Abstract
The way cells are organized within a tissue dictates how they sense and respond to extracellular signals, as cues are received and interpreted based on expression and organization of receptors, downstream signaling proteins, and transcription factors. Part of this microenvironmental context is the result of forces acting on the cell, including forces from other cells or from the cellular substrate or basement membrane. However, measuring forces exerted on and by cells is difficult, particularly in an in vivo context, and interpreting how forces affect downstream cellular processes poses an even greater challenge. Here, we present a simple method for monitoring and analyzing forces generated from cell collectives. We demonstrate the ability to generate traction force data from human embryonic stem cells grown in large organized epithelial sheets to determine the magnitude and organization of cell–ECM and cell–cell forces within a self-renewing colony. We show that this method can be used to measure forces in a dynamic hESC system and demonstrate the ability to map intracolony protein localization to force organization.
Keywords: Traction force, Human embryonic stem cells, Self-organization, Mechanotransduction, Monolayer stress microscopy, Epiblast
1. Introduction
Coordinated cell movements are required in physiological processes important to development, growth, and disease states, including embryogenesis, adult stem cell differentiation, wound repair, and metastasis. The ability of a cell to determine when and how to move in the context of a tissue is dependent on its ability to sense and respond to extracellular cues, which include the forces supplied by neighboring cells and the rigidity and composition of the extracellular matrix. An extrinsic force can cause dramatic changes in cell state, for example, shear stress caused by blood flow can affect endothelial cell proliferation, apoptosis, migration, and gene expression [1]. But merely altering the properties of the substrate to which cells are adhered also causes significant changes to the cell, as cells apply forces to their substrates and respond to the resistance that they sense [2]. The resulting changes include reorganization of the cytoskeleton and cellular adhesions, including those at cell–cell and cell–ECM boundaries. These changes can feedback to stabilize a morphological transition, as modulation of ECM properties that affect cell–ECM forces directly alters cell–cell tension [3]. As adhesions form and break down, their associated proteins are stabilized or degraded, which can affect downstream signaling pathways and lead to a transcriptional response. Though we know much about how cells acting alone sense, process, and transmit mechanical signals, less is understood about the forces that regulate cells working as collectives in the physiological context of a tissue.
Cell reorganization in the context of a tissue is a primary component of embryonic development, as cells must be precisely localized within the complex embryo as their fate is specified. Work in model organisms has defined specific examples of forces driving the cellular rearrangements of embryogenesis, for example, mechanical stimuli are required for epidermal elongation in Caenorhabditis elegans [4], mechanical stretching of Drosophila wing discs promotes cell proliferation during development [5], and mechanotransduction through cell–cell adhesion is a driver of Xenopus gastrulation [6]. In all these cases, actomyosin organization and rearrangement play a role, particularly in terms of coordinating the cell–cell versus cell–ECM adhesion forces that are coupled to actomyosin networks [7,8]. Actomyosin contractility also plays an instructive role in gastrulation [9–11], when cells within the epiblastic epithelium undergo an EMT to migrate into the primitive streak and form the endoderm and mesoderm germ layers [12,13]. While much is known about the soluble signals that emanate proximal and distal to the streak to drive this process, the extent to which the forces elaborated during maturation of the epiblastic epithelium contribute to gastrulation is unknown. Thus, the way cells are spatiotemporally organized in terms of cell–cell and cell–ECM protein localization and the resulting force maturation in the developing embryo is a relatively unexplored but potentially crucially important component of developmental signaling.
What we do know about the forces present in the developing embryo is largely drawn from experiments in model organisms that use techniques such as compression tests on whole embryos or explanted embryonic tissue to assess deformability [14], laser ablation to locally cut a tissue and measure the resulting tension release [15,16], or FRET-based approaches to track the activation of mechanosignaling proteins such as Rac and Rho during development [17]. A recent novel method to study endogenous forces in living and developing tissues using fluorescently labeled microdroplets has enabled measurements of the cell-generated stresses in the dental mesenchyme of live mice, and is promising for future developmental studies [18]. However, such studies are difficult to perform in the presence of external manipulations in a highly controlled environment.
Recent in vitro approaches permit analysis of how forces are organized in cell collectives, which provides relevant context to the cells in terms of cell–cell and cell–matrix forces. It is becoming increasingly evident that cells in collectives behave differently than single cells in terms of junction assembly and mechanotransduction events [19], so in vitro methods for careful study of specific collective cell properties will allow for tractable systems in which to better understand these emergent phenomena. In one approach known as traction force microscopy, adherent cells are grown on hydrogels containing fluorescent microbeads, whose displacement indicates the force applied by the cell to its substrate. Such work has provided insights into collective cell migration [20,21], heterogenous distribution of physical forces in colonies [22], and epithelial tissue dynamics [23]. FRET-based approaches have also been applied to epithelial collectives to assess transduction of mechanical forces [24] and intercellular tension distribution [25]. Applying these approaches to other cell types of epithelial origin, particularly those that are relevant to developmental processes, has the potential to uncover previously unknown requirements for the elaboration of forces in dictating cell fate and driving differentiation.
Human embryonic stem cells (hESCs) are isolated from the inner cell mass of a blastocyst, and are thought to be the in vitro equivalent of the pluripotent epiblast [26]. We previously showed that hESCs cultured on mechanically deformable polyacrylamide substrates of an appropriate stiffness can recapitulate the structural and morphological organization of an in vivo epiblast, including a columnar epithelium with basally displaced nuclei and well-developed E-cadherin-based adherens junctions with cortical F-actin fibers [27]. Because hESCs cultured in this manner represent an epithelial sheet formed in a context that is relevant to embryonic development, understanding the mechanical properties exerted by these cells as they organize into colonies reminiscent of epiblastic organization could provide insight into the contribution of mechanical forces to embryo formation.
To build on our previously demonstrated ability to establish viable hESC colonies on soft hydrogels [27], we sought to leverage the recently developed techniques described above [20,22,23] to quantify the forces exerted by hESCs grown in colonies as they organized into epithelial sheets. To this end, we recently developed methods for hESC patterning coupled with measurements designed to analyze development and maturation of cell–ECM and cell–cell forces. We describe a method for preparation of poly-acrylamide gels that can be used for force measurements and are robustly compatible with adherence and long-term culture of hESCs. We demonstrate how to plate hESCs in pre-gastrulation-stage embryo-sized colonies of defined geometries onto traction force hydrogels of defined compliance in a way that allows the cells to unrestrictedly adhere and organize, with the ability to model the embryo in terms of size, shape, and composition. With this system, we show how to apply traction force microscopy to measure cell–ECM forces in real time as the colonies mature, and also assess how cell–cell forces develop with a technique called monolayer stress microscopy [20], allowing us to generate a comprehensive picture of how forces develop in a developmentally relevant epithelial model system. Many differentiation protocols that begin with human pluripotent stem cells use monolayer culture as a starting point for differentiation [28–32], and it has been shown that both extrinsic and intrinsic cell–matrix forces affect differentiation in other contexts [33,34], so the ability to quantify and manipulate forces could enhance our understanding of how mechanics is involved in many aspects of development.
This method enables us to observe and measure the cell mechanics that underlie embryonic processes. Embryonic stem cells have distinct mechanical properties, being softer and more sensitive to stress than their differentiated counterparts [35] and presumably than other model epithelial lines such as MDCK or MCF10A cells, systems in which most work has been done to establish principles for collective cell cohesion and organization. We thus describe a tool for understanding the relationship between tissue-scale self-organization and force in a simplified system that bridges the gap between pure tissue culture studies and those performed in embryos. Recent work has indicated that self-organized patterning of hESCs occurs when colony geometry is controlled [36], a result that was attributed to paracrine gradients but may also include a contribution from adhesion forces in these developmental analogs. With our system, we can examine not only how protein and signaling gradients are set up within epiblastic epithelium-like colonies, but also track the cell-intrinsic forces involved in differentiation and developmental processes. Our technique also allows for retroactive superimposition of relevant biomarker expression with traction force maps, enabling the establishment of links between endogenous forces and embryonic epithelial organization in the context of tissue geometries that are widely applicable to live-cell temporal studies of development, differentiation, and migration.
2. Methods
The methods we present here build on our previously published method for preparing and culturing hESCs on ligand modified polyacrylamide gels [27], which was used to study the effects of substrate compliance on hESC differentiation at a population level in which hESCs were plated as single cells or small randomly plated colonies. However, as described above, the organization of cells within tissues underlies collective cell behaviors such as germ layer differentiation during development. This modified method provides improvements in gel casting and the chemistry of ligand conjugation but, more significantly, allows for control of colony size, shape, and position and integrates with current matrix traction force methods to quantify cell matrix and intercellular forces. Importantly, position control provides a means of non-destructively acquiring these forces, allowing end point analysis for correlation of molecular markers and underlying forces. This is accomplished by a set of unique approaches involving custom designed 3D printed culture wells and microscope stage mounts that simplify and streamline the preparation, plating and culture process.
