lear T, Iannone O, Shiva S, Corey C, Rajbhandari S, Jerome J, Chen BB, Mallampalli RK. The Proapoptotic F-box Protein Fbxl7 Regulates Mitochondrial Function by Mediating the Ubiquitylation and Proteasomal Degradation of Survivin. J Biol Chem. 2015;290:11843–11852. doi: 10.1074/jbc.M114.629931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B, Baulida J, Bonilla F, de Herreros AG, Diaz VM. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J Biol Chem. 2010;285:3794–3805. doi: 10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- [200].Zheng H, Du Y, Hua Y, Wu Z, Yan Y, Li Y. Essential role of Fbxl14 ubiquitin ligase in regulation of vertebrate axis formation through modulating Mkp3 level. Cell research. 2012;22:936–940. doi: 10.1038/cr.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. The Journal of cell biology. 2011;194:17–25. doi: 10.1083/jcb.201012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Zhu J, Li K, Dong L, Chen Y. Role of FBXL20 in human colorectal adenocarcinoma. Oncol Rep. 2012;28:2290–2298. doi: 10.3892/or.2012.2065. [DOI] [PubMed] [Google Scholar]
- [203].Takagi H, Setou M, Ito S, Yao I. SCRAPPER regulates the thresholds of long-term potentiation/depression, the bidirectional synaptic plasticity in hippocampal CA3-CA1 synapses. Neural plasticity. 2012;2012:352829. doi: 10.1155/2012/352829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, Inokuchi K, Ohtsuka T, Macgregor GR, Tanaka K, Setou M. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T, Liu M, Jin M, Wu K, Yuan J. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015;29:184–196. doi: 10.1101/gad.252528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Wu W, Ding H, Cao J, Zhang W. FBXL5 inhibits metastasis of gastric cancer through suppressing Snail1. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;35:1764–1772. doi: 10.1159/000373988. [DOI] [PubMed] [Google Scholar]
- [207].Chen Z-W, Liu B, Tang N-W, Xu Y-H, Ye X-Y, Li Z-M, Niu X-M, Shen S-P, Lu S, Xu L. FBXL5-mediated degradation of single-stranded DNA-binding protein hSSB1 controls DNA damage response. Nucleic acids research. 2014;42:11560–11569. doi: 10.1093/nar/gku876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Ruiz JC, Walker SD, Anderson SA, Eisenstein RS, Bruick RK. F-box and leucine-rich repeat protein 5 (FBXL5) is required for maintenance of cellular and systemic iron homeostasis. J Biol Chem. 2013;288:552–560. doi: 10.1074/jbc.M112.426171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell metabolism. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- [210].Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Zhang N, Liu J, Ding X, Aikhionbare F, Jin C, Yao X. FBXL5 interacts with p150Glued and regulates its ubiquitination. Biochem Biophys Res Commun. 2007;359:34–39. doi: 10.1016/j.bbrc.2007.05.068. [DOI] [PubMed] [Google Scholar]
- [213].Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, Diaz VM. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic acids research. 2014;42:1079–1094. doi: 10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Tsutsumi T, Kuwabara H, Arai T, Xiao Y, Decaprio JA. Disruption of the Fbxw8 gene results in pre- and postnatal growth retardation in mice. Mol Cell Biol. 2008;28:743–751. doi: 10.1128/MCB.01665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Tsunematsu R, Nishiyama M, Kotoshiba S, Saiga T, Kamura T, Nakayama KI. Fbxw8 is essential for Cul1-Cul7 complex formation and for placental development. Molecular and cellular biology. 2006;26:6157–6169. doi: 10.1128/MCB.00595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Kim SJ, DeStefano MA, Oh WJ, Wu C.-c., Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Molecular cell. 2012;48:875–887. doi: 10.1016/j.molcel.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Kong C, Samovski D, Srikanth P, Wainszelbaum MJ, Charron AJ, Liu J, Lange JJ, Chen P-I, Pan Z-Q, Su X, Stahl PD. Ubiquitination and degradation of the hominoid-specific oncoprotein TBC1D3 is mediated by CUL7 E3 ligase. PloS one. 2012;7:e46485. doi: 10.1371/journal.pone.0046485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Okabe H, Lee S-H, Phuchareon J, Albertson DG, McCormick F, Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PloS one. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Wang H, Chen Y, Lin P, Li L, Zhou G, Liu G, Logsdon C, Jin J, Abbruzzese JL, Tan T-H, Wang H. The CUL7/F-box and WD repeat domain containing 8 (CUL7/Fbxw8) ubiquitin ligase promotes degradation of hematopoietic progenitor kinase 1. The Journal of biological chemistry. 2014;289:4009–4017. doi: 10.1074/jbc.M113.520106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Fu J, Qiu H, Cai M, Pan Y, Cao Y, Liu L, Yun J, Zhang CZ. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer science. 2013;104:508–515. doi: 10.1111/cas.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Singhal S, Amin KM, Kruklitis R, DeLong P, Friscia ME, Litzky LA, Putt ME, Kaiser LR, Albelda SM. Alterations in cell cycle genes in early stage lung adenocarcinoma identified by expression profiling. Cancer biology & therapy. 2:291–298. doi: 10.4161/cbt.2.3.399. [DOI] [PubMed] [Google Scholar]
- [222].Tetzlaff MT, Bai C, Finegold M, Wilson J, Harper JW, Mahon KA, Elledge SJ. Cyclin F disruption compromises placental development and affects normal cell cycle execution. Molecular and cellular biology. 2004;24:2487–2498. doi: 10.1128/MCB.24.6.2487-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC, Elledge SJ. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].D'Angiolella V, Donato V, Forrester FM, Jeong Y-T, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Klein DK, Hoffmann S, Ahlskog JK, O'Hanlon K, Quaas M, Larsen BD, Rolland B, Rosner HI, Walter D, Kousholt AN, Menzel T, Lees M, Johansen JV, Rappsilber J, Engeland K, Sorensen CS. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat Commun. 2015;6:5800. doi: 10.1038/ncomms6800. [DOI] [PubMed] [Google Scholar]
- [227].Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;28:3157–3166. doi: 10.1038/onc.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Chen X, Sahasrabuddhe AA, Szankasi P, Chung F, Basrur V, Rangnekar VM, Pagano M, Lim MS, Elenitoba-Johnson KS. Fbxo45-mediated degradation of the tumor-suppressor Par-4 regulates cancer cell survival. Cell Death Differ. 2014;21:1535–1545. doi: 10.1038/cdd.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Xu M, Zhu C, Zhao X, Chen C, Zhang H, Yuan H, Deng R, Dou J, Wang Y, Huang J, Chen Q, Jiang B, Yu J. Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget. 2015;6:979–994. doi: 10.18632/oncotarget.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Chiorazzi M, Rui L, Yang Y, Ceribelli M, Tishbi N, Maurer CW, Ranuncolo SM, Zhao H, Xu W, Chan W-CC, Jaffe ES, Gascoyne RD, Campo E, Rosenwald A, Ott G, Delabie J, Rimsza LM, Shaham S, Staudt LM. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3943–3948. doi: 10.1073/pnas.1217271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Samuelson DJ, Hesselson SE, Aperavich BA, Zan Y, Haag JD, Trentham-Dietz A, Hampton JM, Mau B, Chen K-S, Baynes C, Khaw K-T, Luben R, Perkins B, Shah M, Pharoah PD, Dunning AM, Easton DF, Ponder BA, Gould MN. Rat Mcs5a is a compound quantitative trait locus with orthologous human loci that associate with breast cancer risk. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6299–6304. doi: 10.1073/pnas.0701687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Smits BMG, Traun BD, Devries TL, Tran A, Samuelson D, Haag JD, Gould M. An insulator loop resides between the synthetically interacting elements of the human/rat conserved breast cancer susceptibility locus MCS5A/Mcs5a. Nucleic acids research. 2012;40:132–147. doi: 10.1093/nar/gkr610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, Hunter AJ, Cheeseman M, Brown SD. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15:3273–3279. doi: 10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- [235].Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell. 2013;49:1147–1158. doi: 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Rossi M, Duan S, Jeong Y-T, Horn M, Saraf A, Florens L, Washburn MP, Antebi A, Pagano M. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Molecular cell. 2013;49:1159–1166. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Zheng H, Shen M, Zha Y-L, Li W, Wei Y, Blanco Mario A.A., Ren G, Zhou T, Storz P, Wang H-Y, Kang Y. PKD1 Phosphorylation-Dependent Degradation of SNAIL by SCF-FBXO11 Regulates Epithelial-Mesenchymal Transition and Metastasis. Cancer Cell. 2014;26:358–373. