Abstract
MicroRNAs (miRNAs) are small endogenous non-coding RNAs, which play critical roles in cancer development by suppressing gene expression at the post-transcriptional level. In general, oncogenic miRNAs are upregulated in cancer, while miRNAs that act as tumor suppressors are downregulated, leading to decreased expression of tumor suppressors and upregulated oncogene expression, respectively. F-box proteins function as the substrate-recognition components of the SKP1-CUL1-F-box (SCF)-ubiquitin ligase complex for the degradation of their protein targets by the ubiquitin-proteasome system. Therefore F-box proteins and miRNAs both negatively regulate target gene expression post-transcriptionally. Since each miRNA is capable of fine-tuning the expression of multiple target genes, multiple F-box proteins may be suppressed by the same miRNA. Meanwhile, one F-box proteins could be regulated by several miRNAs in different cancer types. In this review, we will focus on miRNA-mediated downregulation of various F-box proteins, the resulting stabilization of F-box protein substrates and the impact of these processes on human malignancies. We provide insight into how the miRNA: F-box protein axis may regulate cancer progression and metastasis. We also consider the broader role of F-box proteins in the regulation of pathways that are independent of the ubiquitin ligase complex and how that impacts on oncogenesis. The area of miRNAs and the F-box proteins that they regulate in cancer is an emerging field and will inform new strategies in cancer treatment.
Keywords: microRNA, F-box protein, cancer
1. Introduction
1.1 F-box proteins in cancer
F-box proteins are a large protein family (69 putative members in humans), which harbor the consensus F-box motif [1–3]. The F-box motif is a protein-protein interaction domain that was first identified in cyclin F (also known as F-box only 1/FBXO1) [4]. The F-box motif-mediated interaction between F-box proteins and Skp1 (S-phase kinase-associated protein 1) is essential for pivotal functions of the Skp1-cullin1-F-box protein (SCF) E3 ligase complexes in regulating the cell cycle, circadian rhythm, cell proliferation and apoptosis [5, 6]. Within the SCF complex, cullin 1 serves as the docking platform to bridge RING-box protein 1 (Rbx1), which recruits the E2 ubiquitin-conjugating enzyme, and Skp1 into the ligase machinery complex. The Skp1-associated F-box proteins can recognize specific degradation motifs (degrons) in their substrates, enabling the SCF ligase complex to modify substrates by attaching ubiquitin moieties, and thereby leading to their degradation by proteasome. Therefore, the variable F-box proteins within the SCF complex determines the specificity of the substrates that are ubiquitinated by the SCF complex [5].
F-box proteins can be organized into three subfamilies based on the presence of specific substrate recognition domains [1]. The FBXW subfamily is comprised of 10 members, including β-TrCP1 (β-transducin repeat-containing protein1, also known as FBXW1), β-TrCP2 (also known as FBXW11) and FBXW7 (also known as FBW7 or CDC4). The FBXW proteins harbor multiple WD40 motifs, which recognize canonical phosphodegrons in their respective substrates. The FBXL subfamily has 22 members, which share clusters of leucine-rich repeat domains. Although systematic characterization of potential FBXL substrates by quantitative proteomic approaches has been recently performed [7], the shared criteria for FBXL substrate recognition have not yet been defined. The remaining 37 F-box proteins comprise the FBXO subfamily where the “O” refers to the functionally uncharacterized “other” domain. Most interestingly, several FBXO proteins, such as FBXO4, FBXO11 and FBXO31, apparently play critical roles in regulating oncogenesis [8–10].
SCF complexes direct specific protein substrates for proteasomal degradation through the attachment of polyubiquitin chains. Therefore, F-box proteins play roles as oncogenes, tumor suppressors, or both in a context-dependent manner, depending on the substrates that they target for degradation, [11]. For example, FBXW7 suppresses tumorigenesis by promoting the degradation of oncogenic proteins, such as cyclin E, MYC, NOTCH and MCL1 [12], and is frequently mutated in cholangiocarcinoma and T cell acute lymphoblastic leukemia (TALL) [13, 14]. Mice with T cell-specific deletion of FBXW7 showed excessive accumulation of MYC accompanied by spontaneous development of thymic lymphoma [15]. An intestine-specific deletion of FBXW7 increased the tumor incidence in mice with the adenomatous polyposis coli mutation, in part due to increased NOTCH1 and JUN protein expression in intestinal cells [16]. Another tumor suppressive F-box protein FBXO11 was found to target BCL-6 (B cell lymphoma 6) for degradation, which is crucial in the pathogenesis of diffused large B cell lymphoma (DLBCL), the most common adult lymphoma [17]. Human DLBCL cell lines and primary cancers harboring mutations or deletion of FBXO11 gene displayed diminished ability to promote BCL-6 degradation, resulting in lymphomagenesis in immunocompromised mice. In contrast, the F-box protein Skp2 has been shown to promote oncogenesis. Overexpression of Skp2 in mammary gland, prostate or T lymphoid drives the development of breast cancer, prostate cancer and T cell lymphoma in mice [19–21]. Skp2 is overexpression in a variety of human cancers [22–26], and may promote oncogenesis through the ubiquitination and degradation of tumor suppressors, such as p21, p27, p57, RBL2 (retinoblastoma-like protein 2) and FOXO1 [11].
In contrast to FBXW7 and Skp2, the roles of β-TrCP1 and β-TrCP2 in modulating tumorigenesis may be context-dependent. β-TrCP1 targets both IκBα (inhibitor of nuclear factor κB) and β-catenin [27–32], leading to NF-κB pathway activation and suppression of the Wnt/β-catenin pathway, respectively. β-TrCP overexpression has been found in colorectal, pancreatic, prostate, breast and gastric cancer [33–35] whereas somatic mutations of β-TrCP that disrupts the E3 ligase activity has been found in some human gastric cancers [36, 37]. The expanding target list of β-TrCP proteins includes both oncogenes (Mcl-1, MYC and cyclin D) and tumor suppressors (p53, PDCD4 and FOXO3a), which further highlights the context-dependent functions of β-TrCP proteins in human cancer [11, 38].