2.1. Casting PA gels embedded with fluorescent beads
Polyacrylamide (PA), to a good approximation, is an elastic material. The elastic modulus, E, in pascals (Pa) is determined by the concentrations of acrylamide and the crosslinker bisacrylamide according to Table 1 and spans the range from soft tissue-like (E < 1 kPa) to stiff dense fibrous tissue ECMs (E = 10–30 kPa) [37]. While the stiffness chosen will generally reflect that which is physiologically or experimentally relevant, at present there are some limitations to the application of this method to the entire range of possible stiffnesses. Most epithelial cells, including hESCs, will not generally develop sufficient contractile forces to distort bead positions for E > 10 kPa. We have also observed that there can be a z component to the force over the colony. This is small relative to xy displacements for 1 kPa > E > 10 kPa, so we make the approximation over this range that it is negligible. For softer matrices (E < 1 kPa), however, it may become more significant, requiring tracking of bead positions in three dimensions, which complicates the mathematical analysis. In our experience the elastic modulus that this applies to is somewhat cell type dependent. For example hESCs, in contrast to the mammary epithelial line MCF10A, relocalize much of their actomyosin contractile apparatus to the apical rather than basal domain, resulting in strong and concerted intercellular contractility. In large mm sized colonies where the hESC colony appears to be anchored primarily at the edges, the result of this coordinated contraction is a significant downward bending of the substrate (tens of microns) for E < 1 kPa. This is less apparent for MCF10A cells. This method does not treat these more complicated cases.
Table 1.
Components of hydrogel solution for PA gels of defined elastic modulus. All rows correspond to 1 ml total volume.
| Elastic Modulus (Pa) |
Final % acrylamide |
Final % Bis- acrylamide |
ddH2O (µl) |
40% acrylamide (µl) |
2% Bis- acrylamide (µl) |
10× PBS (µl) |
1% TEMED (µl) |
1% PPS (µl) |
1/4 dilution of microspheres (µl) |
|---|---|---|---|---|---|---|---|---|---|
| 400 | 3 | 0.05 | 590 | 75 | 25 | 100 | 75 | 75 | 60 |
| 1050 | 3 | 0.1 | 565 | 75 | 50 | 100 | 75 | 75 | 60 |
| 2700 | 7.5 | 0.035 | 485 | 187.5 | 17.5 | 100 | 75 | 75 | 60 |
| 4000 | 7.5 | 0.05 | 477.5 | 187.5 | 25 | 100 | 75 | 75 | 60 |
| 6000 | 7.5 | 0.07 | 467.5 | 187.5 | 35 | 100 | 75 | 75 | 60 |
| 13,800 | 7.5 | 0.15 | 427.5 | 187.5 | 75 | 100 | 75 | 75 | 60 |
| 60,000 | 10 | 0.5 | 190 | 250 | 250 | 100 | 75 | 75 | 60 |
PBS = phosphate-buffered saline, PPS = potassium persulfate. Microspheres are diluted in water.
In this application of PA gels as adhesive surfaces for cell culture the gel is cast between two coverslips, one modified to covalently couple to the polymerizing PA gel that provides a rigid bottom support and the other (top) rendered non-stick that is peeled away following polymerization. Use of small volumes of polymerizing solution keeps the gel thin (~100 µm) and surface tension prevents both spillage out of the side and ideally maintains the gel at a uniform thickness. For MSM applications, fiduciary fluorescent beads are ideally exactly at the surface of the gel-cell interface (the mathematical analysis of cell–matrix traction forces assumes that all forces are exerted in a flat plane parallel to the plane). To achieve this, gels are cast between top and bottom coverslips separated by a thin spacer and the assembly is clamped together at the top of a screw cap centrifuge tube. This assembly can be centrifuged to sediment beads much more rapidly and uniformly to the interface, before the onset of significant polymerization.
Gels are prepared on #1 18 mm round coverslips for both top and bottom. For cleaning and glutaraldehyde modification of the bottom coverslip we refer to our earlier publication [27]. Top coverslips are cleaned by washing for several hours in a 10% dilution of household bleach followed by water washes, 100% ethanol, further water washes then air drying. Top cover-slips are then rendered non-stick by coating with 1 or 2 microliters of Rain-X (available from any automotive supply store) spread over the surface with a pipet tip. The dried Rain-X is spread to a uniform layer by buffing with a lint-free wipe (Kim-wipes). It is helpful to avoid confusion as to which side has been treated to label the side opposite with the word TOP. Top and bottom coverslips can be reused: Top coverslips by repeating the procedure described above. Treatment of bottom coverslips for 1+ days in 10% bleach before repeating the protocol described in [27] is sufficient to remove residual acrylamide. Modified bottom coverslips can be autoclaved to sterilize.
Spacers with an outer diameter of 18 mm and an inner diameter of 14 mm are made with corresponding round hammer-driven punches for soft materials that may be obtained from many tool suppliers (ex McMaster-Carr). Spacers may be made from many types of plastic sheet but for MSM we prefer thin (127 µm or 0.00500 thick; McMaster-Carr, Cat.#855815K102) polycarbonate film. Following punching these are gently sanded with very fine (>600 grit) sandpaper to remove rough edges that may be created during punching. Gently wash with water before use to remove loose sanded material. These may be reused indefinitely but any that become kinked either during preparation or subsequent use should be discarded.
Refer to [27] for a list of materials in Table 1. Many sizes and colors of fluorescent beads may be used. The chosen bead size should be significantly (at least 10-fold) smaller than one cell diameter so several beads underlie each cell, but large enough to provide accurate positioning. In general, smaller beads can be used as the power of the microscope objective increases. In practice we find that 1 µm diameter beads (FluoSpheres, Molecular Probes, Cat# F-8821) are the minimum size if a 4× objective is used because of the possibility of positioning errors, while for a 10× objective, much smaller sizes could be used. These are resuspended and diluted 1:4 in sterile PBS as the working stock used in Table 1. Beads are sonicated for 15 min in an ultrasonic cleaner/water bath (ex Branson Model 2510), then left undisturbed for 30 min to allow larger bead aggregates to sediment. Gel solutions are assembled according to Table 1 excepting potassium persulfate. The bead suspension is added by pipetting the appropriate aliquot from just below the surface of the suspension after standing. The gel solution is briefly sonicated again (5 min) to further break up residual bead clumps. Potassium persulfate (PPS) is dissolved in sterile water and degassed together with the gel solution for at least 30 min under vacuum (house vacuum of 20 ± 5 mmHg is sufficient). Freshly made PPS and degassing are required for efficient polymerization of these thin gels. Following degassing, gel solution and PPS are placed for 5 min on ice to cool. Cooling slows polymerization and allows more time for assembling the gel sandwich.
-
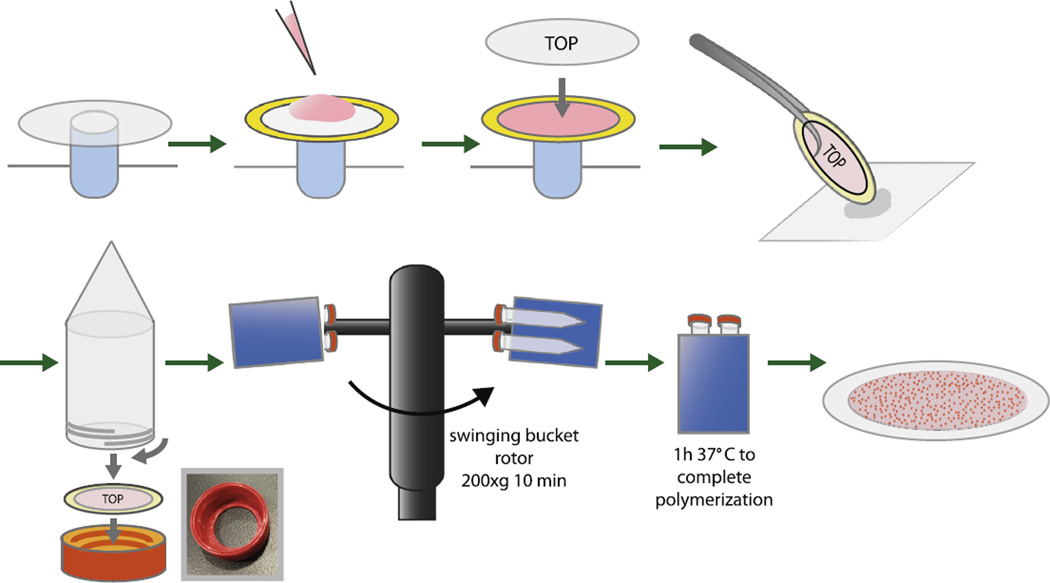
To cast gels the bottom coverslip is placed on a small post (we use a 2 ml capless centrifuge tube wedged into a standard Eppendorf rack and wrapped at the top with Parafilm to render it less slippery). The spacer is placed on the coverslip and 70 µl of the cooled gel solution is added to the center immediately after the appropriate amount of 1% PPS has been added (Fig. 1). Working quickly, the top coverslip (non-stick side down) is laid on the cooled gel solution. The sandwich is picked up at one edge with blunt forceps and the opposite edge is dabbed onto a Kimwipe to drain excess volume, ensuring that the volume of gel solution is relatively constant. The assemblage is then clamped into the centrifuge tube as described below (Fig. 1).
