Abstract
We used two different infection models to investigate the kinetics of the PcpA-dependent pneumococcal disease in mice. In a bacteremic pneumonia model, we observed a PcpA-dependent increase in bacterial burden in the lungs, blood, liver, BAL and spleens of mice at 24-hrs post infection. This PcpA-dependent effect on bacterial burden appeared earlier (within 12-hrs) in the focal-pneumonia model, which lacks bacteremia or sepsis. Histological changes show that the ability of pneumococci to make PcpA was associated with unresolved inflammation in both models of infection. Using our bacteremic pneumonia model we further investigated the effects of PcpA on recruitment of innate immune regulatory cells. The presence of PcpA was associated with increased IL-6 levels, suppressed production of TNF-related apoptosis - inducing ligand (TRAIL) and reduced infiltration of polymorphonuclear cells. The ability of pneumococci to make PcpA negatively modulated both the infiltration and apoptosis of macrophages and the recruitment of myeloid-derived suppressor-like cells (MDSCs). The latter have been shown to facilitate clearance and control of bacterial pneumonia. Taken together, the ability to make PcpA was strongly associated with increased bacterial burden, inflammation and negative regulation of innate immune cell recruitment to the lung tissue during bacteremic pneumonia.
INTRODUCTION
Streptococcus pneumoniae (Sp), an aerotolerant Gram-positive anerobe, is the leading cause of pneumonia, bacteremia, otitis media, sinusitis, and meningitis worldwide. Sp is responsible for about 850,000 deaths per year in children under the age of 5, with its greatest impact in the developing countries [1]. S. pneumoniae normally colonizes individuals asymptomatically and is considered a commensal in the upper respiratory tract. During the disease process, pneumococci are aspirated into the lower respiratory tract, which can lead to the development of pneumonia [2-4]. The exact mechanism of how Sp changes from a state of asymptomatically colonizing the upper airways of individuals to causing pneumonia has not been elucidated.
Several surface antigens associated with the pneumococcus aid in immune evasion [5] during pneumococcal infection. Although the role of capsule and other pneumococcal virulence factors in the host response during infection has been investigated, little is known about the role of PcpA during pneumonia and invasive pneumococcal disease (IPD). PcpA [6-9], a choline binding protein, is considered an excellent candidate for inclusion into a pneumococcal protein vaccine [10]. This surface antigen is not produced under the high Mn2+ concentrations present in the nasopharynx [8] and antibody to PcpA does not protect against colonization [9]. Thus, immunity to it would not have an affect on colonization and thus would not open up a niche for an opportunistic pathogen lacking PcpA. PcpA contains several leucine-rich repeat (LRR) regions, which may contribute to its mechanism of virulence. Previous studies have shown that PcpA contributed to adherence of pneumococci to an immortalized alveolar epithelial cell line in vitro [11,12]; our in vivo and in vitro results reported here identify another possible activity of PcpA, the modulation of innate immunity to the benefit of the pneumococcus.
During the initial stage of pneumococcal pneumonia, ineffective phagocytosis by alveolar macrophages and differential pro-inflammatory cytokine release in lung tissues and airways are observed [13,14]. Pneumococci then rapidly multiply in the alveoli (4-24 hrs), followed by increased influx of neutrophils into the tissue accompanied by elevated cytokine and leukotriene levels in both the airways and lung tissue. The bulk of the damage to the lung is seen during the third stage (24-48 hrs), which presents as alveolar injury and interstitial edema and is marked also by regeneration and resolution. This is the window during which bacteria invade the bloodstream, followed by an increase in alveolar monocyte and lymphocyte activity. If bacterial infection is not sufficiently controlled during the final stage (72-96 hrs), there is further bacterial multiplication, massive tissue damage and high mortality [13-17].
Another hallmark of the immune response during pneumococcal pneumonia is the rapid induction of apoptosis in macrophages [18]. Phagocytosis of apoptotic macrophages is critical for limiting excessive tissue damage and inflammation during infection [19-26]. The inhibition of induction of apoptosis in macrophages during bacterial infections has been shown to decrease bacterial clearance. Recently, Steinwede et al. have shown that pneumococcal infection with serotype 19F strain EF3030 induces production of TRAIL (TNF-α-related apoptosis-inducing ligand) by neutrophils and neutrophil-dependent up-regulation of DR5 (the mouse homolog to human Trail receptor R2) expression on alveolar macrophages [18]. The subsequent binding of TRAIL to the DR5 receptor ultimately led to apoptosis of alveolar macrophages [18]. When alveolar macrophages underwent apoptosis instead of necrosis, the environment of the lungs was more anti-inflammatory than pro-inflammatory which proved to be protective for the host during pneumococcal pneumonia [18].
Recent studies have also provided evidence of the role of immature myeloid-derived suppressor (MDSC)-like cells in the resolution of bacterial pneumonia [27]. Although MDSCs are known for immune suppression via amino acid depletion and cytokine /reactive free radical-mediated modulation of T cell function, MDSCs have recently been implicated in the efferocytosis of apoptotic neutrophils in a Klebsiella pneumoniae infection model, suggesting an association of their infiltration with resolution of inflammation in bacterial pneumonia. Subsets of MDSC-like cells have also recently been identified as important regulators of allergic airway inflammation [28]. But the defined role for MDSCs in resolution of Sp-associated inflammation has not yet been investigated.
In this study, we show that the expression of the virulence factor PcpA had a strong impact on host defense in two distinct models of pneumococcal lung infection. In a mouse model of bacteremic pneumonia, PcpA expression was associated with increased bacterial burden in the lung, blood, spleen, and liver, increased inflammation, increased IL-6 production in the airways and decreased neutrophil and recruitment of MDSCs to the lungs during infection. We also observed a significant decrease in TRAIL production, DR5 expression and apoptosis of macrophages in the presence of PcpA. Taken together, our studies suggest that PcpA is likely to have a role in innate immune modulation during pneumococcal pneumonia.
METHODS AND MATERIALS
Mice
All mice used in this study were CBA/CaHN-Btkxid/J (CBA/N) (The Jackson Laboratory Bar Harbor, ME). Female mice were purchased at 6-8 weeks of age and rested for 7-14 days before use in specific pathogen free facilities at The University of Alabama at Birmingham (UAB). All studies were carried under the supervision of veterinarians using protocols approved by the Institutional Animal Care and Use Committee of UAB.
Bacterial strains, media, and growth conditions
S. pneumoniae strains TIGR4, EF3030 and their respective pcpA− mutants, JEN11 and JEN18 [8] were grown to an OD600~ 0.3 at 37°C in Todd-Hewitt broth with 0.5% yeast extract (Fisher Scientific, Pittsburgh, PA) (THY) or on blood agar plates (Becton Dickinson, Sparks, MD.). Previous expression studies have shown that an OD600 ~0.3 yields optimal PcpA expression. All infection stocks were started from plates of −80°C parent stocks of each strain in 10% glycerol. PcpA expression of TIGR4, EF3030 and their respective mutants was assayed by staining with polyclonal anti-PcpA antibody, and flow cytometry analysis was performed. Flow cytometry analysis [11,29] confirmed expression of PcpA by both WT strains and its absence in the mutant strains. Supplementary Fig 1 shows representative data for TIGR4 and EF3030 and their pcpA mutants JEN11 and JEN18 respectively. The wild type and mutants were grown in THY or blood agar plates with the addition of erythromycin (Fisher Scientific) for growth of the mutants at a concentration of 0.3 μg/mL. The list of wild type and mutant bacterial strains as well as the plasmid constructs is provided in Supplementary Figure 1. The construction of the mutant strains has been described previously [8].
Pneumococci were grown in high and low Mn2+ THY for these studies. PcpA is under control of PsaR, a manganese dependent repressor and therefore pneumococci were grown in low Mn2+ THY to induce expression of PcpA prior to infection [8] [9]. High Mn2+ THY is defined as regular Todd-Hewitt with 0.5% yeast extract. Low Mn2+ THY was made by depleting manganese out of the media and adding it back at a known concentration. Low Mn2+ was prepared according to manufacturer’s instructions with Chelex-100 [2%wt/vol] (Sigma Aldrich, St. Louis, MO.) being added prior to autoclaving. After autoclaving, low Mn2+ solution was stirred overnight at room temperature and then filter-sterilized. Prior to use, ZnCl2, MgCl2, CaCl2 and FeSO4 were added at a concentration of 1mM each and MnSO4 (Fisher Scientific) was added to a concentration of 0.1μM. Infection stocks were made by growing strains to 0.3 OD600, and storing bacteria in 10% glycerol in cryovials at −80°C. At least two weeks after freezer stocks were made, CFU in a representative vial were quantitated by serial dilution and plating on blood agar.