1.2 MicroRNAs in cancer
MicroRNAs (miRNAs) are a family of small non-coding RNAs (19~24 nucleotides) that are highly conserved throughout evolution, and negatively regulate target gene by suppressing translation and/or inducing mRNA decay [39]. Intergenic miRNA genes are transcribed by RNA polymerase II as long, capped and polyadenylated transcripts [40]. These primary miRNA transcripts, which vary from several hundred to several thousand nucleotides, are then processed into precursor miRNAs by a nuclear microprocessor complex consisting of Drosha and DGCR8 (DiGeorge syndrome critical region 8, also known as Pasha) [41]. Alternatively, precursor-miRNAs can also result from RNA splicing, when the miRNA sequence resides in protein-coding gene introns and is co-transcribed with the mRNA of the host gene. Precursor miRNAs are transported into the cytoplasm by exportin 5, and cleaved into mature double-stranded miRNAs by cytoplasmic type III RNase Dicer. The miRNA duplexes are then loaded into the RNA-induced silencing complex (RISC), where the RNA duplexes are unwound and the passenger strands are removed, yielding the functional mature RISC [42]. The overall function of RISC-loaded miRNAs is to post-transcriptionally repress target genes by inhibiting ribosome-dependent protein translation and/or destabilizing mRNA expression. The seed region (2–7 nucleotides) of miRNAs plays an essential role in recognizing complementary sequences (so called MRE, miRNA recognition element) within target mRNA, which are typically located in the 3′ untranslated region (3′-UTR) [39]. However, MREs within 5′-UTRs or open reading frames of target mRNAs have also been reported, which could effectively mediate miRNA-dependent gene suppression [43, 44].
MiRNAs play pivotal roles in modulating cancer initiation and progression [45]. MiRNA genes are found frequently located within cancer-associated genomic regions, which are amplified or deleted in different human cancers. Accordingly, miRNAs can act either as oncogenes or tumor suppressors, depending on the genes being repressed in particular cancer types. Moreover, one miRNA can be either tumor-suppressive or oncogenic depending on the cellular context and different cancer types. Various F-box proteins are negatively regulated by miRNAs, which adds another layer of regulation to the versatile roles of these proteins in governing the pathogenesis of human malignancies.
2. MiRNA-dependent regulation of F-box proteins and its role in cancer
2.1 MiRNA targeting of FBXW7
Since FBXW7 is a critical tumor suppressor that regulates cell proliferation, apoptosis, cell cycle and differentiation in various human cancers, it is not surprising that a variety of oncogenic miRNAs have been reported to promote tumorigenesis by targeting FBXW7. For example, miR-27a was reported to negatively regulate FBXW7 expression in primary mouse erythroblasts leading to increased stability and aberrant activation of cyclin E [46]. In colon cancer cells, FBXW7 was identified as a direct target of miR-27a, which was frequently co-amplified with miR-23a in primary tumors and cancer stem cells from stage I/II colorectal cancer patients [47]. MiR-27a upregulation promoted colon cancer cell proliferation, which likely involved increased cyclin E levels due to suppression of FBXW7 expression by miR-27a. Similar observations were also made in human lung cancer specimens and pediatric B cell acute lymphoblastic leukemia cells [48, 49]. Upregulation of miR-27a by SV40 small T antigen was found to play a critical role in malignant transformation of normal human bronchial epithelial cells. In these cells, FBWX7 was identified as a primary miR-27a target, whose downregulation resulted in increased stability of c-Myc, c-Jun, cyclin E and NOTCH1 and subsequently in accelerated cell-cycle progression [48]. Interestingly, miR-27a-dependent FBXW7 suppression may be cell cycle phase specific, i.e., promoting G1-S phase transition by suppressing FBXW7 and relieving FBXW7-mediated suppression of cyclin E [49]. Taken together, these findings indicate that FBXW7 downregulation likely plays a major role in miR-27a-promoted cancer progression.
A number of studies have established that FBXW7 is a target of miR-223. For example, protein levels of FBXW7 negatively correlate with miR-223 expression in gastric tumor tissue [50]. Similarly, an inverse correlation between FBXW7 and miR-223 was found in patient samples from T-ALL [51] and esophageal squamous cell carcinoma [52], and associated with poor prognosis. Relatively high miR-223 expression has been found in hepatocellular carcinoma, T-ALL, ovarian cancer, gastric cancer, esophageal squamous cell carcinoma and bladder cancer [53], suggesting that miR-223 may play multifaceted roles in oncogenesis. Moreover, miR-223 could also serve as a downstream effector of other oncogenes, such as TAL1, NOTCH1 and NF-κB, to diminish the tumor suppressive function of FBXW7, leading to T-ALL progression [54, 55]. In addition, miR-223-dependent FBXW7 repression may also contribute to the reduced sensitivity of gastric cancers to trastuzumab and cisplatin [56, 57]. Interestingly, downregulating miR-223 by treatment with the tyrosine kinase inhibitor genistein increased FBXW7 expression, resulted in inhibited cell growth, decreased invasion, and increased apoptosis in pancreatic cancer cells [58]. Nevertheless, whether the miR-223/FBXW7 axis will be an effective therapeutic target for cancer treatment requires further study.