Critical: these gels should be made quickly and in small batches to ensure that they are centrifuged prior to gel solidification.
To make the coverslip-spacer-gel sandwich clamp we used a 3D printer (UPrint Plus, 254 µm layer resolution, Stratasys) to print an ABS modified version of the cap on a 15 ml conical bottom centrifuge tube (Corning Cat# 430052) with a thread compatible with the tube (Fig. 1, photograph inset). The Autodesk Inventor file as well as the corresponding STL file for this and all other printed parts referred to in this article is available on request. As described below, this modified cap is multifunctional, serving both as clamp and as a component of the cell culture well/microscope stage.
-
Working quickly, the gel sandwich is coaxed into the bottom of the inverted cap with top coverslip up. Once seated in the cap base the 15 ml conical tube is inverted and screwed in until it gently but firmly squeezes the gel sandwich assembly. The tube is turned upright and centrifuged for 10 min at room temperature at 200g using a swinging bucket rotor (Fig. 1). This sediments the beads to the interface of gel and top coverslip. After centrifugation the tube assembly is placed upright in a stand and transferred to a humidified environment at 37 °C for 1 h to polymerize.
Critical: always use a swinging bucket rotor so tubes are not spun at an angle, to ensure even bead distribution and keep tubes perfectly upright after centrifugation while gels are solidifying.
Following polymerization the clamp assembly is unscrewed and the sandwich is carefully removed. The top coverslip is removed by prying using the edge of a razor blade. The spacer is removed and the bottom coverslip with attached gel is dipped briefly in ethanol and then placed gel side up in a petri dish containing sterile 0.9% (w/v) NaCl. Placing the gel sandwich in a petri dish of cold water for 5–10 min is helpful for removal of the top coverslip. Gels can be stored for long periods at 4 °C in 0.9% (w/v) NaCl but adding NaN3 to 2 mM is advisable to prevent growth of microorganisms.
Fig. 1.
Diagram of casting method for preparing traction force gels with planar fluorescent bead layer. See text for details. Inset photograph shows 3D–printed cassette used to house polyacrylamide traction force gels.
2.2. Functionalizing PA gels with ECM ligands
The chemistry of functionalizing the surface of PA gels with ECM ligands is similar to the method we published previously [27]. Briefly, sites of unsaturation that remain on the surface of the gel provide points of covalent attachment for a thin protein reactive layer polymerized on the surface. In our previous method this was a free radical UV photoinitiated copolymer of bisacrylamide and a linear heterobifunctional compound with acrylic acid and N-hydroxysuccinimide ester functionalities at opposite ends (N6) whose synthesis we described [27]. This chemistry allowed sufficient levels of ECM ligands to be immobilized on the gel surface such that cell adhesion was reasonably robust and cell spreading was dependent on PA elastic modulus.
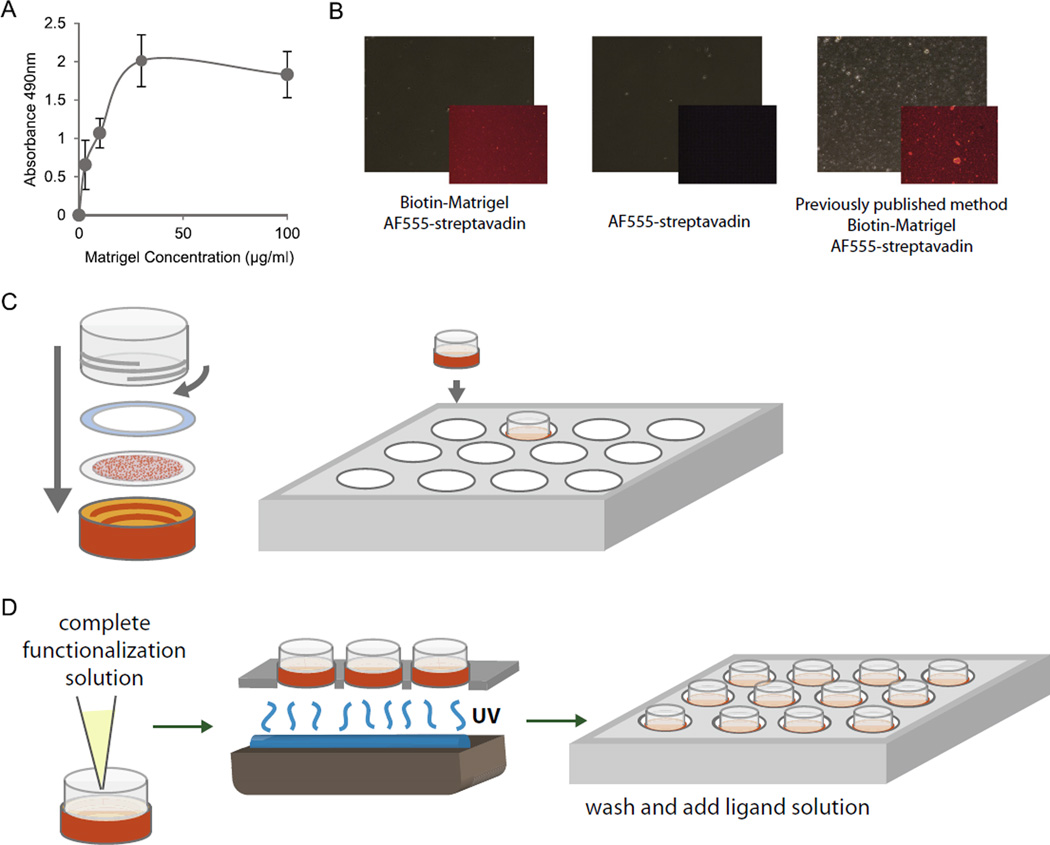
Since our previous publication, multifunctional methacrylates were shown to serve as robust adhesive surfaces for the culture of hESCs and can physically absorb proteins, providing additional mechanisms for ECM-ligand attachment [38]. While these materials, used as solid polymers, were limited in their range of potential compliances, their chemistry was compatible with our copolymerizing system. A combination of the addition of dilute concentrations of di(trimethylolpropane) tetraacrylate and a 10-fold reduction in N6 concentrations was found to maintain robust ECM-ligand surface attachment (Fig. 2A), and promote the adherence and survival of hESCs with similar dependencies of cell spreading as a function of elastic modulus. Importantly though, this new formulation was less prone to the formation of macroscopic copolymeric deposits than our previous work [27], producing a more even and uniform distribution of ligand (Fig. 2B). In our hands (unpublished observations), this new formulation has been successfully applied to many ligands (Fibronectin, Collagen I, Laminin 1,1,1, and Matrigel) and many different cell types, including neural stem cells, endodermal progenitor cells, mouse ESCs, mammary epithelial cell lines, neural tumor cell lines, mammary tumor cells, primary mouse hepatocytes, primary fibroblasts, and primary mammary epithelial cells.
Fig. 2.
Functionalization of traction force gels compatible with multi-day culture of hESCs. (A) Relative PA gel surface bound biotin labeled Matrigel proteins as a function of total solution phase Matrigel concentration for the modified chemistry described herein. Biotin-labeled proteins were indirectly detected by streptavidin-horseradish peroxidase complex in an ELISA format using O-phenylenediamine-H2O2 as substrate with endpoint product measured by absorption at 490 nm. (B) Phase images of gel surfaces (large panels) and corresponding distribution of PA gel surface-bound biotin labeled Matrigel proteins detected with Alexa555 labeled streptavidin (insets). Leftmost two panels show specific (left panel; with Matrigel) and non-specific (middle panel; without Matrigel) streptavidin binding for the modified chemistry described herein compared to specific binding for the earlier chemistry described in [27] (right panel). Note the discrete co-polymeric deposits of N6 and bisacrylamide in this particular example of the latter compared to the more consistent uniform distribution obtained with the modified chemistry. (C) Traction force gels are re-inserted into cassettes and put into multi-well plates for functionalization. (D) Functionalization solution containing N6, Irgacure, and tetramethacrylate is added to gels before undergoing UV treatment and subsequent washes prior to plating cells.