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Jin Y, Shenoy AK, Doernberg S, Chen H, Luo H, Shen H, Lin T, Tarrash M, Cai Q, Hu X, Fiske R, Chen T, Wu L, Mohammed KA, Rottiers V, Lee SS, Lu J. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer letters. 2015;362:70–82. doi: 10.1016/j.canlet.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Jeong Y-T, Cermak L, Guijarro MV, Hernando E, Pagano M. FBH1 protects melanocytes from transformation and is deregulated in melanomas. Cell cycle (Georgetown, Tex.) 2013;12:1128–1132. doi: 10.4161/cc.24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Zhang Y, Long J, Lu W, Shu X-O, Cai Q, Zheng Y, Li C, Li B, Gao Y-T, Zheng W. Rare coding variants and breast cancer risk: evaluation of susceptibility Loci identified in genome-wide association studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:622–628. doi: 10.1158/1055-9965.EPI-13-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Laulier C, Cheng A, Huang N, Stark JM. Mammalian Fbh1 is important to restore normal mitotic progression following decatenation stress. DNA repair. 2010;9:708–717. doi: 10.1016/j.dnarep.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Lawrence CL, Jones N, Wilkinson CR. Stress-induced phosphorylation of S. pombe Atf1 abrogates its interaction with F box protein Fbh1. Current biology : CB. 2009;19:1907–1911. doi: 10.1016/j.cub.2009.09.044. [DOI] [PubMed] [Google Scholar]
- [244].Lockwood WW, Chandel SK, Stewart GL, Erdjument-Bromage H, Beverly LJ. The novel ubiquitin ligase complex, SCF(Fbxw4), interacts with the COP9 signalosome in an F-box dependent manner, is mutated, lost and under-expressed in human cancers. PLoS One. 2013;8:e63610. doi: 10.1371/journal.pone.0063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Baumann U, Fernández-Sáiz V, Rudelius M, Lemeer S, Rad R, Knorn A-M, Slawska J, Engel K, Jeremias I, Li Z, Tomiatti V, Illert A-L, Targosz B-S, Braun M, Perner S, Leitges M, Klapper W, Dreyling M, Miething C, Lenz G, Rosenwald A, Peschel C, Keller U, Kuster B, Bassermann F. Disruption of the PRKCD–FBXO25–HAX-1 axis attenuates the apoptotic response and drives lymphomagenesis. Nature Medicine. 2014;20:1401–1409. doi: 10.1038/nm.3740. [DOI] [PubMed] [Google Scholar]
- [246].Jang JW, Lee WY, Lee JH, Moon SH, Kim CH, Chung HM. A novel Fbxo25 acts as an E3 ligase for destructing cardiac specific transcription factors. Biochem Biophys Res Commun. 2011;410:183–188. doi: 10.1016/j.bbrc.2011.05.011. [DOI] [PubMed] [Google Scholar]
- [247].Teixeira FR, Manfiolli AO, Soares CS, Baqui MM, Koide T, Gomes MD. The F-box protein FBXO25 promotes the proteasome-dependent degradation of ELK-1 protein. J Biol Chem. 2013;288:28152–28162. doi: 10.1074/jbc.M113.504308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- [249].Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- [251].Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nature genetics. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- [252].Weng AP, Ferrando AA, Lee W, Morris J.P.t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- [253].Siu KT, Xu Y, Swartz KL, Bhattacharyya M, Gurbuxani S, Hua Y, Minella AC. Chromosome instability underlies hematopoietic stem cell dysfunction and lymphoid neoplasia associated with impaired Fbw7-mediated cyclin E regulation. Mol Cell Biol. 2014;34:3244–3258. doi: 10.1128/MCB.01528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- [255].Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle. 2011;10:3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, Zou C, Ellis B, Sciurba FC, Zhang Y, Mallampalli RK. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol. 2013;14:470–479. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Li Y, Hao B. Structural basis of dimerization-dependent ubiquitination by the SCF(Fbx4) ubiquitin ligase. The Journal of biological chemistry. 2010;285:13896–13906. doi: 10.1074/jbc.M110.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Barbash O, Lee EK, Diehl JA. Phosphorylation-dependent regulation of SCFFbx4 dimerization and activity involves a novel component, 14-3-3? Oncogene. 2011;30:1995–2002. doi: 10.1038/onc.2010.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Koo J, Yue P, Gal AA, Khuri FR, Sun S-Y. Maintaining glycogen synthase kinase-3 activity is critical for mTOR kinase inhibitors to inhibit cancer cell growth. Cancer research. 