FBXW7 is targeted by other oncogenic miRNAs, such as miR-92a. In cervical cancer cells, miR-92a is upregulated and targets FBXW7, which enhances cell cycle transition from G1 to S phase and promotes cancer cell proliferation [59]. In colon cancer cells, miR-182 and miR-503 were sequentially upregulated and cooperatively targeted FBXW7 for gene suppression [60]. Moreover, miR-182 could be upregulated by sterol regulatory element-binding proteins (SREBPs), and SREBP-induced miR-182 increased SREBP nuclear accumulation by downregulating FBXW7 [61], suggesting that a SREBP/miR-182 positive feedback loop may regulate intracellular lipid homeostasis and pathogenesis of liver cancer.
FBXW7 is targeted by other miRNAs whose functions in tumorigenesis are more context-dependent. The MRE within the 3′-UTR of FBXW7 is recognized by miR-25, directly repressing FBXW7 levels in gastric cancer and prostatic small cell neuroendocrine carcinoma cells [62, 63], which promotes rapid proliferation and aggressive behavior of these cancer cells. Interestingly, miR-25 upregulation in prostatic small cell neuroendocrine carcinoma cells was driven by mutated p53, which lead to increased Aurora kinase A expression due to miR-25 suppression of FBXW7 levels [63]. Along with other cell cycle promoting oncogenes (e.g. c-Myc, c-Jun, cyclin E) downregulated by FBXW7, increased Aurora kinase A levels enhanced cancer cell cycle progression and accelerated growth. In addition, the suppression of FBXW7 by miR-25a also promoted reprograming of mouse somatic cells into induced pluripotent stem cells, where Klf5 is a downstream target of FBXW7 and appears to play a critical role in this process [64]. Whether suppression of FBXW7 by miR-25 also contributes to maintenance and self-renewal of cancer stem-like cells needs further exploration. In contrast to the oncogenic function of miR-25 in gastric cancer [62], ovarian cancer [65] and esophageal squamous cell carcinoma [66], miR-25 was found to be downregulated in human colon cancer and thyroid carcinoma [67, 68]. It will be interesting to determine whether FBXW7 levels are altered in colon cancer and thyroid carcinoma, which exhibit decreased miR-25 expression, and the role of FBXW7 in these cancers.
2.2 MiRNA targeting of FBXO11
In contrast to the well-characterized substrate-recognition mechanisms of FBXW proteins, how FBXO proteins select their substrate for SCF-mediated ubiquitination and proteasomal degradation is relatively unclear. Nevertheless, recent studies have revealed the FBXO proteins, FBXO4 and FBXO11, play important roles in modulating cancer development, although their substrate-recognition mechanisms may vary [5, 11].
Deletion or mutation of FBXO11 was found in ~20% of DLBCL patients, indicating that FBXO11 is a tumor suppressor in DLBCL. Oncoprotein BCL6, whose upregulation drives DLBCL development, was identified as a substrate of FBXO11 for SCF-mediated degradation [9]. Besides, FBXO11 mutations were observed in colon cancer, lung cancer, ovarian cancer, head and neck squamous cell carcinoma, as well as Burkitt lymphoma, implying that FBXO11 may act as a tumor suppressor in several cancers [5]. Interestingly, the oncogenic miRNA miR-21 was found to directly suppress expression of FBXO11, resulting in increased BCL6 expression, in melanoma, prostate cancer and glioma [69, 70]. Knockdown of FBXO11 expression attenuated the tumor suppressive effect of miR-21 silencing in a melanoma xenograft model, demonstrating that FBXO11 is a critical miR-21 target gene. An inverse correlation between miR-21 and FBXO11 expression was observed in glioma patients, and high FBXO11 level was associated with lower tumor grade and better patient survival. These findings strongly support that FBXO11 suppression is a critical mechanism mediating the oncogenic activity of miR-21 in melanoma, glioma and prostate cancer progression.
FBXO11 interacts with Cdt2 (Cdc10-dependent transcript 2; also known as DTL and DCAF2) and promotes its proteasomal degradation [71, 72]. Cdt2 is a component of Cul4-RING protein ubiquitin ligase (CRL4) complex, which promotes ubiquitination and degradation of the cell cycle regulator p21 (CDKN1A) and SET domain-containing lysine methyltransferase 8 (SETD8). Cdt2 degradation controls cell-cycle exit timing by FBXO11, while FBXO11-promoted SETD8 stabilization may enhance cancer cell migration and decrease apoptosis [71, 73]. These observations raise the possibility that FBXO11 could promote cancer progression in specific stages and/or in certain cancers. In the majority of breast cancer patient samples that harbor a FBXO11 gene alteration, amplification and upregulation of FBXO11 was observed. Moreover, FBXO11 suppression by miR-621 increased the sensitivity to chemotherapy in breast cancers by enhancing p53-mediated apoptosis [74]. It was reported that FBXO11 could inhibit p53 transactivity by promoting p53 neddylation [75]. Therefore, FBXO11 suppression by miR-621 may enhance p53-mediated apoptosis in breast cancer cells by repressing 53 neddylation and/or inhibiting SETD8-promoted p53 destabilization. However, FBXO11 was also shown to inhibit breast cancer metastasis by promoting Snail degradation, which inhibited epithelial-mesenchymal transition [76]. Taken together these findings suggest that, while FBXO11 may promote oncogenesis in specific cancer types and stages, overall FBXO11 likely functions as a tumor suppressor in many cancers. It remains to be further explored that how miRNAs modulates FBXO11 protein levels in specific cancer types/stages and the role of the miRNA/FBXO11 pathway in the development of particular cancers.