We present here this modified method, which preserves the cellular response to bulk gel compliance and provides significant advantages, including low cost, scalability and reproducibility. In addition to the modification in the chemistry we also present changes in the logistics of surface functionalization that take advantage of the modified 3D printed caps described above.
Bottom coverslips with 14 mm concentric PA gels are removed from storage and placed gel side up on a small non-slip post (See Protocol 2.1 Step 4).
Using hammer driven round punches, compressible, biocompatible gaskets of 18 mm outer diameter and 14 mm inner diameter are prepared from soft thin silicone film (McMaster-Carr, Cat#86435K45 0.0200 thick, Durometer 20A or 35A). These are washed in water and sterilized by autoclaving and are placed on the concentric outer ring of bare glass that remains after disassembly of the gel sandwich described above (Fig. 2C).
-
Bottom coverslip and gasket are placed in the 3D printed cap gel side up. A 15 ml Corning conical bottom centrifuge tube can be modified to form the walls of the functionalization/culture well. The tube is cut below the 14 ml mark with a small saw, filed as level as possible to the 14 ml mark and gently sanded. This is screwed into the base, compressing the gasket against the glass edge, forming a liquid tight seal and a gel-bottomed culture well. Alternatively, we designed and 3D printed an ABS modification of this part that in addition contains internal guides that more accurately center and solidly hold the plating guides described below (Protocol 2.3). Microscopic pores between extruded ABS layers in the inner wall of this part must be sealed post printing either by applying acetone to the surface or by exposing to acetone vapor to ensure that it is liquid tight. Having the gel fixed in the base of the vessel facilitates ease of subsequent handling as well as providing the means of imaging unstressed bead positions accurately prior to plating cells (Protocol 2.4).
Critical: maintaining sterility of gel chambers requires washing caps and gaskets in 10% bleach overnight, or autoclaving these materials before each use. Once the gel is reassembled, all steps should be performed in a laminar flow hood suited for tissue culture.
The gel assembly is placed into one well of a modified standard 12 well tissue culture plate and filled with 1 ml of 0.9% (w/v) NaCl to prevent gel drying while solutions are prepared for functionalization. The bottoms of the wells of this plate are drilled out using an 11/1600 spade bit. This leaves an edge of several mm at the base on which the gel well assembly rests with a wide enough opening to permit complete UV illumination from below (Step 6).
Using Table 2 as a guide and calculating 0.5 ml solution per gel, sterile water, 0.5 M HEPES pH 6, ethanol and 0.2% (w/v) bisacrylamide are combined and degassed under vacuum for 20 min. 10 min before degassing is finished, 0.9% NaCl is aspirated and gels are washed first with 1 ml of sterile water and then with 1 ml of 70% (v/v) ethanol for 5 min. Gels should not be exposed to ethanol for long periods as swelling and/or dehydration may irreversibly deform the surface.
After degassing the mixture in Step 5, N6 is dissolved to 0.6 mg/ml in ethanol and added, along with the appropriate volume of 3% (w/v) Irgacure 2959 (Ciba) and 0.2% (v/v) di (trimethylolpropane) tetraacrylate (Sigma Cat#408360), both prepared as stocks in ethanol. Ethanol is aspirated from the gels and 0.5 ml of functionalizing solution is added. Gels are then exposed to UV light for 10 min to copolymerize N6, bisacrylamide and di(trimethylolpropane) tetraacrylate (Fig. 2D). We use a Spectroline model EN-180 handheld UV source with long wavelength (365 nm) peak emission, invert the source and place the plate-gel assembly on top so the gels are illuminated from below. We refer the reader to our earlier article concerning synthesis, stability, and storage and handling of N6 and the substitution of the commercially available compound acrylic acid N-hydroxysuccinimide ester (Sigma Cat#A8060) for N6 [27].
After functionalizing, the plate gel assembly is placed on ice, the solution is aspirated and gels are washed in succession 5 min per wash 1 ml per gel with gentle agitation two times with cold, sterile 25 mM HEPES pH 6 and two times with cold, sterile 0.9% (w/v) NaCl.
After aspiration of the last wash gels are incubated with the ECM ligand of choice. Several buffers are possible for this step but should be amine free and alkaline in pH. A good general choice for a number of ECM ligands including fibronectin, laminin1,1,1, collagen I, collagen II and Matrigel (BD Biosciences Cat#354234) is 0.1 M HEPES pH 8, 0.1 M NaCl. Gels are incubated in 0.5 ml/gel cold sterile buffer with the appropriate concentration of ECM ligand overnight at 4 °C. Concentrations of ECM ligands vary from 10 to 200 µg/ml. We refer again to our earlier article for a discussion of the variation of immobilized ligand density, and parameters of cell adherence including spreading and generation of single cell traction forces as a function of the solution phase ligand concentration [27]. The modification in chemistry presented here does not significantly change these recommendations. For hESCs we use a mix of 200 µg/ml Matrigel and 25 µg/ml rat tail collagen I.
-
After immobilizing, ECM ligand gels are incubated for 30 min with 1 ml/gel sterile 50 mM HEPES, 0.1 M glycine pH 8 at room temperature then washed four times over 1 h with 1 ml/gel sterile 1x PBS.
TIP: thorough washing of gels is required to quench remaining unreacted N6 and other functionalization components.
Finally, gels are placed in a cell culture incubator for 1–2 days in 1 ml/gel DMEM:F12 containing 250 µg/ml Fungizone, 100u/ml Penicillin, 100 u/ml Streptomycin and 50 µg/ml Gentamycin. These washes remove undesirable toxic agents/byproducts and the incubation in media ensures that the gels are sterile prior to use.
Table 2.
Components of functionalization solution for PA gels. All volumes are listed as microliter amounts.
| ddH2O | 0.5 M HEPES NaOH pH = 6.0 |
100% ethanol |
0.2% Bis- acrylamide |
0.2% tetramethacrylate in ethanol |
0.03% N6 in 50% ethanol |
3% Irgacure in ethanol |
Total volume |
|---|---|---|---|---|---|---|---|
| 433.3 | 100 | 65 | 50 | 10 | 333.3 | 8.33 | 1000 |
2.3. Plating epithelial cells as collectives
Modeling the forces that underlie cell movements in coherent epithelial monolayers, as may occur during wound healing or in the large scale movements associated with embryonic development, necessitates methods for preparing cellular monolayers. The distribution of these forces is sensitive to the overall size and shape of the monolayer and can influence the developmental fate of cells [39], so some control of these parameters is also desirable. Many disaggregated epithelial cells, including hESCs, will regenerate such monolayers rapidly if plated at high densities on an adherent substrate, but the final monolayer geometry in standard tissue culture is unpredictable. Micropatterning approaches that spatially control adherent and non-adherent regions allow precise control of monolayer shape and size but may be difficult to implement and do not permit study of the forces underlying outward epithelial monolayer migration. A simple solution to these demands is to plate cells at high density using a mask of defined geometry that restricts access of plated cells to a small region of the adherent surface [40].
We present here an implementation of this approach using 3D printed plating guides that act like a funnel, guiding cells to a defined location on an adherent PA surface. Guides are removed after cell adherence, maintaining the overall size and geometry of the funnel outlet as coherent epithelial monolayers reform over 12–24 h. With these guides we can also control the final location of the monolayer on the gel, which permits acquisition of unstressed bead images prior to rather than after plating cells. This allows forces to be recovered non-destructively, permitting use of these monolayers for subsequent molecular analysis. This provides a means of mapping cell and molecular correlates to the underlying forces without having to design and employ cells expressing often difficult-to-generate real time molecular reporters.
-
Fig. 3A shows an image of a 3D printed plating guide within the cap cassette assembly. The guide was constructed using an Alaris Objet24 printer (Stratasys) out of Vero White Plus employing PolyJet printing technology with z layer resolution of 28 µm and xy resolution of 42 µm. Alternative printing materials may be used, though this finer resolution is required for some of the smaller features of the guide. The guide is made as two parts to minimize support material and is screwed together by miniature machine screws.
TIP: removing all traces of support material from guides by scraping and washing is crucial to survival of plated cells, as support material was found to be cytotoxic.