FACS analysis of PcpA expression
The expression of PcpA in different manganese conditions was assessed through flow cytometry. TIGR4, EF3030, JEN11 and JEN18 were grown up in low and high Mn2+ media to an OD600=0.2, diluted back to an OD600=0.1 and grown up to OD600=0.3. Bacterial cultures were then aliquoted into 250μL samples and spun at 8000rpm for 5 minutes. After removal of the supernatant, the pellet was resuspended in 100μL of primary antibody diluted 1:100 in 1% BSA in PBS. Cells were incubated with polyclonal anti-rabbit PcpA for 30 minutes at room temperature (RT).
After incubation, cells were washed with 750μL of 1% BSA in PBS, spun down and supernatant was removed. The pellet was resuspended in 100μL of FITC-labeled secondary antibody diluted 1:100 in 1% BSA in PBS and underwent incubation for 30 minutes at RT. After incubation, cells were washed with 1% BSA in PBS, spun down and supernatant was removed. Pellets were resuspended in 125μL of 1% BSA in PBS and 125μL of 2-4% paraformaldehyde in a FACS tube. Samples were analyzed within two days using a BD LSR II. Expression of PcpA was described as fold change in the mean fluorescence intensity (MFI) of the sample over MFI of the negative control, which were cells that had only the secondary antibody (FITC-labeled goat anti-rabbit) added to them. FlowJo (Tree star, Inc. Ashland, OR) was used to analyze dot plots and statistics on MFI of samples.
Detecting PcpA expression in vivo
To determine in vivo expression of PcpA, a PcpA capture-ELISA assay was carried out as follows. Microtiter 96-well plates (NUNC Weisbaden, Germany) were coated with an optimized combination of 3 mouse monoclonal antibodies (mAb) 20.10, 163, and 2.16, all of isotype IgG1) to PcpA (Widener RW, Grubbs JA, King JD, Schachern P, Swords WE, Briles DE. In preparation). The plates were coated with 2 ug/mL of total mAb in 1×PBS (Fisher Scientific, Pittsburg, PA) and incubated at 4°C overnight. As negative controls each assay contained mAb-coated wells to which no sample was added. To evaluate the specificity of PcpA detection, PcpA-uncoated wells were included. After coating, plates were washed three times with ELISA wash buffer (1×PBS with 0.05% Tween 20). Each well was blocked with 150uL of 1% BSA/0.2% casein in 1×PBS for 1hr and then washed three times with ELISA wash buffer. Samples of BAL from infected mice and a standard with a known concentration of rPcpA were then added to the plate and incubated at room temperature for one hour and washed three times with ELISA wash buffer. Plates were next incubated at room temperature for one hour with rabbit anti-PcpA IgG and then washed three times with ELISA wash buffer. Plates were then incubated at room temperature for one hour with biotin-conjugated donkey anti-rabbit immunoglobulin IgG (H+L) (Southern Biotechnology Associates, Birmingham, AL) and then washed three times with ELISA wash buffer. Plates were then incubated at room temperature for one hour with strepavidin-alkaline phosphatase (Southern Biotechnology Associates, Birmingham AL) and washed three times with ELISA wash buffer. The plates were next developed with p-nitrophenyl phosphate (Sigma, St. Louis, MO) and absorbance was read at 405 nm. Data was reported as ng PcpA/ml.
Lung Infection and in vivo mouse studies
The bacteremic pneumonia model was established by intratracheally infecting 8-12 week old male CBA/N mice with the capsular type 4 pneumococcal strain, TIGR4. This model is characterized by severe lung infection, development of bacteremia and sepsis within 24 hours, and mortality within 48 hrs post infection.
The focal pneumonia model was established by intratracheally infecting 8-12 week old male CBA/N mice with the capsular type 19F pneumococcal strain, EF3030. This model is characterized by a self-resolving of lung infection generally lacking any detectable bacteremia [30].
Mice were anesthetized with isoflurane (Henry Schein Animal Health, Dublin, OH). After anesthetization, mice were intratracheally (i.t.) infected with between 3.0-5.0×106 CFUs of TIGR4, EF3030, or their respective pcpA− mutants grown in low manganese in 40μL of lactated ringer’s solution (Hospira, Lakeforest, IL). At 12, 24, 36 and 96-hrs post infection mice were euthanized, and bronchoalveolar lavage fluid (BAL), lung tissue, blood, liver, and spleen were harvested. Prior to whole lungs being harvested, the trachea was cut at the top of the larynx and lungs were lavaged with 1mL of lactated ringer’s solution. Lungs, spleen, and liver were then harvested and homogenized in 1mL of ringer’s in stomacher bags. The BAL and homogenized tissue samples were serially diluted and plated on blood agar plates with gentamicin (Lonza, Walkersville, MD) or erythromycin. Plates were incubated at 37°C under 5% CO2 overnight and used to calculate CFU recovery.
Lung Histology
8-12 week old CBA/N mice were infected as previously described and were euthanized at 6, 12, 24, and 48-hr time points after infection. Prior to removal of the lung tissues, 1mL of 10% formalin solution (Fisher Scientific) was introduced into the lungs via the cannulated trachea. Inflated lungs were surgically removed, immediately submerged into 10% formalin and after fixation embedded in paraffin. Lung sections were stained with hematoxlyin-eosin (Fisher Scientific), mounted, and examined microscopically with a 20× objective. Lung sections were scored for inflammation and congestion. Mouse lung tissues were fixed in formalin, processed, sectioned (5 μm thick) and stained with hematoxylin-eosin. Each lung lobe tissue was examined microscopically by a pathologist for the presence of inflammatory infiltrate and congestion, and further graded. Inflammation was characterized by the presence of lymphocytic infiltrates. Grade 1 – scattered lymphocytes in perivascular and peribronchial area. Grade 2 – dense lymphocytes in perivascular and peribronchial area. Grade 3 – diffuse lymphocytic infiltrate involving portion of a lobe. Congestion was characterized by dense alveoli with blood-filled capillaries. Grade 1 – focal congestion (less than half of lobe). Grade 2 – diffuse congestion (more than half of lobe).
Harvesting and Isolation of Murine Lung Immune Cells
After bacterial challenge as described in the in vivo mouse studies, CBA/N mice were anesthetized using 50-100μL of a 1:1 ketamine (VETone, UAB Veterinary Care)/xylazine (Lloyd laboratories, Shenandoah, IA) mixture. Mice were euthanized by cardiac perfusion with 5-7 mLs of chilled 1× PBS (Fisher Scientific) followed by bilateral thoracotomy. The BAL was produced by lavaging lungs 4× with 1 mL of chilled PBS. The first 1mL of lavage fluid collected was centrifuged and the clarified supernatant was used for quantitating CFU recovery and measuring cytokine production. We enumerated infiltrating immune cells recruited to the airways during infection by pooling cellular components from all the lavage aspirates. The whole lungs were then harvested, placed in complete Iscove’s media (Mediatech Inc., Manassas, VA), and minced into fine pieces. Collagenase B (Roche Diagnostics, Indianapolis, IN) and DNAse (Sigma-Aldrich, St. Louis, MO) were added to the minced lung mixture and incubated at 37° C for 30 minutes as described before [28]. After incubation, digested lung tissue extract was then strained through a 40μm cell strainer as previously described [28,31].
Flow Cytometry Analyses of Lung Immune Cells
Both the BAL and lung homogenates were spun down at 1400rpm for 5 minutes at 4°C. Supernatants were removed and the pellet was washed 2× with 1mL of 1× PBS. Samples were then blocked with anti- CD16/ CD32 (2.4G2, Thermo Fisher scientific, Waltham, MA) in 1mL of 3% BSA (Fisher Scientific) in PBS for 30 minutes at 4 °C. Cells were stained with rat- anti-mouse antibodies for CD11b (M1/70, eBioscience, San Diego, CA), Ly6G (Gr-1) (RB6-8C5, eBioscience, San Diego, CA), Ly6C (HK1.4, eBioscience, San Diego, CA), F4/80 (BM8, eBioscience, San Diego, CA), and Annexin V+ (eBioscience, San Diego, CA). Lung cells and BAL were first gated on live cells. Lung and BAL neutrophils were identified as Gr-1+CD11b+Ly-6G+F4/80−. Alveolar macrophages were identified as cells that were CD11b+CD11c+F4/80+. PMNs where defined as cells that were CD11b+, Ly-6G+, F4/80−. MDSCs were identified as Gr-1+CD11b+Ly-6C+F4/80+ cells. Samples were fixed with 2-4% paraformaldehyde (Fisher Scientific) and data acquisition for flow cytometry was performed using a BD LSRII Flow Cytometer. Flow cytometry data were imported and differences in cell recruitment were analyzed using Flowjo (Tree Star, Inc. Ashland, OR). Absolute cell numbers were calculated as Total cells in tissue x % gated x % positive.