2014;74:2555–2568. doi: 10.1158/0008-5472.CAN-13-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Pontano LL, Aggarwal P, Barbash O, Brown EJ, Bassing CH, Diehl JA. Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Molecular and cellular biology. 2008;28:7245–7258. doi: 10.1128/MCB.01085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Lee EK, Lian Z, D'Andrea K, Letrero R, Sheng W, Liu S, Diehl JN, Pytel D, Barbash O, Schuchter L, Amaravaradi R, Xu X, Herlyn M, Nathanson KL, Diehl JA. The FBXO4 Tumor Suppressor Functions as a Barrier to BrafV600E-Dependent Metastatic Melanoma. Molecular and Cellular Biology. 2013;33:4422–4433. doi: 10.1128/MCB.00706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Chandrasekaran S, Tan TX, Hall JR, Cook JG. Stress-stimulated mitogen-activated protein kinases control the stability and activity of the Cdt1 DNA replication licensing factor. Mol Cell Biol. 2011;31:4405–4416. doi: 10.1128/MCB.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Rizzardi LF, Coleman KE, Varma D, Matson JP, Oh S, Cook JG. CDK1-dependent inhibition of the E3 ubiquitin ligase CRL4CDT2 ensures robust transition from S Phase to Mitosis. J Biol Chem. 2015;290:556–567. doi: 10.1074/jbc.M114.614701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Abbas T, Keaton M, Dutta A. Regulation of TGF-β signaling, exit from the cell cycle, and cellular migration through cullin cross-regulation: SCF-FBXO11 turns off CRL4-Cdt2. Cell cycle (Georgetown, Tex.) 2013;12:2175–2182. doi: 10.4161/cc.25314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai S-C, Zhu W, Nakajima H, Nakajima HO, Field LJ, Wang R, Pan Z-Q. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Molecular cell. 2008;30:403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Lin P, Fu J, Zhao B, Lin F, Zou H, Liu L, Zhu C, Wang H, Yu X. Fbxw8 is involved in the proliferation of human choriocarcinoma JEG-3 cells. Molecular biology reports. 2011;38:1741–1747. doi: 10.1007/s11033-010-0288-7. [DOI] [PubMed] [Google Scholar]
- [269].Cen G, Ding H-H, Liu B, Wu W-D. FBXL5 targets cortactin for ubiquitination-mediated destruction to regulate gastric cancer cell migration. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:8633–8638. doi: 10.1007/s13277-014-2104-9. [DOI] [PubMed] [Google Scholar]
- [270].Dragoi A-M, Swiss R, Gao B, Agaisse H. Novel strategies to enforce an epithelial phenotype in mesenchymal cells. Cancer research. 2014;74:3659–3672. doi: 10.1158/0008-5472.CAN-13-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [272].Fung TK, Siu WY, Yam CH, Lau A, Poon RYC. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. The Journal of biological chemistry. 2002;277:35140–35149. doi: 10.1074/jbc.M205503200. [DOI] [PubMed] [Google Scholar]
- [273].Li J, D'Angiolella V, Seeley ES, Kim S, Kobayashi T, Fu W, Campos EI, Pagano M, Dynlacht BD. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature. 2013;495:255–259. doi: 10.1038/nature11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes & development. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Kim J-H, Kim J, Kim D-H, Ryu G-H, Bae S-H, Seo Y-S. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic acids research. 2004;32:2287–2297. doi: 10.1093/nar/gkh534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. The Journal of cell biology. 2009;186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Jeong Y-T, Rossi M, Cermak L, Saraf A, Florens L, Washburn MP, Sung P, Schildkraut CL, Schildkraut C, Pagano M. FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. The Journal of cell biology. 2013;200:141–149. doi: 10.1083/jcb.201209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Fugger K, Chu WK, Haahr P, Kousholt AN, Beck H, Payne MJ, Hanada K, Hickson ID, Sørensen CS. FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress. Nature communications. 2013;4:1423. doi: 10.1038/ncomms2395. [DOI] [PubMed] [Google Scholar]
- [279].Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, Logothetis CJ, Hung MC, Zhang S, Lin HK. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Shuvalov O, Petukhov A, Daks A, Fedorova O, Ermakov A, Melino G, Barlev NA. Current Genome Editing Tools in Gene Therapy: New Approaches to Treat Cancer. Curr Gene Ther. 2015;15:511–529. doi: 10.2174/1566523215666150818110241. [DOI] [PubMed] [Google Scholar]