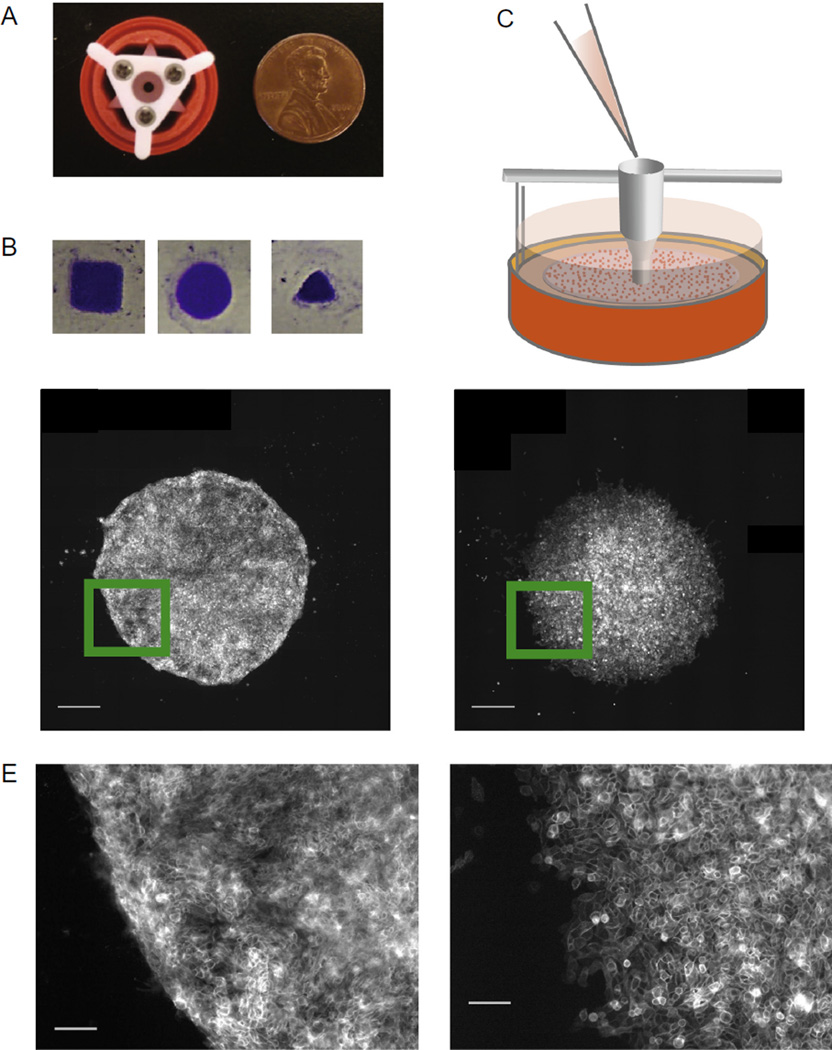
The plating guide is designed to center the epithelial monolayer with respect to the walls of a cut 15 ml centrifuge tube (Step 3 Protocol 2.2) but the best centering and stability are obtained with the ABS printed replacement. Guides may be printed with many sizes and shape of outlet and the resulting monolayer adopts these shapes and sizes for at least 1–2 days, as we demonstrate with colonies of varied geometry (Fig. 3B). Those we use throughout the article are circles with diameters of 1.8 mm (Fig. 3A).
Unstressed bead images may be collected either before or after cell plating. The setup required for collection of unstressed bead images before plating is described in Method 2.4.
-
Before plating cells, plating guides are sterilized by immersion in 70% ethanol and dried under laminar flow. The guides are placed funnel side down in wells and dropped from a low height to gently rest against the gel (Fig. 3C).
TIP: to form a continuous column of liquid within the channel of the guide and avoid trapping air, it is helpful to turn the guide upside down and pipet media into the guide channel base before placing the guide in the well and adding the cell suspension to the top of the liquid column.
The guide should be placed upright on the gel to minimize tilting, but it is important to note that for softer gels (E < 400 Pa) the weight of the guide may permanently deform the gel in a way that partially compromises acquisition of unstressed bead images prior to plating. These deformations are usually confined to where the outside of the guide rests against the gel and show up as a characteristic arc shape that is on the order of 1 mm from the colony edge. It is thus distal from cells and can be ignored.
For a 1.8 mm diameter circle, 5 µl of a suspension of 4 × 106 cells/ml (20,000 cells) is sufficient to form a coherent monolayer in 12–24 h. Before resuspending at high density, we pass a more dilute cell suspension through a porous filter (70 µm) to remove cell aggregates, resulting in more uniform plating densities. The cell suspension is best added using sterile tips designed for gel loading as they have a long narrow tip that can fit into the top of the guide channel without bumping the sides. The tip is inserted at the top of the guide below the level of the liquid column and cells are expelled with the pipettor, being careful not to expel any air. The gel/guide assembly is then returned to the incubator and cells are allowed to plate and adhere for 1–2 h, after which the guide is lifted vertically out of the well. Sideways motions of the guide should be minimized to avoid damaging the gel and to avoid liquid flows which may disturb the sometimes loosely adhered cells. The resulting colonies maintain their circular shape for several days on soft gels (Fig. 3D), though differences can be seen in the edge morphology of colonies made up of cells of different epithelial origin (Fig. 3E).
We refer to our earlier publication for the maintenance and culture of hESCs [27], which discusses the requirement for the Rho kinase inhibitor Y27632 for adhesion of hESCs especially to soft gels. Here, when we plate hESCs as described above, they reform monolayers within 12–16 h in the presence of 10 µM Y27632 even on the softest gels, after which time Y27632 can be removed stepwise without triggering extensive cell death. Half of the media is removed and replaced with an equal volume of fresh pre-warmed primary mouse embryo fibroblast conditioned media, and cells are allowed to adapt for a couple of hours before this is repeated 3 times throughout one day. Once the monolayer is formed, cells are ready for acquisition of stressed bead images either as a single time point or as part of a time sequence and may incorporate experimental treatments including drug treatment or induction of differentiation.
Fig. 3.
Plating unrestricted cell colonies of arbitrary geometry. (A) Photograph of 3D–printed plating guide sitting within gel cassette used to create adherent colonies of arbitrary geometry on polyacrylamide gels. (B) Images of crystal-violet stained hESC colonies created using different shaped plating guides. (C) Diagram of funnel-shaped plating guide resting on traction force gel for cell seeding. (D) Image of mature hESC colony growing in self-renewal media (left) and MCF10A colony (right), both on a 1000 Pa gel. Scale bars = 500 µm. (E) Magnified image of colonies shown in (D) to show colony edge morphology. Scale bars = 100 µm.
2.4. Acquisition of bead images
Stressed and unstressed bead images are acquired using an inverted widefield epifluorescence microscope equipped with a computer controlled motorized microscope stage with xyz linear encoders for accurate positioning, equipped with sample temperature, humidity and gas control. The printed stage insert we designed has a single gas port leading into a chamber, for which a low gas flow of a blended gas mix of 5% CO2 95% air regulated by a rotameter (Cole Parmer Model PMR1-010291 is one such low flow alternative) is sufficient to maintain adequate pH control of standard tissue culture media. The gas is bubbled through sterile water maintained at 37 °C and small reservoirs of sterile water are included in the chamber to maintain chamber humidity and prevent osmotic changes due to evaporation. In our setup the entire microscope stage, objectives and condenser are enclosed in a Plexiglass box with forced air temperature feedback control (In Vivo Scientific) to maintain temperature at 37 °C. With this arrangement cells can be imaged for at least several days. The microscope should be equipped with motorized high speed excitation and emission filter wheels and a high quality cooled CCD camera. We are using a Nikon Eclipse TE2000-U inverted epifluorescent microscope with Prior Scientific xyz positioning stage and high speed filter wheels and a Roper Scientific monochrome cooled CCD camera. Software control is via Nikons NIS Elements.