In vivo depletion of MDSCs
The effects of in vivo depletion of MDSCs were examined in the bacteremic pneumonia model. After bacterial challenge with TIGR4 and JEN11 strains as described in the in vivo mouse studies, CBA/N mice received anti-Gr1 antibody (150 μg/100μl) [32] or IgG control (BioXCell, West Lebanon, NH) intraperitoneally 3 hours after infection. At 24 hours after infection, BAL fluid and lung homogenates were harvested from mice and CFU were quantitated as described before.
TRAIL, TNF-α, and IL-6 ELISA
The BAL fluid from the in vivo mouse experiments taken at various time points were used in ELISA experiments to quantitate IL-6, TNF-α, and TRAIL levels in the airways of CBA/N mice in our bacteremic and focal pneumonia models. IL-6, TNF-α, and TRAIL levels were measured using IL-6 Quantikine ELISA (R & D Systems, Minneapolis, MN), and TNF-α Quantikine ELISA (R & D Systems, Minneapolis, MN), and TRAIL-R1 ELISA (MyBioSource, SanDiego, CA) following manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using Graphpad Prism software. All animal experiments contained 5-7 mice per group. Each figure represents the pooled data from at least two independent experiments. Differences in the median between two groups were compared by the Mann-Whitney two-sample rank-test. P-values of <0.05 were considered significant.
RESULTS
The PcpA-dependent effect on bacterial burden in the lung does not become apparent until 12 to 24 hours post infection
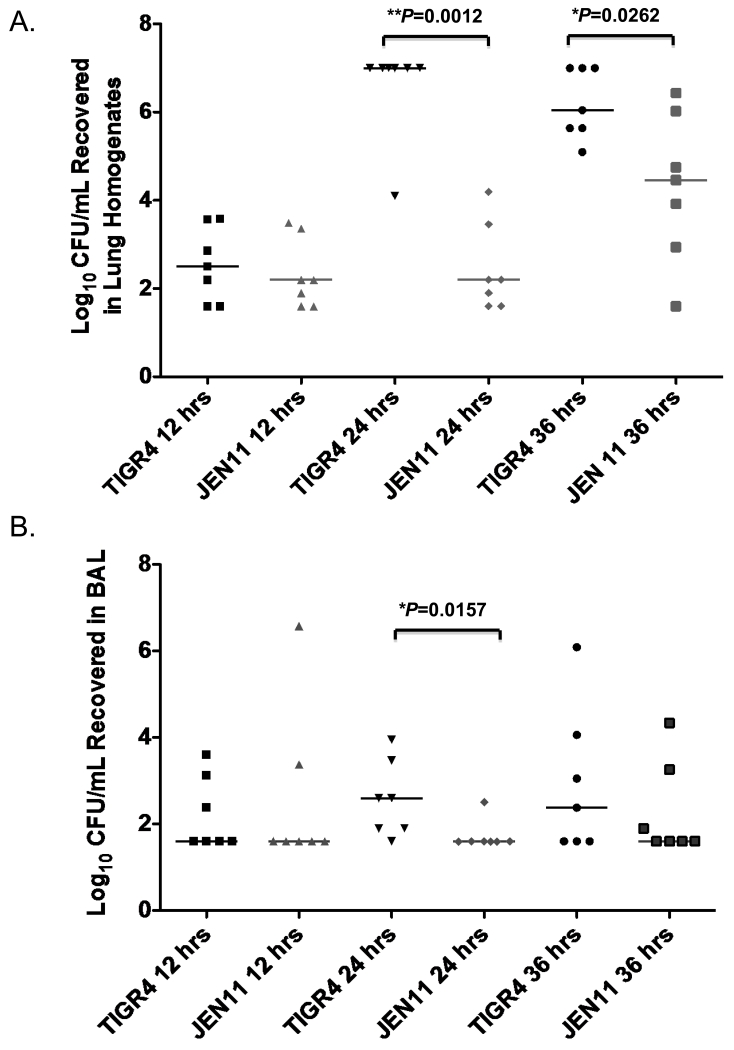
We utilized two models of lung infection, septic pneumonia and focal pneumonia, to investigate the effects of PcpA expression in vivo. In the septic pneumonia model, CBA/N mice were intratracheally infected with 106 CFUs of TIGR4 (serotype 4) or JEN11 (a pcpA mutant of TIGR4) grown in low Mn2+ THY. Sp are thought to generally be a commensal of the human nasopharynx. The exact mechanism of how this microorganism transitions from the nasopharynx where it asymptomatically colonizes to a more invasive site is unknown. The concentration of manganese within the mucosa tends to be in the micromolar range [33] and has been measured in the saliva at 36 μM. In the internal sites such as the blood and lungs the concentration of manganese is in the nanomolar range [33]. The lung and blood have lower concentrations of manganese and PcpA expression is de-repressed in this particular environment allowing PcpA to be a virulence factor in invasive infection in the lung and blood [9]. Our preliminary studies indicated that within the first 12-hrs, the numbers of CFU in the lung, blood, spleen, and liver remained low and were not effected by mutations in pcpA. In this model WT TIGR4 pneumococci usually produced lung inflammation and damage, and from 40 to 72 hours most of the mice became moribund or died. At the 12-hr time point, we looked at the effects of PcpA on the bacterial burden recovered in the lung tissue of pneumococcal infected mice and found no significant differences in CFUs recovered between TIGR4 (PcpA+) and JEN11 (PcpA−) (Figure 1A). We also found that at 12 hours after infection, there was no significant difference between WT and pcpA− TIGR4 pneumococci in terms of bacterial load in the airways (Figure 1B). However at 24-hrs after pneumococcal challenge we saw a significant increase (**P=0.0012 and 0.0157) in bacterial burden in both the lungs and airways in the presence of PcpA (Figure 1A and B). This difference remained significant in the lung tissue at 36-hrs post infection (*P=0.0262) but not in the BAL (Figure 1A and B).
Figure 1. The PcpA-dependent effect on bacterial burden in the bacteremic pneumonia model occurs late in infection.
(A) At 12, 24, and 36 hrs post infection, bacterial burdens were quantified in the lung homogenates of mice infected with 1.0× 106 CFU of TIGR4 or JEN11 (**P=0.0012 for 24 hrs and *P=0.0262 for 36 hrs). (B) After 12, 24, and 36 hrs post infection, bacterial burdens were quantified in the BAL of mice infected with 1.0× 106 CFU of TIGR4 or JEN11 (*P=0.0157 for 24 hrs). The horizontal bars indicate the medians. These data are representative of three independent experiments.