2.3 MiRNA targeting of the F-box protein Skp2
The oncogenic F-box protein Skp2 is also tightly controlled by a variety of tumor suppressive miRNAs. In an inducible mouse model, Skp2 was identified as a miR-203 target gene, and Skp2 suppression was found to be essential for miR-203 to promote cell cycle exit and inhibit long-term cell proliferation [77]. Skp2 promotes ubiquitination and subsequent degradation of the cell cycle regulators, p21 and p27 [78, 79], whose downregulation accelerates cell proliferation and reduces cell senescence. MiR-203-mediated Skp2 suppression likely enhanced the stability of p21/p27, leading to cell cycle arrest and increased senescence. The ectopic expression of Skp2 alone is sufficient to rescue cell cycle exit induced by miR-203 [77]. Skp2 was also identified as a major target for the tumor suppressive miRNA miR-7, whose downregulation led to increased p27 and cell growth arrest [80]. Similarly, the tumor suppressive activity of miR-340 in non-small cell lung carcinoma (NSCLC) cells was associated with increased p27 levels, which is due in part to miR-340-dependent suppression of Skp2 [81]. Skp2 overexpression and consequent p27-suppression has been associated with the aggressiveness in NSCLC [82]. In accordance, miR-340 levels were found to inversely correlated with clinical staging in NSCLC patients [81], suggesting that miR-340-mediated Skp2 downregulation plays a critical role in regulating NSCLC progression. Furthermore, Skp2-suppression may also contribute to the premetastatic lung niche for cancer metastasis. It was shown that miR-30 family was downregulated in pre-metastatic lungs, whereas fibroblast-derived miR-30 attenuated melanoma cell lung metastasis by stabilizing pulmonary vessels [83]. Skp2 was identified as a direct target gene of the miR-30 family in lung fibroblasts. Interestingly, overexpression of Skp2 increased pulmonary vascular permeability and promoted lung metastasis of B16 melanoma cells, indicating that miR-30-depedent Skp2 suppression may have a critical role in antagonizing lung metastasis in melanoma patients.
2.4 MiRNA targeting of the F-box protein β-TrCP1
As already discussed, the roles of β-TrCP1/2 in cancer pathogenesis are highly context-dependent [11, 38]. Similarly, miRNAs function in context-dependent manner in the development of various cancers [45, 84]. Therefore, the pathological significance of miRNA-mediated β-TrCP suppression in cancer development depends on the particular substrate of SCFβ-TrCP, as well as the specific cancer type.
In human monocytic lymphoma U937 cells, β-TrCP1 promotes the degradation of the Sp1 transcription factor, resulting in decreased transcription of ADAM17 [85]. Downregulation of β-TrCP mediated by miR-183 increased ADAM17 expression by stabilizing Sp1, which correlates with increased TNFα/NF-κB signaling and enhanced cancer cell survival. ADAM17 promotes the shedding of cell surface-anchored TNFα thereby increasing the levels of soluble TNFα, which is required for TNFα-induced activation of NF-κB canonical pathway. NF-κB signaling plays important roles in promoting the progression of hematopoietic malignancies. Thus, miR-183 may promote oncogenesis in U937 cells, while β-TrCP1 has tumor suppressor activity, by regulating TNFα/NF-κB signaling. Likewise, β-TrCP1 may also play an anti-tumorigenic role by promoting TAZ degradation and inhibiting the Hippo pathway in NSCLC cells. Oncogenic miR-135b was found to be upregulated in NSCLC cells lines as well as in NSCLC patient samples [86]. Moreover, high miR-135b expression correlated with poor overall survival in NSCLC patients. β-TrCP1 was identified as a direct miR-135b target that regulates the Hippo pathway in the NSCLC cells. Downregulation of β-TrCP1 by miR-135b led to increased TAZ stability and enhanced transactivation of Hippo pathway target genes, which may contribute to NSCLC progression and metastasis. Interestingly, DNA methylation and oncogenic NF-κB signaling in NSCLC cells regulate miR-135b expression. It will be interesting to determine whether β-TrCP1-modulated Sp1/ADAM17/TNFα axis is also involved in regulating NF-κB activation in NSCLC cells. Nevertheless, NF-κB signaling can be directly regulated by IκB degradation mediated by β-TrCP1, which is required for NF-κB activation. The direct impact of miR-183/miR-135b-mediated β-TrCP1 suppression on NF-κB signaling in cancer cells requires further examination.
2.5 MiRNA targeting of other F-box proteins
Besides F-box proteins with well-characterized roles in cancer development, several F-box proteins are targeted by miRNAs whose roles in tumorigenesis are less understood. MiR-218 downregulates FBXW8 in human JEG-3 choriocarcinoma cells and promotes cell proliferation [87]. Since FBXW8 mediates the ubiquitination and degradation of p27 [88], the miR-218 mediated decrease of FBXW8 would be expected to lead to increased p27 levels, as well as impaired cell cycle arrest and enhanced JEG-3 cell proliferation.
In cytogenetically normal acute myeloid leukemia (CN-AML) patients, intronic miR-3151 was co-amplified with the host oncogene BAALC [89]. High miR-3151 expression was associated with shorter disease-free as well as overall survival in CN-AML patients. FBXL20 was identified as a direct target of miR-3151, although the pathological function of miR-3151-mediated FBXL20 suppression is unclear. FBXL20 was recently reported to promote ubiquitination and degradation of vacuolar protein-sorting 34 (Vps34), which is a critical regulator of autophagy and receptor degradation [90]. Interestingly, FBXL20 transcription is regulated by p53, which is also a miR-3151 target [91]. The multiple-layered downregulation of FBXL20 by miR-3151 suggest that FBXL20 may serve as a critical target for miR-3151 to promote oncogenesis.