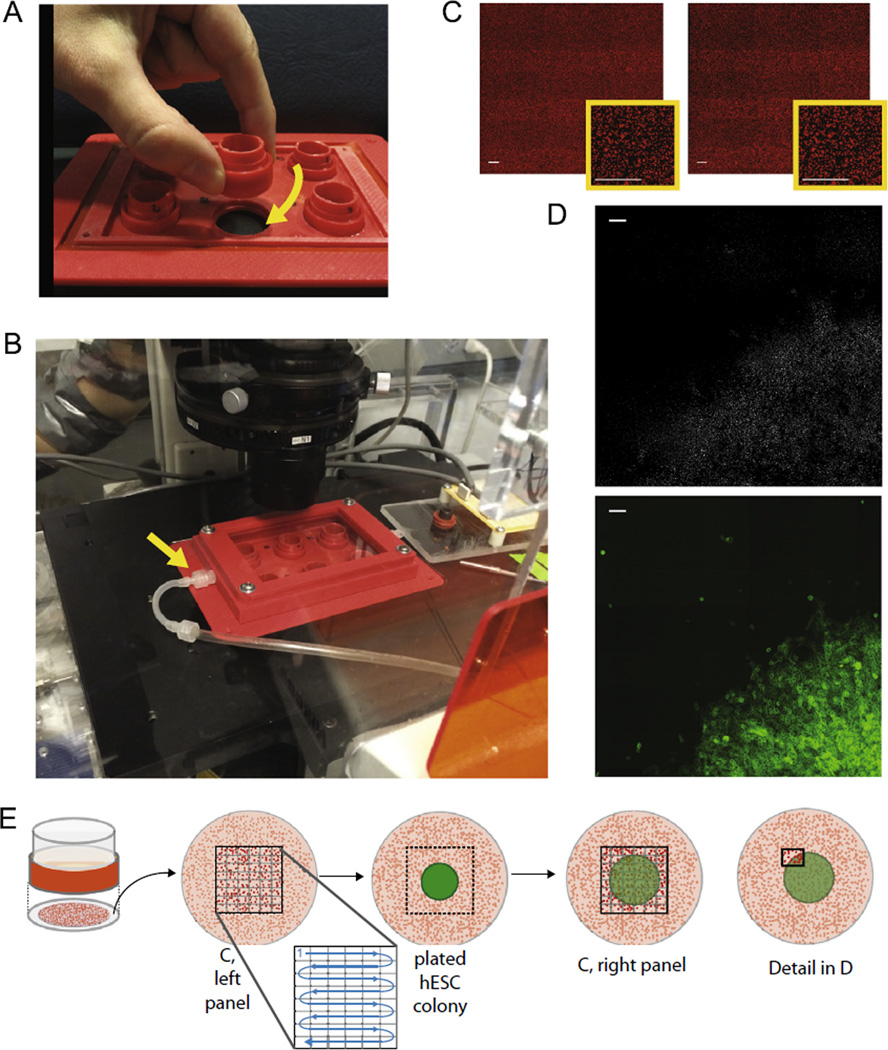
To collect unstressed bead images before plating, it is a precondition that gels maintain a fixed position in x and y with respect to the positioning system of the microscope stage and do not undergo rotation when transported to and from the microscope. This was done by 3D printing a stage insert that could double as a tissue culture plate in a standard incubator, into which the gel assembly can be tightly screwed (Fig. 4A). After screwing gels into the stage, 0.7 ml/gel media is added to the wells and a 3D printed environmental chamber is screwed into the microscope stage to isolate the gels. This assembly is then fitted tightly into the motorized microscope stage and images of unstressed beads are acquired (Fig. 4B).
Epithelial monolayers are initially plated on the order of mm and this size will usually increase over time. If unstressed bead images are collected before plating, some allowance must also be made for random deviation in the position of the plating guides that changes the exact position of the colony on the gel. With the printed parts we describe here we find that this deviation is usually on the order of a few hundred microns at most. The size of the unstressed bead field that must be acquired will depend on these considerations. For the 1.8 mm diameter hESC colonies that we plated here using our guides we find that imaging an approximately 4 mm by 4 mm square of unstressed beads prior to plating provides a generous margin of error.
-
To image this large area requires multiple overlapping fields of view (FOV) that are stitched together post acquisition. The number of required FOV depends on the size of the overall image, the degree of overlap, the magnification of the objective, and the chip size of the CCD camera. We used a Nikon 10× objective (Plan Apo NA 0.45 and 4 mm working distance), which gives an unbinned pixel size of 0.65 µm/pixel and an overall camera FOV of 904.8 µm by 676 µm for our Roper Scientific CoolSnap HQ Camera. We designed a simple Excel spreadsheet calculator that has as fixed input camera FOV (in µm) and as parameters desired FOV fractional overlap in x and y and desired total scan length in x and y (both in µm) as well as the desired number of overlapping FOV in x and y. We iteratively change these parameters to optimize the overall scan. We use a Fiji plugin to stitch images together, which has a single overlap input for both x and y, requiring us to fit to a desired FOV fractional overlap that is fixed for both x and y. With the configuration described above, a 6 × 8 = 48 FOV image covers 4.040 mm × 3.957 mm from edge to edge with 0.3 as fractional overlap. A second Excel spreadsheet calculator then takes as input scan distance and fractional overlap together with a point defined by the stage positioning system to calculate a series of row by row stage positions that accomplishes these scan parameters with the defined point as the center of the image. For each well of our stage insert this point is defined by the stage positioning system when the camera FOV is centered on the outlet of the plating guide inserted into that well with the stage insert fixed in the microscope stage. Corresponding scan points are input into the xy stage positioning system of the NIS elements interface, and this saved set of points can then be offset to any arbitrary stage position to define sets of points for other wells. The scanned stitched image covers an area centered on the point at which the epithelial monolayer will subsequently be plated. For each xy position the optimum focus for the beads is set and the position recorded, then the image is acquired automatically for this set of scan points. Individual images are exported as tiffs and stitched together using the Fiji plugin Grid Collection Stitching [41] with subpixel resolution. With our custom printed stages and gel mounts the gel surface is orthogonal or nearly orthogonal to the optical axis. Typical deviations are on the order of 10 microns (in z) over 4 mm in x or y or 1.7 micron per FOV in x and 1.3 micron per FOV in y. These small changes in z per FOV mean that the relevant beads are nearly always within the focal plane of the 10× objective with resulting improvements in positional accuracy.
Critical: if focal plane deviates significantly across the image due to a canted or uneven gel such that there are FOVs with regions of beads that are not perfectly in focus, this gel will likely be unsuitable for image analysis and processing.
Once cells are plated as described above (Method 2.3), the stage insert is returned to the microscope stage, and for each well the stage is moved to the initial scan point as defined above and the beads are brought into focus. The first unstressed bead image recorded earlier is opened in NIS elements and the xy position of the live image is adjusted so that its FOV matches that of the unstressed image. The first scan point in our example is very far from plated cells so there is no distortion of the two images due to contractile cellular forces that would complicate this alignment. This new stage position is then used to offset all of the previous scan points, and each position is visited to update the z position for the best focal plane and the image is acquired.
Following acquisition of scanned bead images again (after plating) these bead images are stitched using Grid Collection Stitching and before (unstressed) and after (stressed) stitched images are registered. Good registration can be obtained using the Fiji plugin TurboReg [42] treating the images as rigid bodies aided by the large area of unstressed beads surrounding the colony in both images. Rigid registration is recommended to prevent any artifactual bead deformations that might result for those methods that allow for elastic deformations in the registration process.
How the bead movements defined by these registered images are converted to cell–cell and cell–matrix vector force fields has been recently covered in detail [40] and will not be restated here.
Fig. 4.
Stage insert and microscope setup for gathering traction force images prior to plating cells. (A) Photograph of 6-well assembly for culture and microscopy of consistently centered colonies on traction force gels. Cassettes have threads to screw firmly into stage insert. (B) Fully assembled stage insert with environmental control lid inserted into microscope. Arrow indicates connection to CO2 input line. (C) Bead images before (left) and after (right) plating cells. All scale bars = 100 µm. (D) The subtracted image indicating changes in bead location from large bead plots shown in (C). Bottom panel displays corresponding cell location via a MUC1-venus membrane marker. Scale bars = 100 µm. (E) Diagram showing where and when bead images are taken in the context of the gel and cell colony. Images are taken sequentially across a grid before and after colonies are plated.
3. Results and discussion
Using the methods described here, one can generate detailed maps of cell–matrix traction forces from hESCs seeded in mm-sized circular monolayers on bead-containing polyacrylamide gels of a range of elastic moduli from physiologically soft to supraphysiologically hard. We showed previously that on soft but not stiff matrices, these monolayers form a columnar pseudostratified epithelia resembling the in vivo pregastrulation epiblast [27]. Here, we add control of monolayer size and shape, demonstrating geometries that are similar to those of the embryo at a relevant developmental stage. Recently it was shown that similar sized and shaped colonies of hESCs undergo a complex and radially organized pattern of germ layer differentiation when treated with high concentrations of BMP4 [36], suggesting that developmental self-organization may emerge in the collective dynamics of large cellular aggregates. In addition to biochemical gradients, it is likely that cell–cell and cell–matrix mechanical forces contribute to these large tissue scale reorganizations [43], but little is known about such forces in the context of developmental biology. To address this, we have developed an approach to measure dynamic mechanical forces, which can be applied to studies of germ layer differentiation and developmental patterning. While other approaches have demonstrated that mechanical signals affect stem cell differentiation [44,45] and embryonic development [4–6], our platform allows for correlation between specific developmental fates and the associated tissue-level forces. By demonstrating not only the ability to alter the mechanical signals presented to cells but also to non-destructively measure the cell-intrinsic forces and analyze the spatial distribution of molecular markers, we show that this simple system can be applied to study the forces associated with developmental processes.