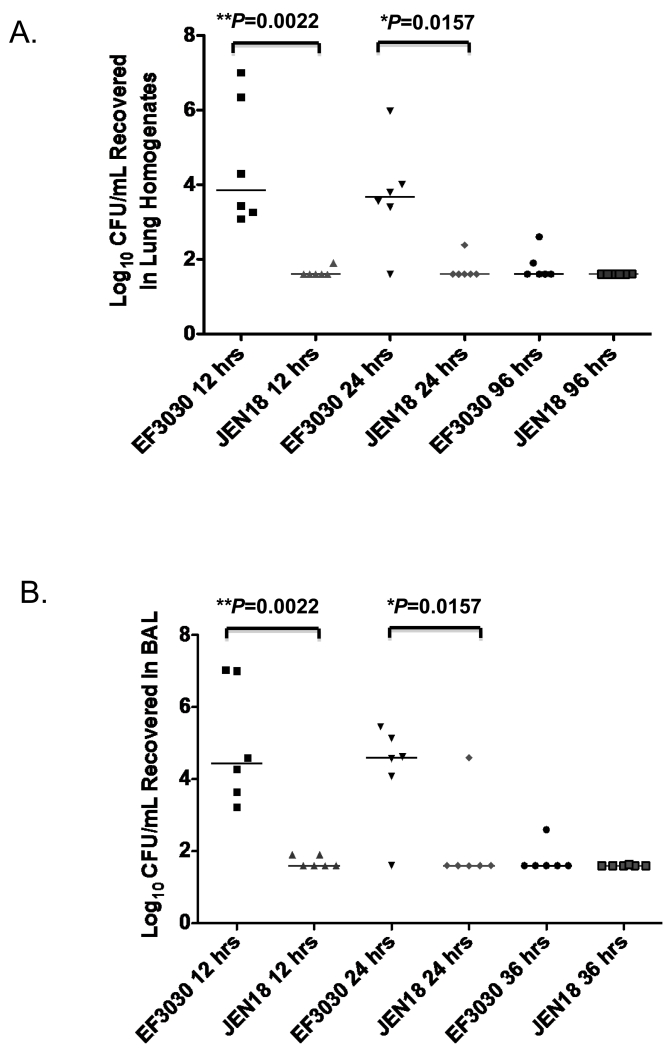
The focal lung infection model in which CBA/N mice were infected i.t. with EF3030 strain produced a self-resolving focal pneumonia infection with only rare incidences of bacteremia [30]. It is characteristic of 19F strains like EF3030 not to cause sepsis [34] and when present in the lung, to be restricted to the lungs and upper respiratory tract [30]. Because the lung disease was less severe in this model, we were able to analyze the lungs at later time points than with the TIGR4 infections. We infected CBA/N mice i.t. infected with 106 CFUs of EF3030 (WT) or JEN18 (pcpA− mutant) and harvested the lungs and BAL at 12, 24 and 96-hrs post infection. Surprisingly, the PcpA-dependent effect on bacterial burden of EF3030 in the lungs appeared within the first 12-hrs after challenge with Sp. At the 12 hr time point we saw greater clearance of bacteria from the lung tissue (**P=0.0022) and the BAL (**P=0.0022) in the absence of PcpA (Figure 2A and B). This difference between EF3030 and its pcpA mutant remained significant at 24-hrs (*P=0.0157) but not at 96-hrs post infection (Figure 2A and B) as the infections were largely resolved by that time. Even though the course of disease with both pathogens was very different, our data revealed a consistent effect with higher CFU levels with each WT strain compared to their pcpA mutants suggesting that this effect was PcpA dependent.
Figure 2. The PcpA-dependent effect on bacterial burden in the focal pneumonia model occurs earlier than in the bacteremic model.
(A) After 12, 24, and 96 hrs post infection, bacterial burdens were quantified in the lung homogenates of mice infected with 1.0× 106 CFU of EF3030 or JEN18 (**P=0.0022 for 12 hrs and *P=0.0157 for 24 hrs). (B) After 12, 24, and 96 hrs post infection, bacterial burdens were quantified in the BAL of mice infected with 1.0× 106 CFU of EF3030 or JEN18 (**P=0.0022 for 12 hrs and *P=0.0157 for 24 hrs). The horizontal bars indicate the medians. These data are representative of two independent experiments.
The effect of PcpA on survival in infected mice
There was no effect of the pcpA mutation on the survival time of EF3030 since mice do not die at the challenge dose used, but have a self-resolving infection in the lungs. All mice infected with TIGR4 died. The median survival time of 5 mice infected with TIGR4 was 29.5 hours and the longest surviving mouse died at 68 hours. For JEN11-infected mice the median survival was 168 hours. One of the five mice died at 101 hours and 4 died at 168 hr (P value = 0.0075, Mann Whitney test).
The expression of PcpA is associated with significant increases in CFUs recovered in the blood and the spleen and liver
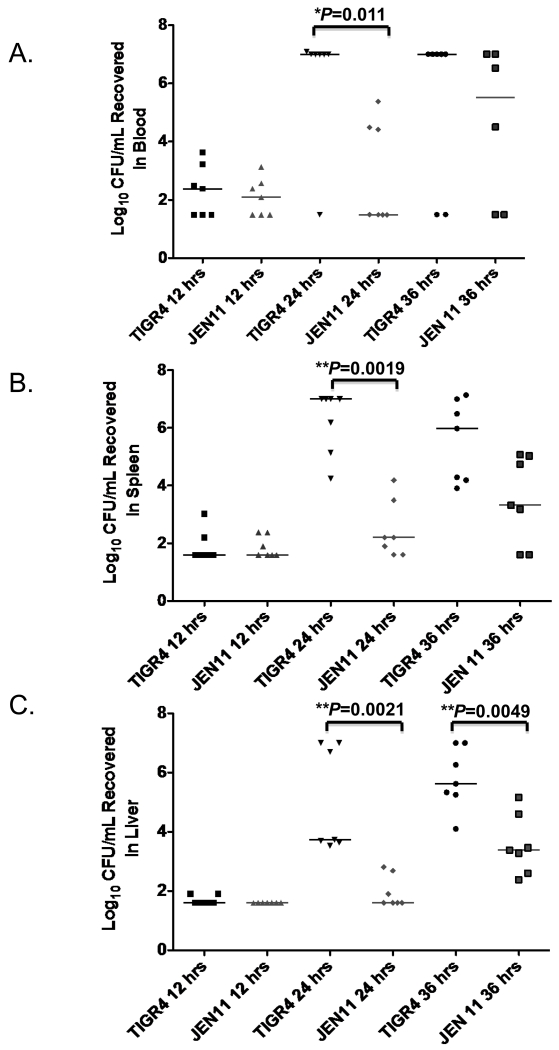
To more completely understand PcpA’s role during infection we looked at its effects on levels of the bacteria in the lungs to blood, liver, and spleen. CBA/N mice were infected i.t. with 106 CFUs of TIGR4 or JEN11 (pcpA−) in our bacteremic pneumonia model and the bacterial burdens in the blood, liver, and spleen were quantitated at 12, 24, and 36-hrs post infection. Twenty-four hours after pneumococcal challenge, in the presence of PcpA expression, we observed a significant increase in CFUs recovered from blood (*P=0.0111), spleen (***P=0.0019), and liver (***P=0.0021) (Figure 3A, B, and C). These observations were consistent with the findings at the same time points in the lung tissue and BAL of these same mice (Figure 1). Thus, PcpA expression strongly enhanced the levels of pneumococci in the lungs, which may have accounted for their higher levels in the blood, liver, and spleen.
Figure 3. The PcpA-dependent effect on bacterial burden in the blood, spleen, and liver appears by 24 hrs post infection.
(A) After 12, 24, and 36 hrs post infection, bacterial burden was quantified in the blood of mice infected with 1.0× 106 CFU of TIGR4 or JEN11 (*P=0.0111 for 24 hrs). (B) After 12, 24, and 36 hrs post infection, bacterial burden was quantified in the spleen of mice infected with 1.0× 106 CFU of TIGR4 or JEN11 (***P=0.0019 for 24 hrs). (C) After 12, 24, and 36 hrs post infection, bacterial burden was quantified in the liver of mice infected with 1.0× 106 CFU of TIGR4 or JEN11 (***P=0.0021 for 24 hrs and **P=0.0049 for 36 hrs). The horizontal bars indicate the medians. These data are representative of three independent experiments.
As expected, virtually no CFUs were noted in the blood, spleen and liver of mice in the focal pneumonia model at any time point (data not shown). Only one mouse out of 36 infected with EF3030, or its pcpA− mutant JEN18, yielded any detectable CFU and these were seen only in the liver (log 3.3 CFU) at the 12-hour time-point (data not shown). We further validated in vivo PcpA expression using a PcpA capture ELISA assay. As shown in Supplemental Fig. 2, PcpA was detected in the BAL of mice infected with TIGR4 and EF3030 pneumococci but not in the BAL of mice infected with the mutants Jen11 or Jen 18. PcpA is not detectable by our PcpA ELISA in lysates of Jen11 or Jen18. Neither is it detectable on the surface of these strains of pneumococci with rabbit anti-PcpA sera and secondary FITC-labeled goat-anti-rabbit antiserum by flow cytometry when these strains are grown in normal THY media or low-manganese THY. It is measurable by these methods on the TIGR4 and EF3030 background strains.