FBXL5 has been identified as a potential direct target of miR-290-295 miRNA cluster that is enriched in mouse embryonic stem cells and promotes stem cell survival [92][93]. FBXL5 may promote epithelial-to-mesenchymal transition (EMT) by mediating E-cadherin degradation [94], consistent with a role for FBXL5 in regulating cancer metastasis. However, Snail1, an EMT-promoting transcription factor, was reported to be degraded by FBXL5-dependent pathway in the nucleus [95]. All in all, the biological significance of miR-290-295 cluster-regulated EMT in metastasis and its impact on cancer stem-like cell survival in human cancer warrants further investigation.
The muscle-specific F-box protein FBXO32 reportedly regulates apoptosis and functions as a tumor suppressor [96, 97]. FBXO32 was epigenetically silenced in ovarian cancers and esophageal squamous cell carcinomas, whereas reconstitution of FBXO32 expression inhibited ovarian cancer proliferation [98, 99]. Intriguingly, oncogenic miR-19a/b was shown to suppress FBXO32 expression in cardiomyocytes [100]. MiR-19a/b are members of oncogenic miR-17-92a cluster, which is amplified in a variety of cancers [101]. Whether miR-19a/b-dependent suppression of FBXO32 plays a role in the oncogenic function of miR-17-92a cluster in various cancers remained to be further explored.
3. Conclusions and perspectives
In general, F-box proteins and miRNAs negatively regulate target gene expression post-transcriptionally (see Table 1). Through recognizing specific degrons, F-box proteins direct polyubiquitination on specific target proteins by the SCF ligase complex and thereby promoting subsequent proteasomal degradation. Similarly, a miRNA recognizes specific MREs in target mRNAs that is complimentary to its seed region, thereby guiding the loading of the target mRNA into RISC for translation inhibition and mRNA decay. Most F-box proteins or miRNAs regulate multiple substrates, which play multifaceted pathophysiological roles in human diseases. Therefore, the function of F-box proteins or miRNAs in oncogenesis should be considered in the context of the specific substrates suppressed by the F-box protein/miRNA and in the specific cancer (Table 1).
Table 1.
MiRNA-dependent F-box protein suppression and its impact on cancer
| F-box protein | Targeting miRNA | Cancer/cell type specificity | Impact of F-box protein suppression |
|---|---|---|---|
| FBXW7 | miR-25 | gastric cancer | oncogenic |
| prostatic small cell neuroendocrine carcinoma | oncogenic | ||
| miR-27a | colon cancer lung cancer pediatric B-ALL |
oncogenic | |
| miR-92a | cervical cancer | oncogenic | |
| miR-182 | colon cancer | oncogenic | |
| miR-223 | gastric cancer T-ALL esophageal squamous cell carcinoma |
oncogenic | |
| miR-503 | colon cancer | oncogenic | |
| FBXO11 | miR-21 | prostate cancer melanoma glioma |
oncogenic |
| miR-621 | breast cancer | tumor suppressive | |
| Skp2 | miR-7 | CHO cells | tumor suppressive |
| miR-30 | lung fibroblasts | ||
| miR-203 | skin cancer | ||
| miR-340 | NSCLC | ||
| β-TrCP1 | miR-183 | monocytic lymphoma | oncogenic |
| miR-135b | NSCLC | ||
| FBXW8 | miR-218 | choriocarcinoma | oncogenic |
| FBXL20 | miR-3151 | CN-AML | oncogenic |
| FBXL5 | miR-290-295 | mouse ESCs | unknown |
| FBXO32 | miR-19a/b | ovarian cancer | oncogenic |
| FBXL11 | miR-19b | HEK293 | unknown |
In this review, we focused on miRNA-mediated downregulation of F-box proteins, the consequent stabilization of F-box protein substrates and their impact on human malignancies (Figure 1). Although this multi-layered regulation is discussed generally in the framework of one miRNA targeting one F-box protein thereby stabilizing one specific substrate of the F-box protein, each miRNA has the ability to suppress multiple target genes simultaneously, even multiple F-box proteins, which synergistically contribute to the overall impact on particular cancer cells. Moreover, each F-box protein likely is regulated by several upstream miRNAs, and could regulate multiple downstream substrates. The phenotypes observed in cancer cells by modulating one miRNA: F-box protein pair may reflect the collective outcome of altered expression of multiple target genes and/or several signaling pathways, which synergistically or antagonistically regulate oncogenesis and cancer progression. Considering the continuingly expanding lists of substrates of major oncogenic/tumor suppressive F-box proteins, future studies may provide a more comprehensive understanding of the complex responses in cancer cells as the consequences of altering one specific miRNA or F-box protein, as well as reveal unappreciated functions of the particular miRNA/F-box protein in modulating cancer development.
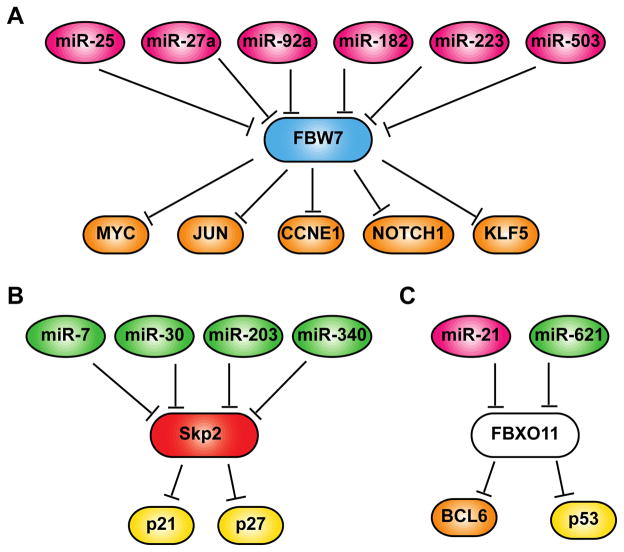
Figure 1.