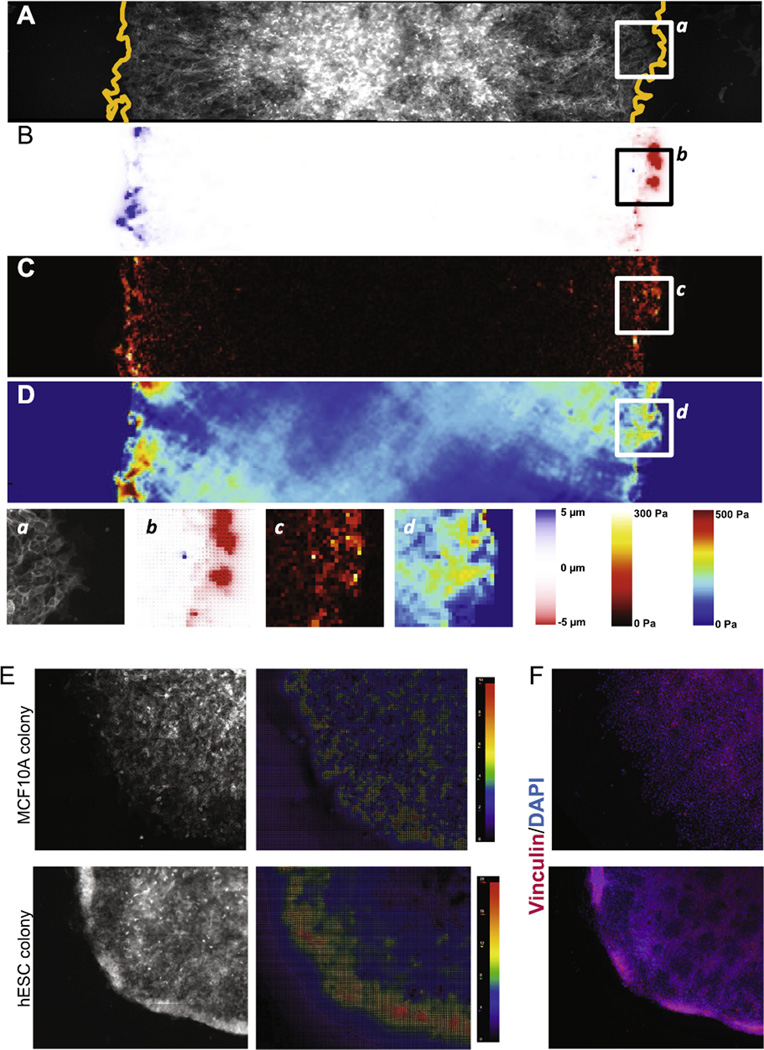
By way of demonstration of the feasibility of our approach we show the results from a horizontal cross-section through a single hESC colony (Fig. 5A) seeded on a polyacrylamide gel of E = 1000 Pa. The stitched unstressed bead image was acquired prior to plating cells and acquisition of stressed bead images. Registration of the two images yields a map of displacements (Fig. 5B), which can be converted to displacement vectors by Particle Image Velocimetry (PIV). This displacement map is utilized to reconstruct cell–ECM tractions by assuming a linear-elastic substrate and applying the unconstrained Fourier transformed traction cytometry method (Fig. 5C) [40,46,47]. Because these tractions must be balanced throughout the cell layer according to Newton’s laws, cell–cell forces can be computed by using a finite element approach (Fig. 5D) [20,40]. We show that cell–matrix and cell–cell stresses are strongest around the edges of the colony and predominately point inwards, and are low or virtually absent within the center (Fig. 5C).
Fig. 5.
Force organization in hESC colonies. (A) Fluorescent image of a cross-section of an hESC colony plated on a 1000 Pa gel growing in self-renewal media. Cells express MUC1-venus membrane marker. Dashed line indicates the limits of the colony. (B) Colony map showing gel deformations in the x direction. (C) High-resolution traction force map resulting from the calculation using the Boussinesq algorithm for infinite gel substrate of finite thickness. (D) Intercellular and intracellular stresses σxx. (a–d) Magnification of the colony edge showing cells, deformations, tractions and stresses. (E) Fluorescent image of the edge of an MCF10A (left) and an hESC colony (right) and the associated particle image velocimetry map depicting the direction and magnitude of bead movements from the unstressed versus stressed conditions. Both colonies are on 1000 Pa gels. (F) Immunofluorescence images of colony edges shown in (E).
Our prior work suggested that hESC colonies are collectively highly contractile [27] but the magnitude of this relative to other epithelia was unclear. In Fig. 5E we show a simple comparison of an hESC colony to a similar sized colony of a normal polarized epithelial cell line, MCF10A, acquired by our approach and analyzed by PIV using the PIV plugin for FIJI [48]. Cell matrix stresses are much higher at the edges of hESC colonies than those at MCF10A colony edges, which are lower and more diffusely distributed through the colony.
While we do not have sufficient experimental data to determine the underlying molecular basis for this difference we conjecture that this is the result of differences in the organization of the actomyosin contractile apparatus between these two epithelia. On softer substrates, hESCs organize the contractile apparatus as circumferential bands at the apical surface near well-developed E-cadherin-based adherens junctions [27], resulting in strong intercellular contractions that seem to be coordinated over most of the extent of the colony. Apical constriction such as this is known to initiate cell ingression during gastrulation [49]. In the unattached state, coordinated apical contractions cause a concave curling up of the colony edges, as precedes topological closure and embryoid body formation of detached colony fragments. When attached, this upward curling force is likely resisted by strong edge attachments, leading to large deformations of the gel at the edges. In the case of MCF10A cells, more of the actomyosin contractile apparatus is located at the basal domain with the consequence that intercellular contraction is lower and groups of cells are likely coordinated over shorter ranges (10–100 s of microns). Subsequent fixing and staining of the hESC and MCF10A colonies shows that vinculin is more strongly concentrated at the edges of hESC colonies and proximal to the highest stresses (Fig. 5F). This suggests that vinculin expression and/or localization may also underlie or result from some of these differences. In addition, this illustrates the potential utility of our method to obtain molecular correlates to the underlying cellular mechanics.
Aspects of our methods have been published elsewhere to achieve similar ends, but we believe that our comprehensive protocol is optimally suited to culture of hESC colonies on traction force gels of low elastic modulus. Other approaches for depositing beads uniformly near the cell-gel interface have been published, including using a two-layer approach [50] or positively charging the top coverslip to coat it with beads [51]. While we have not compared these approaches side-by-side, they have predominantly been applied to single cell traction force microscopy of stiffer gels. However, for the high quality colony-wide traction data we require, gels must be defect-free and uniform over a large area. We find, particularly for soft gels, that hydrophobic as opposed to charged top coverslips are best suited for this application as they are more reliably removed without tearing or deforming the underlying gel. Our method (2.1) permits consistent production of gels of uniform thickness and low compliance with beads at the cell-gel interface, at least for beads of ⩾0.5 µm. However, it is possible that for applications requiring small beads, centrifugation may be inadequate to sediment the beads, so other approaches may be required. Consistent gel preparation and cell plating at low stiffness are important because the epithelial cell behaviors that are most relevant to physiological processes occur in vivo in the context of tissues, which typically have an elastic modulus between 100 and 5000 Pa [52], with developmentally relevant stiffnesses at the low end of this range.
Beyond our technical substrate improvements, our methods also include enhanced control over biophysical parameters which we couple with the ability to correlate forces with protein expression. We can mimic any formation of adherent cell collective that is found in vivo, and analyze how forces develop with addition of relevant growth factors or inhibitors. While we have observed that hESCs maintain pluripotency across multiple passages on soft gels when plated as single cells or in circular colonies, others have demonstrated radial segregation of developmental markers in large hESC colonies that are induced to differentiate [36], exposing patterns that would not be elaborated in single cells. This indicates that geometry and regional heterogeneity can contribute to differentiation potential, and it is possible that beyond soluble signals provided to cells, differences in force organization could underlie this heterogeneity. Substrate stiffness has been shown to influence stem cell differentiation [44,45], so the endogenous forces that cells in a collective exert on each other are also likely to play a role, a hypothesis that can be rigorously tested by adapting the methods described here to include differentiation protocols.
Beyond differentiation and development, additional applications of these methods include cell biology studies involving cell collectives of epithelial origin that present a particular behavior related to cell motility or mechanics. For example, collective cell motility could be analyzed in detail and correlated to expression and localization of cell polarity proteins, or reorganization of cells within a colony could be monitored after addition of an EMT initiating factor such as TGFβ. This method to reproducibly generate high-density colony-wide traction force maps and compare regions of traction and cell stresses to protein expression is robust and widely applicable to address outstanding questions about relationships between force generation and cell behavior.
Acknowledgments
We would like to acknowledge funding from CIRM grant RB5-07409 and CIRM grant TR3-05542. J.N.L. acknowledges NIH TMEN grant U54 CA163155 and L.P. acknowledges CIRM training grant TG2-01153.