The expression of PcpA promotes higher levels of lung inflammation
The above studies led us to conclude that PcpA had no significant effects in vivo during the first few hours of lung infection, at least in the case of infection with TIGR4 strain. We then evaluated whether PcpA expression modulated overall inflammation in the lungs of mice during pneumococcal infection using our bacteremic pneumonia model in which CBA/N mice were i.t. challenged with 106 CFUs of TIGR4 or JEN11. To ensure that the time course of PcpA-mediated effects on inflammation was adequately studied, we examined 6, 12, 24 and 48-hour time points. This time frame allowed us to observe when mice were first developing signs of pneumonia before they start to become septic at about 40 hours. Studies of later time points were not possible because many of the mice infected i.t. with TIGR4 started to die shortly after 48 hours post infection. At a sublethal dose of 5×105 CFU of TIGR4, we did not detect any sign of infection and inflammation within either group (data not shown).
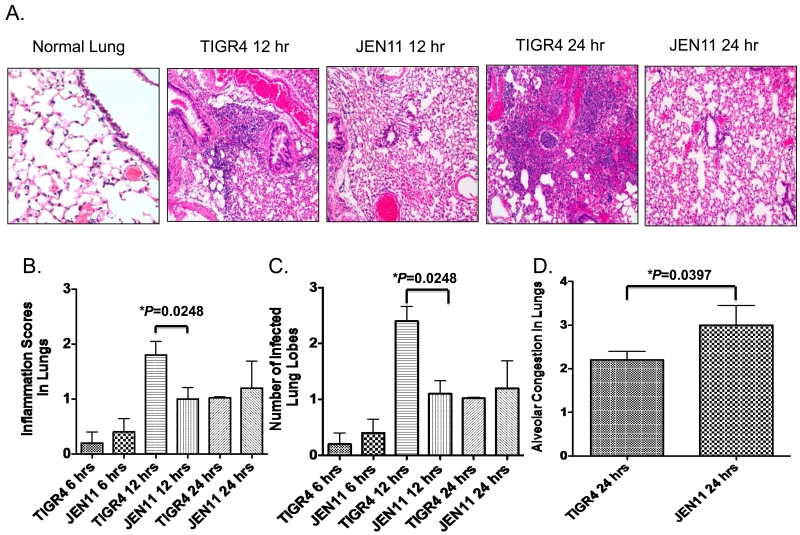
However, using a challenge dose of 1.0 × 106 CFU/mouse, histological evidence of lung infection became apparent (Figure 4A). At the 6 hr time point inflammation was apparent, but there were no observable differences between the mice infected with the WT and mutant TIGR4 pneumococci (Figure 4B). By 12-hrs, lung infection was more pronounced in the mice infected with TIGR4 than those infected with the pcpA mutant of TIGR4 (Figure 4). This was apparent from the higher histologic grade of inflammation (increased cellular infiltrates containing mainly lymphocytes and neutrophils) (Figure 4B), and the presence of more inflamed lung lobes/lung when the infection was with TIGR4 versus its pcpA mutant (Figure 4C, *P=0.0248). At 24-hrs, the WT-infected mice continued to have more cellular infiltrates than the pcpA mutant-infected mice but the difference was not statistically significant (Figure 4B). At 24 hours there was also no longer a difference in numbers of lobes infected by TIGR4 versus its pcpA mutant (Figure 4C).
Figure 4. PcpA expression promotes more inflammation and alveolar congestion in the lungs during bacteremic pneumonia.
(A) CBA/N mice were infected with 1.0×106 CFUs of TIGR4 or JEN11. After 12 and 24 hrs post infection, H&E stained lung section were analyzed for markers of leukocytes in alveoli and bronchi as a marker inflammation and thickening of alveolar septa by intrusion of blood as a marker of congestion. (B) After 6, 12, and 24 hrs post infection, lung sections were analyzed and designated inflammation scores from CBA/N mice infected with 1.0×106 CFUs of TIGR4 or JEN11 (*P=0.0248 for 12 hrs). (C) After 6, 12, and 24 hrs post infection, lung sections were analyzed for number of infected lung lobes from CBA/N mice infected with 1.0×106 CFUs of TIGR4 or JEN11 (*P=0.0248 for 12 hrs). (D) After 24 hrs post infection, alveolar congestion of CBA/N mice infected with 1.0×106 CFUs of TIGR4 or JEN11 were analyzed (*P=0.0397). The bars indicate average scores with standard error of the mean. These data are representative of three independent experiments.
Another parameter we used to evaluate the effects of PcpA expression on lung disease during pneumonia was measurement of alveolar congestion, defined as the thickening and filling with blood of alveolar septa usually occurring during the initial stages of pneumonia. Despite the increased cellular infiltration at 12 and possibly at 24hrs (Figure 4B) in the presence of PcpA, there was significantly less alveolar congestion at 24 hours with WT than the pcpA mutant challenge strain (*P=0.0397) (Figure 4 D).
The presence of PcpA expression was associated with decreased TRAIL production and increased IL-6 and TNF-α production in the lungs of mice
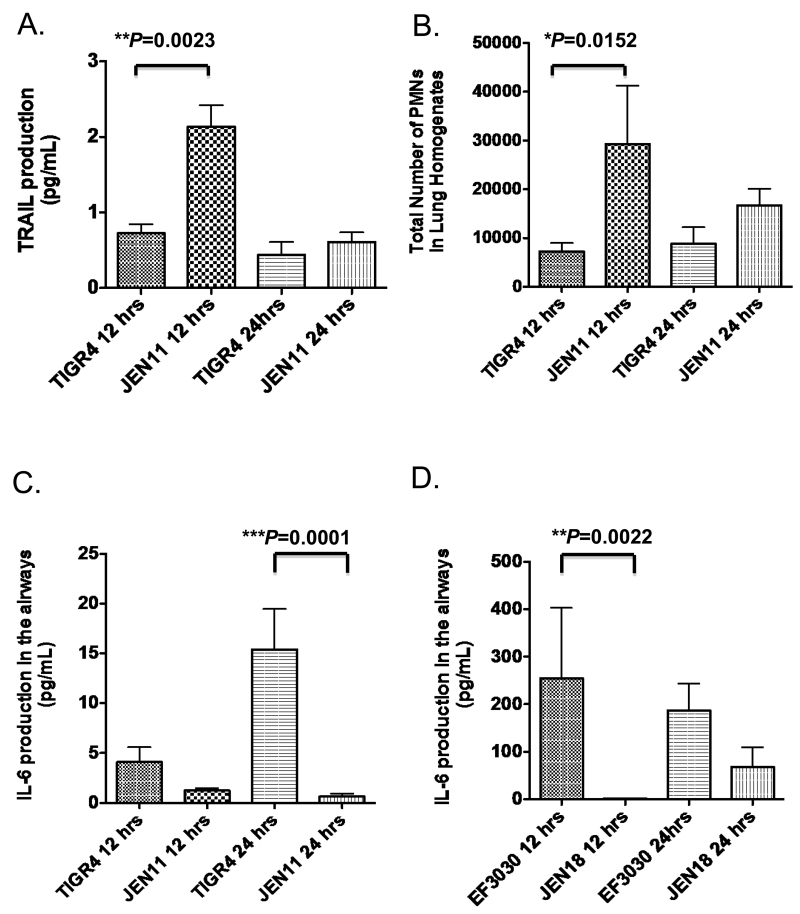
TRAIL is indicative of a potential for triggering anti-inflammatory apoptosis in macrophages [18]. Since we saw an increase in inflammation in the presence of PcpA, we then wanted to determine if the reduced lung inflammation in infections with the pcpA mutant was associated with levels of TRAIL in their lungs. Such an association could be indicative of differences in the potential for triggering anti-inflammatory apoptosis of macrophages in the presence or absence of PcpA. By 12 hrs post infection, in the absence of PcpA (in JEN11 infected mice) there were significantly higher levels of TRAIL (**P=0.0023) in the airways than in mice infected with TIGR4 (Figure 5A). At 24 hrs, the levels of TRAIL were reduced to baseline in the airways in both two groups. As polymorphonuclear (PMN) cells have been reported to be significant contributors of TRAIL, we quantitated the absolute numbers of infiltrating PMNs in the lung tissue. The peak of TRAIL production in the airways appears to positively correlate with the peak of PMN recruitment in the lung tissue (Figure 5B), which are consistent with previous studies using WT pneumococci [18].
Figure 5. PcpA expression significantly decreases PMN-induced TRAIL production and leads to higher levels of the pro-inflammatory cytokine IL-6.