MicroRNA-mediated suppression of F-Box proteins and their impact on cancer progression. A. Oncogenic miRNAs, such as miR-25, miR-27a, miR-92a, miR-182, miR-223 and miR-503, could promote cancer development by suppressing FBW7, thereby promoting proliferation and enhancing survival of cancer cells through stabilizing FBW7 substrates including c-Myc, c-Jun, cyclin E1, NOTCH1 and Klf5. B. Tumor suppressive miRNAs, such as miR-7, miR-30, miR-203 and miR-340, may inhibit cancer cell growth by suppressing Skp2, resulting in increased cell cycle checkpoint regulators p21 and p27. C. Both oncogenic miR-21 and tumor suppressive miR-621 can target FBXO11 in different cancer types. In melanoma, prostate cancer and glioma cells, miR-21-dependent FBXO11 suppression leads to increased Bcl-6 level and enhanced cancer cell growth. In contrast, miR-621 downregulates FBXO11 in breast cancer cells, resulting in increased p53 activity and enhanced response to chemotherapies.
Although the major function of F-box proteins is to regulate SCF-dependent ubiquitination of specific substrates, certain F-box proteins play roles in addition to their E3 ligase component. For instance, FBXL10/FBXL11 harbor a JmjC motif as well as the F-box domain, which functions as histone demethylase [102, 103]. By demethylation of lysine36 in histone H3, FBXL10/FBXL11 promoted cell proliferation by epigenetically inhibiting p15Ink4b transcription [102]. However, FBXL10-repressed ribosomal RNA transcription may antagonize the aggressive behavior of brain tumors [103]. Several lines of evidence support that FBXL10/FBXL11 play important roles in cancer development in a context-dependent manner by the epigenetic regulation of gene transcription. Interestingly, FBXL10 reportedly regulates the transcription of tumor suppressive miRNAs, miR-101 and let-7b, which may regulate cell senescence and proliferation by EZH2 suppression [104]. On the other hand, miR-19b suppresses FBXL11 expression, which increases NF-κB signaling [105]. One can envision that further exploration of the mutual regulation between miRNAs and FBXL10/FBXL11 may reveal more unexpected regulatory mechanisms orchestrated by miRNAs and F-box proteins in cancer development.
Our current knowledge from recent studies has demonstrated the critical and complex roles of miRNA-mediated regulation of F-box proteins in the pathogenesis and progression of human cancer. With recent advances in therapeutic strategies using F-box proteins or miRNAs as drug targets, this growing body of knowledge may soon be translated into novel approaches to treat cancer. Since F-box proteins and miRNAs function in a context-dependent manner, it is likely that genomic profiling of each cancer type and individual patient may ultimately be required for designing the most effective therapeutic approaches targeting specific F-box proteins and or miRNAs as a personalized medicine strategy in cancer patients.
Acknowledgments
We apologize to investigators whose important contributions and original articles were not cited in this review due to space limitation. The work in the Wu laboratory has been supported by NIH R01CA149251 and American Cancer Society (RSG-13-186-01-CSM). The work in the Pfeffer laboratory has been supported by NIH R01CA133322, Department of Defense (W81XWH-11-1-0533) and the Muirhead Chair Endowment at the University of Tennessee Health Science Center.
Abbreviations
- Skp1
S-phase kinase-associated protein 1
- SCF
Skp1-cullin1-F-box protein
- Rbx1
RING-box protein 1
- β-TrCP1
β-transducin repeat-containing protein1
- FBXL
F-box and leucine-rich repeat proteins
- FBXL
F-box and leucine-rich repeat proteins
- RBL2
retinoblastoma-like protein 2
- IκBα
inhibitor of nuclear factor κB
- DGCR8
DiGeorge syndrome critical region 8
- RISC
RNA-induced silencing complex
- T-ALL
T cell acute lymphoblastic leukemia
- MRE
miRNA recognition element
- 3’-UTR
3’ untranslated region
- BCL6
B cell lymphoma 6
- Cdt2
Cdc10-dependent transcript 2
- SETD8
SET domain-containing lysine methyltransferase 8
- NSCLC
non-small cell lung carcinoma
- CN-AML
cytogenetically normal acute myeloid leukemia
Footnotes
Conflict of interest: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes & development. 2004;18:2573–80. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358, e1. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 3.Skaar JR, Pagan JK, Pagano M. SnapShot: F box proteins I. Cell. 2009;137:1160–e1. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–74. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nature reviews Molecular cell biology. 2013;14:369–81. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 7.Tan MK, Lim HJ, Bennett EJ, Shi Y, Harper JW. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Molecular cell. 2013;52:9–24. doi: 10.1016/j.molcel.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaites LP, Lee EK, Lian Z, Barbash O, Roy D, Wasik M, et al. The Fbx4 tumor suppressor regulates cyclin D1 accumulation and prevents neoplastic transformation. Molecular and cellular biology. 2011;31:4513–23. doi: 10.1128/MCB.05733-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–3. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–5. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nature reviews Cancer. 2014;14:233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–73. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer research. 2007;67:9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 14.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–71. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. The Journal of experimental medicine. 2007;204:2875–88. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–41. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 17.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–3. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5934–41. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, et al. Role of the F-box protein Skp2 in lymphomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2515–20. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umanskaya K, Radke S, Chander H, Monardo R, Xu X, Pan ZQ, et al. Skp2B stimulates mammary gland development by inhibiting REA, the repressor of the estrogen receptor. Molecular and cellular biology. 2007;27:7615–22. doi: 10.1128/MCB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, et al. Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate. Cancer research. 2003;63:1583–8. [PubMed] [Google Scholar]