References
- 1.Li Y-SJ, Haga JH, Chien S. Molecular basis of the effects of shear stress on vascular endothelial cells. J. Biomech. 2005;38:1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. http://dx.doi.org/10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. http://dx.doi.org/10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. 2011;108:4708–4713. doi: 10.1073/pnas.1011123108. http://dx.doi.org/10.1073/pnas.1011123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. http://dx.doi.org/10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 5.Aegerter-Wilmsen T, Heimlicher MB, Smith AC, de Reuille PB, Smith RS, Aegerter CM, et al. Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development. 2012;139:3221–3231. doi: 10.1242/dev.082800. http://dx.doi.org/10.1242/dev.082800. [DOI] [PubMed] [Google Scholar]
- 6.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev. Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. http://dx.doi.org/10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005;232:685–694. doi: 10.1002/dvdy.20334. http://dx.doi.org/10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Gonzalez R, de M, Simoes S, Röper J-C, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. http://dx.doi.org/10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pouille P-A, Ahmadi P, Brunet A-C, Farge E. Mechanical signals trigger Myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci. Signal. 2009;2:ra16. doi: 10.1126/scisignal.2000098. http://dx.doi.org/10.1126/scisignal.2000098. [DOI] [PubMed] [Google Scholar]
- 10.Brouzés E, Farge E. Interplay of mechanical deformation and patterned gene expression in developing embryos. Curr. Opin. Genet. Dev. 2004;14:367–374. doi: 10.1016/j.gde.2004.06.005. http://dx.doi.org/10.1016/j.gde.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Brouzés E, Supatto W, Farge E. Is mechano-sensitive expression of twist involved in mesoderm formation? Biol Cell Auspices Eur. Cell Biol. Organ. 2004;96:471–477. doi: 10.1016/j.biolcel.2004.04.009. http://dx.doi.org/10.1016/j.biolcel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr. Biol. 2005;15:R213–R228. doi: 10.1016/j.cub.2005.03.016. http://dx.doi.org/10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Shook DR, Keller R. Epithelial type, ingression, blastopore architecture and the evolution of chordate mesoderm morphogenesis. J. Exp. Zool. B Mol. Dev. Evol. 2008;310:85–110. doi: 10.1002/jez.b.21198. http://dx.doi.org/10.1002/jez.b.21198. [DOI] [PubMed] [Google Scholar]
- 14.Moore SW, Keller RE, Koehl MA. The dorsal involuting marginal zone stiffens anisotropically during its convergent extension in the gastrula of Xenopus laevis. Dev. Camb. Engl. 1995;121:3131–3140. doi: 10.1242/dev.121.10.3131. [DOI] [PubMed] [Google Scholar]
- 15.Hutson MS, Tokutake Y, Chang M-S, Bloor JW, Venakides S, Kiehart DP, et al. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. http://dx.doi.org/10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- 16.Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr. Biol. 2005;15:2208–2221. doi: 10.1016/j.cub.2005.11.064. http://dx.doi.org/10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 17.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat. Protoc. 2011;6:1835–1846. doi: 10.1038/nprot.2011.395. http://dx.doi.org/10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
- 18.Campàs O, Mammoto T, Hasso S, Sperling RA, O’Connell D, Bischof AG, et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods. 2014;11:183–189. doi: 10.1038/nmeth.2761. http://dx.doi.org/10.1038/nmeth.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. http://dx.doi.org/10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 20.Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 2011;10:469–475. doi: 10.1038/nmat3025. http://dx.doi.org/10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol. 2015;17:276–287. doi: 10.1038/ncb3115. http://dx.doi.org/10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 22.Trepat X, Fredberg JJ. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011;21:638–646. doi: 10.1016/j.tcb.2011.06.006. http://dx.doi.org/10.1016/j.tcb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bazellières E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, et al. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. http://dx.doi.org/10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. http://dx.doi.org/10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim JY, Moeller J, Hart KC, Ramallo D, Vogel V, Dunn AR, et al. Spatial distribution of cell–cell and cell-ECM adhesions regulates force balance while maintaining E-cadherin molecular tension in cell pairs. Mol. Biol. Cell. 2015 doi: 10.1091/mbc.E14-12-1618. http://dx.doi.org/10.1091/mbc.E14-12-1618. mbc.E14–12–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. http://dx.doi.org/10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 27.Lakins JN, Chin AR, Weaver VM. Exploring the link between human embryonic stem cell organization and fate using tension-calibrated extracellular matrix functionalized polyacrylamide gels. Methods Mol. Biol. Clifton N.J. 2012;916:317–350. doi: 10.1007/978-1-61779-980-8_24. http://dx.doi.org/10.1007/978-1-61779-980-8_24. [DOI] [PubMed] [Google Scholar]
- 28.van den Berg CW, Elliott DA, Braam SR, Mummery CL, Davis RP. Differentiation of human pluripotent stem cells to cardiomyocytes under defined conditions. Methods Mol. Biol. Clifton N.J. 2015 doi: 10.1007/7651_2014_178. http://dx.doi.org/10.1007/7651_2014_178. [DOI] [PubMed] [Google Scholar]
- 29.Wattanapanitch M, Klincumhom N, Potirat P, Amornpisutt R, Lorthongpanich C, U-pratya Y, et al. Dual small-molecule targeting of SMAD signaling stimulates human induced pluripotent stem cells toward neural lineages. PLoS One. 2014;9:e106952. doi: 10.1371/journal.pone.0106952. http://dx.doi.org/10.1371/journal.pone.0106952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhou J, Fang Z, Jiang M, Chen X. Noggin versus basic fibroblast growth factor on the differentiation of human embryonic stem cells. Neural Regener. Res. 2013;8:2171–2177. doi: 10.3969/j.issn.1673-5374.2013.23.007. http://dx.doi.org/10.3969/j.issn.1673-5374.2013.23.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills JA, Paluru P, Weiss MJ, Gadue P, French DL. Hematopoietic differentiation of pluripotent stem cells in culture. Methods Mol. Biol. Clifton N.J. 2014;1185:181–194. doi: 10.1007/978-1-4939-1133-2_12. http://dx.doi.org/10.1007/978-1-4939-1133-2_12. [DOI] [PubMed] [Google Scholar]
- 32.Banda E, Grabel L. Directed differentiation of human embryonic stem cells into neural progenitors. Methods Mol. Biol. Clifton N.J. 2014 doi: 10.1007/7651_2014_67. http://dx.doi.org/10.1007/7651_2014_67. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y-K, Chen CS. Cell adhesion and mechanical stimulation in the regulation of mesenchymal stem cell differentiation. J. Cell Mol. Med. 2013;17:823–832. doi: 10.1111/jcmm.12061. http://dx.doi.org/10.1111/jcmm.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reilly GC, Engler AJ. Intrinsic extracellular matrix properties regulate stem cell differentiation. J. Biomech. 2010;43:55–62. doi: 10.1016/j.jbiomech.2009.09.009. http://dx.doi.org/10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Chowdhury F, Na S, Li D, Poh Y-C, Tanaka TS, Wang F, et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9:82–88. doi: 10.1038/nmat2563. http://dx.doi.org/10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. http://dx.doi.org/10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells RG. Tissue mechanics and fibrosis. Biochim. Biophys. Acta. 1832;2013:884–890. doi: 10.1016/j.bbadis.2013.02.007. http://dx.doi.org/10.1016/j.bbadis.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat. Mater. 2010;9:768–778. doi: 10.1038/nmat2812. http://dx.doi.org/10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. http://dx.doi.org/10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serra-Picamal X, Conte V, Sunyer R, Muñoz JJ, Trepat X. Mapping forces and kinematics during collective cell migration. In: Paluch EK, editor. Methods Cell Biol. Academic Press; 2015. pp. 309–330. (Chapter 17) [DOI] [PubMed] [Google Scholar]
- 41.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. http://dx.doi.org/10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. Publ. IEEE Signal Process. Soc. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 43.Davidson LA. Epithelial machines that shape the embryo. Trends Cell Biol. 2012;22:82–87. doi: 10.1016/j.tcb.2011.10.005. http://dx.doi.org/10.1016/j.tcb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. http://dx.doi.org/10.1016/S1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. http://dx.doi.org/10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, et al. Physical forces during collective cell migration. Nat. Phys. 2009;5:426–430. http://dx.doi.org/10.1038/nphys1269. [Google Scholar]
- 47.Butler JP, Tolic´-Nørrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. - Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. http://dx.doi.org/10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 48.Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. http://dx.doi.org/10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. http://dx.doi.org/10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- 50.Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. J. Neurosci. 2001;21:6159–6169. doi: 10.1523/JNEUROSCI.21-16-06159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knoll SG, Ali MY, Saif MTA. A novel method for localizing reporter fluorescent beads near the cell culture surface for traction force microscopy. J. Vis. Exp. JoVE. 2014 doi: 10.3791/51873. http://dx.doi.org/10.3791/51873. 51873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. http://dx.doi.org/10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]