CBA/N mice were infected with WT or the pcpA inactivated mutant, lung homogenates and BAL were harvested at 12, 24, and 96 hrs post infection. (A) After 12 and 24 hrs post infection, TRAIL production in the airways after infection was assayed by ELISA in CBA/N mice that were infected with TIGR4 or JEN11 (**P=0.0023 for 12 hrs). (B) After 12 and 24 hrs post infection, total number of PMN in the LH were quantified (***P=0.0152 for 12 hrs). (C) After 12 and 24 hrs post infection, IL-6 production in the airways after infection was assayed by ELISA in mice infected with TIGR4 or JEN11(***P=0.0001 for 12 hr) (D) After 12 and 24 hrs post infection, IL-6 production in the airways after infection with EF3030 or JEN18 (**P=0.0022 for 12 hr). The bars indicate averages with standard error of the mean. These data are representative of two independent experiments.
IL-6 and TNF-α are two of the major pro-inflammatory cytokines that play a role in the pathogenesis of pneumococcal infections in mouse models [13]. IL-6 and TNF-α levels are elevated during early infection in the airways and these levels are reported to increase between 4 and 24 hrs post infection [13]. We determined whether the decreased TRAIL expression in WT infections would be associated with higher levels of IL-6 and TNF-α. Such a result might be expected in a pro-inflammatory environment observed in the WT infection groups. At 24 hrs post infection, in the presence of PcpA, significantly higher levels of IL-6 (Figure 5C ***P=0.0001), and TNF-α (*P=0.028, Supplementary Figure 3) production was observed in the airways of CBA/N mice in the bacteremic model. We observed the same trend in the airways of infected mice in the focal pneumonia model. Mice infected with EF3030 (WT) had higher levels of IL-6 (**P=0.0022) in their airways at 12 hrs than the mutant infected mice (Figure 5D). The increased bacterial burden and the resulting lung inflammation may contribute to the increased IL-6 levels in the airways of mice infected with EF3030. The peak of IL-6 production also positively correlated with peak inflammation scores in histological experiments in the lungs of CBA/N mice. Taken together, in the airways of mice infected with pneumococci expressing PcpA, there were significantly lower levels of TRAIL indicative of less apoptotic potential and significantly higher levels of IL-6 and TNF-α production indicative of pro-inflammatory airway milieu.
The presence of PcpA resulted in decreased numbers of apoptotic macrophages and efferocytotic MDSCs in the lungs of CBA/N mice during pneumococcal lung infection
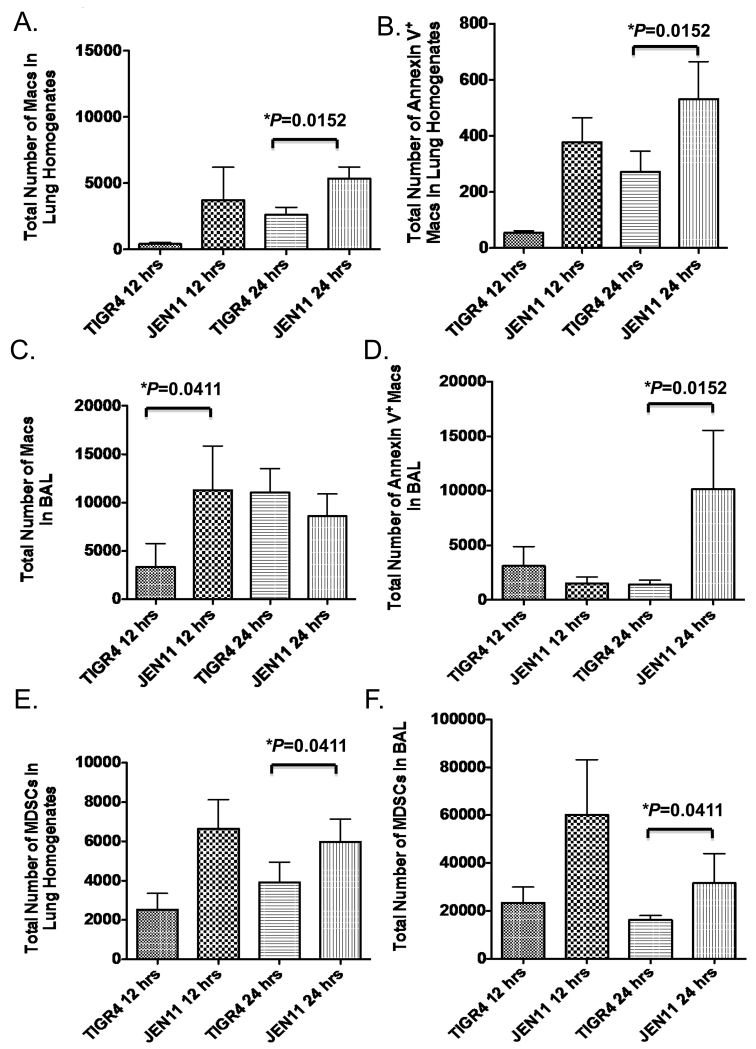
Apoptosis of alveolar macrophages in the lung after pneumococcal challenge is a hallmark of the disease process [18,21-24,35]. The induction of apoptosis in alveolar macrophages limits the inflammatory environment of the infected lungs and promotes tissue repair. Because we observed a significant decrease in TRAIL production in the presence of PcpA, we wanted to see if that translated into a decrease in apoptotic immune cells in the lung tissue and airways during pneumococcal pneumonia. At 12 hrs post infection, CBA/N mice infected with TIGR4 had reduced infiltration of total macrophages and apoptotic cells as measured by Annexin V+ staining in lung homogenates compared to JEN11-infected mice, although the reduction was not statistically significant (Figure 6A and B). By 24 hours, there were significantly more total macrophages (*P=0.0152) and apoptotic cells (*P=0.0152) in the lungs of mutant infected mice compared to the lungs of TIGR4-infected mice (Figure 6A and B). In the airways, CBA/N mice infected with TIGR4 had a significantly reduced infiltration of macrophages at 12 hrs (*P=0.0411) but equivalent numbers at 24 hrs compared to JEN11-infected mice (Figure 6C). As observed in the lungs, a significantly higher number (*P=0.0152) of apoptotic cells was observed in the airways of mutant-infected mice by the 24 hr time point (Figure 6D). We also observed that PcpA expression reduced significantly the total numbers of MDSCs recruited to the lung tissue and airways (*P=0.0411) (Figure 6E and F). At 24 hrs post infection, WT-infected mice had significantly fewer total numbers of DR5 positive (*P=0.0152) macrophages in the BAL (data not shown). Taken together, the presence of PcpA in the lungs of CBA/N mice in our bacteremic pneumonia model leads to suppression of immune regulatory cell recruitment and reduction of apoptotic cells making it more likely to promote an exacerbated pro-inflammatory environment.
Figure 6. PcpA Expression leads to apoptosis of macrophages and decreased MDSC recruitment to the airways during bacteremic pneumonia.
CBA/N mice were infected with TIGR4 and JEN11, lung homogenates and BAL were harvested at 12 and 24 hrs post infection (A) Total number of Macs in the LH (*P=0.0152 for 12 hrs). (B) Total number of Annexin V+ macrophages in the lung homogenates (*P=0.0152 for 12 hrs). (C) Total number of Macs in the LH (*P=0.0411 for 24 hrs). (D) Total number of Macs in the BAL (*P=0.0152 for 12 hrs). (E) Total number of MDSCs in the LH (*P=0.0411 for 24 hrs). (F) Total number of MDSCs in the BAL (*P=0.0411 for 24 hrs). The bars indicate averages with standard error of the mean. These data are representative of two independent experiments.