- 22.Seki R, Ohshima K, Fujisaki T, Uike N, Kawano F, Gondo H, et al. Prognostic significance of S-phase kinase-associated protein 2 and p27kip1 in patients with diffuse large B-cell lymphoma: effects of rituximab. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2010;21:833–41. doi: 10.1093/annonc/mdp481. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: a novel potential therapeutic target for prostate cancer. Biochimica et biophysica acta. 2012;1825:11–7. doi: 10.1016/j.bbcan.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose AE, Wang G, Hanniford D, Monni S, Tu T, Shapiro RL, et al. Clinical relevance of SKP2 alterations in metastatic melanoma. Pigment cell & melanoma research. 2011;24:197–206. doi: 10.1111/j.1755-148X.2010.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuler S, Diersch S, Hamacher R, Schmid RM, Saur D, Schneider G. SKP2 confers resistance of pancreatic cancer cells towards TRAIL-induced apoptosis. International journal of oncology. 2011;38:219–25. [PubMed] [Google Scholar]
- 26.Radke S, Pirkmaier A, Germain D. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene. 2005;24:3448–58. doi: 10.1038/sj.onc.1208328. [DOI] [PubMed] [Google Scholar]
- 27.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes & development. 1999;13:270–83. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 29.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Current biology : CB. 1999;9:207–10. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, et al. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. The EMBO journal. 1999;18:2401–10. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, et al. Ubiquitin-dependent degradation of IkappaBalpha is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3859–63. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, et al. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Molecular cell. 1999;3:527–33. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 33.Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. Journal of the National Cancer Institute. 2004;96:1161–70. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 34.Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, et al. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer research. 2005;65:1316–24. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–36. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 36.Kim CJ, Song JH, Cho YG, Kim YS, Kim SY, Nam SW, et al. Somatic mutations of the beta-TrCP gene in gastric cancer. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2007;115:127–33. doi: 10.1111/j.1600-0463.2007.apm_562.x. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh T, Katoh M. Expression profiles of betaTRCP1 and betaTRCP2, and mutation analysis of betaTRCP2 in gastric cancer. International journal of oncology. 2001;18:959–64. [PubMed] [Google Scholar]
- 38.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nature reviews Drug discovery. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 42.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 43.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao Z, et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene. 2015 doi: 10.1038/onc.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Sengupta T, Kukreja L, Minella AC. MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. The Journal of biological chemistry. 2010;285:34439–46. doi: 10.1074/jbc.M110.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, et al. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer discovery. 2012;2:540–53. doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, Zhang B, et al. Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene. 2011;30:3875–86. doi: 10.1038/onc.2011.103. [DOI] [PubMed] [Google Scholar]
- 49.Lerner M, Lundgren J, Akhoondi S, Jahn A, Ng HF, Akbari Moqadam F, et al. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell cycle. 2011;10:2172–83. doi: 10.4161/cc.10.13.16248. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Guo Y, Liang X, Sun M, Wang G, De W, et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. Journal of cancer research and clinical oncology. 2012;138:763–74. doi: 10.1007/s00432-012-1154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nature genetics. 2011;43:673–8. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. British journal of cancer. 2012;106:182–8. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. Journal of internal medicine. 2013;274:215–26. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. The Journal of experimental medicine. 2013;210:1545–57. doi: 10.1084/jem.20122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, et al. Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:2324–35. doi: 10.1038/leu.2014.133. [DOI] [PubMed] [Google Scholar]
- 56.Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. International journal of cancer Journal international du cancer. 2015;136:1537–45. doi: 10.1002/ijc.29168. [DOI] [PubMed] [Google Scholar]
- 57.Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. Journal of experimental & clinical cancer research : CR. 2015;34:28. doi: 10.1186/s13046-015-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma J, Cheng L, Liu H, Zhang J, Shi Y, Zeng F, et al. Genistein down-regulates miR-223 expression in pancreatic cancer cells. Current drug targets. 2013;14:1150–6. doi: 10.2174/13894501113149990187. [DOI] [PubMed] [Google Scholar]
- 59.Zhou C, Shen L, Mao L, Wang B, Li Y, Yu H. miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochemical and biophysical research communications. 2015;458:63–9. doi: 10.1016/j.bbrc.2015.01.066. [DOI] [PubMed] [Google Scholar]
- 60.Li L, Sarver AL, Khatri R, Hajeri PB, Kamenev I, French AJ, et al. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. The Journal of pathology. 2014;234:488–501. doi: 10.1002/path.4407. [DOI] [PubMed] [Google Scholar]
- 61.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell metabolism. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y, et al. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-3510-3. [DOI] [PubMed] [Google Scholar]
- 63.Li Z, Sun Y, Chen X, Squires J, Nowroozizadeh B, Liang C, et al. p53 Mutation Directs AURKA Overexpression via miR-25 and FBXW7 in Prostatic Small Cell Neuroendocrine Carcinoma. Molecular cancer research : MCR. 2015;13:584–91. doi: 10.1158/1541-7786.MCR-14-0277-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu D, Davis MP, Abreu-Goodger C, Wang W, Campos LS, Siede J, et al. MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming of mouse fibroblast cells to iPSCs. PloS one. 2012;7:e40938. doi: 10.1371/journal.pone.0040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncology reports. 2012;27:594–8. doi: 10.3892/or.2011.1530. [DOI] [PubMed] [Google Scholar]
- 66.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochemical and biophysical research communications. 2012;421:640–5. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 67.Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer letters. 2013;335:168–74. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 68.Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, et al. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. The Journal of clinical endocrinology and metabolism. 2012;97:E710–8. doi: 10.1210/jc.2011-3068. [DOI] [PubMed] [Google Scholar]
- 69.Yang CH, Pfeffer SR, Sims M, Yue J, Wang Y, Linga VG, et al. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. The Journal of biological chemistry. 2015;290:6037–46. doi: 10.1074/jbc.M114.632125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug development research. 2015 doi: 10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]
- 71.Abbas T, Mueller Adam C, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 Promotes Cdt2 Ubiquitylation and Degradation and Regulates Pr-Set7/Set8-Mediated Cellular Migration. Molecular cell. 2013;49:1147–58. doi: 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossi M, Duan S, Jeong Y-T, Horn M, Saraf A, Florens L, et al. Regulation of the CRL4Cdt2 Ubiquitin Ligase and Cell-Cycle Exit by the SCFFbxo11 Ubiquitin Ligase. Molecular cell. 2013;49:1159–66. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. The EMBO journal. 2012;31:110–23. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. 2015 doi: 10.1038/onc.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. The Journal of biological chemistry. 2007;282:1797–804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng H, Shen M, Zha YL, Li W, Wei Y, Blanco MA, et al. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer cell. 2014;26:358–73. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jackson SJ, Zhang Z, Feng D, Flagg M, O’Loughlin E, Wang D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140:1882–91. doi: 10.1242/dev.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11324–9. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature cell biology. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez N, Gallagher M, Lao N, Gallagher C, Clarke C, Doolan P, et al. MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including Skp2 and Psme3. PloS one. 2013;8:e65671. doi: 10.1371/journal.pone.0065671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34:3240–50. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osoegawa A, Yoshino I, Tanaka S, Sugio K, Kameyama T, Yamaguchi M, et al. Regulation of p27 by S-phase kinase-associated protein 2 is associated with aggressiveness in non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:4165–73. doi: 10.1200/JCO.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 83.Qi F, He T, Jia L, Song N, Guo L, Ma X, et al. The miR-30 Family Inhibits Pulmonary Vascular Hyperpermeability in the Premetastatic Phase by Direct Targeting of Skp2. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:3071–80. doi: 10.1158/1078-0432.CCR-14-2785. [DOI] [PubMed] [Google Scholar]
- 84.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews Drug discovery. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu WH, Chang LS. Suppression of Akt/Foxp3-mediated miR-183 expression blocks Sp1-mediated ADAM17 expression and TNFalpha-mediated NFkappaB activation in piceatannol-treated human leukemia U937 cells. Biochemical pharmacology. 2012;84:670–80. doi: 10.1016/j.bcp.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 86.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nature communications. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 87.Shi D, Tan Z, Lu R, Yang W, Zhang Y. MicroRNA-218 inhibits the proliferation of human choriocarcinoma JEG-3 cell line by targeting Fbxw8. Biochemical and biophysical research communications. 2014;450:1241–6. doi: 10.1016/j.bbrc.2014.06.094. [DOI] [PubMed] [Google Scholar]
- 88.Lin P, Fu J, Zhao B, Lin F, Zou H, Liu L, et al. Fbxw8 is involved in the proliferation of human choriocarcinoma JEG-3 cells. Molecular biology reports. 2011;38:1741–7. doi: 10.1007/s11033-010-0288-7. [DOI] [PubMed] [Google Scholar]
- 89.Eisfeld AK, Marcucci G, Maharry K, Schwind S, Radmacher MD, Nicolet D, et al. miR-3151 interplays with its host gene BAALC and independently affects outcome of patients with cytogenetically normal acute myeloid leukemia. Blood. 2012;120:249–58. doi: 10.1182/blood-2012-02-408492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T, et al. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes & development. 2015;29:184–96. doi: 10.1101/gad.252528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eisfeld AK, Schwind S, Patel R, Huang X, Santhanam R, Walker CJ, et al. Intronic miR-3151 within BAALC drives leukemogenesis by deregulating the TP53 pathway. Science signaling. 2014;7:ra36. doi: 10.1126/scisignal.2004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lichner Z, Pall E, Kerekes A, Pallinger E, Maraghechi P, Bosze Z, et al. The miR-290-295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation; research in biological diversity. 2011;81:11–24. doi: 10.1016/j.diff.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 93.Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, Dennis LM, et al. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS genetics. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dragoi AM, Swiss R, Gao B, Agaisse H. Novel strategies to enforce an epithelial phenotype in mesenchymal cells. Cancer research. 2014;74:3659–72. doi: 10.1158/0008-5472.CAN-13-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic acids research. 2014;42:1079–94. doi: 10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14440–5. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo W, Zhang M, Shen S, Guo Y, Kuang G, Yang Z, et al. Aberrant methylation and decreased expression of the TGF-beta/Smad target gene FBXO32 in esophageal squamous cell carcinoma. Cancer. 2014;120:2412–23. doi: 10.1002/cncr.28764. [DOI] [PubMed] [Google Scholar]
- 99.Chou JL, Su HY, Chen LY, Liao YP, Hartman-Frey C, Lai YH, et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Laboratory investigation; a journal of technical methods and pathology. 2010;90:414–25. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song DW, Ryu JY, Kim JO, Kwon EJ, Kim do H. The miR-19a/b family positively regulates cardiomyocyte hypertrophy by targeting atrogin-1 and MuRF-1. The Biochemical journal. 2014;457:151–62. doi: 10.1042/BJ20130833. [DOI] [PubMed] [Google Scholar]
- 101.Olive V, Li Q, He L. mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunological reviews. 2013;253:158–66. doi: 10.1111/imr.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nature structural & molecular biology. 2008;15:1169–75. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–13. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 104.Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, et al. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. The Journal of biological chemistry. 2011;286:33061–9. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic acids research. 2012;40:8048–58. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]