In vivo depletion of MDSCs by anti-Gr1 antibody had no effect on WT-infected mice but significantly increased the bacterial burden in pcpA−mutant-infected mice
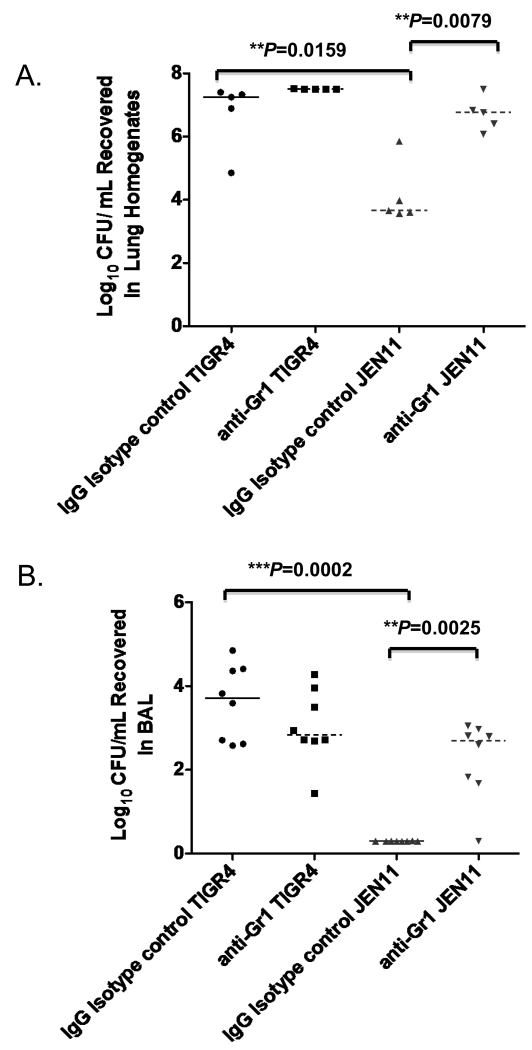
To further investigate whether a significant increase in innate immune cell recruitment, specifically MDSCs, in the lungs of pcpA− mutant infected CBA/N mice were contributing to clearance of the pneumococci, and to determine if monocytic or granulocytic MDSC contribute to the clearance of pneumococci, 3 hours after infection we instilled anti-Gr-1 or an IgG isotype control antibody intraperitoneally in mice infected with either WT pneumococci or the pcpA− mutant. Gr-1 is a GPI-myeloid differentiation marker (also known as Ly6G) whose cellular expression is restricted to monocytes in the bone and neutrophils in the peripheral organs [36]. MDSC are heterogeneous immature myeloid cells including immunosuppressive monocytic and granulocytic subsets. Anti-Gr-1 antibody administration depletes both monocytic and granulocytic subsets of MDSCs [36]. CBA/N mice were sacrificed 24 hrs post infection; lungs were first lavaged to collect CFU counts in the BAL, and lungs were homogenized to quantitate the CFU in the lung homogenates. At 24 hrs, we found that WT-infected CBA/N mice that were given the IgG isotype control had a significant increase in bacterial burden in the lung tissue (*P=0.0159) and airways (*P=0.0002) compared to PcpA− mutant infected mice (Figure 7A and B). When we used anti-Gr-1 antibody to deplete MDSCs in the lungs of WT-infected mice, we found that anti-Gr-1 administration had no significant effects on the bacterial burden in the lung tissue and BAL (Figure 7A and B). Interestingly, in the mutant JEN11 strain-infected mice, depletion of MDSCs led to a significant increase in bacterial burden in both the lung tissue and airways (**P=0.0079 and **P=0.0025) (Figure 7A and B). These data suggest that the significant increases in MDSCs that are recruited to the lungs in the absence of PcpA are responsible for the increased bacterial clearance that is observed during infection with the mutant strain.
Figure 7. In the absence of PcpA, MDSC depletion using anti-Gr-1 antibody leads to a significant increase in lung bacterial burden during bacteremic pneumonia.
CBA/N mice were instilled with anti-Gr-1 or an IgG isotype control antibody and subsequently infected with TIGR4 and JEN11. (A) Lung homogenates (**P=0.0079) and (B) BAL (**P=0.0025) were harvested at 24 hrs post infection and bacterial burden were quantified. The bars indicate averages with standard error of the mean. These data are representative of two independent experiments.
DISCUSSION
PcpA is a virulence factor in the lung and the blood; however, the direct mechanism of virulence has not yet been identified. For this reason we chose to study the role of PcpA during pneumococcal lung disease in two distinct mouse models.
It was previously shown that the expression of PcpA significantly enhanced adherence to A549 cells in vitro but had no biologically relevant role in initial attachment in a mouse model for pneumococcal pneumonia in vivo [11,12]. The observation of Glover et al. that the pcpA− mutant in the EF3030 background was significantly less recovered compared to the WT strain in the blood 7 days post infection [9], was consistent with a the late role of PcpA in infection, that was observed in the present studies. The pathogenesis of pneumococcal pneumonia involves five stages [13-17,37]. The first stage of the immune response during pneumococcal pneumonia is marked by rapid apoptosis of alveolar macrophages and differential pro-inflammatory cytokine release in lung tissues. The failure to induce apoptosis in macrophages leads to a highly inflammatory environment. Our studies shown here suggest that the presence of PcpA has negative effects on the first two stages of the innate immune response during pneumococcal pneumonia. Therefore, we chose to study the role of PcpA during pneumococcal infection in order to tease apart when its expression is critical for virulence in pneumonia. Our current studies have identified a strong association of PcpA expression with increased bacterial burden in the lung tissue, BAL, blood, spleen, and liver by 24-hrs post infection in our bacteremic pneumonia model. By 36 hrs, we observed that mice infected with the pcpA− mutant appear to have a similar bacterial load in each site, leading us to conclude that the increase in mutant pneumococci at that time point is sufficient enough to induce a successful innate immune response. The PcpA-dependent effect on bacterial burden occurring later on in infection seems to suggest that PcpA’s role during pneumococcal pneumonia has little to do with attachment to the lung epithelium and more with modulating the host innate immune response.
We evaluated PcpA’s role in inflammation (where we define it as an influx of neutrophils and lymphocytes) and alveolar congestion after pneumococcal lung infection. In our studies we observed a PcpA-dependent effect on inflammation and alveolar congestion in the lungs during bacteremic pneumonia starting at 12-hrs post infection. This association suggests that the absence of PcpA is protective to the lung tissue and promotes tissue repair earlier than WT-infected mice. We predict that this virulence factor thus promotes lung damage and increased bacterial burden.
We investigated the role of PcpA in recruitment of key players in the innate immune response to bacterial infections. We found that in the presence of pcpA-expressing pneumococci there were significantly fewer professional phagocytes including macrophages and neutrophils aiding in bacterial clearance in the lungs of WT-infected mice. We report here also our novel observation that PcpA suppresses infiltration of MDSCs. We predict that in this environment created by PcpA activity on innate immunity, pneumococci can more rapidly multiply resulting in collateral lung injury and increased burden. MDSCs are a heterogenous population of innate cells. These immature myeloid cells suppress host immune responses through various mechanisms including production of large amounts of reactive oxygen and nitrogen species, immunoregulatory cytokines such as TGF-β and IL-10 and upregulation of immune regulatory enzymatic pathways including inducible nitric oxide synthase and arginase. Although they were discovered first in the microenvironment of tumors, more recently, MDSCs have been shown to play an integral role in the innate immune response during bacterial infection. Those studies showed that lung MDSCs appeared with a delayed kinetics in response to Klebsiella pneumoniae infection [27]. These cells produced IL-10, which was associated with enhanced efferocytosis of apoptotic PMNs. Our data suggests that expression of PcpA is associated with dysregulation of innate immune mechanisms including reduced recruitment of MDSCs, PMNs and macrophages to the lung tissue. Further investigations are necessary to elucidate the direct mechanism of PcpA-dependent suppression of infiltration of MDSCs.
Although PcpA is produced in low amounts by pneumococci it has been shown to be a virulence factor in the lung and blood. Based on our present results, it seems likely that the mode of action of antibody to PcpA could be inhibition of PcpA’s deleterious effect on innate immunity. PcpA may therefore be a useful candidate for incorporation in a vaccine against invasive pneumococcal infection.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank specifically, Janice King, Joanetha Y. Hale and Tong Huan Jin for their technical assistance.
Footnotes
This work was supported in part by National Institutes of Health grants AI021458 (D.B.), P30AI027767(D.B.), F31AI10259 (M.W.), FAMRI YCSA Faculty Award ( J.D.), PACCM funds, and Sanofi Pasteur.
REFERENCES
- 1.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 4.Weiser JN. The pneumococcus: why a commensal misbehaves. J Mol Med (Berl) 2010;88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann S, Hammerschmidt S. Versatility of pneumococcal surface proteins. Microbiology. 2006;152:295–303. doi: 10.1099/mic.0.28610-0. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Beato AR, Lopez R, Garcia JL. Molecμar characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol Lett. 1998;164:207–214. doi: 10.1111/j.1574-6968.1998.tb13087.x. [DOI] [PubMed] [Google Scholar]
- 7.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston JW, Briles DE, Myers LE, Hollingshead SK. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect Immun. 2006;74:1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover DT, Hollingshead SK, Briles DE. Streptococcus pneumoniae surface protein PcpA elicits protection against lung infection and fatal sepsis. Infect Immun. 2008;76:2767–2776. doi: 10.1128/IAI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks WA, Chang LJ, Sheng X, Hopfer R, Team PPRS Safety and immunogenicity of a trivalent recombinant PcpA, PhtD, and PlyD1 pneumococcal protein vaccine in adults, toddlers, and infants: A phase I randomized controlled study. Vaccine. 2015;33:4610–4617. doi: 10.1016/j.vaccine.2015.06.078. [DOI] [PubMed] [Google Scholar]
- 11.Khan MN, Sharma SK, Filkins LM, Pichichero ME. PcpA of Streptococcus pneumoniae mediates adherence to nasopharyngeal and lung epithelial cells and elicits functional antibodies in humans. Microbes Infect. 2012;14:1102–1110. doi: 10.1016/j.micinf.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker MMWR, Coats M, Mirza S, Glover D, Myers L, Ochs M, Briles DE. The effect of PcpA on adherence to A549 epithelial cells may not explain its virulence role in mouse lung infections. PLOS one. 2014 [Google Scholar]
- 13.Gillespie SH, Balakrishnan I. Pathogenesis of pneumococcal infection. J Med Microbiol. 2000;49:1057–1067. doi: 10.1099/0022-1317-49-12-1057. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie SH, McWhinney PH, Kibbler CC. Pneumococcal bacteraemia. Lancet. 1991;337:376. doi: 10.1016/0140-6736(91)91020-u. [DOI] [PubMed] [Google Scholar]
- 15.Bergeron Y, Ouellet N, Deslauriers AM, Simard M, Olivier M, et al. Cytokine kinetics and other host factors in response to pneumococcal pulmonary infection in mice. Infect Immun. 1998;66:912–922. doi: 10.1128/iai.66.3.912-922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cauwels A, Wan E, Leismann M, Tuomanen E. Coexistence of CD14-dependent and independent pathways for stimulation of human monocytes by gram-positive bacteria. Infect Immun. 1997;65:3255–3260. doi: 10.1128/iai.65.8.3255-3260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun JS, Novak R, Gao G, Murray PJ, Shenep JL. Pneumolysin, a protein toxin of Streptococcus pneumoniae, induces nitric oxide production from macrophages. Infect Immun. 1999;67:3750–3756. doi: 10.1128/iai.67.8.3750-3756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinwede K, Henken S, Bohling J, Maus R, Ueberberg B, et al. TNF-related apoptosis-inducing ligand (TRAIL) exerts therapeutic efficacy for the treatment of pneumococcal pneumonia in mice. J Exp Med. 2012;209:1937–1952. doi: 10.1084/jem.20120983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton JC. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol. 1996;4:103–106. doi: 10.1016/0966-842X(96)81526-5. [DOI] [PubMed] [Google Scholar]
- 20.Rubins JB, Charboneau D, Fasching C, Berry AM, Paton JC, et al. Distinct roles for pneumolysin’s cytotoxic and complement activities in the pathogenesis of pneumococcal pneumonia. Am J Respir Crit Care Med. 1996;153:1339–1346. doi: 10.1164/ajrccm.153.4.8616564. [DOI] [PubMed] [Google Scholar]
- 21.Dockrell DH, Marriott HM, Prince LR, Ridger VC, Ince PG, et al. Alveolar macrophage apoptosis contributes to pneumococcal clearance in a resolving model of pulmonary infection. J Immunol. 2003;171:5380–5388. doi: 10.4049/jimmunol.171.10.5380. [DOI] [PubMed] [Google Scholar]
- 22.Knapp S, Leemans JC, Florquin S, Branger J, Maris NA, et al. Alveolar macrophages have a protective antiinflammatory role during murine pneumococcal pneumonia. Am J Respir Crit Care Med. 2003;167:171–179. doi: 10.1164/rccm.200207-698OC. [DOI] [PubMed] [Google Scholar]
- 23.Maus UA, Srivastava M, Paton JC, Mack M, Everhart MB, et al. Pneumolysin-induced lung injury is independent of leukocyte trafficking into the alveolar space. J Immunol. 2004;173:1307–1312. doi: 10.4049/jimmunol.173.2.1307. [DOI] [PubMed] [Google Scholar]
- 24.Winter C, Taut K, Srivastava M, Langer F, Mack M, et al. Lung-specific overexpression of CC chemokine ligand (CCL) 2 enhances the host defense to Streptococcus pneumoniae infection in mice: role of the CCL2-CCR2 axis. J Immunol. 2007;178:5828–5838. doi: 10.4049/jimmunol.178.9.5828. [DOI] [PubMed] [Google Scholar]
- 25.Taut K, Winter C, Briles DE, Paton JC, Christman JW, et al. Macrophage Turnover Kinetics in the Lungs of Mice Infected with Streptococcus pneumoniae. Am J Respir Cell Mol Biol. 2008;38:105–113. doi: 10.1165/rcmb.2007-0132OC. [DOI] [PubMed] [Google Scholar]
- 26.Hahn I, Klaus A, Janze AK, Steinwede K, Ding N, et al. Cathepsin G and neutrophil elastase play critical and nonredundant roles in lung-protective immunity against Streptococcus pneumoniae in mice. Infect Immun. 2011;79:4893–4901. doi: 10.1128/IAI.05593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poe SL, Arora M, Oriss TB, Yarlagadda M, Isse K, et al. STAT1-regulated lung MDSC-like cells produce IL-10 and efferocytose apoptotic neutrophils with relevance in resolution of bacterial pneumonia. Mucosal Immunol. 2013;6:189–199. doi: 10.1038/mi.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshane J, Zmijewski JW, Luther R, Gaggar A, Deshane R, et al. Free radical-producing myeloid-derived regulatory cells: potent activators and suppressors of lung inflammation and airway hyperresponsiveness. Mucosal Immunol. 2011;4:503–518. doi: 10.1038/mi.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniels CC, Briles TC, Mirza S, Hakansson AP, Briles DE. Capsule does not block antibody binding to PspA, a surface virulence protein of Streptococcus pneumoniae. Microb Pathog. 2006;40:228–233. doi: 10.1016/j.micpath.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, et al. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis. 2003;188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 31.Werner JL, Gessner MA, Lilly LM, Nelson MP, Metz AE, et al. Neutrophils produce interleukin 17A (IL-17A) in a dectin-1- and IL-23-dependent manner during invasive fungal infection. Infect Immun. 2011;79:3966–3977. doi: 10.1128/IAI.05493-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mundy-Bosse BL, Lesinski GB, Jaime-Ramirez AC, Benninger K, Khan M, et al. Myeloid-derived suppressor cell inhibition of the IFN response in tumor-bearing mice. Cancer Res. 2011;71:5101–5110. doi: 10.1158/0008-5472.CAN-10-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chicharro JL, Serrano V, Urena R, Gutierrez AM, Carvajal A, et al. Trace elements and electrolytes in human resting mixed saliva after exercise. Br J Sports Med. 1999;33:204–207. doi: 10.1136/bjsm.33.3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briles DE, Crain MJ, Gray BM, Forman C, Yother J. A strong association between capsular type and mouse virulence among human isolates of Streptococcus pneumoniae. Infect Immun. 1992;60:111–116. doi: 10.1128/iai.60.1.111-116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubins JB, Charboneau D, Fasching C, Berry AM, Paton JC, et al. Distinct roles for pneumolysin’s cytotoxic and complement activities in pathogenesis of pneumococcal pneumonia. Am J Respir Crit Care Med. 1996;153:1339–1346. doi: 10.1164/ajrccm.153.4.8616564. [DOI] [PubMed] [Google Scholar]
- 36.Pruijt JF, Verzaal P, van Os R, de Kruijf EJ, van Schie ML, et al. Neutrophils are indispensable for hematopoietic stem cell mobilization induced by interleukin-8 in mice. Proc Natl Acad Sci U S A. 2002;99:6228–6233. doi: 10.1073/pnas.092112999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillespie SH. Aspects of pneumococcal infection including bacterial virulence, host response and vaccination. J Med Microbiol. 1989;28:237–248. doi: 10.1099/00222615-28-4-237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.